Abstract
Introduction
An intricate balance with our intestinal microbes is pivotal to health. A key interface of host-microbial interactions occurs in the mucus covering the intestinal epithelial surface. In the colon, the mucus layer serves as a barrier inhibiting direct epithelial contact with the dense population of microbes. Defects in this system are a hallmark of colitis. The mucus layer is structurally dependent on the polymeric mucin MUC2, and it is synthesized by goblet cells (GCs), specialized secretory cells classically viewed as a homogenous cell type. Studies identifying divergent functional features in GC subpopulations, including differential mucus biosynthesis rate and responses to bacteria, suggests GC populations may in fact be heterogeneous.
Rationale
In the present study, we characterized intestinal GC expression diversity, and defined how a specific GC subtype, localized in the intercrypt surface epithelium, functionally contributes to the formation of the mucus barrier.
Results
Using mCherry-MUC2 transgenic mice to sort and isolate GCs, we generated transcriptomic and proteomic profiles to characterize the GC expression landscape in both the small intestine and colon. Single cell transcriptomic analysis revealed several unique GC clusters in each tissue regions, which segregated into two separate trajectories. One trajectory had enriched expression of known GC-specific genes (e.g., Clca1, Fcgbp), and was designated as canonical GCs. Conversely, the other trajectory was enriched for expression of genes typically associated with enterocytes (e.g., Dmbt1, Gsdmc4), which we designated as non-canonical GCs.
In the colon, the most differentiated GCs are the high mucus turnover cells localized to the surface epithelium between crypts and were thus designated as intercrypt GCs (icGCs). These cells had distinct expression profiles from crypt-resident GCs, thus we investigated their role in forming the mucus barrier. We exploited the lectin binding features of mucus to resolve the three-dimensional organization of mucus in live tissue explants. Results demonstrated that icGCs secreted distinct mucus that filled the spatial regions between mucus plumes secreted from crypt openings. The intercrypt mucus was impenetrable to bacteria-sized beads; however, it was more penetrable to smaller molecules compared to crypt plume mucus. Penetrable surface mucus may be important for absorption of ions, and other compounds, while denser mucus within the crypt compartment contributes to the shielding of the stem cell niche.
Both mucus subtypes appear to be important for the overall protective function of mucus, as a barrier impenetrable to bacteria was formed by the mixed net-like organization of intercrypt and crypt plume mucus. A mouse model with dysfunctional icGCs lacking normal intercrypt mucus exhibited an inadequate mucus barrier and was more susceptible to both chemically-induced and spontaneous (age-dependent) colitis, thus arguing for the indispensable role of icGCs in maintaining a functional mucus barrier. Furthermore, biopsies from patients with ulcerative colitis, also in remission, exhibited increased GC shedding and reduced icGC numbers. These characteristics was associated with structural defects in the mucus barrier, including gaps in the intercrypt mucus that exposed areas of the surface epithelium.
Conclusion
The current study identifies GCs as a heterogeneous population of cells with diverse functional features that indicate a dynamic cellular system. GCs at different locations along the crypt-surface axis contribute to a functional mucus barrier that protects the epithelium from microorganisms. The icGCs possess a unique role in mucus organization, where their malfunction is associated with colitis in both mice and humans.
Keywords: Single cell RNA sequencing, Spdef, Muc2, mucin, epithelium
Diversity of GC expression and function
Intestinal GCs can be divided into several distinct clusters that form canonical and non-canonical trajectories, and gene expression profiles outline their topographical position. The specialized surface GC type, the icGCs, secrete mucus crucial for forming a protective barrier between bacteria and the epithelium. Depletion of icGCs impairs mucus function, a phenomenon observed in ulcerative colitis (UC) patients.

The intestinal mucus layer, a vital element of epithelial protection, is produced by goblet cells. Intestinal goblet cells are assumed to be a homogenous cell type. Herein, we delineated their specific gene and protein expression profiles and identified several distinct goblet cell populations forming two differentiation trajectories. One distinct subtype, the intercrypt goblet cells (icGCs), located at the colonic luminal surface, produced mucus with properties that differed from mucus secreted by crypt residing goblet cells. Mice with defective icGCs had increased sensitivity to chemically induced colitis and manifested spontaneous colitis with age. Furthermore, alterations in mucus and reduced number of icGCs were observed in patients with both active and remissive ulcerative colitis, highlighting the importance of icGCs in maintaining functional protection of the epithelium.
Introduction
Mucus secreted from the intestinal epithelium forms a continuous layer throughout the intestinal tract (1, 2). A primary function of mucus is to protect the epithelium against luminal microbes. In the distal colon, this is achieved by an inner mucus layer on the tissue surface, which creates a physical barrier between the bacteria and the epithelium (3-5). The inner mucus layer is modified to form an expanded outer mucus that is more accessible to bacteria (6); mucus along the whole intestine facilitates fecal transport (6). In the small intestine, a single layer of permeable mucus fortified by antimicrobial peptides protects the epithelium (1, 2).
The primary structural component of the mucus, MUC2, is a large multimeric and heavily O-glycosylated protein. The exposed terminal glycans form an outer surface on MUC2 that can be recognized by bacteria and lectins (7). Upon secretion, oligomers of MUC2 and other mucus components form a sieve-like structure, allowing passage of small molecules whilst hindering translocation of microorganisms. To date, a comprehensive understanding of mucus organization remains to be elucidated.
The vital importance of a functional mucus barrier is demonstrated by mice lacking Muc2, which develop spontaneous colitis (8). Furthermore, several observations have correlated mucus defects with the pathogenesis of ulcerative colitis (UC) and with experimental mouse models of colitis (9-11). Although mucus disruption is observed prior to intestinal inflammation and colitis induced by dextran sodium sulfate (DSS) in mice (12), it remains uncertain whether mucus dysfunction in humans is a cause or consequence of colitis-associated inflammation.
Mucus constituents, including MUC2, are secreted by goblet cells (GCs), and their high expression and complex structure put a high demand on the cellular biosynthesis machinery. GCs express specific proteins involved in these processes as AGR2 and ERN2 (13, 14) (all gene and protein names are listed in Table S1). As with all intestinal epithelial cells, GCs are derived from crypt-residing intestinal stem cells. GCs differentiation is promoted by several transcription factors, including ATOH1 and SPDEF, whereas they are negatively regulated by HES1 (15-18). Global single cell analysis of the intestinal epithelium has often classified GCs as a single population (19-21). However, a subpopulation of GCs only detected in active UC has recently been described in humans (22), suggesting dynamic differentiation of GCs during inflammation.
In the present study, we characterized GC transcript and protein profiles in both small and large intestine. Single cell analysis identified several clusters, forming two distinct differentiation trajectories. GCs in the intercrypt region of the colonic surface epithelium, intercrypt GC (icGCs), possessed a unique transcript profile and produced distinct, more penetrable mucus that filled the spatial regions between mucus plumes secreted from crypt-resident GCs. Spdef−/− mice showed secretory defects in the icGC population and altered mucus architecture. These changes contribute to increased susceptibility to DSS-induced colitis, as well as increased incidence of spontaneous colitis. In a majority of UC patients, both in remission (UCr) and with active disease (UCa), a reduced number of icGCs and an altered mucus organization was observed. Thus, intestinal GCs are remarkably heterogeneous, with colonic icGCs constituting an essential subpopulation for normal formation and function of the mucus barrier.
Goblet cell specific transcript and protein expression
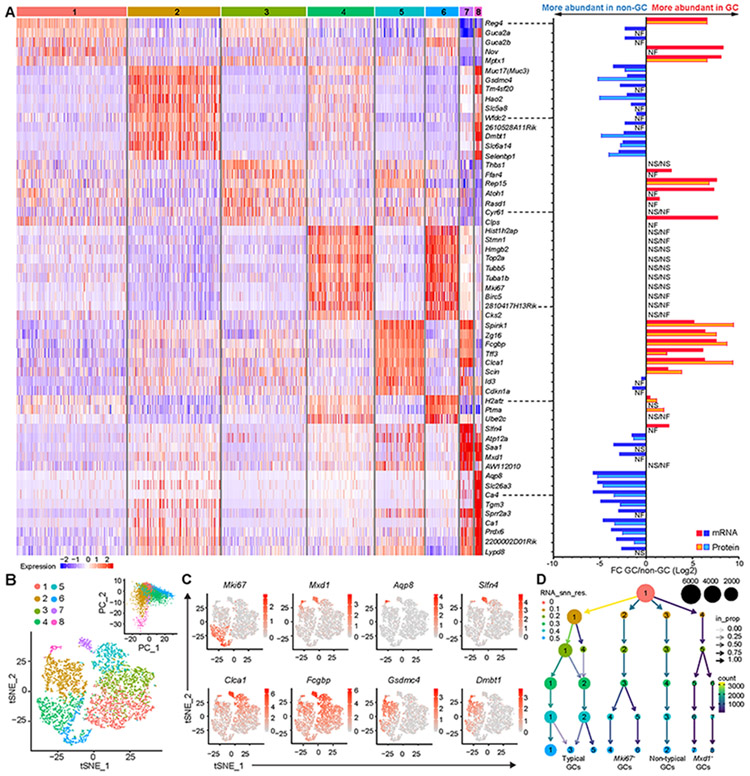
We isolated mCherry-MUC2 positive cells by FACS from the distal colon and the most distal small intestine of transgenic RedMUC298trTg mice (23) (Fig. S1, A and B). GCs comprised 17% and 5% of viable epithelial cells in the colon and small intestine, respectively (Fig. S1C). Bulk RNA sequencing (RNA-seq) and proteomics by mass spectrometry were performed comparing GCs to the non-GC pool (Table S2 and S3). Principal Component Analysis (PCA) grouped samples based both on cell type and origin (Fig. S1D). Protein and mRNA showed good overlap and Pearson’s correlation coefficient comparing mRNA and protein was 0.57 in the colon and 0.63 in the small intestine (Fig. S1, E and F). Comparison of GC and non-GC populations identified approximately 500 proteins and 800 transcripts significantly enriched in GCs (Fig. 1, A and B). Many of the GC enriched genes were shared between the small intestine and colon, but some showed site-specificity (Fig. 1C). Several known mucus components such as Muc2 (endogenous), Clca1, Fcgbp and Zg16 were among the 50 most abundant GC enriched genes and proteins. Gene Ontology (GO) enrichment analysis primarily classified the GC expression profile into secretory related processes but also identified lipid metabolism, immune response, proteolysis, and protein glycosylation (Figs. 1D and S2).
Fig. 1. Multi-omics analysis of purified GCs from the distal colon (DC) and the distal small intestine (Si8).
(A) Volcano plots of transcriptome and proteome in GC and non-GC populations. (B) Venn diagram indicating the number of genes and proteins associated with each population using fold change (FC) values ≥ 2 and adjusted-p value ≤ 0.05 (protein) or ≤ 0.01 (mRNA). (C) Heat maps of the 50 genes and proteins most differentially expressed in the GCs compared to non-GCs. (D) Biological processes upregulated in GCs according to Gene Ontology (GO) enrichment analysis. Four replicates (pool of two mice per replicate).
Goblet cells comprise several different clusters
To study the intrinsic variability of GCs under basal conditions, we performed single cell RNA-seq (scRNA-seq) on sorted GCs from RedMUC298trTg mice. Colonic GCs grouped into eight different clusters (Fig. 2, A and B, and Table S4). Clusters 6 and 4 were enriched for the proliferation marker Mki67, whereas clusters 7 and 8 were enriched for Mxd1, a gene expressed by fully differentiated cells (24). Mxd1+ cells were further separated by more specific markers, Slfn4 and Aqp8. Furthermore, GCs showed two main gene expression profiles; Clusters 1, 3, 5, and 7 exhibited a higher expression of typical GC-associated genes such as Clca1 and Fcgbp (1, 25), whereas clusters 2 and 8 were enriched in expression of Gsdmc4, Dmbt1 or Aqp8; all more abundant in the non-GC population according to bulk RNA-seq (Fig. 2, A and C). PCA analysis showed a higher diversity within the clusters associated with non-GC genes and with proliferation (Fig. 2B). A cluster tree revealed four main groups of GCs: typical, proliferative (Mki67+), non-typical, and fully differentiated (Mxd1+) (Fig. 2D).
Fig. 2. Different clusters identified among the Muc2+ cells in the distal colon by single-cell analysis.
(A) Left panel: Heat map of up to 10 enriched genes per cluster; Right panel: mRNA and protein FC of GC to non-GC genes displayed in the heat map. (B) tSNE plot of colonic GCs grouped in eight clusters and principal component analysis (PCA) of cluster diversity. (C) tSNE plots highlighting expression and distribution of selected genes. (D) Cluster tree showing the four main subgroups of GCs during the clustering process. Two replicates (two mice per replicate) were combined for the analysis. FC: Fold change. NS: Not significant; NF: Not found.
Small intestinal GCs grouped into nine clusters (Fig. S3, A and B, and Table S4) when Muc2 expressing Paneth cells were removed (26) (Fig. S4). Proliferative genes were abundant in clusters 2 and 9 (Mki67+). Fully differentiated GCs (Mxd1+) expressed Ido1 and were further divided into clusters 5 and 8. Typical GC-associated genes (Clca1 and Fcgbp) were most highly expressed in clusters 1, 3, 5 and 8, while cluster 4 showed a more pronounced non-GC profile (Sis, Dmbt1, Gsdmc4) and was more diverse in the PCA (Fig. S3, A to D). Expression of typical Paneth cell products such as Defa24 and Lyz1 was also noted in several clusters, but highest in cluster 3 and 6 (Fig. S3, A and C).
Maturation trajectories of intestinal goblet cells
To investigate whether GC populations follow distinct trajectories throughout their differentiation and maturation, we performed a pseudotime analysis (Table S5). Colonic GCs revealed two trajectories originating from proliferative cells (Mki67+) at the lower region of the crypt. Typical GC markers (Atoh1, Fcgbp and Clca1) were enriched in one trajectory designated as canonical GCs. Expression of these genes were found in Muc2-expressing cells along the crypt (Fig. 3, A to C). A second trajectory comprised cells with a non-GC profile and was designated non-canonical GCs as markers (Hes1, Gsdmc4 and Dmbt1) for this trajectory were more abundant in Muc2-negative cells (Fig. 3, A to C). The terminals of the canonical and non-canonical GC trajectories ended in two small populations of Mxd1+ GCs highly expressing Slfn4 or Aqp8 that were localized at the surface epithelium (Fig. 3, A to C). Similar results were obtained analyzing individual datasets, and the same GC expression pattern of non-canonical GC markers, as seen in RedMUC298trTg mice, was also observed in wild type (WT) mice (Fig. S5, A and B). These trajectories were further supported by expression of different transcription factors. Specifically, Atoh1 was found in most GCs, especially in the canonical GCs, whereas Hes1 was expressed in some GCs along the crypt following the non-canonical trajectory (Fig. 3, B and C).
Fig. 3. Pseudotime analysis unveils two trajectories of GCs in the distal colon.
(A) Trajectory and Partition-based graph abstraction of colonic GCs. Two replicates (2 mice per replicate) were combined. (B) Expression of selected genes along the described trajectories. (C) mRNA-in situ hybridization of genes enriched in different clusters counterstained with Muc2, Epcam for cell borders and Hoechst for DNA. Scale bars = 25 μm. (D) tSNE plot, scatter plot of the bulk RNA-seq data of genes significantly enriched in the canonical and non-canonical GC populations, and biological processes upregulated in canonical GCs compared to non-canonical GCs according to GO enrichment analysis. (E) tSNE plots highlighting expression of sentinel GC signaling genes and genes dysregulated during inflammation.
We next compared functions of genes associated with canonical and non-canonical GC trajectories in the colon (Table S6). Gene ontology (GO) enrichment analysis of differentiated colonic GCs (Mki67−) revealed that processes related to secretion, glycosylation, and endoplasmic reticulum stress were enriched in the canonical GCs, whereas non-canonical GCs demonstrated enrichment of genes associated with lipid and amino acid metabolism, detoxification, and intestinal absorption (Fig. 3D). Expression levels of Il18 and Nlrp6, genes associated with sentinel GCs (senGCs) (23), were higher in the non-canonical population. Wfdc2 and Areg, genes whose expression is altered with colitis (22, 27), were also enriched in the non-canonical GCs (Fig. 3E).
Both pooled and individual datasets from the distal small intestine reveal a trajectory starting from the proliferative pool of GCs that leads to a branching structure (Figs. S6, A and B, and S5C). Unlike colon, this trajectory was dominated by canonical GCs with a later bifurcation made up of non-canonical GCs. The most mature canonical GCs present at the tip of the villi, were marked by the expression of Ido1. Defa24+ cells were spread along the canonical trajectory and identified in several GCs of the crypt, whereas Lyz1 was limited to fewer GCs (Fig. S6, B to D)
Intercrypt goblet cells
The two colonic clusters of fully differentiated GCs (cluster 7 and 8, Mxd1+) expressed many genes in common, but differed as cluster 8 expressed several genes shared with non-GC epithelial cells (Fig. 2, A and B). Markers for these GCs were only detected at the surface epithelium in the intercrypt area (Fig. 3C). Functional profiling of these cells compared to non-proliferative crypt GCs revealed increased expression of genes involved in cellular response to stress and bacteria, likely related to their luminal facing location (Fig. 4A and B, S7A, and Table S6). Additionally, processes such as programmed cell death and negative regulation of differentiation were upregulated as a consequence of their late stage of differentiation. They were also enriched in protein transport-associated genes (Fig. 4, A and C). Several genes involved in protein folding and quality control (Agr2, Pdia4-6) were expressed at lower levels in these cells compared to crypt GCs (Fig. 4D).
Fig. 4. Gene expression in icGCs.
(A) Biological processes upregulated in the surface located GCs (Mxd1+) compared to crypt-residing GCs according to GO enrichment analysis. (B, C, D) Violin plots of gene expression related to response to bacteria (B) protein transport (C) and processing of Muc2 (D) in the different clusters. (E) Staining with WGA, UEA1, the nonO-glycosylated Muc2 precursor and Hoechst for DNA in a colonic cross section. Arrowheads indicate crypt-opening GC (yellow) and an icGC (pink). Scale bars = 20 μm, and 10 μm in magnification; n = 24. (F) Mouse colonic section stained with AB-PAS or mature Muc2 with icGCs indicated (pink arrowheads). Epithelial surface (dashed line), scale bars = 50 and 20 μm (left and right panel in AB-PAS). (G) Violin plots of mucus gene expression in the different clusters. (H) Violin and tSNE plots showing expression of Spdef in GCs.
The possibility to use lectin staining to differentiate GCs at different locations in the epithelium was determined. The wheat germ agglutinin (WGA) lectin (N-acetylglucosamine/sialic acid-binding) detected crypt-residing GCs in fixed tissue, whereas the Ulex europaeus agglutinin I (UEA1) (fucose-binding) readily detected GCs of the upper crypt and surface epithelium (Fig. 4E). Lectin specificity is dependent on the combined effect of the glycosyltransferases expressed and glycan presentation, differential lectin staining could thus not be fully explained by glycosyltransferase expression (Fig. S7B). UEA1+/WGA− GCs were restricted to the intercrypt surface epithelium. Accordingly, these cells were designated as intercrypt GCs (icGCs). In addition to specific lectin reactivity, icGCs were also strongly stained by antiserum to the nonO-glycosylated Muc2 precursor found in the endoplasmic reticulum (Fig. 4E). However, they were barely detectable using AB/PAS or immunostaining of mature Muc2, which instead primarily labelled the theca in crypt-residing GCs (Fig. 4F). This difference was likely due to the rapid secretion and reduced storage of mature Muc2 in icGCs compared to crypt-residing GCs(28).
The icGCs, as defined by their staining, shape and location include all intercrypt surface GCs, but appear to be primarily comprised of the most differentiated canonical GCs (cluster 7, high Slfn4 expression) in line with their high expression of mucus associated genes, such as Muc2, Clca1 and Fcgbp (Fig. 4G). They show a functional profile with response to stress, cell differentiation, apoptosis and protein transport (Fig. S7C). For example, expression of the constitutive secretory regulator Stxbp1 was high, while the regulated secretory equivalent Stxbp2 was predominantly expressed in crypt GCs. Higher expressions of the endoplasmic reticulum stress related gene Ern2 combined with altered expression of proteins involved in folding indicated functional alterations in protein biosynthesis (Fig. 4D), and a subset of icGCs also expressed the immunomodulatory gene Ido1 (Fig. S7, D and E). Although icGCs mainly consist of canonical GCs, the contribution of non-canonical surface GCs (cluster 8) cannot be excluded. The transcription factor Spdef, thought to drive terminal GC differentiation, was expressed by most GCs, but was found to be more abundant in the canonical linage clusters, including icGCs (Figs. 4H, and S7F).
Unlike in the colon, expression of the icGC marker Slfn4 was not restricted to GCs in the distal small intestine, suggesting that cells identical to icGCs are absent in the distal small intestine. Nevertheless, the cluster of Mxd1+/Ido1+ GCs present at the villi could potentially represent a similar subset; however, additional research is required for confirmation (Fig. S8).
Colonic mucus organization and properties
Using fluorescent-conjugated UEA1 and WGA, either alone or in combination, to stain secreted mucus in flushed colonic tissue ex vivo, revealed two distinctive staining patterns: WGA stained plume-like structures positioned on top of crypt openings (crypt plumes), whereas UEA1 primarily stained mucus material filling the space between plumes (intercrypt mucus) (Figs 5, A and B, and S9, A to C, and Movie S1). Intensity histograms and low co-localization coefficients in tissue proximate mucus confirmed that UEA1 and WGA primarily stained different material (Figs. 5C, and S9, D and E). Isosurface mapping demonstrated that UEA1-postive intercrypt mucus emerged from icGCs, whereas WGA-positive material emerged from the crypt (Fig. 5D). The UEA1- and WGA-stained fractions intermixed on top of the plumes and merged to form a dense net-like arrangement at the mucus surface (Fig. 5B), as demonstrated by an increased co-localization coefficient in the mix zone and at the surface (Fig. S9E). Mucus not subjected to flushing had a staining pattern mirroring that observed in flushed mucus; however, the mix zone appeared thicker, and the plumes smaller (Fig. S9F). Formations resembling the crypt-covering mucus plumes could also be observed by staining of fixed colonic tissue sections (Fig. S9G). However, the mucus plumes were also intensely stained by UEA1, indicating different lectin binding depending on method. Consequently, the observed mucus organization observed ex vivo is also likely present in vivo.
Fig. 5. Colonic mucus organization and penetrability.
(A) UEA1 and WGA-stained flushed mouse colonic mucus in x/z-projection with marked (white arrowheads) crypt-openings. Color code arrowheads match images in B. (B) x/y projections at different z-stacks levels as indicated in A. Magnification of the mixed zone mucus (yellow arrowheads mark WGA and UEA1 fusion points); n = 30. (C) Intensity profile of UEA1 and WGA along a straight line in tissue proximate mucus (Fig. S9). Normalized raw data in pale color, crypt-openings (arrows); n = 17. (D) Isosurface overlay of UEA1 and WGA-stained mucus secreted from icGCs (pink arrowheads) or a crypt-opening (yellow arrowhead). (E) x/y and x/z projections of 0.2 μm fluorescent beads in WGA-stained colonic mucus. Areas with low bead concentration (dashed line), underlying crypt-openings (purple spheres), z-position for x/y projections (arrowhead); n = 6. Scale bars: 50 μm (B, E), 25 μm (A, B magnification) and 20 μm (D).
Bacteria-sized 1 μm beads settled on the mucus surface (Fig. S10A). On the contrary, 0.2 μm beads penetrated the mucus surface and intercrypt mucus and reached the epithelium. However, crypt plume mucus excluded even the 0.2 μm beads (Fig. 5E, and S10B). To rule out that intrinsic properties of the beads affected their distribution, FITC-conjugated dextran with a molecular mass of 2,000 kDa (FD2000) was used. Similar to the 0.2 μm beads FD2000 penetrated down to the epithelium in the intercrypt regions but did not fully penetrate the crypt plumes (Fig. S10C). These findings suggest that the crypt plumes have more stringent barrier properties compared to intercrypt mucus.
Inducing ion and liquid flow by forskolin treatment, or reducing it using Cftr-deficient mice did not dramatically alter the overall mucus structure, although the volume of the crypt plumes appeared to increase with forskolin and decrease with impaired Cftr activity (Fig. S10, D and E). The ability of the crypt plumes to exclude 0.2 μm beads remained with deficient Cftr function. Thus, liquid secretion does not seem to determine overall mucus structure.
Altered icGCs in Spdef−/− mice
The transcription factor Spdef is thought to be involved in the terminal differentiation of GCs (16, 17), and as it is expressed in the most differentiated GCs, the icGCs, we investigated GC effects in Spdef deficient mice. Colonic GCs from 6 week-old Spdef−/− (crossed with RedMUC298trTg) mice showed altered expression of several GC-cluster marker genes, primarily in the canonical linage, but also in the proliferative cells, compared to WT GCs (Fig. 6A). Furthermore, several of the genes either ≥2-fold up- or down-regulated in Spdef-deficient compared to WT GCs were more abundant in the icGCs, but genes encoding the main mucus components (5) were unaltered (Fig. 6B, S11A, and Table S7). Many icGC genes were markedly reduced in Spdef−/− mice, whereas Ido1, expressed by few icGCs, initially showed a slight increase in expression and later expanded to more cells (Figs. 6 A and B, and S11, B and C). Thus, the absence of Spdef induce several alteration but appeared to have a pronounced effect on icGCs.
Fig. 6. icGC alterations in Spdef−/− colon.
(A) Fold change (FC) expression of single GC cluster (color bars) marker genes in Spdef−/− versus WT with adjusted p-values. (B) FC expression among the previously defined single cell clusters (left) of the most up- and down-regulated genes in Spdef−/− compared to WT GCs obtained by bulk RNA seq with adjusted p-values (right). (C) UEA1, cell membrane and DNA staining of whole-mounted colonic tissue from 7 and 13 week-old WT and Spdef−/− mice. Magnified areas (white boxes), apical surface of icGCs (arrowheads), scale bars = 20 μm. (D) Quantification of the number of icGCs per mm2 in whole-mounted colons from WT and Spdef−/− mice. Mean ± SD, analyzed by 2-way ANOVA with Sidak’s multiple comparison test; n= 4 - 8. (E) TEM micrographs of icGCs in WT and Spdef−/− colon with magnifications. Scale bars = 2 μm; n = 4. (F) SEM scans of the icGCs and the epithelial surface in WT and Spdef−/− distal colon. Identified GC openings (red arrowheads), scale bars = 1 μm (upper) 20 μm (lower), n = 4. (G) In vivo labeling for 2 and 4 h by GalNAz detected by Alkyne-TAMRA on colonic sections of 13-week old WT and Spdef−/− mice. Scale bar = 20 μm; n = 4.
Lectin-based investigation of icGCs in colonic whole-mounted tissue from 7 and 13 week-old Spdef−/− mice showed normal icGC and crypt numbers, but revealed age-dependent icGC morphological alterations (Figs. 6, C and D, and S12, A to C). At 7 weeks of age Spdef−/− icGCs appeared largely unaffected, whereas in 13 week-old Spdef−/− mice the apical membrane staining of icGCs was more intense, and they contained less UEA1 stained material. Transmission electron microscopy (TEM) further revealed gross morphological changes in icGCs, which had increased number of microvilli at the apical membrane, enlarged ER, and reduced theca size, in line with previous observations (17), whereas other GCs appeared normal (Figs. 6E, and S12D). This can explain the difference in staining patterns observed (Fig 4F).The alterations of the apical membrane in icGCs of Spdef−/− mice were even more evident by scanning electron microscopy (SEM), resulting in quantitatively lower number (Fig. 6F and S12E).
To investigate defects in mucus secretion and turnover, in vivo labeling of O-glycans with N-azidoacetylgalactosamine (GalNAz) was performed (28). Mucus turnover in icGCs and crypt GCs of Spdef−/− mice was similar to WT mice at 13 weeks of age; however, Spdef−/− icGCs, but not crypt GCs, contained less mature mucus compared to their WT counterparts (Figs. 6G, and S12 F and G). The combined results reveal dramatic effects on icGCs in Spdef−/− mice.
Altered mucus phenotype and susceptibility to colitis in Spdef−/− mice
We next investigated the contribution of icGCs to mucus layer organization and function. At 13 weeks of age, Spdef−/− mice largely lacked distinct intercrypt mucus. UEA1 instead primarily stained the WGA-positive crypt plumes (Fig. 7, A and B), which was further confirmed with both isosurface overlay and intensity histogram analysis (Fig. 7, C and D, and S13A). Closer examination of the Spdef−/− mucus surface mesh indicated that it mainly originated from the crypt plumes and appeared less dense than in WT explants (Fig. 7B and 5B). Using immunostaining for the Muc2 nonO-glycosylated precursor as an icGC marker, UEA1-stained material could be detected in icGCs in 13 week-old Spdef−/− mice, indicating that lectin reactivity of mucus produced by Spdef−/− icGCs was retained but decreased (Fig. S13B).
Fig. 7. Age-dependent loss of intercrypt mucus and colitis in Spdef−/− mice.
(A) UEA1- and WGA-stained Spdef−/− colonic mucus in x/z-projection. Crypt-openings (white arrowheads), crypt plumes (dashed line). Color code arrowheads match images in B. (B) x/y projections at different z-stack levels as indicated in A. Magnification of mucus from the crypt plumes expanding to form the mucus surface mesh (arrowhead); n = 12. (C) Isosurface overlay of UEA1 and WGA. Underlying crypt-openings marked by green spheres. (D) Intensity profile of UEA1 and WGA in tissue proximate mucus (Fig. S12). Normalized raw data (pale color), crypt openings (arrows); n = 3. (E) 1 μm beads on WT and Spdef−/− colonic mucus. Mucus surfaces (dashed lines), beads at tissue (arrowhead); n = 4 - 5. (F) Normalized bead penetrability; n = 4 - 5. (G) Bacteria FISH, UEA1 and WGA in sections from WT and Spdef−/− colon; n = 7. (H) Colitis score of Spdef−/− and WT mice treated with 3% DSS; n = 3 - 13 (I) Histological colitis score in 26 week-old WT and Spdef−/− mice. Normal score range of 6-19 week-old WT (n = 14, data not shown) was set as the colitis cut-off (dashed line). (J) AB-PAS stained colonic sections from 26 week-old WT and Spdef−/− mice. Infiltrating inflammatory cells (arrowheads); n = 7. (K) Bacteria FISH in sections from 26 week-old WT and Spdef−/− animals. Epithelial surface (dashed line), bacteria at the epithelium (arrowheads); n = 4. Scale bars = 50 μm. * in images indicate shrinkage artifacts due to fixation. Graphs shows mean ± SD analyzed by 2-way ANOVA with Tukey’s multiple comparison test (F and H) or unpaired Student’s t-test (I).
Mucus penetrability by 1 μm beads was unaltered in 7 week-old Spdef−/− mice compared to WT but the mucus was found to be thinner and more penetrable in 13 week old animals (Fig. 7E and F, and S13C). Nevertheless, crypt-openings remained protected by the plumes (Fig. S13D). A deficient mucus barrier in 13 week-old Spdef−/− animals was confirmed by fluorescent in situ hybridization (FISH). Gut-residing bacteria were separated from the epithelium by a continuous mucus layer in WT and 7 week old Spdef−/−mice, whereas defects in the mucus layer was observed in the older Spdef−/− mice (Fig. 7G).
Spdef−/− and WT mice at 7 weeks of age responded similarly to DSS-induced colitis, whereas colitis was more severe in the 13 week old Spdef−/− mice (Fig. 7H, and S13E). Notably, Spdef−/− mice required euthanasia at 6 days of DSS treatment according to ethical guidelines (Fig. S13F). Additionally, at 26 weeks of age almost all Spdef−/− mice exhibited signs of spontaneous colitis by histopathological examination of colonic tissue sections (Fig. 7, I and J, and fig. S13G). FISH revealed bacteria in proximity to the epithelial surface and at the bottom of the crypts in Spdef−/− mice, whereas bacteria were clearly separated from the tissue in WT colon (Fig. 7K). These results indicated that there is an age-dependent loss of colonic mucus barrier integrity, in Spdef−/− mice where icGC abnormalities are likely a contributing factor
Fewer icGCs and mucus alterations in ulcerative colitis patients
In human sigmoid colon tissue sections, icGCs were strongly stained by the Lycopersicon esculentum lectin (LEL) (N-acetylglucosamine-binding), but stained only weakly or negative for Lotus tetragonolobus lectin (LTL) (fucose-binding). Conversely, crypt-residing GCs were intensely stained by LTL and largely negative or weakly stained for LEL. The WGA lectin stained GCs throughout the crypt, with increasing intensity towards the icGCs. Thus, human icGCs could be broadly distinguished as WGA+/LEL+/LTL− GCs (Fig. 8A). Staining of the nonO-glycosylated MUC2 precursor was as prominent in human colonic icGCs as in the mouse (Fig. 8B).
Fig. 8. Mucus alterations and loss of icGCs in UC patiets.
(A, B) Lectin (A) and nonO-glycosylated MUC2 immunostaining (B) in a human sigmoid colon cross section. Arrowheads: crypt-opening GC (yellow) and icGC (pink); n = 12. (C) Whole-mounted colonic tissue from control (Ctrl) and UC patients. The outer border of the crypt (dashed yellow line). (D, E, F) Number of icGCs per crypt (D), number of GCs (normalized to the relative area for each segment) at different relative positions along the crypt center-border axis (E), and GC shedding score (F) in Ctrl, UCr and UCa colonic biopsies; n= 4 - 9. (G) GC shedding (gray arrowheads) in a fixed whole-mounted UCr biopsy. Position for x/z projection (pink arrowhead). (H) Biopsies from human sigmoid colon stained ex vivo with LTL, WGA and LEL. Crypt plumes (dashed lines), level of corresponding projection (arrowhead), n = 9. (I) Mucus penetrability of 1.0 and 0.2 μm fluorescent beads. Mucus without 0.2 μm beads (dashed line), crypt-openings (arrowheads); n = 3. (J) Mucus defect scores images in biopsies from UC patients. Mucus gaps (orange arrowhead), crypt openings (white arrowheads), and z-positions for x/y projections (color coded arrowheads). (K) Mucus defect score in Ctrl, UCr and UCa patients. Scale bars = 50 μm. Bars indicate median ± interquartile range. Statistical analysis was performed by Kruskal-Wallis analysis with Dunn’s multiple comparison test.
Staining for T-antigen (Galβ1-3GalNAc) by Jacalin, which labels all GCs, was used for GC detection in whole-mounted sigmoid colon biopsies from healthy controls and UC patients (Fig. 8C, and Table S8). Individual GCs were mapped and assigned as crypt-opening GCs or icGCs depending on their location (Fig. S14A). A tendency towards fewer observable GCs per crypt was noted in both UCr and UCa patients compared to controls, where the reduction of icGCs was further pronounced (Fig. 8D, and S14B). This difference was not due to smaller crypt surface area (Fig. S14C). The frequency of icGCs at different relative positions along the crypt center-to-border axis was used to identify crypt-opening and intercrypt zones, based on the presence of icGCs in each area (Fig. S14D). The number of GCs at intercrypt zone was reduced in UCr and UCa patients whereas the number of GCs in the crypt-opening zone was unaltered (Fig. 8E). The difference in number of GC was especially pronounced at the outer border of the crypt (Fig. 8E and S14E). These results indicate that icGCs were lost in the intercrypt zone in UCa and UCr patients.
GC shedding was quantified as mucus-containing cells above the epithelial surface and was scored from 0 to 3 (Figs. 8, F and G, S14F, and Table S9). Both UCr and UCa samples showed more GC shedding. There was a negative correlation between the number of icGCs and the GC shedding score, indicating that the reduced number of icGCs was a consequence of increased cell shedding (Fig. S14K).
The ex vivo mucus organization of healthy human colonic biopsies was similar to that observed in mouse, where crypt covering plumes primarily stained with LTL, and intercrypt mucus stained with both LEL and WGA (Fig. 8H, and Movie S2). The bead penetration assay showed similar results with 1 μm beads settling on the mucus surface, while 0.2 μm beads penetrated the intercrypt mucus (Fig. 8I).
Analysis of biopsies from UC patients revealed a loss of intercrypt mucus in a majority of the patients (Fig. 8J, and Movie S3). Some patients with absent intercrypt mucus maintained a fairly intact mucus surface, whereas others had a very weak mucus surface staining. Overall both UCr and UCa specimens had more discernable mucus defects than controls, based on scores grading the presence/absence of intercrypt mucus and the density of surface mucus from 0 (normal) to 5 (all mucus lost) (Figs. 8, J and K, S14G, and Table S10). Jacalin did not stain the mucus gaps in UC, but gave more pronounced mucus plume staining than in control patients (Fig. S14H). Furthermore, no increase in Jacalin or T-antigen binding Peanut agglutinin (PNA) staining was observed in icGCs in colonic sections from UC (Fig. S14I).
Plotting the mucus defect score to the number of icGCs showed separate clustering of control and UC patients (Fig. S14J), with a trend towards a positive correlation between the mucus defect score and GC shedding score (Fig. S14K). No correlations could be found between available clinical information (number of active episodes during the last two years, time to last active episode, medication), mucus and icGC scores (data not shown).
Our findings revealed that many UC patients with active disease and in remission have reduced numbers of icGCs, increased GC shedding, and altered mucus organization.
Discussion
The protective intestinal mucus is produced by GCs, and their mRNA and protein expression profiles were now determined. Overall, GCs have cellular pathways that focus on protein production, folding, glycosylation, vesicle transport, and secretion, highlighting mucus production as their main task. Yet, in contrast to the current understanding, single cell analysis of GCs identified distinctive canonical and non-canonical differentiation trajectories, comprising several GC clusters in both small and large intestine. While both trajectories were clearly identifiable in the colon, the non-canonical trajectory was less prominent in the distal small intestine. The non-canonical GCs trajectory is characterized by transcripts in common with enterocytes. Enterocyte markers can also be observed in the GC clusters of other single cell datasets of all intestinal epithelial cells when carefully examined (19, 21, 22, 29).The expression of genes involved in the activation of senGCs, as well as genes dysregulated during colitis are found in non-canonical GCs suggesting that these cells could be crucial for maintaining intestinal homeostasis (22, 27, 30). The GC diversity identified indicate an unpredicted degree of plasticity within the GC population.
In the small intestine, epithelial-derived microbial defense mechanisms are largely attributed to the Paneth cells; however we observed GCs with a defense profile, expressing antimicrobial genes indicating that GCs are a more intrinsic part of the innate immune system than previously considered. The co-secretion of antimicrobials and mucus may represent a strategy to increase the concentration of defense proteins close to the epithelium.
The icGCs located at the colonic surface epithelium are the most differentiated GCs. They possess a distinct morphology, a specific gene expression profile, are detected by specific lectin combinations and nonO-glycosylated Muc2 antiserum, and rapidly produce and secrete intercrypt mucus. Based on this we propose that icGCs are a novel, functionally distinct GC subtype, in a similar fashion to the previously described senGCs (23). The high expression of several mucus genes as well as protein transport-associated genes in icGCs is in line with the rapid biosynthesis and secretion of mucus previously observed in these cells (28). Additionally, Stxbp1, enriched in icGCs, has been associated with baseline secretion in lung GCs, whereas Stxbp2, which is more prominent in crypt GCs, is associated with regulated secretion (31). This suggests that secretion by icGCs is continuous at baseline, while secretion from crypt GCs is more responsive to stimulation (32). Furthermore, enhanced mucus production increases the cellular demand of protein folding. However, several chaperones, including the mucin biosynthesis specific Agr2 (13), were expressed at lower levels in icGCs, which might indicate that the continuous secretion from icGCs is different from "plume secretion" from crypt GCs. The increased expression of the GC specific gene Ern2, regulating Muc2 mRNA levels, also indicate different control in these cells (14).
Ex vivo visualization of mucus on live biopsies provided new insight into mucus organization, revealing different properties of the mucus emerging from icGCs and crypt GCs. The stratified mucus layer seen in immunostainings of fixed colonic tissue likely represents the net-like structures seen at the mucus surface ex vivo. The mucus plumes and intercrypt mucus likely correspond to the most recently secreted material, originally described as more MUC2-intense and often associated with the tissue surface in sections (2, 5). The contrasting appearance of mucus by the two methods might arise from different conditions in fixed and live tissue, especially shrinkage upon fixation of the highly hydrated mucus and static pressure exerted by the luminal content.
The enhanced barrier function of the mucus plumes covering the crypt openings provides an extra barrier against microbial intruders, protecting the stem cell niche at the bottom of the crypt. More penetrable intercrypt mucus secreted from icGCs at the epithelial surface interconnects the crypt plumes and while it provides a barrier to microbes, it may enable liquid, ion and nutrient uptake, consistent with the absorptive functions of the surface epithelium. These findings may also explain why motile bacteria can penetrate the mucus at these specific locations (33). Although yet to be resolved, the delineating features of intercrypt mucus and crypt plumes may be a result of cell-specific forms of MUC2, or co-secretion of other factors known to modulate mucus properties, such as proteases, cross-linkers, ions, or fluid volumes (34-37).
The effect of Spdef deficiency was prominent in icGCs, based on altered gene expression, morphology and reduced mucus production and secretion of the surface epithelium. These mice spontaneously developed colitis, but the intercrypt mucus defects were evident prior to the onset of inflammation,supporting a fundamental role of intercrypt mucus. Nevertheless, contributing effects of Spdef ablation on other GCs or cells causing additional functional alterations cannot be ruled out.
Previous studies have shown that patients with UC have mucus barrier defects; however, such impairments are primarily described in the context of active disease (9, 30). Herein, we have shown that a majority of UC patients, not only those with active disease but also those in remission, have alterations in their mucus organization. The explants from UC patients exhibited enhanced GC shedding on the surface epithelium and reduced icGC numbers, resulting in structural defects in the mucus barrier such as gaps in the intercrypt mucus and exposed areas of surface epithelium. Taken together, alterations of icGCs and intercrypt mucus may represent an early and sustained indicator of disease that could contribute to disease initiation.
The results from mouse and human distal colon suggest comparable mucus organization at the epithelial surface between species. The intercrypt mucus contributes to the mucus barrier organization and is important for limiting bacterial penetration. The common denominator identified in inflammatory initiation is the reduced number or function of icGCs resulting in local mucus barrier defects. The mechanisms likely differ between mice and humans and is more diverse among patients. The experimental limitations to specifically target icGCs and the few patients possible to include preclude detailed understanding, but pave the way for further studies and potential treatment strategies.
In summary, in-depth mRNA/protein expression profiling of intestinal GCs unveils striking heterogeneity in this cell population, which points to a markedly nuanced orchestration of the protective mucus system in the gut. Pseudotime analysis not only identified the anticipated canonical trajectory, but also revealed an additional non-canonical path consistent with a complex differentiation network. Plasticity has been observed in several studies of the intestinal stem cell-proliferative cell niche (38), and in post-mitotic cells (39). Our results support dynamic shaping of the GC population that may reflect both intraepithelial communication and influence from environmental cues. We further identified icGCs as a new functionally distinct GC subtype localized at the apical surface of the colonic epithelium. The icGCs secrete mucus that differs from mucus produced by crypt-residing GCs, and both are required to provide a protective mucus barrier. Deficient intercrypt mucus secretion or reduced icGC numbers leads to disruption of the mucus layer with an impaired organization at the mucus surface, correlating with increased colitis susceptibility. In UC patients, reduced icGC numbers and deficient mucus is an early marker as observed not only in active disease patients but also during remission.
Material and methods
A full description of the methods can be found in the supplementary materials
Animals
All animal experimental procedures were in compliance with Swedish animal welfare legislation and approved by the Swedish Laboratory Animal Ethical Committee in Gothenburg, Sweden (number: 280-12, 73-15. 74-15, 2285-19 and 2292-19). C57BL/6, RedMUC298trTg mice (23, 40), Spdef−/− (17), crossed to the RedMUC298trTg strain, and Cftrtm1Unc mice (41) were bred in house in SPF conditions and all experiments used age-matched mice of both genders. DSS-colitis was induced by administration of 3% w/v DSS and disease activity index was scored daily (12, 42).
Human samples
Sigmoid colon biopsies were obtained from patients referred for colonoscopy at Sahlgrenska University Hospital (Gothenburg, Sweden) (Table S8) in compliance with the local human research ethical committee (136-12) and the Declaration of Helsinki as described previously (43). Patients with normal intestine, evaluated macroscopically, were included as controls. Patients with full Mayo score 0 were considered as remission, and ≥1 as active UC.
Intestinal epithelial cell isolation and sorting
Intestinal epithelial cells were isolated from freshly collected distal colon and distal small intestine from RedMUC298trTg mice and Spdef−/− RedMUC298trTg. GC and non-GC were sorted with FACS Jazz (Becton Dickinson) according to the presence or absence of mCherry signal with dead cells and doublets removed. The purity of sorted cells was > 96%.
RNA extraction and sequencing
Sorted cells were pelleted and total RNA extracted using RNeasy kit and QIAshredder columns (Qiagen). The cDNA libraries were prepared using the TruSeq Stranded Total RNA Sample Preparation kit with Ribo Zero Gold (Rev. E; Illumina) and sequenced via paired-end with the NextSeq500 platform (Illumina). The reads were mapped against the mouse reference genome mm10 from Ensembl using STAR (44). The number of mapped reads was calculated using HTseq (45) and different expression and statistical analysis with DESeq2 in R (46).
Protein extraction
Sorted cells were ysed and digested with with LysC and trypsin. Samples were analyzed with an EASY-nLC 1000 system connected to a Q-Exactive mass-spectrometer (Thermo Fisher). MS raw files were processed with MaxQuant software (47) by searching against mouse UniProt database combined with an in-house database containing all human and mouse mucin sequences, including mCherry MUC2 (http://www.medkem.gu.se/mucinbiology/databases/) using Label-Free Quantification (LFQ) method. Statistics were analyzed with Welch’s two-sample t-test, and the Benjamini-Hochberg test for adjusted p-values.
Data analysis for RNA-sequencing and mass spectrometry.
Volcano plots were created using EnhacedVolcano (48). The top 50 genes and proteins were clustered using the heatmap.2 function from gplots. Venn diagrams were made with VennDiagram. All these packages were run in R. Fold change ≥ ±2 and adjusted p-value ≤ 0.05 (proteins) or ≤ 0.01 (RNA) were used to select enriched proteins and genes. Gene Ontology enrichement analysis was performed using the DAVID tool (49, 50).
Single cell preparation, sequencing and analysis
GCs form distal colon and distal small intestine were sorted and droplet-based scRNA-seq was performed using the chromium single cell 3’ v2 platform (10x Genomics). Library sequencing was performed using the NextSeq500 platform. The data processing was performed using Cell Ranger software (10x Genomics) and Seurat (51, 52) in R. Immune, enteroendocrine, Paneth cells, and cells with low expression of Muc2 (<3) were removed. We identified 15392 genes across 6123 cells for the colon and 14794 genes across 3552 cells for the small intestine. Statistical analysis was performed using Wilcoxon Rank Sum test and adjusted p-values Bonferroni correction.
Pseudotime analysis was performed using Scanpy (53) in Python resulting in 14447 genes across 3402 cells for the small intestine and 14892 genes across 5936 cells for the colon. GCs were clustered using the Lovain algoritm. Statistically analysis used Wilcoxon Rank Sum test and the adjusted p-values Benjamini-Hochberg test.
Immunostaining, in situ hybridization and colonic whole-mounted tissue
Paraffin-embedded methanol-Carnoy-fixed sections were dewaxed, rehydrated and antigen-retrieved for immunostaining with different combinations of anti-Ido1 (Cell Signaling), anti-Epcam (Abcam), chicken monoclonal anti-Defa24, raised against a well characterized peptide(54), anti-Lysozyme (Genway) or anti-apoMUC2 (detecting the nonO-glycosylated mucin) antibodies (PH497 and PH1900 for mouse and human respectively (55, 56). Bacterial FISH was performed as previously described (57) with counterstaining using fluorescently conjugated lectins. For RNA-in situ hybridization we used the RNA scope technique (Advanced Cell Diagnostics) according to manufacturer’s instruction on formalin-fixed tissue sections. GCs in formalin-fixed whole-mounted tissue from mouse or human biopsies were stained by lectins or an anti-human apoMUC2 and counterstained with CellMask Deep Red plasma membrane stain (Thermo Fisher Scientific). In all cases, DNA was counterstained by Hoechst-34580 (Thermo Fisher Scientific). Images were recorded with an LSM700 Axio Examiner Z.1 confocal microscope (Zeiss) and further processed using the Imaris software (Bitplane) or ImageJ v1.52k (58). Scoring criteria for GC shedding are given in table S9.
GalNAz labeling of mice and turnover of mucus
WT and Spdef−/− mice at 13 weeks of age were labeled with GalNaz by intraperitoneal injection with subsequent detection using by Alkyne-Tetramethylrhodamine (TAMRA) (both from Thermo Fisher Scientific) on fixed colonic tissue sections as previously described (28).
Ex vivo mucus analysis
Procedure, materials and equipment for ex vivo examination of colonic mucus were employed as previously described (43) with visualization of mucus by fluorescently conjugated lectins and investigation of mucus barrier properties using fluorescent beads or FITC-conjugated dextran (2000 kDa). Quantification of bead penetrability was performed as previously described (59, 60). Z-stacks were acquired using LSM700 with a Plan-Apochromat x 20/1.0 DIC water objective (Zeiss) and the ZEN 2010 software. Image analysis was performed using Imaris. Mucus defect scoring was done in a blinded fashion according to table S10.
Electron microscopy
Colonic tissue was excised, flushed and fixed in modified Karnovsky’s fixative, cut and further processed for either TEM or SEM using a Zeiss Leo 912AB Omega electron microscope or a Zeiss Gemini DSM 982 microscope respectively. GCs were manually counted in a blinded fashion for quantification.
Statistical analysis
Statistical analysis and graphical illustrations were performed using GraphPad PRISM 7.03 or 8.3.1 (GraphPad Software). Data are presented as mean ± standard deviation (SD) or median with range, as indicated. Statistical tests were applied as indicated. For all statistical analyses: * p<0.05, ** p<0.01, *** p<0.0001, **** p<0.00001, ns = non-significant.
Supplementary Material
Acknowledgement
We are indebted to the physicians and nurses at GEA of the Sahlgrenska University hospital for help with obtaining the human material. We acknowledge Hans Clevers and J. Whitsett, Cincinnati Children’s Hospital Medical Center and the University of Cincinnati School of Medicine, for providing the Spdef−/− mouse strain. We thank the Genomics, Centre Cellular Imaging and Bioinformatics Core Facilities at the Sahlgrenska academy for their assistance. We especially thank Vanja Borjesson for helping with the pseudotime analysis. The project is part of the NIH funded MIST collaboration where GCH, MEVJ and CLB are members.
Funding:
This work was supported by the Swedish Research Council (2015-03047, 2017-00958, 2019-01134), The Swedish Cancer Foundation (CAN2016/487, 2017/360), The Knut and Alice Wallenberg Foundation (2017-0028), National Institute of Health (U01AI095473, U01AI125926, R37AI32738, the content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH), European Research Council ERC (694181), IngaBritt and Arne Lundberg Foundation (2015-070, 2018-0117), The Hasselblad foundation, Sahlgrenska University Hospital (ALFGBG-724681, ALFGBG-440741), Wilhelm and Martina Lundgren’s Foundation, and the Sahlgrenska Academy.
Footnotes
Competing interest: The authors do not have any competing interest.
Data and materials availability: RNAseq and proteomics data is deposited as stated in material and methods and all other data is available in the paper and supplementary material
Data availability
MS proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE (61) partner repository with the dataset identifier PXD011527. All raw data from the bulk RNA-seq and scRNA-seq are deposited to the GEO database (GSE144436).
References
- 1.Rodriguez-Pineiro AM et al. , Studies of mucus in mouse stomach, small intestine, and colon. II. Gastrointestinal mucus proteome reveals Muc2 and Muc5ac accompanied by a set of core proteins. Am. J. Physiol Gastrointest. Liver Physiol 305, G348–G356 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ermund A, Schutte A, Johansson ME, Gustafsson JK, Hansson GC, Studies of mucus in mouse stomach, small intestine, and colon. I. Gastrointestinal mucus layers have different properties depending on location as well as over the Peyer's patches. Am. J. Physiol Gastrointest. Liver Physiol 305, G341–G347 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergstrom JH et al. , Gram-positive bacteria are held at a distance in the colon mucus by the lectin-like protein ZG16. Proc. Natl. Acad. Sci. U. S. A 113, 13833–13838 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jakobsson HE et al. , The composition of the gut microbiota shapes the colon mucus barrier. EMBO Rep 16, 164–177 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johansson ME et al. , The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl. Acad. Sci. U. S. A 105, 15064–15069 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergstrom K et al. , Proximal colon-derived O-glycosylated mucus encapsulates and modulates the microbiota. Science 370, 467–472 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arike L, Hansson GC, The Densely O-Glycosylated MUC2 Mucin Protects the Intestine and Provides Food for the Commensal Bacteria. J Mol Biol 428, 3221–3229 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van der SM et al. , Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology 131, 117–129 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Johansson ME et al. , Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut 63, 281–291 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bergstrom K et al. , Core 1- and 3-derived O-glycans collectively maintain the colonic mucus barrier and protect against spontaneous colitis in mice. Mucosal. Immunol 10, 91–103 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carvalho FA et al. , Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host. Microbe 12, 139–152 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johansson ME et al. , Bacteria Penetrate the Inner Mucus Layer before Inflammation in the Dextran Sulfate Colitis Model. PLoS. ONE 5, e12238 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park SW et al. , The protein disulfide isomerase AGR2 is essential for production of intestinal mucus. Proc. Natl. Acad. Sci. U. S. A 106, 6950–6955 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsuru A et al. , Negative feedback by IRE1beta optimizes mucin production in goblet cells. Proc. Natl. Acad. Sci. U. S. A 110, 2864–2869 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van der Flier LG, Clevers H, Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu. Rev. Physiol 71, 241–260 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Noah TK, Kazanjian A, Whitsett J, Shroyer NF, SAM pointed domain ETS factor (SPDEF) regulates terminal differentiation and maturation of intestinal goblet cells. Exp. Cell Res 316, 452–465 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gregorieff A et al. , The Ets-Domain Transcription Factor Spdef Promotes Maturation of Goblet and Paneth Cells in the Intestinal Epithelium. Gastroenterology 137, 1333–1345 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Lo YH et al. , Transcriptional Regulation by ATOH1 and its Target SPDEF in the Intestine. Cell Mol Gastroenterol Hepatol 3, 51–71 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haber AL et al. , A single-cell survey of the small intestinal epithelium. Nature 551, 333–339 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herring CA et al. , Unsupervised Trajectory Analysis of Single-Cell RNA-Seq and Imaging Data Reveals Alternative Tuft Cell Origins in the Gut. Cell Syst 6, 37–51 e39 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y et al. , Single-cell transcriptome analysis reveals differential nutrient absorption functions in human intestine. J Exp Med 217, e20191130 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parikh K et al. , Colonic epithelial cell diversity in health and inflammatory bowel disease. Nature 567, 49–55 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Birchenough GM, Nystrom EE, Johansson ME, Hansson GC, A sentinel goblet cell guards the colonic crypt by triggering Nlrp6-dependent Muc2 secretion. Science 352, 1535–1542 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mariadason JM et al. , Gene expression profiling of intestinal epithelial cell maturation along the crypt-villus axis. Gastroenterology 128, 1081–1088 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Rothenberg ME et al. , Identification of a cKit(+) colonic crypt base secretory cell that supports Lgr5(+) stem cells in mice. Gastroenterology 142, 1195–1205 e1196 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stahl M et al. , The Muc2 mucin coats murine Paneth cell granules and facilitates their content release and dispersion. Am J Physiol Gastrointest Liver Physiol 315, G195–G205 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen F et al. , Neutrophils Promote Amphiregulin Production in Intestinal Epithelial Cells through TGF-beta and Contribute to Intestinal Homeostasis. J Immunol 201, 2492–2501 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johansson ME, Fast renewal of the distal colonic mucus layers by the surface goblet cells as measured by in vivo labeling of mucin glycoproteins. PLoS. ONE 7, e41009 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tabula Muris C et al. , Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367–372 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van der Post S et al. , Structural weakening of the colonic mucus barrier is an early event in ulcerative colitis pathogenesis. Gut 68, 2142–2151 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaramillo AM et al. , Different Munc18 proteins mediate baseline and stimulated airway mucin secretion. JCI Insight 4, e124815 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neutra MR, O'Malley LJ, Specian RD, Regulation of intestinal goblet cell secretion. II. A survey of potential secretagogues. Am. J. Physiol 242, G380–G387 (1982). [DOI] [PubMed] [Google Scholar]
- 33.Furter M, Sellin ME, Hansson GC, Hardt WD, Mucus Architecture and Near-Surface Swimming Affect Distinct Salmonella Typhimurium Infection Patterns along the Murine Intestinal Tract. Cell Rep 27, 2665–2678 e2663 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiao F et al. , Slc26a3 deficiency is associated with loss of colonic HCO secretion, absence of a firm mucus layer and barrier impairment in mice. Acta Physiol (Oxf) 211, 161–175 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Nystrom EEL et al. , Calcium-activated Chloride Channel Regulator 1 (CLCA1) Controls Mucus Expansion in Colon by Proteolytic Activity. EBioMedicine 33, 134–143 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gustafsson JK et al. , Bicarbonate and functional CFTR channel are required for proper mucin secretion and link cystic fibrosis with its mucus phenotype. J. Exp. Med 209, 1263–1272 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Recktenwald CV, Hansson GC, The reduction-insensitive bonds of the MUC2 mucin are isopeptide bonds. J. Biol. Chem 291, 13580–13590 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Sousa EMF, de Sauvage FJ, Cellular Plasticity in Intestinal Homeostasis and Disease. Cell Stem Cell 24, 54–64 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Zecchini V, Domaschenz R, Winton D, Jones P, Notch signaling regulates the differentiation of post-mitotic intestinal epithelial cells. Genes Dev 19, 1686–1691 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Svensson F, Lang T, Johansson MEV, Hansson GC, The central exons of the human MUC2 and MUC6 mucins are highly repetitive and variable in sequence between individuals. Sci Rep 8, 17503 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Snouwaert JN et al. , An animal model for cystic fibrosis made by gene targeting. Science 257, 1083–1088 (1992). [DOI] [PubMed] [Google Scholar]
- 42.Cooper HS, Murthy SN, Shah RS, Sedergran DJ, Clinicopathologic study of dextran sulfate sodium experimental murine colitis. Lab Invest 69, 238–249 (1993). [PubMed] [Google Scholar]
- 43.Gustafsson JK et al. , An ex vivo method for studying mucus formation, properties, and thickness in human colonic biopsies and mouse small and large intestinal explants. Am. J. Physiol Gastrointest. Liver Physiol 302, G430–G438 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dobin A et al. , STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anders S, Pyl PT, Huber W, HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tyanova S, Temu T, Cox J, The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc 11, 2301–2319 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Blighe K, Rana S, Lewis M, EnhancedVolcano: Publication-ready volcano plots with enhanced colouring and labeling. https://github.com/kevinblighe (2019). [Google Scholar]
- 49.Huang da W, Sherman BT, Lempicki RA, Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 50.Huang da W, Sherman BT, Lempicki RA, Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37, 1–13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R, Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stuart T et al. , Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902 e1821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolf FA, Angerer P, Theis FJ, SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 19, 15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Szyk A et al. , Crystal structures of human alpha-defensins HNP4, HD5, and HD6. Protein Sci 15, 2749–2760 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Asker N, Axelsson MAB, Olofsson SO, Hansson GC, Dimerization of the human MUC2 mucin in the endoplasmic reticulum is followed by a N-glycosylation-dependent transfer of the mono- and dimers to the Golgi apparatus. J. Biol. Chem 273, 18857–18863 (1998). [DOI] [PubMed] [Google Scholar]
- 56.Hansson GC, Baeckstrom D, Carlstedt I, Klinga-Levan K, Molecular cloning of a cDNA coding for a region of an apoprotein from the 'insoluble' mucin complex of rat small intestine. Biochem. Biophys. Res. Commun 198, 181–190 (1994). [DOI] [PubMed] [Google Scholar]
- 57.Erickson NA et al. , The Goblet Cell Protein Clca1 (Alias mClca3 or Gob-5) Is Not Required for Intestinal Mucus Synthesis, Structure and Barrier Function in Naive or DSS-Challenged Mice. PLoS. ONE 10, e0131991 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schneider CA, Rasband WS, Eliceiri KW, NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schroeder BO et al. , Bifidobacteria or Fiber Protects against Diet-Induced Microbiota-Mediated Colonic Mucus Deterioration. Cell Host Microbe 23, 27–40 e27 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Volk JK et al. , The Nlrp6 inflammasome is not required for baseline colonic inner mucus layer formation or function. J Exp Med 216, 2602–2618 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vizcaino JA et al. , 2016 update of the PRIDE database and its related tools. Nucleic Acids Res 44, 11033 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
MS proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE (61) partner repository with the dataset identifier PXD011527. All raw data from the bulk RNA-seq and scRNA-seq are deposited to the GEO database (GSE144436).