Abstract
In this study, we describe the genomes of two novel candidate species of non-nitrogen fixing Frankia that were isolated from the root nodules of Coriaria nepalensis and Alnus glutinosa, genospecies CN and Ag, respectively. Comparative genomic analyses revealed that both genospecies lack genes essential for nitrogen-fixation and possess genes involved in the degradation of plant cell walls. Additionally, we found distinct biosynthetic gene clusters in each genospecies. The availability of these genomes will contribute to the study of the taxonomy and evolution of actinorhizal symbioses.
Keywords: Frankia, Actinorhizal symbiosis, genome, non-nitrogen-fixing frankiae, biosynthetic gene clusters
Introduction
The actinobacterial genus Frankia consists of a diverse group of filamentous, gram-positive soil bacteria that are able to form root nodules with members of eight plant families representing about 25 genera of woody, dicotyledonous, perennial angiosperms 1-3. Root nodule formation is host plant-specific, and largely correlates with assignments of strains to specific clusters derived from comparative sequence analyses of amplicons of genes such as 16S or 23S rRNA genes 4, nifH genes 5-7, gyrB or glnII 8 and concatenated sequences of some housekeeping genes (dnaA, ftsZ, and pgk) 9, 10. Clusters 1, 2 and 3 represent Frankia strains that are capable of fixing atmospheric nitrogen (N2) in pure culture and in symbiosis, while members of cluster 4 are typically unable to fix N2, with one exception, and are often not able to form root nodules 4, 11.
While comparative sequence analyses of specific gene amplicons have provided evidence for group-specific assignments of isolates, whole genome sequencing has opened a new avenue to more sophisticated classification of species within the genus Frankia. These analyses include both isolates deposited as type strains in culture collections, and uncultured Frankia populations in root nodules of specific host plants described as candidate species. Frankia alni ACN14aT 12 and Frankia torreyi CpI1T 13, as well as Frankia casuarinae CcI3T 12 and Frankia canadensis ARgP5T 14 have been identified in cluster 1, representing cluster 1a, 1c, and 1d, respectively. Related, but distinct to F. candadensis is Candidatus Frankia nodulisporulans AgTrS, and AgUmASt1 and AgUmASH1 15 as well as Candidatus Frankia alpina AiOr, and AvVan 15. Frankia coriariae BMG5.1T 16, Candidatus Frankia datiscae Dg1 17, Candidatus Frankia californiensis Dg2 18 and Candidatus Frankia meridionalis Cppng1 19 represent species in cluster 2, while Frankia elaeagni BMG5.12T 12, Frankia discariae BCU110501T 20, Frankia soli NRRL B-16219T 21 and Frankia irregularis G2T 22 are species in cluster 3. Cluster 4 frankiae are represented by three species, Frankia inefficax EuI1cT 23, Frankia asymbiotica M16386T 24 and Frankia saprophytica CN3T 25.
Using comparative sequence analyses of amplicons of an actinobacteria-specific insertion in the 23S rRNA genes of additional cluster 4 frankiae, we identified several strains clustering together but distinct from type strains of cluster 4 26. These strains included CN4, CN6, CN7, and CNM7 isolated from Coriaria nepalensis 27, AgW1.1 and AgB1.9 isolated from Alnus glutinosa 28, and a yet undescribed strain AgB1.8 obtained from the same location as strain AgB1.9. The goal of this study was to use whole genome sequence analyses to assess the viability of our previous amplicon-based analysis, and thus affirm the potential of these strains for new species descriptions.
Materials and Methods
Sample preparation
Seven Frankia strains previously identified to represent members of cluster 4 (AgW1.1, AgB1.9, AgB1.8, CN4, CN6, CN7, CNM7) were grown in Defined Propionate Medium (DPM) containing propionate and NH4Cl as C and N source, respectively 29, at 30°C for two weeks. Cells were harvested by centrifugation (15,000 x g, 5 min), and cell aggregates homogenized by brief sonication (10s at 20% output in a S-450 sonifier, Branson Ultrasonics, Danbury, CT) 30. After centrifugation, cell pellets were used for DNA extraction using the SurePrepTM Soil DNA Isolation Kit (Fisher Scientific, Houston, TX) with small modifications as described before 31. Extractions of all samples were done in triplicate, and DNA concentrations measured with a Qubit® 2.0 Fluorometer (Life Technologies, Carlsbad, USA). Library preparation and sequencing was performed at the Microbial Genomics Sequencing Center, Pittsburgh, PA, USA using the Illumina tagmentation protocol and the NextSeq Illumina platform (2 x 150 bp).
Genome assembling
Sequence reads were filtered and trimmed using the default settings of fastp 32, and bbduk (https://jgi.doe.gov/data-and-tools/bbtools/bb-tools-user-guide/) was used to remove reads with average % GC < 54. Genomes were assembled using SPAdes 3.13.0 33. Quast was used to check the quality of the assembled genomes.
Comparative genomic analysis
All “Frankia” genomes available in NCBI Genomes (https://www.ncbi.nlm.nih.gov/genome/) in November, 14, 2019 were downloaded for comparative genomic analyses. Anvi'o 34 was used to compare the shared COG functions among Frankia genomes and generate a core genome phylogenetic tree. The core genome phylogenetic tree was generated using anvi-gen-phylogenomic-tree with the concatenated alignment of 113 single-copy core genes (present in all genomes) that resulted in a total alignment length of 40,557 amino acids. Fragments of the 16S rRNA gene were recovered from the genomes using the HMM profile Ribosomal RNAs implemented in anvi'o. 16S rRNA gene sequences were aligned using SINA 35 and sequence identity was calculated with MEGAX 3 36. Pairwise Average Nucleotide Identity (ANI) was calculated with the fastANI software 37. Identification of open reading frames (ORFs) was performed using Prokka using the default settings 38 and KEGG orthologous annotation was performed with kofamKOALA using the default settings 39. We used antiSMASH 5.0 using the default settings 40 to investigate the presence of biosynthetic gene clusters (BGCs) categories across cluster 4 Frankia strains.
Results
Phylogenetic analysis of Frankia spp. isolates
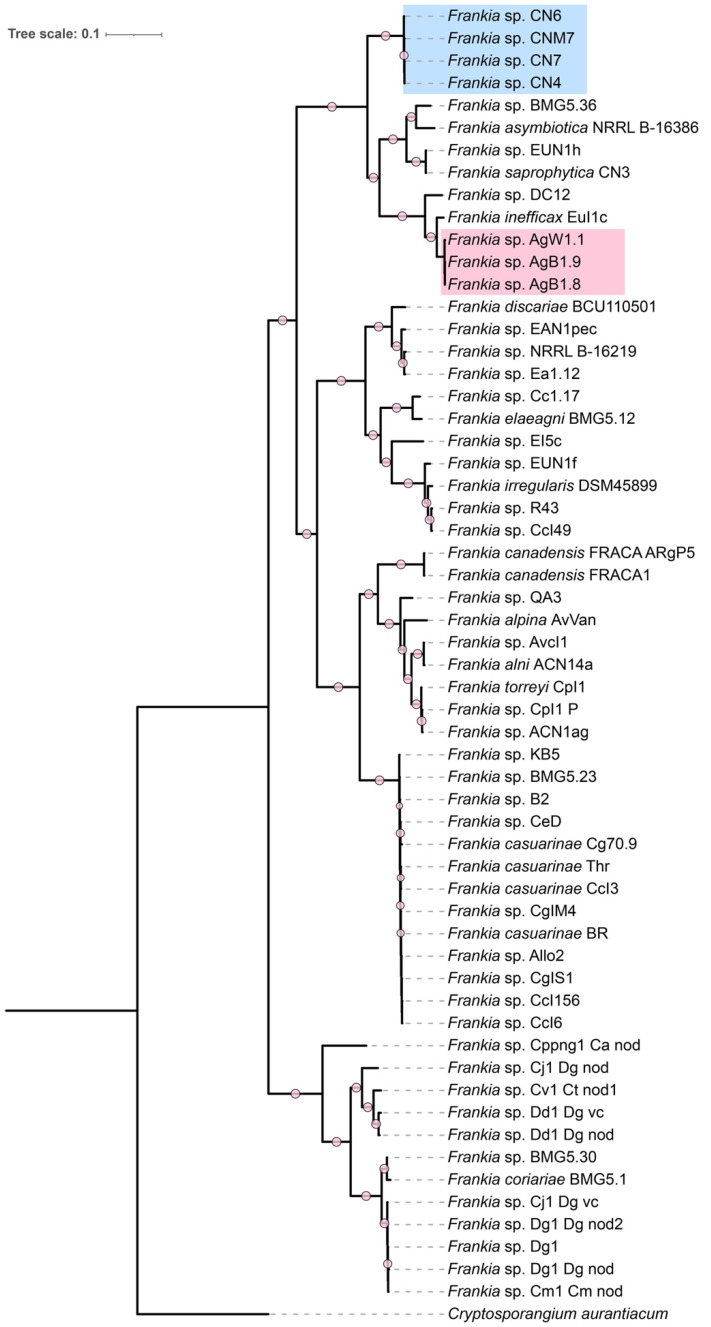
Assembled genomes ranged from 9.8 Mb and 10.7 Mb and the GC content of the genomes was between 71.5 and 72.7% (Table 1). A phylogenetic tree generated from the concatenated alignment of single-copy orthologous proteins present in all 58 Frankia genomes revealed that the closest strains to AgB1.8, AgB1.9, AgW1.1, CNM7, CN4, CN6, CN7 were members of cluster 4, i.e. Frankia saprophytica CN3, Frankia sp. EUN1h, Frankia asymbiotica NRRL B-16386, Frankia sp. BMG5.36, Frankia sp. DC12 and Frankia inefficax EuI1c (Figure 1).
Table 1.
Frankia spp. assembled genomes information.
| Strain | RefSeq assembly accessiom | Number of reads a | Number of scaffolds | N50 scaffold b | Total length | GC (%) c | Protein coding genes |
|---|---|---|---|---|---|---|---|
| AgB1.8 | GCF_016792105.1 | 4,651,340 | 383 | 46,557 | 9,788,199 | 71.67 | 9,155 |
| AgB1.9 | GCF_016786355.1 | 6,564,124 | 278 | 66,606 | 9,788,632 | 71.65 | 9,075 |
| AgW1.1 | GCF_016786345.1 | 3,546,242 | 558 | 35,357 | 9,961,786 | 71.53 | 9,540 |
| CNM7 | GCF_016786385.1 | 3,663,004 | 1,106 | 15,926 | 10,670,374 | 72.72 | 10,336 |
| CN4 | GCF_016786305.1 | 5,509,032 | 607 | 31,737 | 10,707,582 | 72.68 | 9,988 |
| CN6 | GCF_016792085.1 | 6,112,674 | 397 | 48,718 | 10,719,543 | 72.69 | 9,882 |
| CN7 | GCF_016786325.1 | 7,180,944 | 398 | 51,919 | 10,728,937 | 72.68 | 9,843 |
a Number of reads used in the genome assembling after quality control.
b N50 scaffold is the largest length L of which 50% of all nucleotides are in scaffolds of size at least L.
c Total number of G and C nucleotides in the genome, divided by the total length of the genome.
Figure 1.
Maximum likelihood tree from concatenated alignments of 113 single-copy protein (total alignment length of 40,557 amino acids) generated with Anvi'o. Strains sequenced in this study are highlighted. The tree was re-rooted using Cryptoporangium aurantiacum as outgroup. Circles represent branch support values (maximum likelihood bootstrap) ≥ 0.8.
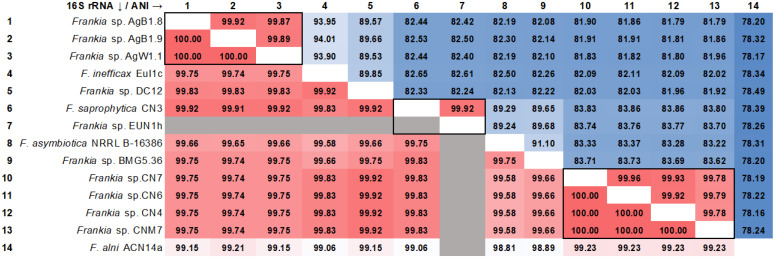
In order to determine if the strains described in this work were potential novel species the average nucleotide identity (ANI) was calculated among all the cluster 4 strains (Figure 2). AgB1.8, AgB1.9, AgW1.1, CNM7, CN4, CN6, CN7 did not show ANI ≥ 95% to any of the reference strains. Strains AgB1.8, AgB1.9, AgW1.1 showed ANI ≥ 99% to each other, which indicates they belong to the same genospecies (genospecies Ag). Strains CNM7, CN4, CN6, CN7 showed ANI ≥ 99% to each other, indicating that they belong to a single genospecies (genospecies CN). Analysis of nearly full length 16S rRNA gene fragment (alignment length 1,202) showed that the identity of cluster 4 frankiae ranged from 99.57% to 100% (Figure 2).
Figure 2.
Heatmap matrix of similarity among genomes of cluster 4 frankiae. Top diagonal values correspond to the pairwise Average Nucleotide Identity (ANI) in percentage. Groups of genomes that fall into the estimated species boundary (ANI ≥ 95%) are delimited with black lines. Bottom diagonal values correspond to the pairwise identities of 16S rRNA genes (alignment length 1,202 bp) in percentage. No 16S rRNA sequence of Frankia sp. EUN1h was found in the genome using the anvi'o pipeline.
Functional analysis of Frankia spp. isolates
We compared the presence of KEGG orthologues (KO) of the two novel genospecies of Frankia Ag and CN with other Frankia strains (Table S2). As expected, nitrogenase complex genes (nif) were absent in all the seven strains sequenced in this study (Table S2). KEGG orthologues common to all cluster 4 genomes and absent in other frankiae included genes involved in the urea transporter system (K11959 to K11963), and the complete tryptophan metabolism pathway (KEGG module M00038). Additionally, KEGG orthologues involved in the biodegradation of xenobiotics were enriched in cluster 4 Frankia genomes when compared to other frankiae (Table S2). 415 KEGG orthologues were found in all 58 Frankia genomes analyzed in this study, including: alkaline phosphatases (K01113), which are involved in solubilization of soil phosphate (Nautiyal et al., 2000, Hahn et al., 2003); adherence proteins (K12510 and K12511), described to be involved in Actinobacillus actinomycetemcomitans host colonization (Kachalany et al. 2001) (Table S2).
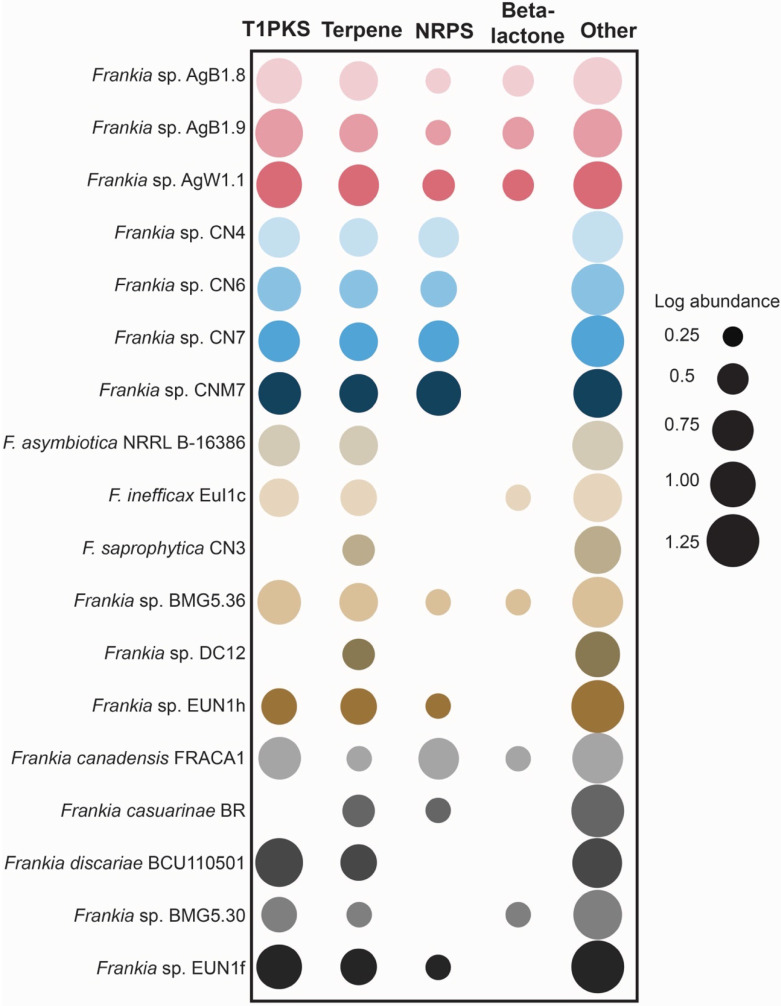
Unique KOs of genomes of Frankia Ag included feruloyl esterase (K09252) and beta-mannosidase (K01192). KOs found in the genomes of Frankia CN genospecies and not found in other cluster 4 genomes included a protein involved nitrate transport (K15577), and a flavohemoglobin (K05916). Additionally, genomes of Frankia Ag genospecies had more Type I polyketide synthase (PKS) and beta-lactone clusters than other cluster 4 Frankia, while Frankia CN genospecies had more non-ribosomal peptide synthetase clusters (NRPS) than other cluster 4 frankiae (Figure 3).
Figure 3.
Abundance of biosynthetic gene cluster (BGCs) categories across selected genomes Frankia strains. The size of the circles represents the log of total number of clusters of each BGC category in each genome. T1PKS, Type-1 Polyketide synthase; NRPS, Non-ribosomal peptide synthetase.
Discussion
In this study, we compared the genomes of three Frankia strains isolated from Alnus glutinosa and four strains isolated from Coriaria nepalensis with 51 publicly available genomes of Frankia. Phylogenomic and ANI analyses confirmed that these strains are related to cluster 4 frankiae, and indicate that the three strains isolated from Alnus glutinosa (AgB1.8, AgB1.9 and AgW1.1; Ag genospecies) belong to a novel species, as do the four strains from Coriaria nepalensis (CN4, CN6, CN7 and CNM7; CN genospecies). Genome sizes of all strains assembled as Ag and CN genospecies were about 9.8 Mb and 10.7 Mb, respectively. Sizes were comparable to those of other strains representing cluster 4 frankiae, i.e. F. inefficax and F. saprophytica with 8.8 Mb and 10 Mb, respectively, and much larger than genomes of most cluster 1 and 3 frankiae (7.5 Mb to 7.9 Mb), including F. casuarinae (4.9 to 5.6 Mb) and A. nodulisporulans (4.9 Mb), as well as F. coriariae as cluster 2 representative (5.8 Mb) (see 11 for review). An exception was the genome size of F. irregularis, a cluster 3 representative with a genome size similar to our cluster frankiae (9.5 Mb) 22, and of related cluster 3 strains R43 (10.44 Mb) 41 and EAN1pec (9.04 Mb) 42.
Smaller genome sizes in frankiae have been related to genome reductions are associated to reduced saprotrophic potential, but not symbiotic potential, while larger genomes created through genome expansions, often through duplication of genes involved in introducing substrates into central metabolic pathways, allowed frankiae to exploit a large variety of environments 17, 42. The comparatively large genomes of cluster 3 and cluster 4 Frankia strains therefore suggest an increased saprotrophic potential compared to strains assigned to other clusters. In support of this hypothesis, indigenous cluster 3 frankiae as well as introduced strain EAN1pec have been shown to persist and grow under a broader range of environmental conditions (e.g. plant species, carbon resource or matric potential) than indigenous cluster 1a frankiae or introduced strains ArI3 (cluster 1a) and CcI3 (cluster 1c) 43, 44. The results of these studies have been used to suggest that cluster 3 frankiae represent a group of generalists 44. In addition, cluster 1a frankiae were characteristic populations of later stages in a succession of Frankia populations in soil associated with plant growth and succession. Frankiae of cluster 1b represented by Frankia strain Ag45/Mut15 were characteristic of soils in early stages of plant-mediated organic matter accumulation. Cluster 1c frankiae represented a group of highly-specialized frankiae that depended on the presence of their host plant to grow saprotrophically in soil 44.
Diversity of frankiae in soils vegetated with host plant species such as Alnus glutinosa or non-host plant species such as Betula nigra was found to be restricted to cluster 1 and 3 frankiae, and generally members of cluster 1b dominating 26. Cluster 1b frankiae have been detected as major populations in several studies, with absolute numbers depending on the sampling depth, physicochemical conditions and the vegetation 43, 45, 46. So far, cluster 4 frankiae have been detected as major Frankia population only in prairie soils where they represented about 50% of all frankiae, that included clusters 1b, 2 and 3, in native, restored and cultivated black prairie soil 26. Native and restored prairie were dominated by Sorghastrum nutans (L.) (Indiangrass) and Andropogon gerardii Vitman (Big bluestem), while the cultivated prairie site was adjacent to the native prairie site, but cultivated continuously with corn. These results suggest that grasses are supporting a diverse community of frankiae in prairie soils, with significant positive effects on the abundance of cluster 4 frankiae. Although it has been shown that frankiae can grow in the rhizosphere of grasses 47, it is yet unclear for prairie environments whether these effects are related to rhizosphere effects of dense grass root environments or to organic matter accumulation in top soils.
Isolates representing cluster 4 frankiae have been obtained from a variety of different host plant species, i.e. Coriaria nepalensis, Alnus glutinosa, Elaeagnus umbellata, Datisca cannabina, and Ceanothus americanus (CN3, AgW1.1, EuI1c, DC12 and CaI1) 27, 28, 48-50. Cluster 4 frankiae have also been detected in bioassays highlighting that they form an important fraction of all frankiae in wet soils under A. glutinosa 51, 52. Some progenies of A. glutinosa, however, exhibited natural resistance to infection by these populations with no or only very small nodules formed 28, 53. The failure to detect nodules has therefore resulted in the assumption that many cluster 4 frankiae are not only ineffective with respect to nitrogen-fixation, but also non-infective with respect to root nodule formation. Cluster 4 frankiae could therefore be characterized as largely or even entirely saprotrophic, with isolates obtained from root nodules as surfaces contaminants 54.
Both genospecies CN and Ag lack genes essential for nitrogen-fixation, as observed for cluster 4 species F. inefficax EUI1cT 23 and F. saprophytica CN3T 25, but not for F. asymbiotica M16386T 24. While F. inefficax EUI1cT was observed to form ineffective nodules, none of the other species were able to form nodules on their original host plant. Members of genospecies Ag have been shown to induce small ineffective nodules on their respective host plant A. glutinosa 28 while members of genospecies CN failed to induce nodules on their host plant Coriaria nepalensis 27. Genomes of genospecies Ag strains harbor genes encoding feruloyl esterases and beta-mannosidases that potentially participate in the degradation of plant cell walls as indicated for some plant fungal pathogens, and could therefore be involved in the plant infection process or in nutrient acquisition 55, 56. Genes for both flavohemoglobin and nitrite/nitrate transport proteins are unique for genospecies CN strains potentially involved in the conversion of nitric oxide to nitrate 57. Our analysis supports previous studies that found significant biosynthetic potential of Frankia spp. It has been hypothesized that specialized metabolites, such as the molecules produced by PKS and NRPS gene clusters, might participate in plant-microbial signaling and interactions 58. While our genome studies confirm our previous expectations and affirm the potential of these strains for new species descriptions, the assessment of ecological differences between our genospecies CN and Ag, and described cluster 4 species requires additional experimental data.
Data Summary
1. Genomes of the strains sequenced in this study from Dr. Dittmar Hahn culture collection and were deposited in the National Center for Biotechnology Information (NCBI), under BioProject Number PRJNA680372. Individual RefSeq assembly accession numbers can be found in Table S1.
2. A list of other Frankia genomes utilized in this study can be found in Table S1. All sequences were downloaded from the NCBI Assembly database.
Supplementary Material
Supplementary tables.
Acknowledgments
The authors acknowledge Quinlan J. Carlos-Shanley for writing a script to parse the genome annotation data.
References
- 1.Schwintzer CR, Tjepkema JD. The biology of Frankia and actinorhizal plants. San Diego: Academic Press. 1990.
- 2.Huss-Danell K. Actinorhizal symbioses and their N-2 fixation. New Phytol. 1997;136:375–405. doi: 10.1046/j.1469-8137.1997.00755.x. [DOI] [PubMed] [Google Scholar]
- 3.Benson DR, Silvester WB. Biology of Frankia strains, actinomycete symbionts of actinorhizal plants. Microbiol Rev. 1993;57:293–319. doi: 10.1128/mr.57.2.293-319.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Normand P, Orso S, Cournoyer B, Jeannin P, Chapelon C, Dawson J. et al. Molecular phylogeny of the genus Frankia and related genera and emendation of the family Frankiaceae. Int J Syst Bacteriol. 1996;46:1–9. doi: 10.1099/00207713-46-1-1. [DOI] [PubMed] [Google Scholar]
- 5.Welsh A, Mirza BS, Rieder JP, Paschke MW, Hahn D. Diversity of frankiae in root nodules of Morella pensylvanica grown in soils from five continents. Syst Appl Microbiol. 2009;32:201–210. doi: 10.1016/j.syapm.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 6.Welsh AK, Dawson JO, Gottfried GJ, Hahn D. Diversity of Frankia in root nodules of geographically isolated Arizona alders in central Arizona (USA) Appl Environ Microbiol. 2009;75:6913–6918. doi: 10.1128/AEM.01103-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mirza BS, Welsh A, Rasul G, Rieder JP, Paschke MW, Hahn D. Variation in Frankia populations of the Elaeagnus host infection group in nodules of six host plant species after inoculation with soil. Microb Ecol. 2009;58:384–393. doi: 10.1007/s00248-009-9513-0. [DOI] [PubMed] [Google Scholar]
- 8.Nouioui I, Ghodhbane-Gtari F, Beauchemin NJ, Tisa LS, Gtari M. Phylogeny of members of the Frankia genus based on gyrB, nifH and glnII sequences. Anton Leeuw Int J G. 2011;100:579–587. doi: 10.1007/s10482-011-9613-y. [DOI] [PubMed] [Google Scholar]
- 9.Pozzi AC, Bautista-Guerrero HH, Nouioui I, Cotin-Galvan L, Pepin R, Fournier P. et al. In-planta sporulation phenotype: a major life history trait to understand the evolution of Alnus-infective Frankia strains. Environ Microbiol. 2015;17:3125–3138. doi: 10.1111/1462-2920.12644. [DOI] [PubMed] [Google Scholar]
- 10.Pozzi AC, Bautista-Guerrero HH, Abby SS, Herrera-Belaroussi A, Abrouk D, Normand P. et al. Robust Frankia phylogeny, species delineation and intraspeciesdiversity based on Multi-Locus Sequence Analysis (MLSA) and Single-Locus Strain Typing (SLST) adapted to a large sample size. System Appl Microbiol. 2018;41:311–323. doi: 10.1016/j.syapm.2018.03.002. [DOI] [PubMed] [Google Scholar]
- 11.Gtari M, Nouioui I, Sarkar I, Ghodhbane-Gtari F, Tisa LS, Sen A. et al. An update on the taxonomy of the genus Frankia Brunchorst, 1886, 174(AL) Anton Leeuw. 2019;112:5–21. doi: 10.1007/s10482-018-1165-y. [DOI] [PubMed] [Google Scholar]
- 12.Nouioui I, Ghodhbane-Gtari F, Montero-Calasanz MD, Goker M, Meier-Kolthoff JP, Schumann P. et al. Proposal of a type strain for Frankia alni (Woronin 1866) Von Tubeuf 1895, emended description of Frankia alni, and recognition of Frankia casuarinae sp. nov and Frankia elaeagni sp. nov. Int J Syst Evol Microbiol. 2016;66:5201–5210. doi: 10.1099/ijsem.0.001496. [DOI] [PubMed] [Google Scholar]
- 13.Nouioui I, Ghodhbane-Gtari F, Jando M, Tisa LS, Klenk HP, Gtari M. Frankia torreyi sp. nov, the first actinobacterium of the genus Frankia Brunchorst 1886, 174(AL) isolated in axeniculture. Anton Leeuw. 2019;112:57–65. doi: 10.1007/s10482-018-1131-8. [DOI] [PubMed] [Google Scholar]
- 14.Normand P, Nouioui I, Pujic P, Fournier P, Dubost A, Schwob G. et al. Frankia canadensis sp nov, isolated from root nodules of Alnus incana subspecies rugosa. Int J Syst Evol Microbiol. 2018;68:3001–3011. doi: 10.1099/ijsem.0.002939. [DOI] [PubMed] [Google Scholar]
- 15.Herrera-Belaroussi A, Normand P, Pawlowski K, Fernandez MP, Wibberg D, Kalinowski J. et al. Candidatus Frankia nodulisporulans sp. nov, an Alnus glutinosa-infective Frankia species unable to grow in pure culture and able to sporulate in-planta. Syst Appl Microbiol. 2020;43:126134. doi: 10.1016/j.syapm.2020.126134. [DOI] [PubMed] [Google Scholar]
- 16.Nouioui I, Ghodhbane-Gtari F, Rohde M, Klenk HP, Gtari M. Frankia coriariae sp nov, an infective and effective microsymbiont isolated from Coriaria japonica. Int J Syst Evol Microbiol. 2017;67:1266–1270. doi: 10.1099/ijsem.0.001797. [DOI] [PubMed] [Google Scholar]
- 17.Persson T, Battenberg K, Demina IV, Vigil-Stenman T, Vanden Heuvel B, Pujic P. et al. Candidatus Frankia datiscae Dg1, the actinobacterial microsymbiont of Datisca glomerata, expresses the canonical nod genes nodABC in symbiosis with its host plant. PLoS One. 2015;10:e0127630. doi: 10.1371/journal.pone.0127630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Normand P, Nguyen TV, Battenberg K, Berry AM, Vanden Heuvel B, Fernandez MP. et al. Proposal of 'Candidatus Frankia californiensis', the uncultured symbiont in nitrogen-fixing root nodules of a phylogenetically broad group of hosts endemic to western North America. Int J Syst Evol Microbiol. 2017;67:3706–3715. doi: 10.1099/ijsem.0.002147. [DOI] [PubMed] [Google Scholar]
- 19.Nguyen TV, Wibberg D, Vigil-Stenman T, Berckx F, Battenberg K, Demchenko KN. et al. Frankia-enriched metagenomes from the earliest diverging symbiotic Frankia cluster: they come in teams. Genome Biol Evol. 2019;11:2273–2291. doi: 10.1093/gbe/evz153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nouioui I, Montero-Calasanz MD, Ghodhbane-Gtari F, Rohde M, Tisa LS, Klenk HP. et al. Frankia discariae sp nov.: an infective and effective microsymbiont isolated from the root nodule of Discaria trinervis. Arch Microbiol. 2017;199:641–647. doi: 10.1007/s00203-017-1337-6. [DOI] [PubMed] [Google Scholar]
- 21.Gtari M, Ghodhbane-Gtari F, Nouioui I. Frankia soli sp. nov, an actinobacterium isolated from soil beneath Ceanothus jepsonii. Int J Syst Evol Micr0biol. 2020;70:1203–1209. doi: 10.1099/ijsem.0.003899. [DOI] [PubMed] [Google Scholar]
- 22.Nouioui I, Ghodhbane-Gtari F, Rhode M, Sangal V, Klenk HP, Gtari M. Frankia irregularis sp nov, an actinobacterium unable to nodulate its original host, Casuarina equisetifolia, but effectively nodulates members of the actinorhizal Rhamnales. Int J Syst Evol Microbiol. 2018;68:2883–2890. doi: 10.1099/ijsem.0.002914. [DOI] [PubMed] [Google Scholar]
- 23.Nouioui I, Ghodhbane-Gtari F, Montero-Calasanz MD, Rohde M, Tisa LS, Gtari M. et al. Frankia inefficax sp nov, an actinobacterial endophyte inducing ineffective, non nitrogen-fixing, root nodules on its actinorhizal host plants. Anton Leeuw. 2017;110:313–320. doi: 10.1007/s10482-016-0801-7. [DOI] [PubMed] [Google Scholar]
- 24.Nouioui I, Gueddou A, Ghodhbane-Gtari F, Rhode M, Gtari M, Klenk HP. Frankia asymbiotica sp nov, a non-infective actinobacterium isolated from Morella californica root nodule. Int J Syst Evol Microbiol. 2017;67:4897–4901. doi: 10.1099/ijsem.0.002153. [DOI] [PubMed] [Google Scholar]
- 25.Nouioui I, Ghodhbane-Gtari F, Klenk HP, Gtari M. Frankia saprophytica sp nov, an atypical, non-infective (Nod-) and non -nitrogen fixing (Fix-) actinobacterium isolated from Coriaria nepalensis root nodules. Int J Syst Evol Microbiol. 2018;68:1090–1095. doi: 10.1099/ijsem.0.002633. [DOI] [PubMed] [Google Scholar]
- 26.Ben Tekaya S, Ganesan AS, Guerra T, Dawson JO, Forstner MRJ, Hahn D. Sybr Green- and TaqMan-based quantitative PCR approaches allow assessment of the abundance and relative distribution of Frankia clusters in soils. Appl Environ Microb. 2017;83:e02833–16. doi: 10.1128/AEM.02833-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mirza MS, Hahn D, Akkermans ADL. Isolation and characterization of Frankia strains from Coriaria nepalensis. Syst Appl Microbiol. 1992;15:289–295. [Google Scholar]
- 28.Hahn D, Starrenburg MJC, Akkermans ADL. Variable compatibility of cloned Alnus glutinosa ecotypes against ineffective Frankia strains. Plant Soil. 1988;107:233–243. [Google Scholar]
- 29.Meesters TM, van Genesen ST, Akkermans ADL. Growth, acetylene reduction activity and localization of nitrogenase in relation to vesicle formation in Frankia strains Cc1.17 and Cp1.2. Arch Microbiol. 1985;143:137–142. [Google Scholar]
- 30.Mirza BS, Welsh A, Hahn D. Saprophytic growth of inoculated Frankia sp. in soil microcosms. FEMS Microbiol Ecol. 2007;62:280–289. doi: 10.1111/j.1574-6941.2007.00382.x. [DOI] [PubMed] [Google Scholar]
- 31.Samant S, Sha Q, Iyer A, Dhabekar P, Hahn D. Quantification of Frankia in soils using SYBR Green based qPCR. Syst Appl Microbiol. 2012;35:191–197. doi: 10.1016/j.syapm.2011.12.004. [DOI] [PubMed] [Google Scholar]
- 32.Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i90. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A. Using SPAdes de novo assembler. Curr Protoc Bioinformatics. 2020;70:e102. doi: 10.1002/cpbi.102. [DOI] [PubMed] [Google Scholar]
- 34.Eren AM, Kiefl E, Shaiber A, Veseli I, Miller SE, Schechter MS. et al. Community-led, integrated, reproducible multi-omics with anvi'o. Nature Microbiol. 2021;6:3–6. doi: 10.1038/s41564-020-00834-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pruesse E, Peplies J, Glöckner FO. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 2012;28:1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stecher G, Tamura K, Kumar S. Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol Biol Evol. 2020;37:1237–1239. doi: 10.1093/molbev/msz312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nature Communications. 2018;9:5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 39.Aramaki T, Blanc-Mathieu R, Endo H, Ohkubo K, Kanehisa M, Goto S. et al. KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics. 2019;36:2251–2252. doi: 10.1093/bioinformatics/btz859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY. et al. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucl Acids Res. 2019;47:W81–W7. doi: 10.1093/nar/gkz310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pujic P, Bolotin A, Fournier P, Sorokin A, Lapidus A, Richau KH. et al. Genome sequence of the atypical symbiotic Frankia R43 strain, a nitrogen-fixing and hydrogen-producing actinobacterium. Genome Announc. 2015;3:e01387–15. doi: 10.1128/genomeA.01387-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Normand P, Lapierre P, Tisa LS, Gogarten JP, Alloisio N, Bagnarol E. et al. Genome characteristics of facultatively symbiotic Frankia sp strains reflect host range and host plant biogeography. Genome Res. 2007;17:7–15. doi: 10.1101/gr.5798407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Samant SS, Dawson JO, Hahn D. Growth responses of indigenous Frankia populations to edaphic factors in actinorhizal rhizospheres. System Appl Microbiol. 2015;38:501–505. doi: 10.1016/j.syapm.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 44.Samant S, Dawson JO, Hahn D. Growth responses of introduced Frankia strains to edaphic factors. Plant Soil. 2016;400:123–132. [Google Scholar]
- 45.Samant S, Huo T, Dawson JO, Hahn D. Abundance and relative distribution of Frankia host infection groups under actinorhizal Alnus glutinosa and non-actinorhizal Betula nigra trees. Microb Ecol. 2016;71:473–481. doi: 10.1007/s00248-015-0643-2. [DOI] [PubMed] [Google Scholar]
- 46.Samant S, Amann RI, Hahn D. Evaluation of the 23S rRNA gene as target for qPCR based quantification of Frankia in soils. Syst Appl Microbiol. 2014;37:229–234. doi: 10.1016/j.syapm.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 47.Rönkkö R, Smolander A, Nurmiaho-Lassila EL, Haahtela K. Frankia in the rhizosphere of nonhost plants: A comparison with root-associated nitrogen-fixing Enterobacter, Klebsiella and Pseudomonas. Plant Soil. 1993;153:85–95. [Google Scholar]
- 48.Lechevalier MP, Ruan JS. Physiology and chemical diversity of Frankia spp. isolated from nodules of Comptonia peregrina (L.) Coult. and Ceanothus americanus L. Plant Soil. 1984;78:15–22. [Google Scholar]
- 49.Mirza MS, Janse JD, Hahn D, Akkermans ADL. Identification of atypical Frankia strains by fatty acid analysis. FEMS Microbiol Lett. 1991;83:91–97. [Google Scholar]
- 50.Baker D, Newcomb W, Torrey JG. Characterization of an ineffective actinorhizal microsymbiont, Frankia sp. EuI1 (Actinomycetales) Can J Microbiol. 1980;26:1072–1089. doi: 10.1139/m80-180. [DOI] [PubMed] [Google Scholar]
- 51.Wolters DJ, Akkermans ADL, Van Dijk C. Ineffective Frankia strains in wet stands of Alnus glutinosa L. Gaertn. in the Netherlands. Soil Biol Biochem. 1997;29:1707–1712. [Google Scholar]
- 52.Wolters DJ, Van Dijk C, Zoetendal EG, Akkermans AD. Phylogenetic characterization of ineffective Frankia in Alnus glutinosa (L.) Gaertn. nodules from wetland soil inoculants. Mol Ecol. 1997;6:971–981. doi: 10.1046/j.1365-294x.1997.00265.x. [DOI] [PubMed] [Google Scholar]
- 53.Van Dijk C, Sluimer A. Resistance to an ineffective Frankia strain type in Alnus glutinosa (L.) Gaertn. New Phytol. 1994;128:497–504. doi: 10.1111/j.1469-8137.1994.tb02996.x. [DOI] [PubMed] [Google Scholar]
- 54.Vemulapally S, Guerra T, Hahn D. Localization of typical and atypical Frankia isolates from Casuarina sp. in nodules formed on Casuarina equisetifolia. Plant Soil. 2019;435:385–393. [Google Scholar]
- 55.Xu M, Gao X, Chen J, Yin Z, Feng H, Huang L. The feruloyl esterase genes are required for full pathogenicity of the apple tree canker pathogen Valsa mali. Mol Plant Pathol. 2018;19:1353–1363. doi: 10.1111/mpp.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kubicek CP, Starr TL, Glass NL. Plant cell wall-degrading enzymes and their secretion in plant-pathogenic fungi. Annu Rev Phytopathol. 2014;52:427–451. doi: 10.1146/annurev-phyto-102313-045831. [DOI] [PubMed] [Google Scholar]
- 57.Frey AD, Kallio PT. Bacterial hemoglobins and flavohemoglobins: versatile proteins and their impact on microbiology and biotechnology. FEMS Microbiol Rev. 2003;27:525–545. doi: 10.1016/S0168-6445(03)00056-1. [DOI] [PubMed] [Google Scholar]
- 58.Udwary DW, Gontang EA, Jones AC, Jones CS, Schultz AW, Winter JM. et al. Significant natural product biosynthetic potential of actinorhizal symbionts of the genus Frankia, as revealed by comparative genomic and proteomic analyses. Appl Environ Microbiol. 2011;77:3617–3625. doi: 10.1128/AEM.00038-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables.