Abstract
Background
The 5-methylcytosine (m5C) is one of the important forms of RNA post modification, and its regulatory mechanism in tumors has received increasing attention. However, its potential role in colorectal cancer remains unclear.
Materials and Methods
Here, we systematically investigated the genetic variation and prognostic value of the 14 m5c RNA methylation regulators in colon cancer. The prognostic risk score was constructed using three m5C regulators, which was verified in the GSE17536 (N=177), GSE41258 (N=248) and GSE38832 (N=122) datasets.
Results
The risk score developed from the three-m5C signature represents an independent prognostic factor, which can accurately predict the prognosis of patients with colon cancer in multiple datasets. The cytokine–cytokine receptor interaction and chemokine signaling pathway were significantly enriched in the low-risk score group. Further analysis showed that the three-m5C signature was related to tumor immune microenvironment (TIME), affecting the abundance of tumor-infiltrating immune cells. Especially, patients with low risk score had higher immune score than those with high risk score. In addition, gene set enrichment analysis (GSEA) confirmed that all three regulatory factors are associated with the MAPK/p38 signaling pathway.
Conclusion
In conclusion, our study illustrates that the three-m5C signature may be involved in the regulation of colon cancer immune microenvironment in synergy with the MAPK signaling pathway. Therefore, further studying the three-m5C signature regulatory mechanisms might provide promising targets for improving the responsiveness of colon cancer to immunotherapy.
Keywords: 5-methylcytosine, the three-m5C signature, colon cancer, prognostic, tumor immune microenvironment
Introduction
It was estimated that over 1.9 million new colorectal cancer (CRC) cases and 935,000 deaths occurred in 2020, which account for approximately one out of 10 cancer cases and deaths. CRC maintains the global third ranking on account of incidence, but ranks second in terms of mortality.1 Incidence rates vary significantly between different countries across the world, probably attributing to differences of the level of economic development and access to medical care. Colon cancer serves as the most common subtype of CRC, the 5-year overall survival (OS) rate, especially for patients at advanced stages, is less than 40%, which can be ascribed to post-operative recurrence and metastasis to a great extent.2–4 With the development of treatment methods, such as surgery, radiotherapy, chemotherapy and immunotherapy, the OS rate of colon cancer patients at different stages has improved.5 However, due to the difficulties in early screening, some patients with advanced colon cancer have limited therapeutic effects, leading to an adverse effect on the prognosis.6 The detection and analysis of tumor prognostic markers are of great significance for evaluating tumor progression, predicting the effect of therapeutic regimen and prolonging survival duration.7 By detecting tumor prognostic markers, it is of great significance for screening colon cancer patients who may have low survival and high mortality, which can assist clinical treatment and improve the prognosis of patients.
Since the description of pseudouridine (Ψ) proposed in 1960 lifted the curtain on post-transcriptional modifications of RNA, epigenetic modification has been winning more and more attention from scientific researchers all over the world.8 It is widely acknowledged that mRNA post-transcriptional modifications emerged as a novel epigenetic modification that is promising to evaluate tumor prognosis, which include 5- terminal capping, pre-mRNA splicing, polyadenylation and mRNA export epigenetic mechanisms.9,10 Among them, 5-methylcytosine is a conserved and prevalent mark in RNAs of human. There is evidence suggesting that m5C is widely distributed in a wide variety of RNA species, including cytoplasmic and mitochondrial ribosomal RNAs (rRNAs) and transfer RNAs (tRNAs) as well as messenger RNAs (mRNAs), enhancer RNAs (eRNAs) and a number of non-coding RNAs.11 As has been confirmed by previous studies, m5C modifications of mRNA discovered in 1925 are located in the untranslated regions (UTRs) of mRNA transcripts and ubiquitous in nature, which are involved in a number of biological processes such as protein translational regulation, RNA processing and stress response.12 The C5 methylation of RNA cytosines is determined by the coordinated actions of the three regulators: methyltransferases (writers), RNA-binding proteins (readers), and demethylases (erasers). At present, m5C regulators consist of eight writers (NOL1/NOP2/SUN domain (NSUN) family and the DNA methyltransferase homologue DNMT2), two readers (YBX1 and ALYREF) and four erasers (translocator family (TET) and ALKBH1).13,14 Current studies have shown that these regulators are involved in biological processes such as cell differentiation and apoptosis.15,16 Mutations in the genes encoding these regulators are associated with a variety of human diseases, and changes in expression levels have been observed in many cancers.17 For example, NSUN2 is overexpressed not only in oral and colorectal cancer but also in other cancer tissues, which indicate it represents a possible useful novel biomarker for cancer diagnostics.18 A recent study showed that m5C RNA methylation regulators are closely related to the prognosis and can participate in the regulation of the immune microenvironment in lung squamous cell carcinoma.9 Increasing studies on RNA modifications have pushed its elucidation of mechanism forward a lot, while the existing studies of m5C RNA modifications is not considerable and calling for more attention.
In this study, we aim to assess the power of the m5C RNA methylation regulators to predict the prognosis of colon cancer patients, and explore the biological function associated with the m5C regulators through comprehensive bioinformatical analysis. Based on RNA sequencing data from TCGA dataset, we systematically analyzed the relationship between 14 m5C regulators and prognosis in colon cancer for establishing a risk signature gene set including NSUN6, TRDMT1 and ALKBH1, which showed that the three-m5C signature had a strong ability to predict the prognosis of colon cancer. Using datasets from different cohorts, we further verified that the signature is highly correlated with the prognosis of patients with colon cancer, which confirmed the stability and reliability of the three-m5C signature model. In addition, the signaling pathways involved with the three-m5C signature were further analyzed to explore the survival mechanism relevant to the scoring of the three-m5C signature, which indicated that the three-m5C signature was able to participate in the regulation of tumor immune microenvironment.
Materials and Methods
Datasets
The GDC-TCGA-COAD dataset and the corresponding clinical data were downloaded from the Cancer Genome Browser website of the University of California, Santa Cruz (http://xena.ucsc.edu/), including normal samples (n = 41) and tumor samples (n = 471).19 Among them, 448 colon cancer patients with OS were enrolled for further survival analysis. The GEO: GSE17536, GSE41258 and GSE38832 datasets from GEO database were used as the external validation cohort. The expression profiling of colon cancer patients and survival information were extracted from GEO: GSE17536 (N=177), GSE41258 (N=248) and GSE38832 (N=122) datasets.
Prognostic Model Establishment
To evaluate the prognostic value of m5C RNA methylation regulators, we used random forest algorithm, Least Absolute time-selection Operator (Lasso) Cox regression algorithm and multivariate Cox regression analysis to screen out genes closely related to survival from 14 m5C RNA methylation regulators. Then, we further analyzed the landscape of genetic variation of each m5C RAN methylation regulator. Combined with gene expression, prognostic value, multivariate Cox regression analysis and genetic variation characteristics, the genes most related to the occurrence and development of colon cancer were screened out, based on which a multivariate Cox regression analysis was performed subsequently. Finally, a risk score formula was calculated by taking into account of the expression of optimized genes and correlation estimated via Cox regression coefficients: Risk score = (exp Gene1 * coef Gene1) + (exp Gene2 * coef Gene2) + (exp Gene3 * coef Gene3). This formula was meant to calculate the risk score of each patient in the TGGA dataset. Colon cancer patients were categorized as low risk and high risk in line with the most exact cutoff determined by χ-tile 3.6.1 software.20 The predictive power of the prognostic model was evaluated by Log rank test and Kaplan-Meier survival analysis.
Nomogram Construction and Evaluation
To assess whether the prognostic signature in patients with colon cancer was related to clinical characteristics, we performed a multivariate Cox regression analysis. The multivariate Cox regression model was used to compile nomograms via the rms package. The calibration curve was evaluated graphically by plotting the observation rate and the prediction probability of the histogram. In addition, the consistency index (C-index) was calculated and the receiver operating characteristic (ROC) curve was drawn to evaluate the reliability and stability of the nomograms.
Biological Enrichment Analysis for the Three-m5C Signature Modification Patterns
Using the limma package to compare the gene expression differences between colon cancer tissues and adjacent normal tissues, the differentially expressed genes (DEGs) were determined with the critical of |log fold change| > 1 and P<0.05. Weighted gene co-expression network analysis (WGCNA) is a systems biology method for describing the correlation patterns among genes across microarray samples.21 In order to identify the biological function of the three-m5C signature, WGCNA package was used to identify the genes and gene modules associated with the three-m5C signature. Then, the biological functions of the three-m5C signature related genes were analyzed by Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis through the Clusterprofiler package.
Gene Set Enrichment Analysis of the Three Selected m5C RNA Methylation Regulators
According to the expression level of genes, every sample was classified as either “High” or “Low” groups. Gene set enrichment analysis of genes was conducted by utilizing Clusterprofiler package. The number of permutations were set at “1000”. The |NES| >1, P < 0.05 and q < 0.25 indicated statistically significant.
Immunohistochemistry (IHC)
To examine gene expression in tumors and matched normal tissue, samples from 16 cancer patients were obtained from the First Affiliated Hospital of Zhengzhou University (The study was approved by the Ethics Committee of the First Affiliated Hospital of Zhengzhou, Henan, China (Data 11/08/2020); all participants were informed of the details of the study design and signed the informed consent form.). Paraffin-embedded, formalin-fixed tissue specimens were necessities for IHC. Sections were incubated overnight in a humidified container at 4°C with the primary antibodies of ALKBH1 (1:100, ab126596; Abcam), NSUN6 (1:100, ab214227; Abcam) and TRDMT1 (1:100, ab220175; Abcam). Briefly, after xylol deparaffinization and rehydration in graded alcohols, colorectal cancer arrays were boiled in citrate buffer, pre-incubated with H2O2, and blocked with rabbit or goat serum (DAKO, Glostrup, Denmark). Arrays were then incubated with a primary antibody and next with an HRP-conjugated secondary antibody. Before counterstaining with hematoxylin, diaminobenzidine was supposed to develop the target proteins. The IHC staining was evaluated by applying a scoring system from 0 to 3 (0 = negative or no staining, 1 = weak or low staining; 2 = moderate or intermediate staining; and 3 = strong or high staining), which determined the score for each tissue based on the percentage of positive cells and intensity of staining. The arrays were read by a pathologist with Olympus BX41 microscope.
Statistical Analysis
Most statistical analyses were performed with R version 4.0.4 software (Institute for Statistics and Mathematics, Vienna, Austria; https://www.r-project.org) (Package: edgeR, ggplot2, rms, glmnet, Clusterprofiler, survminer, timeROC). GraphPad Prism 8.0 and SPSS 24.0 (IBM, NY, USA) were also used for statistical analyses. Survival analysis was performed by using a Log rank test. The χ2 test was utilized for analysis on correlation between risk scores and clinical parameters. P < 0.05 was considered statistically significant.
Results
The Expression, Genetic Variation and Prognostic Value of m5C Regulators in Colon Cancer
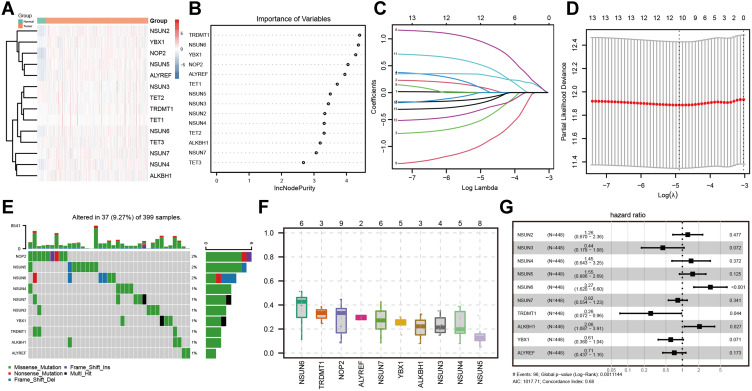
In view of the important role of m5C RNA methylation regulators in tumor progression, we used the GDC-TCGA dataset to comprehensively explore the transcription levels of 14 m5C RNA methylation regulators. The results showed that the expression levels of m5C RNA methylation regulators in colon cancer patients were significantly different from those in the normal control group (Figure 1A). Then, the importance of each gene was calculated by the random forest algorithm, and 13 M5c RNA methylation regulators were screened withIncNodePurity = 3 as the critical value (Figure 1B). In order to better determine the clinical prognostic value of m5C RNA methylation regulators, we used the LASSO Cox regression algorithm for the 13 regulatory factors that were significantly differentially expressed in the TCGA dataset, and obtained variables closely related to survival, including 10 regulatory factors (Figure 1C and D). Besides, we further analyzed the landscape of genetic variation of m5C regulators in colon cancer. Among 399 samples, 37 experienced mutations of m5C regulators, with frequency 9.27%. It was found that the 10 m5C RNA methylation regulators all manifested low mutations and low variant allele frequency (VAF) in colon cancer samples (Figure 1E and F). To comprehensively analyze genes of the most prognostic value among 10 m5C RNA methylation regulators in patients with colon cancer, Cox regression multivariate analysis was further analyzed, which indicated that NSUN6, TRDMT1 and ALKBH1 were significant and strongly correlated with prognosis (Figure 1G). Considering gene expression, survival value and genetic variation characteristics, we constructed an optimal multivariate Cox regression model based on the expression levels of NSUN6, TRDMT1 and ALKBH1. A risk score was calculated for each patient in line with a formula derived from the expression levels of three genes weighted by their regression coefficient: Risk score = (1.0930 * expression of NSUN6) + (−1.8569 * expression of TRDMT1) + (0.4911 * expression of ALKBH1).
Figure 1.
The expression, genetic variation and prognostic value of m5C regulators in colon cancer. (A) the transcription levels of 14 m5C RNA methylation regulators; (B) the importance of each gene calculated by the random forest algorithm; (C) LASSO regression analysis of the 13 m5C RNA methylation regulators; (D) Tenfold cross-validation for tuning the parameter selection in the LASSO regression. The dotted vertical lines represent the optimal values of the tuning parameter (λ) by minimum criteria; the mutations (E) and variant allele frequency (F) of the 10 m5C RNA methylation regulators in colon cancer samples; (G) Cox regression multivariate analysis was used to comprehensively analyze the prognostic value among 10 m5C RNA methylation regulators.
Abbreviation: Lasso, least absolute time-selection operator.
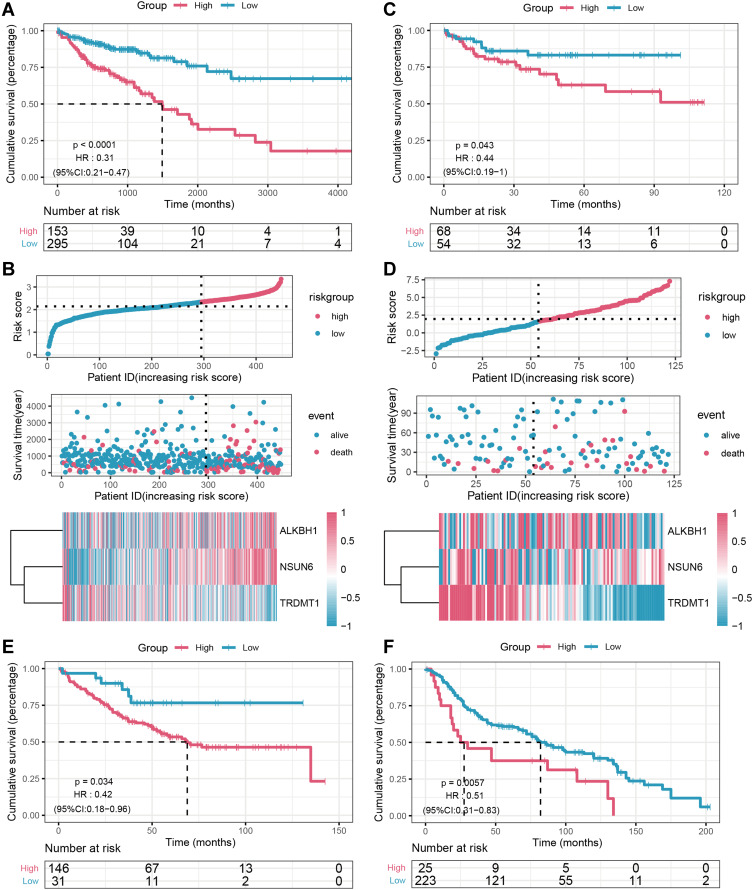
χ-tile 3.6.1 software was used to identify the optimum cut-off value for distinguishing high-risk (n = 153) from low-risk (n = 295) patients, which was estimated to be 2.34 in GDC-TCGA-COAD cohorts. High-risk scores patients had shorter overall survival (OS) (HR = 0.31; 95% CI: 0.21–0.47; P < 0.0001) than patients with low risk scores (Figure 2A). Figure 2B shows the distribution of patient prognostic scores, the survival status and gene expression, and data were ranked according to the prognostic score values for the three-m5C signature. NSUN6 and ALKBH1were associated with high risk and TRDMT1 was turned out to be protective. Patients with high risk scores tended to express NSUN6 and ALKBH1, whereas patients with low risk scores tended to express protective TRDMT1. Patients with high-risk scores had more deaths than low-risk-score ones.
Figure 2.
The prognostic value of the three-m5C signature model from different cohorts. Kaplan–Meier curves for the high and low the risk score patient groups in the GDC_TCGA-COAD cohort (A), GSE38832 (C), GSE17536 (E) and GSE41258 (F). The relationships between the expression of three prognostic genes and the risk score distribution with survival status in the GDC_TCGA-COAD cohort (B), GSE38832 (D) cohorts are shown; the X axis is sorted by the risk scores. Based on the risk score, patients were divided into high-risk and low-risk groups using χ-tile 3.6.1 software.
Validation of the Three-m5C Signature in Multiple GEO Colon Cancer Cohorts
To determine whether the three-m5C signature was robust, the performance of the three-m5C signature was assessed in three independent GEO colon cohorts, which totally consisted of 525 colon patients. The optimum cut-off value for GEO colon cohorts was identified with χ-tile 3.6.1 software, based on the three-m5C signature. For the GSE38832 validation cohort, the three-m5C signature managed to categorize 68 patients into the high-risk group and 54 patients into the low-risk group in terms of disease free survival (DFS) (HR =0.44; 95% CI: 0.19–1; P < 0.05; Figure 2C). The distribution of patient prognostic scores, the survival status and gene expression were ranked according to the prognostic score values for the three-m5C signature in GSE38832 validation cohort, which was basically the same as the results of the training set (Figure 2D). Similar analyses indicated that 146 high-risk patients had poorer OS than 31 low-risk patients in the GSE17536 validation cohort (HR = 0.42; 95% CI: 0.18–0.96; P < 0.05; Figure 2E), and 25 high-risk patients had poorer OS than 287 low-risk patients in the GSE41258 validation cohort (HR = 0.51; 95% CI: 0.31–0.83; P < 0.01; Figure 2F). Similar results were observed in both the training set and the testing set, which suggested that high-risk patients had significantly worse OS or DFS than those who were assigned to the low-risk group according to the three-m5C signature.
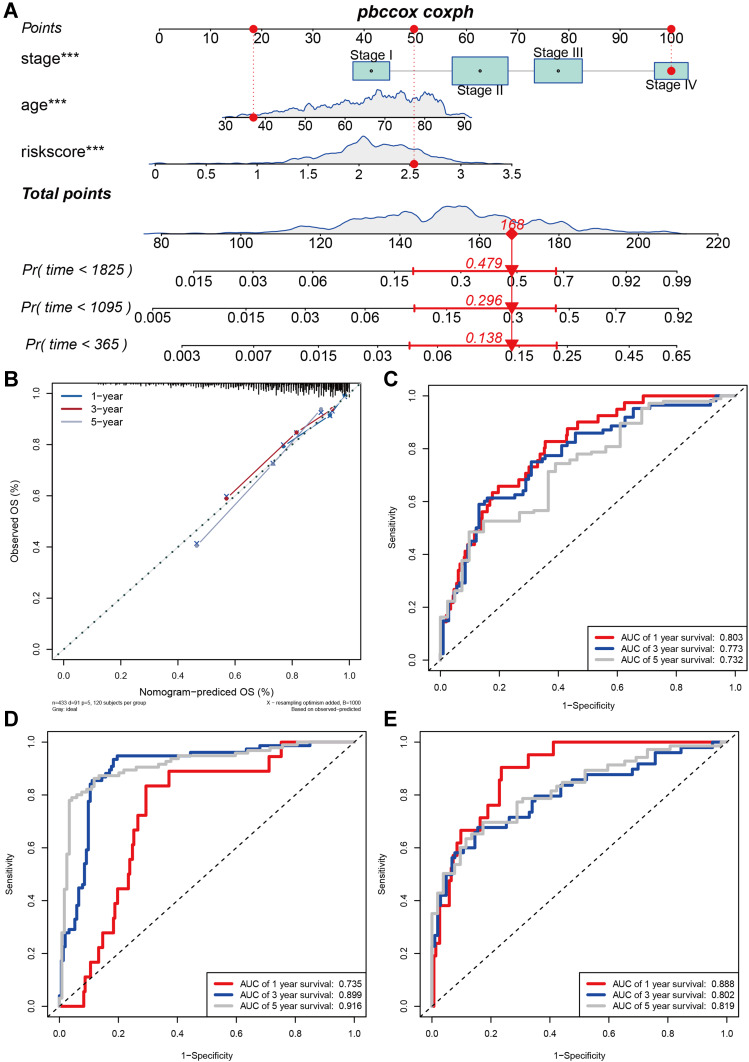
The Construction and Evaluation of a Nomogram Based on the Three-m5C Signature
To explore whether the prognostic value of the three-m5C signature was independent of other clinical factors, the clinical characteristics (including age, sex, clinical stage, lymph node metastasis and distant metastases) between high and low risk-score groups based on the TCGA dataset were further compared, which indicated that there were statistically significant differences in tumor survival state; no significant differences were detected for other clinical features (Table 1). Furthermore, the univariate analysis using TCGA data revealed that age, clinical stage, lymph node metastasis, distant metastases and the risk score were independent prognostic factors for OS of patients with colon cancer (Table 2). Multivariate Cox regression analyses showed that the three-m5C signature could be used as an independent predictor of patients’ survival outcome after being adjusted by clinical characteristics such as age and clinical stage (Table 2). Based on the result of multivariate Cox analysis, a nomogram that integrated the three-m5C signature age and clinical stage, was generated to predict the probability of 1-year, 3-year and 5-year OS for colon cancer patients with the GDC-TCGA-COAD (Figure 3A). Additionally, the calibration curves and the ROC indicated satisfactory discrimination (C-index 0.706) and decent accuracy (Area Under Curve (AUC) of 1-year survival: 0.803; AUC of 3-year survival: 0.773; AUC of 5-year survival: 0.732) (Figure 3B and C). The ROC for the probability of OS at 1, 3 and 5 years were predicted well in the GSE41258 validation cohort (AUC) of 1-year survival: 0.735; AUC of 3-year survival: 0.899; AUC of 5-year survival: 0.916; Figure 3D), and GSE17536 validation cohort (AUC of 1-year survival: 0.888; AUC of 3-year survival: 0.802; AUC of 5-year survival: 0.819; Figure 3E).
Table 1.
Comparison of Clinical Characteristics Between Low-Risk Score Group and High-Risk Score Group in GDC_TCGA_COAD Cohort
| Variable | Case No. (%) | Risk Score | P | |
|---|---|---|---|---|
| High | Low | |||
| Sample | 434 | 217 | 217 | |
| Age (Year) | ||||
| ≥60 | 312 | 147 | 165 | 0.055 |
| <60 | 122 | 70 | 52 | |
| Gender | ||||
| Female | 203 | 101 | 102 | 0.923 |
| Male | 231 | 116 | 115 | |
| Clinical stage | ||||
| I | 75 | 34 | 41 | |
| II | 171 | 82 | 89 | 0.521 |
| III | 126 | 66 | 60 | |
| IV | 62 | 35 | 27 | |
| Lymph node metastasis | ||||
| Yes | 180 | 97 | 83 | |
| No | 254 | 120 | 134 | 0.173 |
| Distant metastases | ||||
| Yes | 62 | 35 | 27 | |
| No | 327 | 155 | 172 | 0.156 |
| Unknown | 45 | 27 | 18 | |
| Survival state | ||||
| Dead | 91 | 60 | 31 | 0.001 |
| Alive | 343 | 157 | 186 | |
Table 2.
Univariate and Multivariate Regression Analyses for Predicting Overall Survival in GDC_TCGA_COAD Cohort
| Variable | Univariate | Multivariate | ||
|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |
| Risk score | 2.7 (1.6–4.4) | <0.001 | 2.4728(1.5451–3.957) | <0.001 |
| Gender | 0.86 (0.56–1.3) | 0.46 | 1.0024(0.6563–1.531) | 0.99 |
| Distant metastases | 1.7 (1.3–2.1) | <0.001 | 1.2291(0.8711–1.734) | 0.24 |
| Lymph node metastasis | 2.6 (1.7–4) | <0.001 | 0.7238(0.393–1.333) | 0.3 |
| Stage | 2.2 (1.8–2.9) | <0.001 | 2.5921(1.8132–3.706) | <0.001 |
| Age | 1 (1–1) | <0.05 | 1.0378(1.0183–1.058) | <0.001 |
Figure 3.
Construction and evaluation of a nomogram based on the three-m5C signature to predict the 1-year, 3-year and 5-year overall survival for colon cancer patients. (A) Nomogram was constructed with the GDC_TCGA-COAD cohorts for predicting the probability of 1-year, 3-year and 5-year OS for colon cancer patients. Calibration (B) and ROC (C) plot of the nomogram for predicting the probability of OS at 1, 3 and 5 years in the GDC_TCGA-COAD cohorts. ROC plot of the nomogram for predicting the probability of OS at 1, 3 and 5 years in the GSE41258 (D) and GSE17536 (E) cohort (***p < 0.001).
Abbreviations: OS, overall survival; ROC, receiver operating characteristic.
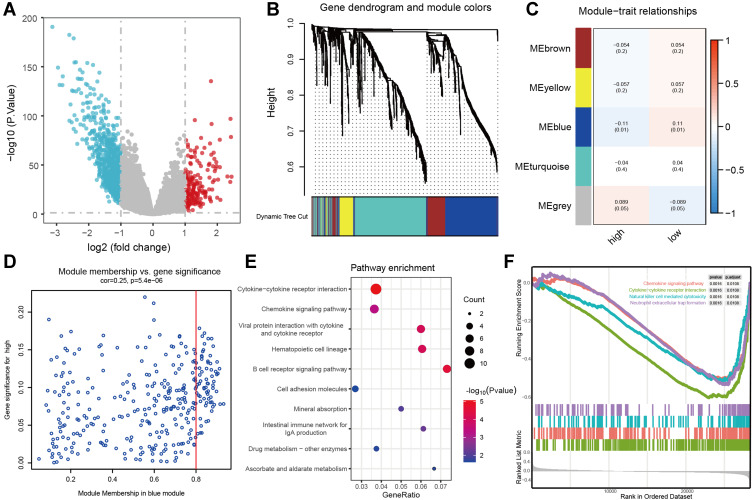
The Biological Characteristics of the Three-m5C Signature in Colon Cancer
With |logFC| > 1, P value < 0.05 as the screening criteria, we firstly obtained 991 differential genes in cancer and normal tissues based on the TCGA dataset (Figure 4A). In order to further understand the molecular mechanisms of the three-m5C signature, we divided colon cancer patients into high-risk group (n = 235) and low-risk score group (n = 236) according to their risk scores. Then, we identified gene modules were related to different modifications of m5C through WGCNA, based on the 991 differential genes. Five gene modules were determined, and high/low-risk score groups matched their corresponding genes (Figure 4B and C). Among them, the blue module was closely relevant to the high/low-risk score groups (Figure 4D). The blue module, containing 323 genes, had been subjected to KEGG pathway enrichment analysis. The results included cytokine−cytokine receptor interaction, chemokine signaling pathway, viral protein interaction with cytokine and cytokine receptor, B cell receptor signaling pathway, etc. (Figure 4E), which are all closely related to the occurrence and development of tumors.22–24 In addition, GSEA was also used to determine the correlated pathways of the three-m5C signatures. Among them, cytokine−cytokine receptor interaction, chemokine signaling pathway, natural killer cell mediated cytotoxicity and neutrophil extracellular trap formation were closely correlated with three-m5C signature, which was consistent with the function pathways of KEGG analysis (Figure 4F). These results suggest that the three-m5C signatures may be involved in the regulation of immune-related pathways.
Figure 4.
The biological characteristics of the three-m5C signature. (A) the volcano plot showing the differentially expressed genes between normal tissue and tumor tissue. (B) Gene dendrogram obtained by average linkage hierarchical clustering. The colour row underneath the dendrogram shows the module assignment determined by the Dynamic Tree Cut, in which 5 modules were identified. (C) Heatmap of the correlation between module eigengenes and the m5C modification patterns. (D) A scatterplot of gene significance for low-risk score group vs module membership in the blue module. (E) the KEGG enrichment analysis of genes in the blue module. (F) GSEA results showing significant enrichment of immune-related phenotype in low-risk score group.
Abbreviations: KEGG, Kyoto Encyclopedia of Genes and Genomes; GSEA, gene set enrichment analysis.
The Relationship Between Immune Infiltrates and the Three-m5C Signature
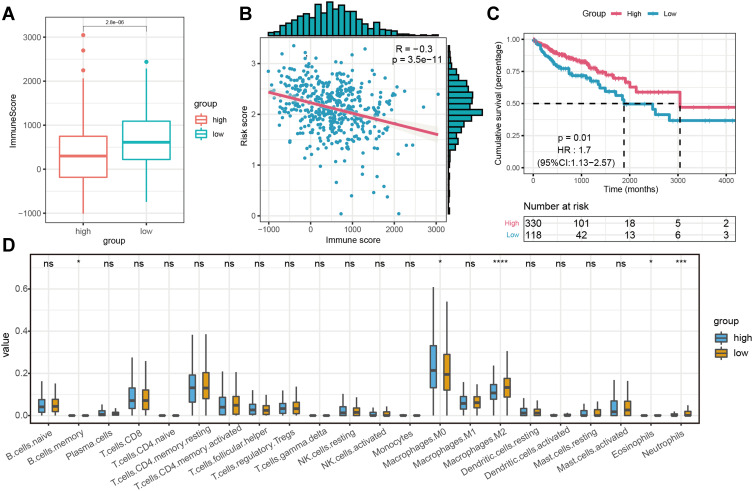
Given KEGG and GSEA enrichment analysis results, we speculated that the three-m5C signature might be closely related to immune infiltrates. In order to study the influence of the three-m5C signature on the TIME of colon cancer, we evaluated the immune score and the level of immune invasion between the high-risk score group and the low-risk score group. Significant differences in immune scores were observed between the two groups, which showed that the low-risk score group with satisfactory prognosis had higher immune scores than the high-risk group (P < 0.001; Figure 5A). Moreover, the immune score of colon cancer patients was significantly negatively correlated with risk score (P < 0.001, R= −0.3; Figure 5B). Based on the optimum cut-off value determined by χ-tile 3.6.1 software, high-immune score patients (n = 330) had longer OS (HR = 1.7; 95% CI: 1.13–2.57; P < 0.05) than patients with low-expression immune score (n = 118) (Figure 5C). Subsequently, we analyzed the proportion of 22 immune cell types between the two groups. Compared with the high-risk score group, there are significantly differences in the infiltration levels of B memory cells, M0 macrophages, M2 macrophages, eosinophils and neutrophils in the low-risk-score group (Figure 5D).
Figure 5.
The Effect on Survival and Immune Infiltration between the high-risk score group and low-risk score group divided by the three-m5C signature in TCGA Cohort. (A) the immunoscore the high-risk score group and low-risk score group; (B) the correlation of immune score and risk score; (C) Kaplan-Meier curves of OS for patients with colon cancer; (D) The infiltrating levels of 22 immune cell types in two groups. (*p < 0.05, ***p < 0.001 and ****p < 0.0001).
Abbreviations: TCGA, The Cancer Genome Atlas; OS, overall survival.
The Prognostic Value of Each Gene in the Three-m5C Signature and the Exploration of Signaling Pathways That They Involve
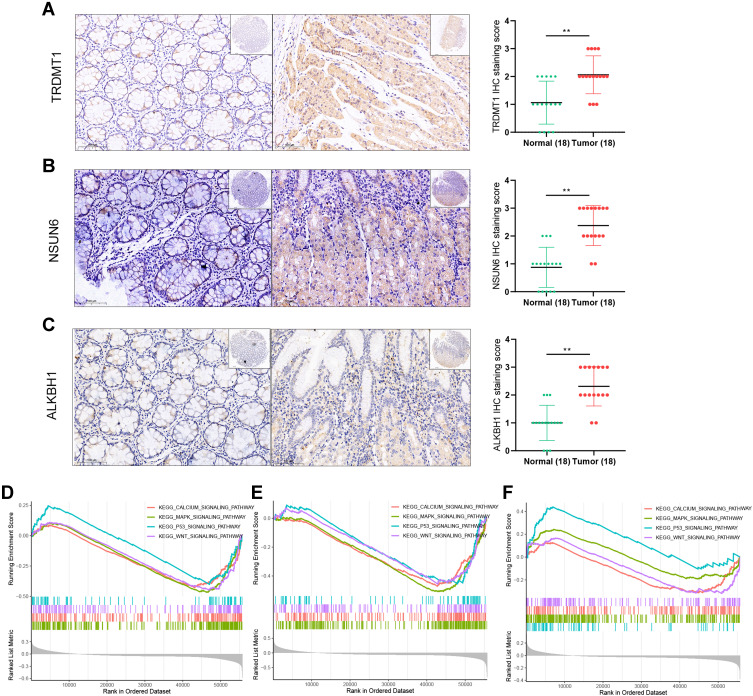
In view of the prognostic value and module function of the three-m5C signature in colon cancer samples, we further explored the expression of the three-m5C signature (TRDMT1, NSUN6 and ALKBH1) in patients with colon cancer, indicating that TRDMT1, NSUN6 and ALKBH1 are differentially expressed in cancer and paracancerous tissues (Figure 6A–C). To understand the importance of each gene in the signature composed of NSUN6, TRDMT1 and ALKBH1, we further explored the signaling pathways that TRDMT1, NSUN6 and ALKBH1 jointly participated in by means of the GSEA. With the screening criteria of |NES| >1, P < 0.05 and q < 0.25, the results indicated that they all participated in 71 signaling pathway, including MAPK signaling pathway, p53 signaling pathway and calcium signaling pathway etc. (Figure 6D–F, Supplementary Table 1). Most of them were all closely related to the occurrence and progression of tumors and the regulation of immune function.25
Figure 6.
The expression and GSEA analysis of the three selected m5C RNA methylation regulators. The expression of TRDMT1 (A), NSUN6 (B) and ALKBH1 (C) in tumors and adjacent no-tumor tissues from patients with colon cancer; GSEA results indicating TRDMT1 (D), NSUN6 (E) and ALKBH1 (F) all participated in MAPK signaling pathway, p53 signaling pathway and calcium signaling pathway (**p < 0.01).
Abbreviations: GSEA, gene set enrichment analysis; TRDMT1, tRNA aspartic acid methyltransferase 1; NSUN6, NOP2/Sun RNA methyltransferase 6; ALKBH1, AlkB homolog 1; MAPK, mitogen-activated protein kinases.
Discussion
As a key post-transcriptional regulator of gene expression programs, RNA modification is closely related to the occurrence and development of many diseases, among which m5C is one of the most common ways of RNA modification.13 m5C is one of the earliest modifications found in a variety of RNAs (mRNA, tRNAs and rRNAs), and has functions such as RNA stability and protein synthesis and translation regulation. In addition, m5C is closely associated with human diseases, and its function involves regulating stem cell stress, cytotoxic stress, mRNA nucleation and gene expression.26 Previous studies have shown that the abnormal expression of RNA m5C methylation regulators can affect the expression of oncogenes and play an important regulatory role in tumorigenesis and development.27
Colon cancer is a common fatal disease, and environmental and genetic factors tend to affect its risk.28 Moreover, data from epidemiological studies show that among adults under 50, the incidence of colon cancer continues to increase.29 In the early diagnosis, surgical resection with or without adjuvant therapy can effectively prolong the survival time of patients with colon cancer; however, the prognosis of advanced patients is usually frustrating.30,31 Therefore, the establishment of a prognostic indicator to accurately identify those patients with poor survival can better guide adjuvant treatment. In this study, based on the GDC-TCGA-COAD cohort, we used the random forest algorithm, LASSO Cox regression algorithm and multivariate Cox regression algorithm to screen out genes closely related to patient survival from 14 m5C regulators. On this basis, we developed a robust prognostic signature, including TRDMT1, NSUN6 and ALKBH1, whose effectiveness was proven on three GEO datasets from different microarray platforms. In order to provide clinicians with a quantitative method to predict the prognosis of colon cancer patients, we constructed a nomogram that integrated the three-m5C signature, age and stage, which was also applicable to the GSE41258 and GSE17536 datasets. It can predict both short-term and long-term survival of colon cancer patients more accurately than a single prognostic factor. Based on the three-m5C regulators, our study is the first to report the feasibility and accuracy of a risk assessment model for determining colon cancer prognosis, which play an important role in prognosis assessment and therapeutic intervention of colon cancer patients.
To explore potential biological function of the three-m5C signature, we identified the three-m5C signature regulation related genes based on the differentially expressed genes between colon cancer and normal tissues. The expression regulation of these genes was affected by m5C modification, and revealed their biological functions was helpful to clarify the occurrence and development mechanism of colon cancer from the perspective of m5C modification. The results of KEGG enrichment analysis of related genes included the cytokine–cytokine receptor interaction, chemokine signaling pathway, viral protein interaction with cytokine and cytokine receptor, B cell receptor signaling pathway etc … Among them, cytokine–cytokine receptor interaction and chemokine signaling pathway also shown in the result of GSEA based on the level of risk score. Low-risk score group showed more activation in cytokine–cytokine receptor interaction, chemokine signaling pathway and other immune signaling pathways which indicated that the cytokine–cytokine receptor and chemokine were highly correlated with the three-m5C signature and the progression of colon cancer. Based on these results, we further compared tumor-infiltrating lymphocyte and immune scores with risk scores. The risk scores based on the three-m5C signature were significantly associated with immune score and immune cell infiltration. Compared with the high-risk score group, there are significantly differences in the infiltration levels of B memory cells, M0 macrophages, M2 macrophages, eosinophils and neutrophils in the low-risk score group. More importantly, the immune score in the high-risk group was significantly lower than that in the low-risk group, indicating that the level of immune infiltration in the tumor microenvironment in the high-risk group was lower. Franck et al have showed that the immunoscore provides a reliable estimate of the risk of recurrence in patients with colon cancer, which can be used to guide clinical treatment and prognosis evaluation.32 These findings suggested that the three-m5C signature was involved in TIME regulation to some extent, which is one of the most important scientific implications of our study.
For the three selected regulators, TRDMT1, NSUN6 and ALKBH1, previous studies have shown that they are all associated with oncogenesis and progression.33 TRDMT1 (tRNA aspartic acid methyltransferase 1), also known as DNMT2, promoting tRNA stability and protein synthesis, participates in cell cycle, apoptosis, autophagy and interleukin levels of many tumors.34,35 However, there are few reports on the correlation between TRDMT1 and colon cancer. NSUN6 (NOP2/Sun RNA methyltransferase 6), is a member of the family of NSUN proteins, which is associated with tRNAs and acts as a tRNA methyltransferase.36,37 Some studies have found that NSUN6 can participate in biological function of tumor progression, such as cell cycle, tumor immune microenvironment and so on.38,39 ALKBH1, a 2-oxoglutarate and Fe (II)-dependent dioxygenase, is essential to multiple cellular processes. Previous studies have shown that ALKBH1 expression promotes tumor metastasis and recurrence in certain tumor types.40 In addition, the GSEA study result indicated that they were also all correlated with calcium signaling pathway, p53 signaling pathway and MAPK signaling pathway, which are all involved in different stages of tumorigenesis and development. Especially, MAPK signal pathway is activated by upstream genomic events and/or activation of multiple signaling events, where the information is merged at this important node pathway. This pathway is strictly regulated by phosphatase and two-way communication with other pathways under normal conditions, such as the mammalian target of protein kinase B/rapamycin (AKT/m-TOR) pathway.41 In addition, there is evidence that MAPK plays a key role in the regulation of immune response, participating in the regulation of innate and adaptive immunity.42 Reviewing the status of ongoing clinical trials investigating MAPK pathway inhibitors, we found that MAPK targeted therapies may collaborate with immune cells, which are consistent with the enrichment analysis result of the three-m5Csignature.43 Therefore, we hypothesize that TRDMT1, NSUN6 and ALKBH1 can jointly act on MAPK signaling pathway, thereby adjusting the level of immune infiltration in colon cancer tumor microenvironment. At present, there are relatively few studies on the signal pathway mediated by three m5C regulators. More relevant studies are needed to reveal the signaling pathways and their physiological and pathological mechanisms of the three m5C regulators both in vitro and in vivo.
Conclusions
In conclusion, this study systematically evaluated prognostic value, the correlation of TIME, and potential regulatory mechanisms of the three-m5C signature in colon cancer. The risk score developed from the three-m5C signature was an independent prognostic indicator of patients with colon cancer. Patients with high-risk score have a lower level of immune infiltration. There is closely relationship between the risk score and immune score in colon cancer. The three-m5C signature might be involved in the regulation of colon cancer immune microenvironment in synergy with the MAPK signaling pathway. Therefore, further studying the three-m5C signature regulatory mechanisms might provide promising targets for improving the responsiveness of colon cancer to immunotherapy. This study provides a basis for further research on the regulatory mechanism of m5c regulators on the immune microenvironment in colon cancer.
Acknowledgments
We thank the reviewers for their insightful comments.
Funding Statement
This work was supported by the National Key Research and Development Program of China (Grant No. 2017YFC0909900). The National Natural Science Foundation of China (Grant No. 82002433), and Science and Technology Project of Henan Provincial.Department of Education (Grant No. 18A320044, 21A320036), Henan Province Medical Science and Technology Research Project Joint Construction Project (Grant No. LHGJ20190003, LHGJ20190055).
Abbreviations
m5C, 5-methylcytosine; TIME, tumor immune microenvironment; GSEA, gene set enrichment analysis; Lasso, least absolute time-selection operator; ROC, receiver operating characteristic; C-index, consistency index; DEGs, differentially expressed genes; WGCNA, weighted gene co-expression network analysis; IHC, immunohistochemistry; VAF, variant allele frequency; OS, overall survival; DFS, disease free survival.
Ethics Approval
This study was approved by the Ethics Committee of the First Affiliated Hospital of Zhengzhou, Henan, China (Data 11/08/2020). Written informed consent was obtained from all patients. The experiments were carried out following the Declaration of Helsinki.
Disclosure
The authors have no conflicts of interest to declare.
References
- 1.Sung H, Ferlay J, Siegel RL. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2.Mejri N, Dridi M, El Benna H, et al. Tumor location impact in stage II and III colon cancer: epidemiological and outcome evaluation. J Gastrointest Oncol. 2018;9(2):263–268. doi: 10.21037/jgo.2017.12.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Favoriti P, Carbone G, Greco M, et al. Worldwide burden of colorectal cancer: a review. Updates Surg. 2016;68(1):7–11. doi: 10.1007/s13304-016-0359-y [DOI] [PubMed] [Google Scholar]
- 4.Mody K, Bekaii-Saab T. Clinical trials and progress in metastatic colon cancer. Surg Oncol Clin N Am. 2018;27(2):349–365. doi: 10.1016/j.soc.2017.11.008 [DOI] [PubMed] [Google Scholar]
- 5.Jahanafrooz Z, Mosafer J, Akbari M, et al. Colon cancer therapy by focusing on colon cancer stem cells and their tumor microenvironment. J Cell Physiol. 2020;235(5):4153–4166. doi: 10.1002/jcp.29337 [DOI] [PubMed] [Google Scholar]
- 6.Argilés G, Tabernero J, Labianca R, et al. Localised colon cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2020;31(10):1291–1305. doi: 10.1016/j.annonc.2020.06.022 [DOI] [PubMed] [Google Scholar]
- 7.Wang X, Duanmu J, Fu X, et al. Analyzing and validating the prognostic value and mechanism of colon cancer immune microenvironment. J Transl Med. 2020;18(1):324. doi: 10.1186/s12967-020-02491-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohn WE. Pseudouridine, a carbon-carbon linked ribonucleoside in ribonucleic acids: isolation, structure, and chemical characteristics. J Biol Chem. 1960;235:1488–1498. doi: 10.1016/S0021-9258(18)69432-3 [DOI] [PubMed] [Google Scholar]
- 9.Pan J, Huang Z, Xu Y. m5C RNA methylation regulators predict prognosis and regulate the immune microenvironment in lung squamous cell carcinoma. Front Oncol. 2021;11:657466. doi: 10.3389/fonc.2021.657466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roundtree IA, Evans ME, Pan T, et al. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169(7):1187–1200. doi: 10.1016/j.cell.2017.05.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edelheit S, Schwartz S, Mumbach MR, et al. Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs. PLoS Genet. 2013;9(6):e1003602. doi: 10.1371/journal.pgen.1003602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He Y, Yu X, Li J, et al. Role of m(5) C-related regulatory genes in the diagnosis and prognosis of hepatocellular carcinoma. Am J Transl Res. 2020;12(3):912–922. [PMC free article] [PubMed] [Google Scholar]
- 13.Nombela P, Miguel-López B, Blanco S. The role of m(6)A, m(5)C and Ψ RNA modifications in cancer: novel therapeutic opportunities. Mol Cancer. 2021;20(1):18. doi: 10.1186/s12943-020-01263-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du E, Li J, Sheng F, et al. A pan-cancer analysis reveals genetic alterations, molecular mechanisms, and clinical relevance of m(5) C regulators. Clin Transl Med. 2020;10(5):e180. doi: 10.1002/ctm2.180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen H, Yang H, Zhu X, et al. m(5)C modification of mRNA serves a DNA damage code to promote homologous recombination. Nat Commun. 2020;11(1):2834. doi: 10.1038/s41467-020-16722-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mei L, Shen C, Miao R, et al. RNA methyltransferase NSUN2 promotes gastric cancer cell proliferation by repressing p57(Kip2) by an m(5) C-dependent manner. Cell Death Dis. 2020;11(4):270. doi: 10.1038/s41419-020-2487-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bohnsack KE, Höbartner C. Eukaryotic 5-methylcytosine (m5C) RNA methyltransferases: mechanisms, cellular functions, and links to disease. Genes (Basel). 2019;10(2):102. doi: 10.3390/genes10020102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okamoto M, Hirata S, Sato S, et al. Frequent increased gene copy number and high protein expression of tRNA (cytosine-5-)-methyltransferase (NSUN2) in human cancers. DNA Cell Biol. 2012;31(5):660–671. doi: 10.1089/dna.2011.1446 [DOI] [PubMed] [Google Scholar]
- 19.Goldman MJ, Craft B. Visualizing and interpreting cancer genomics data via the Xena platform. Nature Biotechnol. 2020;38(6):675–678. doi: 10.1038/s41587-020-0546-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10(21):7252–7259. doi: 10.1158/1078-0432.ccr-04-0713 [DOI] [PubMed] [Google Scholar]
- 21.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 2008;9:559. doi: 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mollica Poeta V, Massara M, Capucetti A, et al. Chemokines and chemokine receptors: new targets for cancer immunotherapy. Front Immunol. 2019;10:379. doi: 10.3389/fimmu.2019.00379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burger JA, Wiestner A. Targeting B cell receptor signalling in cancer: preclinical and clinical advances. Nat Rev Cancer. 2018;18(3):148–167. doi: 10.1038/nrc.2017.121 [DOI] [PubMed] [Google Scholar]
- 24.Coperchini F, Croce L, Marinò M, et al. Role of chemokine receptors in thyroid cancer and immunotherapy. Endocr Relat Cancer. 2019;26(8):R465–r478. doi: 10.1530/erc-19-0163 [DOI] [PubMed] [Google Scholar]
- 25.Liang F, Ren C, Wang J, et al. The crosstalk between STAT3 and p53/RAS signaling controls cancer cell metastasis and cisplatin resistance via the Slug/MAPK/PI3K/AKT-mediated regulation of EMT and autophagy. Oncogenesis. 2019;8(10):59. doi: 10.1038/s41389-019-0165-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Selmi T, Hussain S, Dietmann S, et al. Sequence- and structure-specific cytosine-5 mRNA methylation by NSUN6. Nucleic Acids Res. 2021;49(2):1006–1022. doi: 10.1093/nar/gkaa1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen X, Li A, Sun BF, et al. 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat Cell Biol. 2019;21(8):978–990. doi: 10.1038/s41556-019-0361-y [DOI] [PubMed] [Google Scholar]
- 28.Garrett WS. The gut microbiota and colon cancer. Science. 2019;364(6446):1133–1135. doi: 10.1126/science.aaw2367 [DOI] [PubMed] [Google Scholar]
- 29.Stoffel EM, Murphy CC. Epidemiology and mechanisms of the increasing incidence of colon and rectal cancers in young adults. Gastroenterology. 2020;158(2):341–353. doi: 10.1053/j.gastro.2019.07.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gelibter AJ, Caponnetto S, Urbano F, et al. Adjuvant chemotherapy in resected colon cancer: when, how and how long? Surg Oncol. 2019;30:100–107. doi: 10.1016/j.suronc.2019.06.003 [DOI] [PubMed] [Google Scholar]
- 31.Angell HK, Bruni D. The immunoscore: colon cancer and beyond. Clin Cancer Res. 2020;26(2):332–339. doi: 10.1158/1078-0432.ccr-18-1851 [DOI] [PubMed] [Google Scholar]
- 32.Pagès F, Mlecnik B, Marliot F, et al. International validation of the consensus Immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet. 2018;391(10135):2128–2139. doi: 10.1016/s0140-6736(18)30789-x [DOI] [PubMed] [Google Scholar]
- 33.Subramaniam D, Thombre R, Dhar A, et al. DNA methyltransferases: a novel target for prevention and therapy. Front Oncol. 2014;4:80. doi: 10.3389/fonc.2014.00080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bloniarz D, Adamczyk-Grochala J, Lewinska A, et al. The lack of functional DNMT2/TRDMT1 gene modulates cancer cell responses during drug-induced senescence. Aging. 2021;13(12):15833–15874. doi: 10.18632/aging.203203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Q, Yang H, Zhu X, et al. Integrative analysis of the doxorubicin-associated LncRNA-mRNA network identifies chemoresistance-associated lnc-TRDMT1-5 as a biomarker of breast cancer progression. Front Genet. 2020;11:566. doi: 10.3389/fgene.2020.00566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haag S, Warda AS, Kretschmer J, et al. NSUN6 is a human RNA methyltransferase that catalyzes formation of m5C72 in specific tRNAs. Rna. 2015;21(9):1532–1543. doi: 10.1261/rna.051524.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen YS, Yang WL, Zhao YL, et al. Dynamic transcriptomic m(5) C and its regulatory role in RNA processing. Wiley Interdiscip Rev RNA. 2021;12(4):e1639. doi: 10.1002/wrna.1639 [DOI] [PubMed] [Google Scholar]
- 38.Yang R, Liang X, Wang H, et al. The RNA methyltransferase NSUN6 suppresses pancreatic cancer development by regulating cell proliferation. EBioMedicine. 2021;63:103195. doi: 10.1016/j.ebiom.2020.103195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang Z, Pan J, Wang H, et al. Prognostic significance and tumor immune microenvironment heterogenicity of m5C RNA methylation regulators in triple-negative breast cancer. Front Cell Dev Biol. 2021;9:657547. doi: 10.3389/fcell.2021.657547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H, Zhang Y, Guo Y, et al. ALKBH1 promotes lung cancer by regulating m6A RNA demethylation. Biochem Pharmacol. 2021;189:114284. doi: 10.1016/j.bcp.2020.114284 [DOI] [PubMed] [Google Scholar]
- 41.Burotto M, Chiou VL, Lee JM, et al. The MAPK pathway across different malignancies: a new perspective. Cancer. 2014;120(22):3446–3456. doi: 10.1002/cncr.28864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y, Shepherd EG, Nelin LD. MAPK phosphatases–regulating the immune response. Nat Rev Immunol. 2007;7(3):202–212. doi: 10.1038/nri2035 [DOI] [PubMed] [Google Scholar]
- 43.Shin MH, Kim J, Lim SA, et al. Current insights into combination therapies with MAPK inhibitors and immune checkpoint blockade. Int J Mol Sci. 2020;21(7). doi: 10.3390/ijms21072531 [DOI] [PMC free article] [PubMed] [Google Scholar]