When the coronavirus disease 2019 (COVID-19) pandemic first appeared in December of 2019, the pathophysiological underpinnings of the disease were largely unknown. Scientists, physicians and government institutions from around the globe took an “all-hands on deck” approach with the hope of identifying potential therapies to treat as well as understand the pathophysiology of the disease [1]. Currently, more than 4800 clinical trials listed on clinicaltrials.gov have been performed or proposed around the world, many with subjects from vastly different ethnic and racial backgrounds, as well as different standard-of-care strategies [2]. Despite this effort, apart from monoclonal antibodies, few therapies have emerged as effective treatments of COVID-19; vaccines remain the best approach to control and mitigate the pandemic [3].
Short abstract
Metabolomics changes in COVID-19 predict acute patient outcomes and suggest a role for a bioenergetic crisis. Thus, metabolomics changes in COVID-19 may serve as a biomarker and provide insight into pathogenic mechanisms and pharmacologic targets. https://bit.ly/2XkJeU8
When the coronavirus disease 2019 (COVID-19) pandemic first appeared in December of 2019, the pathophysiological underpinnings of the disease were largely unknown. Scientists, physicians and government institutions from around the globe took an “all-hands on deck” approach with the hope of identifying potential therapies to treat as well as understand the pathophysiology of the disease [1]. Currently, more than 4800 clinical trials listed on clinicaltrials.gov have been performed or proposed around the world, many with subjects from vastly different ethnic and racial backgrounds, as well as different standard-of-care strategies [2]. Despite this effort, apart from monoclonal antibodies, few therapies have emerged as effective treatments of COVID-19; vaccines remain the best approach to control and mitigate the pandemic [3].
Despite the lack of therapeutic successes, we have gained unprecedented insight into the progression of the disease [4]. Among the vast amount of clinical and biological data published over the course of the pandemic, some of the most consistent and exciting results have come from metabolomics profiling of patient serum samples [5]. Metabolomics, a rapidly developing field of research in which metabolites present in tissue or fluids are comprehensively analysed, has deepened our understanding of the pathobiology of multiple disorders, identified predictive biomarkers, and highlighted potential novel therapeutic strategies. Throughout 2020 and 2021, several metabolomics papers focusing on COVID-19 have been published. The earliest reports identified changes in the metabolic pathways for processing lipids, amino acids and carbohydrates in severely ill COVID-19 patients. More recent studies have begun to associate metabolomics changes with symptoms representing specific organ system failures, for example, the acute delirium and post-recovery mental health issues of the nervous system and disruptions of the digestive system [6, 7]. Dysregulation of the kynurenine pathway has been among the most consistent findings as reported in numerous, independent studies [5]. Of potential significance is that many of the metabolomics abnormalities in COVID-19 are similar to those found in sepsis and acute respiratory failure, suggesting a common mechanism leading to an acute bioenergetic crisis [8, 9].
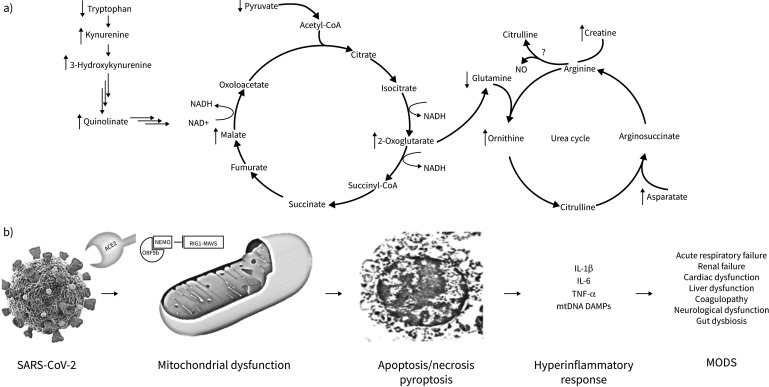
Along these lines, the paper “Metabolomic analyses reveal new stage-specific features of COVID-19” by Jia et al. [10] reports an in-depth, well-designed metabolomics analysis of confirmed COVID-19 patients. The discovery patients (n=63) were subdivided into mild, severe, and recovery groups and independently validated in a second cohort with an additional 90 patients along with 41 non-infected controls. The investigators utilised both broad-spectrum, semi-quantitative mass spectrometry analysis as well as targeted mass spectrometry analysis. The authors found consistent disruptions in glucose metabolism and dysregulation in the TCA and urea cycles which were identified as potential targets for therapeutic intervention (figure 1a). These metabolomics changes are consistent with metabolomics studies in other contexts and point to an underlying bioenergetic crisis as a key pathogenic feature of COVID-19 [5].
FIGURE 1.
Metabolomics changes due to SAR-CoV-2 are predictors of patient acute and long-term outcomes and reflect a bioenergetic crisis likely due to viral hijacking of the mitochondria. a) Metabolomics changes found due to SARS-CoV-2 infections commonly lead to disruption of the kynurenine pathway, TCA cycle and the urea cycle. It is unknown if arginine is converted to citrulline and NO; however, due to the increase in ornithine and creatine, Jia et al. [10] speculate that the urea cycle metabolises arginine instead of the NO producing pathway. b) SARS-CoV-2 is internalised into the cell via the angiotensin converting enzyme 2 (ACE2) receptor. Many viral proteins, including ORF7a and ORF9b locate within the mitochondria. These proteins can inhibit RIG1-MAVS (retinoic acid-inducible gene I-mitochondrial antiviral signalling protein)-dependent interferon signalling, enhance viral replication and disrupt mitochondrial function. This can ultimately lead to cell death via apoptosis, necrosis or pyroptosis, the release of proinflammatory cytokines as well as mitochondrial DAMPs (damage-associated molecular patterns) and ultimately cause multiple organ dysfunction syndrome (MODS). Jia et al. [10]also demonstrate that many of the predictive metabolites altered due to SARS-CoV-2 strongly correlate with proinflammatory cytokines. TNF: tumour necrosis factor; NEMO: nuclear factor κB essential modulator.
Furthermore, the authors were able to correlate changes in interleukin (IL)-1β, tumour necrosis factor (TNF)-α and IL-6 with metabolomics profiles. Not surprisingly, all three cytokines increased with the severity of disease. Although the levels fell during recovery, they were persistently elevated relative to non-infected control patients. Importantly, cytokine levels positively correlated with TCA cycle-related metabolites, including aspartate, creatinine, malate and 2-oxoglutarate. There was also positive correlation with arginine, a key component of the uric acid cycle, which can be converted to either ornithine or nitric oxide and citrulline. The authors posited that the decrease in arginine and increase in ornithine suggests that arginine is metabolised via arginase and the urea cycle as opposed to the nitric oxide cycle. Nonetheless, since nitric oxide could reduce viral RNA production by affecting the spike protein and its primary target, angiotensin converting enzyme 2 (ACE2), clinical trials of the therapeutic gas in COVID-19 are currently underway [11].
The metabolomic findings of Jia et al. [10], like similar studies in sepsis and acute respiratory failure (ARF) [9, 12–15], point to a pivotal role for mitochondrial dysfunction as a driver of COVID-19 outcomes [16]. In this context, the abundance of mitochondrial DNA damage-associated molecular patterns (mtDNA DAMPs), a category of DAMP known to promote cytokine production by both immune and non-immune cells [17, 18], in the plasma of COVID-19 patients is an early predictor of intensive care unit (ICU) admission, need for intubation, and mortality in COVID-19 [19]. Experimental studies have identified multiple pathways by which SARS-CoV-2 evokes mitochondrial dysfunction leading to pathophysiological effects of COVID-19 (figure 1b). For example, SARS-CoV-2 impairs oxidative metabolism and promotes a transition to a glycolytic phenotype in peripheral blood mononuclear cells from COVID-19 patients [20]. This effect may be mediated by ACE2, widely known as the receptor mediating SARS-CoV-2 entry into the cell, which is believed to directly alter mitochondrial function leading to decreased ATP production and activation of NADPH oxidase (NOX) 4 [21, 22]. Increased reactive oxygen species generation associated with NOX4 activation could exert multiple deleterious events, including damaging the mitochondrial genome leading to its fracture into proinflammatory mtDNA DAMPs [23, 24], as well as activating PARP1 causing NAD+ depletion with attendant reduced interferon production, enhanced viral replication and decreased mitophagy [25]. Along with ACE2, proteins encoded by SARS-CoV-2 also may perturb mitochondrial functions. Here, studies on open reading frames ORF-9b and ORF-7a of SARS show that these proteins localise to mitochondria. ORF9b can inactivate the retinoic acid-inducible gene I-mitochondrial antiviral signalling protein (RIG1-MAVS)-dependent interferon signalling pathway by disrupting K63-linked polyubiquitination of nuclear factor κB essential modulator (NEMO) [26]. In SARS, ORF-7a and ORF-8a promote viral replication, while ORF-8a can activate caspase-3 mediated apoptosis [22, 27]. Unlike SARS, the SARS-CoV-2 genome includes ORF-8, which is known to play a role in immune evasion by downregulating major histocompatibility complex (MHC) class Ι, targeting the protein for lysosomal degradation via the beclin-1 autophagy initiation pathway [28]. While the full function of ORF-8 has not been elucidated, it is tempting to speculate that it might adversely affect mitochondria based on the fact that its ability to promote degradation of MHC class I requires a close interaction with beclin 1, the latter of which is known to interact with cardiolipin and mediate mitophagy [29]. The release of the virus, as well as proinflammatory cytokines and mtDNA DAMPs, induces a hyper-inflammatory response [22, 24, 27, 30, 31]. Determining how these bioenergetic metabolomic changes relate to mitochondrial dysfunction could project to new therapies that aim to return the metabolomic profiles back to homeostasis.
One of the more surprising findings was that the greatest number of metabolomics differences between control subjects and COVID-19 patients were in the recovery group, with 98 of 240 metabolites reported as significantly different. This finding may be of particular significance to so-called “Long COVID”, which bears certain similarities to the long-term reduction in quality-of-life (QoL) noted in survivors of other forms of severe illness [32, 33]. It is estimated that 50% of patients admitted to the ICU requiring mechanical ventilation go on to develop post-intensive care syndrome, and up to 80% of survivors of critical illness are readmitted to a nursing home, rehabilitation centre or ICU within 2 years after their initial illness. Half of survivors suffer long-term cognitive decline. Each additional day in the ICU can lead to an 11% loss in muscle mass even after 2 year follow-up. With many of these patients never returning to work, the healthcare system and society will be dealing with these issues many years after the pandemic is abated. Understanding how the metabolomic changes relate to QoL outcomes could potentially identify therapeutic strategies to improve long-term QoL in COVID-19 survivors.
Final strengths of this study were that many of the metabolites identified can predict patient outcomes, were validated in the independent cohort, and had strong overlap with other studies. A large meta-analysis with predictive modelling split between discovery and validation cohorts could provide valuable biomarkers, not only for prediction for SARS-CoV-2 infection, but also may determine whether acute and chronic outcomes in sepsis and ARF display pathways in common with COVID-19.
There were still some limitations in this report. As the authors note, their cohorts were not age-matched due to the fact that the severe cases were primarily observed in the elderly. Larger cohorts would provide further confidence in these results.
Considering the consistent metabolic derangements seen in multiple studies, as well as substantial overlap with metabolomics changes in sepsis and ARF, these results suggest that metabolic biomarkers should be regularly monitored to determine time-dependent changes during evolution of COVID-19. Future therapies could potentially consider how targeted nutraceutical interventions may return the metabolomics profiles back to homeostasis [34]. For example, Jia et al. [10] suggest further investigation into whether type I interferon regulates the urea cycle in infected epithelial cells and whether COVID-19 infection switches the metabolic pathway of glucose metabolism to the urea cycle by reducing nitric oxide production, thereby protecting viral replication. Other therapies that target the kynurenine pathway and the NAD+/NADH ratio may mitigate the bioenergetic crisis and NAD-regulated immune responses [8, 25]. Finally, strategies to suppress mitochondrial oxidant stress or repair oxidative mtDNA damage also have the potential to emerge as therapeutic strategies guided by targeted metabolomics monitoring [23, 35, 36].
Shareable PDF
Acknowledgements
As pointed out in the correspondence by Dr. Milad Zandi, published in this issue of the European Respiratory Journal (https://doi.org/10.1183/13993003.02818-2021), an early view publication of this manuscript suggested that ORF-8a is present in the SARS-CoV-2 genome. This inference has been removed from final published manuscript and clarity added regarding the function of ORF-8 in SARS-CoV-2. We thank Dr. Zandi for pointing out the important differences between the SARS and SARS-CoV-2 genomes.
Footnotes
Author contributions: All authors participated in writing and editing this manuscript, and approved the final manuscript.
Conflict of interest: All authors declare support from the National Institutes of Health for the present manuscript, with no further disclosures.
Support statement: Support was provided by the National Institutes of Health: 5KL2TR003097, NR01933801A1, UL1TR003096, R01HL11361406A1, R01GM127823. Funding information for this article has been deposited with the Crossref Funder Registry.
References
- 1.Park JJH, Mogg R, Smith GE, et al. How COVID-19 has fundamentally changed clinical research in global health. Lancet Glob Health 2021; 9: e711–e720. doi: 10.1016/S2214-109X(20)30542-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Song G, Cheng MQ, Wei XW. Analysis of the WHO ICTRP for novel coronavirus clinical trial registrations. Medicine (Baltimore) 2020; 99: e22840. doi: 10.1097/MD.0000000000022840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tregoning JS, Flight KE, Higham SL, et al. Progress of the COVID-19 vaccine effort: viruses, vaccines and variants versus efficacy, effectiveness and escape. Nat Rev Immunol 2021; 21: 626–636. doi: 10.1038/s41577-021-00592-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh S, Pandey R, Tomar S, et al. A brief molecular insight of COVID-19: epidemiology, clinical manifestation, molecular mechanism, cellular tropism and immuno-pathogenesis. Mol Cell Biochem 2021; 476: 3987–4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hasan MR, Suleiman M, Perez-Lopez A. Metabolomics in the diagnosis and prognosis of COVID-19. Front Genet 2021; 12: 721556. doi: 10.3389/fgene.2021.721556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuperlovic-Culf M, Cunningham EL, Teimoorinia H, et al. Metabolomics and computational analysis of the role of monoamine oxidase activity in delirium and SARS-COV-2 infection. Sci Rep 2021; 11: 10629. doi: 10.1038/s41598-021-90243-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lv L, Jiang H, Chen Y, et al. The faecal metabolome in COVID-19 patients is altered and associated with clinical features and gut microbes. Anal Chim Acta 2021; 1152: 338267. doi: 10.1016/j.aca.2021.338267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Migaud M, Gandotra S, Chand HS, et al. Metabolomics to predict antiviral drug efficacy in COVID-19. Am J Respir Cell Mol Biol 2020; 63: 396–398. doi: 10.1165/rcmb.2020-0206LE [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Langley RJ, Migaud ME, Flores L, et al. A metabolomic endotype of bioenergetic dysfunction predicts mortality in critically ill patients with acute respiratory failure. Sci Rep 2021; 11: 10515. doi: 10.1038/s41598-021-89716-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jia H, Liu C, Li D, et al. Metabolomic analyses reveal new stage-specific features of the COVID-19. Eur Respir J 2022; 59: 2100284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karmouty-Quintana H, Thandavarayan RA, Keller SP, et al. Emerging mechanisms of pulmonary vasoconstriction in SARS-CoV-2-induced acute respiratory distress syndrome (ARDS) and potential therapeutic targets. Int J Mol Sci 2020; 21: 8081. doi: 10.3390/ijms21218081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Langley RJ, Wong HR. Early diagnosis of sepsis: is an integrated omics approach the way forward? Mol Diagn Ther 2017; 21: 525–537. doi: 10.1007/s40291-017-0282-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langley RJ, Tsalik EL, Velkinburgh JC, et al. An integrated clinico-metabolomic model improves prediction of death in sepsis. Sci Transl Med 2013; 5: 195ra195. doi: 10.1126/scitranslmed.3005893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rogers AJ, McGeachie M, Baron RM, et al. Metabolomic derangements are associated with mortality in critically ill adult patients. PLoS ONE 2014; 9: e87538. doi: 10.1371/journal.pone.0087538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langley RJ, Tipper JL, Bruse S, et al. Integrative ‘omic’ analysis of experimental bacteremia identifies a metabolic signature that distinguishes human sepsis from systemic inflammatory response syndromes. Am J Respir Crit Care Med 2014; 190: 445–455. doi: 10.1164/rccm.201404-0624OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shenoy S. Coronavirus (Covid-19) sepsis: revisiting mitochondrial dysfunction in pathogenesis, aging, inflammation, and mortality. Inflamm Res 2020; 69: 1077–1085. doi: 10.1007/s00011-020-01389-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodriguez-Nuevo A, Zorzano A. The sensing of mitochondrial DAMPs by non-immune cells. Cell Stress 2019; 3: 195–207. doi: 10.15698/cst2019.06.190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krysko DV, Agostinis P, Krysko O, et al. Emerging role of damage-associated molecular patterns derived from mitochondria in inflammation. Trends Immunol 2011; 32: 157–164. doi: 10.1016/j.it.2011.01.005 [DOI] [PubMed] [Google Scholar]
- 19.Scozzi D, Cano M, Ma L, et al. Circulating mitochondrial DNA is an early indicator of severe illness and mortality from COVID-19. JCI Insight 2021; 6: e143299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ajaz S, McPhail MJ, Singh KK, et al. Mitochondrial metabolic manipulation by SARS-CoV-2 in peripheral blood mononuclear cells of patients with COVID-19. Am J Physiol Cell Physiol 2021; 320: C57–C65. doi: 10.1152/ajpcell.00426.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi TT, Yang FY, Liu C, et al. Angiotensin-converting enzyme 2 regulates mitochondrial function in pancreatic beta-cells. Biochem Biophys Res Commun 2018; 495: 860–866. doi: 10.1016/j.bbrc.2017.11.055 [DOI] [PubMed] [Google Scholar]
- 22.Singh KK, Chaubey G, Chen JY, et al. Decoding SARS-CoV-2 hijacking of host mitochondria in COVID-19 pathogenesis. Am J Physiol Cell Physiol 2020; 319: C258–C267. doi: 10.1152/ajpcell.00224.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuck JL, Obiako BO, Gorodnya OM, et al. Mitochondrial DNA damage-associated molecular patterns mediate a feed-forward cycle of bacteria-induced vascular injury in perfused rat lungs. Am J Physiol Lung Cell Mol Physiol 2015; 308: L1078–L1085. doi: 10.1152/ajplung.00015.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ganji R, Reddy PH. Impact of COVID-19 on mitochondrial-based immunity in aging and age-related diseases. Front Aging Neurosci 2020; 12: 614650. doi: 10.3389/fnagi.2020.614650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Habeichi NJ, Tannous C, Yabluchanskiy A, et al. Insights into the modulation of the interferon response and NAD+ in the context of COVID-19. Int Rev Immunol 2021; in press [ 10.1080/08830185.2021.1961768]. doi: 10.1080/08830185.2021.1961768 [DOI] [PubMed] [Google Scholar]
- 26.Wu J, Shi Y, Pan X, et al. SARS-CoV-2 ORF9b inhibits RIG-I-MAVS antiviral signaling by interrupting K63-linked ubiquitination of NEMO. Cell Rep 2021; 34: 108761. doi: 10.1016/j.celrep.2021.108761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen CY, Ping YH, Lee HC, et al. Open reading frame 8a of the human severe acute respiratory syndrome coronavirus not only promotes viral replication but also induces apoptosis. J Infect Dis 2007; 196: 405–415. doi: 10.1086/519166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Chen Y, Li Y, et al. The ORF8 protein of SARS-CoV-2 mediates immune evasion through down-regulating MHC-Iota. Proc Natl Acad Sci USA 2021; 118: e2024202118. doi: 10.1073/pnas.2024202118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dudek J. Role of cardiolipin in mitochondrial signaling pathways. Front Cell Dev Biol 2017; 5: 90. doi: 10.3389/fcell.2017.00090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi CS, Qi HY, Boularan C, et al. SARS-coronavirus open reading frame-9b suppresses innate immunity by targeting mitochondria and the MAVS/TRAF3/TRAF6 signalosome. J Immunol 2014; 193: 3080–3089. doi: 10.4049/jimmunol.1303196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prasun P. COVID-19: a mitochondrial perspective. DNA Cell Biol 2021; 40: 713–719. doi: 10.1089/dna.2020.6453 [DOI] [PubMed] [Google Scholar]
- 32.Jaffri A, Jaffri UA. Post-intensive care syndrome and COVID-19: crisis after a crisis? Heart Lung 2020; 49: 883–884. doi: 10.1016/j.hrtlng.2020.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chippa V, Aleem A, Anjum F. Post Acute Coronavirus (COVID-19) Syndrome. Treasure Island, StatPearls, 2021. [PubMed] [Google Scholar]
- 34.James PT, Ali Z, Armitage AE, et al. The role of nutrition in COVID-19 susceptibility and severity of disease: a systematic review. J Nutr 2021; 151: 1854–1878. doi: 10.1093/jn/nxab059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan YB, Pastukh VM, Gorodnya OM, et al. Enhanced mitochondrial DNA repair resuscitates transplantable lungs donated after circulatory death. J Surg Res 2020; 245: 273–280. doi: 10.1016/j.jss.2019.07.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fock EM, Parnova RG. Protective effect of mitochondria-targeted antioxidants against inflammatory response to lipopolysaccharide challenge: a review. Pharmaceutics 2021; 13: 144. doi: 10.3390/pharmaceutics13020144 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This one-page PDF can be shared freely online.
Shareable PDF ERJ-02417-2021.Shareable (275.3KB, pdf)