Summary
The brain has a remarkable but underappreciated capacity to limit memory formation and expression. The term memory suppressor gene was coined in 1998 as an attempt to explain emerging reports that some genes appeared to limit memory. At that time, only a handful of memory suppressor genes were known, and they were understood to work by limiting cAMP-dependent consolidation. In the intervening decades, almost 100 memory suppressor genes with diverse functions have been discovered that affect not only consolidation but also acquisition and forgetting. Here we highlight the surprising extent to which biological limits are placed on memory formation through reviewing the literature on memory suppressor genes. In this review, we present memory suppressors within the framework of their actions on different memory operations: acquisition, consolidation, and forgetting. This is followed by a discussion of the reasons why there may be a biological need to limit memory formation.
Keywords: Memory enhancement, acquisition, consolidation, forgetting, behavioral flexibility, memory accuracy
Introduction
Molecular and cellular studies of learning and memory over the last five decades have focused nearly exclusively on functions that are required for normal learning and memory. The general approach has been to apply insults to certain molecules or cells to see if this compromises these processes. For instance, genetic approaches that were employed early identified learning and memory mutants in Drosophila, including the well-known mutant, rutabaga. Rutabaga was found to encode a Ca2+/calmodulin responsive adenylyl cyclase (Levin et al., 1992; Livingstone et al., 1984), a critical molecule supporting many different forms of learning across phyla by acting as a molecular coincidence detector between the stimuli that are associated (Gervasi et al., 2010; Tomchik and Davis, 2009). A second classic example of a molecule required for normal memory formation is the N-methyl-d-aspartate (NMDA) ionotropic glutamate receptor, which is crucial for the formation of many different types of memory (Morris, 2013). This receptor also acts as a molecular coincidence detector, sensing the coincident release of neurotransmitter from a presynaptic neuron and the depolarization of the postsynaptic neuron (Bliss and Collingridge, 1993). The cumulative research over the last five decades has identified hundreds of genes and the molecules they encode that are essential for normal learning and memory processes.
However, there is a yin for every yang. Recent research has revealed that the brain is also designed with processes that constrain memory formation, such that reducing the activity of these molecules enhances learning and/or memory performance. Genes that encode molecules limiting memory in a wildtype animal are termed ‘memory suppressor genes’ by analogy to tumor suppressor genes (Abel et al., 1998). Thus, the normal function of these molecules is to suppress a process involved in the formation, storage or recall of memory. The opposite of a memory suppressor gene is a memory enhancer gene, which increases memory formation when its activity is increased. A notable example of a memory enhancer is the NMDA receptor subunit NR2B. Overexpression of the NR2B subunit in the mouse forebrain enhances long-term memory (LTM) in a variety of tasks, including fear conditioning, novel object recognition, and Morris water maze performance, paralleled by increased hippocampal long-term potentiation (Tang et al., 1999); whereas reduction of the NR2B subunit impairs spatial memory and long-term potentiation (Clayton et al., 2002).
The earliest memory suppressor molecules identified include repressors of CREB, such as ApCREB2 in Aplysia. Inhibiting ApCREB2 activity by injecting anti-ApCREB2 antiserum into Aplysia sensory neurons allows the formation of long-term facilitation (LTF) after only a single application of serotonin, whereas LTF is normally produced only with multiple applications of serotonin. Thus, ApCREB2 suppresses the formation of LTF by repressing CREB function (Bartsch et al., 1995). Studies of CREB repressors such as ATF4 in the mouse (Chen et al., 2003; Yin et al., 1994) provide cross-species support that negative regulators of CREB function as memory suppressor molecules.
Only recently has there been focused and systematic research on memory suppressor genes. This focus is important for several reasons. First, a deep understanding of memory suppressors will pinpoint the neurophysiological processes that limit memory formation. This will reveal the number and type of molecular control points that limit learning and memory. Arguably, understanding the negative regulators of memory is as equally important as understanding the positive ones. Second, identifying memory suppressors is relevant for understanding enhanced and compromised memory that occurs in the human population. Savant syndrome, for example, is characterized by remarkable memory capacity, often occurring in individuals with autism spectrum disorder (ASD) (Treffert, 2009). Conversely, post-traumatic stress disorder (PTSD) presents as maladaptive, abnormally strong memories – potentially arising from the dysregulation of memory suppressor genes and/or their products. Third, the products of memory suppressor genes offer targets for designing drugs that could act as cognitive enhancers, since identifying antagonists of molecular targets is often easier than identifying agonists.
The formation of memory that becomes retrievable over short- and long-periods of time can be conceptualized as four basic operations: (1) the initial acquisition or encoding of information, (2) the consolidation of memory into a more stable form resistant to disruption, (3) the active forgetting of memory, and (4) the retrieval of memory (Figure 1). In principle, memory suppressors could function by limiting one or more of these four operations. Only the first three – acquisition, consolidation, and forgetting – have been explored using molecular genetics approaches because retrieval remains relatively intractable. Here, we provide a comprehensive review of memory suppressors within the framework of the first three memory operations. Many of the memory suppressors identified in the scientific literature could not be placed into this framework due to the lack of data required for assignment. Nevertheless, we have included them in Supplemental Tables to serve as a comprehensive resource, grouping the genes and gene products into three categories. Table S1 contains suppressors of early memory (effects found at the earliest time point tested within 4hr of training). These are likely to include suppressors of acquisition as well as suppressors of early memory. Table S2 includes genes that suppress late memory (no effects at the earliest time point tested but affecting time points thereafter). This category would contain suppressors of consolidation or promoters of forgetting. Table S3 summarizes memory suppressors that could not be categorized into either of the first two groups. We identify memory suppressors from the enhancement of behavioral memory when the suppressor is inhibited, and we provide a mechanistic basis for the memory enhancement when it is known. Finally, this is followed by a discussion of the reasons why memory suppressors exist.
Figure 1: Memory Operations.
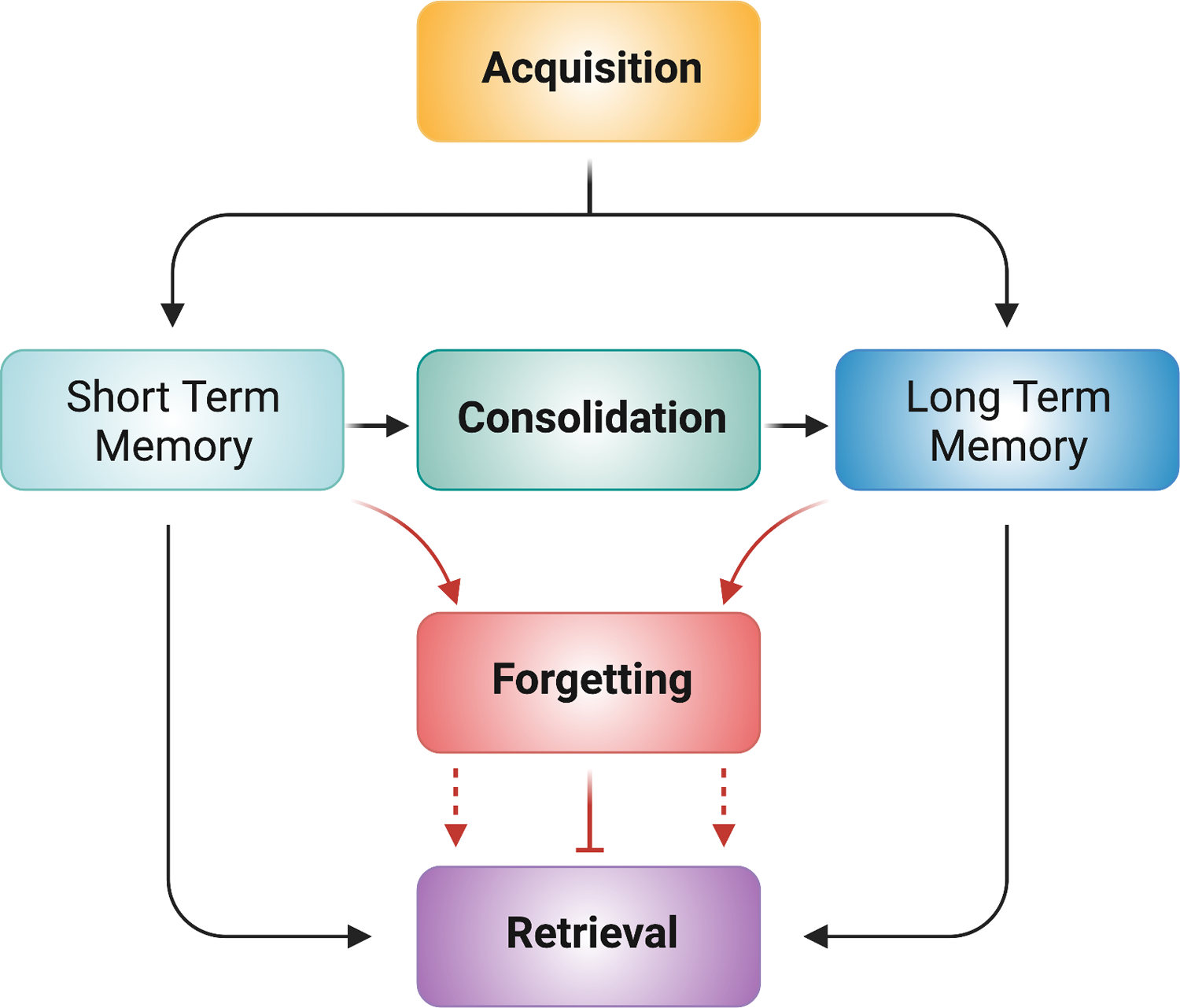
Acquisition includes the molecular, cellular, and circuit processes during learning that encode memory. Short-term memory is unstable and prone to disruption. Consolidation stabilizes memories into a long-term form for subsequent retrieval. Short-term or long-term memory can be altered by forgetting processes (red lines), leading to a reduction in retrieval (dotted red lines with arrow). Alternatively, transient forgetting processes can temporarily block retrieval (solid red line with blocked end). Retrieval allows for the access of information stored in short- and long-term memory.
Suppressors of Acquisition
We define suppressors of acquisition as those genes or molecules that, when inhibited, result in enhanced memory performance when the animal is tested immediately after training. In the Drosophila literature, memory acquisition is frequently measured using multiple, short conditioning trials to monitor how memory performance builds as a function of training trial number (Beck et al., 2000). Moreover, by altering the training trial number across groups to normalize performance immediately after training, researchers have been able to dissociate effects on acquisition from those on memory consolidation or forgetting (Figure 2). However, most rodent learning paradigms measure memory performance hours to weeks after training, so only a few of the memory suppressors identified in mouse studies could definitively be assigned as suppressors of acquisition. Nevertheless, it is likely that many of the early or unassigned genes and their products in Table S1 and S3 act on acquisition.
Figure 2: Normalizing for differences in memory acquisition.
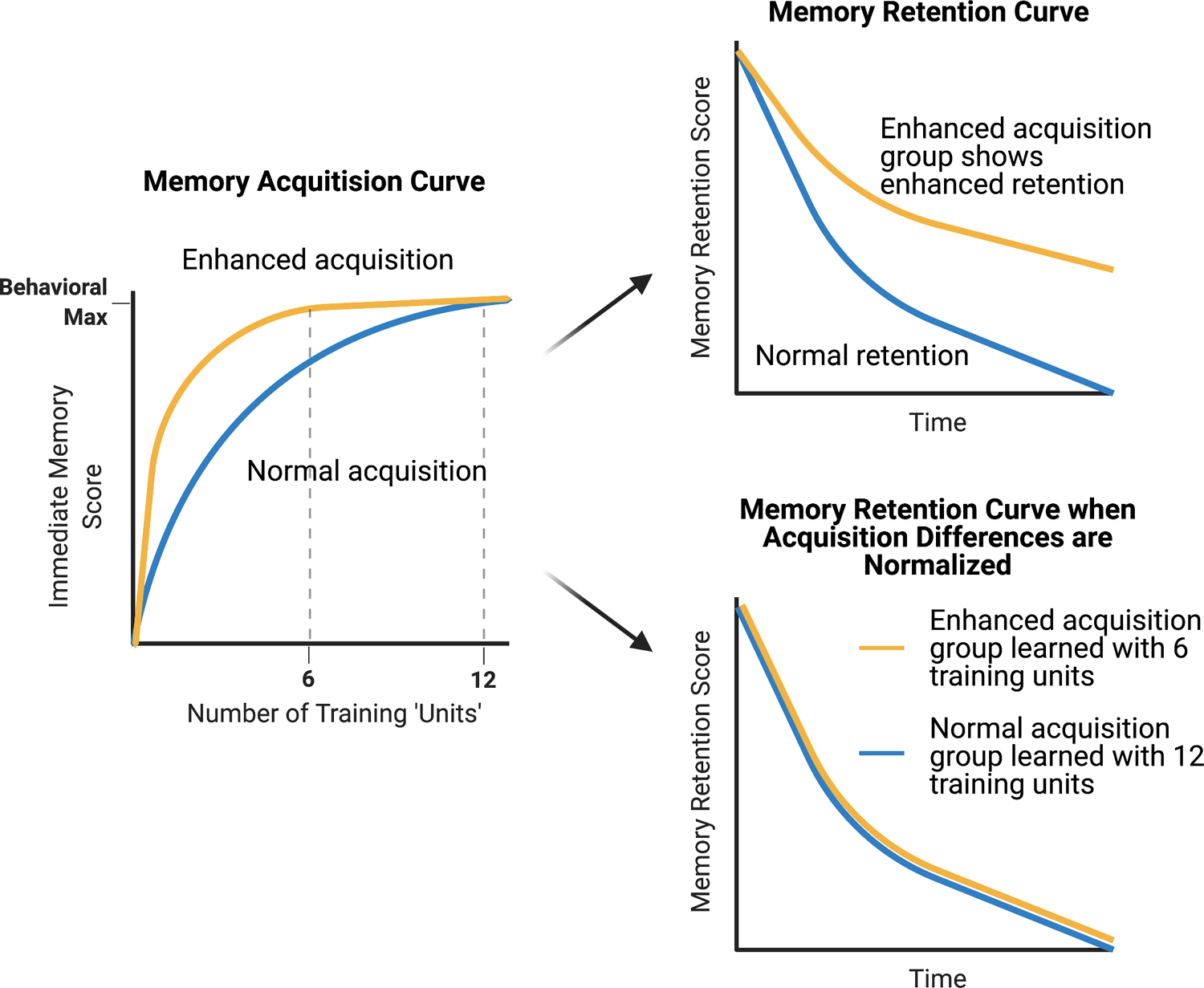
Reducing memory suppressors of acquisition results in enhanced memory acquisition scores (yellow line) compared to wildtype animals (blue line) when short, multiple training sessions (‘Units’) are applied (left graph). Both groups often reach the same measured level of behavioral performance with sufficient training due to ceiling levels on performance in the memory tasks employed. When these groups are tested for later memory, they often exhibit enhanced retention scores across time (upper right graph). However, a retention curve obtained when groups reach ceiling levels of performance does not discriminate between enhanced acquisition, enhanced consolidation, or reduced forgetting. To tease these scenarios apart, the two groups are trained with different numbers of training ‘units’, normalizing for differences in acquisition before testing for memory retention (lower right graph). If the retention scores align with one another, this molecule is designated a suppressor of acquisition. If retention scores remain distinct, the molecule is assigned as having a role in acquisition as well as consolidation or active forgetting.
GABAergic inhibition limits the potency of learned stimuli
Genes that encode molecules of GABAergic inhibitory systems function as suppressors of acquisition. The γ-amino butyric acid (GABA)A receptors are ligand-gated chloride channels that, upon binding of GABA neurotransmitter, hyperpolarize neurons – reducing their ability to be depolarized and fire action potentials. Resistance to dieldrin (Rdl), which codes for a subunit of the GABAA receptor, offers the earliest report of a suppressor of acquisition in Drosophila (Liu et al., 2007; Liu et al., 2009). Rdl is highly expressed in the antennal lobes and mushroom body neurons (MBn; Liu et al., 2007), areas known be important for Drosophila olfactory processing and memory (Davis, 2004) and which are analogous to the mammalian olfactory bulb and primary olfactory cortex (Murthy, 2011). Rdl overexpression in the MBn impairs aversive olfactory conditioning, while RNAi-mediated reduction of Rdl in the MBn enhances the performance of flies in both aversive and appetitive olfactory conditioning tasks (Liu et al., 2007; Liu et al., 2009). Interestingly, Rdl knockdown enhances memory acquisition using short, multiple training sessions like those described in Figure 2, but produces aligned memory retention scores with control flies when knockdown flies are normalized for the enhanced acquisition (Liu et al., 2007). Examining MBn activity in response to olfactory stimuli using the optical Ca2+ reporter GCaMP indicates that Rdl overexpression reduces, while Rdl knockdown enhances, odor induced MBn activity (Liu et al., 2007). These observations led to the conclusion that reducing GABAA receptor function by knocking down Rdl increases the MBn response to odors, which serves as the conditioned stimulus, leading to increased acquisition. In other words, reducing GABAA receptor increases the potency of the odors used for conditioning as experienced by the MBn (Figure 3).
Figure 3: Memory Suppressors of Acquisition.
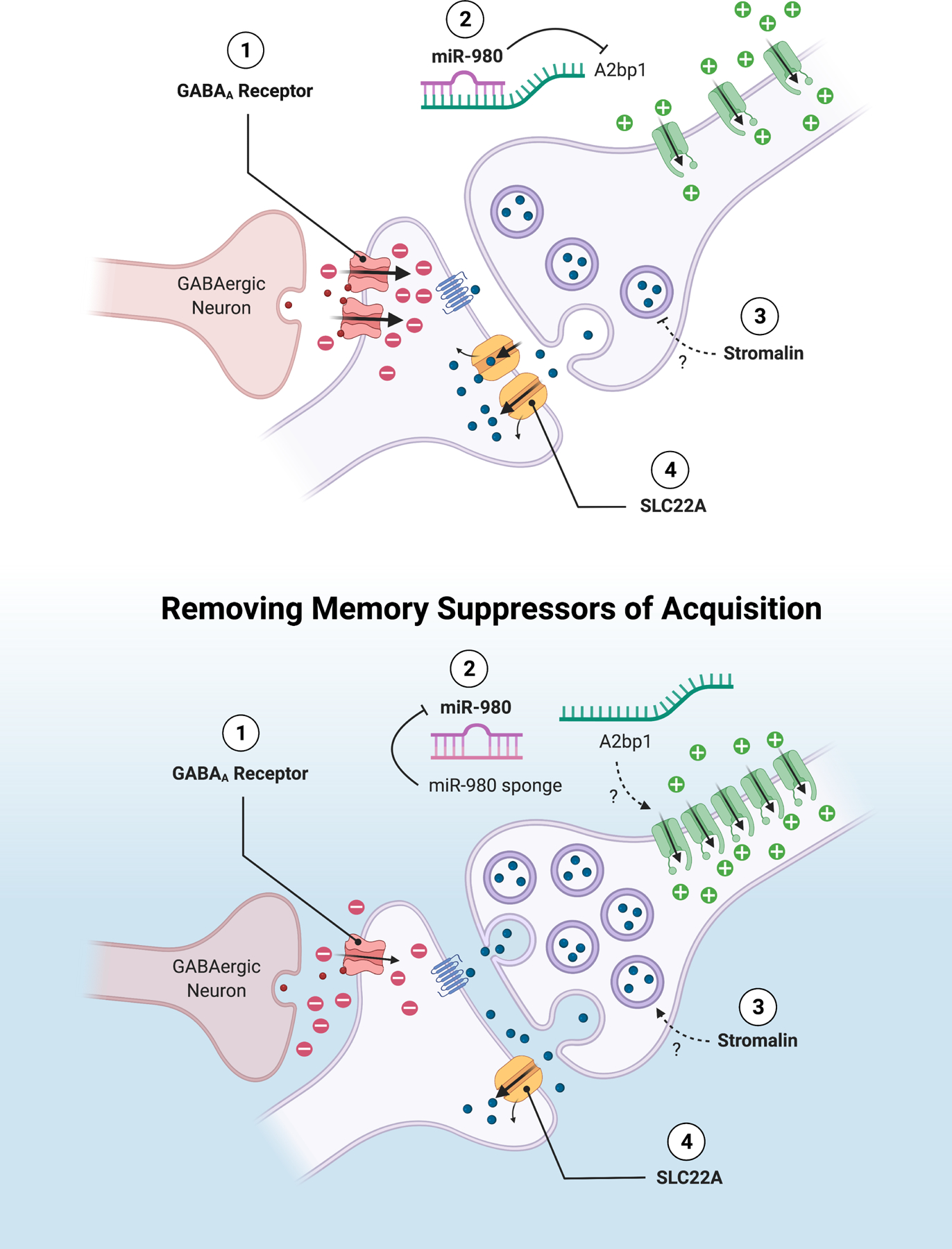
Suppressors of acquisition function in several ways. (1) Inhibiting the neural representation of stimulus strength in the circuit through GABAergic receptor function. Both GABAA and GABAB receptors suppress acquisition. (2) Capping neuronal excitability by miR-980 inhibition of A2bp1. (3) Limiting the number of synaptic vesicles through Stromalin regulation of gene expression. (4) Reducing the neurotransmitter persistence in the synapse by the transporter SLC22A. These suppressors of acquisition function by controlling neuronal activity or synaptic strength in the memory-relevant neural circuit.
Studies of GABAA receptor function in the mouse support this general conclusion. GABAA receptor subunit α5 (GABRAα5) knockout (KO) mice acquire spatial memory in the matching-to-place version of the Morris water maze faster than control mice (Collinson et al., 2002). This subunit exhibits preferential expression in the mouse hippocampus, a brain area known to be required for spatial and contextual memory. The same study discovered that the hippocampal neurons in the KO mice exhibit increased paired pulse facilitation, indicating they have a higher amplitude of excitatory synaptic potentials compared to controls. Thus, in both Drosophila and mice, reducing inhibitory signaling through GABAA receptors leads specifically to enhanced memory acquisition.
Genes encoding metabotropic GABAB receptors also function as acquisition suppressors in Drosophila. Reducing levels of the GABA-B-R3 receptor in the dopamine neurons (DAn) responsible for conveying the sugar reward during olfactory appetitive conditioning results in enhanced acquisition of the reward memory (Yamagata et al., 2021). Inhibiting GABA release from the upstream neuron by reducing glutamic acid decarboxylase 1 (Gad1) and the vesicular GABA transporter (VGAT) also enhance appetitive memory. Interestingly, these flies exhibit a higher learning maximum, rather than reaching the same ceiling asymptotic level as observed with many other acquisition suppressors. Calcium imaging revealed that GABA-B-R3 acts to limit the responses of reward DAn synapses to sugar ingestion (Yamagata et al., 2021). Thus, the hyperpolarization resulting from both GABAA and GABAB receptors limit acquisition to balance the strength of memory formed with the stimuli used in olfactory conditioning.
Several other mouse studies have also reported GABAergic genes as memory suppressors, although it is not clear whether they suppress acquisition since measures of immediate memory post-training were not recorded. Knockout of GABAA receptor subunits GABRA4 (Fan et al., 2020), GABRAα2 (Engin et al., 2015), GABRAβ2 (Parker et al., 2011), and heterozygous knockouts of the GABA transporter GAT1 (Shi et al., 2012) were all reported to lead to enhanced performance in a variety of memory tasks including aversive and appetitive conditioning and spatial memory in the Morris water maze.
MicroRNAs that constrain the excitability of neurons
MicroRNAs (miRs) are small, non-coding RNAs that regulate gene expression through translational repression and/or through mRNA degradation (Bushati and Cohen, 2007). To identify novel miRs that participate in learning and memory processes, a Drosophila miR screen was conducted using a “microRNA sponge” approach to individually inhibit more than 130 miRs, preventing them from binding and silencing their mRNA targets (Busto et al., 2015). This screen identified several miRs that act as memory suppressor genes. One miR identified from the screen, miR-980, was shown to suppress memory acquisition.
MiR-980 inhibition in all neurons, and various subsets of neurons involved in aversive olfactory conditioning (except for GABAergic neurons), led to increased memory scores in the aversive olfactory conditioning paradigm, while miR-980 overexpression in MBn impaired memory (Guven-Ozkan et al., 2016). When memory acquisition and retention were assayed, miR-980 inhibition in all neurons was shown to increase memory scores in both experiments. However, when memory retention was tested after normalizing for the increased memory acquisition of miR-980 knockdown flies (Figure 2), intermediate memory scores (3hrs) were found to be higher than the control, suggesting that miR-980 may independently constrain memory acquisition and alter consolidation and/or forgetting processes (Guven-Ozkan et al., 2016). To determine how a reduction in miR-980 might alter neuron function leading to increased memory expression, the authors examined calcium responses of the MBn to odor. The results indicated that reduced miR-980 led to a stronger odor response in these neurons. Furthermore, current-clamp recordings of the projection neurons that deliver odor information to the MBn, showed that miR-980 inhibition increases firing frequency compared to control neurons or neurons that overexpress miR-980. Thus, miR-980 suppresses memory acquisition by limiting the excitability of neurons (Figure 3). The gene, A2bp1, was identified in this study as a major mediator of the observed miR-980 effects. A2bp1 is an autism and epilepsy-susceptibility gene functioning principally via a role in RNA alternative splicing (Lee et al., 2009). The conclusion that miR-980 functions by limiting neuronal excitability parallels results obtained by reducing GABA receptor function.
Transcriptional regulation that limits the potency of unconditioned stimuli
Stromalin is a member of the highly conserved cohesion complex that is best known for its role in supporting proper chromosome segregation during mitosis and in regulating gene expression (Peters et al., 2008). A memory suppressor function of Stromalin was first identified in a large, RNAi behavioral screen using Drosophila (Walkinshaw et al., 2015). RNAi-mediated depletion of Stromalin in MBn or DAn enhanced memory, while overexpression had no effect (Phan et al., 2019). Further experiments (Figure 2) showed that Stromalin knockdown specifically increases memory acquisition. The authors focused on the role of Stromalin in DAn to identify the cellular mechanisms of Stromalin’s memory suppression effects. DAn in the Drosophila aversive olfactory conditioning paradigm are known to encode the aversive or foot-shock information. RNAi-mediated reduction of Stromalin increases communication between the DAn and their downstream MBn partners, determined by measuring cAMP increases in the MBn in response to DAn activation, while not affecting the calcium responses of the DAn itself to the shock stimulus. Cellular studies revealed that this occurred because of an increase in the number of synaptic vesicles in Stromalin knockdown DAn compared to controls. The important conclusion drawn from this study is that Stromalin suppresses memory acquisition by limiting the number of synaptic vesicles in the DAn that convey the unconditioned stimuli (Figure 3).
In an unexpected twist, the authors found that Stromalin’s memory suppressor function occurs not during adulthood, when olfactory memory was assayed, but during the development of 3rd instar larvae (Phan et al., 2019). This observation was made possible using tools that allow the experimenter to control the timing of RNAi-mediated knockdown. Due to its developmental function that becomes apparent in adult memory tests, the mechanism by which the gene limits synaptic vesicle number was hypothesized to be through the developmental regulation of gene expression from cohesin’s known role in gene expression.
One other gene has been found to function during development to limit adult memory formation. Diaphanous homologous protein 1 (Diap1) is an actin polymerization regulator, whose reduction in utero using RNAi or overexpression of miR-9 results in adult mice that display improved performance in an auditory fear conditioning task 48hrs after training (Lin et al., 2017). Unlike Stromalin, Diap1’s effects are not due to enhancements in memory acquisition, but on LTM. Diap1 appears to limit dendritic branching and synapse formation of developing cortical neurons. Thus, the attractive explanation for the experimental results is that this memory suppressor gene constrains adult memory by restricting the potential to form synaptic connections during development.
Limiting neurotransmitter function at the synapse
DmSLC22A (CG7442) was also identified from the aforementioned RNAi screen (Walkinshaw et al., 2015). Based on its homology to the mammalian SLC22A family of transporter proteins, it was predicted to be an organic cation transporter, but no substrates were known when its memory suppressor function was discovered. In vitro experiments identified DmSLC22A as a transporter for choline and acetylcholine (Figure 3; Gai et al., 2016). RNAi-mediated depletion of DmSLC22A in MBn or DAn increases memory scores, whereas overexpression in the MBn impairs memory performance of flies in the aversive olfactory conditioning paradigm. Since the projection neurons that provide olfactory input into the MBn are cholinergic and SLC22A localizes to the dendrites of the MBn, the gene product appears to function by limiting the persistence of acetylcholine at the projection neuron-MBn synapse (Figure 3).
In summary, the known suppressors of acquisition function by: (1) limiting the potency of stimuli through circuit inhibition, (2) capping neuronal excitability, (3) limiting the availability or release of neurotransmitter, and (4) limiting neurotransmitter persistence/function in the synapse (Figure 3).
Suppressors of Consolidation
Consolidation is the process of converting newly formed memories that are initially sensitive to disruption into long-lasting memories that are stable and resistant to interference (McGaugh and Alpern, 1966; Quinn and Dudai, 1976; Scoville and Milner, 2000; Squire and Davis, 1981). The strength and repetition of the stimuli being learned are the major factors that influence consolidation (Carew et al., 1972; Cepeda et al., 2006; Tully et al., 1994). Thus, consolidation provides a filter to save highly consequential events and/or events likely to be encountered again. Suppressors of consolidation prevent inconsequential information or spurious associations from being encoded into LTM.
Consolidation suppressors limit cAMP-signaling and CREB regulated gene expression
Early cellular studies with Aplysia and genetic studies with Drosophila established that the cAMP signaling pathway is critical for acquisition and short-term memory (Brunelli et al., 1976; Byers et al., 1981; Livingstone et al., 1984; Scholz and Byrne, 1988). Research with these same model systems subsequently established that the normal function of CREB is required for consolidation of LTM and the plasticity associated with it (Dash et al., 1990; Kaang et al., 1993; Tully et al., 1994). A suppressor of consolidation associated with CREB function was identified from studies with Aplysia. When neutralizing antibodies to the endogenous CREB inhibitor, ApCREB2, are injected into sensory neurons, a single pulse of serotonin, which is normally insufficient to drive long-term changes in plasticity, is able to produce long-term facilitation (Bartsch et al., 1995). Similar results were obtained using RNAi to knockdown ApCREB2 (Lee et al., 2003). Inhibition of ATF4 in the mouse, a homologue of ApCREB2, was found to enhance spatial memory and long-term potentiation (LTP) (Chen et al., 2003). These and other related studies have established that CREB-dependent transcription is required for LTM and that suppressors of CREB prevent the consolidation of short-term memory (STM) into LTM (Figure 4).
Figure 4: Memory Suppressors of Consolidation.
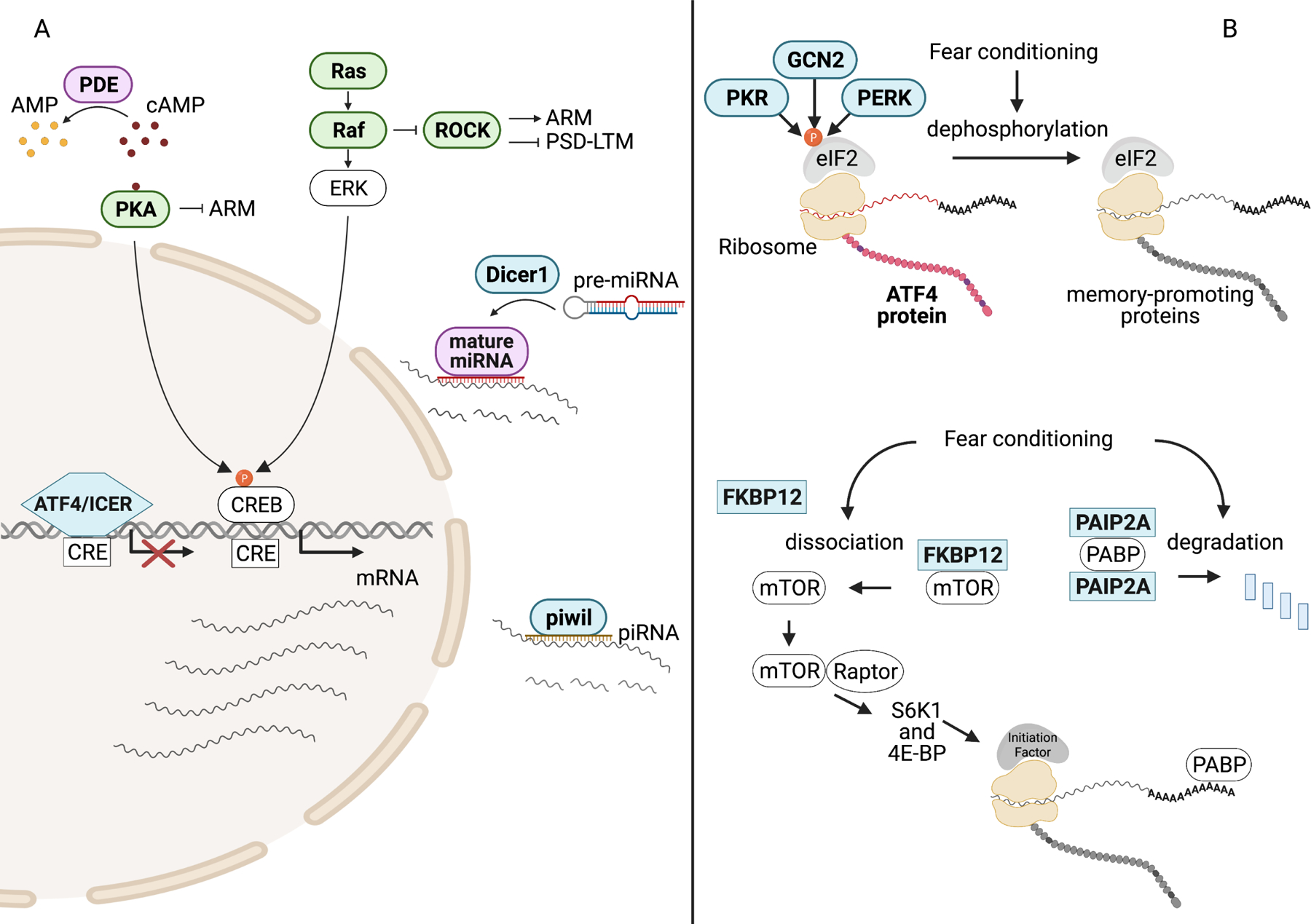
Memory suppressor genes are in bold and color-coded green (fly), blue (mouse), and purple (both mouse and fly). A) PDEs regulate cAMP levels by converting cAMP to 5’-AMP. Activation of PKA by cAMP promotes consolidation through the phosphorylation of CREB but also inhibits consolidated ARM in Drosophila through an unknown mechanism. In Drosophila, ROCK activity enhances ARM and inhibits PSD-LTM. ROCK activity is suppressed by ERK-independent Ras signaling. Phosphorylated CREB binds CRE and drives transcription of mRNA required for memory formation. Proteins that inhibit CREB-dependent transcription, like ATF4 and ICER, suppress consolidation. Following transcription, memory-relevant mRNA can be reduced by small, non-coding RNAs (miRNA, piRNA) that either prevent translation or promote mRNA degradation. Dicer is required for the processing of pre-miRNA and piwil is required for piRNA-mediated mRNA degradation.
B) Regulation of translation factors by inhibitory proteins through phosphorylation or protein-protein interactions prevents ribosomal translation of mRNA to protein. Phosphorylation of the alpha subunit of eIF2 by PKR, GCN2, and PERK promotes translation of ATF4 and suppresses the synthesis of memory-promoting proteins. Fear conditioning stimulates dephosphorylation of eIF2α thereby facilitating consolidation. Fear conditioning also promotes consolidation by disinhibiting mTOR and PABP. FKBP12 association with mTOR prevents mTOR/raptor complex formation and subsequent control of translation regulators by mTOR/raptor. PAIP2A complex formation with PABP prevents PABP association with mRNA poly(A) tails. Fear conditioning drives the degradation of PAIP2A, freeing PABP to facilitate translation through its direct interaction with mRNA.
The conceptual model for protein-synthesis dependent consolidation described above supports the notion that natural constraints on CREB activity, such as the activity of the ApCREB2 gene/protein, or ATF4, limit consolidation. Indeed, the CREB repressor ICER (Mioduszewska et al., 2003) meets the definition of a memory suppressor gene and it achieves this suppression by altering the CREB-activity threshold for memory consolidation. In mice, loss of ICER enhanced fear memory after weak but not strong training (Kojima et al., 2008). This indicates that ICER acts to prevent weak events from being encoded into LTM. Consistent with this model, overexpression of ICER in the forebrain impaired long-term fear memory but not memory measured one hour after training. Interestingly, elevated cAMP drives the expression of ICER (Mioduszewska et al., 2003), indicating that the same signals that promote the activation of CREB also lead to increased CREB-suppressor activity.
How does CREB promote consolidation and how might CREB suppression prevent it? Several studies have found that CREB modulates basal neuronal excitability and the development of LTP. Mice that lack CREB display impaired fear and spatial LTM and a failure to develop hippocampal LTP (Bourtchuladze et al., 1994). Conversely, overexpression of CREB in the amygdala increases neuronal excitability (Yiu et al., 2014, Zhou et al., 2009) and LTM after suboptimal fear conditioning (Josselyn et al., 2001). Similar increases in neuronal excitability occur following CREB overexpression in the hippocampus (Barco et al., 2002; Gruart et al., 2012) and the rat nucleus accumbens (Dong et al., 2006). Expression of CREB in random subsets of mouse amygdala neurons increases encoding of fear memory in those neurons relative to neurons without ectopic CREB expression (Han et al., 2007, Han et al., 2009, Zhou et al., 2009). Together, these data suggest that CREB’s role in consolidation may occur by increasing neuronal excitability.
Since CREB abundance and activation provide a threshold for determining the probability that a memory undergoes CREB-dependent consolidation (Figure 4), one would anticipate that genes and gene products that limit this activation, besides the ATF4/ICER proteins discussed above, would function as consolidation suppressors. There exist two obvious control points at which memory suppressors might function. The first is through the negative regulation of cAMP levels from the activity of 3’,5’-cyclic phosphodiesterases (PDEs) and the second through negative regulators of PKA (protein kinase A), a cAMP-dependent kinase that stimulates CREB transcriptional activity (Figure 4).
The Drosophila dnc gene encodes a cAMP PDE with four mammalian orthologs known as PDE4a-d (Byers et al., 1981; Chen et al., 1986; Davis and Kiger, 1981). Consistent with a memory suppressor function, Scheunemann et al (2018) reported that a partial reduction of dnc activity in serotonergic projection neurons (SNP) neurons facilitates LTM following subthreshold training. In wildtype flies, the dnc PDE prevents SPN cAMP elevation during a single round of training. However, after multiple spaced training cycles, the PDE is inhibited and elevated cAMP levels promote SPN activity, which ultimately drives LTM consolidation. Nevertheless, the role of dnc as a memory suppressor gene is difficult to cleanly unpack since it seems to have neuron-specific roles in memory formation and the complete loss-of-function of dnc throughout the organism causes a profound impairment in memory formation (Dudai et al., 1976).
The function of PDE4s as memory suppressors has more robust support from mammalian studies. Injection of the PDE4 inhibitor rolipram (Nemoz et al., 1985; Henkel-Tiggs and Davis, 1990) into the brains of mice enhances contextual long-term fear memory but has no effect on STM (Barad et al., 1998). In addition, rolipram injection specifically during the consolidation period enhances object recognition memory (Rutten et al., 2006) and rescues age-related memory impairments in object recognition (Wimmer et al., 2020). Genetic studies support the role of PDE4s as consolidation suppressors. PDE4a KO mice exhibit enhanced passive avoidance (Hansen et al., 2014) and PDE4d KO and hippocampal knockdown enhances spatial memory and novel object recognition (Li et al., 2011). Interestingly, inhibition of PDE2 during consolidation promotes novel object location memory when combined with suboptimal doses of a PDE4 inhibitor (Paes et al., 2021), suggesting that multiple PDEs constrain consolidation. Indeed, PDE8b PDE1b, and PDE7 have been reported to suppress multiple types of memory. (McQuown et al., 2019; Tsai et al., 2012, McQuown et al., 2021).
The evidence is less robust for a potential role of PKA inhibitors in consolidation and indeed, some evidence is contrary to this point of view. Eukaryotic cells express a family of PKA inhibitors (PKA-I), that include PKIα, β1, β2 and γ (Chen and Sabatini, 2021). Although no systematic studies have been performed to determine whether any of these molecules function as memory suppressors, significant reductions in PKIα mRNA levels were observed following neuronal stimulation in the rat hippocampus, raising the possibility that learning relieves PKIα inhibition of PKA and promotes subsequent PKA-dependent consolidation (de Lecea et al., 1998). Contrary to the well-established role for PKA in protein synthesis dependent-consolidation, Horiuchi et al (2008) reported that a partial loss of PKA activity in the MBn leads to enhanced protein synthesis-independent consolidated memory (Horiuchi et al., 2008), and this partial-loss-of function rescues age-related impairments in consolidated memory with no effect on learning (Yamazaki et al., 2010).
Small, noncoding RNAs function as consolidation suppressors by regulating protein product abundance
Cellular systems that silence RNA expression act to restrict consolidation (Figure 4). Disrupting the function of miRs as well as piRNA, another type of small, non-coding RNA that represses target mRNA translation and transcript degradation (Huang et al., 2017), enhances memory. The enzyme Dicer1 is required for the conversion of mature miRNA to pre-miRNA (Bartel, 2007) and loss of Dicer1 in the mouse brain enhances conditioned fear memory and alters mRNA levels of memory-related genes (Fiorenza et al., 2016; Konopka et al., 2010). There are three piwi-like (piwil) genes (O’Donnell and Boekel, 2007) in the mouse that participate in piRNA-mediated mRNA degradation, two of which serve redundantly as memory suppressors. Hippocampal knockdown of piwil1 and piwil2 simultaneously in adult mice enhances long-term contextual fear memory, while leaving behavior unaltered during the training phase (Leighton et al., 2019).
The specific piRNAs that mediate piwil-dependent memory suppression remain unknown. However, progress has been made in identifying the miRNAs that suppress consolidation. In Drosophila, miR-92a, a highly conserved miR across species (Yuva-Aydemir et al., 2015), was identified as a memory suppressor gene in a miR memory screen (Busto et al., 2015). Inhibition of miR-92a in the MBn has no effect on learning but enhances one form of consolidated memory known as anesthesia-resistant memory (ARM) (Quinn and Dudai, 1976). This form of consolidated memory is erased by a cold shock to the flies within one hour after acquisition and is distinct from protein synthesis-dependent consolidation. One gene regulated by miR-92a that appears to be largely responsible for ARM consolidation encodes the motor protein Khc73 (Guven-Ozkan et al., 2020), whose overexpression also enhances ARM. These observations implicate protein transport in the process of consolidation, which is consistent with the earlier discovery that consolidation is enhanced by the overexpression of kinesin heavy chain in Aplysia sensory neurons (Puthanveetil et al., 2008).
MiR-182 levels are rapidly reduced in the mouse amygdala after auditory fear conditioning, suggesting that this miR may keep consolidation at bay until released by miR-182 degradation (Griggs et al., 2013). Consistent with this proposition, overexpression of miR-182 in the lateral amygdala reduces the expression of the plasticity promoting proteins, Rac1 and cortactin, and impairs LTM but not STM after cued fear conditioning. It seems likely that miR-182 is a consolidation suppressor, but this provisional conclusion is based on overexpression experiments. A miR-182 disruption experiment is required to satisfy the defining criteria for a consolidation suppressor.
Consolidation suppressors that constrain protein synthesis
Translation of pre-existing and newly synthesized transcripts is required for protein synthesis-dependent (PSD) consolidation of LTM (Flexner et al., 1962; Hernandez and Abel, 2008), making the proteins that regulate translation ideal consolidation control points. The translation initiation factor eIF2α is one such control point whose activity is regulated by several memory suppressor genes. eIF2α is rapidly dephosphorylated following contextual fear conditioning in the mouse hippocampus, derepressing its activity. This derepression promotes memory formation, since mice with phosphorylation-deficient eIF2α display enhanced spatial memory, fear memory, taste memory, and elevated late phase-LTP (Costa-Mattioli et al., 2007). Phosphorylation of eIF2α suppresses memory formation via two distinct mechanisms. First, eIF2-dependent production of memory-supporting proteins is decreased (Boye and Grallert, 2020). Second, the translation of the CREB inhibitor ATF4 is increased (Lu et al., 2004; Vattem and Wek, 2004). The memory suppressor genes PKR, GCN2, and PERK all code for kinases that phosphorylate eIF2α at the S51 residue, thereby suppressing eIF2α-dependent consolidation and synaptic plasticity (Costa-Mattioli et al., 2005; Sharma et al., 2018; Zhu et al., 2011).
Other than translation initiation factors, memory suppressors operate at several other points in the translation pathway. The first is through Poly(A)-binding protein (PABP). PABP interacts with and promotes the translation of memory relevant mRNAs (Gray et al., 2000), but is inhibited by interactions with poly(A) binding protein interacting protein 2 (PAIP2A). Fear conditioning drives the degradation of PAIP2A, freeing PABP to form a complex with mRNA and translation initiation factors (Khoutorsky et al., 2013). PAIP2A+/− mice exhibit normal memory 1 hour after fear conditioning but have significantly elevated memory 1 day later. In mice with the complete loss of PAIP2A, 24-hour memory is enhanced after weak training, L-LTP forms more easily and PABP binding to memory related mRNAs is increased. Second, mTOR (mechanistic target of rapamycin) is required for fear memory consolidation through its complex formation with the protein Raptor (Jobim et al., 2012). Fear conditioning promotes the formation of this complex in the hippocampus, overcoming mTOR inhibition by the protein FKBP12 (Hoeffer et al., 2008) in the mouse forebrain enhances contextual long-term fear memories without altering STM. FKB12 knock out also increases hippocampal L-LTP. But the enhanced L-LTP is eliminated by protein synthesis inhibition, supporting the proposition that FKBP12 suppresses consolidation by controlling mTOR-dependent protein translation. The net effect of mTOR activation is the regulation S6K1 and 4E-BP, which modulate the activity of translation initiation factors (Hoeffer and Klann, 2010).
Ras pathway signaling limits consolidation
Ras signaling affects processes throughout the neuron and regulates synaptic plasticity through both pre- and post-synaptic mechanisms (Curtis and Finkbeiner, 1999; Kushner et al., 2005; Platenik et al., 2000; Xing et al., 1996). Although mammalian Ras activating mutations can lead to either enhancement or impairment of memory depending on the conditions (Fasano and Brambilla, 2011; Kushner et al., 2005), Ras is generally held to be required for PSD- LTM (Mazzucchelli and Brambilla, 2000), in other words, a LTM facilitator.
The positive influence of mammalian Ras on LTM is at least partly due to its effects on learning-induced protein synthesis through the downstream kinase ERK (Peng et al., 2010). However, Ras also has an ERK-independent role in both protein synthesis-dependent and independent consolidated memory in Drosophila. In this organism, ERK-independent Ras signaling through Raf suppresses protein synthesis-independent ARM and facilitates PSD-LTM (Noyes et al., 2020). Knockdown of Ras in the MBn enhances ARM without altering acquisition, but strongly reduces or eliminates PSD-LTM. Prior behavioral studies had shown that ARM and PSD-LTM are antagonistic (Isabel et al., 2004; Placais et al., 2012). Thus, Ras function within its signaling systems provides a molecular explanation for the antagonism between ARM and PSD-LTM. PSD-LTM in Drosophila is analogous to PSD-LTM in mammalian systems, but it remains unclear how ARM relates to mammalian memory. However, many genes that modulate ARM also regulate mammalian memory, including Ras, Raf (Fasano and Brambilla, 2011; Noyes et al., 2020), Rho Kinase (ROCK) (Huentelman et al., 2009; Noyes et al., 2020), CDC42 (Kim et al., 2014; Zhang et al., 2016) and PDE4/dnc (Bolger, 2017; Scheunemann et al., 2012; Scheunemann et al., 2018). Therefore, we speculate that genes discovered in Drosophila that promote or inhibit ARM may prove to be suppressors or facilitators, respectively, of mammalian memory. For instance, Drosophila ROCK activity enhances consolidated ARM, while its pharmacological inhibition in mice improves memory and rescues memory deficits in disease models (Koch et al., 2018).
Thus, there are multiple mechanisms for filtering information to be consolidated into LTM. The major mechanisms include the control of protein synthesis for PSD-LTM by regulating gene expression through transcription factors such as CREB, or by controlling protein translation through small, inhibitory RNAs and/or translation initiation factors (Figure 4). An offshoot of this involves Ras signaling for ARM in Drosophila, whose relationship with mammalian consolidation is currently unknown.
Suppressors that Act on Forgetting
Forgetting is the temporary or permanent inability to retrieve a previously acquired memory. Although forgetting is often viewed as an impediment, as seen in aging or neurodegenerative diseases, it is a critical process for selecting and maintaining those memories that will drive advantageous behavior. For example, behavioral flexibility requires an individual to forget prior information that would prevent updating of memories with new information. This can be observed experimentally using reversal learning paradigms, in which an organism previously trained to associate a stimulus with particular valence must later learn to associate the same stimulus with a neutral or opposite valence (Izquierdo et al., 2017). Forgetting is also critical for memory generalization, the process that allows memories of specific situations to be used to make predictions about similar, but non-identical situations (Robertson, 2018). In this process, forgetting causes the loss of memory details and allows the memory to be retrieved using broad similarities rather than the details present during acquisition. The identification of memory suppressor genes has played a central role as an entrée to the molecular mechanisms for active forgetting.
Enhanced consolidation and impaired forgetting both result in increased memory performance at time points after acquisition. There are several observations that support the categorization into either one of the two categories. (1) Impaired forgetting results in slower memory decay over time, whereas enhanced consolidation should produce a normal memory decay rate. (2) In Drosophila, many of the studies on forgetting have focused on labile memory in the absence of PSD-consolidation. (3) Effects that occur outside the generally accepted time window for consolidation (e.g., ~1 day for PSD-LTM) suggest that consolidation is not affected.
AMPA receptor mediated forgetting
Across species, synaptic strength is efficiently modulated by changes in synaptic AMPA (α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid) receptor (AMPAR) levels (Turrigiano and Nelson, 1998). Insertion of additional AMPARs into post-synaptic sites after learning increases synaptic strength and underlies the formation of some types of memory. Conversely, internalization of surface AMPAR reduces the functional connection between potentiated synapses and is proposed to be one mechanism for forgetting (Figure 5A; Hardt et al., 2014). Some evidence supports this proposal. Postsynaptic levels of the AMPAR subunit Glu2a in the amygdala positively correlate with memory strength after fear conditioning (Migues et al., 2010) and inhibiting hippocampal Glu2a internalization following training suppresses the forgetting of episodic memory (Migues et al., 2016). Although there is some debate on the issue, there is evidence that PKMζ-dependent maintenance of hippocampal memory is achieved by sustaining surface levels of synaptic AMPARs (Migues et al., 2010).
Figure 5: Memory Suppressors that Act on Forgetting.
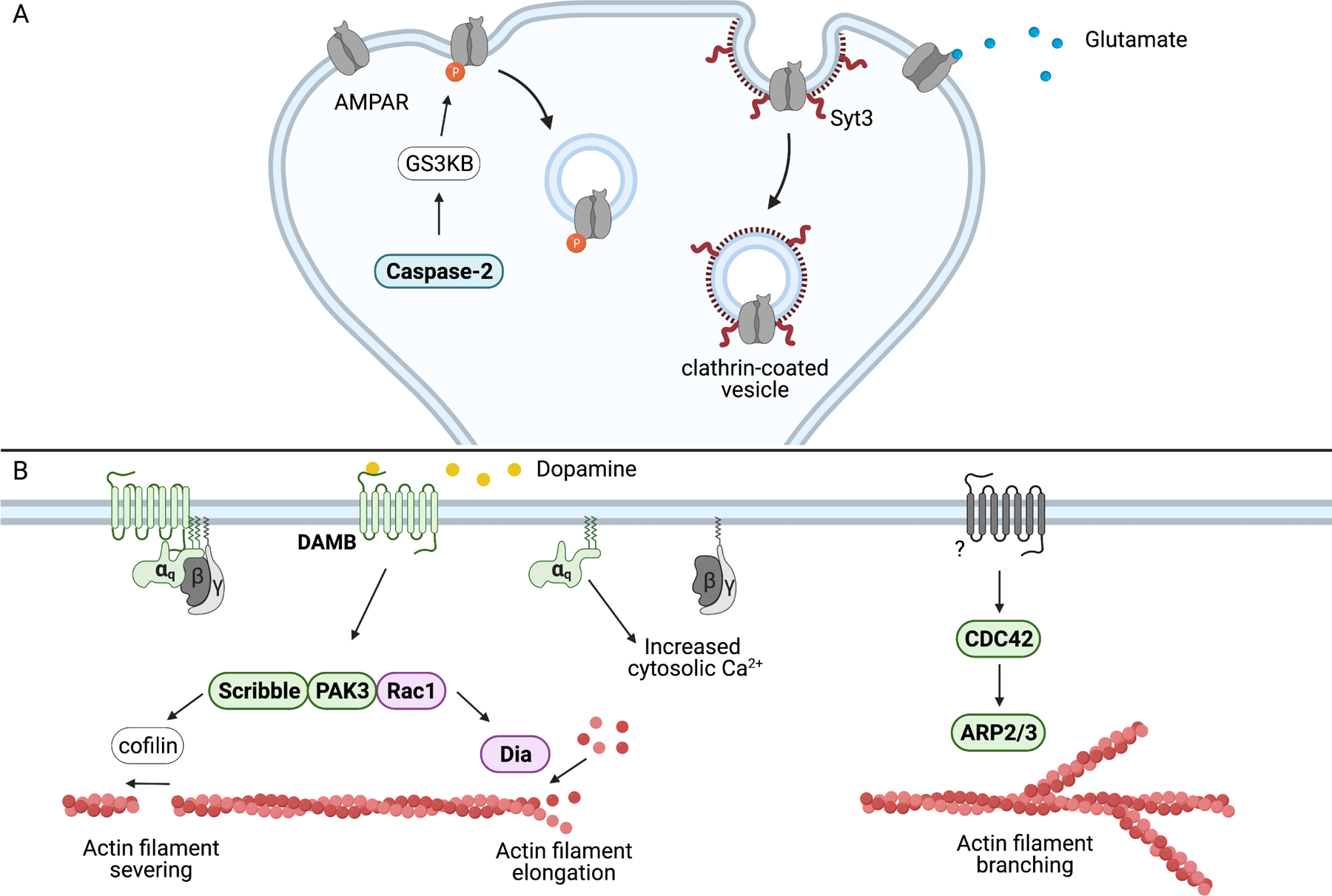
Memory suppressor genes are in bold and color-coded green (fly), blue (mouse), and purple (both mouse and fly). A) Caspase-2 activity initiates a multi-step signaling cascade, starting with the cleavage of Rictor in the protein complex mTORC2. This results in the activation of the kinase AKT followed by the kinase GSK3β. Activated dendritic GSK3β phosphorylates the AMPAR subunit GluA2, inducing AMPAR internalization and a weakening of synaptic strength. Syt3 also regulates AMPAR internalization through GluA2, driving clathrin-mediated endocytosis of AMPARs.
B) Dopamine-driven active forgetting in Drosophila is mediated by the Gαq-coupled DAMB receptor. DAMB signals downstream to a protein complex that includes Rac1. Rac accelerates forgetting in both mice and Drosophila. In Drosophila, Rac1 propagates the forgetting signal to cofilin that induces actin filament severing and to dia that promotes actin filament elongation. CDC42 drives forgetting of consolidated memory through ARP2/3, a protein that controls the branching of actin filaments. Upstream activators involved in CDC42-facilitated forgetting are not known.
Two genes have recently been identified that regulate memory stability presumably through AMPAR internalization. First, reducing expression of the memory suppressor gene, Caspase-2, slows the forgetting of spatial memory and impairs the internalization of hippocampal AMPARs by reducing GSK3β activity (Xu et al., 2019). Caspase-2 is also required for the formation of LTD, which is formed by the internalization of AMPARs from the post-synaptic membrane (Malenka, 2003). The second AMPAR-regulating gene to affect memory stability, syt3, does not fit the working definition of a memory suppressor, as there is no evidence that loss of the gene function alters memory decay. However, syt3 KO mice are impaired in a Morris water maze reversal-learning task (Awasthi et al., 2019). These mice fail to forget the previously learned position of a platform after they are repeatedly trained to the platform’s new position. Like Caspase-2, Syt3 promotes the endocytosis of AMPAR and is required for LTD formation. Thus, syt3 may also be a memory suppressor gene functioning by facilitating the endocytosis of AMPARs.
Dopamine signaling and active forgetting
Research using Drosophila has revealed that dopamine (DA), in addition to being critical for the acquisition of memories, is an intrinsic signal that drives forgetting. The ongoing activity of a subset of DAn is sufficient to cause significant forgetting of labile memories (Berry et al., 2012). Inhibiting these neurons prolongs memory retention while their activation quickly impairs memory performance. These same neurons are required for normal associative olfactory learning and their activation can be substituted in place of an unconditioned stimulus to form an “artificial” memory (Aso and Rubin, 2016; Claridge-Chang et al., 2009). This seemingly contradictory role of DA in learning and forgetting is achieved through two different DA receptors. The D1-like receptor, dda1, is responsible for acquisition, registering the DA signal produced by the unconditioned stimulus (Kim et al., 2007; Tomchik and Davis, 2009). Loss of dda1 in the MBn eliminates associative olfactory memory (Kim et al., 2007). DA-induced forgetting is mediated by DAMB (Berry et al., 2012). Although learning is relatively normal in DAMB mutants, labile memory decay is significantly reduced. Dopamine signaling through DAMB elicits Gαq-dependent increases in cytosolic Ca2+ and reduction of MBn Gαq results in impaired forgetting (Figure 5B; Himmelreich et al., 2017). Interestingly, Drosophila DAns may promote memory decay through co-neurotransmitters. Nitric oxide produced in DAns during aversive conditioning signals through guanylate cyclase in MBn, resulting in reduced memory retention (Aso et al., 2019). This possibility is supported by an earlier study showing that RNAi knockdown of a subunit of guanylyl cyclase enhances memory (Walkinshaw et al., 2015).
Evidence has emerged linking the mammalian dopaminergic system to forgetting. Post-training injection of a D1 dopamine receptor antagonist in the rat hippocampus after training enhances 7-day food and cocaine-place conditioning memory (Diaz et al., 2019), raising the possibility that the D1 receptor drives forgetting in mammals. Recent work has tested the role of rat D1 receptors in retrieval-induced forgetting, a phenomenon in which retrieval of a memory promotes the forgetting of other related memories. Pharmacological inhibition of prefrontal cortex D1 receptors eliminates retrieval-induced forgetting while activation promotes it (Gallo et al., 2021). Dopamine has also been suggested to contribute to retrieval-induced forgetting in humans. Study subjects with an allele for catechol-O-methyltransferase (COMT) associated with higher prefrontal cortex dopamine levels display higher levels of retrieval-induced forgetting (Wimber et al., 2011) despite exhibiting normal or enhanced performance in other cognitive tasks (Savitz et al., 2006). Intriguingly, dopamine promotes the transient blockade of aversive LTM retrieval in Drosophila (Sabandal et al., 2021). This temporary forgetting is dependent on MBn DAMB, which appears to mediate stimuli-induced dopamine release from upstream dopaminergic neurons.
Scribble, first identified as a gene of interest in a memory suppressor screen (Walkinshaw et al., 2015), was later discovered to function downstream of DAMB (Cervantes-Sandoval et al., 2016). Knockdown of Scribble in the MBn recapitulates the DAMB mutant forgetting phenotype and results from epistasis experiments suggest that the Scribble protein propagates the forgetting signaling from DAMB to downstream proteins. Importantly, these studies provided a link between DAMB and the previously established role for Rac1 in active forgetting (Shuai et al., 2010). Current evidence suggests that Scribble acts as a scaffolding protein, physically interacting with Rac and other Rac signaling molecules (Cervantes-Sandoval et al., 2016).
Small G-proteins, including Rac1 and CDC42, drive active forgetting
The role of Rac1, a Rho family GTPase, in forgetting has been demonstrated in mice for several types of memory. Hippocampal Rac1 activity is increased following contextual fear conditioning and pharmacological inhibition of this activity enhanced the conditioned fear memory (Jiang et al., 2016; Lv et al., 2019). Importantly, hippocampal injection of the Rac1 inhibitor was delivered after training, indicating that Rac1 activity has a post-acquisition role in producing the increased memory performance. Lv et al (2019) also showed that the expression of a dominant negative (DN) or constitutively active (CA) form of Rac1 in the hippocampus enhanced and impaired long-term contextual fear memory, respectively, with neither insult altering acquisition. Hippocampal inhibition of Rac1 reduces forgetting of episodic memory tested by novel object recognition (Liu et al., 2016) and in a social discrimination paradigm (Liu et al., 2018) , while Rac1 activation accelerates memory decay in these same tasks. However, Rac1 has been reported to have roles in other memory forming operations depending on the nature of the genetic insult, the neurons that are affected, and the task being learned (Gao et al., 2015; Haditsch et al., 2009; Oh et al., 2010).
The process of Rac-dependent active forgetting is conserved across animal phyla. Drosophila MBn expression of dominant-negative (DN) Rac slows the forgetting of labile memory produced by associative olfactory conditioning while constitutively active (CA) Rac increases forgetting (Shuai et al., 2010). Like Scribble, which scaffolds Rac to downstream signaling components (Cervantes-Sandoval et al., 2016), MBn Rac also regulates behavioral flexibility observed in reversal learning and retroactive interference experiments (Shuai et al., 2010).
While Rac is responsible for active forgetting of labile memories, another small G-protein, Cdc42, drives the forgetting of consolidated memories. A single cycle of aversive olfactory conditioning causes an increase in Cdc42 activity. Inhibition of this activity in the MBn through expression of DN Cdc42 impairs the forgetting of ARM (Zhang et al., 2016). After massed training, consolidated ARM is elevated at later time points compared to single cycle training but initial ARM levels are similar in both training paradigms. Massed training prevents the increase in Cdc42 activity observed following single cycle training. This suggests consolidated ARM is formed at similar levels regardless of the type of training, but the type of training alters the forgetting of ARM through Cdc42 activity.
What molecular processes are altered by Rac1 and Cdc42 activation to cause forgetting? The discovery of Rac1 and Cdc42 as regulators of forgetting implicates actin dynamics as the process that may mediate forgetting. Rac1 and Cdc42 are known to modulate dendritic spine morphology, cellular projections and presynaptic organization via control over actin dynamics (Chen et al., 2012; Duman et al., 2015), consistent with the possibility that undoing structural changes that occur during acquisition may underlie active forgetting. In Drosophila, SCAR/WAVE and dia were found to be downstream from Rac, mediating Rac-induced forgetting of labile memories (Gao et al., 2019). The same paper concluded WASp and ARP2/3 are downstream from Cdc42 and mediate Cdc42–induced forgetting of consolidated ARM. In each case, RNAi knockdown of the individual genes led to enhanced memory after acquisition, with epistasis experiments revealing the relative position of each component in the active forgetting pathways. These four genes are well-established regulators of the actin cytoskeleton (Kovar, 2006; Miki and Takenawa, 2003; Rotty et al., 2013).
It remains unknown how these proteins undo distinct types of memory but their distinct roles in actin dynamics offers a path forward. Dia promotes unbranched actin filaments (Kovar, 2006), while Arp2/3 can nucleate branched filaments (Rotty et al., 2013). But the specific changes in actin dynamics that underlie forgetting may be specific to the developmental stage, behavioral task, and/or organism being studied. Arp2/3 was found to have the opposite effect on associative memory in C elegans. In this organism, the memory suppressor gene mushai-1 reduces the expression Arp2/3, thereby facilitating forgetting (Hadziselimovic et al., 2014). Interestingly, the mouse version of dia, Diap1, is also a memory suppressor gene. Knockdown of Diap1 in the medial prefrontal cortex produced enhanced conditioned fear memory in adult mice (Lin et al., 2017). However, this effect was produced by development-specific knockdown, while Drosophila dia regulates forgetting through increased activity in the adult brain, presumably initiated during acquisition.
In summary, molecules that participate in active forgetting have been identified as memory suppressor genes from genetic screens for enhanced memory performance along with candidate gene approaches. The general principles learned to date include: (1) memory suppressor genes encode molecules that participate in the endocytosis of neurotransmitter receptors so as to reduce excitatory synaptic input, and (2) certain DAn have the responsibility to remove memories by activating specific DA receptors on postsynaptic engram cells, initiating a signaling cascade that seems to terminate in the activation of small G-proteins and the rearrangement of the cytoskeleton in the engram cells. Both mechanisms are attractive as forgetting mechanisms, the first to simply reduce the receptive state of the engram cell and the second to modify or eliminate the structural changes at the synapse that occur with acquisition and consolidation (Figure 5).
Discussion
The literature discussed above indicates that memory suppressors exist, and they function in at least three of the basic operations that underlie memory formation: acquisition, consolidation, and forgetting. In addition, some of the mechanisms by which they constrain memory formation are summarized in the subtitles above. Future investigations will undoubtedly reveal deeper insights into the various mechanisms that the brain utilizes to suppress memory formation and perhaps elucidate other important principles beyond those discussed here. But a general and important question is why do memory suppressors exist? Why should the brain be designed with biological limits for memory formation?
Information overload?
One possibility is that memory suppressors may reduce information overload, allowing the brain to function efficiently only with the essential and important information that it needs for optimal evolutionary fitness of its host. This explanation based on limited capacity is intuitively appealing. However, there are rare and exceptional outliers among us that seemingly have remarkable memories in the apparent absence of neurodevelopmental disorders or brain injury that throw this possibility into question (Brandt and Bakker, 2018). Theoretical and computational modeling of memory capacity also suggest that saturation of memory capacity is unlikely to be a concern (Richards and Frankland, 2017). Therefore, although this explanation cannot be eliminated, there exist sound reasons to search for alternatives.
Allow for behavioral flexibility?
A second possibility is that memory suppression provides for behavioral flexibility in changing environments, allowing individuals to suppress memories that underlie behaviors that are not adaptive or advantageous. A recent review nicely discusses this idea in the context of forgetting (Richards and Frankland, 2017), but the concept can also be applied to suppressors of other memory operations.
For instance, reversal experiments using flies impaired in forgetting show that they have difficulty adjusting their behavior to the altered contingencies, that is, to new rules or environmental conditions (Berry et al., 2012; Cervantes-Sandoval et al., 2016; Shuai et al., 2010). Such observations extend to mice carrying a conditional knockout of LTM suppressor FK506-binding protein 12 or knockdown of WT1 (Hoeffer et al., 2008; Mariottini et al., 2019). Therefore, the normal function of active forgetting molecules may facilitate the ability of animals to update memories to new situations.
Extinction experiments with many different memory suppressors (Rin1, GABRAα5, DREAM, WT1, JIP1-JNK, and Tet2) also reveal that reducing their function limits behavioral flexibility (Bliss et al., 2010; Engin et al., 2015; Fontan-Lozano et al., 2009; Mariottini et al., 2019; Morel et al., 2018; Zengeler et al., 2019). As one specific example, inhibiting the function of the memory suppressor gene, calcineurin (Malleret et al., 2001), impairs the extinction of fear memories (Havekes et al., 2008). The repeated exposure of wildtype mice after acquisition to the context originally learned but without the associated foot shock extinguishes their fear responses. Mice defective in calcineurin function fail to extinguish their fear responses, exposing their lack of behavioral flexibility. However, inhibiting some memory suppressor genes, including NHe5, STEP, Sharp1&2, Cdk5, Hdac2, and PKR, enhances both memory and behavioral flexibility as measured by reversal or extinction experiments (Chen et al., 2017; Hawasli et al., 2007; Morris et al., 2013; Shahmoradi et al., 2015; Venkitaramani et al., 2011; Zhu et al., 2011). Why some memory suppressors promote behavioral flexibility while others suppress flexibility remains a mystery. However, such behavior would be consistent with these memory suppressors functioning through acquisition or through limiting the lability of memory.
Promote accurate association memories?
A third possibility is that memory suppressors promote the formation of more accurate associations. Memory suppressor molecules are predicted to allow associations only for those things that reach a threshold for acceptance as an authentic association, reducing the probability of forming relationships between items that are unrelated or only tangentially related. Three types of experimental data support this idea: (1) Conditional knockout mice of GABRAα5, an acquisition suppressor, produces a deficiency in latent inhibition of auditory fear conditioning. Latent inhibition refers to the phenomenon that organisms are less likely to associate familiar stimuli with an unconditioned stimulus compared to novel stimuli. It is often approached experimentally by pre-exposing the organism to a conditioned stimulus prior to the association trial. This leads to a decrease in the strength of association, presumably reflecting a reduced fidelity of the relationship between the familiar conditioned stimulus and the unconditioned stimulus. The GABRAα5 conditional knockout mice form memories that are similar in strength with or without preexposure to the conditioned stimulus (Engin et al., 2015). Thus, these mice fail to devalue the conditioned stimulus after pre-exposure, leading to an erroneously strong association with the unconditioned stimulus. (2) Loss of function in two memory suppressor genes, Dicer1 and GABRA4, prolongs the duration of trace conditioning (Fan et al., 2020; Konopka et al., 2010). Trace conditioning occurs when the conditioned stimulus precedes the unconditioned stimulus with a gap in time. Longer gaps in time between the conditioned stimulus and unconditioned stimulus increases the probability that the two events are unrelated. (3) The GABRAα5 conditional knockout mice exhibit difficulties in pattern separation between two similar contexts compared to control animals, such that they erroneously exhibit a fear response to a context similar, but not identical, to the trained context (Engin et al., 2015). Similarly, reduction of GABAergic neuron function and GABAB receptor GABA-B-R3 in flies leads to difficulties in odor discrimination (Lin et al., 2014; Yamagata et al., 2021). These are errors in stimulus generalization due to the loss of function of a memory suppressor gene.
Promote social behaviors?
Dysfunction of some memory suppressors also result in disrupted social behavior. Inhibition of two GABAA receptor subunits, GABRAα5 and GABRA4, produced altered sociability in mice. The GABRA4 knockout mice exhibit impaired social recognition memory (Fan et al., 2020) and GABRAα5 knockout mice display reduced social behaviors, making fewer social contacts with conspecifics as adults and producing fewer ultrasonic vocalizations as pups (Zurek et al., 2016). In flies, the knockdown of Rdl alters the mating behavior of females (Ishimoto and Kamikouchi, 2020). Similarly, knockout of the M1 muscarinic receptor impairs social recognition (Anagnostaras et al., 2003), STEP knockout increases dominance behaviors and impaires social memory (Blazquez et al., 2019; Venkitaramani et al., 2011), while Lrfn2 knockout decreases social interest and increases social avoidance (Morimura et al., 2017). Sociability is argued to increase fitness (Hawkley and Capitanio, 2015).
A speculative aspect of this explanation extends to human behavior. Humans with savant syndrome are characterized by having significant mental disability but exceling in at least one skill, such as in music, art, calendar counting, mathematics, or in mechanical or spatial skills (Treffert, 2009). Interestingly, regardless of the type of skill they possess, savants always exhibit extraordinary memory (Treffert, 2009). Individuals with savant syndrome usually have some type of neurodevelopmental disorder but can also acquire the syndrome after brain injury. Approximately 50% of savants have autism spectrum disorder (ASD), which is characterized by abnormal social behavior and/or social information processing, and it has been estimated that 10% of individuals with ASD have savant-like skills (Treffert, 2009). Such observations follow the conceptual thread of an increased memory capacity associated with decreased sociability. Nevertheless, some memory suppressors also seem to function as suppressors of social behavior. Np65 knockout mice (Li et al., 2019) and HCN1 knockdown mice display significantly elevated levels of social interaction while also exhibiting enhanced spatial and object recognition memory (Amuti et al., 2016; Li et al., 2019; Nolan et al., 2004; Silveira Villarroel et al., 2018). This indicates the existence of genetic suppressors of sociability, a research area that may have implications for ASD.
Conclusions
Research on memory suppressors has thus far revealed they function at most operations of memory tested; acquisition, consolidation and forgetting. This organization provides a conceptual framework for future studies. The number of suppressors described in the literature (Table S1–S3) strengthens the case that there is a strong biological need to limit and balance memory formation, but additional research is required to provide an exhaustive list of all their mechanisms of action. In addition, the question of why memory suppressors exist needs further exploration. Currently, there is only limited evidence to support the possibilities that they provide behavioral flexibility, facilitate accurate memories, or modulate social behaviors.
Supplementary Material
Supplemental Table 1: Comprehensive list of early memory suppressor genes. The data in this table contains memory suppressor genes that act on early memory (effects found at the earliest time point tested within 4hr of training). This category may include both suppressors of acquisition and suppressors of early memory.
Manipulation Abbreviations:
KO = knockout
cKO = conditional knockout
KD = knockdown
WT= wild type
Behavioral Test Abbreviations:
AFC = Auditory Fear Conditioning
CFC = Contextual Fear Conditioning
MWM = Morris Water Maze
NOR = Novel Object Recognition
NOL= Novel Object Location
RAM= Radial Arm Maze
Neuron Function Abbreviations:
LTP = Long-Term Potentiation
STP= Short-Term Potentiation
LTD= Long-Term Depression
EPSC = Excitatory Postsynaptic Current
mEPSC = miniature Excitatory Postsynaptic Current
fEPSC= field Excitatory Postsynaptic Current
eEPSP= extracellular Excitatory Postsynaptic Potential
IPSC = Inhibitory Postsynaptic Current
mIPSC = miniature Inhibitory Postsynaptic Current
Supplemental Table 2: Comprehensive list of late memory suppressor genes. The data in the table contains memory suppressor genes that act on late memory. These genes have no effects at the earliest time point tested but effects on memory thereafter. This category likely contains suppressors of consolidation and/or forgetting.
Manipulation Abbreviations:
KO = knockout
cKO = conditional knockout
KD = knockdown
WT= wild type
Behavioral Test Abbreviations:
AFC = Auditory Fear Conditioning
CFC = Contextual Fear Conditioning
MWM = Morris Water Maze
NOR = Novel Object Recognition
NOL= Novel Object Location
RAM= Radial Arm Maze
Neuron Function Abbreviations:
LTP = Long-Term Potentiation
STP= Short-Term Potentiation
LTD= Long-Term Depression
EPSC = Excitatory Postsynaptic Current
mEPSC = miniature Excitatory Postsynaptic Current
fEPSC= field Excitatory Postsynaptic Current
eEPSP= extracellular Excitatory Postsynaptic Potential
IPSC = Inhibitory Postsynaptic Current
mIPSC = miniature Inhibitory Postsynaptic Current
Supplemental Table 3: Comprehensive list of undefined memory suppressor genes. The data in the table contains memory suppressor genes that act on an undefined memory operation. These memory suppressors could not be placed into either of the early or late memory genes defined above due to insufficient data.
Manipulation Abbreviations:
KO = knockout
cKO = conditional knockout
KD = knockdown
WT= wild type
Behavioral Test Abbreviations:
AFC = Auditory Fear Conditioning
CFC = Contextual Fear Conditioning
MWM = Morris Water Maze
NOR = Novel Object Recognition
NOL= Novel Object Location
RAM= Radial Arm Maze
Neuron Function Abbreviations:
LTP = Long-Term Potentiation
STP= Short-Term Potentiation
LTD= Long-Term Depression
EPSC = Excitatory Postsynaptic Current
mEPSC = miniature Excitatory Postsynaptic Current
fEPSC= field Excitatory Postsynaptic Current
eEPSP= extracellular Excitatory Postsynaptic Potential
IPSC = Inhibitory Postsynaptic Current
mIPSC = miniature Inhibitory Postsynaptic Current
Acknowledgements
We thank three anonymous referees for their extremely valuable comments. Research in the authors’ labs was supported by NIH grant R35NS097224 to R.L.D. and a Natural Sciences and Engineering Research Council grant RGPIN-2020–04009 to A.P.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
REFERENCES
- Abel T, Martin KC, Bartsch D, and Kandel ER (1998). Memory suppressor genes: inhibitory constraints on the storage of long-term memory. Science 279, 338–341. [DOI] [PubMed] [Google Scholar]
- Alexander JC, McDermott CM, Tunur T, Rands V, Stelly C, Karhson D, Bowlby MR, An WF, Sweatt JD, and Schrader LA (2009). The role of calsenilin/DREAM/KChIP3 in contextual fear conditioning. Learn Mem 16, 167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amuti S, Tang Y, Wu S, Liu L, Huang L, Zhang H, Li H, Jiang F, Wang G, Liu X, et al. (2016). Neuroplastin 65 mediates cognitive functions via excitatory/inhibitory synapse imbalance and ERK signal pathway. Neurobiol. Learn. Mem 127, 72–83. [DOI] [PubMed] [Google Scholar]
- Anagnostaras S, Murphy G, Hamilton S, Mitchell S, Rahnama N, Nathanson N, and Silva A (2002). Selective cognitive dysfunction in acetylcholine M1 muscarinic receptor mutant mice. J. Cognitive Neurosci, 137–137. [DOI] [PubMed]
- Aso Y, Ray RP, Long X, Bushey D, Cichewicz K, Ngo TT, Sharp B, Christoforou C, Hu A, Lemire AL, et al. (2019). Nitric oxide acts as a cotransmitter in a subset of dopaminergic neurons to diversify memory dynamics. Elife 8:e49257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awasthi A, Ramachandran B, Ahmed S, Benito E, Shinoda Y, Nitzan N, Heukamp A, Rannio S, Martens H, Barth J, et al. (2019). Synaptotagmin-3 drives AMPA receptor endocytosis, depression of synapse strength, and forgetting. Science 363, 44–+. [DOI] [PubMed] [Google Scholar]
- Barad M, Bourtchouladze R, Winder DG, Golan H, and Kandel E (1998). Rolipram, a type IV-specific phosphodiesterase inhibitor, facilitates the establishment of long-lasting long-term potentiation and improves memory. Proc. Natl. Acad. Sci. USA 95, 15020–15025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barco A, Alarcon JM, and Kandel ER (2002). Expression of constitutively active CREB protein facilitates the late phase of long-term potentiation by enhancing synaptic capture. Cell 108, 689–703. [DOI] [PubMed] [Google Scholar]
- Barrett GL, Reid CA, Tsafoulis C, Zhu WM, Williams DA, Paolini AG, Trieu J, and Murphy M (2010). Enhanced spatial memory and hippocampal long-term potentiation in p75 neurotrophin receptor knockout mice. Hippocampus 20, 145–152. [DOI] [PubMed] [Google Scholar]
- Bartel DP (2007). MicroRNAs: genomics, biogenesis, mechanism, and function (Reprinted from Cell, vol 116, pg 281–297, 2004). Cell 131, 11–29 [DOI] [PubMed] [Google Scholar]
- Bartsch D, Ghirardi M, Skehel PA, Karl KA, Herder SP, Chen M, Bailey CH, and Kandel ER (1995). Aplysia CREB2 represses long-term facilitation: relief of repression converts transient facilitation into long-term functional and structural change. Cell 83, 979–992. [DOI] [PubMed] [Google Scholar]
- Beck CD, Schroeder B, and Davis RL (2000). Learning performance of normal and mutant Drosophila after repeated conditioning trials with discrete stimuli. J. Neurosci 20, 2944–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit CE, Bastianetto S, Brouillette J, Tse Y, Boutin JA, Delagrange P, Wong T, Sarret P, and Quirion R (2010). Loss of quinone reductase 2 function selectively facilitates learning behaviors. J. Neurosci 30, 12690–12700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry JA, Cervantes-Sandoval I, Nicholas EP, and Davis RL (2012). Dopamine is required for learning and forgetting in Drosophila. Neuron 74, 530–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorge MD, Hildrestrand GA, Scheffler K, Suganthan R, Rolseth V, Kusnierczyk A, Rowe AD, Vagbo CB, Vetlesen S, Eide L, et al. (2015). Synergistic actions of Ogg1 and Mutyh DNA glycosylases modulate anxiety-like behavior in mice. Cell Rep 13, 2671–2678. [DOI] [PubMed] [Google Scholar]
- Blazquez G, Castane A, Saavedra A, Masana M, Alberch J, and Perez-Navarro E (2019). Social memory and social patterns alterations in the absence of STriatal-enriched protein tyrosine phosphatase. Front. Behav. Neurosci 12, 317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliss JM, Gray EE, Dhaka A, O’Dell TJ, and Colicelli J (2010). Fear learning and extinction are linked to neuronal plasticity through Rin1 signaling. J. Neurosci. Res 88, 917–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliss TV, and Collingridge GL (1993). A synaptic model of memory: long-term potentiation in the hippocampus. Nature 361, 31–39. [DOI] [PubMed] [Google Scholar]
- Bolger GB (2017). The PDE4 cAMP-specific phosphodiesterases: targets for drugs with antidepressant and memory-enhancing action. Adv. Neurobiol 17, 63–102. [DOI] [PubMed] [Google Scholar]
- Bourtchuladze R, Frenguelli B, Blendy J, Cioffi D, Schutz G, and Silva AJ (1994). Deficient long-term-memory in mice with a targeted mutation of the camp-responsive element-binding protein. Cell 79, 59–68. [DOI] [PubMed] [Google Scholar]
- Boye E, and Grallert B (2020). eIF2 alpha phosphorylation and the regulation of translation. Curr. Genet 66, 293–297. [DOI] [PubMed] [Google Scholar]
- Brandt J, and Bakker A (2018). Neuropsychological investigation of “the amazing memory man.” Neuropsychology 32, 304–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridi M, Schoch H, Florian C, Poplawski SG, Banerjee A, Hawk JD, Porcari GS, Lejards C, Hahn CG, Giese KP, et al. (2020). Transcriptional corepressor SIN3A regulates hippocampal synaptic plasticity via Homer1/mGluR5 signaling. Jci. Insight 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunelli M, Castellucci V, and Kandel ER (1976). Synaptic facilitation and behavioral sensitization in Aplysia - possible role of serotonin and cyclic-amp. Science 194, 1178–1181. [DOI] [PubMed] [Google Scholar]
- Bushati N, and Cohen SM (2007). microRNA functions. Annu. Rev. Cell Dev. Biol 23, 175–205. [DOI] [PubMed] [Google Scholar]
- Busto GU, Guven-Ozkan T, Fulga TA, Van Vactor D, and Davis RL (2015). microRNAs That promote or inhibit memory formation in Drosophila melanogaster. Genetics 200, 569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers D, Davis RL, and Kiger JA (1981). Defect in cyclic-AMP phosphodiesterase due to the dunce mutation of learning in Drosophila melanogaster. Nature 289, 79–81. [DOI] [PubMed] [Google Scholar]
- Carew TJ, Pinsker HM, and Kandel ER (1972). Long-term habituation of a defensive withdrawal reflex in Aplysia. Science 175, 451–4. [DOI] [PubMed] [Google Scholar]
- Caughey S, Harris AP, Seckl JR, Holmes MC, and Yau JLW (2017). Forebrain-specific transgene rescue of 11 beta-HSD1 associates with impaired spatial memory and reduced hippocampal brain-derived neurotrophic factor mRNA levels in aged 11 beta-HSD1 deficient mice. J. Neuroendocrinol 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cepeda NJ, Pashler H, Vul E, Wixted JT, and Rohrer D (2006). Distributed practice in verbal recall tasks: a review and quantitative synthesis. Psychol. Bull 132, 354–380. [DOI] [PubMed] [Google Scholar]
- Cervantes-Sandoval I, Chakraborty M, MacMullen C, and Davis RL (2016). Scribble scaffolds a signalosome for active forgetting. Neuron 90, 1230–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Muzzio IA, Malleret G, Bartsch D, Verbitsky M, Pavlidis P, Yonan AL, Vronskaya S, Grody MB, Cepeda I, et al. (2003). Inducible enhancement of memory storage and synaptic plasticity in transgenic mice expressing an inhibitor of ATF4 (CREB-2) and C/EBP proteins. Neuron 39, 655–669. [DOI] [PubMed] [Google Scholar]
- Vronskaya S, Grody MB, Cepeda I, et al. (2003). Inducible enhancement of memory storage and synaptic plasticity in transgenic mice expressing an inhibitor of ATF4 (CREB-2) and C/EBP proteins. Neuron 39, 655–669. [DOI] [PubMed] [Google Scholar]
- Chen C, Wirth A, and Ponimaskin E (2012). Cdc42: an important regulator of neuronal morphology. Int. J. Biochem. Cell B 44, 447–451. [DOI] [PubMed] [Google Scholar]
- Chen CN, Denome S, and Davis RL (1986). Molecular analysis of cDNA clones and the corresponding genomic coding sequences of the Drosophila dunce+ gene, the structural gene for cAMP phosphodiesterase. Proc. Natl. Acad. Sci. USA 83, 9313–9317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Shu S, Chen YT, Liu Z, Yu LJ, Yang LX, Xu Y, and Zhang MJ (2019). AIM2 deletion promotes neuroplasticity and spatial memory of mice. Brain Res. Bull. 152, 85–94. [DOI] [PubMed] [Google Scholar]
- Chen X, Wang X, Tang L, Wang J, Shen C, Liu J, Lu S, Zhang H, Kuang Y, Fei J, et al. (2017). Nhe5 deficiency enhances learning and memory via upregulating Bdnf/TrkB signaling in mice. Am. J. Med. Genet. B. Neuropsychiatr. Genet 174, 828–838. [DOI] [PubMed] [Google Scholar]
- Chen XJ, Wang XY, Tang LY, Wang JJ, Shen CL, Liu JB, Lu SY, Zhang HX, Kuang Y, Fei J, et al. (2017a). Nhe5 deficiency enhances learning and memory via upregulating Bdnf/TrkB signaling in mice. Am. J. Med. Genet B 174, 828–838. [DOI] [PubMed] [Google Scholar]
- Chen Y, and Sabatini BL (2021). The kinase specificity of protein kinase inhibitor peptide. Front Pharmacol 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC, Ma YL, Lin CH, Cheng SJ, Hsu WL, and Lee EHY (2017b). Galectin-3 negatively regulates hippocampus-dependent memory formation through inhibition of integrin signaling and galectin-3 phosphorylation. Front Mol. Neurosci 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claridge-Chang A, Roorda RD, Vrontou E, Sjulson L, Li HY, Hirsh J, and Miesenbock G (2009). Writing memories with light-addressable reinforcement circuitry. Cell 139, 405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton DA, Mesches MH, Alvarez E, Bickford PC, and Browning MD (2002). A hippocampal NR2B deficit can mimic age-related changes in long-term potentiation and spatial learning in the Fischer 344 rat. J. Neurosci 22, 3628–3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole CJ, Mercaldo V, Restivo L, Yiu AP, Sekeres MJ, Han JH, Vetere G, Pekar T, Ross PJ, Neve RL, et al. (2012). MEF2 negatively regulates learning-induced structural plasticity and memory formation. Nat. Neurosci 15, 1255–U1121. [DOI] [PubMed] [Google Scholar]
- Collinson N, Kuenzi FM, Jarolimek W, Maubach KA, Cothliff R, Sur C, Smith A, Otu FM, Howell O, Atack JR, et al. (2002). Enhanced learning and memory and altered GABAergic synaptic transmission in mice lacking the alpha 5 subunit of the GABA(A) receptor. J. Neurosci 22, 5572–5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Mattioli M, Gobert D, Harding H, Herdy B, Azzi M, Bruno M, Bidinosti M, Ben Mamou C, Marcinkiewicz E, Yoshida M, et al. (2005). Translational control of hippocampal synaptic plasticity and memory by the eIF2 alpha kinase GCN2. Nature 436, 1166–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Mattioli M, Gobert D, Stern E, Gamache K, Colina R, Cuello C, Sossin W, Kaufman R, Pelletier J, Rosenblum K, et al. (2007). eIF2 alpha phosphorylation bidirectionally regulates the switch from short- to long-term synaptic plasticity and memory. Cell 129, 195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis J, and Finkbeiner S (1999). Sending signals from the synapse to the nucleus: possible roles for CaMK, Ras/ERK, and SAPK pathways in the regulation of synaptic plasticity and neuronal growth. J. Neurosci. Res 58, 88–95. [PubMed] [Google Scholar]
- Dash PK, Hochner B, and Kandel ER (1990). Injection of the camp-responsive element into the nucleus of Aplysia sensory neurons blocks long-term facilitation. Nature 345, 718–721. [DOI] [PubMed] [Google Scholar]
- Davis RL (2004). Olfactory learning. Neuron 44, 31–48. [DOI] [PubMed] [Google Scholar]
- Davis RL, and Kiger JA (1981). Dunce mutants of Drosophila melanogaster: mutants defective in the cyclic AMP phosphodiesterase enzyme system. J. Cell. Biol 90, 101–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lecea L, Criado JR, Rivera S, Wen W, Soriano E, Henriksen SJ, Taylor SS, Gall CM, and Sutcliffe JG (1998). Endogenous protein kinase A inhibitor (PKI alpha) modulates synaptic activity. J. Neurosci. Res 53, 269–278. [DOI] [PubMed] [Google Scholar]
- Dere E, De Souza-Silva MA, Topic B, Spieler RE, Haas HL, and Huston JP (2003). Histidine-decarboxylase knockout mice show deficient nonreinforced episodic object memory, improved negatively reinforced water-maze performance, and increased neo- and ventro-striatal dopamine turnover. Learn Mem 10, 510–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhaka A, Costa RM, Hu HL, Irvin DK, Patel A, Kornblum HI, Silva AJ, O’Dell TJ, and Colicelli J (2003). The RAS effector RIN1 modulates the formation of aversive memories. J. Neurosci 23, 748–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz FC, Hernandez MA, Capella T, and Medina JH (2019). Dopamine Neurotransmission in the Ventral Tegmental Area Promotes Active Forgetting of Cocaine-Associated Memory. Mol Neurobiol 56, 6206–6217. [DOI] [PubMed] [Google Scholar]
- Dong Y, Green T, Saal D, Marie H, Neve R, Nestler EJ, and Malenka RC (2006). CREB modulates excitability of nucleus accumbens neurons. Nat. Neurosci 9, 475–477. [DOI] [PubMed] [Google Scholar]
- Dudai Y, Jan YN, Byers D, Quinn WG, and Benzer S (1976). Dunce, a mutant of Drosophila deficient in learning. Proc. Natl. Acad. Sci. USA 73, 1684–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duman JG, Mulherkar S, Tu YK, Cheng JXX, and Tolias KF (2015). Mechanisms for spatiotemporal regulation of Rho-GTPase signaling at synapses. Neurosci. Lett 601, 4–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engin E, Sigal M, Benke D, Zeller A, and Rudolph U (2020). Bidirectional regulation of distinct memory domains by alpha 5-subunit-containing GABA(A) receptors in CA1 pyramidal neurons. Learn Memory 27, 423–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engin E, Zarnowska ED, Benke D, Tsvetkov E, Sigal M, Keist R, Bolshakov VY, Pearce RA, and Rudolph U (2015). Tonic inhibitory control of dentate gyrus granule cells by alpha5-containing GABAA receptors reduces memory interference. J. Neurosci 35, 13698–13712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C, Gao Y, Liang G, Huang L, Wang J, Yang X, Shi Y, Drager UC, Zhong M, Gao, et al. (2020). Transcriptomics of Gabra4 knockout mice reveals common NMDAR pathways underlying autism, memory, and epilepsy. Mol. Autism 11, 13–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasano S, and Brambilla R (2011). Ras-ERK signaling in behavior: old questions and new perspectives. Front Behav. Neurosci 5, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorenza A, Lopez-Atalaya JP, Rovira V, Scandaglia M, Geijo-Barrientos E, and Barco A (2016). Blocking miRNA biogenesis in adult forebrain neurons enhances seizure susceptibility, fear memory, and food intake by increasing neuronal responsiveness. Cereb. Cortex 26, 1619–1633. [DOI] [PubMed] [Google Scholar]
- Flexner JB, Flexner LB, Stellar E, Roberts RB, and Haba GDL (1962). Inhibition of protein synthesis in brain and learning and memory following puromycin. J. Neurochem 9, 595–605. [DOI] [PubMed] [Google Scholar]
- Fontan-Lozano A, Romero-Granados R, del-Pozo-Martin Y, Suarez-Pereira I, Delgado-Garcia JM, Penninger JM, and Carrion AM (2009). Lack of DREAM protein enhances learning and memory and slows brain aging. Curr. Biol 19, 1332–1332. [DOI] [PubMed] [Google Scholar]
- Gai YC, Liu Z, Cervantes-Sandoval I, and Davis RL (2016). Drosophila SLC22A transporter is a memory suppressor gene that influences cholinergic neurotransmission to the mushroom bodies. Neuron 90, 581–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo FT, Saad MBZ, Morici JF, Miranda M, Anderson MC, Weisstaub NV, and Bekinschtein P (2021). Dopamine modulates adaptive forgetting in medial prefrontal cortex. bioRxiv [DOI] [PMC free article] [PubMed]
- Gao QQ, Yao WQ, Wang JJ, Yang T, Liu C, Tao YZ, Chen YJ, Liu X, and Ma L (2015). Post-training activation of Rac1 in the basolateral amygdala is required for the formation of both short-term and long-term auditory fear memory. Front Mol. Neurosci 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Shuai YC, Zhang XC, Peng YW, Wang LZ, He J, Zhong Y, and Li Q (2019). Genetic dissection of active forgetting in labile and consolidated memories in Drosophila. Proc. Natl. Acad. Sci. USA 116, 21191–21197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genoux D, Haditsch U, Knobloch M, Michalon A, Storm D, and Mansuy IM (2002). Protein phosphatase 1 is a molecular constraint on learning and memory. Nature 418, 970–975. [DOI] [PubMed] [Google Scholar]
- Gervasi N, Tchenio P, and Preat T (2010). PKA dynamics in a Drosophila learning center: coincidence detection by rutabaga adenylyl cyclase and spatial regulation by dunce phosphodiesterase. Neuron 65, 516–529. [DOI] [PubMed] [Google Scholar]
- Gouzi JY, Bouraimi M, Roussou IG, Moressis A, and Skoulakis EMC (2018). The Drosophila receptor tyrosine kinase Alk constrains long-term memory formation. J. Neurosci 38, 7701–7712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray NK, Coller JM, Dickson KS, and Wickens M (2000). Multiple portions of poly(A)-binding protein stimulate translation in vivo. Embo J 19, 4723–4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griggs EM, Young EJ, Rumbaugh G, and Miller CA (2013). MicroRNA-182 regulates amygdala-dependent memory formation. J. Neurosci 33, 1734–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruart A, Benito E, Delgado-Garcia JM, and Barco A (2012). Enhanced cAMP response element-binding protein activity increases neuronal excitability, hippocampal long-term potentiation, and classical eyeblink conditioning in alert behaving mice. J. Neurosci 32, 17431–17441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J, Nieland TJF, Zhou Y, Wang XY, Mazitschek R, et al. (2009). HDAC2 negatively regulates memory formation and synaptic plasticity. Nature 459, 55–U58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guven-Ozkan T, Busto GU, Jung JY, Drago I, and Davis RL (2020). miR-92a suppresses mushroom body-dependent memory consolidation in Drosophila. Eneuro 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guven-Ozkan T, Busto GU, Schutte SS, Cervantes-Sandoval I, O’Dowd DK, and Davis RL (2016). MiR-980 is a memory suppressor microRNA that regulates the autism-susceptibility gene A2bp1. Cell Rep 14, 1698–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haditsch U, Leone DP, Farinelli M, Chrostek-Grashoff A, Brakebusch C, Mansuy IM, McConnell SK, and Palmer TD (2009). A central role for the small GTPase Rac1 in hippocampal plasticity and spatial learning and memory. Mol. Cell. Neurosci 41, 409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadziselimovic N, Vukojevic V, Peter F, Milnik A, Fastenrath M, Fenyves BG, Hieber P, Demougin P, Vogler C, de Quervain DJF, et al. (2014). Forgetting is regulated via musashi-mediated translational control of the Arp2/3 complex. Cell 156, 1153–1166. [DOI] [PubMed] [Google Scholar]
- Han JH, Kushner SA, Yiu AP, Cole CJ, Matynia A, Brown RA, Neve RL, Guzowski JF, Silva AJ, and Josselyn SA (2007). Neuronal competition and selection during memory formation. Science 316, 457–460. [DOI] [PubMed] [Google Scholar]
- Han JH, Kushner SA, Yiu AP, Hsiang HL, Buch T, Waisman A, Bontempi B, Neve RL, Frankland PW, and Josselyn SA (2009). Selective Erasure of a Fear Memory. Science 323, 1492–1496. [DOI] [PubMed] [Google Scholar]
- Hansen RT, Conti M, and Zhang HT (2014). Mice deficient in phosphodiesterase-4A display anxiogenic-like behavior. Psychopharmacology 231, 2941–2954. [DOI] [PubMed] [Google Scholar]
- Hardt O, Nader K, and Wang YT (2014). GluA2-dependent AMPA receptor endocytosis and the decay of early and late long-term potentiation: possible mechanisms for forgetting of short- and long-term memories. Philos. T. R. Soc. B 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havekes R, Nijholt IM, Visser AK, Eisel UL, and Van der Zee EA (2008). Transgenic inhibition of neuronal calcineurin activity in the forebrain facilitates fear conditioning, but inhibits the extinction of contextual fear memories. Neurobiol. Learn Mem 89, 595–598. [DOI] [PubMed] [Google Scholar]
- Hawasli AH, Benavides DR, Nguyen C, Kansy JW, Hayashi K, Chambon P, Greengard P, Powell CM, Cooper DC, and Bibb JA (2007). Cyclin-dependent kinase 5 governs learning and synaptic plasticity via control of NMDAR degradation. Nat. Neurosci 10, 880–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkley LC, and Capitanio JP (2015). Perceived social isolation, evolutionary fitness and health outcomes: a lifespan approach. Phil. Trans. R. Soc. B 370, 20140114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkel-Tigges J, and Davis RL (1990). Rat Homologs of the Drosophila-Dunce Gene Code for Cyclic-Amp Phosphodiesterases Sensitive to Rolipram and Ro-20–1724. Mol Pharmacol 37, 7–10. [PubMed] [Google Scholar]
- Hernandez PJ, and Abel T (2008). The role of protein synthesis in memory consolidation: Progress amid decades of debate. Neurobiol Learn Mem 89, 293–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelreich S, Masuho I, Berry JA, MacMullen C, Skamangas NK, Martemyanov KA, and Davis RL (2017). Dopamine receptor DAMB signals via Gq to mediate forgetting in Drosophila. Cell Rep 21, 2074–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffer CA, and Klann E (2010). mTOR signaling: at the crossroads of plasticity, memory and disease. Trends Neurosci 33, 67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffer CA, Tang W, Wong H, Santillan A, Patterson RJ, Martinez LA, Tejada-Simon MV, Paylor R, Hamilton SL, and Klann E (2008). Removal of FKBP12 enhances mTOR-Raptor interactions, LTP, memory, and perseverative/repetitive behavior. Neuron 60, 832–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi J, Yamazaki D, Naganos S, Aigaki T, and Saitoe M (2008). Protein kinase A inhibits a consolidated form of memory in Drosophilae. Proc. Natl. Acad. Sci. USA 105, 20976–20981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn KE, Xu B, Gobert D, Hamam BN, Thompson KM, Wu CL, Bouchard JF, Uetani N, Racine RJ, Tremblay ML, et al. (2012). Receptor protein tyrosine phosphatase sigma regulates synapse structure, function and plasticity. J. Neurochem 122, 147–161. [DOI] [PubMed] [Google Scholar]
- Hsu YT, Li J, Wu D, Sudhof TC, and Chen L (2019). Synaptic retinoic acid receptor signaling mediates mTOR-dependent metaplasticity that controls hippocampal learning. Proc. Natl. Acad. Sci. USA 116, 7113–7122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XW, Toth KF, and Aravin AA (2017). piRNA Biogenesis in Drosophila melanogaster. Trends Genet 33, 882–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huentelman MJ, Stephan DA, Talboom J, Corneveaux JJ, Reiman DA, Gerber JD, Barnes CA, Alexander GE, Reiman EM, and Bimonte-Nelson HA (2009). Peripheral delivery of a ROCK inhibitor improves learning and working memory. Behav. Neurosci 123, 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Sawatari E, Hisamoto N, Kitazono T, Teramoto T, Fujiwara M, Matsumoto K, and Ishihara T (2013). Forgetting in C. elegans is accelerated by neuronal communication via the TIR-1/JNK-1 pathway. Cell Rep 3, 808–819. [DOI] [PubMed] [Google Scholar]
- Irvine EE, Drinkwater L, Radwanska K, Al-Qassab H, Smith MA, O’Brien M, Kielar C, Choudhury AI, Krauss S, Cooper JD, et al. (2011). Insulin receptor substrate 2 is a negative regulator of memory formation. Learn Mem 18, 375–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isabel G, Pascual A, and Preat T (2004). Exclusive consolidated memory phases in Drosophila. Science 304, 1024–1027. [DOI] [PubMed] [Google Scholar]
- Ishimoto H, and Kamikouchi A (2020). A feedforward circuit regulates action selection of pre-mating courtship behavior in female Drosophila. Curr. Biol 30, 396–407.e4. [DOI] [PubMed] [Google Scholar]
- Izquierdo A, Brigman JL, Radke AK, Rudebeck PH, and Holmes A (2017). The neural basis of reversal learning: an updated perspective. Neuroscience 345, 12–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon D, Song I, Guido W, Kim K, Kim E, Oh U, and Shin HS (2008). Ablation of Ca2+ channel beta 3 subunit leads to enhanced N-methyl-D-aspartate receptor-dependent long term potentiation and improved long term memory. J. Biol. Chem 283, 12093–12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon D, Yang YM, Jeong MJ, Philipson KD, Rhim H, and Shin HS (2003). Enhanced learning and memory in mice lacking Na+/Ca2+ exchanger 2. Neuron 38, 965–976. [DOI] [PubMed] [Google Scholar]
- Jiang LZ, Mao RR, Zhou QX, Yang YX, Cao J, Ding YQ, Yang Y, Zhang X, Li LJ, and Xu L (2016). Inhibition of Rac1 activity in the hippocampus impairs the forgetting of contextual fear memory. Mol. Neurobiol 53, 1247–1253. [DOI] [PubMed] [Google Scholar]
- Jobim PFC, Pedroso TR, Christoff RR, Werenicz A, Maurmann N, Reolon GK, and Roesler R (2012). Inhibition of mTOR by rapamycin in the amygdala or hippocampus impairs formation and reconsolidation of inhibitory avoidance memory. Neurobiol. Learn Mem 97, 105–112. [DOI] [PubMed] [Google Scholar]
- Josselyn SA, Shi CJ, Carlezon WA, Neve RL, Nestler EJ, and Davis M (2001). Long-term memory is facilitated by cAMP response element-binding protein overexpression in the amygdala. J. Neurosci 21, 2404–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaang BK, Kandel ER, and Grant SGN (1993). Activation of cAMP-responsive genes by stimuli that produce long-term facilitation in Aplysia sensory neurons. Neuron 10, 427–435. [DOI] [PubMed] [Google Scholar]
- Kelly MP, Logue SF, Brennan J, Day JP, Lakkaraju S, Jiang LX, Zhong XT, Tam M, Rizzo SJS, Platt BJ, et al. (2010). Phosphodiesterase 11A in brain is enriched in ventral hippocampus and deletion causes psychiatric disease-related phenotypes. Proc. Natl. Acad. Sci. USA 107, 8457–8462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoutorsky A, Yanagiya A, Gkogkas CG, Fabian MR, Prager-Khoutorsky M, Cao RF, Gamache K, Bouthiette F, Parsyan A, Sorge RE, et al. (2013). Control of Synaptic Plasticity and Memory via Suppression of Poly(A)-Binding Protein. Neuron 78, 298–311. [DOI] [PubMed] [Google Scholar]
- Kim IH, Wang H, Soderling SH, and Yasuda R (2014). Loss of Cdc42 leads to defects in synaptic plasticity and remote memory recall. Elife 3: e02839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YC, Lee HG, and Han KA (2007). D-1 dopamine receptor dDA1 is required in the mushroom body neurons for aversive and appetitive learning in Drosophila. J Neurosci 27, 7640–7647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch JC, Tatenhorst L, Roser AE, Saal KA, Tonges L, and Lingor P (2018). ROCK inhibition in models of neurodegeneration and its potential for clinical translation. Pharmacol. Therapeut 189, 1–21. [DOI] [PubMed] [Google Scholar]
- Kojima N, Borlikova G, Sakamoto T, Yamada K, Ikeda T, Itohara S, Niki H, and Endo S (2008). Inducible cAMP early repressor acts as a negative regulator for kindling epileptogenesis and long-term fear memory. J. Neurosci 28, 6459–6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka W, Kiryk A, Novak M, Herwerth M, Parkitna JR, Wawrzyniak M, Kowarsch A, Michaluk P, Dzwonek J, Arnsperger T, et al. (2010). MicroRNA loss enhances learning and memory in mice. J. Neurosci 30, 14835–14842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar DR (2006). Molecular details of formin-mediated actin assembly. Curr. Opin. Cell Biol 18, 11–17. [DOI] [PubMed] [Google Scholar]
- Kushner SA, Elgersma Y, Murphy GG, Jaarsma D, Hojjati MR, Cui YJ, LeBoutillier JC, Marrone DF, Choi ES, De Zeeuw CI, et al. (2005). Modulation of presynaptic plasticity and learning by the H-ras/extracellular signal-regulated kinase/synapsin I signaling pathway. J. Neurosci 25, 9721–9734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Kim H, Lee YS, and Kaang BK (2003). Overexpression and RNA interference of Ap-cyclic AMP-response element binding protein-2, a repressor of long-term facilitation, in Aplysia kurodai sensory-to-motor synapses. Neurosci. Lett 337, 9–12. [DOI] [PubMed] [Google Scholar]
- Lee JA, Tang ZZ, and Black DL (2009). An inducible change in Fox-1/A2BP1 splicing modulates the alternative splicing of downstream neuronal target exons. Genes Dev 23, 2284–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SE, Simons SB, Heldt SA, Zhao ML, Schroeder JP, Vellano CP, Cowan DP, Ramineni S, Yates CK, Feng Y, et al. (2010). RGS14 is a natural suppressor of both synaptic plasticity in CA2 neurons and hippocampal-based learning and memory. Proc. Natl. Acad. Sci. USA 107, 16994–16998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leighton LJ, Wei W, Marshall PR, Ratnu VS, Li X, Zajaczkowski EL, Spadaro PA, Khandelwal N, Kumar A, and Bredy TW (2019). Disrupting the hippocampal Piwi pathway enhances contextual fear memory in mice. Neurobio. Learn Mem 161, 202–209. [DOI] [PubMed] [Google Scholar]
- Levin LR, Han PL, Hwang PM, Feinstein PG, Davis RL, and Reed RR (1992). The Drosophila Learning and Memory Gene Rutabaga Encodes a Ca2+/Calmodulin-Responsive Adenylyl Cyclase. Cell 68, 479–489. [DOI] [PubMed] [Google Scholar]
- Li HH, Liu YT, Gao XQ, Liu LF, Amuti S, Wu DD, Jiang F, Huang L, Wang GY, Zeng JJ, et al. (2019). Neuroplastin 65 modulates anxiety- and depression-like behavior likely through adult hippocampal neurogenesis and central 5-HT activity. Febs J 286, 3401–3415. [DOI] [PubMed] [Google Scholar]
- Li YF, Cheng YF, Huang Y, Conti M, Wilson SP, O’Donnell JM, and Zhang HT (2011). Phosphodiesterase-4D Knock-Out and RNA Interference-Mediated Knock-Down Enhance Memory and Increase Hippocampal Neurogenesis via Increased cAMP Signaling. J. Neurosci 31, 172–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin AC, Bygrave AM, de Calignon A, Lee T, and Miesenbock G (2014). Sparse, decorrelated odor coding in the mushroom body enhances learned odor discrimination. Nat. Neurosci 17, 559–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Ponnusamy R, Widagdo J, Choi JA, Ge W, Probst C, Buckley T, Lou M, Bredy TW, Fanselow MS, et al. (2017). MicroRNA-mediated disruption of dendritogenesis during a critical period of development influences cognitive capacity later in life. Proc. Natl. Acad. Sci. USA 114, 9188–9193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Buchanan ME, Han KA, and Davis RL (2009). The GABAA receptor RDL suppresses the conditioned stimulus pathway for olfactory learning. J. Neurosci 29, 1573–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Krause WC, and Davis RL (2007). GABA(A) receptor RDL inhibits Drosophila olfactory associative learning. Neuron 56, 1090–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YL, Du SW, Lv L, Lei B, Shi W, Tang YK, Wang LZ, and Zhong Y (2016). Hippocampal activation of Rac1 regulates the forgetting of object recognition memory. Curr. Biol 26, 2351–2357. [DOI] [PubMed] [Google Scholar]
- Liu YL, Lv L, Wang LZ, and Zhong Y (2018). Social isolation induces Rac1-dependent forgetting of social memory. Cell Rep 25, 288–295. [DOI] [PubMed] [Google Scholar]
- Livingstone MS, Sziber PP, and Quinn WG (1984). Loss of calcium/calmodulin responsiveness in adenylate cyclase of rutabaga, a Drosophila learning mutant. Cell 37, 205–215. [DOI] [PubMed] [Google Scholar]
- Lu PD, Harding HP, and Ron D (2004). Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J Cell Biol 167, 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv L, Liu YL, Xie JX, Wu Y, Zhao JJ, Li Q, and Zhong Y (2019). Interplay between alpha 2-chimaerin and Rac1 activity determines dynamic maintenance of long-term memory. Nat. Commun 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malenka RC (2003). Synaptic plasticity and AMPA receptor trafficking. Ann. Ny. Acad. Sci 1003, 1–11. [DOI] [PubMed] [Google Scholar]
- Malleret G, Haditsch U, Genoux D, Jones MW, Bliss TV, Vanhoose AM, Weitlauf C, Kandel ER, Winder DG, and Mansuy IM (2001). Inducible and reversible enhancement of learning, memory, and long-term potentiation by genetic inhibition of calcineurin. Cell 104, 675–686. [DOI] [PubMed] [Google Scholar]
- Malmevik J, Petri R, Knauff P, Brattas PL, Akerblom M, and Jakobsson J (2016). Distinct cognitive effects and underlying transcriptome changes upon inhibition of individual miRNAs in hippocampal neurons. Sci. Rep 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamiya T, Yamada K, Miyamoto Y, Konig N, Watanabe Y, Noda Y, and Nabeshima T (2003). Neuronal mechanism of nociceptin-induced modulation of learning and memory: involvement of N-methyl-D-aspartate receptors. Mol. Psychiatr 8, 752–765. [DOI] [PubMed] [Google Scholar]
- Mariottini C, Munari L, Gunzel E, Seco JM, Tzavaras N, Hansen J, Stern SA, Gao V, Aleyasin H, Sharma A, et al. (2019). Wilm’s tumor 1 promotes memory flexibility. Nat. Commun 10, 3756–x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurice T, Strehaiano M, Simeon N, Bertrand C, and Chatonnet A (2016). Learning performances and vulnerability to amyloid toxicity in the butyrylcholinesterase knockout mouse. Behav. Brain Res 296, 351–360. [DOI] [PubMed] [Google Scholar]
- Mazzucchelli C, and Brambilla R (2000). Ras-related and MAPK signalling in neuronal plasticity and memory formation. Cell Mol Life Sci 57, 604–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaugh JL, and Alpern HP (1966). Effects of electroshock on memory: amnesia without convulsions. Science 152, 665–666. [DOI] [PubMed] [Google Scholar]
- McQuade JM, Vorhees CV, Xu M, and Zhang JH (2002). DNA fragmentation factor 45 knockout mice exhibit longer memory retention in the novel object recognition task compared to wild-type mice. Physiol. Behav 76, 315–320. [DOI] [PubMed] [Google Scholar]
- McQuown S, Xia SZ, Baumgartel K, Barido R, Anderson G, Dyck B, Scott R, and Peters M (2019). Phosphodiesterase 1b (PDE1B) regulates spatial and contextual memory in hippocampus. Front Mol. Neurosci 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuown S, Paes D, Baumgärtel K, Prickaerts J, and Peters M (2021). Pharmacological inhibition of phosphodiesterase 7 enhances consolidation processes of spatial memory. Neurobiology of learning and memory, 177, 107357. [DOI] [PubMed] [Google Scholar]
- Migues PV, Hardt O, Wu DC, Gamache K, Sacktor TC, Wang YT, and Nader K (2010). PKM zeta maintains memories by regulating GluR2-dependent AMPA receptor trafficking. Nat. Neurosci 13, 630–U147. [DOI] [PubMed] [Google Scholar]
- Migues PV, Liu LD, Archbold GEB, Einarsson EO, Wong J, Bonasia K, Ko SH, Wang YT, and Hardt O (2016). Blocking synaptic removal of GluA2-containing AMPA receptors prevents the natural forgetting of long-term memories. J. Neurosci 36, 3481–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H, and Takenawa T (2003). Regulation of actin dynamics by WASP family proteins. J. Biochem 134, 309–313. [DOI] [PubMed] [Google Scholar]
- Mioduszewska B, Jaworski J, and Kaczmarek L (2003). Inducible cAMP early repressor (ICER) in the nervous system - a transcriptional regulator of neuronal plasticity and programmed cell death. J. Neurochem 87, 1313–1320. [DOI] [PubMed] [Google Scholar]
- Miyake A, Takahashi S, Nakamura Y, Inamura K, Matsumoto S, Mochizuki S, and Katou M (2009). Disruption of the ether-a-go-go K+ channel gene BEC1/KCNH3 enhances cognitive function. J. Neurosci 29, 14637–14645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MD, Cushman J, Chandra D, Homanics GE, Olsen RW, and Fanselow MS (2010). Trace and contextual fear conditioning is enhanced in mice lacking the alpha 4 subunit of the GABA(A) receptor. Neurobiol. Learn Mem 93, 383–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel C, Sherrin T, Kennedy NJ, Forest KH, Avcioglu Barutcu S, Robles M, Carpenter-Hyland E, Alfulaij N, Standen CL, Nichols RA, et al. (2018). JIP1-Mediated JNK Activation Negatively Regulates Synaptic Plasticity and Spatial Memory. J. Neurosci 38, 3708–3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimura N, Yasuda H, Yamaguchi K, Katayama K, Hatayama M, Tomioka NH, Odagawa M, Kamiya A, Iwayama Y, Maekawa M, et al. (2017). Autism-like behaviours and enhanced memory formation and synaptic plasticity in Lrfn2/SALM1-deficient mice. Nat. Commun 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RGM (2013). NMDA receptors and memory encoding. Neuropharmacology 74:32–40. [DOI] [PubMed] [Google Scholar]
- Morris MJ, Mahgoub M, Na ES, Pranav H, and Monteggia LM (2013). Loss of histone deacetylase 2 improves working memory and accelerates extinction learning. J. Neurosci 33, 6401–6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy GG, Fedorov NB, Giese KP, Ohno M, Friedman E, Chen R, and Silva AJ (2004). Increased neuronal excitability, synaptic plasticity, and learning in aged Kv beta 1.1 knockout mice. Curr. Biol 14, 1907–1915. [DOI] [PubMed] [Google Scholar]
- Murthy VN (2011). Olfactory maps in the brain. Annu Rev Neurosci 34, 233–58. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Manabe T, Watanabe M, Mamiya T, Ichikawa R, Kiyama Y, Sanbo M, Yagi T, Inoue Y, Nabeshima T, et al. (2001). Enhancement of hippocampal LTP, reference memory and sensorimotor gating in mutant mice lacking a telencephalon-specific cell adhesion molecule. Eur. J. Neurosci 13, 179–189. [DOI] [PubMed] [Google Scholar]
- Nemoz G, Prigent AF, Moueqqit M, Fougier S, Macovschi O, and Pacheco H (1985). Selective inhibition of one of the cyclic-AMP phosphodiesterases from rat brain by the neurotropic compound rolipram. Biochem. Pharmacol 34, 2997–3000. [DOI] [PubMed] [Google Scholar]
- Nishiyama H, Knopfel T, Endo S, and Itohara S (2002). Glial protein S100B modulates long-term neuronal synaptic plasticity. Proc. Natl. Acad. Sci. USA 99, 4037–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan MF, Malleret G, Dudman J Poplawski T, Buhl DL, Santoro B, Gibbs E, Vronskaya S, Buzsaki G, Siegelbaum SA, Kandel ER, et al. (2004). A behavioral role for dendritic integration: HCN1 channels constrain spatial inputs to distal dendrites memory and plasticity at of CA1 pyramidal neurons. Cell 119, 719–732. [DOI] [PubMed] [Google Scholar]
- Noyes NC, Walkinshaw E, and Davis RL (2020). Ras acts as a molecular switch between two forms of consolidated memory in Drosophila. Proc. Natl. Acad. Sci. USA 117, 2133–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell KA, and Boekel JD (2007). Mighty piwis defend the germline against genome intruders. Cell 129, 37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh D, Han S, Seo J, Lee JR, Choi J, Groffen J, Kim K, Cho YS, Choi HS, Shin H, et al. (2010). Regulation of synaptic Rac1 activity, long-term potentiation maintenance, and learning and memory by BCR and ABR Rac GTPase-activating proteins. J. Neurosci 30, 14134–14144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paes D, Xie KQ, Wheeler DG, Zook D, Prickaerts J, and Peters M (2021). Inhibition of PDE2 and PDE4 synergistically improves memory consolidation processes. Neuropharmacology 184. [DOI] [PubMed] [Google Scholar]
- Pan B, Wang W, Zhong P, Blankman JL, Cravatt BF, and Liu QS (2011). Alterations of endocannabinoid signaling, synaptic plasticity, learning, and memory in monoacylglycerol lipase knock-out mice. J. Neurosci 31, 13420–13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JG, Wanat MJ, Soden ME, Ahmad K, Zweifel LS, Bamford NS, and Palmiter RD (2011). Attenuating GABA(A) receptor signaling in dopamine neurons selectively enhances reward learning and alters risk preference in mice. J. Neurosci 31, 17103–17112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S, Zhang Y, Zhang JN, Wang H, and Ren BX (2010). ERK in learning and memory: a review of recent research. Int. J. Mol. Sci 11, 222–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, Tedeschi A, and Schmitz J (2008). The cohesin complex and its roles in chromosome biology. Genes Dev 22, 3089–3114. [DOI] [PubMed] [Google Scholar]
- Phan A, Thomas CI, Chakraborty M, Berry JA, Kamasawa N, and Davis RL (2019). Stromalin constrains memory acquisition by developmentally limiting synaptic vesicle pool size. Neuron 101, 103–118 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Placais PY, Trannoy S, Isabel G, Aso Y, Siwanowicz I, Belliart-Guerin G, Vernier P, Birman S, Tanimoto H, and Preat T (2012). Slow oscillations in two pairs of dopaminergic neurons gate long-term memory formation in Drosophila. Nat. Neurosci 15, 592–599. [DOI] [PubMed] [Google Scholar]
- Platenik J, Kuramoto N, and Yoneda Y (2000). Molecular mechanisms associated with long-term consolidation of the NMDA signals. Life Sci 67, 335–364. [DOI] [PubMed] [Google Scholar]
- Poplawski SG, Garbett KA, McMahan RL, Kordasiewicz HB, Zhao H, Kennedy AJ, Goleva SB, Sanders TH, Motley ST, Swayze EE, et al. (2020). An antisense oligonucleotide leads to suppressed transcription of Hdac2 and long-term memory enhancement. Mol. Ther-Nucl. Acids 19, 1399–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthanveettil SV, Monje FJ, Miniaci MC, Choi YB, Karl KA, Khandros E, Gawinowicz MA, Sheetz MP, and Kandel ER (2008). A New Component in Synaptic Plasticity: Upregulation of Kinesin in the Neurons of the Gill-Withdrawal Reflex. Cell 135, 960–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn WG, and Dudai Y (1976). Memory phases in Drosophila. Nature 262, 576–577. [DOI] [PubMed] [Google Scholar]
- Rappaport AN, Jacob E, Sharma V, Inberg S, Elkobi A, Ounallah-Saad H, Pasmanik-Chor M, Edry E, and Rosenblum K (2015). Expression of quinone reductase-2 in the cortex is a muscarinic acetylcholine receptor-dependent memory consolidation constraint. J. Neurosci 35, 15568–15581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards BA, and Frankland PW (2017). The persistence and transience of memory. Neuron 94, 1071–1084. [DOI] [PubMed] [Google Scholar]
- Robertson EM (2018). Memory instability as a gateway to generalization. Plos. Biol 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romberg C, Yang SJ, Melani R, Andrews MR, Horner AE, Spillantini MG, Bussey TJ, Fawcett JW, Pizzorusso T, and Saksida LM (2013). Depletion of perineuronal nets enhances recognition memory and long-term depression in the perirhinal cortex. J. Neurosci 33, 7057–7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotty JD, Wu CY, and Bear JE (2013). New insights into the regulation and cellular functions of the ARP2/3 complex. Nat. Rev. Mol. Cell Bio 14, 7–12. [DOI] [PubMed] [Google Scholar]
- Rutten K, Prickaerts J, and Blokland A (2006). Rolipram reverses scopolamine-induced and time-dependent memory deficits in object recognition by different mechanisms of action. Neurobiol. Learn Mem 85, 132–138. [DOI] [PubMed] [Google Scholar]
- Sabandal JM, Berry JA, and Davis RL (2021). Dopamine-based mechanism for transient forgetting. Nature 591, 426–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson DJ, Good MA, Skelton K, Sprengel R, Seeburg PH, Rawlins JNP, and Bannerman DM (2009). Enhanced long-term and impaired short-term spatial memory in GluA1 AMPA receptor subunit knockout mice: evidence for a dual-process memory model. Learn Memory 16, 508–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitz J, Solms M, and Ramesar R (2006). The molecular genetics of cognition: dopamine, COMT and BDNF. Genes Brain Behav 5, 311–328. [DOI] [PubMed] [Google Scholar]
- Scheunemann L, Jost E, Richlitzki A, Day JP, Sebastian S, Thum AS, Efetova M, Davies SA, and Schwarzel M (2012). Consolidated and labile odor memory are separately encoded within the Drosophila brain. J. Neurosci 32, 17163–17171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheunemann L, Placais PY, Dromard Y, Schwarzel M, and Preat T (2018). Dunce Phosphodiesterase Acts as a Checkpoint for Drosophila Long-Term Memory in a Pair of Serotonergic Neurons. Neuron 98, 350–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz KP, and Byrne JH (1988). Intracellular injection of cAMP induces a long-term reduction of neuronal K+ currents. Science 240, 1664–1666. [DOI] [PubMed] [Google Scholar]
- Scoville WB, and Milner B (2000). Loss of recent memory after bilateral hippocampal lesions. 1957. J Neuropsychiatry Clin. Neurosci 12, 103–113. [DOI] [PubMed] [Google Scholar]
- Shahmoradi A, Radyushkin K, and Rossner MJ (2015). Enhanced memory consolidation in mice lacking the circadian modulators Sharp1 and-2 caused by elevated Igf2 signaling in the cortex. Proc. Natl. Acad. Sci. USA 112, E3582–E3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V, Ounallah-Saad H, Chakraborty D, Hleihil M, Sood R, Barrera I, Edry E, Chandran SK, de Leon SB, Kaphzan H, et al. (2018). Local inhibition of PERK enhances memory and reverses age-related deterioration of cognitive and neuronal properties. J. Neurosci 38, 648–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Cai Y, Liu G, Gong N, Liu Z, Xu T, Wang Z, and Fei J (2012). Enhanced learning and memory in GAT1 heterozygous mice. Acta Biochim. Biophys. Sin. (Shanghai) 44, 359–366. [DOI] [PubMed] [Google Scholar]
- Shuai Y, Lu B, Hu Y, Wang L, Sun K, and Zhong Y (2010). Forgetting is regulated through Rac activity in Drosophila. Cell 140, 579–589. [DOI] [PubMed] [Google Scholar]
- Shumyatsky GP, Tsvetkov E, Malleret G, Vronskaya S, Hatton M, Hampton L, Battey JF, Dulac C, Kandel ER, and Bolshakov VY (2002). Identification of a signaling network in lateral nucleus of amygdala important for inhibiting memory specifically related to learned fear. Cell 111, 905–918. [DOI] [PubMed] [Google Scholar]
- Silveira Villarroel H, Bompolaki M, Mackay JP, Miranda Tapia AP, Michaelson SD, Leitermann RJ, Marr RA, Urban JH, and Colmers WF (2018). NPY induces stress resilience via downregulation of Ih in principal neurons of rat basolateral amygdala. J. Neurosci 38, 4505–4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slane JM, Lee HS, Vorhees CV, Zhang JH, and Xu M (2000). DNA fragmentation factor 45 deficient mice exhibit enhanced spatial learning and memory compared to wild-type control mice. Brain Res 867, 70–79. [DOI] [PubMed] [Google Scholar]
- Squire LR, and Davis HP (1981). The pharmacology of memory: a neurobiological perspective. Annu. Rev. Pharmacol. Toxicol 21, 323–356. [DOI] [PubMed] [Google Scholar]
- Sterneck E, Paylor R, Jackson-Lewis V, Libbey M, Przedborski S, Tessarollo L, Crawley JN, and Johnson PF (1998). Selectively enhanced contextual fear conditioning in mice lacking the transcriptional regulator CCAAT/enhancer binding protein delta. Proc. Natl. Acad. Sci. USA 95, 10908–10913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan DP, Liu QY, Koshiya N, Gu H, and Alkon D (2006). Enhancement of long-term memory retention and short-term synaptic plasticity in cbl-b null mice. Proc. Natl. Acad. Sci. USA 103, 5125–5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YP, Shimizu E, Dube GR, Rampon C, Kerchner GA, Zhuo M, Liu G, and Tsien JZ (1999). Genetic enhancement of learning and memory in mice. Nature 401, 63–69. [DOI] [PubMed] [Google Scholar]
- Teng LL, Lu GL, Chiou LC, Lin WS, Cheng YY, Hsueh TE, Huang YC, Hwang NH, Yeh JW, Liao RM, et al. (2019). Serotonin receptor HTR6-mediated mTORC1 signaling regulates dietary restriction-induced memory enhancement. Plos. Biol 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomchik SM, and Davis RL (2009). Dynamics of learning-related cAMP signaling and stimulus integration in the Drosophila olfactory pathway. Neuron 64, 510–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treffert DA (2009). The savant syndrome: an extraordinary condition. A synopsis: past, present, future. Philos. Trans. R. Soc. Lond. B. Biol. Sci 364, 1351–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai LCL, Chan GCK, Nangle SN, Shimizu-Albergine M, Jones GL, Storm DR, Beavo JA, and Zweifel LS (2012). Inactivation of Pde8b enhances memory, motor performance, and protects against age-induced motor coordination decay. Genes Brain Behav 11, 837–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tully T, Preat T, Boynton SC, and Del Vecchio M (1994). Genetic dissection of consolidated memory in Drosophila. Cell 79, 35–47. [DOI] [PubMed] [Google Scholar]
- Turrigiano GG, and Nelson SB (1998). Thinking globally, acting locally: AMPA receptor turnover and synaptic strength. Neuron 21, 933–935 [DOI] [PubMed] [Google Scholar]
- Vattem KM, and Wek RC (2004). Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. USA 101, 11269–11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkitaramani DV, Moura PJ, Picciotto MR, and Lombroso PJ (2011). Striatal-enriched protein tyrosine phosphatase (STEP) knockout mice have enhanced hippocampal memory. Eur. J. Neurosci 33, 2288–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkinshaw E, Gai Y, Farkas C, Richter D, Nicholas E, Keleman K, and Davis RL (2015). Identification of genes that promote or inhibit olfactory memory formation in Drosophila. Genetics 199, 1173–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimber M, Schott BH, Wendler F, Seidenbecher CI, Behnisch G, Macharadze T, Bauml KHT, and Richardson-Klavehn A (2011). Prefrontal dopamine and the dynamic control of human long-term memory. Transl Psychiat 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer ME, Blackwell JM, and Abel T (2020). Rolipram treatment during consolidation ameliorates long-term object location memory in aged male mice. Neurobiol. Learn Mem 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LJ, Mellstrom B, Wang HS, Ren M, Domingo S, Kim SS, Li XY, Chen T, Naranjo JR, and Zhuo M (2010). DREAM (downstream regulatory element antagonist modulator) contributes to synaptic depression and contextual fear memory. Mol. Brain 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J, Ginty DD, and Greenberg ME (1996). Coupling of the RAS-MAPK pathway to gene activation by RSK2, a growth factor-regulated CREB kinase. Science 273, 959–963. [DOI] [PubMed] [Google Scholar]
- Xu ZX, Tan JW, Xu HF, Hill CJ, Ostrovskaya O, Martemyanow KA, and Xu BJ (2019). Caspase-2 promotes AMPA receptor internalization and cognitive flexibility via mTORC2-AKT-GSK3 beta signaling. Nat. Commun 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata N, Ezaki T, Takahashi T, Wu H, Tanimoto H (2021). Presynaptic inhibition of dopamine neurons controls optimistic bias. Elife 10:e64907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki D, Horiuchi J, Miyashita T, and Saitoe M (2010). Acute inhibition of PKA activity at old ages ameliorates age-related memory impairment in Drosophila. J. Neurosci 30, 15573–15577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HT, Hou HL, Pahng A, Gu H, Nairn AC, Tang YP, Colombo PJ, and Xia HH (2015). Protein phosphatase-1 inhibitor-2 is a novel memory suppressor. J. Neurosci 35, 15082–15087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin JC, Wallach JS, Del Vecchio M, Wilder EL, Zhou H, Quinn WG, and Tully T (1994). Induction of a dominant negative CREB transgene specifically blocks long-term memory in Drosophila. Cell 79, 49–58. [DOI] [PubMed] [Google Scholar]
- Yiu AP, Mercaldo V, Yan C, Richards B, Rashid AJ, Hsiang HLL, Pressey J, Mahadevan V, Tran MM, Kushner SA, et al. (2014). Neurons Are Recruited to a Memory Trace Based on Relative Neuronal Excitability Immediately before Training. Neuron 83, 722–735. [DOI] [PubMed] [Google Scholar]
- Yuva-Aydemir Y, Xu XL, Aydemir O, Gascon E, Sayin S, Zhou WK, Hong Y, and Gao FB (2015). Downregulation of the host gene jigr1 by miR-92 Is essential for neuroblast self-renewal in Drosophila. Plos. Genet 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zengeler KE, Gettens CP, Smith HC, Caron MM, Zhang X, Howard AH, Boitnott AR, Gogliettino AR, Reda A, Malachowsky BG, et al. (2019). Tet2 negatively regulates memory fidelity
- Zhang XC, Li Q, Wang LZ, Liu ZJ, and Zhong Y (2016). Cdc42-dependent forgetting regulates repetition effect in prolonging memory retention. Cell Rep 16, 817–825. [DOI] [PubMed] [Google Scholar]
- Zhou M, Greenhill S, Huang S, Silva TK, Sano Y, Wu SM, Cai Y, Nagaoka Y, Sehgal M, Cai DJ, et al. (2016). CCR5 is a suppressor for cortical plasticity and hippocampal learning and memory. Elife 5: e20985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Won J, Karlsson MG, Zhou M, Rogerson T, Balaji J, Neve R, Poirazi P, and Silva AJ (2009). CREB regulates excitability and the allocation of memory to subsets of neurons in the amygdala. Nat. Neurosci 12, 1438–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu PJ, Huang W, Kalikulov D, Yoo JW, Placzek AN, Stoica L, Zhou H, Bell JC, Friedlander MJ, Krnjevic K, et al. (2011). Suppression of PKR promotes network excitability and enhanced cognition by interferon-gamma-mediated disinhibition. Cell 147, 1384–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zovkic IB, Paulukaitis BS, Day JJ, Etikala DM, and Sweatt JD (2014). Histone H2A.Z subunit exchange controls consolidation of recent and remote memory. Nature 515, 582–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurek AA, Kemp SW, Aga Z, Walker S, Milenkovic M, Ramsey AJ, Sibille E, Scherer SW, and Orser BA (2016). alpha5GABAA receptor deficiency causes autism-like behaviors. Ann. Clin. Transl. Neurol 3, 392–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1: Comprehensive list of early memory suppressor genes. The data in this table contains memory suppressor genes that act on early memory (effects found at the earliest time point tested within 4hr of training). This category may include both suppressors of acquisition and suppressors of early memory.
Manipulation Abbreviations:
KO = knockout
cKO = conditional knockout
KD = knockdown
WT= wild type
Behavioral Test Abbreviations:
AFC = Auditory Fear Conditioning
CFC = Contextual Fear Conditioning
MWM = Morris Water Maze
NOR = Novel Object Recognition
NOL= Novel Object Location
RAM= Radial Arm Maze
Neuron Function Abbreviations:
LTP = Long-Term Potentiation
STP= Short-Term Potentiation
LTD= Long-Term Depression
EPSC = Excitatory Postsynaptic Current
mEPSC = miniature Excitatory Postsynaptic Current
fEPSC= field Excitatory Postsynaptic Current
eEPSP= extracellular Excitatory Postsynaptic Potential
IPSC = Inhibitory Postsynaptic Current
mIPSC = miniature Inhibitory Postsynaptic Current
Supplemental Table 2: Comprehensive list of late memory suppressor genes. The data in the table contains memory suppressor genes that act on late memory. These genes have no effects at the earliest time point tested but effects on memory thereafter. This category likely contains suppressors of consolidation and/or forgetting.
Manipulation Abbreviations:
KO = knockout
cKO = conditional knockout
KD = knockdown
WT= wild type
Behavioral Test Abbreviations:
AFC = Auditory Fear Conditioning
CFC = Contextual Fear Conditioning
MWM = Morris Water Maze
NOR = Novel Object Recognition
NOL= Novel Object Location
RAM= Radial Arm Maze
Neuron Function Abbreviations:
LTP = Long-Term Potentiation
STP= Short-Term Potentiation
LTD= Long-Term Depression
EPSC = Excitatory Postsynaptic Current
mEPSC = miniature Excitatory Postsynaptic Current
fEPSC= field Excitatory Postsynaptic Current
eEPSP= extracellular Excitatory Postsynaptic Potential
IPSC = Inhibitory Postsynaptic Current
mIPSC = miniature Inhibitory Postsynaptic Current
Supplemental Table 3: Comprehensive list of undefined memory suppressor genes. The data in the table contains memory suppressor genes that act on an undefined memory operation. These memory suppressors could not be placed into either of the early or late memory genes defined above due to insufficient data.
Manipulation Abbreviations:
KO = knockout
cKO = conditional knockout
KD = knockdown
WT= wild type
Behavioral Test Abbreviations:
AFC = Auditory Fear Conditioning
CFC = Contextual Fear Conditioning
MWM = Morris Water Maze
NOR = Novel Object Recognition
NOL= Novel Object Location
RAM= Radial Arm Maze
Neuron Function Abbreviations:
LTP = Long-Term Potentiation
STP= Short-Term Potentiation
LTD= Long-Term Depression
EPSC = Excitatory Postsynaptic Current
mEPSC = miniature Excitatory Postsynaptic Current
fEPSC= field Excitatory Postsynaptic Current
eEPSP= extracellular Excitatory Postsynaptic Potential
IPSC = Inhibitory Postsynaptic Current
mIPSC = miniature Inhibitory Postsynaptic Current


