Abstract
Alleles of the vacuolating cytotoxin gene (vacA) of Helicobacter pylori vary between strains, particularly in the region encoding the signal sequence (which may be type s1 or s2) and the midregion (which may be type m1 or m2). Using a PCR-based typing system developed in the United States, we showed that 36 strains from Asia and South America were all vacA signal sequence type s1; 3 were midregion type m1 and 11 were m2, but 22 could not be typed for the vacA midregion. All strains possessed cagA (cytotoxin-associated gene A), another virulence marker. vacA nucleotide sequence analysis showed that midregion typing failure was due to base substitutions at the primer annealing sites. Using the new sequence data, we developed two new PCR-based vacA midregion typing systems, both of which correctly typed 41 U.S. strains previously typed by the old system and successfully typed all 36 of the non-U.S. strains. All previously untypeable strains were vacA m1, other than one m1/m2 hybrid. In summary, we describe and validate a simple PCR-based system for typing vacuolating cytotoxin (vacA) alleles of H. pylori and show that this system correctly identifies the signal and midregion types of vacA in 77 strains from Asia and North and South America.
The two best-established methods for grading Helicobacter pylori strain virulence are typing of the allele of the vacuolating cytotoxin gene, vacA, and determination of the presence or absence of the cytotoxin-associated gene, cagA (a marker for the cag pathogenicity island) (4). A copy of vacA is present in essentially all strains of H. pylori, but its nucleotide sequence varies between strains. This variation is most marked in the region encoding the signal sequence, which may be type s1 (with subtypes a, b, or c) or type s2, and the midregion, which may be type m1 or type m2 (1, 16). vacA alleles with all possible combinations of signal and midregion types have been found (1, 9). Among strains from the United States, the vacA midregion type of a strain correlates with its ability to induce vacuolation in HeLa cells in vitro and with gastric epithelial damage in vivo, but not with gastric inflammatory cell infiltrate in vivo or with peptic ulceration (2). However, in Taiwan, an association between the m1 type of vacA (the more cytotoxic type) and peptic ulceration has been described (17). In contrast, in the United States, the vacA signal region type correlates with the level of gastric inflammatory cell infiltration (2) and in many but not all studies from the United States and Europe, it also correlates with peptic ulceration (2, 5, 12, 14, 15). In the Far East, this association is not found, as essentially all strains described are vacA signal type s1a or s1c (8, 10, 11, 16, 17). This work on the association of vacA poymorphisms with disease has been hampered by the finding of several groups, especially in the Far East, that the originally described primers used for PCR typing of the vacA midregion do not satisfactorily categorize all strains (8, 10, 11, 12, 14, 17). We aimed to assess the extent of this problem, define the reasons for typing failure, and develop a modified system which would allow simple and accurate vacA typing of all strains.
MATERIALS AND METHODS
H. pylori isolates.
In the first part of the study, we used 36 previously obtained isolates from subjects outside the United States: 8 from dyspeptic patients without ulcers from Lima, Peru; 13 from dyspeptic patients (6 with duodenal ulceration, 5 with gastric ulceration, 1 with both types of ulceration, and 1 with no ulcers) from Yamagucchi, Japan; 9 from dyspeptic patients from Bangkok, Thailand; and 6 from asymptomatic patients from Shandong Province, China. In the standardization of our new PCR-based vacA-typing strategies, we used 41 isolates from patients from Nashville, Tenn., who underwent upper gastrointestinal endoscopy for a variety of indications. We have previously reported on the characterization of the cagA and vacA genotypes of most of these U.S. isolates with our previous systems (2).
vacA and cagA typing using original methodology.
Stock cultures of H. pylori stored at −70°C were regrown under microaerobic conditions (Campypak plus; Becton Dickinson, Cockeysville, Md.), and chromosomal DNA was extracted by lysis with GES (guanadinium thiocyanate, EDTA, Sarkosyl), chloroform extraction, and isopropanol precipitation as described previously (1, 3). The vacA signal and midregion were typed by both PCR and high-stringency colony hybridization with previously described primers and probes (those for the vacA midregion are shown schematically in Fig. 1). vacA signal sequence typing was by PCR with primers VA1-F and VA1-R to categorize this region as type s1 or s2 and the reverse primer VA1-R together with allelic subtype-specific forward primers to subclassify type s1 alleles (the primers used were primer SS1-F for subtype s1a or s1c, whose predicted peptide signal sequences differ by only two amino acids [15], and primer SS3-F for subtype s1b [1, 3]). vacA midregion typing was performed by two separate PCRs with m1-specific primers (primers VA3-F and VA3-R) in the first PCR and m2-specific primers (primers VA4-F and VA4-R) in the second PCR (1, 3) (Fig. 1). The presence of cagA was determined by colony hybridization with a 2,334-bp probe, as described previously (1, 3).
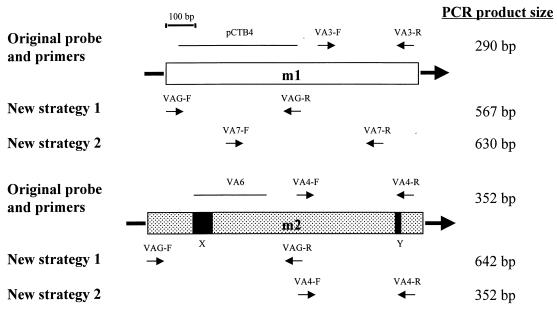
FIG. 1.
Schematic of vacA midregions for the m1 and m2 allelic types, showing the original midregion allelic type-specific typing system (1) and the new PCR strategies. The diagram is drawn to scale except for the sizes of the arrows representing the PCR primers. The boxed area is 910 bp for strain 60190 (coordinates 1395 to 2304 numbered from the adenine of the start codon [8]) and 1,000 bp for strain Tx30a (coordinates 1371 to 2370 [1]). X represents an insertion of 75 bp, and Y represents an insertion of 15 bp; both are present in m2 alleles but not in m1 alleles. The arrows labeled VA3-F and VA3-R and those labeled VA4-F and VA4-R are the allelic type-specific primers used in our original PCR-based system (1). The solid lines labeled pCTB4 and VA6 are the allelic type-specific probes used for hybridization. VAG-F and VAG-R are the primers used in new strategy 1, and they anneal to regions of vacA conserved between m1 and m2 alleles; the types are then differentiated on the basis of PCR product size. Primers VA7-F and VA7-R and primers VA4-F and VA4-R are allelic type-specific primers used together in new strategy 2. Product size depends on which specific primers anneal, and the expected annealing primers are shown below the m1 and m2 regions.
Nucleotide sequencing of vacA regions of selected strains for which PCR typing was unsuccessful.
For selected strains from which vacA midregions could not be typed by our original PCR-based methods, 1.7-kb vacA fragments (coordinates 726 to 2445 bp in the published vacA sequence of strain Tx30a [1]) were amplified by PCR with primers VA10-F (5′-CGCTGAAATCTCTCTTTATG) and VA10-R (5′-CATGCTTTGATTGCCGATAGC). PCR products were subcloned into pT7Blue (Novagen, Madison, Wis.), and then both strands were sequenced with a minimum of twofold redundancy with vector primers and then by primer walking. For selected strains whose vacA signal sequence regions could not be subtyped, we determined the nucleotide sequence of this region by sequencing directly PCR amplicons from primers A1043 (5′-ATTTTACCTTTTTACACATTCTAGCC) and B771 (5′-AGAAGCCCTGAGACCG). Nucleotide sequencing was performed with an automated ABI sequencer.
PCR-based vacA genotyping using new strategies.
Two new strategies were used in this study to classify the midregion types of the vacA alleles. New strategy 1 used primers VAG-F (5′-CAATCTGTCCAATCAAGCGAG) and VAG-R (5′-GCGTCAAAATAATTCCAAGG) (Fig. 1). The PCR protocol was similar to that described previously (1, 3), except that the reaction mixtures were heated initially to 95°C for 90s and then underwent 35 cycles of 30 s at 95°C, 60 s at 56°C, and 90 s at 72°C. After cycling, the products were extended for a further 5 min at 72°C. New strategy 2 used four primers together in one reaction tube: VA7-F (5′-GTAATGGTGGTTTCAACACC) and VA7-R (5′-TAATGAGATCTTGAGCGCT) and the previously described primers VA4-F and VA4-R (1) (Fig. 1). The thermocycling conditions were as described for new strategy 1.
RESULTS
vacA and cagA typing using original methodology.
The vacA signal regions of all 36 non-U.S. isolates were categorized as type s1 by PCR with conserved primers VA1-F and VA1-R (1, 2). Further PCR subtyping with allelic subtype-specific primers was successful for all strains other than two from Peru (Table 1). vacA midregion typing with (separately) m1-specific primers (VA3-F and VA3-R) and m2-specific primers (VA4-F and VA4-R) (Fig. 1) had previously successfully amplified vacA fragments from all 70 U.S. strains tested (1, 2). However, for the strains in the current study, the typing method failed to amplify DNA from 21 of 28 Asian strains, including all 13 Japanese strains tested, and DNA from 1 of 8 Peruvian strains (Table 1). The strains from which DNA was not amplified hybridized weakly with the m1-specific probe (pCTB4) but did not hybridize detectably with the m2-specific probe (VA6) (Fig. 1). All 28 Asian and 6 of 7 Peruvian strains were cagA positive. Thus, vacA alleles from most Asian strains were sufficiently different from those of U.S. strains to prevent effective midregion typing, and alleles from two Peruvian vacA s1 strains were sufficiently different to prevent vacA signal region subtyping.
TABLE 1.
cagA status and vacA genotypes of strains used in this study as assessed by PCR typing
| Country of origin | No. of strains tested | No. of strains
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
cagA status
|
vacA signal sequence type
|
vacA midregion type
|
|||||||||||
| Original method
|
New method
|
||||||||||||
| + Positive | − Negative | s1a or s1ca | s1b | s1, not s1a or -c, or s1b | s2 | m1 | m2 | Neither | m1 | m2 | Neither | ||
| United Statesb | 41 | 31 | 10 | 18 | 13 | 0 | 10 | 15 | 26 | 0 | 15 | 26 | 0 |
| Peru | 8 | 7 | 1c | 5d | 1e | 2f | 0 | 3 | 4 | 1 | 4 | 4 | 0 |
| Japan | 13 | 13 | 0 | 13 | 0 | 0 | 0 | 0 | 0 | 13 | 13 | 0 | 0 |
| Thailand | 9 | 9 | 0 | 9 | 0 | 0 | 0 | 0 | 6 | 3 | 3 | 6 | 0 |
| China | 6 | 6 | 0 | 6 | 0 | 0 | 0 | 0 | 1 | 5 | 4 | 1 | 1g |
| Total | 77 | 66 | 11 | 51 | 14 | 2 | 10 | 18 | 37 | 22 | 39 | 37 | 1 |
The vacA s1 subtyping system that we used was not designed to distinguish between types s1a and s1c.
These U.S. strains were described previously (1) but are included here because they were retyped with our new primers.
This cagA-negative strain was vacA s1 (not s1a or -c, or s1b) and m2.
Nucleotide sequence analysis of two of these alleles showed that they were intermediate between s1a and s1b.
This vacA s1b strain was cagA positive and vacA m1.
One of these strains was cagA positive and vacA m1, and the other was cagA negative and vacA m2. The vacA alleles of these strains were intermediate between s1a and s1b but were more similar to s1b.
The vacA allele of this strain was an m1/m2 hybrid.
Determination by nucleotide sequencing of reasons for vacA midregion typing failure.
To define why PCR-based typing of the vacA midregion was unsuccessful for some strains, we sequenced a 1.7-kb region of vacA containing the primer and probe annealing sites from five of the strains that had not been successfully typed: 90-40 (Peru), 88-29 (Thailand), 88-32 (Thailand), HPK3 (Japan), and Ch2 (China). At the m1 forward primer (VA3-F) annealing site, from 0 to 3 of 19 bases were noncomplementary, whereas at the reverse primer (VA3-R) annealing site, 6 to 7 of 20 bases were noncomplementary. In the 439-bp annealing region for the m1 probe (pCTB4), 9% or fewer of the bases were noncomplementary. Comparative analysis with previously obtained nucleotide sequences from other type m1 vacA alleles showed that these newly sequenced alleles did not form a specific subgrouping within the m1 family, in that they were not related more closely to each other than to other type m1 alleles (7, 13). The annealing region for the 308-bp m2 probe (VA6) included a 75-bp insertion not found in type m1 vacA alleles (Fig. 1). Of the remaining 234 bp to which the m2 probe could potentially anneal, more than 25% of bases were noncomplementary in all cases. These findings explain the annealing of the m1 probe and the absence of annealing of the m2 probe under high-stringency conditions. The final strain, strain Ch2, had a vacA midregion with an m1-like sequence at the forward primer annealing site and an m2-like sequence at the reverse primer annealing site and appeared to be a naturally occurring m1/m2 hybrid. Thus, overall, poor annealing of primers or probes due to sequence divergence between vacA alleles explained PCR and hybridization failures for all but one strain (vacA m1/m2 hybrid strain Ch2).
Design and testing of two new PCR strategies for vacA midregion typing.
Next, so that the vacA alleles of all strains could be typed successfully, we sought to develop new PCR strategies to distinguish between m1 and m2 alleles. We used these strategies to type the 36 non-U.S. strains described above and also (to ensure that our new strategies were valid) 41 strains from the United States previously successfully typed by our previous strategy. For new strategy 1, taking advantage of the conserved m2-specific 75-bp insertion, we designed primers VAG-F and VAG-R to span this region and to anneal to sites conserved among all presently known m1 and m2 vacA alleles (Fig. 1). This permitted sorting of PCR products by size (predicted size of 567 bp for type m1, coordinates 1278 to 1844 from beginning of the open reading frame in strain 60190 [6], and predicted size of 642 bp for type m2, coordinates 1254 to 1895 in strain Tx30a [1]). By use of these primers, PCR amplification of vacA alleles gave, for every strain, a product of one of the two predicted sizes, and for all 58 strains for which previous PCR typing had been successful, the result was the same. All 22 previously untypeable vacA alleles were now typed as m1 with the new primers, including the m1/m2 allele from strain Ch2 (Fig. 2B).
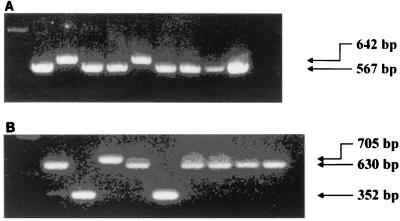
FIG. 2.
One percent agarose gels with ethidium bromide showing examples of PCR-based vacA midregion typing by new strategy 1 (A) and new strategy 2 (B). Details about the PCR primers and expected product sizes are given in the text and in Fig. 1. The PCR products shown were amplified from the following nine strains (in the lanes from left to right), respectively: J238 (from the United States, m1 control), J226 (from the United States, m2 control), Ch2 (from China, m1/m2 hybrid), Ch3 (from China, m1), Ch4 (from China, m2), Ch5 (from China, m1), Ch7 (from China, m1), Ch8 (from China, m1), and 92-24 (from the United States m1 control).
To confirm our results by a different method, we designed a second new PCR-based typing strategy using a new primer pair, VA7-F and VA7-R, to amplify the vacA midregion from type m1 alleles. We then performed a multiplex PCR with these new primers and the original m2 primers (VA4-F and VAF-R) in the same reaction mixture (Fig. 1) using the same PCR conditions that we used in our first strategy. The predicted product sizes were 630 bp for m1 alleles (coordinates 1535 to 2164 in strain 60190 [6], 352 bp for m2 alleles (coordinates 1939 to 2290 in strain Tx30a [1]), 705 bp for m1/m2 alleles (such as Ch2), and 277 bp for m2/m1 alleles (if they existed). When applied to the same set of 77 strains, this second strategy successfully typed vacA alleles from all strains, with results identical to those from the first strategy, with the single exception of those for strain Ch2, which, as predicted, gave a product of 705 bp (Fig. 2B). No other strains gave a product of this size, and no strains gave a product of the size predicted for an m2/m1 allele. In summary, we developed two new PCR-based vacA typing systems which successfully characterized as either m1, m2, or (in one case) m1/m2 all the strains from the United States, Peru, Thailand, China, and Japan tested.
Determination by nucleotide sequence analysis of the reasons for vacA s1 signal sequence subtyping failure in two Peruvian strains.
To deduce why vacA s1 alleles from two Peruvian strains (strains 90-16 and 90-22) could not be subtyped as s1a or s1b, we determined the nucleotide sequences of their vacA signal regions. The vacA alleles from these strains were more similar to s1b alleles than to s1a alleles (data not shown). Because of the difficulty subtyping the s1 vacA alleles of these two Peruvian strains, we similarly obtained the nucleotide sequences of two Peruvian strains classified as s1a by the PCR-based system and the single strain classified as s1b. Nucleotide sequence analysis of the two s1a strains (strains 90-14 and 90-40) revealed that although they had s1a-like sequences at the primer annealing site, 5′ to this their sequences were closer to s1b sequences (data not shown). Nucleotide analysis of the Peruvian s1b strain showed a typical s1b allele. Thus, in summary, all 57 U.S. and Asian strains could be successfully typed as s1a, s1b, or s2. Peruvian strains were successfully typed as s1, but since they were often intermediate between s1a and s1b, this subtyping system was not appropriate for these strains.
DISCUSSION
With the modifications to the vacA midregion typing system described in this paper, the vacA alleles of all strains that we have examined can be typed by just two PCRs: one for typing of the midregion as m1 or m2 and one for typing of the signal sequence as s1 or s2. For midregion typing, we recommend new strategy 1 (Fig. 1 and 2A) as the most direct strategy. For signal sequence typing, we recommend the use of primers VA1-F and VA1-R (1, 3). vacA s1 alleles can be further subtyped by PCR, although this will need validation in individual countries and may not be suitable for all locales (as was found for Peru in this study). Other groups have also improved PCR typing of vacA alleles on the basis of studies with European (12, 14) and international (15, 16) strain collections. The latter strategy uses a PCR approach similar to the one that we have described but, additionally, uses reverse hybridization with oligonucleotide probes immobilized on a nitrocellulose strip. While this confirms specificity, it is unlikely to be convenient for most researchers, unless premade nitrocellulose strips with immobilized oligonucleotides are inexpensively available commercially. The improved typing system described in our present study is easy to use and should facilitate studies to assess which groups of vacA alleles are prevalent in H. pylori strains colonizing different populations around the world. Studies can then be designed for appropriate populations to assess whether H. pylori strains with specific vacA alleles are differently associated with particular diseases.
ACKNOWLEDGMENTS
This work was supported by grants AI 39657 and DK 5707, by a center grant from the National Cancer Institute (CA 68485), and by the Medical Research Service of the U.S. Department of Veterans Affairs. John Atherton is funded by a Clinician Scientist Fellowship from the Medical Research Council (United Kingdom).
REFERENCES
- 1.Atherton J C, Cao P, Peek R M, Tummuru M K R, Blaser M J, Cover T L. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270:17771–17777. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- 2.Atherton J C, Peek R M, Tham K T, Cover T L, Blaser M J. Clinical and pathological importance of heterogeneity in vacA, the vacuolating cytotoxin gene of Helicobacter pylori. Gastroenterology. 1997;112:92–99. doi: 10.1016/s0016-5085(97)70223-3. [DOI] [PubMed] [Google Scholar]
- 3.Atherton J C. Molecular techniques to detect pathogenic strains of Helicobacter pylori. In: Clayton C L, Mobley H L T, editors. Methods in molecular medicine, Helicobacter pylori protocols. Totowa, N.J: Humana Press; 1997. pp. 133–143. [Google Scholar]
- 4.Atherton J C. H. pylori virulence factors. Br Med Bull. 1998;54:105–120. doi: 10.1093/oxfordjournals.bmb.a011662. [DOI] [PubMed] [Google Scholar]
- 5.Basso D, Navaglia F, Brigayo L, Piva M G, Toma A, Greco E, Di Mario F, Galeotti F, Roveroni G, Corsini A, Plebani M. Analysis of Helicobacter pylori vacA and cagA genotypes and serum antibody profile in benign and malignant gastroduodenal diseases. Gut. 1998;43:182–186. doi: 10.1136/gut.43.2.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cover T L, Tummuru M K R, Cao P, Tompson S A, Blaser M J. Divergence of genetic sequences for the vacuolating cytotoxin among Helicobacter pylori strains. J Biol Chem. 1994;269:10566–10573. [PubMed] [Google Scholar]
- 7.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 8.Ito Y, Azuma T, Ito S, Miyaji H, Hirai M, Yamazaki Y, Sato F, Kato T, Kohli Y, Kuriyama M. Analysis and typing of the vacA gene from cagA-positive strains of Helicobacter pylori isolated in Japan. J Clin Microbiol. 1997;35:1710–1714. doi: 10.1128/jcm.35.7.1710-1714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Letley D P, Lastovica A, Louw J A, Hawkey C J, Atherton J C. Allelic diversity of the Helicobacter pylori vacuolating cytotoxin gene in South Africa: rarity of the vacA s1a genotype and natural occurrence of an s2/m1 allele. J Clin Microbiol. 1999;37:1203–1205. doi: 10.1128/jcm.37.4.1203-1205.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maeda S, Ogura K, Yoshida H, Kanai F, Ikenoue T, Kato N, Shiratori Y, Omata M. Major virulence factors, VacA and CagA, are commonly positive in Helicobacter pylori isolates in Japan. Gut. 1998;42:338–343. doi: 10.1136/gut.42.3.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan Z-H, Berg D E, van der Hulst R W M, Su W-W, Raudonikiene A, Xiao S-D, Dankert J, Tytgat G N J, van der Ende A. Prevalence of vacuolating cytotoxin production and distribution of distinct vacA alleles in Helicobacter pylori from China. J Infect Dis. 1998;178:220–226. doi: 10.1086/515601. [DOI] [PubMed] [Google Scholar]
- 12.Rudi J, Kolb C, Maiwald M, Kuck D, Sieg A, Galle P R, Stremmel W. Diversity of Helicobacter pylori vacA and cagA genes and relationship to VacA and CagA protein expression, cytotoxin production, and associated diseases. J Clin Microbiol. 1998;36:944–948. doi: 10.1128/jcm.36.4.944-948.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saitou N, Nei M. The neighbour-joining method; a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 14.Strobel S, Bereswill S, Ballig P, Allgaier P, Sonntag H-G, Kist M. Identification and analysis of a new vacA genotype variant of Helicobacter pylori in different patient groups in Germany. J Clin Microbiol. 1998;36:1285–1289. doi: 10.1128/jcm.36.5.1285-1289.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Doorn L J, Figueiredo C, Rossau R, Jannes G, van Asbroeck M, Sousa J C, Carneiro F, Quint W G V. Typing of Helicobacter pylori vacA gene and detection of cagA gene by PCR and reverse hybridization. J Clin Microbiol. 1998;36:1271–1276. doi: 10.1128/jcm.36.5.1271-1276.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Doorn L-J, Figueiredo C, Sanna R, Pena S, Midolo P, Ng E K W, Atherton J C, Blaser M J, Quint W G V. Expanding allelic diversity of Helicobacter pylori vacA. J Clin Microbiol. 1998;36:2597–2603. doi: 10.1128/jcm.36.9.2597-2603.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H-J, Kuo C-H, Yeh A A M, Chang P C L, Wang W-C. Vacuolating toxin production in clinical isolates of Helicobacter pylori with different vacA genotypes. J Infect Dis. 1998;178:207–212. doi: 10.1086/515600. [DOI] [PubMed] [Google Scholar]