Abstract
We conducted a comprehensive serum transcriptomic analysis to explore the roles of miRNAs in alcoholic hepatitis (AH) pathogenesis and their prognostic significance. Serum miRNA profiling was performed in 15 controls, 20 heavy drinkers without liver disease, and 65 patients with AH and compared to publicly available hepatic miRNA profiling in AH patients. Among the top 26 miRNAs, the expression of miR-30b-5p, miR-20a-5p, miR-146a-5p, and miR-26b-5p were significantly reduced in both serum and liver of AH patients. Pathway analysis of the potential targets of these miRNAs uncovered the genes related to DNA synthesis and cell cycle progression pathways, including RRM2, CCND1, CCND2, MYC, and PMAIP1. We found a significant increase in the protein expression of RRM2, CCND1, and CCND2, but not MYC and PMAIP1 in AH patients who underwent liver transplantation; miR-26b-5p and miR-30b-5p inhibited the 3’-UTR luciferase activity of RRM2 and CCND2, and miR-20a-5p reduced the 3’-UTR luciferase activity of CCND1 and CCND2. During a median follow-up of 346 days, 21% of AH patients died; these patients had higher BMI, MELD, serum miR-30b-5p, miR-20a-5p, miR-146a-5p, and miR-26b-5p than those who survived. Cox regression analysis showed BMI, MELD score, miR-20a-5p, miR-146a-5p, and miR-26b-5p predicted the mortality.
Conclusion:
Patients with AH attempt to deal with hepatocyte injury by down-regulating specific miRNAs and upregulating genes responsible for DNA synthesis and cell cycle progression. Higher expression of these miRNAs, suggestive of a diminished capacity in liver regeneration, predicts short-term mortality in AH patients.
Keywords: hepatitis, alcoholic; microRNAs; cell cycle, prognosis
INTRODUCTION
Alcohol-associated liver disease (ALD) is a complex disorder secondary to excessive alcohol use (1). Its pathogenesis is complex and involves a multi-step process leading to a series of hepatic histopathological changes (2). Patients may present with minimal abnormalities ranging from steatosis to more severe manifestations associated with inflammation and fibrosis (2).
Alcoholic hepatitis (AH) is a clinical syndrome and is characterized by an abrupt rise in serum bilirubin levels, fever, coagulopathy, and liver-related complications (3, 4). AH only occurs in a subset of patients with excessive alcohol use (4). Histologically, these patients have hepatic steatosis and signs of hepatocellular injury/damage as well as inflammatory cell infiltration (5). Once developed, it is associated with significant morbidity and mortality (5–7). It is still elusive why only a subset of excessive alcohol drinkers develop AH and what determines the severity and clinical outcomes of this condition.
The new landscape of human transcriptome along with the identification of non-coding RNAs has discovered their roles in the pathophysiology of human diseases (8, 9). Non-coding RNAs include highly abundant and functionally important RNAs such as microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) (8, 9). MiRNA, a small non-coding RNA molecule containing about 22 nucleotides, functions in RNA silencing and post-transcriptional regulation of gene expression (8). They play a critical role in metabolic regulation through their abilities to regulate gene transcription (8, 9). MiRNA expression patterns are organ- dependent and varies during the disease stage (10, 11). Specific miRNAs are principal regulators that control major cellular functions and involve in disease pathogenesis. Recent studies suggested the involvement of miRNAs in the pathogenesis of hepatic steatosis, viral hepatitis, and non-alcoholic fatty liver disease (12–14). MiRNAs also play role in the development of ALD (14–16). Integrative analysis of the miRNAs and potentially regulated pathways in patients with AH were described, however, the prognostic significance of miRNAs was not reported (17).
The TREAT (Translational Research and Evolving Alcoholic hepatitis Treatment) consortium, consisting of Indiana University, Virginia Commonwealth University, and Mayo Clinic, is supported by the National Institute of Alcohol Abuse and Alcoholism. Its primary objectives are to conduct clinical research in AH and develop novel treatments. As part of the research protocol, a prospective, multi-center observational study of patients with well-characterized AH (cases) and heavy drinkers without evidence of liver disease (controls) was conducted. Serum samples were collected at baseline and patients were prospectively followed for up to 12 months. The objectives of our study were to (i) explore differentially expressed serum miRNAs and their targets in a well-characterized cohort of patients with AH, (ii) identify the potential miRNA-based mechanism underlying the pathogenesis of AH, and (iii) determine the prognostic significance of miRNAs and outcomes in patients with AH.
METHODS
Study cohort
Our study cohort consisted of (i) 15 healthy controls (HC) without a history of heavy alcohol consumption and underlying medical illnesses, (ii) 65 patients with well-characterized AH, and (iii) 17 heavy drinkers (HD) without liver disease. HC patients were recruited from the Roudebush Veterans Administration Medical Center, Indianapolis, IN. AH and HD patients were enrolled as part of the TREAT consortium study (NCT02172898), as previously described (4, 18). The diagnosis of AH in our study was based on clinical evaluation and appropriate laboratory testing as defined by total bilirubin > 3 mg/dL and aspartate aminotransferase (AST) > 50 U/L and history of excessive alcohol use for a minimum of 6 months and within the 6 weeks before the enrollment (3). HD patients were those with a history of excessive alcohol use with no prior history of jaundice, signs of chronic liver disease, or portal hypertension. Demographic data, past medical history, and detailed data collection were collected as previously described (4, 18). In brief, the quantity and pattern of alcohol consumption before enrollment were determined using Time Line Follow-Back (TLFB). Blood samples were collected for complete blood counts, comprehensive metabolic panel, hepatic panel, and coagulation tests. The tests were performed at the local clinical laboratories at each site. Serum samples were stored at −80°C for future use. This study was approved by the Institutional Review Board at the respective institutions and all participants have signed informed consent before enrollment.
Human liver tissues of patients with AH
Human liver tissues from patients with AH who underwent liver transplantation as well as normal controls were provided by the Department of Surgery at John Hopkins Hospital under the funding from the NIAAA (R24AA025017, Clinical resources for alcoholic hepatitis investigators).
miRNA profiling, Bioinformatic analysis:
Please refer to detailed information in Supplementary materials.
Western blot analysis
Cells or liver tissues were lysed with RIPA buffer (Millipore, MA) for 10–15 min on ice. The lysis was centrifuged for 15 min at 12,000 rpm. The supernatants were transferred to a new tube and the protein concentration was determined using Pierce BCA Protein Assay Kit (ThermoFisher, MA). The total protein of 30ug from each sample was loaded for the western blot analysis, as previously described (19). The antibodies for CCND1 (Cat #2978), CCND2 (Cat #3741), MYC (Cat #18583), PMAIP1 (Cat #14766), and GAPDH (Cat #5174) were purchased from Cell Signaling Technology (Danvers, MA); RRM2 was purchased from Millipore (Cat # MABC154MI, Burlington, MA) and actin was from Santa Cruz biotechnology (Cat # sc-47778 HRP, Dallas, TX). The quantifications were performed using Image J (Version 1.52n).
Immunohistochemistry staining
Liver tissues were collected and fixed in 10% formalin on shaking device for 24–48 hours, paraffin-embedded, and sliced into 5 μm sections for the following immunohistochemistry staining, according to the standard protocol. Antibodies against CCND2 (Cell Signaling Technology, Cat #3741), RRM2 (Cell Signaling Technology, Cat #65939), and CCND1 (Cell Signaling Technology, Cat #55506) were used to stain the paraffin sections, and visualized with DAB Peroxidase Substrate Kit (Vector Laboratories, CA). The images were taken by Olympus BX41 microscope (Olympus, NY). The quantifications were performed using Image J (Version 1.52n).
Luciferase assay
The 3’-UTR constructs of CCND1, CCND2, and RRM2 were purchased from Applied Biological Materials (British Columbia, Canada). The mutant constructs of CCND1 and RRM2 3’-UTRs were generated by Synbio-tech Inc. (Kaihsiung City, Taiwan). The CCND2 3’-UTR were constructed using Q5 Site-Directed Mutagenesis Kit from New England Biolabs (NEB, Ipswich, MA). The 3’-UTR sequences and mutant sites were provided in Supplementary Fig. S6. For the experiment, 25ng of 3’-UTR constructs or mutants and 25ng of Renilla plasmids were co-transfected with 50 nM of miRNA mimics (Sigma, MO) for 24 hours in HepG2 or HC-04 cells. The luciferase activities were detected using Dual-Luciferase Reporter Assay from Promega (Madison, WI) following the instructions (20).
Quantitative PCR (qPCR)
Total RNA was isolated using TRIzol Reagent (Invitrogen, CA) and cDNA synthesis was performed with High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, CA) for mRNA and miRCURY LNA RT kit (Qiagen, Germany). Quantitative PCR was performed with iTaq Universal SYBR Green Supermix (Bio-Rad, CA) for mRNA and miRCURY LNA SYBR Green PCR kit (Qiagen, Germany) for microRNAs. The primers were provided in Supplementary Table 3. The primers for miRNAs were purchased from miRCURY LNA miRNA PCR assay (Qiagen, Germany). Each quantitative PCR analysis was run in triplicates. The relative ratio of the indicated genes was normalized to internal control as indicated. The correlations were analyzed using GraphPad Prism (Version 8.2).
Statistical analysis
Basic descriptive statistics were presented as mean, standard deviations (SD), and frequencies (percentages). Appropriate comparison tests including the chi-square test for categorical variables, and Student’s t-test or Analysis of Variance for continuous variables were used. To determine the prognostic significance of variables including miRNAs, the survival data were univariately assessed with Kaplan–Meier survival estimates, and log-rank statistics were utilized for comparison of univariate survival curves. Overall survival was estimated as the interval from the date of enrollment to the time of death or the end of the study on July 31, 2018. None of our patients underwent liver transplantation. We utilized a correlation matrix of selected serum miRNAs with the highest expression to determine their associations before variable selection into the survival model. The evaluation of predictors of survival was conducted using the Cox proportional hazards regression analysis with risk estimates reported as hazard ratios (HR) with corresponding 95% confidence intervals (95% CI). A p-value < 0.05 was considered statistically significant. ROC (Receiver Operating Characteristic) curve analysis was applied to test the prognostic performance of the model for end-stage liver disease (MELD) score and miRNAs. The optimal cut-point values for MELD score and miRNAs were obtained to demonstrate good discriminating powers between those who were deceased and alive. In this study, the optimal cut-point value was defined as its absolute difference between sensitivity and specificity was minimum. All analyses were performed with SAS software version 9.4 (SAS Institute, Cary, NC).
RESULTS
Baseline demographics and clinical characteristics of the study cohort.
Detailed baseline demographics and clinical characteristics of the study cohort are shown in Table 1. There were no differences in mean age (p=0.59) and body mass index (BMI, p=0.60) among groups. Among AH cases, 54% were male and 65% were White. Total drinks in the last 30 days prior to the recruitment, as expected, were significantly higher in HD (368.5±300 standard drinks) and AH (293.4±256.7 standard drinks) groups when compared to controls (1.3±1.3, p=0.0001). As previously reported, total drinks were significantly lower in AH cases compared to HD. Patients with AH had the highest levels of total bilirubin, AST, alanine aminotransferase (ALT), and creatinine (all with p<0.01). The average MELD score among patients with AH was 22.1.
Table 1:
Baseline demographic and clinical characteristics of the study cohort
| Healthy Controls (n=15) |
Heavy drinkers without liver disease (n=17) |
Alcoholic hepatitis (n=65) |
p-value | |
|---|---|---|---|---|
| Age | 44.5±12.8 | 44.1±9.5 | 46.8±11.9 | 0.59 |
| Gender (male, %) | 33% | 45% | 54% | 0.006 |
| Race (White, %) | 53% | 95% | 65% | 0.002 |
| Body mass index (Kg/m2) | 30.1±8.6 | 31.0±8.5 | 29.1±6.8 | 0.60 |
| Drinks last 30 day period | 1.3±1.3 | 368.5±300 | 293.4±256.7^,$ | 0.0001 |
| Hemoglobin (g/dl) | 14.1±1.5 | 13.1±1.5 | 10.5±2.1^,$ | 0.0001 |
| White cell counts (cells/mm3) | 6.1±1.8 | 6.1±2.3 | 11.6±7.9^,$ | 0.001 |
| Platelet counts (cells/mm3) | 242.3±46.1 | 245.7±55.8 | 154.1±100.2^,$ | 0.0001 |
| INR# | - | 0.9±0.3 | 1.6±0.5$ | 0.001 |
| Creatinine (mg/dl) | 0.9±0.2 | 0.7±0.1 | 1.3±1.2 | 0.05 |
| Total bilirubin (mg/dl) | 0.4±0.2 | 0.6±0.2^ | 15.7±2.3^,$ | 0.0001 |
| ALT (U/L) | 23.3±7.2 | 27.5±13.4 | 74.5±70.1^,$ | 0.001 |
| AST (U/L) | 23.9±7.2 | 26.5±8.8 | 143.3±80.1^,$ | 0.0001 |
| Alkaline phosphatase (U/L) | 67.6±12.8 | 73.4±27.0 | 193.0±146.9^,$ | 0.0001 |
| Albumin (g/dl) | 4.3±0.3 | 3.7±0.5^ | 2.9±0.5^,$ | 0.0001 |
| Total Protein (g/dl) | 6.9±0.7 | 6.4±0.5 | 5.7±0.8 | 0.0001 |
| MELD score# | N/A | 6.0±0.1 | 22.1±10.7$ | 0.0001 |
compared to healthy controls,
compared to excessive drinkers without liver disease,
using student T-test for the analysis
Alterations of serum miRNA profiles in patients with alcoholic hepatitis (AH) compared to healthy control (HC) and heavy drinker (HD) groups.
We performed serum miRNA microarray profiling and the heatmap of the top 100 most significantly altered miRNAs among 3 groups is shown in Fig. 1A. Heatmap analysis revealed a similar pattern of miRNA expression levels between HC and HD groups, but not AH. We next employed Sparse Partial Least Squares-Discriminant Analysis (sPLS) with two (2D)- and 3-dimension (3D) plots to demonstrate the differentiation of serum miRNA profiles in our study cohort (Fig. 1B and 1C). sPLS analysis revealed that there were some partial segregations of the differentially expressed miRNA between HC and HD in the 2D plot and 3D plot (Fig. 1B–1C). The top 25 differentially expressed miRNAs in AH compared to HC and HD were illustrated in Fig. 1D and we selected the top 10 with the most differentially expressed miRNAs based on the p-value for further analysis (Fig. 1E and Fig. 2). Additional analysis of the miRNA profiles specifically to healthy controls and AH patients was illustrated in Supplementary Fig. S1.
Fig. 1. Statistical analysis of serum miRNAs profile among healthy controls (HC, n=15), heavy drinkers without liver disease (HD, n=17), and patients with alcoholic hepatitis (AH, n=65).
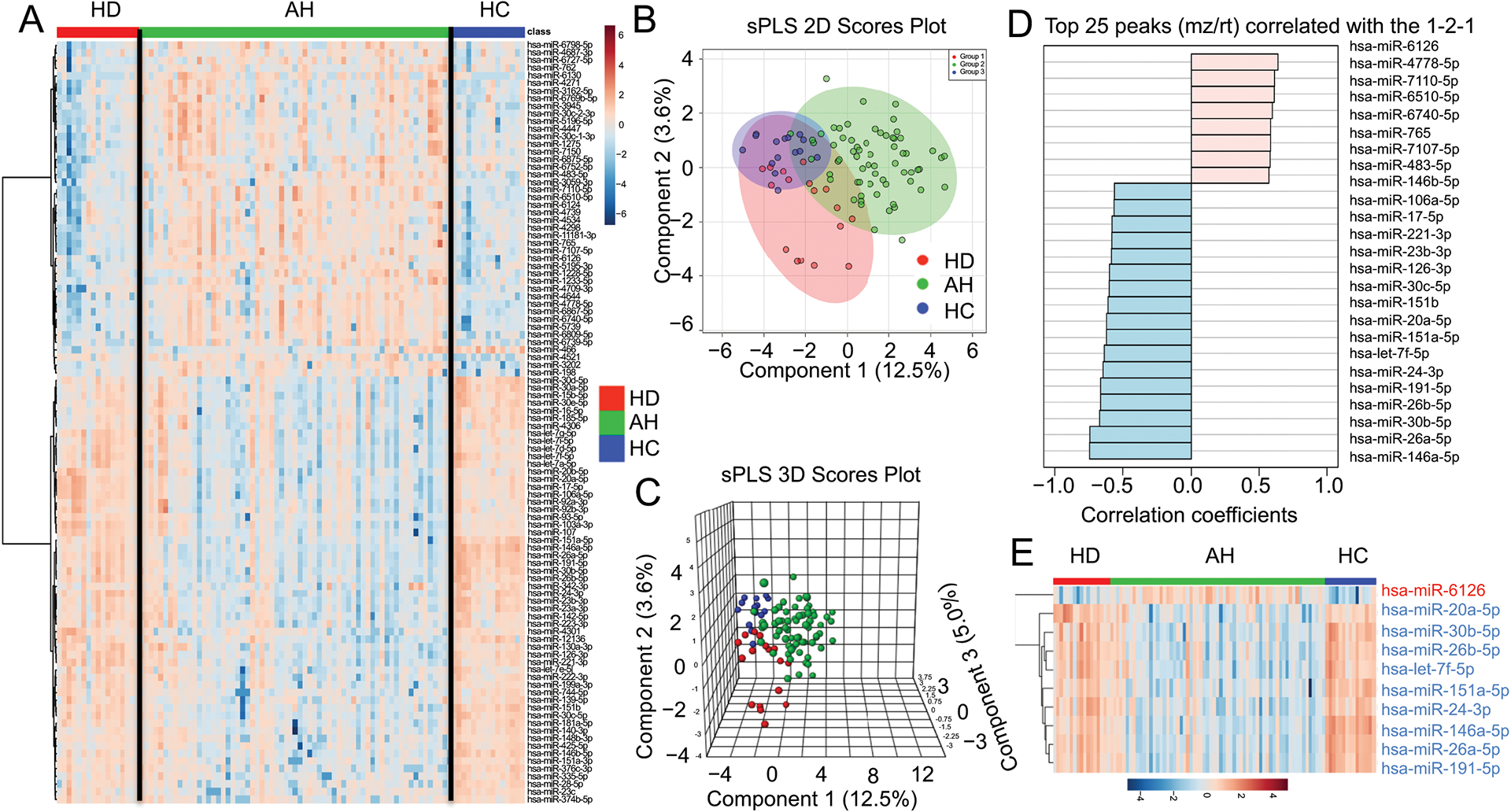
(A) Heatmap of the top 100 most significantly altered miRNAs. (HD: n=17; AH: n=65; HC: n=15). (B–C) 2D (B) and 3D (C) Score plot using sparse Partial Least Squares-Discriminant Analysis (sPLS-DA) between the selected components. The explained variances are shown in the brackets. (D) The top 25 peaks of miRNAs which are correlated with the low-high-low expression pattern in HD-AH-HC groups. (E) Heatmap analysis of the 10 significantly differential serum miRNAs among HC, HD, and AH.
Fig. 2. The relative expression level of the indicated miRNAs based on the microarray reads.
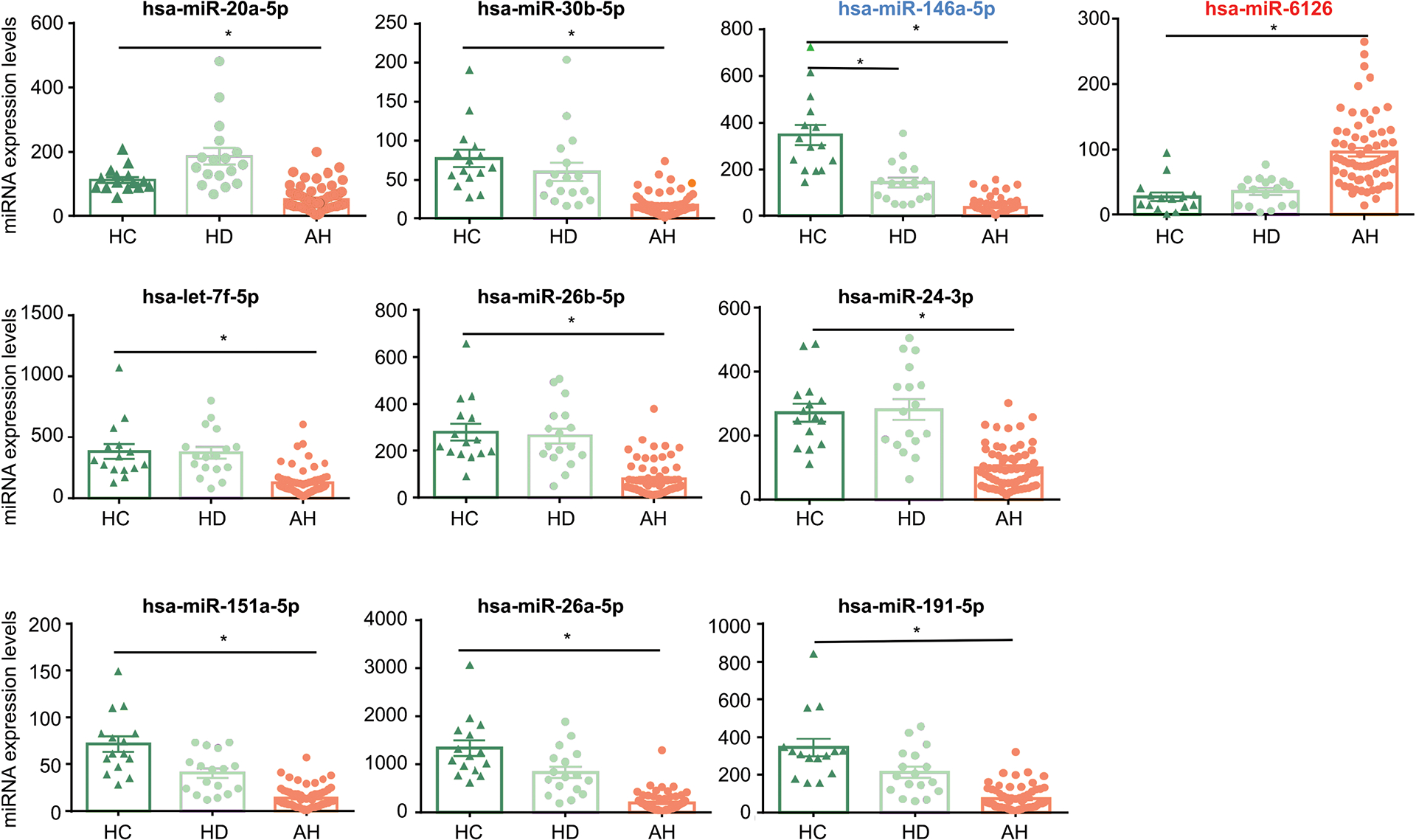
Only Hsa-miR-6126 (in red) was significantly increased in the AH group compared to the HC group. The remaining miRNAs were significantly decreased in patients with AH. Hsa-miR-146a-5p-5p (in blue) was reduced in both HD and AH groups. *: p<0.05 vs HC.
Each dot in Fig. 2 indicated the expression of miRNA from an individual sample. Consistent with the heatmap analysis, we found the serum level of miRNA-6126 significantly increased in the AH group compared to that in HC and HD (Fig. 2, first row, right panel). The remaining serum miRNAs were significantly lower in patients with AH compared to that in HC and HD.
Bioinformatic analysis of the potential gene targets of significantly altered serum miRNAs in patients with AH
We utilized miRNet (www.mirnet.ca) as the tool to identify pathways, potential gene targets, as well as miRNA-target interaction networks(21). We analyzed all the potential targets of the top 10 most significantly altered miRNAs, as shown in Fig. 1E and Fig. 2. The pathway analysis of all identified gene targets revealed that they primarily belong to the P53 signaling and cell cycle pathway (Fig. 3A). The miRNA-target interaction network was illustrated in Fig. 3B; the orange circles representing the targets belong to the P53 signaling pathway.
Fig. 3. Bioinformatic analysis of the potential targets of significantly differential expression of top 10 miRNAs.
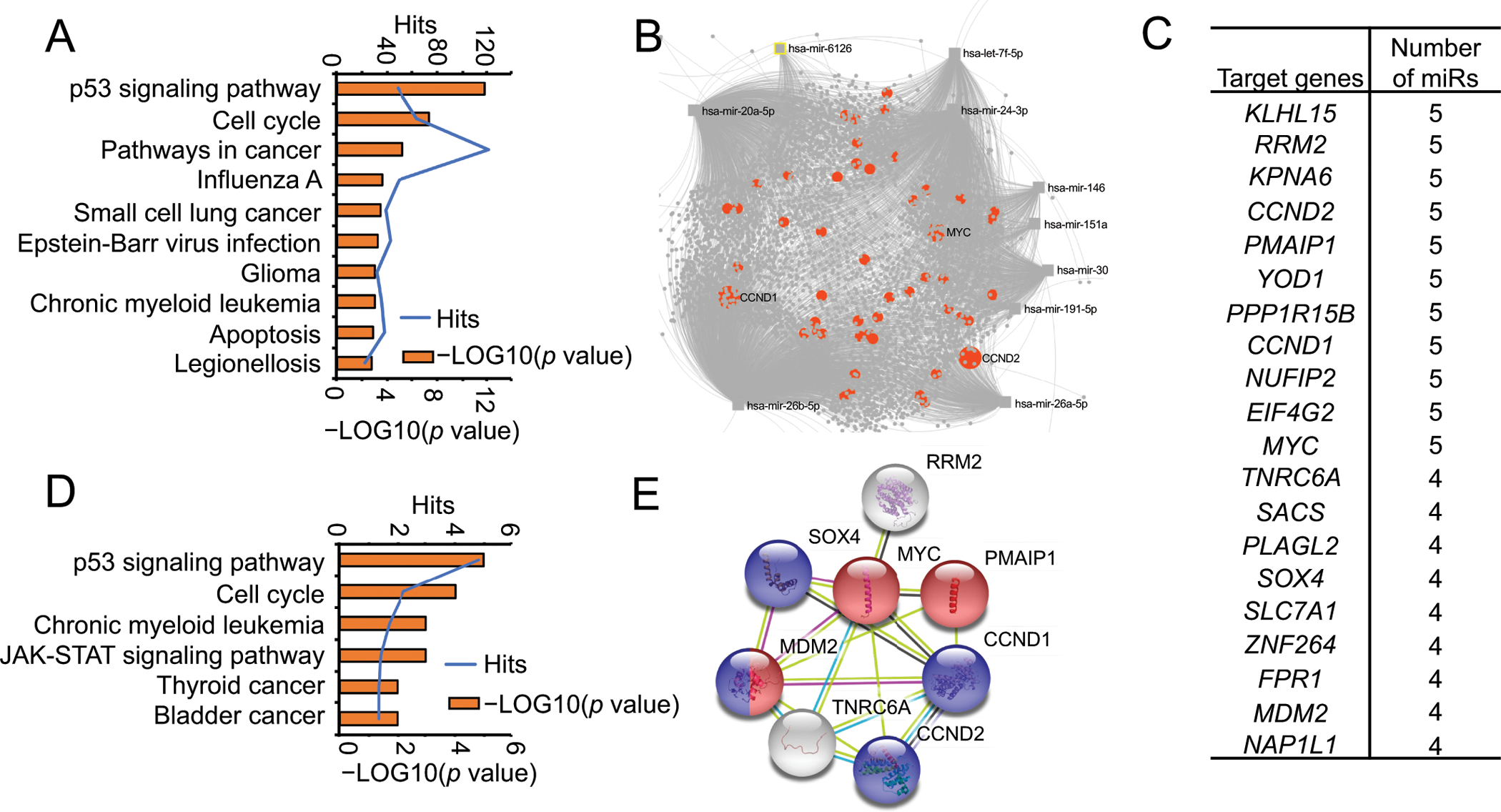
(A) Pathway analysis of all the potential targets of the top 10 significantly differential miRNAs in Fig. 1E. The column sorted with the −LOG10 (p value), the blue line representing the hits in each pathway. (B) The representing network of the top 10 significantly differential miRNAs and their targets. The orange dots represent the targets that belong to the p53 signaling and cell cycle pathway. (C–E) The list of gene targets was shared by at least 4 miRNAs. The pathway analysis of these potential gene targets (C) provided in (D). The networks among these targets were presented in (E). The genes with red balls belong to the regulation of response to DNA damage stimulus (GO:2001020), as the genes with blue balls belong to the regulation of G1/S transition of the mitotic cell cycle (GO:2000045).
It is known that each miRNA can regulate multiple target genes and that each target gene may also be regulated by more than one miRNAs. To increase the probability of selecting specific target genes for further analysis, we identified a set of target genes that were regulated by at least 4 miRNAs among the top 10 most significantly altered miRNAs in the serum of patients with AH (Fig. 3C). We next performed additional pathway analysis specifically to these gene targets and found that they again mainly belong to the P53 signaling and cell cycle pathway (Fig. 3D). The STRING database (https://string-db.org) provided useful computational predictions by integrating publicly available sources (22). Using this database, as expected, 8 of these target genes (Fig. 3C) shared a tight connection with the functions related to the regulation of response to DNA damage stimulus and G1/S transition of the mitotic cell cycle (Fig. 3E). The following 5 genes, RRM2 (ribonucleotide reductase regulatory subunit M2), CCND1 (cyclin D1), CCND2 (cyclin D2), PMAIP1 (phorbol-12-myristate-13-acetate-induced protein 1), and MYC (MYC proto-oncogene) were regulated by 5 miRNAs (Fig. 3C). These genes were not only targeted by the top differentially expressed miRNAs (Fig. 1E and Fig. 2), but they also belonged to the top two pathways, based on the STRING database. The full list of the miRNAs which targeted these 5 genes is provided in Supplementary Fig. S2B and S3A.
Identifying differentially expressed serum miRNAs and hepatic miRNAs in patients with AH
The liver is the major organ responsible for metabolizing alcohol and is affected by alcohol-induced tissue injury. To determine whether the differentially expressed serum miRNAs also occurred in the liver tissues, we examined publicly available hepatic miRNA profiling in the liver of patients with AH (GSE59492)(15) and the top 100 dysregulated serum miRNAs in our patients samples (Fig. 1A). We compared the significantly changed miRNAs in those liver samples with the top 100 dysregulated miRNAs in our blood samples using Venn diagram Supplementary Fig. S2A. A total of 26 miRNAs (Supplementary Fig. S2A) were differentially expressed in both serum and liver of patients with AH. Of these 26 miRNAs, 4 miRNAs (miR-30b-5p, miR-20a-5p, miR-146a-5p, and miR-26b-5p) were in the top 10 most differentially expressed serum miRNAs as shown in Fig. 1E and Fig. 2.
Validation of selected hepatic and serum miRNAs and their target genes
The expression of 4 hepatic miRNAs, miR-30b-5p, miR-20a-5p, miR-146a-5p, and miR-26b-5p, which we identified from the miRNA profiling data (GSE59492) (15), is shown in Fig. 4A. These 4 miRNAs are predicted to target the following genes as shown in Fig. 3C, 3E, Supplementary Fig. S2B, and S3A, RRM2, CCND1, CCND2, PMAIP1, and MYC. We next hypothesized that the reduction of these 4 hepatic miRNAs could lead to an increase in the protein levels of their respective targets. We performed western blot analyses from the liver of healthy controls and patients with AH and found a significant increase in the level of CCND2 (~4 fold), RRM2 (~4 fold), and CCND1 (~2 fold), but not PMAIP1 (Fig. 4B). We were not able to detect the protein expression of MYC in our samples (Supplementary Fig. S3C). To validate our western blot analyses, we next performed the immunohistochemical staining and confirmed an increase in the level of CCND2, RRM2, and CCND1 in patients with AH (Fig. 4D and Supplementary Fig. S3D). Taken together, our data suggested that RRM2, CCND1, and CCND2 are the target of our candidate miRNAs.
Fig. 4. The expression levels of the selected miRNAs and their targets in the liver of patients with AH.
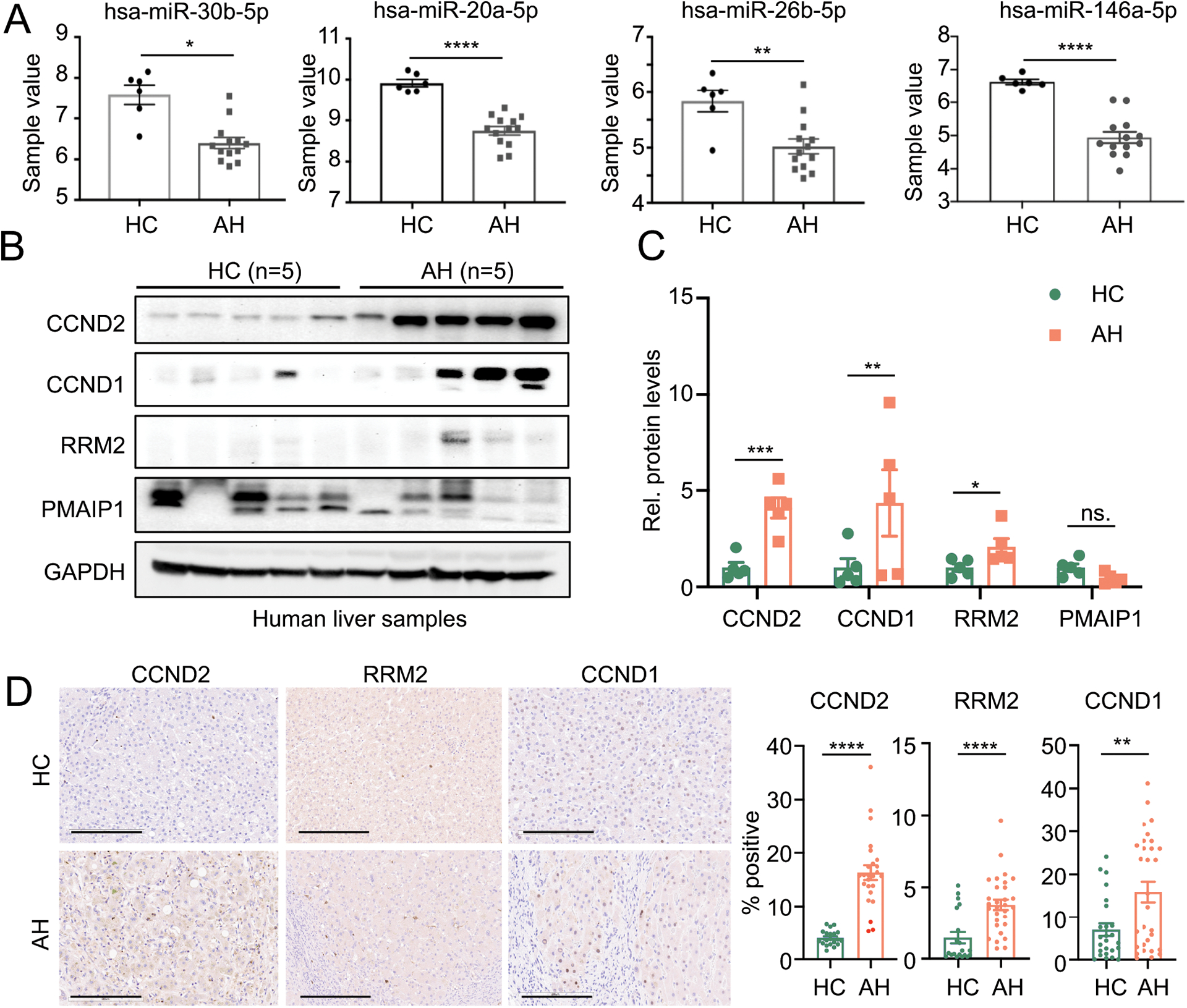
(A) The selected miRs levels in control and AH liver samples from published RNA-seq dataset (GSE59492). (B–C) The representing western blot images and quantifications of selected targets using 5 samples in each group. (D) The representing IHC images and quantification of CCND2, RRM2 and CCND1. Scale bar: 300μm. *p<0.05; **p<0.01; ***p <0.001; ****p <0.0001 vs HC.
Next, we determined the predicted 3’-UTR specific binding sites of RRM2, CCND1, and CCND2 genes for each miRNA using TargetScan (www.targetscan.org) and miRanda database (www.microrna.org) (Fig. 5A and Supplementary Fig. S4–S5). A dose-dependent inhibition of the protein level of these target genes by their respective miRNAs was observed in Fig. 5B. To further confirm that these genes are the target of each miRNA, we constructed the 3’-UTRs with wild-type and mutated binding sites into a luciferase report vector (Supplementary Fig. S6). The luciferase reporter assay to determine the luciferase activity changes of targets 3’UTR by miR-30b-5p, miR-20a-5p, and miR-26b-5p was then performed. We did not conduct experiments with miR-146a-5p as it was previously reported that CCND1 and CCND2 are its targets (23). We found that miR-26b-5p and miR-30b-5p inhibited the 3’-UTR luciferase activity of RRM2 and CCND2 and miR-20a-5p reduced the 3’-UTR luciferase activity of CCND1 and CCND2 (Supplementary Fig. S7). The site mutation on the binding sites of the CCND2 or RRM2 3’-UTR completely blocked the inhibition by miR-26a or miR-30b-5p on luciferase activity. Additionally, the mutation on the binding site 1, but not 2, on the CCND1 3’-UTR ameliorated the inhibition of luciferase activity by miR-20a-5p (Supplementary Fig. S6 and Fig. 5C).
Fig. 5. Validation of the predicted targets.
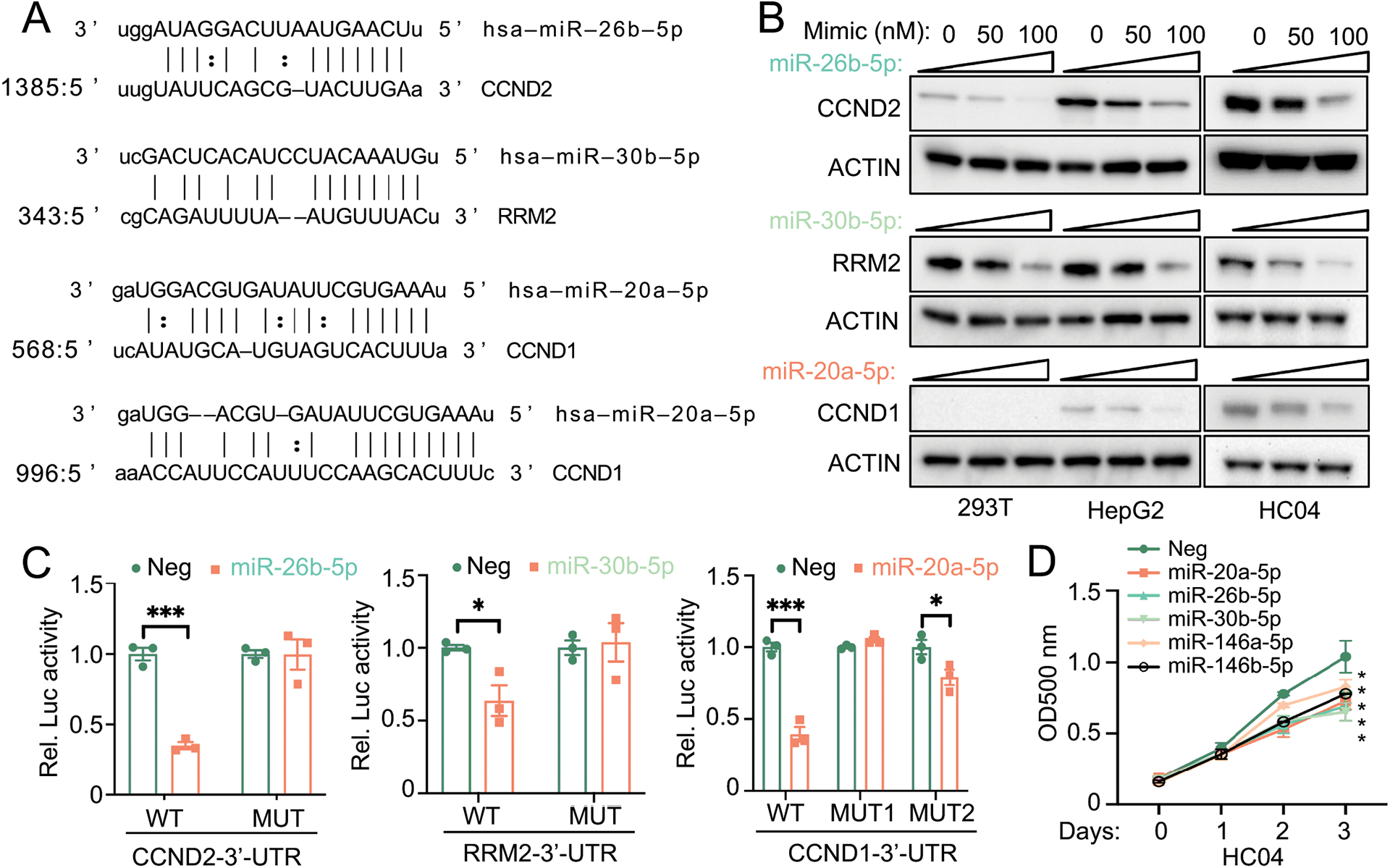
(A) The selected miRNAs and the potential binding sites on the 3’-UTR of their target genes (B) Dose-dependent inhibition of miRNAs on the protein expression of their target genes. Indicated miRNA mimics were transfected into HEK293T, HepG2, or HC04 cells at 50 nM or 100 nM for 24 hrs. Cells were then collected for western blot analysis for respective proteins. (C) The relative luciferase activity of the targets 3’-UTR and 3’-UTR with mutant binding sites after the transfection with miRNA mimics (50 nM), as indicated in HC04 cells. *p<0.05; **p<0.01; ***p<0.001 vs Neg (negative control mimic). (D) The MTS assay in HC04 cells after the transfection of indicated miR mimics (50 nM). *p<0.05 vs Neg.
The functionality of hepatic and serum miRNAs on cellular proliferation
To determine the functional relevance and confirm that selected hepatic and serum miRNAs facilitate cellular proliferation, we next performed an MTS assay by transfecting HC-04 cells with respective miRNA mimics. We found the ability of miR-30b-5p, miR-20a-5p, miR-146a-5p, miR-146b-5p, and miR-26b-5p in inhibiting cellular proliferation (Fig. 5D). We also determined the expression of several hepatic genes in patients with AH, which are previously reported to be associated with liver proliferation and regeneration. We observed a significant increase in the expression of CCND2, SOX4 (SRY-Box Transcription Factor 4), SOX9, EPCAM (epithelial cell adhesion molecule), and MDM2 (mouse double minute 2) (Supplementary Fig. S8). We also found that the hepatic expression of miR-26b-5p and miR-30b-5p was inversely associated with that of CCND1 in AH patients (Supplementary Fig. S9).
The only one of the upregulated miRNAs in the top 10 significantly changed serum miRNAs among HC, HD and AH was miR-6126 (Fig. 2). This miRNA was not part of the publicly available hepatic miRNA profiling, GSE59492 (15), that we used in our analysis in Supplementary Fig. S2. We determined its expression in the liver of patients with AH and found that the miR-6126 levels were increased by 3 folds in AH liver samples compared to HC livers. (Supplementary Fig. S10A). The miR-6126 was recently reported to be involved in oncogenic processes in ovarian cancer cells(24). However, the function of this miRNA in the liver is not known. We analyzed the pathway enrichment of miR-6126’s target genes predicted by TargetScan (www.targetscan.org) and found that several enriched pathways were related to cell growth, cellular proliferation, and liver generation, such as signaling pathways regulating pluripotency of stem cells and FOXO (forkhead box O) signaling (Supplementary Fig. S10B). The top 20 genes in association with these pathways belonged to the PIK3 (phosphoinositied 3-kinases) family and WNT pathway (Supplementary Fig. S10C). We generated stable HC-04 cells overexpressing miR-6126 and performed MTS assay and immunofluorescence staining for proliferating cell nuclear antigen, PCNA, a widely used marker for cellular proliferation(25). We found that overexpression of miR-6126 significantly promoted cellular proliferation (Supplementary Fig. S10D–E). Interestingly, the level of miR-6126 is also associated with serum AST (Supplementary Fig. S11).
Implications and prognostic significance of serum miRNAs in patients with AH
During the median follow-up period of 346 days, 14 (21%) AH patients died. The baseline demographic and characteristics of patients who died or survived were shown in Table 2. There were no differences in age, gender, race, and serum level of miR-6126 in both groups. Those who died during the follow-up had a higher BMI (32.5 vs. 27.2 kg/m2, p=0.007) and MELD score (28.6 vs. 20.4, p=0.009) compared to those who were alive. Interestingly, the serum level of miR-30b-5p (24.1 vs. 16.0, p=0.05), miR-20a-5p (77.4 vs. 47.4, p=0.009), miR-146a-5p (58.9 vs. 34.6, p=0.02), and miR-26b-5p (115.6 vs. 69.4, p=0.03) was significantly higher in AH patients who died. The association between the level of these serum miRNAs and MELD score is shown in Supplementary Fig. S12A–E. Because these miRNAs shared similar target genes (Fig. 3), we thus conducted the correlation matrix to determine their correlations (Supplementary Table 1) before the analysis with the Cox proportional hazards model to determine its prognostic significance on survival outcomes. As expected, a significant correlation was observed among our selected miRNAs (Supplementary Table 1); univariate Cox regression analysis was then utilized. We found that BMI (Hazard ratios, HR 1.108, p=0.004), baseline MELD score (HR 1.05, p=0.01), miR-20a-5p (HR 1.013, p=0.01), miR-146a-5p (HR 1.01, p=0.04), and miR-26b-5p (HR 1.005, p=0.05) were the predictor of mortality in patients with AH (Supplementary Table 2).
Table 2:
Baseline demographic and clinical characteristics of patients with alcoholic hepatitis who were deceased or alive at the end of the study
| Alcoholic hepatitis – deceased (n=14) |
Alcoholic hepatitis - alive (n=51) |
p-value | |
|---|---|---|---|
| Age | 48.3±11.2 | 45.5±11.3 | 0.42 |
| Gender (male, %) | 71% | 71% | 0.95 |
| Race (White, %) | 100% | 86% | 0.33 |
| Body mass index (Kg/m2) | 32.5±7.0 | 27.2±6.2 | 0.007 |
| Drinks per last 30 day period | 216.8±174.3 | 313.5±282.5 | 0.24 |
| Hemoglobin (g/dl) | 10.4±1.7 | 10.8±2.3 | 0.64 |
| White cell counts (cells/mm3) | 10.5±5.9 | 11.2±7.3 | 0.76 |
| Platelet counts (cells/mm3) | 127.3±51.7 | 155.2±96.1 | 0.30 |
| INR | 2.1±0.5 | 1.6±0.6 | 0.007 |
| Creatinine (mg/dl) | 1.5±0.9 | 1.3±1.4 | 0.64 |
| Total bilirubin (mg/dl) | 23.2±16.0 | 12.4±13.8 | 0.01 |
| ALT (U/L) | 60.9±32.2 | 75.5±72.5 | 0.46 |
| AST (U/L) | 143.1±78.7 | 143.9±86.5 | 0.97 |
| Alkaline phosphatase (U/L) | 193.1±156.1 | 178.1±121. | 0.70 |
| Albumin (g/dl) | 2.8±0.6 | 3.0±0.6 | 0.31 |
| Total Protein (g/dl) | 6.0±1.2 | 5.9±0.8 | 0.67 |
| MELD score | 28.6±8.5 | 20.4±10.7 | 0.009 |
| miR_30b | 24.1± 11.4 | 16.0± 11.0 | 0.05 |
| miR-20a-5p | 77.4± 46.1 | 47.4± 34.2 | 0.009 |
| miR-146a-5p | 58.9±49.4 | 34.6±28.1 | 0.02 |
| miR_26b | 115.6± 86.5 | 69.4 ± 64.3 | 0.03 |
| miR_6126 | 112.4±57.6 | 91.5± 52.5 | 0.20 |
Next, we determined the relationship between the variables identified from the Cox regression model and survival outcomes by dichotomizing the variables based on the optimal cut-off to demonstrate good discriminating powers between those who died and survived. The optimal cut-point value was defined as its absolute difference between sensitivity and specificity was minimum based on the ROC curve analysis. The Kaplan-Meier curve survival analyses showed that a MELD score cut-off at 24 (p=0.01) and miR-20a-5p cut-off at 57 (p=0.003) were significant predictors of overall survival. We also observed the trend for miR-146a-5p (cut-off at 33, p=0.09) and miR-26b-5p (cut-off at 71, p=0.07) in association with the survival outcome in patients with AH (Fig. 6). Lastly, the serum level of miR-6126 was not associated with the survival outcome (Supplementary Table 2 and Supplementary Fig. S12F).
Fig. 6.
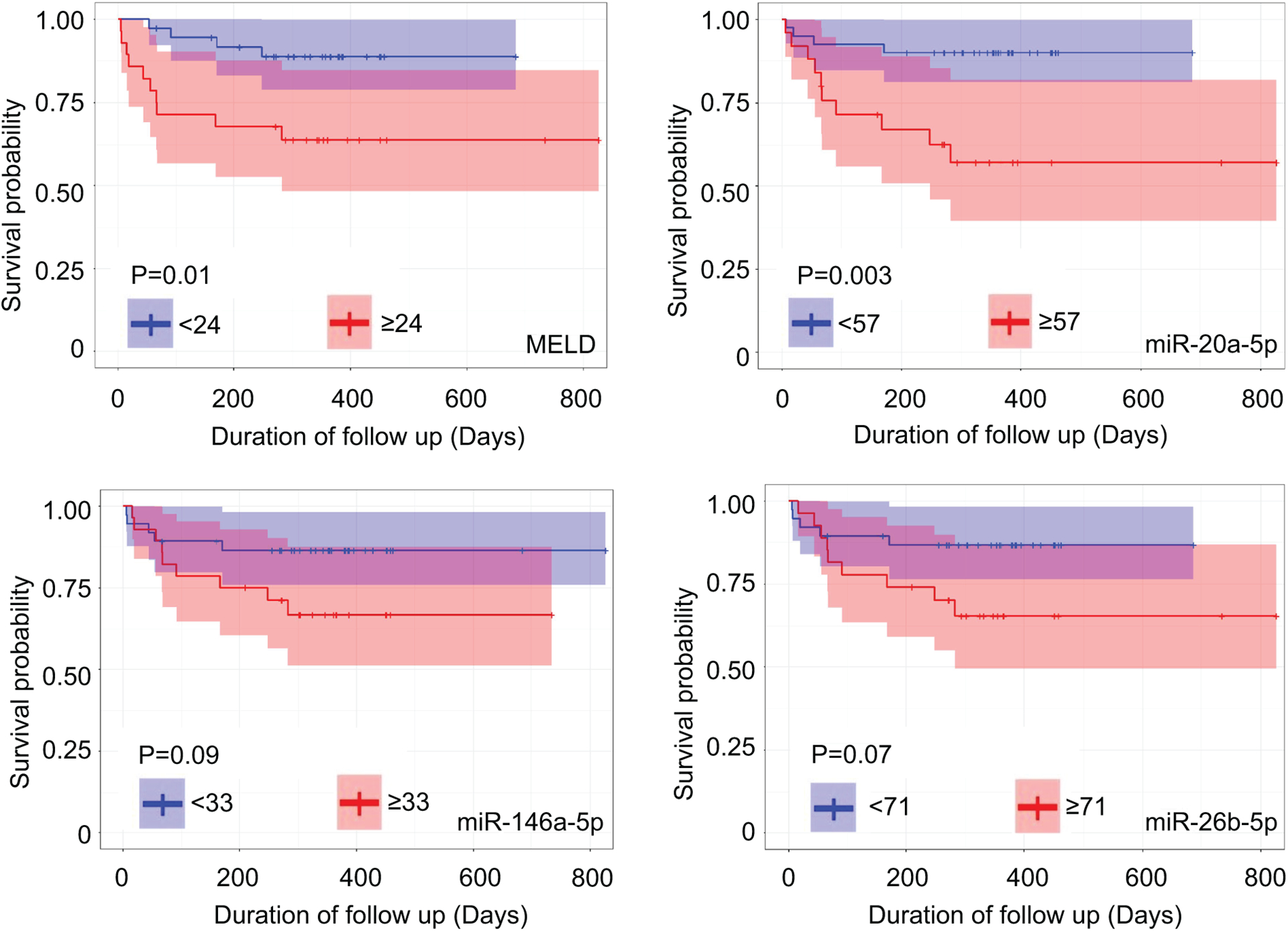
Kaplan-Meier curve survival analyses of MELD score, miR-20a-5p, miR-146a-5p, and miR-26b-5p and their appropriate cut-offs on survival outcome in patients with AH.
Discussion
MiRNAs are endogenous, small non-coding, single-stranded RNAs, 18–24 nucleotides in length, that regulate target gene expression after transcription (26). MiRNAs bind to mRNAs by partial base pairing (partial complementarity) notably at the 3’-UTRs and thereby interfere with the process of translation, repressing translation, or inducing mRNA degradation (27, 28). MiRNAs regulate several biological processes in almost all cell types and specific miRNAs are principal regulators for cellular functions and are involved in disease pathogenesis (13, 14).
Excessive alcohol consumption leads to adverse health outcomes affecting several organ systems. In the liver, excessive alcohol use can cause a spectrum of histopathological changes spanning from simple steatosis, steatohepatitis, advanced fibrosis, and cirrhosis (1). The underlying mechanisms of alcohol‐induced liver injury are multifaceted, involving the alterations in lipid metabolism, immune responses, and epigenetic regulation (29–31); all of these processes are under the control of miRNAs (14, 32). While hepatic steatosis occurs almost universally among excessive drinkers, advanced liver disease notably AH develops only in a subset of heavy drinkers. Because miRNAs are implicated in multiple pathways underlying ALD pathogenesis, we hypothesized that a unique set of miRNAs, which are differentially expressed in excessive drinkers with AH but not in those without AH, is important in driving the development of AH. These distinct miRNAs can be served as surrogates in predicting clinical outcomes. To address this hypothesis, we comprehensively analyzed and established a miRNA signature in the serum specifically expressed in healthy controls, heavy drinkers without liver disease, and those with AH. Serum miRNA signature which was uniquely expressed in AH was validated in the liver of patients with AH to explore their functions and prognostic significance.
Our results demonstrated the serum miRNA signature that is exclusively expressed in excessive drinkers with AH when compared to healthy controls and heavy drinkers without AH; this set of miRNAs is likely implicated in the development of AH among excessive drinkers. Using the selection criteria to identify the top 10 differentially expressed serum miRNAs, we found that they share similar target genes that are responsible for DNA damage and the G1/S transition of the mitotic cell cycle. Among these miRNAs, 4 miRNAs, miR-30b-5p, miR-20a-5p, miR-146a-5p, and miR-26b-5p; have the differential expression in the liver of patients with AH, based on the publicly available hepatic miRNA profiling (15), in analogy to that in the serum. Interestingly, these 4 miRNAs are previously shown to be associated with cellular differentiation and proliferation (23, 33–35). MiR-30b-5p also regulates insulin sensitivity and is associated with the presence of hepatic steatosis (36). To further our understanding of the functional significance of these miRNAs, we found that RRM2, CCND1, and CCND2 are their gene targets. RRM2 converts ribonucleotides to deoxynucleotides which is required for DNA polymerization and repair. It plays an important role in DNA synthesis and cell growth (37). CCND1 and CCND2 encode the protein cyclin D1 and cyclin D2, the members of the Cyclin protein family. Cyclin D1 and D2 are important regulators of the G1 to S phase of cell cycle progression(38). We performed in vitro experiments and observed the ability of these 4 miRNAs in inhibiting cellular proliferation. In addition to miR-30b-5p, miR-20a-5p, miR-146a-5p, and miR-26b-5p, we found a high abundance of miR-6126 in the serum of patients with AH. The function of this miRNA is related to cellular proliferation, as confirmed by the MTS assay and immunofluorescence staining for PCNA. However, unlike miR-30b-5p, miR-20a-5p, miR-146a-5p, and miR-26b-5p, the level of miR-6126 was not an independent predictor of survival in our study cohort.
The “Yin and Yang” of hepatocyte division either activation or inhibition can be attributed to underlying hepatocyte injury from alcohol-induced oxidative stress and inflammation (39, 40). Our results suggested that patients with AH attempt to deal with hepatocyte injury/death by down-regulating hepatic miR-30b-5p, miR-20a-5p, miR-146a-5p, and miR-26b-5p expression and compensatory stimulating hepatocyte division through the activation of their target genes, RRM2, CCND1, and CCND2. In a normal liver, most cells are in the resting G0 phase with a small proportion of hepatocytes in the other phases of the cell cycle (41). Much has been studied on the underlying mechanism driving hepatocyte injury, however, little is known about the effect of alcohol on the hepatocyte repairing process specifically in patients with AH. The liver is a highly regenerative tissue with the capability of rapid regeneration after injury, as illustrated by an increase in the expression of several genes which are involved in liver regeneration such as CCND2, SOX4, SOX9, EPCAM, and MDM2. While we did not observe an induction in mRNA level of CCND1 and RRM2 (Supplementary Fig. 8), their protein expression is markedly increased in AH patients (Fig. 4B and C), suggesting the post-transcriptional regulations of CCND1 and RRM2 by selected miRNAs(42). There is a substantial increase in S-phase DNA synthesis in the hepatocytes among patients with AH (41, 43). The increased expression of liver progenitor cell markers has been reported in these patients and a similar observation is also observed in ethanol-fed rodents with liver injury (41, 43). When we carefully analyzed the significance of these miRNAs on the outcome of patients with AH, we found that those who died had significantly higher expression of these miRNAs compared to that of those who survived. The upstream signaling or molecular mechanisms leading to the differential inhibition of these selected miRNAs and survival outcome should be further explored. It is plausible that the process to inhibit these miRNAs may be impaired resulting in an inappropriate compensatory mechanism to stimulate hepatocyte division in patients who died during the follow-up. We observed a dose-dependent inhibition of the protein levels involving in DNA synthesis and cell cycle progression by these miRNAs; suggesting that those who died may have a diminished capacity in cellular DNA synthesis and cell-cycle progression.
Our work also provides additional insights on the use of miRNAs as the diagnostic or prognostic markers in patients with AH. Increased serum level of miR-122 and miR-155 has been reported in mouse models of ALD (8, 44). Serum miRNA-192 and miR-30a are significantly increased in AH patients when compared to healthy controls (45). Interestingly, we did not find these miRNAs as the most differentially expressed miRNAs in our dataset. It is plausible that our miRNA selection was based on the significantly altered miRNAs not only in healthy controls but also in excessive drinkers without liver disease.
In conclusion, our miRNA profiling study has led us to the discovery of a unique set of miRNA panel in the serum and liver of patients with AH. These miRNAs exert their functions through several target genes which are primarily involved in DNA synthesis and cell cycle progression, an important process in hepatocyte regeneration. The balance between hepatocyte injury, cellular DNA synthesis, and cell cycle progression is an important factor driving hepatic function, liver failure, and mortality in patients with AH. Our results suggest that hepatocyte proliferation is inhibited leading to worsening liver failure (indicated by high MELD score) and short-term mortality in these patients. The treatment strategy for patients with AH should therefore be a combination therapy not only targeting inflammation but also promoting hepatocyte proliferation (or regeneration) (40). At present, there are few prognostic markers in AH patients (46). We found, as previously reported, that the MELD score is associated with mortality in patients with AH (47). Our results also provide a new avenue on the potential use of a panel of miRNAs with specific biological functions related to cell cycle progression and DNA synthesis as the prognostic markers for these patients. Future studies to validate the prognostic ability of our miRNAs in AH and determine if the combination of data between MELD score and miRNAs is better in predicting outcomes should be explored.
Supplementary Material
Fig. 7. Schematic diagram on the significance of selected miRNAs in AH pathogenesis.
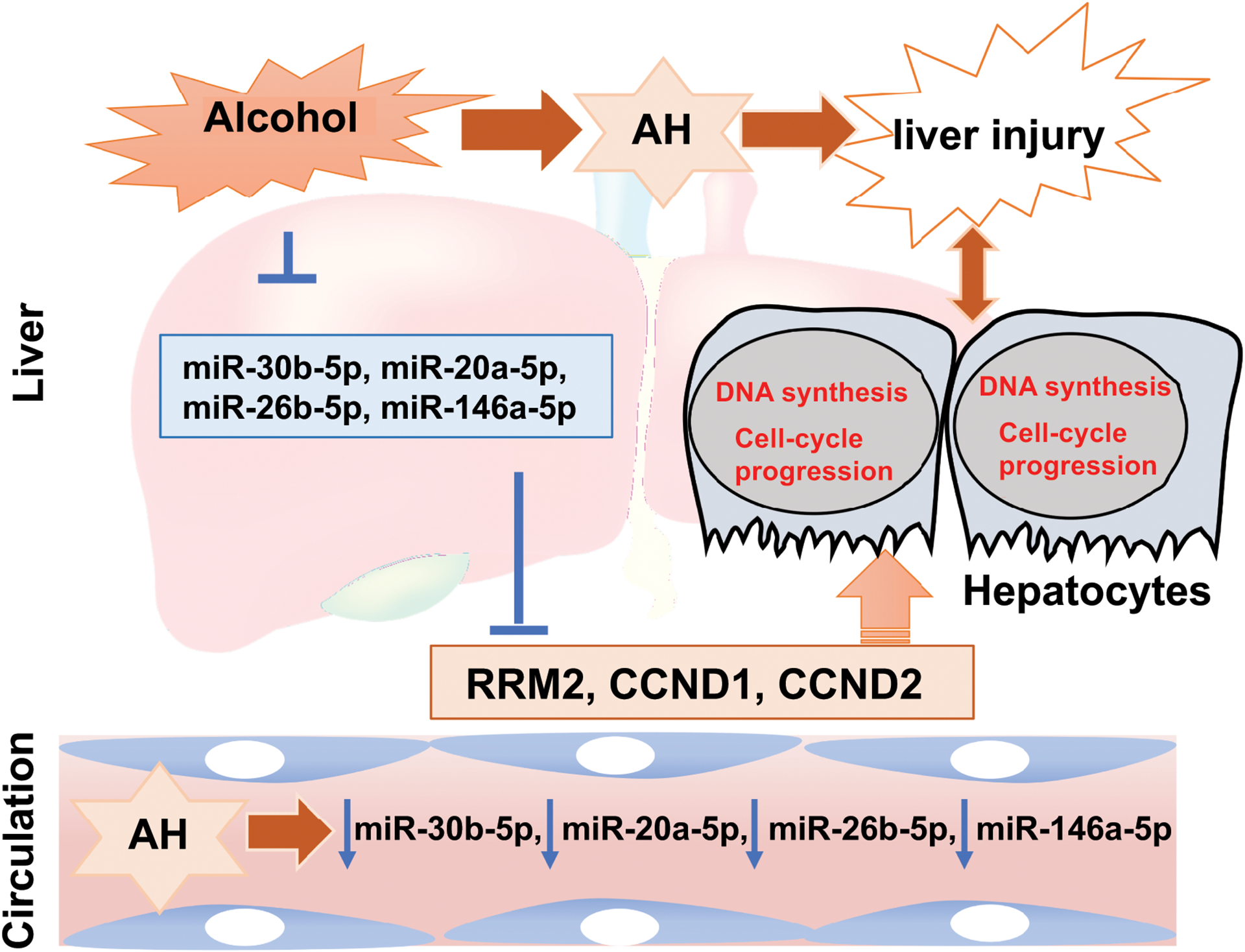
Hepatic and serum expression of miR-30b-5p, miR-20a-5p, miR-26b-5p, and miR-146a-5p are significantly reduced in patients with alcoholic hepatitis. These miRNAs regulate genes, RRM2, CCND1, and CCND2, which are involved in DNA synthesis and cell-cycle progression, in response to liver injury. Higher expression of these miRNAs, suggestive of a diminished capacity in DNA synthesis and cell-cycle progression, predicts short-term mortality in AH patients.
Source of Funding
This study is supported by The Translational Research and Evolving Alcoholic Hepatitis Treatment (TREAT) Consortium is supported by the National Institute on Alcohol Abuse and Alcoholism (NIAAA) (grants 5U01AA021883-04, 5U01AA021891-04, 5U01AA021788-04, 5U01AA021840-04) and partly supported by R01DK107682, R01AA025208, U01 AA026917, UH2/UH3 AA026903, I01CX000361, U01AA026817, Showalter Scholar, and Dean’s Scholar for Medical Research (to SL); and K01 AA026385 and the Ralph W. and Grace M. Showalter Research Trust (to ZY).
Abbreviations:
- AH
Alcoholic hepatitis
- ALD
Alcoholic liver disease
- HD
Heavy drinker without liver disease
- BMI
Body mass index
- CCND1
Cyclin D1
- CCND2
Cyclin D2
- EPCAM
Epithelial cell adhesion molecule
- LncRNAs
Long non-coding RNAs
- MDM2
Mouse double minute 2
- MELD
Model for end-stage liver disease
- MYC
MYC proto-oncogene
- PMAIP1
Phorbol-12-myristate-13-acetate-induced protein 1
- RRM2
Ribonucleotide reductase regulatory subunit M2
- SOX
SRY-box transcription factor
- TLFB
Timeline follow back
- TREAT
the Translational Research and Evolving Alcoholic hepatitis Treatment
Footnotes
Conflict of interest: None
References:
- 1.Liangpunsakul S, Haber P, McCaughan GW. Alcoholic Liver Disease in Asia, Europe, and North America. Gastroenterology 2016;150:1786–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sozio MS, Liangpunsakul S, Crabb D. The role of lipid metabolism in the pathogenesis of alcoholic and nonalcoholic hepatic steatosis. Semin Liver Dis 2010;30:378–390. [DOI] [PubMed] [Google Scholar]
- 3.Crabb DW, Bataller R, Chalasani NP, Kamath PS, Lucey M, Mathurin P, McClain C, et al. Standard Definitions and Common Data Elements for Clinical Trials in Patients With Alcoholic Hepatitis: Recommendation From the NIAAA Alcoholic Hepatitis Consortia. Gastroenterology 2016;150:785–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liangpunsakul S, Puri P, Shah VH, Kamath P, Sanyal A, Urban T, Ren X, et al. Effects of Age, Sex, Body Weight, and Quantity of Alcohol Consumption on Occurrence and Severity of Alcoholic Hepatitis. Clin Gastroenterol Hepatol 2016;14:1831–1838 e1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mandrekar P, Bataller R, Tsukamoto H, Gao B. Alcoholic hepatitis: Translational approaches to develop targeted therapies. Hepatology 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chayanupatkul M, Liangpunsakul S. Alcoholic hepatitis: a comprehensive review of pathogenesis and treatment. World J. Gastroenterol 2014;20:6279–6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thursz MR, Richardson P, Allison M, Austin A, Bowers M, Day CP, Downs N, et al. Prednisolone or pentoxifylline for alcoholic hepatitis. N Engl J Med 2015;372:1619–1628. [DOI] [PubMed] [Google Scholar]
- 8.Szabo G, Bala S. MicroRNAs in liver disease. Nat. Rev. Gastroenterol. Hepatol 2013;10:542–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takahashi K, Yan I, Haga H, Patel T. Long noncoding RNA in liver diseases. Hepatology 2014;60:744–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics 2007;8:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ludwig N, Leidinger P, Becker K, Backes C, Fehlmann T, Pallasch C, Rheinheimer S, et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res 2016;44:3865–3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pirola CJ, Fernandez Gianotti T, Castano GO, Mallardi P, San Martino J, Mora Gonzalez Lopez Ledesma M, Flichman D, et al. Circulating microRNA signature in non-alcoholic fatty liver disease: from serum non-coding RNAs to liver histology and disease pathogenesis. Gut 2015;64:800–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han S, Zhang T, Kusumanchi P, Huda N, Jiang Y, Liangpunsakul S, Yang Z. Role of microRNA-7 in liver diseases: a comprehensive review of the mechanisms and therapeutic applications. J Investig Med 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang T, Yang Z, Kusumanchi P, Han S, Liangpunsakul S. Critical Role of microRNA-21 in the Pathogenesis of Liver Diseases. Front Med (Lausanne) 2020;7:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blaya D, Coll M, Rodrigo-Torres D, Vila-Casadesus M, Altamirano J, Llopis M, Graupera I, et al. Integrative microRNA profiling in alcoholic hepatitis reveals a role for microRNA-182 in liver injury and inflammation. Gut 2016;65:1535–1545. [DOI] [PubMed] [Google Scholar]
- 16.Li M, He Y, Zhou Z, Ramirez T, Gao Y, Gao Y, Ross RA, et al. MicroRNA-223 ameliorates alcoholic liver injury by inhibiting the IL-6-p47(phox)-oxidative stress pathway in neutrophils. Gut 2017;66:705–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yao J, Cheng Y, Zhang D, Fan J, Zhao Z, Li Y, Jiang Y, et al. Identification of key genes, MicroRNAs and potentially regulated pathways in alcoholic hepatitis by integrative analysis. Gene 2019;720:144035. [DOI] [PubMed] [Google Scholar]
- 18.Liangpunsakul S, Beaudoin JJ, Shah VH, Puri P, Sanyal AJ, Kamath PS, Lourens SG, et al. Interaction between the patatin-like phospholipase domain-containing protein 3 genotype and coffee drinking and the risk for acute alcoholic hepatitis. Hepatol Commun 2018;2:29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Z, Tsuchiya H, Zhang Y, Lee S, Liu C, Huang Y, Vargas GM, et al. REV-ERBalpha Activates C/EBP Homologous Protein to Control Small Heterodimer Partner-Mediated Oscillation of Alcoholic Fatty Liver. Am J Pathol 2016;186:2909–2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Z, Tsuchiya H, Zhang Y, Hartnett ME, Wang L. MicroRNA-433 inhibits liver cancer cell migration by repressing the protein expression and function of cAMP response element-binding protein. J Biol Chem 2013;288:28893–28899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan Y, Xia J. miRNet-Functional Analysis and Visual Exploration of miRNA-Target Interactions in a Network Context. Methods Mol Biol 2018;1819:215–233. [DOI] [PubMed] [Google Scholar]
- 22.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li YL, Wang J, Zhang CY, Shen YQ, Wang HM, Ding L, Gu YC, et al. MiR-146a-5p-5p inhibits cell proliferation and cell cycle progression in NSCLC cell lines by targeting CCND1 and CCND2. Oncotarget 2016;7:59287–59298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanlikilicer P, Rashed MH, Bayraktar R, Mitra R, Ivan C, Aslan B, Zhang X, et al. Ubiquitous Release of Exosomal Tumor Suppressor miR-6126 from Ovarian Cancer Cells. Cancer Res 2016;76:7194–7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bologna-Molina R, Mosqueda-Taylor A, Molina-Frechero N, Mori-Estevez AD, Sanchez-Acuna G. Comparison of the value of PCNA and Ki-67 as markers of cell proliferation in ameloblastic tumors. Med Oral Patol Oral Cir Bucal 2013;18:e174–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ambros V The functions of animal microRNAs. Nature 2004;431:350–355. [DOI] [PubMed] [Google Scholar]
- 27.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Brien J, Hayder H, Zayed Y, Peng C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front Endocrinol (Lausanne) 2018;9:402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao B, Ahmad MF, Nagy LE, Tsukamoto H. Inflammatory pathways in alcoholic steatohepatitis. J Hepatol 2019;70:249–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang Y, Zhang T, Kusumanchi P, Han S, Yang Z, Liangpunsakul S. Alcohol Metabolizing Enzymes, Microsomal Ethanol Oxidizing System, Cytochrome P450 2E1, Catalase, and Aldehyde Dehydrogenase in Alcohol-Associated Liver Disease. Biomedicines 2020;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao B, Bataller R. Alcoholic liver disease: pathogenesis and new therapeutic targets. Gastroenterology 2011;141:1572–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han S, Zhang T, Kusumanchi P, Huda N, Jiang Y, Liangpunsakul S, Yang Z. Role of microRNA-7 in liver diseases: a comprehensive review of the mechanisms and therapeutic applications. J Investig Med 2020;68:1208–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Q, Liu S, Zhang J, Ma X, Dong M, Sun B, Xin Y. Roles and regulatory mechanisms of miR-30b-5p in cancer, cardiovascular disease, and metabolic disorders (Review). Exp Ther Med 2021;21:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ai F, Zhang Y, Peng B. miR-20a-5p regulates proliferation, differentiation and apoptosis in P19 cell model of cardiac differentiation by targeting Smoothened. Biol Open 2016;5:1260–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang N, Zhou X, Zhao M, Zhao D, Zhu Z, Li S, Yang H. Down-regulation of microRNA-26b modulates non-small cell lung cancer cells chemoresistance and migration through the association of PTEN. Acta Biochim Biophys Sin (Shanghai) 2015;47:530–538. [DOI] [PubMed] [Google Scholar]
- 36.Dai LL, Li SD, Ma YC, Tang JR, Lv JY, Zhang YQ, Miao YL, et al. MicroRNA-30b regulates insulin sensitivity by targeting SERCA2b in non-alcoholic fatty liver disease. Liver Int 2019;39:1504–1513. [DOI] [PubMed] [Google Scholar]
- 37.Lee B, Ha SY, Song DH, Lee HW, Cho SY, Park CK. High expression of ribonucleotide reductase subunit M2 correlates with poor prognosis of hepatocellular carcinoma. Gut Liver 2014;8:662–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alao JP. The regulation of cyclin D1 degradation: roles in cancer development and the potential for therapeutic invention. Mol Cancer 2007;6:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sancho-Bru P, Altamirano J, Rodrigo-Torres D, Coll M, Millan C, Jose Lozano J, Miquel R, et al. Liver progenitor cell markers correlate with liver damage and predict short-term mortality in patients with alcoholic hepatitis. Hepatology 2012;55:1931–1941. [DOI] [PubMed] [Google Scholar]
- 40.Gao B, Shah VH. Combination therapy: New hope for alcoholic hepatitis? Clin Res Hepatol Gastroenterol 2015;39 Suppl 1:S7–S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Apte UM, McRee R, Ramaiah SK. Hepatocyte proliferation is the possible mechanism for the transient decrease in liver injury during steatosis stage of alcoholic liver disease. Toxicol Pathol 2004;32:567–576. [DOI] [PubMed] [Google Scholar]
- 42.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 2008;9:102–114. [DOI] [PubMed] [Google Scholar]
- 43.Dubuquoy L, Louvet A, Lassailly G, Truant S, Boleslawski E, Artru F, Maggiotto F, et al. Progenitor cell expansion and impaired hepatocyte regeneration in explanted livers from alcoholic hepatitis. Gut 2015;64:1949–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bala S, Szabo G. MicroRNA Signature in Alcoholic Liver Disease. Int J Hepatol 2012;2012:498232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Momen-Heravi F, Saha B, Kodys K, Catalano D, Satishchandran A, Szabo G. Increased number of circulating exosomes and their microRNA cargos are potential novel biomarkers in alcoholic hepatitis. J Transl Med 2015;13:261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moreno C, Mueller S, Szabo G. Non-invasive diagnosis and biomarkers in alcohol-related liver disease. J Hepatol 2019;70:273–283. [DOI] [PubMed] [Google Scholar]
- 47.Dunn W, Jamil LH, Brown LS, Wiesner RH, Kim WR, Menon KV, Malinchoc M, et al. MELD accurately predicts mortality in patients with alcoholic hepatitis. Hepatology 2005;41:353–358. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


