Abstract
The c-Myc oncoprotein is a transcription factor which is a critical regulator of cellular proliferation. Deregulated expression of c-Myc is associated with many human cancers, including Burkitt's lymphoma. The c-Myc protein is normally degraded very rapidly with a half-life of 20 to 30 min. Here we demonstrate that proteolysis of c-Myc in vivo is mediated by the ubiquitin-proteasome pathway. Inhibition of proteasome activity blocks c-Myc degradation, and c-Myc is a substrate for ubiquitination in vivo. Furthermore, an increase in c-Myc stability occurs in mitotic cells and is associated with inhibited c-Myc ubiquitination. Deletion analysis was used to identify regions of the c-Myc protein which are required for rapid proteolysis. We found that a centrally located PEST sequence, amino acids 226 to 270, is necessary for rapid c-Myc degradation, but not for ubiquitination. Also, N-terminal sequences, located within the first 158 amino acids of c-Myc, are necessary for both efficient c-Myc ubiquitination and subsequent degradation. We found that c-Myc is significantly stabilized (two- to sixfold) in many Burkitt's lymphoma-derived cell lines, suggesting that aberrant c-Myc proteolysis may play a role in the pathogenesis of Burkitt's lymphoma. Finally, mutation of Thr-58, a major phosphorylation site in c-Myc and a mutational hot spot in Burkitt's lymphoma, increases c-Myc stability; however, mutation of c-Myc is not essential for stabilization in Burkitt's lymphoma cells.
The c-myc proto-oncogene belongs to a family of related genes which includes L-myc, N-myc, s-myc, and B-myc (reviewed in reference 31). The c-myc gene encodes a short-lived nuclear phosphoprotein which is a central regulator of cell growth. Expression of c-myc is induced by mitogenic signals and is suppressed by growth-inhibitory signals. Constitutive c-myc expression inhibits exit from the cell cycle and prevents differentiation (31, 46). Moreover, c-Myc activity is sufficient to drive quiescent cells into the cell cycle in the absence of growth factors, but also induces apoptosis when survival factors are missing (17, 19, 28). Homozygous inactivation of c-myc in fibroblasts severely diminishes their rate of proliferation by prolonging both the G1 and G2 phases of the cell cycle (47). Thus, c-Myc is a potent and critical promoter of cellular proliferation. Consistent with this fact, deregulated c-myc expression is very common in cancer. Activation of c-myc by proviral insertion, gene amplification, and chromosomal translocation has been described in a variety of tumors from several species, including humans. Furthermore, overexpression of c-myc in transgenic mice results in tumor development (46, 63).
The c-Myc protein is a transcription factor of the basic helix-loop-helix–leucine zipper (bHLH-LZ) class. Two regions of c-Myc are critical for biological function—the N-terminal transactivation/repression domain and the C-terminal bHLH-LZ DNA binding domain. c-Myc can activate the transcription of specific E-box-containing genes as a heterodimeric complex with its partner protein, Max (23, 31). Additionally, c-Myc can specifically repress gene transcription (13, 20). Although the precise mechanism remains poorly understood, c-Myc is thought to exert its biological effects by regulating the expression of target genes involved in cellular proliferation (16, 20).
The expression of c-myc is tightly regulated at many different levels. In addition to transcriptional initiation and attenuation, c-myc expression is regulated posttranscriptionally at the levels of mRNA stability, translation, and protein stability (46, 63). Indeed, a remarkable feature of the c-Myc protein is its very short half-life, usually 20 to 30 min (27, 36, 41, 51, 73). A number of short-lived transcription factors, including c-Myb (8), c-Jun (69), c-Fos (66), p53 (45), and E2F (10, 29, 33), among others, are degraded by the ubiquitin-proteasome pathway. In this pathway, ubiquitin polypeptides are covalently attached to lysine residues of the target protein by the concerted action of at least three enzymes: the ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a ubiquitin-protein ligase (E3). Upon multiubiquitination, substrate proteins are targeted for rapid proteolysis by the 26S proteasome (11, 12, 71).
In this report, we present an analysis of c-Myc proteolysis. We show that c-Myc proteolysis is mediated by the ubiquitin-proteasome pathway in vivo. We also demonstrate that two regions of the c-Myc protein are important for rapid degradation, a central PEST sequence unnecessary for c-Myc ubiquitination, and an N-terminal region required for efficient ubiquitination. Furthermore, we show that c-Myc is stabilized in a number of Burkitt's lymphoma cell lines, suggesting that defective c-Myc proteolysis by the ubiquitin pathway may play a role in lymphomagenesis.
MATERIALS AND METHODS
Cell lines.
COS-7 cells were obtained from S. Brandt (Vanderbilt University, Nashville, Tenn.). The human colon carcinoma cell line COLO320 was obtained from M. Groudine (Fred Hutchinson Cancer Research Center, Seattle, Wash.). NIH 3T3 cells were obtained from American Type Culture Collection (ATCC). COS-7, COLO320, and NIH 3T3 cells were maintained in Dulbecco's modified Eagle medium (DMEM; GibcoBRL) containing 10% calf serum (Hyclone). The avian bursal lymphoma cell line Bk3A was obtained from M. Linial (Fred Hutchinson Cancer Research Center) and was maintained in DMEM containing 10% tryptose phosphate broth (GibcoBRL), 5% calf serum (Hyclone), and 1% heat-inactivated chick serum (GibcoBRL). The Burkitt's lymphoma cell lines CA46, Daudi, JD38, KK124, JLPc119, Namalwa, and p3HR1 were obtained from M. Zajac-Kaye (National Cancer Institute, Bethesda, Md.); DW6 (also called B7CL) and ST486 (also called B3CL) were obtained from C. Dang (Johns Hopkins University, Baltimore, Md.); Ramos and Walker were obtained from R. Eisenman (Fred Hutchinson Cancer Research Center); Jijoye was obtained from ATCC. All Burkitt's cell lines were maintained in RPMI 1640 medium (GibcoBRL) containing 10% fetal calf serum (Hyclone). All cell lines were maintained at 37°C in a humidified 5% CO2 atmosphere.
Cell treatments.
The specific protease inhibitors ALLN (N-acetyl-l-leucinyl-l-leucinyl-l-norleucinal; Sigma), ALLM (N-acetyl-l-leucinyl-l-leucinyl-methioninal; Sigma), E-64 [trans-epoxysuccinyl-l-leucylamido-(4-guanidino)butane; Boehringer Mannheim], and lactacystin (BIOMOL) were dissolved in dimethyl sulfoxide (DMSO). COS-7 or COLO320 cells at a density of ∼5 × 106 cells per 10-cm-diameter dish were treated with the indicated protease inhibitor at the indicated concentration for 2 h prior to pulse-chase analysis. The final concentration of DMSO in the tissue culture medium was 0.5%. To examine the accumulation of ubiquitin conjugates, Bk3A cells at a density of ∼5 × 105 cells/ml were treated with ALLN at 100 μM for the indicated amount of time prior to Western blot analysis.
To block cells in mitosis, Bk3A cells at a density of ∼5 × 105 cells/ml were treated with nocodazole (Sigma) at 500 ng/ml for 24 h. Cells were then treated for the indicated amount of time with cycloheximide (Sigma) at 50 ng/ml to examine protein turnover, or with ALLN at 100 μM, prior to Western blot analysis.
Plasmids.
The simian virus 40 (SV40) promoter-driven expression plasmids containing wild-type human c-myc (pSV-myc) or the indicated deletion mutants (67) contain the SV40 origin of replication allowing for overexpression in COS cells and were a gift from C. Dang (Johns Hopkins University). The construction of the cytomegalovirus (CMV) promoter-driven murine c-myc expression plasmid, CMV-c-Myc2, has been described previously (26). The murine c-myc cDNA has a mutation at the CUG upstream initiation site to prevent synthesis of the c-Myc1 protein. In addition, the murine c-Myc penultimate glycine is mutated to arginine which allows for selective immunoprecipitation of the exogenously expressed murine c-Myc protein with our avian-specific antibody (anti-av-Myc12C) from COS, murine, and human cells (26). Single-stranded DNA from CMV-Myc2 was used to create the deletion or point mutant constructs described in the text by site-directed mutagenesis according to the method of Kunkel (37). All mutations were verified by sequencing. To create the CMV-driven c-MycS expression plasmids, the BbvII-XbaI fragment from CMV-Myc2, or a deletion-mutated derivative, was subcloned into pcDNA3 (Invitrogen), which results in removal of the c-Myc2 initiation site. The CMV-hemagglutinin (HA) epitope-tagged ubiquitin expression plasmid (pMT123) has been described previously (69) and was a gift from G. Kato (Johns Hopkins University).
Cell transfections.
For transfection of COS-7 cells, cells were plated at a density of 2.5 × 106 per 10-cm-diameter dish the day prior to transfection and were transfected with the indicated plasmids for 4 to 6 h by the calcium phosphate method as previously described (65). For transfection of COLO320 cells, cells were plated at a density of 4 × 106 cells per 10-cm-diameter dish the day prior to transfection and were transfected for 6 h by liposome-mediated transfection (Lipofectamine; Life Technologies) according to the manufacturer's instructions. For transfection of NIH 3T3 cells, cells were plated at a density of 2.5 × 106 per 10-cm-diameter dish the day prior to transfection and were transfected with the indicated plasmids as described for COLO320 cells. Cells were harvested approximately 48 h posttransfection for analysis of c-Myc. CA46 cells were transfected with 15 μg of CMV-Myc2 by using DMRIE-C reagent (Life Technologies) according to the manufacturer's instructions followed by selection in medium containing 800 μg of G418 per ml (Geneticin; GibcoBRL) for 3 weeks.
Antibodies.
The affinity-purified rabbit polyclonal c-Myc antibodies anti-MycFL (against full-length murine c-Myc) and avian-specific anti-av-Myc12C (against C-terminal avian c-Myc peptide) were generated as previously described (65). The c-Myc mouse monoclonal antibody C-33 was purchased from Santa Cruz Biotechnology. Mouse monoclonal anti-HA antibody (12CA5) was obtained from A. Reynolds (Vanderbilt University).
Immunoprecipitation.
Radiolabeled cells were lysed in cold Ab lysis buffer (25) with 20 U of aprotinin per ml followed by sonication. The amount of radiolabel incorporated into cellular proteins was determined by precipitation with 10% trichloroacetic acid (TCA). Equivalent amounts of TCA-precipitable counts from each cellular lysate were adjusted to equal volumes with Ab buffer, precleared with Staphylococcus aureus membranes (Pansorbin; Calbiochem), and clarified by centrifugation. Lysates were then incubated with 5 μg of the indicated c-Myc antibody for 1 to 2 h at 4°C, and the immune complexes were precipitated with Pansorbin for 20 min. The immunoprecipitates were washed three times with radioimmunoprecipitation assay (RIPA) buffer (25) and incubated at 95°C for 3 min in Laemmli sample buffer. Samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with 10% polyacrylamide gels followed by autoradiography. High-range protein molecular weight markers (GibcoBRL) were used as standards in each SDS-PAGE gel.
Western blot analysis.
Cells were lysed in RIPA buffer with 1 mM phenylmethylsulfonyl fluoride (PMSF), 20 U of aprotinin per ml, and 5 mM N-ethylmaleimide (NEM) followed by sonication. Protein concentrations of cellular lysates were determined using the DC protein assay (Bio-Rad). Equal amounts of lysate (100 μg of total protein) were boiled for 3 min in Laemmli sample buffer and subjected to 10% polyacrylamide SDS-PAGE. High-range molecular weight markers (Rainbow markers; Amersham) were used as standards in each SDS-PAGE gel. Proteins were then electrophoretically transferred to nitrocellulose membranes (Protran; Schleicher & Schuell) followed by blocking in milk buffer (3% milk in Tris-buffered saline [TBS]; pH 7.5) and incubation with the indicated primary antibody in milk buffer overnight at 4°C. The membranes were washed with TBS (pH 7.5), and proteins were detected by incubation with horseradish peroxidase-conjugated donkey anti-rabbit immunoglobulin G (IgG) (Jackson Laboratories) or goat anti-mouse IgG (Amersham) secondary antibodies, as appropriate, followed by enhanced chemiluminescence (ECL; Amersham) according to the manufacturer's instructions.
Pulse-chase experiments.
COS-7 or COLO320 cells were plated at a density of 2 × 106 or 4 × 106 cells per 10-cm-diameter dish, respectively, the day prior to pulse-chase analysis. Transiently transfected COS-7 or NIH 3T3 cells were split 1:4 or 1:5 into 10-cm-diameter dishes 24 h after transfection and were subjected to pulse-chase analysis 24 h later. For the pulse-chase, the cells were washed with phosphate-buffered saline (PBS) and labeled with 200 to 500 μCi of [35S]methionine/cysteine (Trans35S-label; ICN) per plate in methionine/cysteine-free DMEM (GibcoBRL) at 37°C for 10 min. After labeling, the cells were washed with PBS and chased with complete DMEM containing 10% calf serum at 37°C for the indicated amount of time. Where indicated, specific protease inhibitors were included in the chase medium. For the Burkitt's lymphoma cells, the cells were grown to a density of ∼5 × 105 cells/ml. Cells (4 × 107) were labeled with 1 mCi of Trans35S-label and were chased in RPMI 1640 medium containing 10% fetal calf serum for the indicated amount of time. A total of 107 cells were collected at each time point. Cells were subsequently subjected to immunoprecipitation analysis as described above. Band intensities were measured by scanning densitometry (Un-Scan-It; Silk Scientific) of autoradiographs.
In vivo ubiquitination assay.
COS-7 or COLO320 cells were transiently transfected with 4 μg of CMV-ubiquitin-HA and/or 2 μg of the indicated c-Myc expression plasmid, and the cells were split 1:4 into 10-cm-diameter dishes 24 h posttransfection. Where indicated, the cells were treated with ALLN (100 μM) as described in the text. The cells were lysed in Ab lysis buffer with 1 mM PMSF, 20 U of aprotinin per ml, and 5 mM NEM followed by sonication. Cellular lysates were equalized for total protein concentration, and immunoprecipitation was performed as described above, except that lysates were incubated with 2 μg of the indicated c-Myc antibody overnight at 4°C. Immunoprecipitates were subjected to 10% polyacrylamide SDS-PAGE, transferred to nitrocellulose, and blotted with 12CA5 at 1 to 5 μg/ml, and protein-ubiquitin conjugates were detected by ECL as described above. In some cases, blots were “stripped” as suggested by the manufacturer (Amersham) and reprobed with another antibody as indicated.
RESULTS
Proteolysis of c-Myc by the ubiquitin-proteasome pathway.
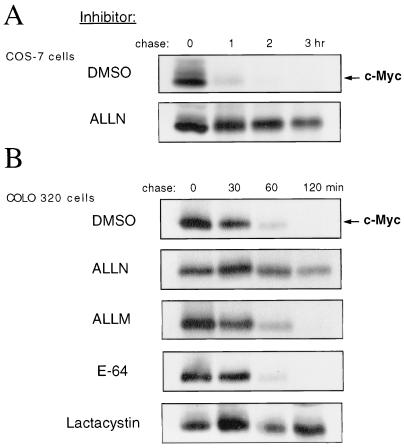
The c-Myc protein has a very short half-life, usually ∼20 to 30 min (27, 36, 41, 51, 73), relative to other cellular proteins. Because the ubiquitin-proteasome pathway is responsible for the degradation of many short-lived transcription factors, we sought to determine whether this proteolytic pathway is involved in the rapid degradation of c-Myc in vivo. We first investigated whether c-Myc degradation is dependent on the activity of the 26S proteasome. COS-7 cells were treated with ALLN, a peptide aldehyde that inhibits the proteasome (53), or with the solvent DMSO as a control. To determine the rate of c-Myc protein turnover in these cells, pulse-chase experiments were performed followed by immunoprecipitation analysis of c-Myc. In cells treated with ALLN, turnover of endogenous c-Myc was markedly inhibited (Fig. 1A). Similar results were observed with several different cell lines, including COLO320 (Fig. 1B), HeLa, and the avian bursal lymphoma cell lines 243 and Bk3A (data not shown). Since ALLN not only is a potent inhibitor of the proteasome but also is inhibitory to other proteases, including calpains (72), we examined the effects of additional protease inhibitors on c-Myc turnover in COLO320 cells by pulse-chase analysis. As shown in Fig. 1B, turnover of c-Myc was blocked in cells treated with either ALLN or with lactacystin, an inhibitor specific for the proteasome (21). Conversely, neither ALLM, a peptide aldehyde which is more inhibitory to calpain than to the proteasome (53), nor E-64 [trans-epoxysuccinyl-l-leucylamido-(4-guanidino)butane], an inhibitor of lysosomal cysteine proteases (6), had a significant effect on c-Myc degradation (Fig. 1B). These results suggest that proteolysis of c-Myc is proteasome dependent.
FIG. 1.
Inhibition of proteasome activity inhibits c-Myc turnover. (A) COS-7 cells were treated with either DMSO or ALLN (100 μM) for 2 h and pulse-chased in the presence of DMSO or ALLN. The cells were labeled with [35S]methionine/cysteine for 10 min (pulse) and incubated in unlabeled complete medium for the indicated times (chase). Cell lysates were prepared, equalized for total labeled protein, and c-Myc was immunoprecipitated with anti-MycFL, followed by analysis by SDS-PAGE and autoradiography. (B) COLO320 cells were treated with DMSO, ALLN (100 μM), ALLM (100 μM), E-64 (50 μM), or lactacystin (50 μM) for 2 h, pulse-chased in the presence of the respective compound, and analyzed as described for panel A. Lactacystin was not included in the chase medium because it is an irreversible inhibitor of the proteasome.
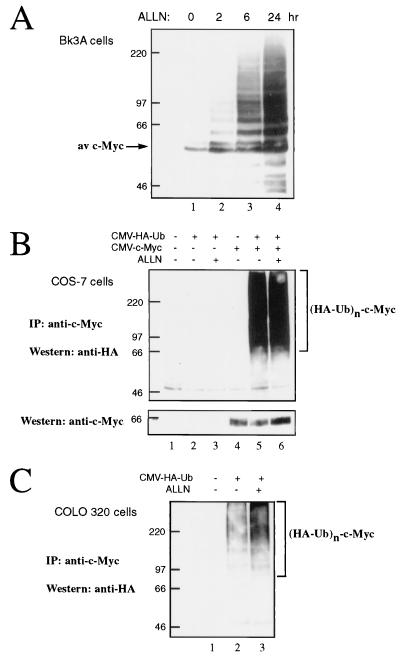
Substrates of the ubiquitin-proteasome pathway are conjugated to ubiquitin, and the formation of a multiubiquitin chain targets them for proteasomal degradation. The hypothesis that c-Myc is degraded by this pathway predicts that prolonged inhibition of the proteasome would lead to the accumulation of multiubiquitinated forms of c-Myc. To test this prediction, Bk3A cells were treated with the proteasome inhibitor ALLN for up to 24 h. Western blot analysis with anti-c-Myc revealed the accumulation of a ladder of upper-molecular-weight c-Myc forms (Fig. 2A) indicative of ubiquitination. These slower-migrating c-Myc forms were evident after 2 h of treatment (lane 2) and accumulated to high levels after 6 and 24 h of treatment with ALLN (lanes 3 and 4).
FIG. 2.
c-Myc is ubiquitinated in vivo. (A) Bk3A cells were left untreated or treated with ALLN (100 μM) for 2, 6, or 24 h as indicated. Cell lysates were prepared and analyzed by Western blotting using anti-av-Myc12C. (B) COS-7 cells were transiently transfected with expression plasmids encoding murine c-Myc (CMV–c-Myc2 [2 μg]) and HA-tagged ubiquitin (CMV-HA-Ub [4 μg]) as indicated. Forty-six hours later, cells were left untreated or treated with ALLN (100 μM) for 2 h as indicated. Cell lysates were subjected to immunoprecipitation (IP) with anti-av-Myc12C and subsequently analyzed by Western blotting with anti-HA antibody 12CA5 to detect c-Myc–HA–ubiquitin conjugates. The blot was stripped and reprobed with the monoclonal antibody C33 to detect c-Myc expression (lower panel). (C) COLO320 cells were transiently transfected with CMV-HA-Ub (4 μg) as indicated. Forty-six hours later, cells were left untreated or treated with ALLN (100 μM) for 2 h as indicated. Cell lysates were subjected to immunoprecipitation with anti-MycFL, followed by Western blot analysis with 12CA5 to detect c-Myc–HA–ubiquitin conjugates.
To directly assess whether c-Myc is a substrate for ubiquitination in vivo, we used an expression plasmid encoding HA epitope-tagged ubiquitin. This plasmid was transiently transfected into COS-7 cells together with an expression plasmid encoding murine c-Myc. Forty-six hours after transfection, cells were left untreated or treated with the proteasome inhibitor ALLN for 2 h. Cell lysates were subjected to immunoprecipitation with a c-Myc antibody specific for the exogenously expressed c-Myc followed by Western blotting for the HA epitope. As shown in Fig. 2B, an HA-immunoreactive upper-molecular-weight ladder of bands was observed in c-Myc immunoprecipitates derived from cells cotransfected with HA-ubiquitin and c-Myc (lanes 5 and 6), but not from mock-transfected cells (lane 1) or cells transfected with either HA-ubiquitin (lanes 2 and 3) or c-Myc (lane 4) alone. The HA-reactive species migrated above where unmodified c-Myc migrated at 65 kDa (see Fig. 2B, lower panel). These c-Myc-ubiquitin conjugates were easily detectable even in the absence of proteasome inhibition (Fig. 2B, lane 5).
To verify that the ubiquitination of c-Myc is not merely a consequence of exogenous overexpression, we sought to confirm that endogenous c-Myc is also a substrate for ubiquitination. Thus, COLO320 cells were transiently transfected with HA-ubiquitin and cells were treated as described above. The endogenous human c-Myc was immunoprecipitated with anti-c-Myc followed by Western blot analysis with anti-HA. As shown in Fig. 2C, HA-reactive upper-molecular-weight species were observed in c-Myc immunoprecipitates from cells transfected with HA-ubiquitin (lanes 2 and 3), but not from mock-transfected cells (lane 1). As expected, c-Myc-ubiquitin conjugates were more readily detectable from cells treated with the proteasome inhibitor ALLN than from untreated cells (compare lanes 2 and 3). Taken together, these results demonstrate that c-Myc is ubiquitinated and that c-Myc degradation occurs via the ubiquitin-proteasome pathway.
c-Myc regions required for rapid proteolysis and ubiquitination.
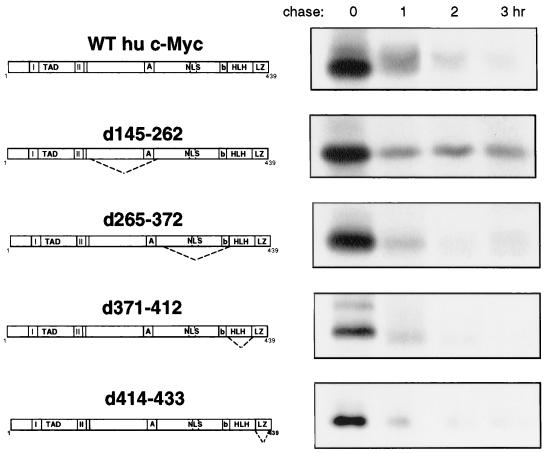
We next wanted to determine what regions of the c-Myc protein are responsible for targeting c-Myc for rapid degradation by the ubiquitin-proteasome pathway in vivo. To this end, we utilized a large panel of expression plasmids encoding c-Myc with various deletion mutations throughout the c-Myc protein. We began by examining the requirement of central and C-terminal regions for c-Myc proteolysis. Plasmids encoding wild-type (WT) human c-Myc or various c-Myc deletion mutants (diagrammed in Fig. 3) were transiently transfected into COS-7 cells, and the rate of protein turnover was analyzed in pulse-chase experiments. Deletion of various C-terminal regions spanning amino acids 265 to 433 had no significant effect on c-Myc turnover (Fig. 3). Smaller deletion mutations within this region (d265–317 and d312–368) were also tested, and likewise, c-Myc turnover was unaffected (data not shown). The C-terminal portion of c-Myc contains the basic region (b; amino acids 355 to 368), HLH motif (amino acids 368 to 410), and LZ domain (amino acids 411 to 439), which are required for dimerization with Max and DNA binding. Also, the c-Myc nuclear localization signals are contained within this region, the main nuclear localization signal being located at amino acids 320 to 328 (31). Because removal of these sequences did not inhibit turnover of c-Myc, it appears that dimerization with Max, binding to DNA, or nuclear localization is not required for rapid c-Myc degradation. One mutant, c-Myc d145–262, consistently showed partial stabilization in pulse-chase experiments (Fig. 3). Although the central region of c-Myc is dispensable for c-Myc function (67), this observation suggested it may contain sequences which play a role in c-Myc proteolysis. Indeed, the 145 to 262 deletion removes a conserved central acidic region (A; amino acids 242 to 261) and most of a PEST sequence in c-Myc.
FIG. 3.
Deletion analysis of c-Myc protein turnover. COS-7 cells were transiently transfected with 2 μg of an expression plasmid encoding wild-type human (WT hu) c-Myc (pSV-myc) or c-Myc with the indicated deletion mutation. Forty-eight hours after transfection, cells were subjected to pulse-chase analysis as described in the legend to Fig. 1A, and c-Myc was immunoprecipitated with anti-MycFL.
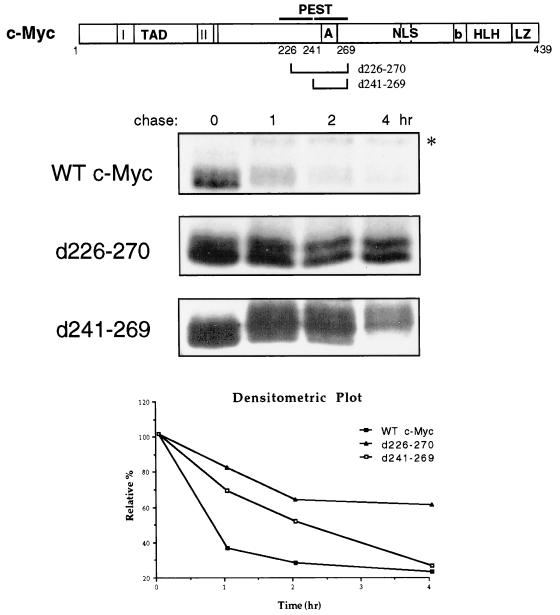
PEST sequences are sequences enriched in proline (P), glutamic acid (E), serine (S), threonine (T), and aspartic acid (D), which occur with high frequency in short-lived proteins (52, 54). c-Myc contains PEST sequences spanning amino acids 207 to 269 (diagrammed in Fig. 4) with the “highest score” PEST sequence being amino acids 241 to 269 (54). To investigate the importance of this PEST region for c-Myc degradation, PEST deletion mutants of murine c-Myc (d241–269 and a slightly larger deletion, d226–270) were analyzed in pulse-chase experiments along with wild-type c-Myc in COS-7 cells. As shown in Fig. 4, the d241–269 deletion mutation partially stabilized whereas d226–270 substantially stabilized c-Myc. In contrast, a deletion of amino acids 226 to 241 had no effect on c-Myc turnover (data not shown). These results suggest that a PEST sequence specified within amino acids 226 to 270 is required for efficient c-Myc proteolysis and removal of this entire region is necessary to confer a substantial increase in protein stability. It is worth noting that overexpressed c-Myc was generally somewhat more stable (on average, half-life of ∼40 min) relative to the turnover of endogenous c-Myc in this cell line (half-life, 20 to 30 min [see Fig. 1A]). This increased stability is partially due to the fact that during the chase, a small proportion of c-Myc shifts to a larger stable form (marked with an asterisk in Fig. 4) which migrates approximately 6 kDa higher than unmodified c-Myc, and we do not yet know what this apparent modification is. We believe the slightly prolonged half-life of c-Myc is likely a consequence of the high level of exogenous c-Myc expression in these experiments. Nonetheless, c-Myc was still degraded relatively rapidly, and our ability to discern an increase in stability, as with c-Myc d226–270 (half-life, ∼5 h), was unaffected.
FIG. 4.
Deletion of a PEST sequence stabilizes c-Myc. COS-7 cells were transiently transfected with 2 μg of an expression plasmid encoding wild-type (WT) murine c-Myc (CMV–c-Myc2) or c-Myc with the indicated deletion mutation. Forty-eight hours after transfection, the cells were subjected to pulse-chase analysis as described in the legend to Fig. 1A followed by immunoprecipitation of exogenous c-Myc proteins with anti-av-Myc12C. The data obtained from densitometric analysis of the experiment shown above were expressed as the relative percentage of the amount of c-Myc protein at the zero time point and plotted as a function of time.
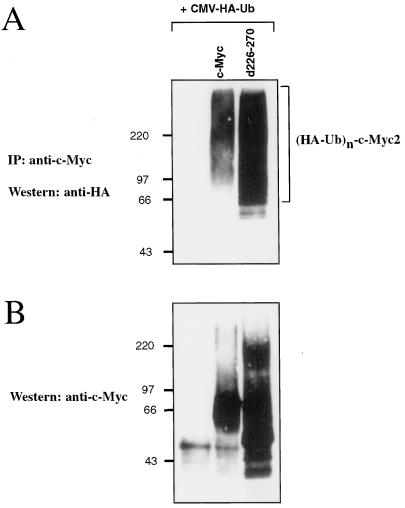
Having demonstrated that c-Myc is degraded by the ubiquitin-proteasome pathway, we reasoned that the inability of c-Myc d226–270 to be rapidly degraded may be due to an inability of this mutant protein to be ubiquitinated. To test this hypothesis, c-Myc d226–270, or wild-type c-Myc, was cotransfected into COS-7 cells with HA-ubiquitin, exogenous c-Myc proteins were specifically immunoprecipitated, and immunoprecipitates were analyzed by Western blotting for HA. Unexpectedly, c-Myc d226–270 consistently displayed a higher level of ubiquitination relative to wild-type c-Myc (Fig. 5A, compare lanes 2 and 3). When c-Myc protein expression was analyzed by Western blotting for c-Myc, we observed that upper-molecular-weight forms of c-Myc d226–270, indicative of ubiquitin conjugates, had accumulated (Fig. 5B, lane 3). These results indicate that the PEST sequence is necessary for rapid c-Myc proteolysis at a step which comes after ubiquitination. A block to c-Myc degradation by the proteasome, but not to ubiquitination, would explain the accumulation of ubiquitinated forms of c-Myc d226–270.
FIG. 5.
PEST sequence of c-Myc is not required for ubiquitination. (A) COS-7 cells were transiently transfected with 4 μg of a plasmid encoding HA-tagged ubiquitin (CMV-HA-Ub) and 2 μg of a plasmid encoding wild-type murine c-Myc or the c-Myc PEST deletion mutant d226–270 as indicated. Forty-eight hours after transfection, cell lysates were prepared. Exogenous c-Myc proteins were immunoprecipitated with anti-av-Myc12C and subsequently analyzed by Western blotting using anti-HA antibody 12CA5 to detect c-Myc–HA–ubiquitin conjugates. (B) The blot shown in panel A was stripped and reprobed with anti-av-Myc12C to examine c-Myc expression.
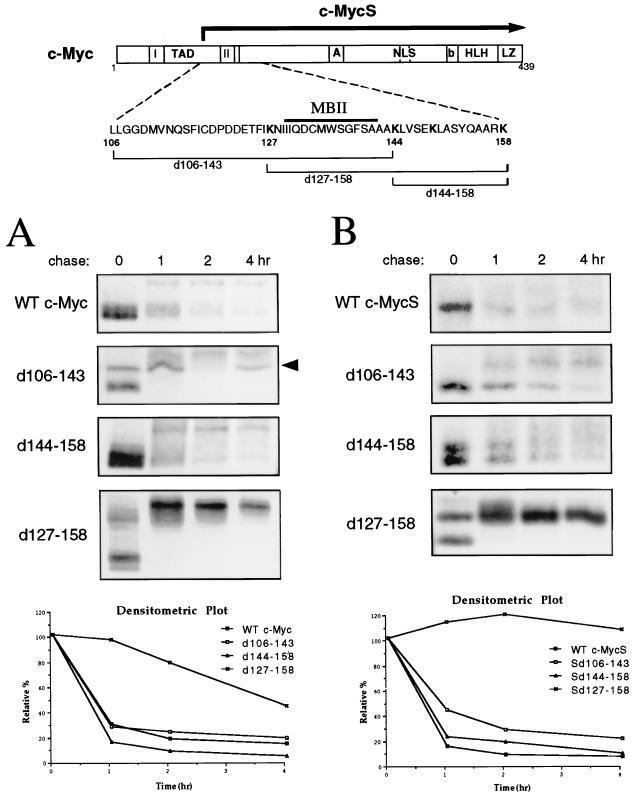
We next examined the requirement of N-terminal regions for rapid c-Myc proteolysis in search of a region or regions which might target c-Myc for ubiquitination. The N terminus contains the c-Myc transactivation domain (TAD; amino acids 1 to 143), which includes two highly conserved regions known as Myc homology boxes, MBI and MBII, the latter of which is essential for c-Myc biological activity (31, 75). We have previously shown that c-MycS, a downstream-initiated alternative translational form of c-Myc which lacks the first approximately 100 amino acids compared with full-length c-Myc, is rapidly degraded in vivo (65). In addition, c-MycS is ubiquitinated to a similar extent compared with c-Myc (data not shown). This suggests that the N-terminal 100 amino acids of c-Myc are not required for rapid proteolysis. Also, we have shown that B-Myc, a c-Myc homologue that is highly homologous (68% identity) to the N terminus of c-Myc (2, 3), has a short in vivo half-life (M. A. Gregory, Q. Xiao, G. A. Cornwall, B. Lutterbach, and S. R. Hann, unpublished data). The only region c-Myc, c-MycS, and B-Myc have in common is a sequence (amino acids 110 to 158; shown in the diagram in Fig. 6) which encompasses MBII (amino acids 130 to 142 in murine c-Myc). We thus reasoned that this region of c-Myc may contain a necessary degradation signal. To test this idea, deletion mutants of murine c-Myc, d106–143, d127–158, and d144–158, or wild-type c-Myc, were transfected into COS-7 cells and examined by pulse-chase analysis. Only the deletion of amino acids 127 to 158 significantly affected the stability of c-Myc (Fig. 6A). Similar results were obtained when these deletion mutants were expressed in HeLa cells (data not shown). c-Myc d127–158 displayed considerable stabilization (half-life, 2.5 h) relative to wild-type c-Myc (half-life, 35 min); unexpectedly, however, it completely shifted to a slower-migrating form (∼12 kDa larger) during the chase. This large shift in mobility appears to be due to phosphorylation because it can be removed by treatment with phosphatase (data not shown). Although c-Myc d127–158 was more stable, it was still mostly degraded over the course of the pulse-chase (Fig. 6A), and none of the mutants tested in this assay showed a demonstrable difference in its ability to be ubiquitinated compared with wild-type c-Myc (data not shown).
FIG. 6.
Deletion of N-terminal sequences stabilizes c-Myc. (A) COS-7 cells were transiently transfected with 2 μg of an expression plasmid encoding wild-type (WT) murine c-Myc or the indicated c-Myc deletion mutant. Forty-eight hours after transfection, the cells were subjected to pulse-chase analysis as described in the legend to Fig. 1A, followed by immunoprecipitation of exogenous c-Myc proteins with anti-av-Myc12C. The arrow indicates a nonspecific background band. The data obtained from densitometric analysis of the experiment shown above were expressed as the relative percentage of the amount of c-Myc protein at the zero time point and plotted as a function of time. (B) COS-7 cells were transiently transfected with 2 μg of an expression plasmid encoding wild-type murine c-MycS or the indicated c-MycS deletion mutant. Pulse-chase analysis, immunoprecipitation of c-MycS, and densitometric plotting were carried out as described for panel A.
Although the first 100 amino acids of c-Myc are not required for rapid c-Myc turnover, we reasoned that they might contribute to c-Myc's instability. Indeed, in a recent report by Flinn et al., it was shown that both MBI (amino acids 45 to 63) and MBII can independently destabilize a heterologous protein (22). To test our hypothesis, we generated c-MycS expression plasmids containing the deletion mutations described above. Thus, these deletion mutants, c-MycS d106–143, d127–158, and d144–158, lack the first 100 amino acids of c-Myc, including MBI, in addition to the respective MBII region deletion. These constructs were transfected into COS-7 cells and subjected to pulse-chase analysis. As shown in Fig. 6B, deletion of amino acids 106 to 143 partially stabilized c-MycS, and this small increase in stability was reproducibly observed (data not shown). c-MycS d127–158, however, was largely stabilized, and, in fact, turnover of c-MycS d127–158 was virtually undetectable in this assay. This effect was not solely due to the removal of MBI and MBII, since c-Myc d106–143, which also lacks these sequences, was only minimally stabilized. As with full-length c-Myc, the d127–158 mutation caused c-MycS to shift into a slower-migrating form (∼3 kDa larger) (Fig. 6B). Because the shift in mobility could be removed with phosphatase treatment, it may be due to hyperphosphorylation of c-Myc as a consequence of defective proteolysis. Indeed, phosphopeptide mapping experiments have revealed that c-MycS d127–158, in comparison with wild-type c-MycS, becomes phosphorylated at several new sites which have not previously been observed in c-Myc peptide maps (data not shown). The results presented above suggest that amino acids 127 to 158 play an essential role in rapid c-Myc proteolysis and that upstream sequences, within the first 100 amino acids of c-Myc, additionally contribute.
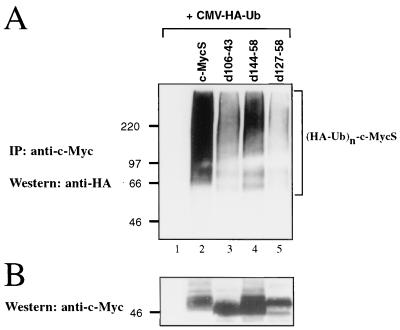
To determine if the N-terminal regions of c-Myc which are required for rapid proteolysis are required for ubiquitination, the c-MycS deletion mutants described above, or wild-type c-MycS, were coexpressed with HA-ubiquitin in COS-7 cells and the exogenous c-Myc proteins were immunoprecipitated. Western blotting for HA showed that ubiquitination of c-MycS d127–158 was significantly impaired (Fig. 7A, lane 5) compared with wild-type c-MycS (lane 2), as was ubiquitination of c-MycS d106–143 (lane 3), although to a lesser extent. Western blotting for c-Myc showed approximately equal levels of expression of the various c-MycS forms (Fig. 7B). The highly stable c-MycS d127–158 did not seem to accumulate to high levels (lane 5), as would be expected; however, RNA levels were not measured, and there may have been changes in mRNA stability or translation. Although these results support a role for N-terminal sequences in targeting c-Myc for ubiquitination and subsequent degradation, it appears these sequences are not absolutely required for ubiquitination. Both c-MycS d106–143 and the more stable c-MycS d127–58 appear to remain partially ubiquitinated (Fig. 7A, lanes 3 and 5). Thus, there still might be additional sequences in c-Myc which can target the protein for ubiquitination, albeit inefficiently.
FIG. 7.
Deletion of N-terminal sequences inhibits c-Myc ubiquitination. (A) COS-7 cells were transiently transfected with 4 μg of a plasmid encoding HA-tagged ubiquitin (CMV-HA-Ub) and 2 μg of a plasmid encoding wild-type murine c-MycS or the indicated c-MycS deletion mutant. Forty-eight hours after transfection, cell lysates were prepared. Exogenous c-MycS proteins were immunoprecipitated with anti-av-Myc12C and subsequently analyzed by Western blotting with anti-HA antibody 12CA5 to detect c-MycS–HA–ubiquitin conjugates. (B) The blot shown in panel A was stripped and reprobed with anti-av-Myc12C to examine c-MycS expression.
Degradation and ubiquitination of c-Myc are blocked in mitosis.
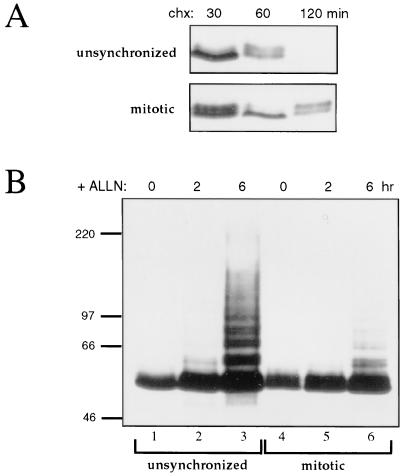
An interesting question is whether c-Myc proteolysis by the ubiquitin-proteasome pathway is merely a constitutive process or is regulated. A previous study provided evidence that c-Myc may be stabilized in mitosis (40). In order to directly test this hypothesis, Bk3A cells were blocked in mitosis by nocodazole treatment and compared with unsynchronized cycling Bk3A cells. The stability of c-Myc was assessed by monitoring c-Myc levels by Western blot analysis after treatment of cells with cycloheximide to inhibit protein synthesis. As shown in Fig. 8A, turnover of c-Myc was rapid in unsynchronized cells, but was significantly slower in mitotic cells. The amount of c-Myc turnover observed in the nocodazole-treated cells might, at least partially, be accounted for by a population of cells not blocked in mitosis. To determine if ubiquitination of c-Myc is compromised in mitosis, unsynchronized or mitotic Bk3A cells were treated with ALLN for up to 6 h to block proteasome activity. c-Myc was then examined by Western blot for the accumulation of ubiquitin conjugates. In unsynchronized cells, upper-molecular-weight forms of c-Myc, indicating ubiquitin conjugation, rapidly accumulated to a high level by 6 h of proteasome inhibition (Fig. 8B, lane 3). In mitotic cells, however, the accumulation of ubiquitinated forms was largely blocked (compare lanes 3 and 6) indicating that c-Myc ubiquitination is abrogated in mitosis. Importantly, these results demonstrate a direct correlation between the ability of c-Myc to be ubiquitinated and its ability to be degraded, which is expected if c-Myc proteolysis is dependent on the ubiquitin pathway.
FIG. 8.
Degradation and ubiquitination of c-Myc are inhibited in mitotic cells. (A) Bk3A cells were blocked in mitosis by treatment with nocodazole for 24 h. To examine the rate of c-Myc turnover, unsynchronized, or nocodazole-treated cells were treated with cycloheximide (chx) for the indicated amount of time, followed by Western blot analysis with anti-av-Myc12C to detect endogenous c-Myc. (B) Unsynchronized or mitosis-arrested Bk3A cells were treated the proteasome inhibitor ALLN (100 μM) for the indicated amount of time and analyzed by Western blotting with anti-av-Myc12C.
Stabilization of c-Myc in Burkitt's lymphoma cell lines.
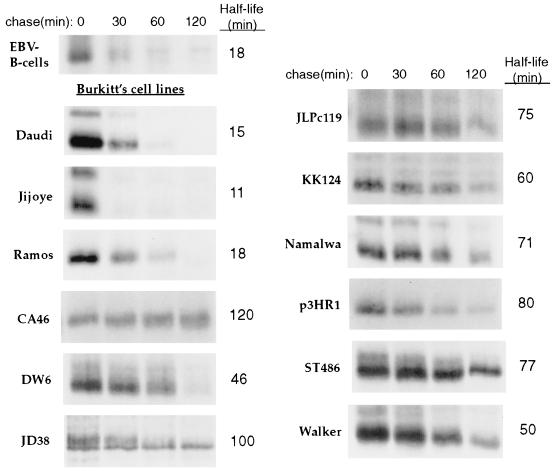
It was previously observed that c-Myc is stabilized in several human glioma cell lines (61). Additionally, a prolonged N-Myc half-life was observed in a human neuroblastoma cell line (15), thus leading to the suggestion that aberrant Myc protein degradation may contribute to oncogenesis. In order to determine if c-Myc stabilization occurs in another tumor cell type, we examined c-Myc protein turnover in several Burkitt's lymphoma cell lines. We chose to study Burkitt's lymphoma because it is clear that c-Myc plays a role in this cancer. Burkitt's lymphoma is a human B-cell lymphoma in which the c-myc gene is translocated to one of the heavy or light chain Ig gene loci resulting in constitutive c-myc expression (44). Turnover of c-Myc was analyzed in 12 Burkitt's lymphoma cell lines in pulse-chase experiments. As a control, c-Myc turnover in Epstein-Barr virus (EBV)-immortalized B cells was also analyzed. As shown in Fig. 9, c-Myc was significantly stabilized in 9 of the 12 Burkitt's lines relative to EBV-immortalized B cells. In these nine cell lines (CA46, DW6, JD38, JLPc119, KK124, Namalwa, p3HR1, ST486, and Walker), the half-life of c-Myc ranged from 46 to 120 min, which is significantly prolonged (approximately two- to sixfold longer) compared with the c-Myc half-life in EBV-immortalized B cells (18 min) and the 20- to 30-min c-Myc half-life observed in other cell lines (e.g., see Fig. 1). Only the Daudi, Jijoye, and Ramos cell lines displayed a normal short c-Myc half-life (15, 11, and 18 min, respectively). These results suggest that stabilization of c-Myc may play a role in the pathogenesis of Burkitt's lymphoma.
FIG. 9.
c-Myc is stabilized in Burkitt's lymphoma cell lines. EBV-immortalized B cells or the indicated Burkitt's lymphoma cell line were subjected to pulse-chase analysis as described in the legend to Fig. 1A, followed by immunoprecipitation of c-Myc with anti-MycFL. The half-lives were determined by a logarithmic analysis of the data obtained from densitometric scanning and the values shown are an average from two or more independent experiments. A representative pulse-chase experiment is shown.
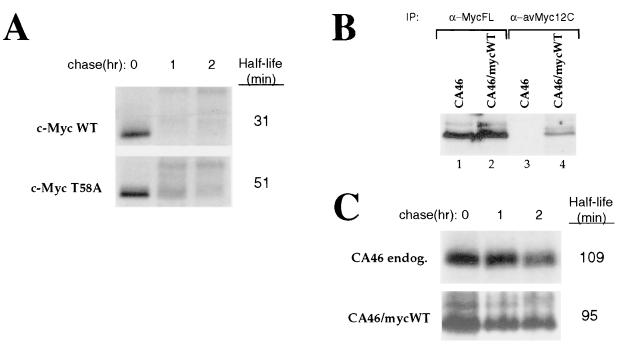
Mutations in the N-terminal domain of c-Myc are commonly observed in Burkitt's lymphoma, with Thr-58 being a hot spot for mutation (1, 4, 7, 76). Because Thr-58 is a major site of phosphorylation in c-Myc (43), this suggests the possibility that phosphorylation of this residue regulates c-Myc stability. To examine this possibility, NIH 3T3 cells were transiently transfected with an expression plasmid encoding either wild-type c-Myc or c-Myc with Thr-58 mutagenized to nonphosphorylatable Ala and subjected to pulse-chase analysis. As shown in Fig. 10A, c-Myc T58A was moderately more stable than wild-type c-Myc (approximately 1.6-fold). Similar results were obtained using COS-7 cells where c-Myc T58A was approximately 1.7-fold more stable than wild-type c-Myc (data not shown). These data suggest that phosphorylation of Thr-58 facilitates rapid c-Myc proteolysis and that mutations at or near Thr-58 may contribute to the increased stability of c-Myc in Burkitt's lymphoma cells.
FIG. 10.
Mutation of Thr-58 increases c-Myc stability. (A) NIH 3T3 cells were transiently transfected with 2 μg of a plasmid encoding wild-type (WT) murine c-Myc or c-Myc with a Thr-58-to-Ala mutation (T58A). Forty-eight hours after transfection, the cells were subjected to pulse-chase analysis as described in the legend to Fig. 1A followed by immunoprecipitation of exogenous c-Myc proteins with anti-av-Myc12C. The half-life values were determined by a logarithmic analysis of the data obtained from densitometric scanning and the values shown are an average from two independent experiments. (B) CA46 cells were stably transfected with a plasmid encoding wild-type murine c-Myc as described in Materials and Methods (CA46/mycWT). c-Myc expression in untransfected CA46 and CA46/mycWT was analyzed by immunoprecipitation (IP) with either anti-(α)-MycFL or anti-av-Myc12C. (C) CA46 or CA46/mycWT cells were subjected to pulse-chase analysis followed by immunoprecipitation with either anti-MycFL (for CA46) or anti-av-Myc12C (for CA46/mycWT). endog., endogenous. The half-life values were determined as described for panel A.
In order to determine if mutation of c-Myc fully accounts for the observed increase in c-Myc stability in Burkitt's lymphoma cells, we wished to examine the stability of wild-type c-Myc in the context of a Burkitt's lymphoma cell line in which endogenous mutant c-Myc is stabilized. To this end, CA46 cells, the cell line in which c-Myc was found to be the most stable (Fig. 9), were stably transfected with wild-type c-myc. Immunoprecipitation analysis using anti-MycFL, which recognizes both the endogenous and exogenous c-Myc, revealed that c-Myc levels were not substantially higher in CA46/mycWT compared with untransfected CA46 cells (Fig. 10B, compare lanes 1 and 2), demonstrating that expression of exogenous wild-type c-Myc is comparable to that of endogenous c-Myc. Anti-av-Myc12C, directed against the C terminus of the exogenous c-Myc, immunoprecipitated c-Myc in CA46/mycWT (Fig. 10B, lane 4) but not in untransfected CA46 cells (lane 3), confirming the specificity of this antibody for exogenous c-Myc. Untransfected CA46 and CA46/mycWT cells were used in pulse-chase experiments followed by immunoprecipitation analysis of endogenous or exogenous c-Myc. As shown in Fig. 10C, exogenous wild-type c-Myc in CA46/mycWT cells displayed a significantly prolonged half-life (95 min) similar to that of endogenous mutant c-Myc in CA46 cells. These results suggest that the increased stability of c-Myc in CA46 cells is not primarily due to c-Myc mutation indicating that there are additional, cellular mechanisms which can operate to stabilize c-Myc in Burkitt's lymphoma cells.
DISCUSSION
In recent years, it has become clear that ubiquitin-mediated proteolysis is a key mechanism for regulating critical proteins involved with cell growth (18, 35). In this report, we have demonstrated that c-Myc is degraded by the ubiquitin-proteasome pathway in vivo. Our results are in agreement with recent reports, published while this work was in progress, implicating the ubiquitin-proteasome pathway in the degradation of c-Myc in vivo (24, 57). As in these studies, we have found that rapid proteolysis of c-Myc depends on the activity of the 26S proteasome. Furthermore, we have shown that endogenous c-Myc is a substrate for ubiquitination and that multiubiquitinated forms of c-Myc rapidly accumulate upon proteasome inhibition. Additionally, we have shown that there is a direct correlation between c-Myc ubiquitination and degradation because both of these processes are inhibited in mitotic cells. These data strongly argue that c-Myc proteolysis occurs via the ubiquitin-proteasome pathway.
In our attempts to identify the regions of c-Myc which are required for its rapid degradation, we have found that C-terminal sequences (amino acids 265 to 439) are unnecessary. Because this region contains the c-Myc nuclear localization signals and the bHLH-LZ domain, we suggest that nuclear localization, dimerization with Max, and DNA binding are unimportant for rapid c-Myc degradation. A centrally located PEST sequence (amino acids 226 to 270 of murine c-Myc), however, appears to be critical since removal of this sequence substantially blocked c-Myc degradation. PEST sequences are found in numerous short-lived proteins and are thought to target proteins for rapid degradation via the proteasome, although the exact mechanism is unclear (52). In many cases, PEST sequences act as conditional proteolytic signals. For example, phosphorylation of a PEST sequence by casein kinase II (CKII) appears to promote the degradation of Drosophila Cactus (39) and its mammalian counterpart, IκBα (38, 48, 59). The PEST sequence of c-Myc could also be regulated by phosphorylation as c-Myc is a substrate for phosphorylation by CKII in vitro at sites which fall within the PEST region (amino acids 240 to 262) (42). However, phosphopeptide mapping experiments have not confirmed that c-Myc is phosphorylated in vivo within this region (M. A. Gregory and S. R. Hann, unpublished data).
How do PEST sequences target proteins for degradation? A prevalent theory is that PEST sequences, or the kinase target sites located therein, target the protein for ubiquitination and thus degradation by the proteasome, although substantial evidence is lacking. Surprisingly, we have found that although the PEST sequence of c-Myc (amino acids 226 to 270) is required for rapid degradation, it is not required for ubiquitination; in fact, removal of this sequence caused the accumulation of ubiquitinated c-Myc. We conclude that the PEST sequence of c-Myc plays a necessary role in a step which comes between c-Myc ubiquitination and its subsequent degradation by the proteasome. Thus, the c-Myc PEST sequence would seem to be necessary but not sufficient for rapid degradation. Similar observations have been made regarding the PEST sequences of other substrates of the ubiquitin-proteasome pathway. For example, the PEST sequences of IκBα and yeast Cln2 were found to be necessary for rapid proteolysis, but were insufficient by themselves in destabilizing a heterologous protein (9, 56). The precise role of the PEST region in c-Myc proteolysis is open to speculation. Brown et al. have suggested that the high degree of negative charge conferred by acidic PEST residues may be required for unfolding of a protein substrate after docking with the proteasome (9). Our results with c-Myc would be consistent with this hypothesis. A less likely possibility is that the PEST sequence of c-Myc is important for efficient recognition by the proteasome and ubiquitination could serve to alter the protein conformation such that the PEST region is exposed.
We also have demonstrated a role for N-terminal sequences in targeting c-Myc for ubiquitin-mediated proteolysis. Our previous studies showed that c-MycS, which lacks the N-terminal 100 amino acids of c-Myc, is rapidly degraded (65). This suggests that the N-terminal 100 amino acids, which includes the highly conserved MBI, do not contain an essential degradation signal, but does not exclude the possibility that this region might contribute to c-Myc's instability. In this report, we have shown that removal of a specific sequence encompassing highly conserved MBII, amino acids 127 to 158 of murine c-Myc, significantly inhibited c-Myc degradation. However, when this sequence was removed from c-MycS, degradation was largely blocked and ubiquitination was inhibited. This effect was not due solely to the removal of MBII (amino acids 130 to 142) as a deletion of amino acids 106 to 143 only partially inhibited c-MycS ubiquitination and degradation. From these results, we conclude that the c-Myc N terminus contains multiple regions involved in targeting c-Myc for ubiquitination and rapid degradation, including a primary region encompassing MBII (amino acids 127 to 158) and a secondary region(s) within the first 100 amino acids.
Additional evidence supporting a role for N-terminal sequences in Myc proteolysis comes from our studies of the B-Myc protein. B-Myc, a 170-amino-acid protein (murine B-Myc) which is highly homologous to the N-terminal 158 amino acids of c-Myc (3), is rapidly degraded in vivo by the ubiquitin-proteasome pathway (M. A. Gregory, Q. Xiao, G. A. Cornwall, B. Lutterbach, and S. R. Hann, unpublished data). Furthermore, an N-terminal fragment of c-Myc consisting of the first 167 amino acids is also ubiquitinated and rapidly degraded (M. A. Gregory and S. R. Hann, unpublished data). This suggests that the c-Myc N terminus contains sufficient information to target c-Myc for ubiquitination and degradation, at least in context of a smaller protein. As neither of these small Myc proteins contain a discernible PEST sequence, we conclude the PEST region is only necessary for the degradation of larger Myc proteins, which would be consistent with the unfolding hypothesis previously discussed.
It is interesting that neither of the two deletions, d106–143 or d144–158, which remove either half of the d127–158 sequence mimicked the substantial stabilizing effect of d127–158 on c-MycS. This could be explained if the d127–158 sequence contained multiple elements capable of recruiting the ubiquitination machinery. An alternative possibility might be that this region contains multiple lysine residues which can serve as the acceptor sites for ubiquitin. Indeed, d127–158 removes all the lysines from this region of c-Myc (K127, K144, K149, and K158 [see Fig. 6]). The only two other lysines in the c-Myc N terminus occur at amino acids 51 and 52, within MBI, which are lacking in c-MycS. Thus, in the absence of MBI, the MBII region lysines could be critical sites for ubiquitination. Most of these lysines are evolutionarily conserved in a c-Myc and are present in other Myc family members. Although specific lysines are usually not required, in the case of IκB for example, two specific lysines are necessary target sites for ubiquitination (58).
Two recent reports have also investigated the regions of c-Myc involved in proteolysis. Flinn et al. (22) demonstrated that both MBI and MBII can act as autonomous degradation signals in yeast cells and observed that removal of MBI and MBII caused substantial accumulation of c-Myc in mammalian cells. Our results seem to conflict with the last observation, since we did not observe significant accumulation of c-MycS d106–143, which lacks both MBI and MBII, in COS-7 cells. Additionally, our pulse-chase analysis showed that c-MycS d106–43 is only minimally stabilized, leading us to conclude that N-terminal sequences other than just MBI and MBII play an important role in c-Myc proteolysis (discussed above). Salghetti et al. (57) localized a necessary signal for ubiquitin-mediated degradation to the N-terminal 147 amino acids of c-Myc based on deletional analysis and showed that multiple sequence elements from this region can act as degradation signals when fused to a heterologous protein. Our results are partially in conflict with their report. They found that removal of the first 94 amino acids was sufficient to considerably stabilize c-Myc whereas we have shown that c-MycS, which lacks this sequence, is rapidly degraded similar to full-length c-Myc. Also, they found that deletion of the N-terminal 147 amino acids both stabilized c-Myc and abrogated detectable ubiquitination. This deletion mutation is highly similar to our d106–143 mutation of c-MycS, which we've shown only partially inhibits ubiquitination and degradation. These inconsistencies might be explained by differences in the experimental systems used. Nonetheless, the two aforementioned reports and our results all converge on the conclusion that multiple regions within the c-Myc N terminus can signal ubiquitination and degradation. As the N-terminal domain of c-Myc is essential for c-Myc biological function, the prospect that this region contains multiple elements which signal ubiquitin-mediated degradation is intriguing. It is interesting to speculate that c-Myc's ability to recruit the ubiquitination machinery may be critical for c-Myc function (57).
It is obvious that the rapid rate of c-Myc protein degradation allows tight regulation of c-myc function. In addition, several observations suggest that c-Myc proteolysis itself is regulated. First, a twofold increase in the rate of c-Myc degradation was observed during the postcommitment phase of murine erythroleukemia cell differentiation (64). Secondly, it was recently shown that Ras stabilizes c-Myc in rat REF52 fibroblasts (60). Thirdly, we have shown in this report that c-Myc overexpression can lead to moderate protein stabilization. Fourth, we also have demonstrated that c-Myc degradation is inhibited in mitosis. Lastly, c-Myc is stabilized in some tumor cells (discussed below). Thus, it seems that ubiquitin-mediated proteolysis of c-Myc is not merely a constitutive process but is regulated.
The abundance of key growth regulatory proteins is frequently altered in cancer cells. For example, elevated Myc expression due to gene amplification is observed in a wide range of human tumors (46). It is reasonable to speculate that aberrant or deregulated proteolysis of Myc might be an alternative or additional mechanism of oncogenic activation. Indeed, stabilization of c-Myc has been observed in human glioma cell lines (61) and N-Myc stabilization was seen in a neuroblastoma cell line lacking N-myc amplification (15). We have shown that c-Myc is significantly stabilized (two- to sixfold, half-lives, 46 to 120 min) in a large number of Burkitt's lymphoma-derived cell lines leading us to conclude that defective c-Myc proteolysis may contribute to the uncontrolled proliferation of Burkitt's lymphoma cells. Additionally, Ruf et al. recently demonstrated that c-Myc was stabilized severalfold (half-life, 120 min) in the Akata cell line, but not in two other Burkitt's lymphoma cell lines examined (55). In Burkitt's lymphoma, c-myc is translocated to one of the Ig loci resulting in constitutive, although not necessarily elevated, c-myc expression (63, 68). Therefore, c-Myc protein stabilization could provide a means for high-level constitutive c-Myc expression in this lymphoma.
Mutations in c-Myc are common in Burkitt's lymphoma and in AIDS-associated lymphomas; these mutations frequently occur in the N-terminal transactivation domain clustering around Thr-58, a highly conserved and functionally important phosphorylation site in MBI (1, 4, 7, 14, 76). Mutation of Thr-58 increases c-Myc's activity in transformation assays, suggesting that phosphorylation at Thr-58 transduces a negative growth signal (30, 49). Interestingly, the three Burkitt's cell lines in which c-Myc turnover was normal (Daudi, Jijoye, and Ramos) have been shown to express c-Myc with a wild-type sequence in the N-terminal domain (1, 50, 74). Of the nine Burkitt's cell lines in which c-Myc was stabilized, c-Myc was found to be mutated in the N-terminal domain in five of them, CA46, DW6, JLPc119, p3HR1, and ST486 (1, 4, 7, 32, 62) and of these five, three have c-Myc mutations at Pro-57 (CA46, p3HR1, and ST486), a mutation which abolishes Thr-58 phosphorylation (32). Also, the half-life of c-Myc was significantly extended in two additional Burkitt's cell lines which have c-Myc mutations at Thr-58 (J. Niklinski, G. F. Claassen, M. A. Gregory, F. J. Kaye, C. J. Allegra, S. R. Hann, and M. Zajac-Kaye, unpublished data). These observations suggest the possibility that Thr-58 phosphorylation facilitates rapid c-Myc proteolysis and that decreased proteolysis of c-Myc in Burkitt's lymphoma may be the result of mutation within the c-Myc N-terminal domain at or near Thr-58. In support of this idea, we've shown that a Thr-58-to-alanine mutation moderately stabilizes ectopic c-Myc in NIH 3T3 and COS-7 cells. Similarly, Salghetti et al. observed stabilization of c-Myc in human U2OS cells with a Thr-58-to-alanine mutation, as well as with other mutations which occur in Burkitt's lymphoma (57). However, c-MycS, which lacks Thr-58, retains a normal rapid half-life (discussed above). This suggests that the absence of the N-terminal domain precludes the requirement of Thr-58 phosphorylation for rapid c-Myc proteolysis.
Although mutation of c-Myc is apparently sufficient to moderately stabilize the protein, it does not appear to be essential for c-Myc stabilization in Burkitt's lymphoma. In this report, we have shown that ectopic wild-type c-Myc has a significantly prolonged half-life similar to that of endogenous mutant c-Myc in CA46 Burkitt's lymphoma cells. Furthermore, mutations were not detected in the N-terminal domain of c-Myc in three of the Burkitt's cell lines (KK124, Namalwa, and Walker) (1, 7) in which we've shown that c-Myc is stabilized. It is also worth noting that the neuroblastoma cell line NBL-S, in which stabilization of the highly related c-Myc homologue N-Myc was observed (15), expresses only wild-type N-Myc (34). From these observations, we conclude that there are additional or alternative mechanisms, other than c-Myc mutation, that stabilize c-Myc in Burkitt's lymphoma. For example, there may be a defect in the pathway which mediates c-Myc proteolysis, such as alteration of an E2 or E3 enzyme which directs c-Myc ubiquitination.
ACKNOWLEDGMENTS
We are very grateful to Chi Dang, Gregory Kato, Robert Eisenman, Albert Reynolds, and Maria Zajac-Kaye for sharing cell lines and reagents. We thank Gisela Claassen, Bart Lutterbach, Gail Cornwall, and Rebecca Chinery for critical review of the manuscript. We also thank Jingyu Shi for technical assistance.
This work was supported by U.S. Public Health Service grants CA47399 and CA78888 from the National Cancer Institute.
REFERENCES
- 1.Albert T, Urlbauer B, Kohlhuber F, Hammerson B, Eick D. Ongoing mutations in the N-terminal domain of c-Myc affect transactivation in Burkitt's lymphoma cell lines. Oncogene. 1994;9:759–763. [PubMed] [Google Scholar]
- 2.Asker C, Steinitz M, Andersson K, Sumegi J, Klein G, Ingvarsson S. Nucleotide sequence of the rat Bmyc gene. Oncogene. 1989;4:1523–1527. [PubMed] [Google Scholar]
- 3.Asker C E, Magnusson K P, Piccoli S P, Andersson K, Klein G, Cole M D, Wiman K G. Mouse and rat B-myc share amino acid sequence homology with the c-myc transcriptional activator domain and contain a B-myc specific carboxy terminal region. Oncogene. 1995;11:1963–1969. [PubMed] [Google Scholar]
- 4.Axelson H, Henriksson M, Wang Y, Magnusson K P, Klein G. The amino-terminal phosphorylation sites of C-MYC are frequently mutated in Burkitt's lymphoma lines but not in mouse plasmacytomas and rat immunocytes. Eur J Cancer. 1995;31A:2099–2104. doi: 10.1016/0959-8049(95)00449-1. [DOI] [PubMed] [Google Scholar]
- 5.Bader J P, Hausman F A, Ray D A. Intranuclear degradation of the transformation-inducing protein encoded by avian MC29 virus. J Biol Chem. 1986;261:8303–8308. [PubMed] [Google Scholar]
- 6.Barrett A J, Kembhavi A A, Brown M A, Kirschke H, Knight C G, Tamai M, Hanada K. l-trans-Epoxysuccinyl-leucylamido(4-guanidino)butane (E-64) and its analogues as inhibitors of cysteine proteinases including cathepsins B, H, and L. Biochem J. 1982;201:189–198. doi: 10.1042/bj2010189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhatia K, Huppi K, Spangler G, Siwarski D, Iyer R, Magrath I. Point mutations in the c-Myc transactivation domain are common in Burkitt's lymphoma and mouse plasmacytomas. Nat Genet. 1993;5:56–61. doi: 10.1038/ng0993-56. [DOI] [PubMed] [Google Scholar]
- 8.Bies J, Wolff L. Oncogenic activation of c-Myb by carboxyl-terminal truncation leads to decreased proteolysis by the ubiquitin-26S proteasome pathway. Oncogene. 1997;14:203–212. doi: 10.1038/sj.onc.1200828. [DOI] [PubMed] [Google Scholar]
- 9.Brown K, Franzoso G, Baldi L, Carlson L, Mills L, Lin Y-C, Gerstberger S, Siebenlist U. The signal response of IκBα is regulated by transferable N- and C-terminal domains. Mol Cell Biol. 1997;17:3021–3027. doi: 10.1128/mcb.17.6.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campanero M R, Flemington E K. Regulation of E2F through ubiquitin-proteasome-dependent degradation: stabilization by the pRB tumor suppressor protein. Proc Natl Acad Sci USA. 1997;94:2221–2226. doi: 10.1073/pnas.94.6.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ciechanover A. The ubiquitin-proteasome proteolytic pathway. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- 12.Ciechanover A. The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J. 1998;17:7151–7160. doi: 10.1093/emboj/17.24.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Claassen G F, Hann S R. Myc-mediated transformation: the repression connection. Oncogene. 1999;18:2925–2933. doi: 10.1038/sj.onc.1202747. [DOI] [PubMed] [Google Scholar]
- 14.Clark H M, Yano T, Otsuki T, Jaffe E S, Shibata D, Raffeld M. Mutations in the coding region of c-MYC in AIDS-associated and other aggressive lymphomas. Cancer Res. 1994;54:3383–3386. [PubMed] [Google Scholar]
- 15.Cohn S L, Salwen H, Quasney M W, Ikegaki N, Cowan J M, Herst C V, Kennett R H, Rosen S T, DiGiuseppe J A, Brodeur G M. Prolonged N-myc protein half-life in a neuroblastoma cell line lacking N-myc amplification. Oncogene. 1990;5:1821–1827. [PubMed] [Google Scholar]
- 16.Cole M D, McMahon S B. The Myc oncoprotein: a critical evaluation of transactivation and target gene regulation. Oncogene. 1999;18:2916–2924. doi: 10.1038/sj.onc.1202748. [DOI] [PubMed] [Google Scholar]
- 17.Eilers M, Schirm S, Bishop J M. The c-Myc protein activates transcription of the alpha prothymosin gene. EMBO J. 1991;10:133–141. doi: 10.1002/j.1460-2075.1991.tb07929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elledge S J, Harper J W. The role of protein stability in the cell cycle and cancer. Biochim Biophys Acta. 1998;1377:M61–M70. doi: 10.1016/s0304-419x(98)00005-5. [DOI] [PubMed] [Google Scholar]
- 19.Evan G I, Wyllie A H, Gilbert C S, Littlewood T D, Land H, Brooks M, Waters C M, Penn L Z, Hancock D C. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- 20.Facchini L M, Penn L Z. The molecular role of Myc in growth and transformation: recent discoveries lead to new insights. FASEB J. 1998;12:633–651. [PubMed] [Google Scholar]
- 21.Fenteany G, Standaert R F, Lane W S, Choi S, Corey E J, Schreiber S L. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- 22.Flinn E M, Magnus C, Busch C, Wright A P H. myc boxes, which are conserved in myc family proteins, are signals for protein degradation via the proteasome. Mol Cell Biol. 1998;18:5961–5969. doi: 10.1128/mcb.18.10.5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grandori C, Eisenman R. Myc target genes. Trends Biochem Sci. 1997;22:177–181. doi: 10.1016/s0968-0004(97)01025-6. [DOI] [PubMed] [Google Scholar]
- 24.Gross-Mesilaty S, Reinstein E, Bercovich B, Tobias K E, Schwartz A L, Kahana C, Ciechanover A. Basal and human papillomavirus E6 oncoprotein-induced degradation of Myc proteins by the ubiquitin pathway. Proc Natl Acad Sci USA. 1998;95:8058–8063. doi: 10.1073/pnas.95.14.8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hann S R, Abrams H D, Rohrschneider L R, Eisenman R N. Proteins encoded by v-Myc and c-Myc oncogenes: identification and localization in acute leukemia transformants and bursal lymphoma cell lines. Cell. 1983;34:789–798. doi: 10.1016/0092-8674(83)90535-4. [DOI] [PubMed] [Google Scholar]
- 26.Hann S R, Dixit M, Sears R C, Sealy L. The alternatively initiated c-Myc proteins differentially regulate transcription through a noncanonical DNA-binding site. Genes Dev. 1994;8:2441–2452. doi: 10.1101/gad.8.20.2441. [DOI] [PubMed] [Google Scholar]
- 27.Hann S R, Eisenman R N. Proteins encoded by the human c-myc oncogene: differential expression in neoplastic cells. Mol Cell Biol. 1984;4:2486–2497. doi: 10.1128/mcb.4.11.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harrington E A, Bennett M R, Fanidi A, Evan G I. c-Myc-induced apoptosis in fibroblasts is inhibited by specific cytokines. EMBO J. 1994;13:3286–3295. doi: 10.1002/j.1460-2075.1994.tb06630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hateboer G, Kerkhoven R M, Shvarts A, Bernards R, Beijersbergen R L. Degradation of E2F by the ubiquitin-proteasome pathway: regulation by retinoblastoma family proteins and adenovirus transforming proteins. Genes Dev. 1996;10:2960–2970. doi: 10.1101/gad.10.23.2960. [DOI] [PubMed] [Google Scholar]
- 30.Henriksson M, Bakardjiev A, Klein G, Luscher B. Phosphorylation sites mapping in the N-terminal domain of c-Myc modulate its transforming potential. Oncogene. 1993;8:3199–3209. [PubMed] [Google Scholar]
- 31.Henriksson M, Luscher B. Proteins of the Myc network: essential regulators of cell growth and differentiation. Adv Cancer Res. 1996;68:109–182. doi: 10.1016/s0065-230x(08)60353-x. [DOI] [PubMed] [Google Scholar]
- 32.Hoang A T, Lutterbach B, Lewis B C, Yano T, Chou T-Y, Barrett J F, Raffeld M, Hann S R, Dang C V. A link between increased transforming activity of lymphoma-derived MYC mutant alleles, their defective regulation by p107, and altered phosphorylation of the c-Myc transactivation domain. Mol Cell Biol. 1995;15:4031–4042. doi: 10.1128/mcb.15.8.4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hofmann F, Martelli F, Livingston D M, Wang Z. The retinoblastoma gene product protects E2F-1 from degradation by the ubiquitin-proteasome pathway. Genes Dev. 1996;10:2949–2959. doi: 10.1101/gad.10.23.2949. [DOI] [PubMed] [Google Scholar]
- 34.Hogarty M D, Brodeur G M. Wild-type sequence of MYCN in neuroblastoma cell lines. Int J Cancer. 1999;80:630–631. doi: 10.1002/(sici)1097-0215(19990209)80:4<630::aid-ijc24>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 35.Isaksson A, Musti A M, Bohmann D. Ubiquitin in signal transduction and cell transformation. Biochim Biophys Acta. 1996;1288:F21–F29. doi: 10.1016/0304-419x(96)00011-x. [DOI] [PubMed] [Google Scholar]
- 36.King M W, Roberts J M, Eisenman R N. Expression of the c-myc proto-oncogene during development of Xenopus laevis. Mol Cell Biol. 1986;6:4499–4508. doi: 10.1128/mcb.6.12.4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin R, Beauparlant P, Makris C, Meloche S, Hiscott J. Phosphorylation of IκBα in the C-terminal PEST domain by casein kinase II affects intrinsic protein stability. Mol Cell Biol. 1996;16:1401–1409. doi: 10.1128/mcb.16.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Z, Galindo R L, Wasserman S A. A role for CKII phosphorylation of the cactus PEST domain in dorsoventral patterning of the Drosophila embryo. Genes Dev. 1997;11:3413–3422. doi: 10.1101/gad.11.24.3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luscher B, Eisenman R N. Mitosis-specific phosphorylation of the nuclear oncoproteins Myc and Myb. J Cell Biol. 1992;118:775–784. doi: 10.1083/jcb.118.4.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lüscher B, Eisenman R N. c-myc and c-myb protein degradation: effect of metabolic inhibitors and heat shock. Mol Cell Biol. 1999;8:2504–2512. doi: 10.1128/mcb.8.6.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luscher B, Kuenzel E A, Krebs E G, Eisenman R N. Myc proteins are phosphorylated by casein kinase II. EMBO J. 1989;8:1111–1119. doi: 10.1002/j.1460-2075.1989.tb03481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lutterbach B, Hann S R. Hierarchical phosphorylation at N-terminal transformation-sensitive sites in c-Myc protein is regulated by mitogens and in mitosis. Mol Cell Biol. 1994;14:5510–5522. doi: 10.1128/mcb.14.8.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Magrath I. The pathogenesis of Burkitt's lymphoma. Adv Cancer Res. 1990;55:133–270. doi: 10.1016/s0065-230x(08)60470-4. [DOI] [PubMed] [Google Scholar]
- 45.Maki C G, Huibregtse J M, Howley P M. In vivo ubiquitination and proteasome-mediated degradation of p53. Cancer Res. 1996;56:2649–2654. [PubMed] [Google Scholar]
- 46.Marcu K B, Bossone S A, Patel A J. Myc function and regulation. Annu Rev Biochem. 1992;61:809–860. doi: 10.1146/annurev.bi.61.070192.004113. [DOI] [PubMed] [Google Scholar]
- 47.Mateyak M K, Obaya A J, Adachi S, Sedivy J M. Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 1997;8:1039–1048. [PubMed] [Google Scholar]
- 48.McElhinny J A, Trushin S A, Bren G D, Chester N, Paya C V. Casein kinase II phosphorylates IκBα at S-283, S-288, S-293, and T-291 and is required for its degradation. Mol Cell Biol. 1996;16:899–906. doi: 10.1128/mcb.16.3.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pulverer B J, Fischer K, Vousden K, Littlewood T, Evan G, Woodgett J R. Site-specific modulation of c-Myc cotransformation by residues phosphorylated in vivo. Oncogene. 1994;9:59–70. [PubMed] [Google Scholar]
- 50.Rabbitts T H, Hamlyn P H, Baer R. Altered nucleotide sequences of a translocated c-myc gene in Burkitt lymphoma. Nature. 1983;306:760–765. doi: 10.1038/306760a0. [DOI] [PubMed] [Google Scholar]
- 51.Ramsay G, Evan G I, Bishop J M. The protein encoded by the human proto-oncogene c-myc. Proc Natl Acad Sci USA. 1984;81:7742–7746. doi: 10.1073/pnas.81.24.7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rechsteiner M, Rogers S W. PEST sequences and regulation by proteolysis. Trends Biochem Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- 53.Rock K L, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg A L. Inhibitors of the proteasome block degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 54.Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- 55.Ruf I K, Rhyne P W, Yang H, Borza C M, Hutt-Fletcher L M, Cleveland J L, Sample J T. Epstein-Barr virus regulates c-MYC, apoptosis, and tumorigenicity in Burkitt lymphoma. Mol Cell Biol. 1999;19:1651–1660. doi: 10.1128/mcb.19.3.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Salama S R, Hendricks K B, Thorner J. G1 cyclin degradation: the PEST motif of yeast Cln2 is necessary, but not sufficient, for rapid protein turnover. Mol Cell Biol. 1994;14:7953–7966. doi: 10.1128/mcb.14.12.7953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Salghetti S E, Kim S Y, Tansey W P. Destruction of Myc by ubiquitin-mediated proteolysis: cancer-associated and transforming mutations stabilize Myc. EMBO J. 1999;18:717–726. doi: 10.1093/emboj/18.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scherer D C, Brockman J A, Chen Z, Maniatis T, Ballard D W. Signal-induced degradation of IκBα requires site-specific ubiquitination. Proc Natl Acad Sci USA. 1995;92:11259–11263. doi: 10.1073/pnas.92.24.11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schwarz E M, van Antwerp D, Verma I M. Constitutive phosphorylation of IκBα by casein kinase II occurs preferentially at serine 293: requirement for degradation of free IκBα. Mol Cell Biol. 1996;16:3554–3559. doi: 10.1128/mcb.16.7.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sears R, Leone G, Degregori J, Nevins J R. Ras enhances Myc protein stability. Mol Cell. 1999;3:169–179. doi: 10.1016/s1097-2765(00)80308-1. [DOI] [PubMed] [Google Scholar]
- 61.Shindo H, Tani E, Matsumuto T, Hashimoto T, Furuyama J. Stabilization of c-myc protein in human glioma cells. Acta Neuropathol. 1993;86:345–352. doi: 10.1007/BF00369446. [DOI] [PubMed] [Google Scholar]
- 62.Showe L C, Ballantine M, Nishikura K, Erikson J, Kaji H, Croce C M. Cloning and sequencing of a c-myc oncogene in a Burkitt's lymphoma cell line that is translocated to a germ line alpha switch region. Mol Cell Biol. 1985;5:501–509. doi: 10.1128/mcb.5.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spencer C A, Groudine M. Control of c-Myc regulation in normal and neoplastic cells. Adv Cancer Res. 1991;56:1–48. doi: 10.1016/s0065-230x(08)60476-5. [DOI] [PubMed] [Google Scholar]
- 64.Spotts G D, Hann S R. Enhanced translation and increased turnover of c-myc proteins occurs during differentiation of murine erythroleukemia cells. Mol Cell Biol. 1990;10:3952–3964. doi: 10.1128/mcb.10.8.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Spotts G D, Patel S V, Xiao Q, Hann S R. Identification of downstream-initiated c-Myc proteins which are dominant-negative inhibitors of transactivation by full-length c-Myc proteins. Mol Cell Biol. 1997;17:1459–1468. doi: 10.1128/mcb.17.3.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stancovski I, Gonen H, Orian A, Schwartz A L, Ciechanover A. Degradation of the proto-oncogene product c-Fos by the ubiquitin proteolytic system in vivo and in vitro: identification and characterization of the conjugating enzymes. Mol Cell Biol. 1995;15:7106–7116. doi: 10.1128/mcb.15.12.7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stone J, de Lange T, Ramsay G, Jakobovits E, Bishop J M, Varmus H, Lee W. Definition of regions in human c-myc that are involved in transformation and nuclear localization. Mol Cell Biol. 1987;7:1697–1709. doi: 10.1128/mcb.7.5.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Taub R, Moulding C, Battey J, Murphy W, Vasicek T, Lenoir G M, Leder P. Activation and somatic mutations of the translocated c-myc gene in Burkitt lymphoma cells. Cell. 1984;36:339–348. doi: 10.1016/0092-8674(84)90227-7. [DOI] [PubMed] [Google Scholar]
- 69.Treier M, Staszewski L M, Bohmann D. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the δ domain. Cell. 1994;78:787–798. doi: 10.1016/s0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- 70.Tsurumi C, Ishida N, Tamura T, Kakizuka A, Nishida E, Okumura E, Kishimoto T, Inagaki M, Okazaki K, Sagata N, Ichihara A, Tanaka K. Degradation of c-Fos by the 26S proteasome is accelerated by c-Jun and multiple protein kinases. Mol Cell Biol. 1995;15:5682–5687. doi: 10.1128/mcb.15.10.5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Varshavsky A. The ubiquitin system. Trends Biochem Sci. 1997;22:383–387. doi: 10.1016/s0968-0004(97)01122-5. [DOI] [PubMed] [Google Scholar]
- 72.Wang K K W. Developing selective inhibitors of calpain. Trends Pharmacol Sci. 1990;11:139–142. doi: 10.1016/0165-6147(90)90060-L. [DOI] [PubMed] [Google Scholar]
- 73.Waters C M, Littlewood T D, Hancock D C, Moore J P, Evan G I. c-myc protein expression in untransformed fibroblasts. Oncogene. 1991;6:797–805. [PubMed] [Google Scholar]
- 74.Wiman K G, Clarkson B, Hayday A C, Saito H, Tonegawa S, Hayward W S. Activation of a translocated c-myc gene: role of structural alterations in the upstream region. Proc Natl Acad Sci USA. 1984;81:6798–6802. doi: 10.1073/pnas.81.21.6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xiao Q, Claassen G, Shi J, Adachi S, Sedivy J, Hann S R. Transactivation-defective c-MycS retains the ability to regulate proliferation and apoptosis. Genes Dev. 1998;12:3803–3808. doi: 10.1101/gad.12.24.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yano T, Sander C A, Clark H M, Dolezal M V, Jaffe E S, Raffeld M. Clustered mutations in the second exon of the MYC gene in sporadic Burkitt's lymphoma. Oncogene. 1993;8:2741–2748. [PubMed] [Google Scholar]