Abstract
Immune checkpoint blockade (ICB) therapy revolutionized cancer treatment, but many patients with impaired MHC-I expression remain refractory. Here, we combined FACS-based genome-wide CRISPR screens with a data-mining approach to identify drugs that can upregulate MHC-I without inducing PD-L1. CRISPR screening identified TRAF3, a suppressor of the NF-κB pathway, as a negative regulator of MHC-I but not PD-L1. The Traf3-knockout (Traf3-KO) gene expression signature is associated with better survival in ICB-naive cancer patients and better ICB response. We then screened for drugs with similar transcriptional effects as this signature and identified SMAC mimetics. We experimentally validated that the SMAC mimetic birinapant upregulates MHC-I, sensitizes cancer cells to T-cell-dependent killing, and adds to ICB efficacy. Our findings provide preclinical rationale for treating tumors expressing low MHC-I expression with SMAC mimetics to enhance sensitivity to immunotherapy. The approach used in this study can be generalized to identify other drugs that enhance immunotherapy efficacy.
Keywords: TRAF3, MHC-I, PD-L1, birinapant, immune checkpoint blockade
Introduction
Immune checkpoint blockade (ICB), primarily anti-PD-1, anti-PD-L1, and anti-CTLA-4, has been shown to induce remarkable response across different cancer types in many clinical studies (1–8). However, only a minority of patients responds to ICB, and resistance can develop in patients who initially respond (9). The mechanisms of resistance have been under extensive investigation. Multiple cancer cell-intrinsic mechanisms have been identified, including low neoantigen load (2,3,10,11), loss of antigen presentation (12,13), loss of HLA heterozygosity (14,15), impaired response or prolonged exposure to interferon-gamma (IFNγ) (16–19), activation of certain oncogenic pathways/signatures (20–22), as well as other mechanisms (22,23). Among these ICB-resistance mechanisms, insufficient antigen presentation to activate T cells comprises a large proportion of patients (24–26). Major histocompatibility complex class I (MHC-I), the key component in antigen presentation, presents intracellular peptide antigens to the cell surface for recognition by the specific CD8+ T cells (27). Loss of MHC-I by genetic mutation or epigenetic silencing impairs T-cell recognition and activation, and compromises antitumor activity. Furthermore, ICB treatment of CD8+ T cells in the absence of antigen stimulation can induce, rather than alleviate, T cell dysfunction (28). Therefore, enhancing MHC-I levels in cancer cells, especially in cancers with low baseline MHC-I levels, is a promising strategy to improve ICB efficacy.
Genes encoding the MHC-I components (for example HLA-A/B/C and B2M) are expressed widely yet are under tight regulation (29). In tumors, interferon gamma (IFNγ) secreted by tumor-infiltrating T cells can induce the expression of the MHC-I component genes through the JAK-STAT pathway (30) and boost the CD8+ T-cell-dependent cancer cell elimination (31). However, recent reports showed that IFNγ can also exert immunosuppressive functions, partially through its induction of immune checkpoint genes such as CD274 (encoding PD-L1) to inhibit T cell activation and promote T cell dysfunction (19,32,33). In addition to IFNγ, multiple other cytokines were reported to upregulate both MHC-I and PD-L1 with opposite effects on tumor immunity (31). Therefore, we sought to identify gene perturbations and drugs that specifically induce MHC-I expression without increasing PD-L1 expression.
CRISPR screening is a powerful technology with increasing popularity for target identification in cancer and immunology, and fluorescence-activated cell sorting (FACS)-based CRISPR screens have previously identified regulators of MHC-I or PD-L1 (34–36). However, it is difficult to identify factors that regulate MHC-I but not PD-L1 based on existing data, due to variations in sorting strategies and limitations in statistical power. Likewise, several exciting studies have reported chemotherapy or targeted-therapy with immunomodulatory effects by regulating MHC-I and/or PD-L1 (37–40), but a systematic approach to identifying drugs that preferentially modulate MHC-I has proven elusive.
In this study, we employed a CRISPR screening approach with dual-marker FACS sorting to identify factors that decouple the regulation of MHC-I and PD-L1. The experimentally validated target was used to generate a KO differential expression signature. Using this signature, we analyzed transcriptome data from drug perturbation studies to identify drugs that regulate MHC-I but not PD-L1. Finally, we validated the effect of the identified drug to enhance ICB response in a T-cell-dependent manner in vivo.
Results
An integrated workflow to identify drugs with MHC-I-modulatory effects
Analysis of bulk tumor gene expression data from The Cancer Genome Atlas (TCGA) revealed a strong positive correlation between genes encoding MHC-I and PD-L1 in multiple cancer types (Figure S1A). Furthermore, analysis of Cancer Cell Line Encyclopedia (CCLE) cell line gene expression data in vitro revealed a similar trend (Figure S1B). Despite the overall positive correlation, the ratio of MHC-I over PD-L1 expression is positively correlated with patient overall survival in the majority of cancer types, although TCGA tumors were not treated with immunotherapy (Figure S1C).
To search for drugs that can specifically upregulate MHC-I without inducing PD-L1, we designed an integrated experimental and computational workflow, which consists of 5 steps (Figure S1D). First, we performed genome-wide CRISPR screens with dual-marker sorting to identify candidate genes whose KO can upregulate MHC-I without upregulating PD-L1. Second, we performed experiments on the identified gene candidates to validate the MHC-I-specific effect and test the sensitivity to ICB therapy. Third, we characterized the target KO gene expression signature in vitro. Next, we searched the Gene Expression Omnibus (GEO) to identify drugs with differential gene expression signature similar to that from the candidate gene KO. Finally, we validated the drug for its immunomodulatory effects in vitro and in vivo.
CRISPR screen identifies TRAF3 as a negative regulator of MHC-I
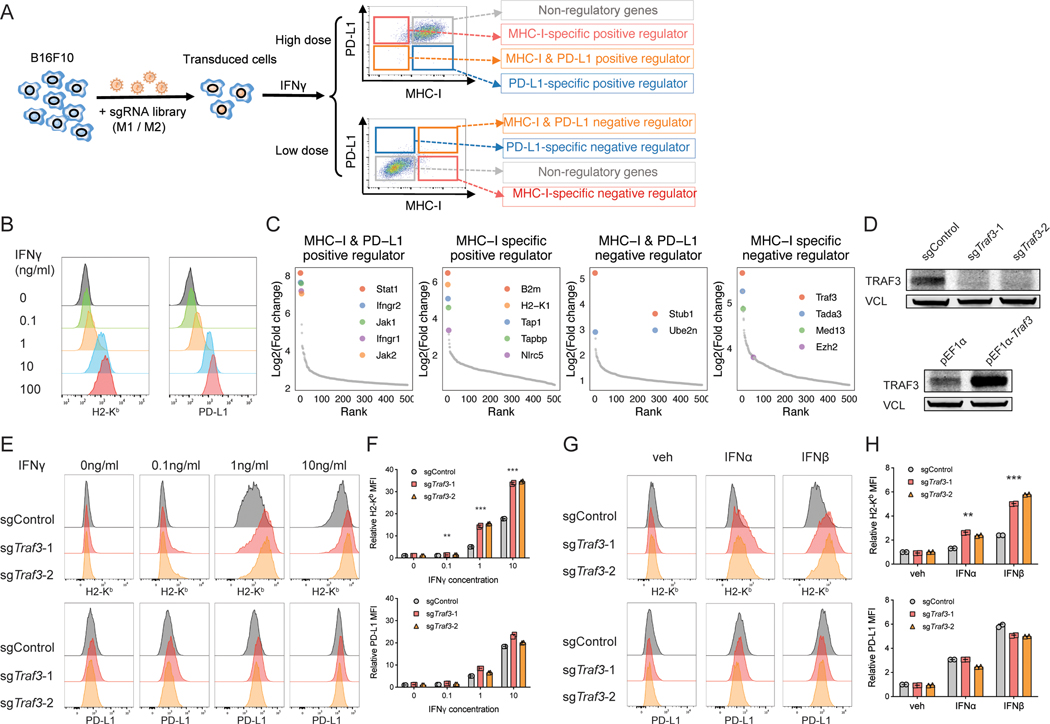
To systematically identify regulators of MHC-I and PD-L1, we performed FACS-based genome-wide CRISPR screens using the mouse melanoma cell line B16F10 (Figure 1A), which can be induced by IFNγ to express MHC-I and PD-L1. Low dose (0.1ng/ml) IFNγ showed minimal induction of MHC-I and PD-L1 and enabled us to identify negative regulators whose deficiency can increase MHC-I and/or PD-L1 protein level (Figure 1B). In comparison, high dose (10ng/ml) IFNγ strongly induced MHC-I and PD-L1 and enabled us to identify the positive regulators of MHC-I and/or PD-L1 protein (Figure 1B). We transduced the B16F10 cells with our mouse genome-wide CRISPR KO library, expanded the transduced cells, and treated the cells with either a low or high dose of IFNγ for 2 days. We then performed FACS to isolate the MHC-IhiPD-L1hi, MHC-IhiPD-L1lo, MHC-IloPD-L1hi, and MHC-IloPD-L1lo subpopulations and compared the sgRNA frequencies in each subpopulation with the unsorted population (Figure 1A). There was a large decrease in the number of detected sgRNAs in the regulator-enriched quadrants compared to the unsorted population, indicating strong selections in these subpopulations (Figure S2A).
Figure 1. CRISPR screens identify novel regulators of MHC-I.
(A) Workflow of using CRISPR screens to identify the positive or negative regulators of MHC-I and/or PD-L1. We transduced B16F10 mouse melanoma cells with an in-house-designed genome-wide sgRNA library, expanded the transduced cells, and stimulated the cells with 0.1ng/ml (low dose) or 10ng/ml IFNg (high dose) for different levels of MHC-I/PD-L1 induction. We then performed FACS to isolate the MHC-IhiPD-L1hi, MHC-IhiPD-L1lo, MHC-IloPD-L1hi, and MHC-IloPD-L1lo sub-populations. We amplified and sequenced the gRNAs in these sub-populations as well as the bulk pre-sorting population to identify genes that were enriched in each sorted sub-population. MHC-I and/or PD-L1 regulators are expected to be enriched in the sub-populations as indicated. (B) Titration of IFNγ concentration to test its effect on MHC-I and PD-L1 expression. Histogram of H2-Kb and PD-L1 levels assessed by flow cytometry following two-day treatment with different concentrations of IFNγ. (C) CRISPR screen reveals known and novel candidate regulators of MHC-I and/or PD-L1. Ranked dot plots of gene enrichment in each sorted sub-population compared to the unsorted population are shown. The X axis shows the rank of each gene, and the Y axis shows the log2 enrichment of sgRNAs for each gene in the indicated sub-population compared to the unsorted population. (D) Immunoblot of B16F10 cells with indicated genotypes shows good efficiency of Traf3 knockout or overexpression. (E-F) Validation of TRAF3 as a negative regulator of MHC-I with IFNγ induction. B16F10 cells transduced with control sgRNA or sgTraf3 were cultured for 48 hours, with IFNγ concentrations as indicated, and then assessed on their MHC-I and PD-L1 levels. (E) Typical histogram of H2-Kb and PD-L1 FACS plot of control or Traf3-deficient B16F10 cells in each treatment condition. (F) Quantification of median fluorescence intensity (MFI) of H2-Kb or PD-L1 from (E). Values are normalized to the sgControl group with vehicle treatment. (**P < 0.01, ***P < 0.001; Two-way ANOVA with Benjamini-Hochberg post test comparing sgTraf3 and sgControl in each condition). (G-H) Validation of TRAF3 as a negative regulator of MHC-I with type-I IFN induction. B16F10 cells transduced with control sgRNA or sgTraf3 were cultured for 48 hours with treatment as indicated, and then assessed on their MHC-I and PD-L1 levels. 500U/ml IFNα or IFNβ was used. (G) Typical histogram of H2-Kb and PD-L1 FACS plot of control or Traf3-deficient B16F10 cells in each treatment condition. (H) Quantification of MFI of H2-Kb or PD-L1 from (G). Values are normalized to the sgControl group with vehicle treatment. (**P < 0.01, ***P < 0.001; Two-way ANOVA with Benjamini-Hochberg post test comparing sgTraf3 and sgControl in each condition).
We used the MAGeCK (41,42) (Figure 1C, Figure S2B, Table S1) and RIGER (43) (Figure S2C) computational algorithms to analyze the CRISPR screen data. Comparison of the MHC-IloPD-L1lo subpopulation with the unsorted population in the 10ng/ml IFNγ cohort identified known positive regulators of both MHC-I and PD-L1, including Jak/Stat and Ifngr genes (Figure 1C). Comparison of the MHC-IhiPD-L1lo subpopulation with the unsorted population in the 10ng/ml IFNγ cohort identified Cd274 (encoding PD-L1), as expected, as the top PD-L1-specific positive regulator (Figure S2B). Comparison of the MHC-IhiPD-L1hi subpopulation with the unsorted population in the 0.1ng/ml IFNγ cohort identified novel candidate common negative regulators, including Stub1 and Ube2n (Figure 1C). Stub1 was reported to downregulate PD-L1 (35), so its role in regulating MHC-I merits further study.
The comparisons most central to the goals of this study are those differentially regulating MHC-I and PD-L1 (MHC-IloPD-L1hi and MHC-IhiPD-L1lo). Comparison of the MHC-IloPD-L1hi subpopulation with the unsorted population in the 10ng/ml IFNγ condition revealed genes known to be involved in MHC-I transcription and assembly, including B2m, Nlrc5, Tap1, Tap2, Tapbp, and H2-K1, which were ranked among the top MHC-I-specific positive regulators (Figure 1C). Comparison of the MHC-IhiPD-L1lo subpopulation with the unsorted population in the 0.1ng/ml IFNγ condition identified previously reported (Ezh2) (36,44) and novel candidate negative regulators of MHC-I, including Traf3, Tada3, and Med13 (Figure 1C). TRAF3, the top-ranking putative MHC-I-specific negative regulator in our screens, is an adaptor protein involved in multiple signaling pathways leading to NF-κB activation (45). NF-κB is known to regulate MHC-I component genes by directly binding to their enhancers (46). A recent study in neuroblastoma also implicated TRAF3 as mediating the MHC-I repressive effects of N4BP1 on MHC-I (47). These previous studies comport with our screen results that TRAF3 is a negative regulator of MHC-I.
We then validated the effect of TRAF3 perturbation on MHC-I and PD-L1 levels using CRISPR-mediated single gene KO of Traf3 with two different sgRNAs, and overexpression of Traf3 (Figure 1D). As expected, Traf3-KO increased the expression of MHC-I, but not PD-L1, in response to various doses of IFNγ (Figure 1E-F). Conversely, over-expression of Traf3 specifically decreased the expression of MHC-I, but not PD-L1, in response to IFNγ (Figure S2D). To test whether MHC-I is upregulated by cell-extrinsic autocrines, we compared the sgControl or sgTraf3 cells cultured in conditioned media from Traf3-WT or Traf3-KO cells. We did not detect a significant difference in MHC-I expression between different media conditions (Figure S2E). Furthermore, we compared the co-culture of GFP+ WT B16F10 cells with GFP- Traf3-WT or Traf3-KO B16F10 cells, and observed no significant difference in MHC-I expression level in GFP+ WT cells between different co-culture conditions either (Figure S2F). These data suggest that Traf3-KO specifically upregulates MHC-I in a cell-autonomous manner.
To test the generality of this regulation, we measured the effect of Traf3 deficiency in the context of induction by other cytokines besides IFNγ. We found that Traf3 deficiency also led to specific upregulation of MHC-I in response to TNFα and type I interferons (Figure 1G-H, Figure S2G-H). We also assessed the role of TRAF3 in MHC-I regulation in an independent mouse cell line (Figure S2I-M), the colorectal cancer line CT26. Despite higher baseline MHC-I expression in CT26, Traf3-KO also led to specific upregulation of MHC-I with or without cytokine treatments (Figure S2I-M). We further tested the function of TRAF3 in human cancer cells, and found that its KO can preferentially upregulate MHC-I in multiple human lines (Figure S2N-Q), including SKMEL5 (melanoma), K028 (melanoma), HT29 (colorectal cancer), and MCF7 (breast cancer). These results support the general role of TRAF3 in suppressing MHC-I in human and mouse cancer cell lines.
TRAF3 depletion activates MHC-I through NF-κB
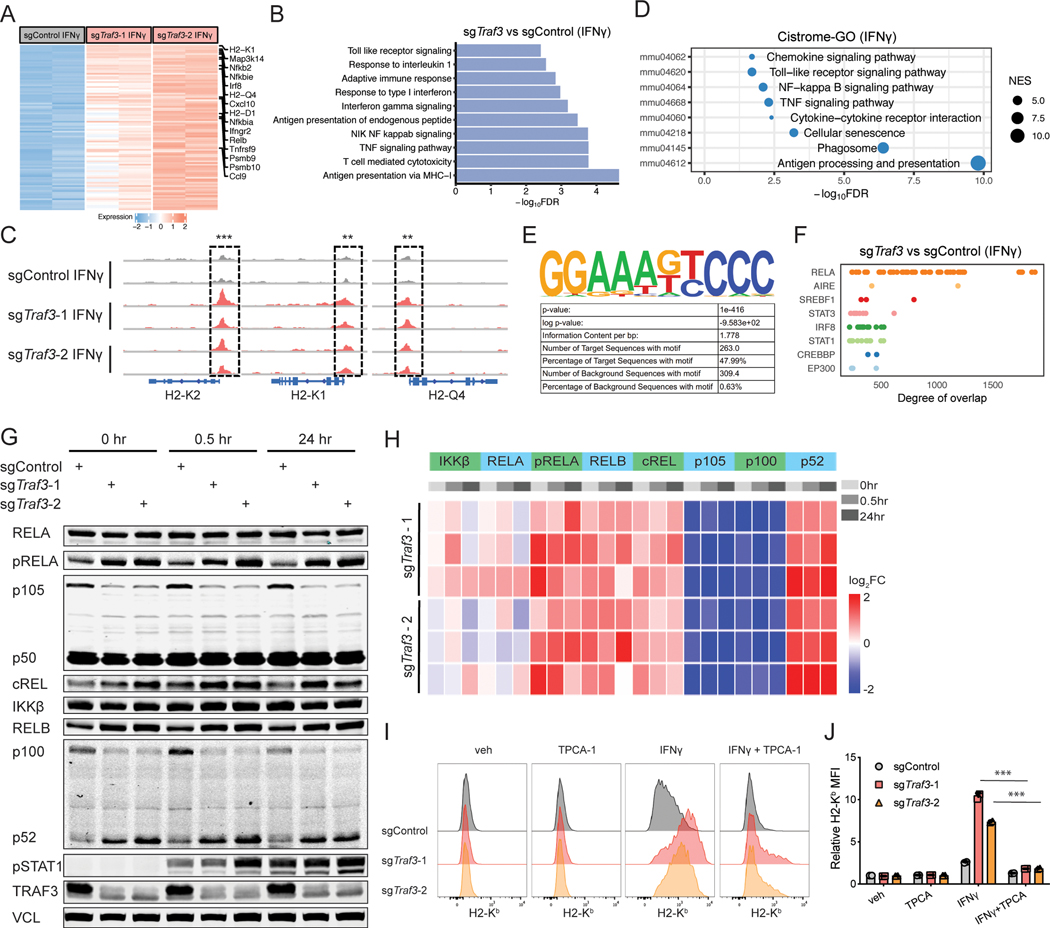
To study the mechanism by which TRAF3 negatively regulates MHC-I levels, we performed RNA-seq and ATAC-seq on Traf3-WT or -KO B16F10 cells that were treated with vehicle or 1ng/ml IFNγ. As expected, RNA-seq revealed multiple MHC-I components, such as H2-K1 and H2-D1, among the top upregulated genes in the Traf3-KO cells under baseline condition or after IFNγ induction (Figure 2A, Figure S3A, Table S2). Other immune-related genes were also upregulated, including the pro-inflammatory chemokines Cxcl10 and Ccl9 and components of the NF-κB pathway (Figure 2A, Figure S3A, Table S2). By contrast, Traf3-KO did not alter the transcription of Cd274 (PD-L1) (Table S2). Gene Ontology (GO) analysis shows that genes upregulated upon Traf3 knockout were enriched in antigen presentation, the NF-κB pathway, and TNF-mediated signaling (Figure 2B, Figure S3B). This is consistent with previous reports that TRAF3 is a negative regulator of NF-kB in response to various extracellular signals (48–50).
Figure 2. TRAF3 deficiency upregulates MHC-I through NF-κB.
(A) RNA-seq of B16F10 cells transduced with sgControl or sgTraf3 shows upregulation of MHC-I-related genes in the absence of Traf3. Heatmap of differential expression of genes induced by TRAF3 deficiency with IFNγ treatment. (B) GSEA enrichment analysis of upregulated pathways (GO biological pathway) in sgTraf3 cells compared to sgControl cells with IFNγ treatment. Multiple pathways, such as antigen presentation and NF-kB signaling, were upregulated by the deletion of Traf3. (C) ATAC-seq of Traf3-normal or -deficient B16F10 cells revealed that TRAF3 deficiency leads to higher chromatin accessibility near genes encoding components of the MHC-I complex. (D) Cistrome-GO analysis of the more accessible regions in sgTraf3 compared to sgControl cells with IFNγ treatment. (E) Cistrome toolkit analysis of ATAC-seq data revealed that DNA-binding sites of RELA were more open in the Traf3-deficient cells. (F) Enrichment of motifs in the accessible chromatin regions specific to Traf3-deficient cells. The top enriched motifs (RELA) is shown. (G) Typical immunoblot of NF-κB signaling components in Traf3-normal or -deficient B16F10 cells in response to IFNγ induction. B16F10 cells transduced with sgControl or sgTraf3 were induced by 1ng/ml IFNγ for 0, 0.5, or 24 hours, and then harvested for immunoblot. Data from 1 typical experiment out of 3 biological replicates is shown. (H) Quantification of immunoblot signals from panel (G) based with 3 biological replicates. TRAF3 deletion leads to upregulated NF-kB signaling. (I-J) B16F10 cells transduced with control sgRNA or sgTraf3 were treated with vehicle control, IFNγ (1ng/ml), and/or TPCA-1 (1μM) for 48 hours, and then assessed on their MHC-I and PD-L1 levels. (I) Typical histogram of H2-Kb and PD-L1 FACS plot of control or Traf3-deficient B16F10 cells in each treatment condition. (J) Quantification of MFI of H2-Kb or PD-L1 from (I). Values are normalized to sgControl group with vehicle treatment. (***P < 0.001; Two-way ANOVA with Benjamini-Hochberg post test comparing IFNγ and IFNγ+TPCA groups).
Consistent with the RNA-seq data, ATAC-seq showed that DNA accessibility is higher in Traf3-KO cells in the promoter regions of MHC-I components (Figure 2C, Figure S3C, Table S2). We also performed Cistrome GO analysis (51) of the ATAC-seq data, which ranks each gene’s regulatory potential with nearby peaks’ strength and distance, and then examines the enrichment of GO pathways on the top ranking genes. Genes with increased regulatory potential in Traf3-KO cells showed significant enrichment of antigen processing and presentation, TNF signaling pathway, and NF-κB signaling pathway (Figure 2D, Figure S3D). In addition, HOMER motif enrichment analysis (52) found NF-κB family motif to be the most enriched in the sites with increased chromatin accessibility upon Traf3 KO (Figure 2E). Furthermore, we used the CistromeDB Toolkit function (53,54) to identify transcription factors (TFs) whose published binding sites have the highest overlap with our differential ATAC-seq peaks, quantified by GIGGLE score (55). This analysis found binding sites of RELA, a component of NF-κB, to be the most enriched in the increased accessible sites in Traf3-KO cells (Figure 2F, Figure S3E). Other TFs with enriched binding sites, such as AIRE, STAT3, IRF8, and STAT1 (Figure 2F), also are known to be involved in immune response.
The NF-κB pathway is involved in multiple biological processes, such as immune response, developmental processes, cell proliferation, and cell death (56–58). The NF-κB family of TFs can be activated by “canonical” or “non-canonical” signaling pathways, which differ from each other in upstream regulators, TF components, and signaling kinetics (59). The canonical pathway predominantly involves RELA, cREL, and NFKB1 (p105/p50), whereas the non-canonical pathway mainly involves RELB and NFKB2 (p100/p52). Immunoblot of Traf3-WT or Traf3-KO cells with IFNγ treatment revealed that TRAF3 deficiency led to activation of both canonical and non-canonical NF-κB pathways, with increased phospho-RELA, RELB, cREL, and p52 (the active form of NFKB2) (Figure 2G-H). Analysis of existing ChIP-seq datasets in Cistrome DB (53,54) revealed that both canonical and non-canonical NF-κB TFs can bind to the promoter/enhancers of MHC-I in human and mouse cells (60–62) (Figure S3F). In addition, consistent with the reported role of TRAF3 as an adaptor for cIAP-mediated degradation of NIK (48,49), an activator of the non-canonical NF-κB pathway, TRAF3-deficient cells showed higher NIK levels (Figure S3G). To test whether the NF-κB pathway underlies the upregulation of MHC-I in the absence of TRAF3, we treated Traf3-KO cells with NF-κB inhibitors, TPCA-1 and IKK-16, and tested whether the upregulation of MHC-I in Traf3-KO cells could be reversed. Indeed, the ability for IFNγ to upregulate MHC-I was drastically attenuated when Traf3-KO cells were treated with TPCA-1 (Figure 2I-J) or IKK-16 (Figure S3H-I). This finding is consistent with recent studies that activation of NF-κB-related pathways can induce MHC-I expression in melanoma cells (63,64). Taken together, these data suggest that TRAF3 deficiency leads to upregulation of MHC-I through elevated NF-κB activity.
TRAF3 deficiency facilitates T-cell-driven killing of cancer cells
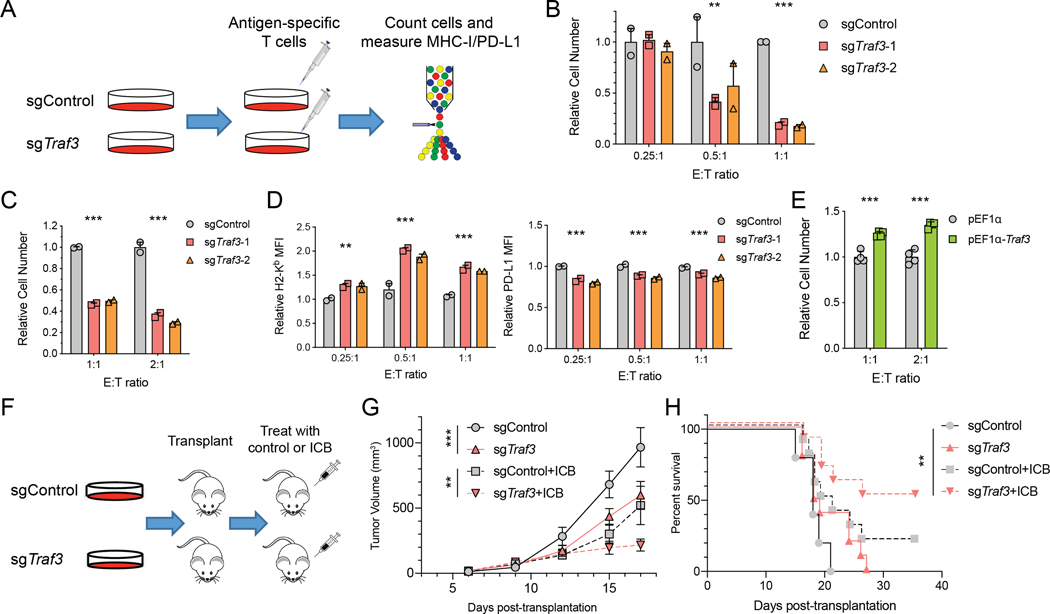
Because TRAF3 loss upregulated MHC-I but not PD-L1 in response to IFNγ, we hypothesized that it may also render cancer cells more susceptible to T-cell driven cytotoxicity. We tested this hypothesis by co-culturing B16F10 cells with either Pmel-1 T cells, which have low-affinity TCR for an endogenous melanoma antigen, or OT-I T cells, which have high-affinity for an exogenously introduced ovalbumin antigen (Figure 3A). Traf3-KO resulted in fewer viable cancer cells and higher MHC-I expression than Traf3-WT after co-culture in both models (Figure 3B-D, Figure S4A-E). In the co-culture with Pmel-1 T cells, the killing efficiency in WT B16F10 cells is low at baseline due to the low expression of MHC-I. Therefore, it is common to pre-treat cancer cells with IFNγ, which boosts MHC-I expression, prior to co-culture with T cells in this model. We used this method and observed significantly more efficient killing of Traf3-KO cells (relative cell number in Figure 3C, raw killing% in Figure S4C). Furthermore, we found that the Traf3-KO B16F10 cells could be efficiently killed by Pmel-1 T cells even without IFNγ pre-treatment (relative cell number in Figure S4A, raw killing% in Figure S4D), and exhibited a substantial induction of MHC-I expression (Figure S4E). Conversely, over-expression of TRAF3 resulted in reduced sensitivity of cancer cells to T-cell-driven cytotoxicity (relative cell number in Figure 3E, raw killing% in Figure S4F).
Figure 3. TRAF3 deletion sensitizes cancer cells to T-cell-driven cytotoxicity.
(A) Workflow of testing the role of TRAF3 in regulating the response of cancer cells to T-cell-driven cytotoxicity through in vitro co-culture. Traf3-normal or -deficient B16F10 cells were cultured with antigen-specific CD8+ T cells (Pmel-1 or OT-I) for 1–3 days. Cell number and cell surface MHC-I/PD-L1 expression were quantified by FACS. (B-C) Relative B16F10 cell number after co-culture with (B) OT-I T cells or (C) Pmel-1 T cells at different E:T ratios revealed a higher sensitivity of Traf3-deficient B16F10 cells to T-cell-mediated cytotoxicity. For co-culture with Pmel-1 T cells, B16F10 cells were pre-treated with 1ng/ml IFNγ for 12 hours prior to the co-culture. The bar plots present the relative cell number in each group, normalized to the cell number in the sgControl group in each E:T condition. Mean ± s.d. and individual replicate values are shown for each group. (**P < 0.01, ***P < 0.001; Two-way ANOVA with Benjamini-Hochberg post test comparing sgTraf3 and sgControl in each condition). (D) Relative MFI of H2-Kb or PD-L1 of B16F10 cells co-cultured with OT-I T cells. Values are normalized to the sgControl group in E:T = 0.25 condition. Mean ± s.d. and individual replicate values are shown for each group. (**P < 0.01, ***P < 0.001; Two-way ANOVA with Benjamini-Hochberg post test comparing sgTraf3 and sgControl in each condition). (E) Relative B16F10 cell number after co-culture with Pmel-1 T cells at different E:T ratios revealed a lower sensitivity of Traf3-overexpressing B16F10 cells to T-cell-mediated cytotoxicity. The bar plots present the relative cell number in each group, normalized to the cell number in the pEF1α group in each E:T condition. Mean ± s.d. and individual replicate values are shown for each group. (**P < 0.01, ***P < 0.001; Two-way ANOVA with Benjamini-Hochberg post test comparing pEF1α-Traf3 and pEF1α in each condition). (F) Workflow of testing the role of TRAF3 in modulating the ICB response in vivo. 4×105 B16F10 cells (Traf3-normal or -deficient) were transplanted subcutaneously into syngeneic recipient mice. Starting on Day 6 post-transplantation, we treated the recipients with control IgG or combination ICB every 3rd day for a total of 4 doses. We monitored tumor size and recipient survival. (G) Longitudinal tumor size of sgControl or sgTraf3 tumors treated by control IgG or ICB. Mean ± s.e.m. is shown for each group at each time point. (**P < 0.01, ***P < 0.001; Two-way ANOVA with Benjamini-Hochberg post test comparing sgTraf3 and sgControl in each condition). (H) Kaplan-Meier curves of recipients of sgControl or sgTraf3 tumors treated by control IgG or ICB. The sgTraf3 cohort with ICB treatment survived significantly longer than the other groups. (**P < 0.01; Log-rank test with Benjamini-Hochberg adjustment of multiple comparisons).
We further tested the effect of Traf3 KO on tumor response to ICB treatment in vivo. Specifically, we transplanted Traf3-WT or -KO B16F10 cancer cells in vivo, treated the recipient mice with control IgG or combined anti-PD-1/anti-CTLA-4 therapy, and monitored disease progression over time (Figure 3F). Consistent with the in vitro co-culture results, Traf3-KO cancer cells showed a better response to ICB treatment than Traf3-WT cells in vivo, and recipients of Traf3-KO cancer cells survived significantly longer upon ICB treatment (Figure 3G-H). Interestingly, Traf3-KO tumors also grew more slowly than TRAF3-normal tumors even without ICB (Figure 3G). In addition, depletion of CD8 T cells by anti-CD8 treatment ablated the slower tumor growth caused by Traf3-KO, with control IgG or ICB treatment (Figure S4G). This indicated that the effect of Traf3-KO on tumor growth is dependent on the presence of CD8 T cells. To provide further support of our experimental observations, we analyzed multiple published co-culture or in vivo CRISPR screens. In general, Traf3-KO tended to sensitize cancer cells to T-cell-driven cytotoxicity in those studies (Figure S4H), in agreement with our finding. In contrast, in NK-cell co-culture CRISPR screens, Traf3-KO seemed to desensitize cancer cells to NK-cell-killing (Figure S4I), consistent with the role of MHC-I in inhibiting NK cell cytotoxicity (65).
Traf3-KO signature is correlated with higher MHC-I in primary patient samples
Next we evaluated whether TRAF3 function was correlated with MHC-I expression or T cell infiltration in clinical cohorts where bulk tumor RNA-seq has been performed. Because TRAF3 regulates downstream TF activity, rather than directly using TRAF3 expression itself to measure TRAF3 functional deficiency, we derived a Traf3-KO gene expression signature from RNA-seq (Figure S5A, Table S3). Specifically, we used the top differential genes from TRAF3-deficiency as signature genes, and evaluated the Traf3-KO signature score in each clinical tumor RNA-seq sample by a weighted sum of signature gene expression. We further used it to assess the association of TRAF3 deficiency with MHC-I antigen presentation, cytotoxic T cell infiltration, overall survival, and immunotherapy response in clinical cohorts as appropriate.
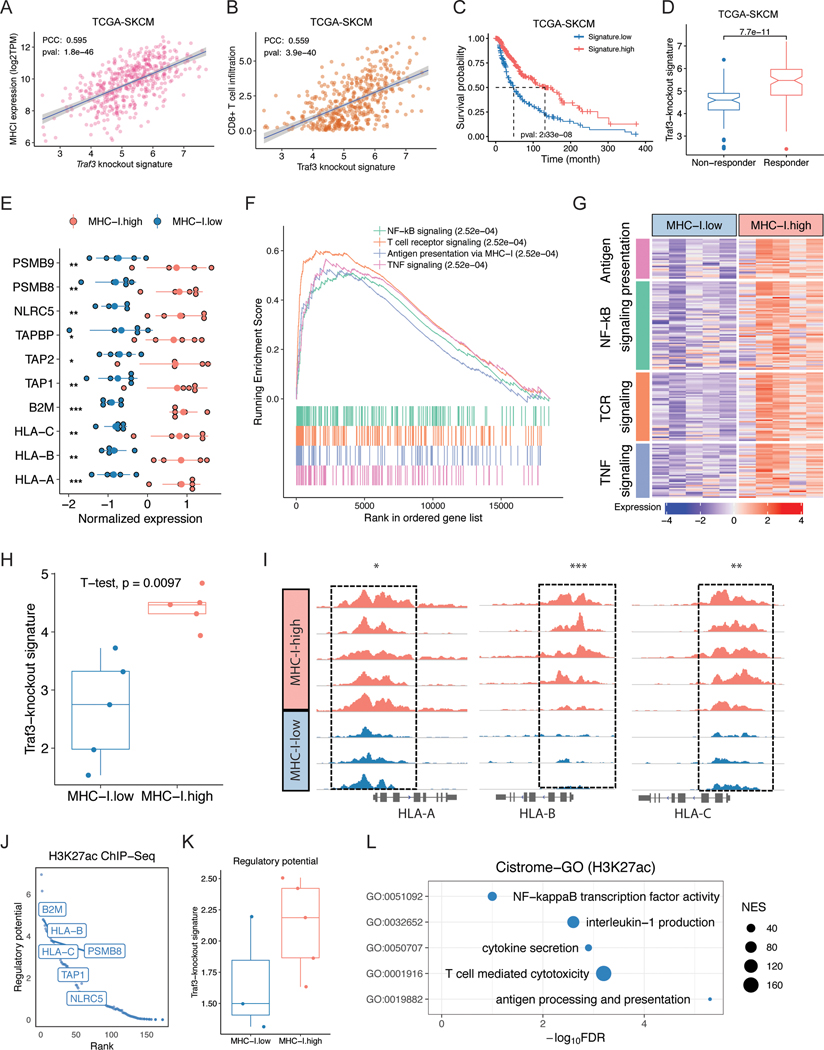
We first examined TCGA tumors, where baseline anti-tumor immunity can influence prognosis, although patients were not treated with ICB (66,67). We found the Traf3-KO signature level to be positively correlated with MHC-I expression, CD8+ T cell infiltration, and patient survival in melanoma (Figure 4A-C). We also applied an orthogonal algorithm TIDE (66), which we developed to predict ICB response based on expression signatures of T cell dysfunction and exclusion, to TCGA tumors as if they were to be treated with ICB. The predicted responders also showed higher Traf3-KO signature score than the predicted non-responders (Figure 4D). Furthermore, we observed positive correlation of Traf3-KO signature and MHC-I expression, CD8 T-cell infiltration, and patient survival in multiple other cancer types in TCGA (Figure S6A-D). Finally, to assess the MHC-I-specific effect of Traf3-KO, we created a restricted Traf3-KO signature to include just the differential genes in the MHC-I component genes. This MHC-I-restricted Traf3-KO signature still shows positive correlation with patient survival in many cancer types in TCGA (Figure S6E). These results suggest that the MHC-I-induction effect by TRAF3 inhibition is associated with better clinical outcome.
Figure 4. Traf3-knockout signature is correlated with higher MHC-I in primary patient samples.
(A-C) Traf3 knockout signature is positively correlated with (A) MHC-I expression, (B) CD8+ T cell infiltration, and (C) patient survival in the TCGA SKCM dataset. PCC: Pearson correlation coefficient. (D) TIDE-predicted ICB responders showed higher Traf3-knockout signature values in the TCGA SKCM dataset. (E) Expression of MHC-I component or related genes in MHC-I-high or -low samples. (F) GSEA of differentially expressed genes in MHC-I-high versus MHC-I-low samples. (G) MHC-I-high samples show higher expression of genes involved in antigen presentation, NF-κB signaling, TNF signaling, and Toll-like receptor pathways. (H) Traf3-knockout signature score in MHC-I-high or MHC-I-low RNA-seq samples. (I) H3K27ac ChIP-seq results for the HLA-A/B/C loci in MHC-I-high or MHC-I-low samples. (J) Top 200 genes with the higher regulatory potential value in MHC-I-high versus MHC-I-low H3K27ac ChIP-seq samples. (K) Traf3-knockout signature score of H3-K27ac ChIP-seq regulatory potential values in MHC-I-high or -low samples. (L) Cistrome-GO enrichment analysis of H3K27ac ChIP-seq peaks with stronger signal in MHC-I-high compared to MHC-I-low samples.
In addition to publicly available databases, we also collected primary samples from treatment-naive melanoma patients treated at the Dana-Farber Cancer Institute. Assessment of MHC-I levels by immunohistochemistry (IHC) of the formalin-fixed paraffin-embedded (FFPE) samples identified MHC-I-high and MHC-I-low tumors, from which we macrodissected cancer cell sections for RNA-seq and H3K27 acetylation FitAc-seq (68) to examine the molecular features associated with MHC-I expression. RNA-seq results on MHC-I components (HLA-A, HLA-B, HLA-C, and B2M) and other genes in the same pathway were consistent with the IHC results (Figure 4E). Examination of the differentially expressed genes revealed 673 upregulated and 425 downregulated genes in the MHC-I-high compared to the MHC-I-low samples, with the upregulated genes being enriched in NF-κB and TNF signaling pathways (Figure 4F-G, Table S4). Indeed, MHC-I-high samples showed a significantly higher Traf3-KO signature score (Figure 4H). Consistent with the RNA-seq data, ChIP-seq revealed higher levels of H3K27ac near MHC-I and higher regulatory potential of MHC-I-related genes (Figure 4I-J, Table S4). The regulatory potential of Traf3-KO signature genes was also higher in the MHC-I-high samples (Figure 4K). Cistrome GO analysis of the differentially active chromosomal regions from ChIP-seq revealed that NF-κB binding sites are more active in the MHC-I-high samples (Figure 4L). These results suggest that the negative regulation of MHC-I by TRAF3 is also reflected in the primary melanoma tumors from patients.
Traf3-KO signature is correlated with better ICB response
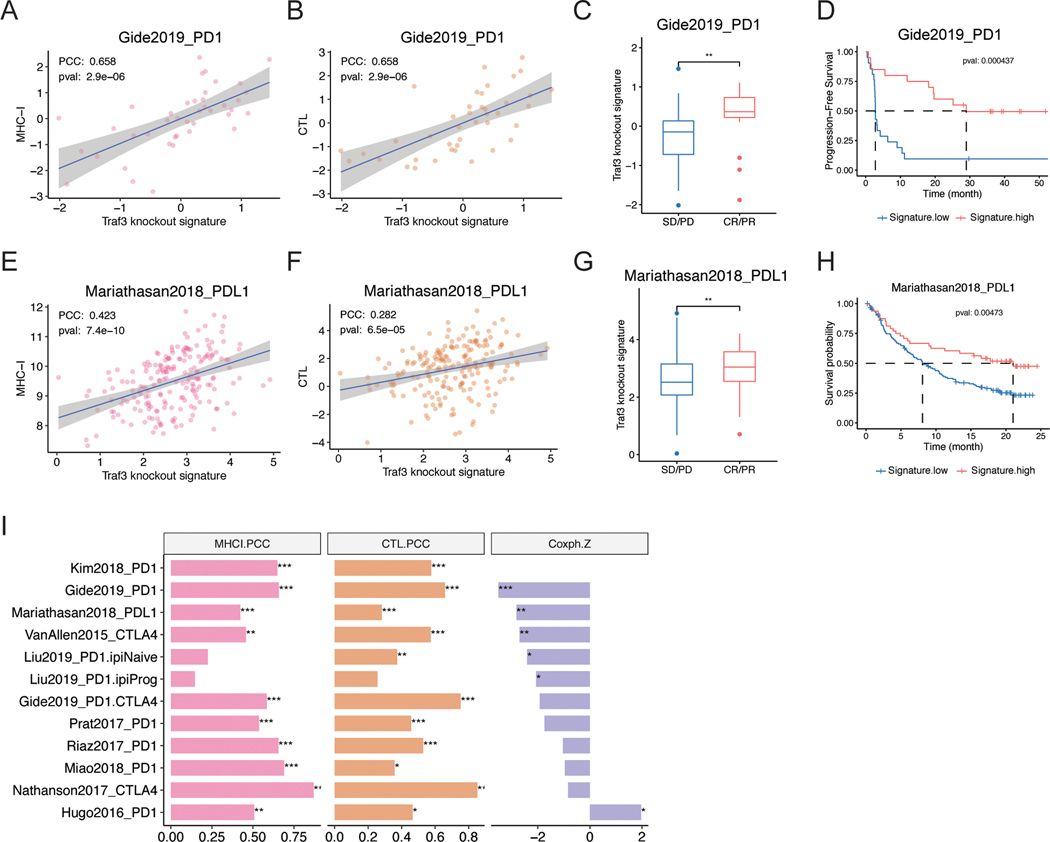
In patients treated with immunotherapy, antigen expression and/or presentation levels is known to be a critical factor of treatment response and an issue addressed in multiple response biomarker studies (69,70). Higher MHC-I expression is associated with better ICB response in most published ICB cohorts (Figure S7A). Based on the role of TRAF3 in MHC-I regulation, we tested the correlation between TRAF3 functional deficiency and immunotherapy response in the published ICB treatment cohorts. We examined the RNA-seq data of pre-treatment biopsies in a recent melanoma cohort with 63 biopsies treated with anti-PD-1 monotherapy and 57 treated with combined anti-PD-1 and anti-CTLA-4 (71). We confirmed that the Traf3-KO signature was positively correlated with tumor MHC-I, cytotoxic T-cell infiltration, ICB response, and progression-free survival (Figure 5A-D). Because the MHC-I-TCR (T-Cell Receptor) interaction is a conserved mechanism for T-cell recognition of cancer cells, we expect the Traf3-KO signature to be associated with ICB response in various cancer types.
Figure 5. Traf3-knockout signature is correlated with better response to ICB.
(A-D) Traf3 knockout signature is positively correlated with (A) MHC-I expression, (B) intratumoral CTL infiltration, (C) ICB response, and (D) overall survival and progression-free survival in patients treated by anti-PD-1 or combined anti-PD-1 and anti-CTLA-4 in the Gide et al. (71) study in melanoma. (E-H) Traf3 knockout signature is positively correlated with (E) MHC-I expression, (F) intratumoral CTL infiltration, (G) ICB response, and (H) overall survival and progression-free survival in patients treated by anti-PD-1 or combined anti-PD-1 and anti-CTLA-4 in the Mariathasan et al. (72) study in urothelial carcinoma. (I) Traf3 knockout signature is positively correlated with MHC-I expression and cytotoxic T cell infiltration and negatively correlated with survival hazard in most ICB treatment clinical trials.
We therefore applied our findings to another study with ~300 urothelial carcinoma biopsies (72), the largest published cohort with available expression data. Again, Traf3-KO signature was positively correlated with tumor MHC-I, cytotoxic T cell infiltration, ICB response, and progression-free survival (Figure 5E-H). Furthermore, the complete or MHC-I-restricted Traf3-KO signature showed similar trend (Table S5, Figure 5I, Figure S7B-E) in most published ICB clinical studies across different cancer types. Together, our data support the general relevance of Traf3-KO signature in predicting ICB response.
Identification of a SMAC mimetic, birinapant, as a regulator of MHC-I to enhance ICB efficacy
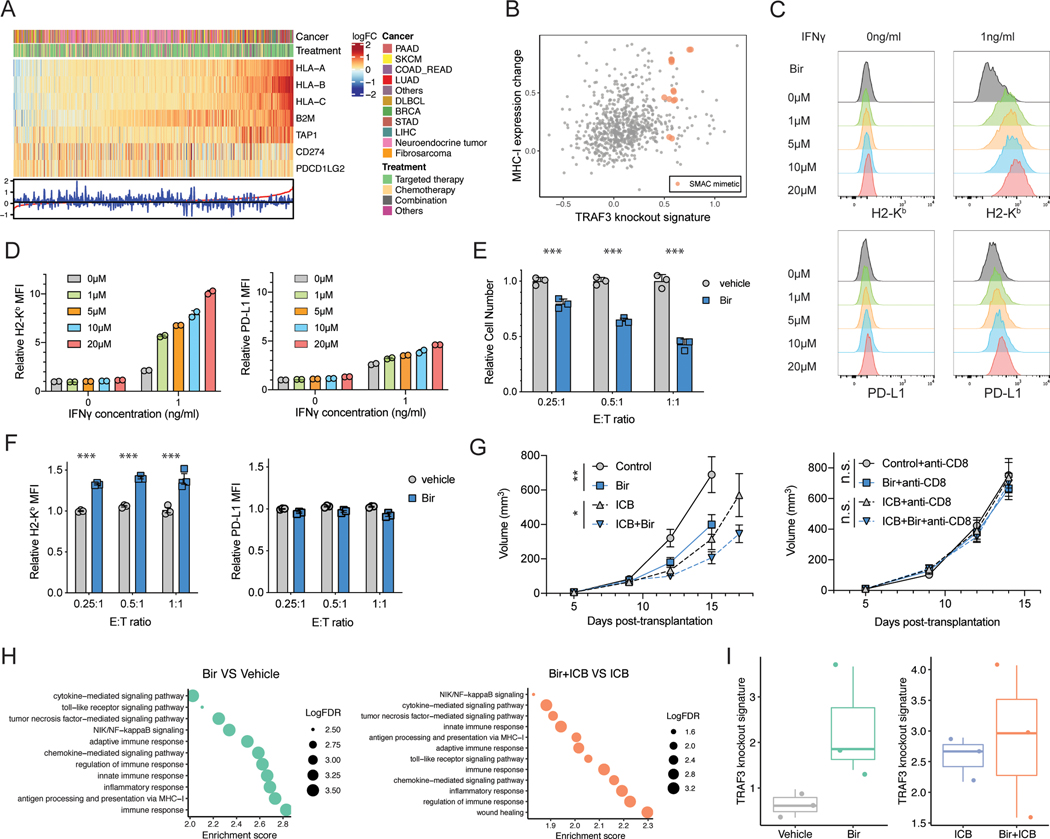
Based on the effect of Traf3-KO on MHC-I levels and sensitivity to T-cell-driven killing, we hypothesized that drug treatment that mimics the transcriptional effect of Traf3-KO may achieve similar outcomes. We searched existing transcriptomic studies on drug treatment effects in the GEO database, with differential expression data involving over 200 targeted-/chemo-therapy drugs. Different drug treatments showed distinct transcriptional regulation of MHC-I components and PD-L1/PD-L2 (Figure 6A, Figure S8A, Table S6). We computed the Traf3-KO signature score of each drug treatment and reassuringly, it is significantly positively correlated with MHC-I expression (Figure S8B). Focusing on drugs that increase Traf3-KO signature and MHC-I expression, we found several drugs previously reported to upregulate MHC-I, including the CDK4/6 inhibitor palbociclib (38) and the EZH2 inhibitor EPZ6438 (73) (Table S6). Furthermore, we found multiple SMAC (Second Mitochondria-derived Activator of Caspase) mimetics as novel candidates for MHC-I-specific regulation (Figure 6B). SMAC mimetics are a family of pro-apoptotic compounds that can inhibit the cIAP/XIAP E3 ligases, leading to activated NF-κB activity. We therefore examined the immunomodulatory effects of a SMAC mimetics, birinapant, using in vitro and in vivo assays.
Figure 6. Birinapant can specifically upregulate MHC-I and add to the efficacy of ICB treatment.
(A) Heatmap of the regulation of transcription of MHC-I components and PD-L1/PD-L2 in each drug treatment condition. Data are ordered by the magnitude of MHC-I regulation from the most upregulated (left) to the most downregulated (right). (B) Characterization of each drug treatment for effect on MHC-I expression and Traf3-knockout signature value. (C) Flow cytometry of B16F10 cells treated by IFNg and/or different concentrations of the SMAC mimetic birinapant (Bir). Typical histograms of H2-Kb and PD-L1 levels in each condition are shown. (D) The relative MFI of samples in (C). Mean ± s.d. is shown for each group. Values are normalized to the mean MFI at 0ng/ml IFNγ 0μM birinapant. (E) Relative B16F10 cell number after co-culture with OT-I T cells at different E:T ratios revealed a higher sensitivity of B16F10 cells to T-cell-mediated cytotoxicity under birinapant treatment. The bar plots present the relative cell number in each group, normalized to the cell number in the vehicle treatment group in each E:T condition. Mean ± s.d. and individual replicate values are shown for each group. (***P < 0.001; Two-way ANOVA with Benjamini-Hochberg post test). (F) Relative MFI of H2-Kb or PD-L1 of B16F10 cells co-cultured with OT-I T cells. Values are normalized to the vehicle treatment group in E:T = 0.25 condition. Mean ± s.d. and individual replicate values are shown for each group. (***P < 0.001; Two-way ANOVA with Benjamini-Hochberg post test comparing vehicle and SMAC treatment groups in each condition). (G) Longitudinal tumor size of tumors under different treatments. Mean ± s.e.m. is shown for each group at each time point. (**P < 0.01, ***P < 0.001; Two-way ANOVA with Benjamini-Hochberg post test). (H) GSEA of the bulk tumor RNA-seq data evaluating the gene sets that were upregulated in response to SMAC mimetic. (I) Quantification of Traf3 knockout signature in each treatment group revealed a trend toward upregulation in response to birinapant treatment.
We first tested the effect of birinapant treatment on MHC-I levels in mouse B16F10 and CT26 cells. Indeed, birinapant treatment specifically upregulated MHC-I in response to IFNγ treatment, with minimal effect on PD-L1 (Figure 6C-D, Figure S8C). Furthermore, birinapant can also specifically upregulate MHC-I in multiple human cancer cell lines (Figure S8D-G). Similar to the effect of Traf3-KO, birinapant treatment led to reduced inactive forms of NF-κB (p100 and p105) and increased active form of NF-κB (p52, Figure S8H). Consistent with the reported role of SMAC mimetics to induce cIAP degradation (74,75), we found lower cIAP and higher NIK levels in birinapant-treated cells (Figure S8I). When added to the cancer cells co-cultured with OT-I T cells, birinapant resulted in specific upregulation of MHC-I in cancer cells and facilitated better cancer-cell killing by T cells (Figure 6E-F). We further tested whether this immunomodulatory effect of birinapant functions through TRAF3 in cancer cells by using Traf3-WT or -KO cancer cells co-cultured with antigen-specific T cells, treated by vehicle or birinapant. TRAF3-deficient cancer cells showed comparable high levels of MHC-I expression and sensitivity to T-cell-mediated cytotoxicity as birinapant-treated cells, and were not further sensitized by birinapant (Figure S8J-K). These data suggest that birinapant’s effect on MHC-I expression is dependent on the presence of TRAF3.
We further tested the effect of birinapant in vivo, by treating established B16F10 melanoma tumors with birinapant and/or combination anti-PD-1/CTLA-4. We observed moderately reduced tumor growth in vivo with birinapant treatment alone, and further reduced tumor progression with combination therapy of birinapant and ICB (Figure 6G, left panel). In addition, CD8 T cell depletion by anti-CD8 treatment ablated the slower tumor growth caused by birinapant treatment (Figure 6G, right panel), indicating the effect of birinapant treatment on tumor growth to be CD8 T-cell-dependent. We performed RNA-seq of the bulk tumor in each treatment group to identify the pathways regulated by birinapant. Indeed, birinapant-upregulated genes, whether or not with ICB treatment, are involved in inflammatory response, innate/adaptive immune response, chemokine/cytokine-mediated signaling, NF-κB signaling, and MHC-I antigen presentation (Figure 6H, Table S7). Inference of tumor immune infiltration by TIMER (76,77) predicted a trend for higher infiltration of dendritic cells and CD8+ T cells, though the difference is not statistically significant (Figure S8L-M). In addition, birinapant treatment increased the Traf3-KO signature in the tumor (Figure 6I, left panel), consistent with our in silico prediction and in vitro epistatic experiment showing that SMAC mimetic treatment phenocopies Traf3-KO. In contrast, we did not observe a significant difference in Traf3-KO signature between Bir+ICB and ICB groups (Figure 6I, right panel). Since we evaluated tumor RNA-seq after ICB treatment, the cancer cells that show higher Traf3-KO signature were likely more susceptible to immune killing, making it difficult to detect such signature through bulk tumor RNA-seq. This ICB-induced selection for immune-resistant clones was characterized in our recent work applying clonal tracing of cancer cells following ICB treatment (78). Furthermore, the presence of other cell types in the tumor microenvironment might obscure the enrichment of Traf3-KO signature.
Discussion
Immunotherapy, especially ICB, has brought paradigm shifts to cancer treatment and improved patient survival in many cancer types. Its therapeutic effect relies on the recognition of cancer-specific antigens by cytotoxic T cells, and defects in cancer-cell antigen presentation is a major ICB-resistance mechanism. Multiple studies have sought to identify regulators of MHC-I (36,44). However, due to the common regulatory pathways shared by MHC-I and PD-L1, upregulating MHC-I often leads to upregulation of PD-L1, which suppresses anti-tumor immunity. In this study, we established a workflow to identify drugs with MHC-I-specific enhancing effects. We first performed dual-marker FACS-based CRISPR screens to identify regulators of MHC-I and/or PD-L1, and found TRAF3 as an MHC-I-specific negative regulator through its modulation of the NF-κB pathway. TRAF3 depletion sensitized cancer cells to antigen-specific T-cell-driven cytotoxicity. Traf3-KO signature is significantly correlated with higher expression of genes in the antigen presentation pathway, higher cytotoxic T-cell infiltration, and better overall survival in cancer patients from many TCGA cohorts and ICB clinical trials. Furthermore, data mining of public transcriptomic profiles of drug treatment followed by experimental validation identified that birinapant, a SMAC mimetic, phenocopies the effect of TRAF3-deficiency on MHC-I expression and adds to the efficacy of ICB treatment in suppressing tumor growth. Our study identifies a novel treatment strategy that can potentially be used in MHC-I-low cancers to enhance ICB therapeutic efficacy.
TRAF3 has been reported to negatively regulate the NF-κB pathway activity, and multiple ChIP-seq studies revealed NF-κB binding sites in the promoter/enhancer regions of MHC-I components. Consistent with these previous studies, we found that Traf3-KO upregulates MHC-I in an NF-κB-dependent manner. There are no published TRAF3 small molecule inhibitors, so we searched published studies to identify drugs that phenocopy the effect of TRAF3 inhibition. The SMAC mimetic family ranked among the top of the list. SMAC mimetics can trigger the auto-ubiquitination and degradation of cIAPs, thereby promoting NF-κB activation via canonical or non-canonical pathways (49,79). We then experimentally validated that birinapant, a SMAC mimetic, can promote the immunosensitivity of cancer cells through specifically inducing MHC-I expression in a TRAF3/NF-κB-dependent manner. A recent study also showed that SMAC mimetics can synergize with ICB in a CD8 T-cell- and TNFα-dependent manner (80), although SMAC mimetics did not upregulate MHC-I in their glioblastoma model (CT-2A). Our preliminary analyses of published scRNA-seq data from various cancer types (81) found that glioblastoma cells tend to have high expression of MHC-I and NF-κB component genes. We speculate that cells already high in MHC-I and NF-κB might not increase MHC-I when treated with SMAC mimetics. Indeed, in A375 and 786O cell lines with higher baseline MHC-I and NF-κB expression, birinapant also failed to increase MHC-I. These results suggest context-dependent MHC-I regulation and anti-tumor response induced by SMAC mimetics, which merit further mechanistic studies.
In addition to SMAC mimetics, mining the GEO database identified multiple other targeted-therapy or chemotherapy agents with the potential to cause a transcriptional effect similar to TRAF3 inhibition. This comports with multiple other studies (37–39,82,83) in revealing the immunomodulatory effects of chemotherapy and targeted therapies. For example, palbociclib, a CDK4/6 inhibitor reported to induce MHC-I via type-III interferon (38), and EPZ6438, an EZH2 inhibitor reported to induce MHC-I via chromatin modification (36,44), ranked among the top candidates. Regimens containing azacitidine, another top-ranking candidate, were also reported to induce antigen presentation in non-small-cell lung cancer cells (84). Our approach using differential gene expression signatures can be extended to identify drugs with other immunomodulatory effects.
Our study has limitations that future studies could address. First, our finding can potentially assist the treatment of tumors whose MHC-I is suppressed through gene regulatory mechanisms, but not those with genetic deletion of MHC-I components or loss-of-heterozygosity of HLA genes. Nevertheless, the clinical observation in melanoma treated by ICB (13) suggests that a large number of patients can still benefit from this approach. Second, in addition to upregulation of MHC-I, other mechanisms may also contribute to the immune sensitivity of Traf3-KO tumors. For example, Traf3-KO cells had higher expression of chemokines which may facilitate T cell infiltration. Further studies are needed to elucidate such alternative mechanisms and their impact on the immune sensitivity of Traf3-KO tumors. Third, when mining drug treatment data to predict drugs that may upregulate MHC-I through TRAF3/NF-κB, our analysis may be limited by the amount of drug treatment data available in GEO. More publicly available transcriptional profiling data on drug screening would improve the power of our approach.
In summary, we present a workflow integrating CRISPR screens with functional genomics characterization and computational mining of publicly accessible data to identify novel regulators of tumor immunity. We showed that the SMAC mimetic birinapant can phenocopy TRAF3 deficiency and preferentially induce MHC-I expression, leading to enhanced sensitivity of cancer cells to T-cell-driven cytotoxicity. The combination of SMAC mimetic and ICB is likely to benefit patients with a low baseline level of MHC-I expression. This is potentially exciting given the biosafety of profiles of birinapant from multiple early phase clinical studies. Furthermore, our general approach can be applied to identify other drugs with immunomodulatory effects.
Materials and Methods
TCGA cohorts
We downloaded all the TCGA tumor data from the Broad GDAC Firehose, including transcriptomic profiles and clinical information of tumor samples from 33 cancer types. All of the gene expression levels were measured by logarithmic Transcripts Per Kilobase Million (TPM). We downloaded the IHC-derived tumor purity data from the Aran et al. study (85) and we also estimated the tumor purity using the CHAT algorithm (86). For all the tumors, we estimated the immune cell infiltration in the tumor microenvironment using TIMER algorithm, with default parameters and TPM as input. Because of the distinct clinical behavior and molecular profile of cancer subtypes, we divided breast cancer data sets into the PAM50 subtypes of basal, lumA, lumB and Her2 positive. We focused on cancer types where higher MHC-I is associated with better overall survival (CoxPH regression of patient survival proportional hazard and MHC-I expression with a z-score < −0.5) for downstream analyses.
CCLE cohorts
We downloaded the RNA-Seq data of more than 1,000 cancer cell lines from the DepMap Portal, which contains processed gene expression data quantified according to the Genotype-Tissue Expression (GTEx) pipelines.
Flow cytometry
We performed fluorescence-activated cell sorting (FACS) of B16F10 or CT26 cells treated under different conditions as indicated to quantify the level of H2-Kb/H2-Kd and PD-L1. B16F10 or CT26 cells were dissociated by TrypLE treatment, washed with PBS-2%FBS, and incubated with DAPI (1:10,000 dilution), anti-H2-Kb (clone AF6–88.5, BioLegend, 1:400 dilution), and anti-PD-L1 (clone MIH5, BD Biosciences, 1:400 dilution) for 1 hour on ice. Cells were then washed and resuspended in PBS-2%FBS and analyzed on BD LSR-Fortessa instrument. FACS data was then analyzed by Flowjo software.
Cell lines and cell culture
Cell lines B16F10, CT26, SKMEL5, and MCF7 were purchased from ATCC and authenticated using standard short tandem repeat analysis in 2019. Cell line K028 was derived from a melanoma patient under institutional review board–approved protocols. Cell line HT29 was obtained from the CCLE core facility, which obtained them directly from commercial sources and authenticated the lines using standard short tandem repeat analysis. 293FT, B16F10, SKMEL5, K029, and MCF7 were cultured in DMEM supplemented with 10% fetal bovine serum, 1% L-glutamine, and 1% penicillin and streptomycin. CT26 and HT29 were cultured in RPMI supplemented with 10% fetal bovine serum, 1% L-glutamine, and 1% penicillin and streptomycin. All cells were used at low passage numbers and were tested for Mycoplasma using the MycoAlert Mycoplasma Detection Kit (Lonza).
CRISPR screens for regulators of MHC-I
We performed CRISPR screens to identify regulators of MHC-I induced by IFNγ. We first transduced 1×108 B16F10 cells with our M1/M2 mouse genome-wide CRISPR library at MOI of 0.3 to ensure that most transduced cells received only 1 virion. We cultured the transduced cells for 2 days in DMEM full media, selected with 2μg/ml puromycin for 2 days, and cultured for an additional 2 days to allow depletion of residual protein of the targeted genes. We then treated the cells with high or low dosage IFNγ to identify MHC-I regulators.
For identification of positive regulators, we treated the transduced cells with 10ng/ml IFNγ for 2 days, and flow-sorted 2×107 cells for the following populations: MHC-Ihi PD-L1hi, MHC-Ihi PD-L1lo, MHC-Ilo PD-L1hi, MHC-Ilo PD-L1lo. The gRNAs that are present in the MHC-Ilo PD-L1hi population are expected to enrich for MHC-I-specific positive regulators. For identification of negative regulators, we treated the transduced cells with 0.1ng/ml IFNg for 2 days, and flow-sorted 2×107 cells for the following populations: MHC-Ihi PD-L1hi, MHC-Ihi PD-L1lo, MHC-Ilo PD-L1hi, MHC-Ilo PD-L1lo. The gRNAs that are present in the MHC-Ihi PD-L1lo population are expected to enrich for MHC-I-specific negative regulators.
Genomic DNA extraction and gRNA library construction were performed as described before (87). Briefly, we extracted genomic DNA from each sorted population or from the unsorted population by phenol-chloroform extraction, and then performed two rounds of PCR to construct the barcode-indexed sequencing library for each sample. We sequenced each library at ~300X average coverage over the CRISPR library.
Analysis of CRISPR screen
MAGeCK software was used to quantify and test for sgRNA and gene enrichment (41). The sequence reads were trimmed to remove a constant portion of the sgRNA sequences and mapped to the M1/M2 mouse genome-wide CRISPR library with MAGeCK “count” module, which computes read counts for each sgRNA. We used MAGeCK “test” module to identify MHC-I/PD-L1 regulators, for which targeting sgRNA were significantly enriched in the sorted populations compared to the control unsorted populations grown in parallel. MAGeCK returned sgRNA and gene log fold changes (LFCs), which represent the enrichment level of the sgRNAs and genes in each cell population. Since negative selection is meaningless in FACS-based CRISPR screens, we assigned zero log fold-change for genes whose log fold-change were negative. Finally, we calculated the mean log fold-change of genes in M1 and M2 CRISPR screens and visualized the results using functions in R package MAGeCKFlute(42).
We analyzed the data from the cohort treated by 10ng/ml IFNg to identify the positive regulators of MHC-I and/or PD-L1. Specifically, we compared the gRNA distribution between the MHC-Ilo PD-L1lo population and unsorted population to identify positive regulators for both; between the MHC-Ilo PD-L1hi population and the unsorted population to identify MHC-I-specific positive regulators; and between the MHC-Ihi PD-L1lo population and the unsorted population to identify PD-L1-specific positive regulators.
We analyzed the data from the cohort treated by 0.1ng/ml IFNg to identify the negative regulators of MHC-I and/or PD-L1. Specifically, we compared the gRNA distribution between the MHC-Ihi PD-L1hi population and unsorted population to identify negative regulators for both; between the MHC-Ihi PD-L1lo population and the unsorted population to identify MHC-I-specific negative regulators; and between the MHC-Ilo PD-L1hi population and the unsorted population to identify PD-L1-specific negative regulators.
Cloning of screen candidates for validation
We cloned 4 gRNAs of Traf3 into the LentiCRISPRv2-Puro construct to validate the role of TRAF3 in the regulation of MHC-I. We used the cloning protocol as previously established(88,89). Out of the 4 gRNAs, 2 successfully depleted TRAF3 protein level >80%. We used these two gRNAs for our follow-up validation studies.
We also cloned the open reading frame (ORF) of Traf3 into pER1α-Puro vector for over-expression of Traf3 and examination of its role in MHC-I/PD-L1 regulation.
Validation of the role of TRAF3 in MHC-I / PD-L1 regulation
We performed CRISPR-Cas9-mediated deletion of Traf3 in multiple cell lines, including B16F10 and CT26. For each line, we cultured Traf3-normal or -deficient cells with different dosages of IFNγ, TNFα, IFNα, or IFNβ for 2 days to test its regulation of MHC-I and/or PD-L1 with these induction methods.
Virus packaging
Viral packaging was performed as previously described(87). Briefly, we cultured 293FT cells at 40%~50% confluence the day before transfection. Transfection was performed using X-tremeGENE HD (Sigma-Aldrich). For each 10-cm dish, 8 ug of lentivectors, 2.4 ug of pMD2.G, and 6 ug of psPAX2 were added into 2 ml OptiMEM (Life Technologies). 40 ul of X-tremeGENE HD was added to the plasmid mixture. The complete mixture was incubated for 15 min and then added to 1.1 × 107 293FT cells per 10-cm dish. After overnight culture, the media was changed to 8 ml DMEM + 15% FBS for virus collection. We harvested virus at 48hr, 60hr, and 72hr after the start of transfection.
Co-culture of cancer cells and T cells for T cell cytotoxicity assay
Co-culture of cancer cells and T cells was performed as previously described (66). Briefly, B16F10 cells were maintained in complete DMEM media (10% FBS and 50U/ml of Penicillin-Streptomycin). CD8 T cells isolated from spleen and lymph nodes from Pmel-1 or OT-I mice were stimulated with anti-CD3/CD28 beads (ThermoFisher, 11452D) and then cultured in complete RPMI 1640 media (10%FBS, 20mM HEPES, 1mM sodium pyruvate, 0.05mM 2-mercaptoethanol, 2mM L-glutamine, and 50U/ml streptomycin and penicillin, 20ng/ml recombinant mouse IL-2).
To test the sensitivity of cancer cells to T cell-driven cytotoxicity (OT-I or Pmel-1 model), we plated B16F10 cells (sgRosa26, sgTraf3, pEF1a-Empty, or pEF1a-Traf3) at equal density in all wells, and added T cells at ratios to cancer cells. With the OT-I model, we first incubated the B16F10 cells with 1nM SIINFEKL peptide for 2 hours prior to co-culture with T cells. With the Pmel-1 model, B16F10 cells were either pre-treated with 1ng/ml IFNγ overnight or untreated prior to the co-culture. There are 2–4 cell-culture replicates for each condition. After a one-day or three-day co-culture with T cells, we counted the remaining cancer cells by FACS using the precision count beads (BioLegend, 424902). T cells present in these cultures were gated out based on antibodies specific for CD45 (Biolegend, clone 30-F11) or CD8 (BioLegend, clone 53-6.7).
Immunoblot
We treated B16F10 cells (sgRosa26 or sgTraf3) by 1ng/ml IFNγ for 0, 0.5, or 24 hours and lysed cells in RIPA buffer. Lysates were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and immunoblotting was performed as described (90). To visualize NIK levels, cells were pre-treated by 0μM or 5μM MG132 for 4 hours before lysis. Antibodies specific for TRAF3, STAT1, phospho-STAT1(Y705), RELA, RELB, phospho-RELA(S536), cREL, p100/p52, p105/p50, IKK, and NIK were from Cell Signaling Technology, Danvers, MA, USA. Antibody specific for VCL was from Santa Cruz Biotechnology, Dallas, TX, USA. Antibody specific for cIAP was from R&D Systems. Antibodies were used at each manufacturer’s recommended concentrations.
RNA-seq
For comparison of transcription profile between control and Traf3−/− B16F10 cells, we treated sgControl or sgTraf3-transduced B16F10 cells with vehicle control or 1ng/ml IFNγ for 2 days, and extracted total RNA using the miRNeasy Plus Mini Kit (Qiagen, #17004) following the manufacturer’s protocol. For comparison of tumors treated by birinapant and/or ICB, we harvested bulk tumors from recipients in each group on Day 15 post-transplantation and extracted total RNA. Total RNA samples in each experiment were submitted to Novogene Inc. for sequencing. Standard mRNA library preparation kit was used for library preparation. Paired-end 150bp sequencing was performed on Illumina HiSeq 2500. Sequencing reads were mapped to the mm10 genome by RSEM. Statistics for differentially expressed genes were calculated by DESeq2.
Traf3-knockout signature
To study the clinical relevance of TRAF3, we established a Traf3-KO signature by extracting the top 200 upregulated and 200 downregulated genes and taking the normalized DESeq2 wald statistics as weights. The weights of genes are normalized to −1 to 1 range by the equation , where indicates the weight of the gene and indicates the DESeq2 wald statistics of the gene. For each input expression profile, we computed a Traf3-KO signature score to estimate the Traf3-KO level by calculating the weighted sum expression of the signature genes following the equation , where denotes the signature score and denotes the expression level of the gene. Finally, we evaluated the association of TRAF3 deficiency with MHC-I expression, tumor-infiltrating T cells, patient outcome and response to immune checkpoint blockade.
ATAC-seq
For comparison of chromatin accessibility between control and Traf3−/− B16F10 cells, we treated sgControl or sgTraf3-transduced B16F10 cells with vehicle control or 1ng/ml IFNγ for 2 days. ATAC-seq was performed in duplicates (200,000 cells) by the Center for Functional Cancer Epigenetics at DFCI as previously described (78,91). For data analysis, we used Burrows-Wheeler Aligner (BWA) (92) to map sequencing reads to the reference genome and MACS2 (93) for peak calling. DESeq2 (94) was applied to identify the differentially accessible regions between ICB-resistant lines and the parental control line.
Mouse experiments
All mice were housed in standard cage in Dana-Farber Cancer Institute Animal Resources Facility (ARF). All animal procedures were carried out under the ARF Institutional Animal Care and Use Committee (IACUC) protocol and were in accordance with the IACUC standards for the welfare of animals. Wild-type C57BL/6 recipient mice were purchased from Charles River laboratory.
To test the function of TRAF3 in tumor’s response to combination anti-PD-1/anti-CTLA-4 treatment, we transplanted 4×105 sgRosa26 or sgTraf3 B16F10 cells subcutaneously into the right flank of 6–8 week-old female C57BL/6 mice (from Charles River Laboratory). Starting Day 6 post-transplantation, we began treatment with control IgG (clone 2A3, 200μg per mouse in 200ul HBSS buffer) or combination anti-PD-1 (clone 1A12, 100μg per mouse) and anti-CTLA4 (clone 9D9, 100μg per mouse) by intraperitoneal injection every 3rd day for a total of 4 times. We monitored tumor growth along the treatment.
To test the efficacy of ICB and SMAC mimetic combination treatment, we transplanted 4×105 parental B16F10 cells subcutaneously into the left and right flank of 6–8 week-old female C57BL/6 mice (from Charles River Laboratory). Starting Day 5 post-transplantation, we began treatment with vehicle or birinapant (dissolved in 15% captisol, 600μg per mouse) by intraperitoneal injection every 3rd day for a total of 3 times. Starting Day 6 post-transplantation, we began treatment with control IgG (clone 2A3, 200μg per mouse in 200ul HBSS buffer) or combination anti-PD-1 (clone 1A12, 100μg per mouse) and anti-CTLA4 (clone 9D9, 100μg per mouse) by intraperitoneal injection every 3rd day for a total of 3 times. We monitored tumor growth along the treatment and the survival of recipients.
Mice were euthanized using CO2 inhalation. All mouse experiments were performed in compliance with institutional guidelines as approved by the Institutional Animal Care and Use Committees (IACUC) of Dana-Farber Cancer Institute. The maximum tumor diameter permitted under the relevant animal protocols is 20mm, which was not exceeded in any experiment.
Profiling of primary MHC-I-high or MHC-I-low melanoma samples
For comparison of transcription and epigenetic profile between MHC-I-high and MHC-I-low primary melanoma tumors, we performed RNA-seq and FiTAc-seq of human tumor biopsies. Samples were grouped into MHC-I-high or MHC-I-low based on IHC of tumor biopsy from the same sampling.
FiTAc-seq: H3K27Ac ChIP-seq on FFPE samples
FiTAc-seq (68) was used to profile H3K27ac ChIP-seq on FFPE samples. Sample selection was performed with dual-IHC staining assessing tumor enrichment with SOX10 antibody and MHC-I expression status. Accordingly, areas of interest were marked and samples were classified into low or high MHC-I categories. The selected areas of interest were manually macrodissected from 10 unstained sections (10 μm each) per case. Briefly, the macrodissected tissues were washed with xylenes to remove paraffin, rehydrated in an ethanol/water series and incubated overnight with 200 μl of lysis buffer (1%SDS, 50 mM Tris-HCl pH8, 10 mM EDTA) on a thermomixer set at 65C, and shaking at 1000rpm. Next, samples were put on ice, supplemented with protease and deacetylase inhibitors and sonicated with Covaris E220 instrument for 5 min (PIP105, 5%DF, 200cbp) in 1mL AFA Fiber milliTUBEs. Solubilized chromatin (ranging from 450 ng to 3 μg) was pre-cleared, diluted, and immunoprecipitated overnight with 3 μg of H3K27ac antibody (Diagenode, cat#A1723–0041D, lot#C15410196) following a low-input protocol; and finally washed, eluted and DNA purified. Libraries were prepared with Accel-NGS 2S Plus DNA Library Kit (Swift Biosciences, cat. no. 21024) on an automated NGS workstation, amplified for 14 cycles, and sequenced on a Next-seq instrument (PE35).
Immunotherapy trials
We collected 12 cancer patient cohorts with available RNA-Seq data, patient survival, and immunotherapy response from published studies, including more than 900 ICB-treated tumors from 61 gastric cancer, 420 melanoma, 35 renal cell carcinoma, and 348 urothelial cancer. For each cohort, we standardized the transcriptome data across patients by variance stabilizing transformation.
Drug treatment transcriptomic data
By searching for transcriptomic studies on drug perturbation from GEO, we manually collected 719 expression profiles, including both microarray and RNA-seq datasets. Microarray profiles were normalized by the normalizeQuantiles function in limma. For each dataset, we then performed differential expression analysis with limma (for microarray) or DESeq2 (for RNA-seq). Finally, for each drug treatment condition, we got the pattern of gene expression change, which served as the input gene expression signature into signature-based analysis for mining immune-modulating drugs.
Test the effect of SMAC mimetic birinapant on MHC-I expression and sensitivity to T-cell-driven killing
To test the effect of birinapant on MHC-I and PD-L1 expression, we treated B16F10 or CT26 cells with IFNγ (0 or 1ng/ml) and/or birinapant (0, 1, 5, 10, or 20μM) for 48 hours and tested the effects on MHC-I or PD-L1 expression by flow cytometry.
To test the effect of birinapant on sensitivity of cancer cells to T-cell-driven cytotoxicity, we started treatment of B16F10 cells (parental, sgRosa26, or sgTraf3) with vehicle control or 1μM birinapant 24 hours prior to the co-culture and maintained the same condition throughout the co-culture.
Testing the combination efficacy of SMAC mimetic and ICB in vivo is detailed in the section “Mouse experiments”.
Correlation analysis
We took the average log2TPM of HLA-A, HLA-B, HLA-C, and B2M as MHC-I expression, and then tested the correlation between MHC-I and PD-L1 expression by using Pearson’s correlation adjusted by tumor purity estimated by IHC or the CHAT algorithm (85) in 25 cancer types in TCGA. MHC-I and PD-L1 show strong positive correlation in the majority of cancer types, where they also show strong correlation with CD8 T cell infiltration. We also computed the correlation between MHC-I and PD-L1 expression in cancer cell lines, which shows the similar results.
We calculated the Pearson correlation of TRAF3-KO signature score with MHC-I expression and cytotoxic T lymphocyte (CTL) level (average expression of CD8A, CD8B, GZMA, GZMB, and PRF1) in multiple tumor cohorts. The TRAF3-KO signature score shows notable positive correlation with MHC-I expression and CTL level in most of the TCGA cancer types and immunotherapy trials.
Survival analysis
We used the Cox proportional hazards regression analysis to test the association between MHC-I expression and patient survival and selected 16 cancer types, in which the MHC-I expression is positively correlated with patient survival (coxph z-statistics < −0.5), for downstream analysis. To determine the decoupled effect of MHC-I and PD-L1 on patient survival, we tested the association between MHC-I/PD-L1 differential expression and patient survival, and found patients with higher MHC-I/PD-L1 differential expression have longer overall survival in all the 16 cancer types.
The clinical relevance of TRAF3 regulating MHC-I expression is confirmed by testing the association between TRAF3-KO signature and overall survival or progressive survival of patients in TCGA and immunotherapy trials with cox regression.
Gene set enrichment/overrepresentation analysis
All pathway/GO enrichment analysis were performed using the EnrichAnalyze function in MAGeCKFlute R package and the enriched pathways are visualized using functions from MAGeCKFlute.
Cistrome-GO and Toolkit for CistromeDB analysis
We selected top 1000 enriched peaks from ATAC-Seq or H3K27ac ChIP-Seq and entered the BED file into Cistrome-GO website to perform pathway enrichment analysis and Toolkit to identify the transcription factors that have a significant binding overlap with the input peak sets.
Data availability
All sequencing data were uploaded to Gene Expression Omnibus with accession number GSE149826.
Code availability
All codes are available upon request.
Supplementary Material
Significance.
MHC-I loss or downregulation in cancer cells is a major mechanism of resistance to T-cell-based immunotherapies. Our study reveals that birinapant may be used for patients with low baseline MHC-I to enhance ICB response. This represents promising immunotherapy opportunities given the biosafety profile of birinapant from multiple clinical trials.
Highlights.
Novel integrated workflow to identify drugs with desired immunomodulatory effects
Dual-marker CRISPR screens identify candidate gene-specific regulators
Traf3 deletion specifically upregulates MHC-I without inducing PD-L1 in response to various cytokines and sensitizes cancer cells to T-cell-driven cytotoxicity
Traf3 knockout signature is correlated with better survival of cancer patients with or without ICB treatment
The SMAC mimetic birinapant phenocopies Traf3-knockout and sensitizes MHC-I-low melanoma to ICB therapy
Acknowledgements
We thank Drs. Kai Wucherpfennig and Deng Pan for their insightful suggestions on this study.
Additional Information:
Financial support: This study was supported by grants from the NIH (R01CA234018 to XSL, R01AI137337 to BEG, P50CA101942-12 and P50CA206963 to GJF), Breast Cancer Research Foundation (BCRF-19-100 to XSL), Burroughs Wellcome Career Award in Medical Sciences (to BEG), and Sara Elizabeth O’Brien Trust Fellowship (to SG).
Conflict of interest:
G.J.F. has patents/pending royalties on the PD-1/PD-L1 pathway from Roche, Merck MSD, Bristol-Myers-Squibb, Merck KGA, Boehringer-Ingelheim, AstraZeneca, Dako, Leica, Mayo Clinic, and Novartis. G.J.F. has served on advisory boards for Roche, Bristol-Myers-Squibb, Xios, Origimed, Triursus, iTeos, NextPoint, IgM, Jubilant, Trillium and GV20. G.J.F. has equity in Nextpoint, Triursus, Xios, iTeos, IgM, and GV20.
S.J.R. received research funding from Bristol-Myers Squibb, Merck & Co, Kite Pharma/Gilead, and Affimed Pharmaceuticals.
F.S.H. receives research funding from Bristol-Myers Squibb and Novartis; he also consults for Merck, EMD Serono, Novartis, Takeda, Genentech/Roche, Compass Therapeutics, Apricity, Aduro, Sanofi, Pionyr, 7 Hills Pharma, Verastem, Torque, Rheos Kairos, Psioxus Therapeutics, Amgen, and Pieris Pharmaceutical.
M.B. is a consultant to and receives sponsored research support from Novartis. MB is a consultant to and serves on the scientific advisory boards of Kronos Bio, H3 Biomedicine, and GV20 Oncotherapy.
X.S.L. is a cofounder, board member, SAB member, and consultant of GV20 Oncotherapy and its subsidiaries; SAB of 3DMed Care; consultant for Genentech; and stock holder of BMY, TMO, WBA, ABT, ABBV, and JNJ, and received research funding from Takeda and Sanofi.
References
- 1.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371:2189–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372:2509–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ansell SM, Lesokhin AM, Borrello I, Halwani A, Scott EC, Gutierrez M, et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. N Engl J Med. 2015;372:311–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Powles T, Eder JP, Fine GD, Braiteh FS, Loriot Y, Cruz C, et al. MPDL3280A (anti-PD-L1) treatment leads to clinical activity in metastatic bladder cancer. Nature. 2014;515:558–62. [DOI] [PubMed] [Google Scholar]
- 7.Ferris RL, Blumenschein G Jr, Fayette J, Guigay J, Colevas AD, Licitra L, et al. Nivolumab for Recurrent Squamous-Cell Carcinoma of the Head and Neck. N Engl J Med. 2016;375:1856–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McDermott DF, Sosman JA, Sznol M, Massard C, Gordon MS, Hamid O, et al. Atezolizumab, an Anti-Programmed Death-Ligand 1 Antibody, in Metastatic Renal Cell Carcinoma: Long-Term Safety, Clinical Activity, and Immune Correlates From a Phase Ia Study. J Clin Oncol. 2016;34:833–42. [DOI] [PubMed] [Google Scholar]
- 9.Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell. 2017;168:707–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350:207–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. New England Journal of Medicine. 2016;375:819–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodig SJ, Gusenleitner D, Jackson DG, Gjini E, Giobbie-Hurder A, Jin C, et al. MHC proteins confer differential sensitivity to CTLA-4 and PD-1 blockade in untreated metastatic melanoma. Sci Transl Med. 2018;10:1–13. [DOI] [PubMed] [Google Scholar]
- 14.Chowell D, Morris LGT, Grigg CM, Weber JK, Samstein RM, Makarov V, et al. Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science. 2018;359:582–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McGranahan N, Rosenthal R, Hiley CT, Rowan AJ, Watkins TBK, Wilson GA, et al. Allele-Specific HLA Loss and Immune Escape in Lung Cancer Evolution. Cell. 2017;171:1259–71.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao J, Shi LZ, Zhao H, Chen J, Xiong L, He Q, et al. Loss of IFN-γ Pathway Genes in Tumor Cells as a Mechanism of Resistance to Anti-CTLA-4 Therapy. Cell. 2016;167:397–404.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manguso RT, Pope HW, Zimmer MD, Brown FD, Yates KB, Miller BC, et al. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature. 2017;547:413–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patel SJ, Sanjana NE, Kishton RJ, Eidizadeh A, Vodnala SK, Cam M, et al. Identification of essential genes for cancer immunotherapy. Nature. 2017;548:537–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benci JL, Xu B, Qiu Y, Wu TJ, Dada H, Twyman-Saint Victor C, et al. Tumor Interferon Signaling Regulates a Multigenic Resistance Program to Immune Checkpoint Blockade. Cell. 2016;167:1540–54.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sumimoto H, Imabayashi F, Iwata T, Kawakami Y. The BRAF-MAPK signaling pathway is essential for cancer-immune evasion in human melanoma cells. J Exp Med. 2006;203:1651–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature. 2015;523:231–5. [DOI] [PubMed] [Google Scholar]
- 22.Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, et al. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell. 2017;168:542. [DOI] [PubMed] [Google Scholar]
- 23.Peng W, Chen JQ, Liu C, Malu S, Creasy C, Tetzlaff MT, et al. Loss of PTEN Promotes Resistance to T Cell–Mediated Immunotherapy. Cancer Discov. American Association for Cancer Research; 2016;6:202–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu D, Schilling B, Liu D, Sucker A, Livingstone E, Jerby-Amon L, et al. Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat Med. 2019;25:1916–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosenthal R, Cadieux EL, Salgado R, Bakir MA, Moore DA, Hiley CT, et al. Neoantigen-directed immune escape in lung cancer evolution. Nature. 2019;567:479–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garrido F, Aptsiauri N, Doorduijn EM, Garcia Lora AM, van Hall T. The urgent need to recover MHC class I in cancers for effective immunotherapy. Curr Opin Immunol. 2016;39:44–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davis MM, Boniface JJ, Reich Z, Lyons D, Hampl J, Arden B, et al. Ligand recognition by alpha beta T cell receptors. Annu Rev Immunol. 1998;16:523–44. [DOI] [PubMed] [Google Scholar]
- 28.Verma V, Shrimali RK, Ahmad S, Dai W, Wang H, Lu S, et al. PD-1 blockade in subprimed CD8 cells induces dysfunctional PD-1CD38 cells and anti-PD-1 resistance. Nat Immunol. 2019;20:1231–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van den Elsen PJ, Holling TM, Kuipers HF, van der Stoep N. Transcriptional regulation of antigen presentation. Current Opinion in Immunology. 2004;16:67–75. [DOI] [PubMed] [Google Scholar]
- 30.Zhou F. Molecular Mechanisms of IFN-γ to Up-Regulate MHC Class I Antigen Processing and Presentation. International Reviews of Immunology. 2009;28:239–60. [DOI] [PubMed] [Google Scholar]
- 31.Parker BS, Rautela J, Hertzog PJ. Antitumour actions of interferons: implications for cancer therapy. Nat Rev Cancer. 2016;16:131–44. [DOI] [PubMed] [Google Scholar]
- 32.Benci JL, Johnson LR, Choa R, Xu Y, Qiu J, Zhou Z, et al. Opposing Functions of Interferon Coordinate Adaptive and Innate Immune Responses to Cancer Immune Checkpoint Blockade. Cell. 2019;178:933–48.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pai C-CS, Huang JT, Lu X, Simons DM, Park C, Chang A, et al. Clonal Deletion of Tumor-Specific T Cells by Interferon-γ Confers Therapeutic Resistance to Combination Immune Checkpoint Blockade. Immunity. 2019;50:477–92.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burr ML, Sparbier CE, Chan Y-C, Williamson JC, Woods K, Beavis PA, et al. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature. 2017;549:101–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mezzadra R, Sun C, Jae LT, Gomez-Eerland R, de Vries E, Wu W, et al. Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature. 2017;549:106–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burr ML, Sparbier CE, Chan KL, Chan Y-C, Kersbergen A, Lam EYN, et al. An Evolutionarily Conserved Function of Polycomb Silences the MHC Class I Antigen Presentation Pathway and Enables Immune Evasion in Cancer. Cancer Cell. 2019;36:385–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Galluzzi L, Buqué A, Kepp O, Zitvogel L, Kroemer G. Immunological Effects of Conventional Chemotherapy and Targeted Anticancer Agents. Cancer Cell. 2015;28:690–714. [DOI] [PubMed] [Google Scholar]
- 38.Goel S, DeCristo MJ, Watt AC, BrinJones H, Sceneay J, Li BB, et al. CDK4/6 inhibition triggers anti-tumour immunity. Nature. 2017;548:471–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J, Bu X, Wang H, Zhu Y, Geng Y, Nihira NT, et al. Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature. 2018;553:91–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Medina BD, Liu M, Vitiello GA, Seifert AM, Zeng S, Bowler T, et al. Oncogenic kinase inhibition limits Batf3-dependent dendritic cell development and antitumor immunity. J Exp Med. 2019;216:1359–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 2014;15:554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang B, Wang M, Zhang W, Xiao T, Chen C-H, Wu A, et al. Integrative analysis of pooled CRISPR genetic screens using MAGeCKFlute. Nat Protoc. 2019;14:756–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X, et al. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci U S A. 2008;105:20380–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou L, Mudianto T, Ma X, Riley R, Uppaluri R. Targeting EZH2 Enhances Antigen Presentation, Antitumor Immunity, and Circumvents Anti-PD-1 Resistance in Head and Neck Cancer. Clin Cancer Res. 2020;26:290–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Häcker H, Tseng P-H, Karin M. Expanding TRAF function: TRAF3 as a tri-faced immune regulator. Nat Rev Immunol. 2011;11:457–68. [DOI] [PubMed] [Google Scholar]
- 46.Jongsma MLM, Guarda G, Spaapen RM. The regulatory network behind MHC class I expression. Mol Immunol. 2019;113:16–21. [DOI] [PubMed] [Google Scholar]
- 47.Spel L, Nieuwenhuis J, Haarsma R, Stickel E, Bleijerveld OB, Altelaar M, et al. Nedd4-Binding Protein 1 and TNFAIP3-Interacting Protein 1 Control MHC-1 Display in Neuroblastoma. Cancer Res. 2018;78:6621–31. [DOI] [PubMed] [Google Scholar]
- 48.Zarnegar BJ, Wang Y, Mahoney DJ, Dempsey PW, Cheung HH, He J, et al. Noncanonical NF-κB activation requires coordinated assembly of a regulatory complex of the adaptors cIAP1, cIAP2, TRAF2 and TRAF3 and the kinase NIK. Nature Immunology. 2008;9:1371–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vallabhapurapu S, Matsuzawa A, Zhang W, Tseng P-H, Keats JJ, Wang H, et al. Nonredundant and complementary functions of TRAF2 and TRAF3 in a ubiquitination cascade that activates NIK-dependent alternative NF-κB signaling. Nature Immunology. 2008;9:1364–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bista P, Zeng W, Ryan S, Bailly V, Browning JL, Lukashev ME. TRAF3 Controls Activation of the Canonical and Alternative NFκB by the Lymphotoxin Beta Receptor. Journal of Biological Chemistry. 2010;285:12971–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li S, Wan C, Zheng R, Fan J, Dong X, Meyer CA, et al. Cistrome-GO: a web server for functional enrichment analysis of transcription factor ChIP-seq peaks. Nucleic Acids Res. 2019;47:W206–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Molecular Cell. 2010;38:576–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mei S, Qin Q, Wu Q, Sun H, Zheng R, Zang C, et al. Cistrome Data Browser: a data portal for ChIP-Seq and chromatin accessibility data in human and mouse. Nucleic Acids Res. 2017;45:D658–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng R, Wan C, Mei S, Qin Q, Wu Q, Sun H, et al. Cistrome Data Browser: expanded datasets and new tools for gene regulatory analysis. Nucleic Acids Res. 2019;47:D729–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Layer RM, Pedersen BS, DiSera T, Marth GT, Gertz J, Quinlan AR. GIGGLE: a search engine for large-scale integrated genome analysis. Nat Methods. 2018;15:123–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gilmore TD. Introduction to NF-κB: players, pathways, perspectives. Oncogene. 2006;25:6680–4. [DOI] [PubMed] [Google Scholar]
- 57.Vallabhapurapu S, Karin M. Regulation and Function of NF-κB Transcription Factors in the Immune System. Annual Review of Immunology. 2009;27:693–733. [DOI] [PubMed] [Google Scholar]
- 58.Sun S-C. The non-canonical NF-κB pathway in immunity and inflammation. Nat Rev Immunol. 2017;17:545–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hayden MS, Ghosh S. Shared Principles in NF-κB Signaling. Cell. 2008;132:344–62. [DOI] [PubMed] [Google Scholar]
- 60.Zhao B, Barrera LA, Ersing I, Willox B, Schmidt SCS, Greenfeld H, et al. The NF-κB genomic landscape in lymphoblastoid B cells. Cell Rep. 2014;8:1595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Link VM, Duttke SH, Chun HB, Holtman IR, Westin E, Hoeksema MA, et al. Analysis of Genetically Diverse Macrophages Reveals Local and Domain-wide Mechanisms that Control Transcription Factor Binding and Function. Cell. 2018;173:1796–809.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Iannetti A, Ledoux AC, Tudhope SJ, Sellier H, Zhao B, Mowla S, et al. Regulation of p53 and Rb links the alternative NF-κB pathway to EZH2 expression and cell senescence. PLoS Genet. 2014;10:e1004642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Such L, Zhao F, Liu D, Thier B, Le-Trilling VTK, Sucker A, et al. Targeting the innate immunoreceptor RIG-I overcomes melanoma-intrinsic resistance to T cell immunotherapy. Journal of Clinical Investigation. 2020;130:4266–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kalbasi A, Tariveranmoshabad M, Hakimi K, Kremer S, Campbell KM, Funes JM, et al. Uncoupling interferon signaling and antigen presentation to overcome immunotherapy resistance due to JAK1 loss in melanoma. Sci Transl Med. 2020;12:eabb0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–74. [DOI] [PubMed] [Google Scholar]
- 66.Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med. 2018;24:1550–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Scheper W, Kelderman S, Fanchi LF, Linnemann C, Bendle G, de Rooij MAJ, et al. Low and variable tumor reactivity of the intratumoral TCR repertoire in human cancers. Nat Med. 2019;25:89–94. [DOI] [PubMed] [Google Scholar]
- 68.Font-Tello A, Kesten N, Xie Y, Taing L, Varešlija D, Young LS, et al. FiTAc-seq: fixed-tissue ChIP-seq for H3K27ac profiling and super-enhancer analysis of FFPE tissues. Nature Protocols. 2020;15:2503–18. [DOI] [PubMed] [Google Scholar]
- 69.Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N Engl J Med. 2016;375:819–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maude SL, Frey N, Shaw PA, Aplenc R, Barrett DM, Bunin NJ, et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med. 2014;371:1507–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gide TN, Quek C, Menzies AM, Tasker AT, Shang P, Holst J, et al. Distinct Immune Cell Populations Define Response to Anti-PD-1 Monotherapy and Anti-PD-1/Anti-CTLA-4 Combined Therapy. Cancer Cell. 2019;35:238–55.e6. [DOI] [PubMed] [Google Scholar]
- 72.Mariathasan S, Turley SJ, Nickles D, Castiglioni A, Yuen K, Wang Y, et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature. 2018;554:544–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou L, Mudianto T, Ma X, Riley R, Uppaluri R. Targeting EZH2 Enhances Antigen Presentation, Antitumor Immunity, and Circumvents Anti–PD-1 Resistance in Head and Neck Cancer. Clinical Cancer Research. 2020;26:290–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vince JE, Wei-Lynn Wong W, Khan N, Feltham R, Chau D, Ahmed AU, et al. IAP Antagonists Target cIAP1 to Induce TNFα-Dependent Apoptosis. Cell. 2007;131:682–93. [DOI] [PubMed] [Google Scholar]
- 75.Varfolomeev E, Blankenship JW, Wayson SM, Fedorova AV, Kayagaki N, Garg P, et al. IAP Antagonists Induce Autoubiquitination of c-IAPs, NF-κB Activation, and TNFα-Dependent Apoptosis. Cell. 2007;131:669–81. [DOI] [PubMed] [Google Scholar]
- 76.Li B, Severson E, Pignon J-C, Zhao H, Li T, Novak J, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017;77:e108–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gu SS, Wang X, Hu X, Jiang P, Li Z, Traugh N, et al. Clonal tracing reveals diverse patterns of response to immune checkpoint blockade. Genome Biol. 2020;21:263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bertrand MJM, Milutinovic S, Dickson KM, Ho WC, Boudreault A, Durkin J, et al. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol Cell. 2008;30:689–700. [DOI] [PubMed] [Google Scholar]
- 80.Beug ST, Beauregard CE, Healy C, Sanda T, St-Jean M, Chabot J, et al. Smac mimetics synergize with immune checkpoint inhibitors to promote tumour immunity against glioblastoma. Nat Commun. 2017;8:14278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sun D, Wang J, Han Y, Dong X, Zheng R, Ge J, et al. TISCH: a comprehensive web resource enabling interactive single-cell transcriptome visualization of tumor microenvironment. Nucleic Acids Res. 2021;49(D1):D1420–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang H, Christensen CL, Dries R, Oser MG, Deng J, Diskin B, et al. CDK7 Inhibition Potentiates Genome Instability Triggering Anti-tumor Immunity in Small Cell Lung Cancer. Cancer Cell. 2020;37:37–54.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hu-Lieskovan S, Mok S, Homet Moreno B, Tsoi J, Robert L, Goedert L, et al. Improved antitumor activity of immunotherapy with BRAF and MEK inhibitors in BRAF(V600E) melanoma. Sci Transl Med. 2015;7:279ra41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Topper MJ, Vaz M, Chiappinelli KB, DeStefano Shields CE, Niknafs N, Yen R-WC, et al. Epigenetic Therapy Ties MYC Depletion to Reversing Immune Evasion and Treating Lung Cancer. Cell. 2017;171:1284–300.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Aran D, Sirota M, Butte AJ. Systematic pan-cancer analysis of tumour purity. Nat Commun. 2015;6:8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li B, Li JZ. A general framework for analyzing tumor subclonality using SNP array and DNA sequencing data. Genome Biol. 2014;15:473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xiao T, Li W, Wang X, Xu H, Yang J, Wu Q, et al. Estrogen-regulated feedback loop limits the efficacy of estrogen receptor-targeted breast cancer therapy. Proc Natl Acad Sci U S A. 2018;115:7869–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gu S, Sayad A, Chan G, Yang W, Lu Z, Virtanen C, et al. SHP2 is required for BCR-ABL1-induced hematologic neoplasia. Leukemia. 2018;32:203–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Qin Q, Mei S, Wu Q, Sun H, Li L, Taing L, et al. ChiLin: a comprehensive ChIP-seq and DNase-seq quality control and analysis pipeline. BMC Bioinformatics. 2016;17:404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008;9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data were uploaded to Gene Expression Omnibus with accession number GSE149826.