Abstract
The laboratory axolotl (Ambystoma mexicanum) is widely used in biological research. Recent advancements in genetic and molecular toolkits are greatly accelerating the work using axolotl, especially in the area of tissue regeneration. At this juncture, there is a critical need to establish gene and transgenic nomenclature to ensure uniformity in axolotl research. Here, we propose guidelines for genetic nomenclature when working with the axolotl.
Introduction:
The axolotl’s tremendous regeneration ability, large egg size and ex vivo development have attracted generations of researchers to use it in biological research. However, the axolotl’s use was limited in the 20th century due to the lack of genomic information and genetic tools. In the last two decades, major efforts by a number of laboratories have led to the development of new research resources that now allow for resolution of biological processes at the molecular level. These efforts have yielded transcriptome and genome assemblies1–4, transgenic animals5–7, genome editing using CRISPR/Cas9 technology to generate knockout and knock-in animals8–10, implementation of tissue-specific Cre-loxP11,12, virus mediated gene delivery for ectopic gene expression13–16, and tissue-clearing and advanced imaging technologies17–20. With these resources and tools in hand, the axolotl community is expected to enter the phase of exponential growth in coming years, while all researchers will benefit from the unified nomenclature.
Standardized nomenclature of genes, proteins or genetically modified animals is crucial for effective communication among researchers. It also facilitates comparative -omics studies with other organisms. Moreover, nomenclature of a transgenic animal provides essential additional information. For example, it indicates the method and specific genome modification that was used to develop the transgenic and identifies the lab/institute that created the resource. Since the first implementation of standardized gene nomenclature in M. musculus21,22, nomeclature guidelines (detailed in a book written by Wood23) and committees have been introduced in human24,25 and many major model organisms, such as Xenopus26,27, zebrafish28,29, C. elegans30, chicken31, anole32, tunicates33 and planaria - S. mediterranea34.
In the last three years, salamander researchers have met annually to identify critical needs to advance and unify community efforts, including the formation of a gene nomenclature committee. In this early stage of gene annotation, the identification of orthologous axolotl genes is complicated by the limitations of the current assemblies. Additionally, evolutionary processes complicate the identification of orthologous genes, including the nucleotide sequence divergence/convergence, gene duplication and rearrangement of genes within and among chromosomes. As chromosome evolution in the axolotl has been relatively conservative relative to other vertebrate taxa35–37, information from sequence identity and gene order conservation (conserved synteny) can be used to efficiently identify homologous genes in the axolotl genome. Indeed, we expect that many genes in the most recent axolotl genome assembly4 are correctly annotated based on their homology to other vertebrate genes. Nevertheless, at this stage investigators should remain cautious and manually validate gene annotations that seem ambiguous or strange. In following the precedent of other gene nomenclature committees, it is logical to align axolotl gene names and symbols with those in model organism databases to facilitate the inevitable migration of axolotl genome annotations into a unified database format.
Here, we advance a general set of guidelines for describing gene centric information when using the axolotl and other salamanders. These guidelines do not cover the full range of gene and RNA types (miRNA, snRNA, snoRNA) that are annotated in well-curated model organism databases. They will be made available through the Ambystoma genomic stock center (AGSC, https://ambystoma.uky.edu) and axolotl-omics (https://www.axolotl-omics.org), which at present are the two most-widely used web resources for the axolotl community. Regular updates to these guidelines will be made as axolotl genome annotations mature. The current authors of this manuscript will serve as the gene nomenclature committee and Dr. Elspeth Bruford of the HUGO Gene Nomenclatute Committee, which is responsible for naming human genes, will serve as an advisor of this committee. Gene nomenclature guidelines will be expanded in the future if necessary after discussing with the axolotl research community. These guidelines are designed according to FAIR (Findability, Accessibility, Interoperability, and Reusability) principles to maintain consistency with other model organisms wherever possible38. For reader’s ease, we highlight the axolotl as the exemplar salamander species and Prrx1 as the exemplar gene throughout this manuscript to introduce guidelines for the gene nomenclature.
Summary of nomenclature:
Species symbol: Amex
Gene/mRNA/cDNA: Prrx1
Protein: PRRX1
Paralogs: Prrx1 and Prrx2
Transcript variant: Prrx1.1, Prrx1.2
Transgenic line: tgSceI(Mmu.Prrx1:GFPnls-T2a-ERT2-Cre-ERT2)Labcode, tgSceI(Mmu.Prrx1:GFPnls-T2a-ERT2-Cre-ERT2;CAGGS:loxP-GFP-loxP-Cherry)Labcode
Gene knockout: tm(Prrx1153v6D8/153v6D8)Labcode, tm(Prrx1153v6I5/+)Labcode
Gene knock-in: tm(Prrx1t/+:Prrx1-T2a-Cherry)Labcode, tm(Prrx1r/+:Cherry)Labcode, tm(Prrx1r/+:miniPrrx1-T2a-Cherry)Labcode
Detailed description of nomenclature:
Species symbol
-
1
Species abbreviation
Although the axolotl (Ambystoma mexicanum) is one of the most commonly used salamanders, a number of other species such as Ambystoma tigrinum (Tiger salamander), Ambystoma maculatum (Spotted salamander) Notophthalmus viridescens (Eastern newt), and Pleurodeles waltl (Iberian newt) are commonly used for tetrapod tissue regeneration research39. A comparative -omics study often requires identification of orthologous genes across salamander species. We recommend using the first letter denoting the genus name in upper case and the first three letters of the species name in lower case regular font to create the species-specific acronym. For Ambystoma mexicanum, we recommend using Amex as a species symbol. For other ambystomatid species, the same convention would follow, such as Amac (A. maculatum), Aand (A. andersoni), and Acal (A. californiensi). If there is a subspecies, the first letter of the subspecies name can be assigned as a fifth letter in a lower-case italics, and a sixth letter to differentiate between subspecies that share the same first letter of the subspecies name. For example, there are several A. tigrinum subspecies, including A. tigrinum mavortium (Atigma), A. tigrinum melanosticum (Atigme), and A. tigrinum nebulosum (Atign).
Gene/mRNA/cDNA/Protein
-
2
Gene
A gene symbol is described by a series of alphanumeric characters, where the first character is upper-case italics and the rest are lower-case italics. This is in line with the mouse gene nomenclature and it follows current axolotl gene annotations2. For example, a gene Paired related homeobox 1 is referred to as Prrx1.-
2.1Species ambiguityWhen there is an ambiguity about the orthology of a gene from a species, we recommend using a prefix to indicate the species name. The prefixes should not be considered part of the name and hence, a period should be placed between species and the gene name, while an entire phrase should be written in italics, e.g. Amex.Prrx1. When there is no ambiguity of species used for the study, the species symbol must be omitted when describing the gene.
-
2.2Gene, mRNA, cDNAOften a gene, mRNA and cDNA need to be distinguished in a text. In such cases, we recommend putting “gene”, “mRNA” or “cDNA” in regular font in parentheses in front of the gene symbol. Gene, mRNA or cDNA words must be omitted to describe a gene when there is no ambiguity or after describing once in the beginning of the communication.Gene: Prrx1In case of ambiguity between gene, mRNA, cDNA: (gene)Prrx1, (mRNA)Prrx1, (cDNA)Prrx1In case of species ambiguity: (gene)Amex.Prrx1
-
2.1
-
3
Protein
The protein symbol is same as the gene symbol, but in regular (non-italic) font, with all characters in upper-case.
Protein: PRRX1
Note the differences in gene and protein naming convention among commonly used model organisms in Table 1.
Table1:
Gene and protein naming convention in axolotl, zebrafish, Xenopus, anole, mouse, chicken, and human.
Gene nomenclature in axolotl
The chromosome-scale axolotl genome assemblies2,3 made it possible to study not only the coding sequences, but also the evolution and synteny at the whole-genome level and accurate gene annotation is vital for those purposes. However, to ensure accurate and unambiguous communication between scientists, it is crucial to work out a set of rules for how the genes and proteins should be named, how to distinguish paralogs and orthologs, and how to name pseudogenes.
-
4
Orthologs and paralogs
Unlike zebrafish and Xenopus laevis, the axolotl genome was not structured by whole-genome duplication events. However, axolotl-specific (i.e. in-paralogs) as well as salamander-specific (i.e. out-paralogs) gene duplicates are known (Fig. 1a) and can be shared with other salamander taxa 42. In order to define the evolutionary relationship between the genes, it is crucial to define whether the genes in different species derive from a common ancestor (orthologs), or whether the genes in the same species arose from the same ancestor (paralogs) 43. While it is important to standardize the gene names for orthologous genes, it is also important to keep the gene naming compatible to that used across vertebrates, in order to make the analyses of the axolotl data comparable with the large body of available human and mouse datasets. Similar to the human and mouse orthologous gene nomenclature (HUMOT) project 44, we propose to rely on a mixed approach that combines both the orthology information with the synteny and also integrates the data from expert orthology resources. The importance of the synteny can be demonstrated by a following example. Imagine a situation that in the axolotl, a gene homologous to a gene GENEB in another species was duplicated (Fig. 1b), while the copy that is more similar to the ancestral gene (indicated by the exon pattern in Fig. 1b) moved out of the locus. In this case, the copy that stayed in the locus should be annotated as Geneb2, while the one that moved out should be annotated as Geneb1. In contrast, if the copy that moved out is less similar to GENEB than the one that stayed, then the latter should be annotated as Geneb1 and the former as Geneb2. If synteny is not conserved then phylogenetic analysis should be used to identify the relationships between the genes. As these relationships can be complex it is better to deliberately exclude ambiguous orthology information than to propagate incorrect assumptions about the genes.
Fig. 1. Gene nomenclature for duplicated and novel genes.
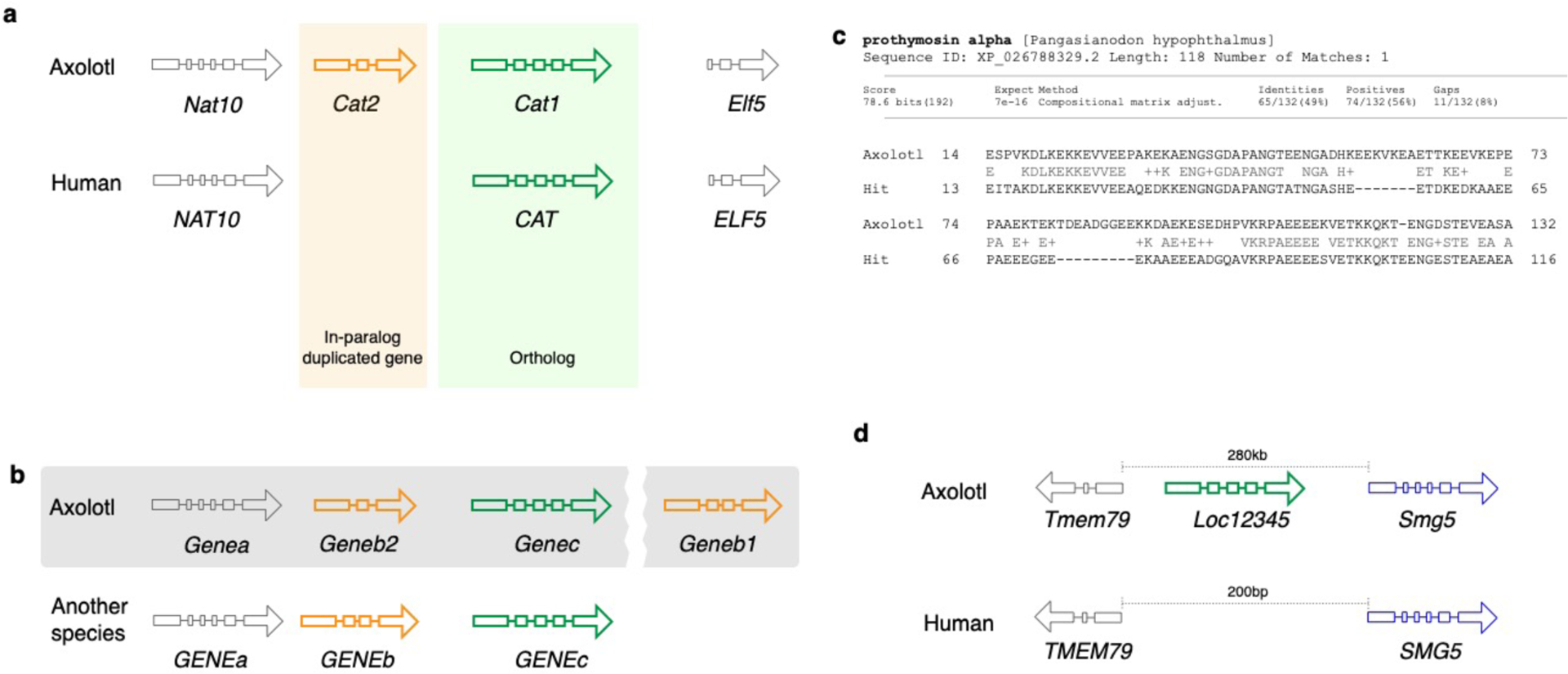
a. Two Cat loci arose by gene duplication. Comparison with the human genome reveals a single copy of CAT in the human genome. Thus, Cat1 and Cat2 are paralogs in the axolotl genome. b. Annotation of orthologs and paralogs. Geneb1 is annotated based on its homology to GENEB in another organism (indicated by the pattern of exons), while a paralogous gene is annotated as Geneb2 since it has a lower sequence similarity. Grey shaded area indicates the chromosome to highlight that Geneb1 is in a different locus c. A putative novel gene family member in axolotl that has some (BLAST e-value 1e-16) similarity to a gene in another organism. d. A putative novel gene that does not have any homologs, but has a long (1941aa) open reading frame.
In the case of paralogous genes, one must be very cautious in order to avoid name collisions across species. Imagine, in the above example, there were also a gene GENEB2 in the same species. However, this GENEB2 is not orthologous to the newly annotated Geneb2 in axolotl. In this case, we suggest using the next available number in the series, following consultation with other gene nomenclature groups. In the outlined example, it would be Geneb3. Hence, we always recommend contacting the axolotl gene nomenclature committee when naming a gene.
-
Paralogs:
Paired related homeobox 1 and Paired related homeobox 2: Prrx1 and Prrx2
-
Duplicated genes:
Catalases - Cat and Cat2
-
5
Novel genes.
While paralogous genes should be treated as described above, potential novel genes should be annotated differently. Ideally, novel genes should be characterized functionally and named based on their function, whenever possible. However, for the vast majority of the novel genes this is not the case. We therefore suggest to examine the orthologous relationships to other functionally characterized or unambiguously annotated genes in axolotl or other organisms. If a novel gene can be assigned to a certain protein family but fails to fulfill the criteria outlined in 4 to be annotated unambiguously, we suggest adding a suffix ‘-like’ to the gene symbol of the closest ortholog based solely on the sequence homology. This annotation may be changed later when its existence is confirmed in the lab and the functional data become available. Finally, if none of those approaches can be applied, the novel gene should be annotated as Locxxxx (Fig. 1d), where xxxx are the NCBI gene IDs. In this case, the gene sequence should be submitted to NCBI first to get the NCBI gene ID, which ensures that this gene symbol is not arbitrary.
Example:
Prothymosin-alpha-like : Prothymosin-alpha-like – Ptal (Fig. 1c) and Loc12345 between Tmem79 and Smg5 (Fig. 1d).-
5.1PseudogenesSome genes may lose their open reading frame (ORF) in the course of the time and, thus, also their function. However, since the gene sequences are still present in the genome, they must be accurately annotated as they may have undesired effects on transgenics, transcript quantification and other analyses that rely on the gene sequence. To stay consistent with the nomenclature guidelines in other organisms and particularly in humans24, we suggest appending the suffix ‘p’ (pseudogene) followed by a number to the gene symbol of the ancestral gene if the gene is processed, while naming unprocessed genes as new members of the family with the suffix ‘p’.
-
5.1
-
6
Transcript variants
We propose that the transcript variant names are formed after the following schema: GeneName.TranscriptNumber. All predicted transcripts of a gene are numbered by the order in which they were annotated. While in organisms with a well-established genome annotation, the nomenclature does not specifically deal with the transcript annotations, we feel that in axolotl the annotation is frequently changed at the moment and therefore it is vital to keep track of which isoforms were proven to be wrong. For example, imagine that a gene has three annotated isoforms, Gene.1, Gene.2 and Gene.3. However, it turns out that Gene.3 is just an artifact. Nevertheless, another isoform, Gene.4, is shown to be very tissue-specific. At this point, it is better to have the isoforms annotated as Gene.1, Gene.2 and Gene.4, to indicate that Gene.3 does not exist and avoid confusion in case any works referred to Gene.3 before it was excluded.
Example:
transcript variants of Prrx1
Prrx1.1, Prrx1.2
-
7
Non-coding transcripts
Similar to the guidelines for the human genome outlined in Seal et al, 202045, we propose that non-coding transcripts are annotated according to their RNA type. MicroRNAs should be annotated as “mir-XXX”, where XXX is the submission ID in the miRbase database46. Transfer RNAs should receive gene names following the pattern tRNA-XXX-YYY- GtRNAdbID, where XXX is the three-letter amino acid code, YYY is the anticodon and GtRNAdbID is the gene ID in GtRNAdb database47. Other classes of non-coding RNAs should be named after consulting the gene naming committee as very little work has been done on non-coding RNAs in axolotl so far and, thus, the exact requirements will be met later.
Similar to the guidelines in other vertebrates, we suggest to use the gene symbol with the suffix ‘-as’ for non-coding transcripts that originate from the same promoter as an annotated protein-coding gene on the opposite strand, e.g. Dio3-as for a non-coding RNA that originates from the Dio3 promoter.
-
8
Mitochondrial genes
In order to stay consistent with the well-annotated species, we propose to use the human annotation of the mitochondrial genes (NCBI Reference Sequence: NC_012920.1) in axolotl. However, in agreement with the remainder of the nomenclature, only the first letter should be capital, e.g. Mtnd2 for the mitochondrially encoded NADH dehydrogenase 2, while the human counterpart would be MTND2.
Chromosomes and aberrations
The axolotl genome is made up of ~32 Gb of DNA, which are distributed across 14 chromosomes2,3,48. Chromosomes are numbered in a descending order based on their size, which was initially determined by meiotic mapping and recombination distances that define linkage groups37,49. Thus, as also seen in the human genome, chromosome ordering by size does not exactly correlate with ordering based upon chromosome base pair length. Further, axolotl chromosomes are divided into short and long arms via centromere. Conventionally, the short arm is called the p arm, while the long arm is called the q arm. For the sake of consistency, we propose to retain this nomenclature for the axolotl chromosome arms.
-
9
Deficiencies, duplication, inversion, insertion, translocation
Chromosomal aberrations are known in every species and the axolotl is no exception. They can be mainly classified into deficiencies, duplication, inversion, insertions and translocations. Although a few chromosomal aberrations have been reported to date42, we anticipate that such annotations will arise in the near future from the analyses of the genome and transcriptome assemblies. We propose to use the following prefix for each of them which is in line with the usage of these terms by the zebrafish community29.
deficiencies, Df
duplication, Dp
inversion, In
insertion, Is
translocation, T
Further, chromosome rearrangements are indicated with the following prefixes in italics, followed by the chromosome aberration details in parentheses and in italics, which in turn is followed by the name of the line in a regular font. Df(Chr#:xxx)lineNN
Transgenic lines and constructs
Since the first successful axolotl transgenics, which were ubiquitous fluorescent reporter expression lines6, transgenesis has made significant progress. In the last decade, a number of transgenic methodologies were successfully implemented in axolotl. This includes the I-SceI mediated transgenesis, Tol2-mediated transgenesis, TALEN-mediated transgenesis and CRISPR/Cas9-mediated transgenesis, all of which allow researchers to perform random transgenesis, generate knock-outs and knock-ins in axolotl5,7–10. With these advances, it is expected that more transgenic animals will be generated in the near future and a standard transgenic nomenclature is, thus, needed to assign the identifier information in a consistent and rigorous manner.
-
10
Random insertion
I-SceI and Tol2- mediated transgenesis utilizes the flanking I-SceI restriction sites or Tol2 transposable elements to the cassette of interest. Co-injection of such construct with the I-SceI meganuclease or Tol2 mRNA/protein allows for random integration of the cassette of interest into the genome. We recommend highlighting such transgenic animals by the tg symbol followed by the method of transgenesis and name of the cassette in parentheses. We also propose separating regulatory elements (enhancer and promoters) and the coding sequence by colons. The name of the full cassette should be written in italics. If a foreign regulatory element is used for making the transgenic animal, then it should be mentioned as a one letter genus and three letter species symbol followed by a period in the nomenclature, such as from mouse - M. musculus (Mmu). If the regulatory element of the axolotl is used then the species information can be omitted. Further, we recommend appending the developer’s lab code as a superscript after the parentheses to indicate the origin of the transgenic animal. In order to obtain the lab code, developer should register their lab/organization with international laboratory code registry (ILAR) (https://www.nationalacademies.org/ilar/lab-code-database).
I-SceI mediated transgenesis
tgSceI(Mmu.Prrx1:GFPnls-T2a-ERT2-Cre-ERT2)Labcode
Tol2 mediated transgenesis
tgTol2(Mmu.Prrx1:GFPnls-T2a-Cre-ERT2)Labcode
Often, transgenic animals are made with more than one cassette. In such instances, we suggest the use of semicolon (;) between two cassettes.
tgSceI(Mmu.Prrx1:GFPnls-T2a-ERT2-Cre-ERT2;CAGGS:loxP-GFP-loxP-Cherry)Labcode
-
11
Knockout lines
Gene mutants are often generated with the aim to perform functional analysis. With the advent of CRISPR/Cas9, it has become relatively easy to generate such lines. Such germline transmitted targeted mutations (tm) should be characterized for insertion or deletion and they should be annotated as I or D, respectively. In addition, the line name should also contain the position of the indel with respect to the start of the gene, the version of the genome and the number of nucleotides that are inserted or deleted. If the generated animals are heterozygous then the wildtype (+) should be mentioned as a second allele.
Example:
tm(Prrx1153v6D8/153v6D8)Labcode, refers to a homozygous deletion of 8 nucleotides at the nucleotide position 153 with respect to the start of Prrx1 in the genome version 6.
tm(Prrx1153v6I5/+)Labcode, refers to a heterozygous insertion of 5 nucleotides at the nucleotide position 153 from the beginning of Prrx1 in the genome version 6.
-
12
Knock-in lines
Similarly, the names of a knock-in animal should contain the name of the gene locus where the transgene is inserted. At the moment only non-homologous end-joining (NHEJ) mediated knock-in possible8 and such transgene knock-ins are generated by targeted mutation (tm) at either the N-terminus or the C-terminus of the endogenous ORF. This, in turn, may either retain or disrupt the native ORF. Similar to the cassette in the random transgenesis, regulatory elements and the coding DNA sequence should be separated by a colon and italicized. When a transgene is inserted at the C-terminus as a tag without disrupting the native coding sequence then it should be written as follows.
tm(Prrx1t/+:Prrx1-T2a-Cherry)Labcode, refers to a heterozygous knock-in at the Prrx1 locus, which retains the native Prrx1 gene structure and allows for tagging (t) at its C-terminus resulting in a fusion of Prrx1 with T2a-Cherry. Similarly, a homozygous knock-in should be labeled as tm(Prrx1t/t:Prrx1-T2a-Cherry)Labcode.
However, the N-terminal insertion of a transgene via the NHEJ disrupts the native gene sequence. Hence, the N-terminus knock-ins are generated in one of the following two ways.
When the native coding sequence is disrupted leading to a heterozygous genotype, tm(Prrx1r/+:Cherry)Labcode, refers to the heterozygous N-terminus knock-in at the Prrx1 locus, which replaces (r) the native gene with Cherry. In this situation, the native gene is not active and hence, these animals are heterozygous knock-outs for Prrx1.
When the native coding sequence is disrupted, but replaced with the cDNA of the native gene, which is also refered to as a mini-gene,
tm(Prrx1r/+:miniPrrx1-T2a-Cherry)Labcode, refers to a heterozygous N-terminus knock-in at the Prrx1 locus, which replaces (r) the native gene with a mini-gene version of Prrx1(cDNA), which is fused to T2a-Cherry. In this situation, since the native gene is replaced by the Prrx1 cDNA, the animals are not considered as knock-outs for Prrx1.
Finally, we want to once again point out that the axolotl gene nomenclature committee should be contacted every time when naming a gene. We envisage that such systemic nomenclature would remove confusion among researchers and serve the entire community.
Acknowledgment:
This system of nomenclature was discussed among salamander community and we thank all our colleagues for their inputs on developing these guidelines. We specially thank Anna Vlasova, Tamsin Jones and Elspeth Bruford for their helpful suggestions regarding homologs and gene naming. Work in the JFF laboratory is supported by grants from National Key R&D Program of China 2019YFE0106700, the Natural Science Foundation of China 31970782, Key-Area Research and Development Program of Guangdong Province 2018B030332001, 2019B030335001. Work in the SRV laboratory is supported by grants from the National Institutes of Health (P40 OD019794, R24OD010435). Work in the EMT laboratory is supported by grants from ERC (AdG 742046) and FWF (Standalone I4353). Work in the PM laboratory is supported by grants from National Institutes of Health - COBRE (5P20GM104318-08) and DFG (429469366).
References:
- 1.Bryant DM, Johnson K, DiTommaso T, et al. A Tissue-Mapped Axolotl De Novo Transcriptome Enables Identification of Limb Regeneration Factors. Cell Rep. Jan 17 2017;18(3):762–776. 10.1016/j.celrep.2016.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nowoshilow S, Schloissnig S, Fei JF, et al. The axolotl genome and the evolution of key tissue formation regulators. Nature. Feb 1 2018;554(7690):50–55. 10.1038/nature25458. [DOI] [PubMed] [Google Scholar]
- 3.Smith JJ, Timoshevskaya N, Timoshevskiy VA, Keinath MC, Hardy D, Voss SR. A chromosome-scale assembly of the axolotl genome. Genome Res. Feb 2019;29(2):317–324. 10.1101/gr.241901.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schloissnig S, Kawaguchi A, Nowoshilow S, et al. The giant axolotl genome uncovers the evolution, scaling, and transcriptional control of complex gene loci. Proc Natl Acad Sci U S A. Apr 13 2021;118(15). 10.1073/pnas.2017176118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khattak S, Murawala P, Andreas H, et al. Optimized axolotl (Ambystoma mexicanum) husbandry, breeding, metamorphosis, transgenesis and tamoxifen-mediated recombination. Nat Protoc. Mar 2014;9(3):529–40. 10.1038/nprot.2014.040. [DOI] [PubMed] [Google Scholar]
- 6.Sobkow L, Epperlein HH, Herklotz S, Straube WL, Tanaka EM. A germline GFP transgenic axolotl and its use to track cell fate: dual origin of the fin mesenchyme during development and the fate of blood cells during regeneration. Dev Biol. Feb 15 2006;290(2):386–97. 10.1016/j.ydbio.2005.11.037. [DOI] [PubMed] [Google Scholar]
- 7.Woodcock MR, Vaughn-Wolfe J, Elias A, et al. Identification of Mutant Genes and Introgressed Tiger Salamander DNA in the Laboratory Axolotl, Ambystoma mexicanum. Sci Rep. Jan 31 2017;7(1):6. 10.1038/s41598-017-00059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fei JF, Lou WP, Knapp D, et al. Application and optimization of CRISPR-Cas9-mediated genome engineering in axolotl (Ambystoma mexicanum). Nat Protoc. Dec 2018;13(12):2908–2943. 10.1038/s41596-018-0071-0. [DOI] [PubMed] [Google Scholar]
- 9.Fei JF, Schuez M, Tazaki A, Taniguchi Y, Roensch K, Tanaka EM. CRISPR-mediated genomic deletion of Sox2 in the axolotl shows a requirement in spinal cord neural stem cell amplification during tail regeneration. Stem Cell Reports. Sep 9 2014;3(3):444–59. 10.1016/j.stemcr.2014.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flowers GP, Timberlake AT, McLean KC, Monaghan JR, Crews CM. Highly efficient targeted mutagenesis in axolotl using Cas9 RNA-guided nuclease. Development. May 2014;141(10):2165–71. 10.1242/dev.105072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fei JF, Schuez M, Knapp D, Taniguchi Y, Drechsel DN, Tanaka EM. Efficient gene knockin in axolotl and its use to test the role of satellite cells in limb regeneration. Proc Natl Acad Sci U S A. Nov 21 2017;114(47):12501–12506. 10.1073/pnas.1706855114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerber T, Murawala P, Knapp D, et al. Single-cell analysis uncovers convergence of cell identities during axolotl limb regeneration. Science. Oct 26 2018;362(6413). 10.1126/science.aaq0681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khattak S, Sandoval-Guzman T, Stanke N, Protze S, Tanaka EM, Lindemann D. Foamy virus for efficient gene transfer in regeneration studies. BMC Dev Biol. May 3 2013;13:17. 10.1186/1471-213X-13-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oliveira CR, Lemaitre R, Murawala P, Tazaki A, Drechsel DN, Tanaka EM. Pseudotyped baculovirus is an effective gene expression tool for studying molecular function during axolotl limb regeneration. Dev Biol. Jan 15 2018;433(2):262–275. 10.1016/j.ydbio.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 15.Roy S, Gardiner DM, Bryant SV. Vaccinia as a tool for functional analysis in regenerating limbs: ectopic expression of Shh. Dev Biol. Feb 15 2000;218(2):199–205. 10.1006/dbio.1999.9556. [DOI] [PubMed] [Google Scholar]
- 16.Whited JL, Tsai SL, Beier KT, et al. Pseudotyped retroviruses for infecting axolotl in vivo and in vitro. Development. Mar 2013;140(5):1137–46. 10.1242/dev.087734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masselink W, Reumann D, Murawala P, et al. Broad applicability of a streamlined ethyl cinnamate-based clearing procedure. Development. Feb 1 2019;146(3). 10.1242/dev.166884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pende M, Vadiwala K, Schmidbaur H, et al. A versatile depigmentation, clearing, and labeling method for exploring nervous system diversity. Sci Adv. May 2020;6(22):eaba0365. 10.1126/sciadv.aba0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duerr TJ, Comellas E, Jeon EK, et al. 3D visualization of macromolecule synthesis. Elife. Oct 14 2020;9. 10.7554/eLife.60354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adrados CS, Yu Q, Bolanos Castro LA, Rodriguez Cabrera LA, Yun MH. Salamander-Eci: An optical clearing protocol for the three-dimensional exploration of regeneration. Dev Dyn. Oct 20 2020. 10.1002/dvdy.264. [DOI] [PubMed] [Google Scholar]
- 21.Davisson MT. Rules and guidelines for nomenclature of mouse genes. International Committee on Standardized Genetic Nomenclature for Mice. Gene. Sep 30 1994;147(2):157–60. 10.1016/0378-1119(94)90060-4. [DOI] [PubMed] [Google Scholar]
- 22.Dunn LCG, Snell H, G. D.. Report of the committee on mouse genetics nomenclature. Journal of Heredity. December 1940 1940;31(12):505–506. 10.1093/oxfordjournals.jhered.a104827. [DOI] [Google Scholar]
- 23.Wood R Trends in Genetics: Genetic nomenclature guide with information on websites.: Cambridge, UK : Elsevier Trends Journals; 1998. [Google Scholar]
- 24.Bruford EA, Braschi B, Denny P, Jones TEM, Seal RL, Tweedie S. Guidelines for human gene nomenclature. Nat Genet. Aug 2020;52(8):754–758. 10.1038/s41588-020-0669-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shows TB, Alper CA, Bootsma D, et al. International system for human gene nomenclature (1979) ISGN (1979). Cytogenet Cell Genet. 1979;25(1–4):96–116. 10.1159/000131404. [DOI] [PubMed] [Google Scholar]
- 26.Xenopus_Gene_Nomenclature_Committee. Gene Nomenclature Guidelines https://wwwxenbaseorg/gene/static/geneNomenclaturejsp 2013.
- 27.Karimi K, Fortriede JD, Lotay VS, et al. Xenbase: a genomic, epigenomic and transcriptomic model organism database. Nucleic Acids Res. Jan 4 2018;46(D1):D861–D868. 10.1093/nar/gkx936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruzicka L, Howe DG, Ramachandran S, et al. The Zebrafish Information Network: new support for non-coding genes, richer Gene Ontology annotations and the Alliance of Genome Resources. Nucleic Acids Res. Jan 8 2019;47(D1):D867–D873. 10.1093/nar/gky1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zebrafish_Nomenclature_Committee. ZFIN Zebrafish Nomenclature Conventions https://wikizfinorg/display/general/ZFIN+Zebrafish+Nomenclature+Conventions 2019.
- 30.Horvitz HR, Brenner S, Hodgkin J, Herman RK. A uniform genetic nomenclature for the nematode Caenorhabditis elegans. Mol Gen Genet. Sep 1979;175(2):129–33. 10.1007/BF00425528. [DOI] [PubMed] [Google Scholar]
- 31.Burt DW, Carre W, Fell M, et al. The Chicken Gene Nomenclature Committee report. BMC Genomics. Jul 14 2009;10 Suppl 2:S5. 10.1186/1471-2164-10-S2-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kusumi K, Kulathinal RJ, Abzhanov A, et al. Developing a community-based genetic nomenclature for anole lizards. BMC Genomics. Nov 11 2011;12:554. 10.1186/1471-2164-12-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stolfi A, Sasakura Y, Chalopin D, et al. Guidelines for the nomenclature of genetic elements in tunicate genomes. Genesis. Jan 2015;53(1):1–14. 10.1002/dvg.22822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reddien PW, Newmark PA, Sanchez Alvarado A. Gene nomenclature guidelines for the planarian Schmidtea mediterranea. Dev Dyn. Nov 2008;237(11):3099–101. 10.1002/dvdy.21623. [DOI] [PubMed] [Google Scholar]
- 35.Smith JJ, Voss SR. Gene order data from a model amphibian (Ambystoma): new perspectives on vertebrate genome structure and evolution. BMC Genomics. Aug 29 2006;7:219. 10.1186/1471-2164-7-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voss SR, Kump DK, Putta S, et al. Origin of amphibian and avian chromosomes by fission, fusion, and retention of ancestral chromosomes. Genome Res. Aug 2011;21(8):1306–12. 10.1101/gr.116491.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Voss SR, Smith JJ, Gardiner DM, Parichy DM. Conserved vertebrate chromosome segments in the large salamander genome. Genetics. Jun 2001;158(2):735–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilkinson MD, Dumontier M, Aalbersberg IJ, et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data. Mar 15 2016;3:160018. 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Joven A, Elewa A, Simon A. Model systems for regeneration: salamanders. Development. Jul 22 2019;146(14). 10.1242/dev.167700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.International_Committee_on_Standardized_Genetic_Nomenclature_for_Mice. Guidelines for Nomenclature of Genes, Genetic Markers, Alleles, and Mutations in Mouse and Rat http://wwwinformaticsjaxorg/mgihome/nomen/geneshtml 2018.
- 41.HUGO_Gene_Nomenclature_Committee. HGNC Guidelines [WWW Document]. HGNC. https://wwwgenenamesorg/about/guidelines/. 2020. [Google Scholar]
- 42.Keinath MC, Timoshevskaya N, Timoshevskiy VA, Voss SR, Smith JJ. Miniscule differences between sex chromosomes in the giant genome of a salamander. Sci Rep. Dec 14 2018;8(1):17882. 10.1038/s41598-018-36209-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koonin EV. An apology for orthologs - or brave new memes. Genome Biol. 2001;2(4):COMMENT1005. 10.1186/gb-2001-2-4-comment1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wright MW, Bruford EA. Human and orthologous gene nomenclature. Gene. Mar 15 2006;369:1–6. 10.1016/j.gene.2005.10.029. [DOI] [PubMed] [Google Scholar]
- 45.Seal RL, Chen LL, Griffiths-Jones S, et al. A guide to naming human non-coding RNA genes. EMBO J. Mar 16 2020;39(6):e103777. 10.15252/embj.2019103777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ambros V, Bartel B, Bartel DP, et al. A uniform system for microRNA annotation. RNA. Mar 2003;9(3):277–9. 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chan PP, Lowe TM. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. Jan 4 2016;44(D1):D184–9. 10.1093/nar/gkv1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Callan HG. Chromosomes and nucleoli of the axolotl, Ambystoma mexicanum. J Cell Sci. Mar 1966;1(1):85–108. [DOI] [PubMed] [Google Scholar]
- 49.Smith JJ, Kump DK, Walker JA, Parichy DM, Voss SR. A comprehensive expressed sequence tag linkage map for tiger salamander and Mexican axolotl: enabling gene mapping and comparative genomics in Ambystoma. Genetics. Nov 2005;171(3):1161–71. 10.1534/genetics.105.046433. [DOI] [PMC free article] [PubMed] [Google Scholar]


