Abstract
Background
PITX2 DNA methylation has been shown to predict outcomes in high-risk breast cancer patients after anthracycline-based chemotherapy. To determine its prognostic versus predictive value, the impact of PITX2 DNA methylation on outcomes was studied in an untreated cohort vs. an anthracycline-treated triple-negative breast cancer (TNBC) cohort.
Material and Methods
The percent DNA methylation ratio (PMR) of paired-like homeodomain transcription factor 2 (PITX2) was determined by a validated methylation-specific real-time PCR test. Patient samples of routinely collected archived formalin-fixed paraffin-embedded (FFPE) tissue and clinical data from 144 TNBC patients of 2 independent cohorts (i.e., 66 untreated patients and 78 patients treated with anthracycline-based chemotherapy) were analyzed.
Results
The risk of 5- and 10-year overall survival (OS) increased continuously with rising PITX2 DNA methylation in the anthracycline-treated population, but it increased only slightly during 10-year follow-up time in the untreated patient population. PITX2 DNA methylation with a PMR cutoff of 2 did not show significance for poor vs. good outcomes (OS) in the untreated patient cohort (HR = 1.55; p = 0.259). In contrast, the PITX2 PMR cutoff of 2 identified patients with poor (PMR >2) vs. good (PMR ≤2) outcomes (OS) with statistical significance in the anthracycline-treated cohort (HR = 3.96; p = 0.011). The results in the subgroup of patients who did receive anthracyclines only (no taxanes) confirmed this finding (HR = 5.71; p = 0.014).
Conclusion
In this hypothesis-generating study PITX2 DNA methylation demonstrated predominantly predictive value in anthracycline treatment in TNBC patients. The risk of poor outcome (OS) correlates with increasing PITX2 DNA methylation.
Keywords: PITX2, DNA methylation, Therapy prediction, Triple-negative breast cancer/neoplasms, Anthracyclines, Chemotherapy, Biomarker, Treatment outcome, Tumor/genetics, Homeodomain proteins/genetics
Introduction
Epigenetics is a breakthrough in patient-tailored medicine, delivering new biomarkers and therapeutic options for several cancer indications including breast cancer, colon cancer, head and neck cancer, and prostate cancer [1, 2].
In the Wnt-signaling pathway, PITX2 regulates the recruitment and stabilization of cytosolic β-catenin, which triggers the transcription of cell cycle regulatory and proliferation genes (e.g., cyclin D1 and c-Myc) and subsequently enhances cell proliferation [3]. Methylation of the PITX2 gene leads to epigenetic silencing of this transcription factor and a subsequent change in molecular signaling.
Significant evidence has accumulated that methylation of the PITX2 promoter gene might serve as a predictive and prognostic biomarker in a variety of cancers [1]. The value of PITX2 DNA methylation for prediction of outcomes after anthracycline-based chemotherapy [4] was confirmed recently with a CE-marked assay using FFPE tumor tissue specimens in ER+, HER2− breast cancer patients [5]. In a pilot study of 56 triple-negative breast cancer (TNBC) patients treated with adjuvant anthracycline-treated chemotherapy, Absmaier et al. [6] reported a potential of PITX2 to predict outcomes after therapy. So far, the question of whether PITX2 DNA methylation has a prognostic and/or predictive value in breast cancer patients remains unanswered.
Considerable misunderstandings exist regarding the definition of and difference between the prognostic and predictive value of a given biomarker. According to Ballman [7] a prognostic biomarker delivers information about the probability of a cancer disease outcome such as disease recurrence, progression, or death independently of treatment. A biomarker is predictive for a specific therapy if the treatment effect is different for biomarker-positive patients compared to biomarker-negative patients. Following these definitions, evidence for the predictive value is obtained if a biomarker identifies patients with poor versus good outcomes in the same clinicopathological patient population only for a specific therapy and not in the same indication with another treatment or no treatment at all.
In this exploratory study using archived tissue specimens, the prognostic versus predictive value of PITX2 was evaluated in 2 TNBC populations by comparing the impact of PITX2 DNA methylation on the outcome (DFS/overall survival [OS]) in an untreated TNBC cohort versus an anthracycline-treated TNBC cohort.
Materials and Methods
Patients, Samples, Study Design and Conduction
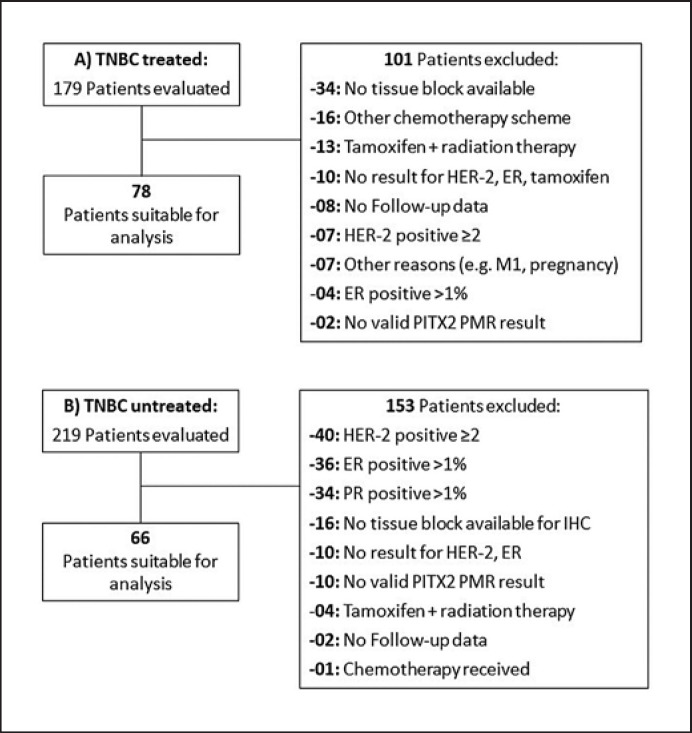
Patient samples and clinical data of 144 TNBC patients from 2 cohorts with availability of clinical follow-up data for at least 18 months and valid PITX2 percent DNA methylation ratio (PMR) values per tissue sample were analyzed. Cohort 1 comprised 66 untreated patients from the Karolinska Institute in Sweden (primary diagnosis in 1971–1976). Cohort 2 comprised 78 patients treated with anthracycline-based chemotherapy (ANT±T) from the Comprehensive Cancer Center of the Technical University of Munich (TUM) in Germany (primary diagnosis in 1996–2009). The main eligibility criteria were as follows: histologically confirmed invasive breast cancer; primary tumor stage pT1, pT2, or pT3; ER-negative, PR-negative, and HER2-negative cancer (TNBC); no endocrine therapy; for cohort 1, no further therapy besides surgery ± radiation; and for cohort 2: surgery followed by standard-of-care adjuvant anthracycline-based chemotherapy (±taxanes, no dose-dense therapy, and no other primary systemic chemotherapy). The CONSORT diagram (Fig. 1) lists all of the patients assessed for eligibility and the reasons for exclusion. For all of the patients, source data verification was performed.
Fig. 1.
The consort diagrams depict the eligibility of the patients and the respective samples that were included in the 2 patient cohorts, i.e., the anthracycline-based chemotherapy treated cohort (A) and the untreated cohort (B), and the reasons for noninclusion.
Clinical Variables Assessed
The ER/PR and HER2 status of all of the samples was assessed by an experienced local pathologist via immunohistochemistry (IHC) using the following cutoffs: ER negativity ≤1%, PR negativity ≤5%, and HER2 IHC status 0 or 1. For most samples of the untreated patients (cohort 1) ER/PR and HER2 status was already available based on an assessment using a standardized method by the Institute of Pathology of the Helmholtz Center Munich. Due to missing values for 21 cases from cohort 1, assessment of the ER/PR/HER2 status was performed at the Research Unit Analytical Pathology of the Helmholtz Center Munich using the same standardized IHC protocol with the cutoffs as described above.
For cohort 2, assessment of the TNBC status was performed by the pathology laboratory of the TUM as a routine diagnostics analysis using the same criteria as for cohort 1. For HER2, an IHC status of 2+ with absence of HER2 amplification analyzed by fluorescence in situ hybridization (FISH) was considered HER2 negative according to national and international guidelines.
Study Evaluation Plan
This study was designed to determine the PITX2 PMR cutoff value in an untreated population (n = 66, prognostic value) and subsequently to investigate this cutoff in a treated population (n = 78, prognostic and predictive value) for statistical significance. If no cutoff was identified in the untreated population, a cutoff was to be determined in the treated population and applied to the other population.
PITX2 DNA-Methylation Assay, DNA Amplification and Input
All primary formalin-fixed paraffin-embedded (FFPE) breast cancer tissue samples were coded to ensure blinding of the operator conducting the PITX2 DNA methylation assay. The PITX2 test (QIAGEN therascreen® PITX2 RGQ PCR Kit) is a quantitative methylation-specific real-time PCR test (qMSP) intended for determination of the PMR in promotor 2 of the PITX2 gene in primary FFPE breast cancer tissue [5, 8]. In order to reduce the tissue input and increase amplifiable DNA copy numbers an adapted therascreen® PITX2 RGQ PCR assay protocol (CEF PITX2 workflow) was used in the present study [9]. Genomic DNA preparation was performed through deparaffinization and lysis of the FFPE tumor tissue section using an Epitect Fast FFPE Lysis Kit (QIAGEN, Hilden, Germany). The lysate was subsequently applied directly for bisulfite conversion using an Epitect Fast DNA Bisulfite Kit (QIAGEN) according to the manufacturer's instruction. Upon cleaning and elution, the bisulfite-converted DNA (bisDNA) was quantified with a QIAxpert spectrophotometer and bisDNA samples were diluted to a final concentration of 6 ng/µL for subsequent qPCR analysis. Samples from patient cohort 1 were very low in DNA content due to age and probably usage of unbuffered formalin for fixation. Therefore, bisDNA was used for preamplification applying the same qPCR reaction mix as used in the therascreen® PITX2 RGQ PCR Kit Handbook [10]. After adding 4 µL of sample bisDNA to 1 well of reaction mix, the following protocol was performed using RGQ software version 2.3.1 on a Rotor-Gene Q real-time PCR cycler: polymerase activation: 2 min at 95°C, followed by 10 cycles with denaturation for 5 s at 95°C and annealing for 5 s at 60°C. PCR reactions were used undiluted as samples for subsequent qPCR analysis and stored at −20°C until further use.
The percent methylation ratio of 3 CpG motifs of the PITX2 gene promoter 2 [8] was quantified by qMSP using the therascreen® PITX2 RGQ PCR Kit, containing qPCR reaction mix, primer, and probes, as well as positive and negative controls. The assay was performed on a Rotor-Gene Q MDx real-time PCR platform (QIAGEN) and automatically assessed by Rotor-Gene AssayManager® software version 2.1 with an installed Gamma plug-in and PITX2 specific assay profile (therascreen_PITX2_FFPE_CE Assay Profile version 1.0.1) for data analysis and quality control [10]. In order to determine the PMR, values were calculated for the methylated and the unmethylated PITX2 DNA methylation status using a modified 2−∆CT method with the following formula: 1/(1 + 2[CTmeth (FAM) − CTunmeth (HEX)]) [10, 11].
Statistical Methods
Disease-free survival (DFS), defined as the time from the primary surgery to the first documented DFS event (local disease recurrence, distant metastasis, life-threatening secondary cancer, and death of any cause) was originally designated as the primary endpoint. However, the imaging technologies applied for follow-up for progression of disease were not comparable in the different decades of tissue sampling in the present study. Therefore, as the results for OS, defined as the secondary endpoint, are regarded as more reliable within the presented study population, OS was selected for hypothesis generation. The date of the primary surgery was considered the follow-up index date. OS was defined as the time from surgery to the documented date of death or, if the patient was still alive, the date of the last follow-up information. The PITX2 cutoff value for OS was established with the maximum-selected log-rank statistic using the maxstat.test function as implemented by the program library maxstat of the program R (R Development Core Team 2012) [12]. Analysis was performed for OS follow-up time censored at 5 and 10 years. Survival curves were calculated according to the incidence function [13, 14]. The log-rank test was used for calculating the respective p values and a two-sided significance level of 5% was considered significant (R software version 3.4.1 [2017] − Single Candle [2017]; The R Foundation for Statistical Computing).
Results
Patients Characteristics and Study Populations
Patient characteristics and the reasons for inclusion and exclusion of the archived patient samples (CONSORT diagrams) are presented in Table 1 and Figure 1, respectively. All of the treated patients (cohort 2) had received anthracycline-based chemotherapy (±docetaxel or paclitaxel) consisting of epirubicin or doxorubicin in combination with cyclophosphamide (EC/AC) or in combination with cyclophosphamide and fluorouracil (FEC). Current national and international guidelines recommend anthracycline-taxane combination regimens, and therefore patients treated with taxanes in addition to anthracyclines were also included in the present study.
Table 1.
Patient cohort characteristics
| Characteristic | Cohort 1: TNBC untreated (n = 66) | Cohort 2: TNBC treated (n = 78) |
|---|---|---|
| Age (years) | ||
| <50 | 15 (23) | 33 (42) |
| ≥50 | 51 (77) | 45 (58) |
| T stage | ||
| T1 (<2.0 cm) | 36 (55) | 33 (42) |
| T2 (2–5cm) | 23 (35) | 40 (51) |
| T3 (>5 cm) | 3 (5) | 5 (6) |
| TX | 4 (6) | 0 (0) |
| Lymph node stage | ||
| NO | 35 (53) | 45 (58) |
| N1 | 17 (26) | 14 (18) |
| N+ | 14 (21) | 19 (24) |
| Grading | ||
| G1 | 8 (12) | 0 (0) |
| G2 | 41 (62) | 6 (8) |
| G3/G4 | 17 (26) | 54 (69) |
| GX | 0 (0) | 18 (23) |
| Histology | ||
| Ductal | 36 (55) | 56 (72) |
| Other | 30 (45) | 22 (28) |
| Radiotherapy | ||
| Yes | 35 (53) | 74 (95) |
| No | 31 (47) | 4 (5) |
| Endocrine therapy | ||
| Yes | 0 (0) | 0 (0) |
| No | 66 (100) | 78 (100) |
| CTx scheme | ||
| ANT | 0 (0) | 50 (64) |
| ANT-T | 0 (0) | 28 (36) |
| UNT | 66 (100) | 0 (0) |
| DFS event | ||
| Yes | 48 (73) | 29 (37) |
| Yes within 10 years | 36 (55) | 28 (36) |
| No | 18 (27) | 49 (63) |
| OS event | ||
| Yes | 37 (56) | 16 (21) |
| Yes within 10 years | 28 (42) | 14 (18) |
| No | 29 (44) | 62 (79) |
Values are presented as numbers (%). ANT, treated with anthracyclines; ANT-T, treated with anthracyclines followed by taxanes; CTx, chemotherapy; UNK, unkown; UNT, untreated (no chemotherapy).
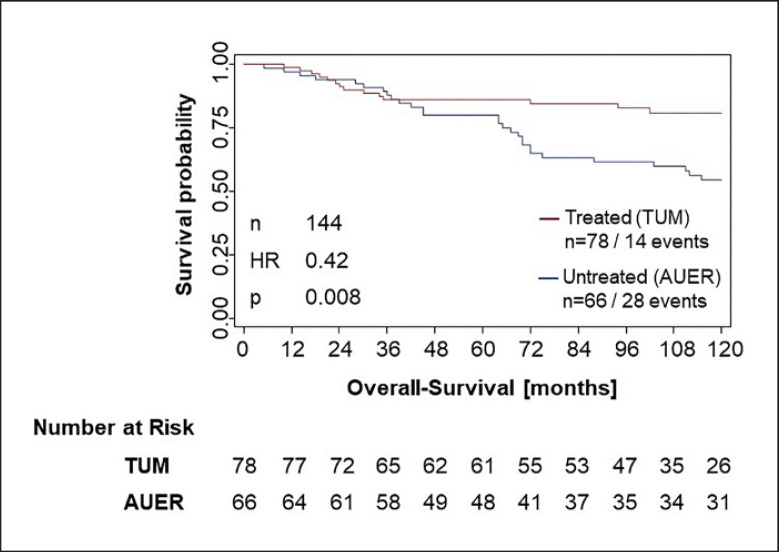
Compared to the untreated population, the anthracycline-treated patient cohort was younger (age <50 years: 42 vs. 23%) and had a higher percentage of T2 stages (51 vs. 35%) and more patients with grading G3/4 (69 vs. 26%). The lymph node status was comparable in both populations (patients with N0: 58 vs. 53%). The different rate of treatment by radiotherapy (95 vs. 53%) was probably due to the higher rate of breast-conserving surgery in the treated population. The untreated TNBC population demonstrated a higher event rate (OS) at 10 years compared to the anthracycline-treated population (28 OS events − 42% event rate vs. 14 OS events − 18% event rate; Fig. 2). The 3-year OS event rate was similar, with 12% in the untreated population versus 14% in the treated population. The OS event rate in the untreated patient population increased to 20% at 5 years and to 42% at 10 years; the treated population developed a lower OS event rate, with 14% at 5 years and 18% at 10 years, assumingly due to the chemotherapy treatment.
Fig. 2.
Kaplan-Meier analysis of the untreated (blue line) vs. the anthracycline-treated (red line) TNBC population. The treated patient cohort showed a significant better OS than the untreated patient cohort.
Based on the different follow-up time intervals, imaging technologies, and analysis methods used during the different time periods of the untreated versus treated cohorts, detection of DFS events was likely subject to bias and therefore not evaluated further.
DNA Quality, Amplification and PITX2 Measurement
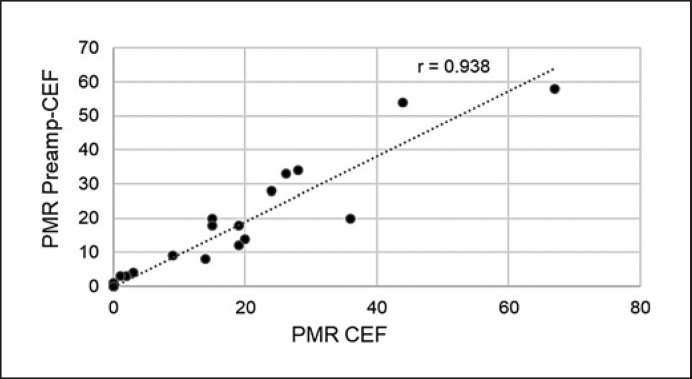
To reduce tissue input and increase amplifiable DNA copy numbers, we applied an adapted therascreen® PITX2 RGQ PCR assay protocol, i.e., CEF PITX2 workflow, which showed a high concordance with the standard therascreen® PITX2 RGQ PCR assay protocol and an increased yield of amplifiable PITX2 copy numbers, allowing a reliable assessment of PITX2 PMR down to a PMR of 2 [9]. In addition, due to the age of the untreated patient cohort 1, preamplification of the DNA was required. The preamplification was performed by 10 PCR cycles applying the PITX2 primers and PCR reaction mixes of the kit. The graph (Fig. 3) shows a high correlation (r = 0.938) between preamplified and non-preamplified PITX2 PMR in an independent set of 20 tumor tissue specimens, confirming data reproducibility. The treated patient cohort 2 was tested for PITX2 PMR also with the CEF workflow, without a preamplification step.
Fig. 3.
Correlation between PMR values generated with or without preamplification. The graph depicts the high correlation (r = 0.938) between PITX2 PMR values obtained with preamplified (y-axis) and nonpreamplified bisulfite-converted DNA (x-axis) from 20 tumor tissue specimens.
PITX2 DNA Methylation and Risk of Recurrence
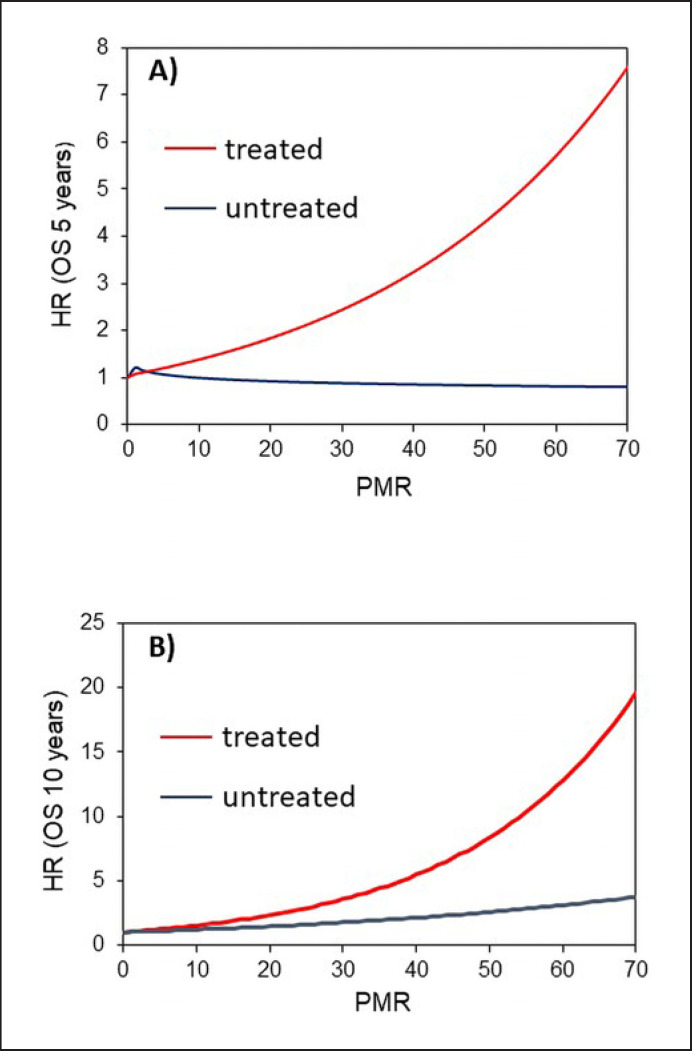
Hypermethylation of PITX2 has been previously demonstrated to confer an increased risk of poor outcomes with respect to DFS in estrogen receptor-positive and HER2-negative patients [5, 15]. The data in the present study show that the risk for OS events at 5-years (blue line in Fig. 4A) did not increase with increasing PITX2 methylation in the untreated TNBC patient cohort 1. At 10 years only a slight increase in OS risk was observed in the untreated patient cohort 1 (blue line in Fig. 4B). In contrast, the risk of OS events in the anthracycline-treated patient cohort 2 significantly increased with increasing PITX2 PMR values not only at 5 years (red line in Fig. 4A) but also at 10 years (red line in Fig. 4B).
Fig. 4.
Increasing PITX2 DNA methylation results and increasing OS risk. A OS at 5 years of follow-up. B OS at 10 years of follow-up. The blue line depicts the untreated patient population and the red line represents the anthracycline-treated patient population.
PITX2 PMR Cutoff Determination in the Two Patient Cohorts
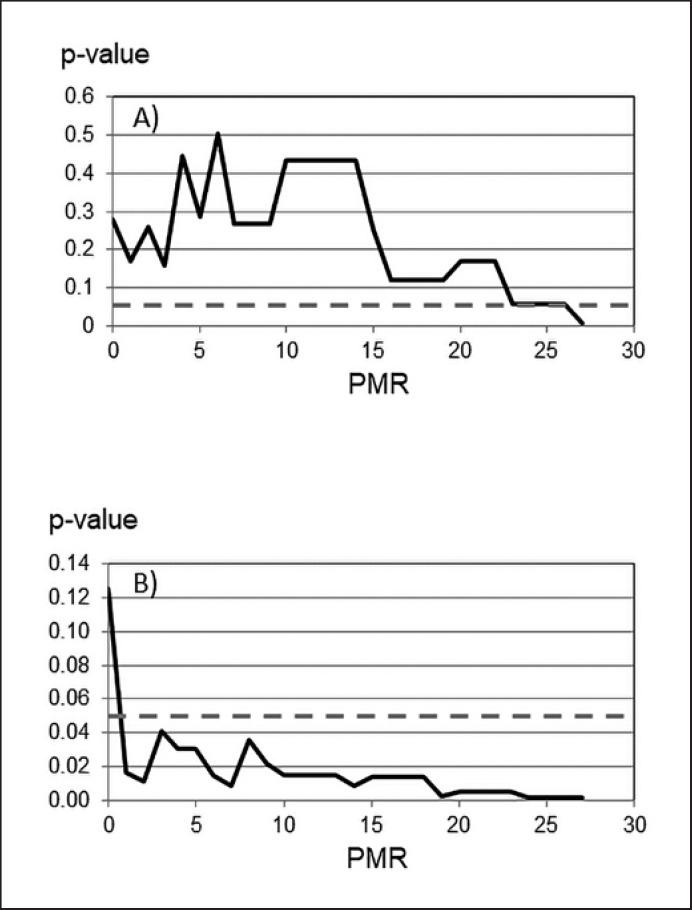
According to the predefined study protocol the PITX2 PMR cutoff value was first determined in the untreated population by log-rank statistics (n = 66, prognostic value). In the 5-year DFS analysis no statistically significant cutoff was found in the untreated population as well as in the treated one. Ten-year DFS analyses revealed cutoffs of 27 in the untreated population and 19 in the treated population (DFS results not shown). Analysis of PITX2 PMR values in the untreated cohort at the 10-year OS follow-up from 0 to 30 resulted in a single significant cutoff point at PMR 27; however, only 2 patients were above this threshold. Therefore, this cutoff was considered to be not clinically relevant and was neglected (Fig. 5A; Table 2).
Fig. 5.
Log-rank statistics to identify the optimal cutoff value for PITX2 DNA methylation. The dashed line represents the p value of significance for OS (p < 0.05). A Untreated patient cohort. B Anthracycline-treated patient cohort.
Table 2.
Statistical significance of 10-year OS as a function of the PITX2 cutoff
| Cutoff applieda | Cohort 1: untreated TNBC |
Cohort 2: ANT-treated TNBC |
||
|---|---|---|---|---|
| 10-year OS HR | p value | 10-year OS HR |
p value | |
| PMR 0 | 1.50 | 0.280 | 2.29 | 0.125 |
| PMR 1 | 0.52 | 0.172 | 3.65 | 0.017 |
| PMR 2 | 1.55 | 0.259 | 3.96 | 0.011 |
| PMR 3–18 | 0.48–2.05 | 0.119–0.505 | 2.98–4.11 | 0.009–0.041 |
| PMR 19 | 2.05 | 0.119 | 5.31 | 0.003 |
| PMR 20–23 | 0.68–2.57 | 0.056–0.169 | 5.44 | 0.005 |
| PMR 24–26 | 2.57 | 0.056 | 6.51 | 0.002 |
| PMR 27 | 4.33 | 0.007 | 6.51 | 0.002 |
≤ vs. > cutoff PMR value applied in cohorts 1 and 2. ANT, anthracycline-based chemotherapy; OS, overall-survival.
Subsequently, we searched for a cutoff value at 10-year OS using log-rank statistics in the treated population (n = 78, prognostic and predictive value). Already at a cutoff of PMR 1, patients with poor versus good outcomes (OS) were identified with a high statistical significance (p < 0.017; Table 2). Significance (p < 0.05) remained valid over all subsequent PMR values tested up to PMR 30 (Fig. 5B; Table 2). It is noteworthy that HR increased with increasing PMR. The optimal separation of the 2 patient cohorts with the highest statistical significance was obtained for a PITX2 cutoff of 2.
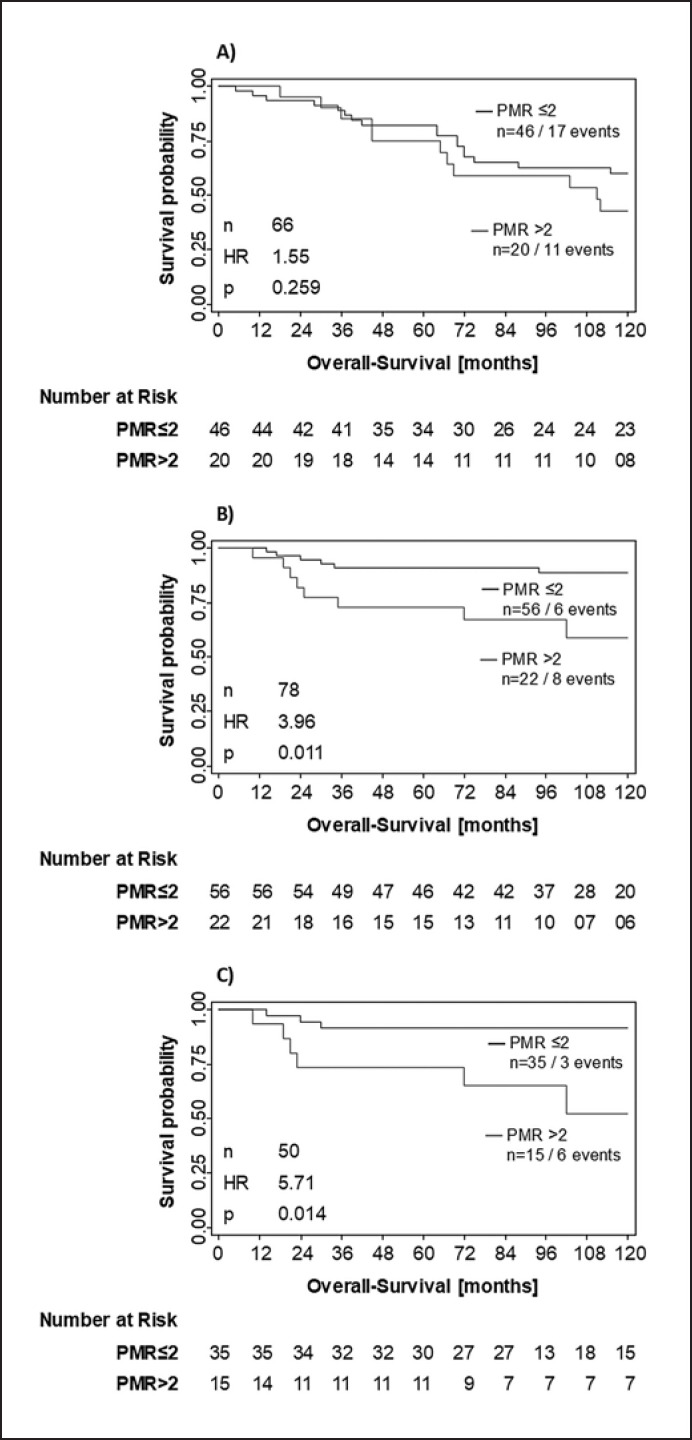
Impact on OS Applying the Statistically Significant PITX2 Cutoff
Applying the PITX2 DNA methylation PMR cutoff value of 2 to the untreated patient population, no difference and no statistical relevance for poor versus good outcomes could be demonstrated for OS (HR = 1.55; p = 0.259; Fig. 6A).
Fig. 6.
Kaplan-Meier analysis applying the PITX2 cutoff value of PMR 2. Untreated patient cohort (A; n = 66), anthracycline-treated patient cohort (B; n = 78), and anthracycline-treated patients who received anthracyclines only (no taxanes added) in their chemotherapy regimens (C; n = 50).
In contrast, the PITX2 PMR cutoff value of 2 identified patient populations with poor versus good survival with a high statistical significance in the anthracycline-treated patient cohort 2 (HR = 3.96; p = 0.011; Fig. 6B). Results in the subgroup of patients who did receive anthracyclines only (no taxanes; n = 50) confirmed this finding (HR = 5.71; p = 0.014; Fig. 6C).
Discussion
The clinical relevance of the PITX2 DNA methylation status in breast cancer has been described in several studies [reviewed in 1]. The precise role of PITX2 DNA methylation in breast carcinogenesis and disease progression, however, remains not fully understood. Whereas Nimmrich et al. [16] described a prognostic role of PITX2 DNA methylation in breast cancer, others have shown a strong correlation with metastasis-free survival in node-negative, tamoxifen-treated breast cancer patients, demonstrating a predictive value [11, 17]. Furthermore, strong evidence has been provided that PITX2 DNA- methylation predicts outcomes in anthracycline-treated high-risk breast cancer patients [4, 5, 15].
PITX2 as homeobox transcription factor plays an important role in general developmental processes and it has been shown to have a strong impact on tumor development, facilitating invasion [18, 19], proliferation [3, 20], and metastasis [21] in different cancer entities. Furthermore, PITX2 has been shown to be also involved in chemo- or radiotherapy resistance [22, 23, 24, 25].
Therefore, the question arises of whether PITX2 DNA methylation has prognostic or predictive value or both. To address this issue, the role of PITX2 DNA methylation was investigated in untreated and treated TNBC patient cohorts. This was only possible because of the availability of a unique untreated patient cohort from the early 1970s. The 20-year time gap of the dates of diagnosis between the patient populations (1971–1976 for the untreated cohort and 1996–2007 for the treated cohort) and the reason why different diagnostic evaluation algorithms were applied (e.g., for staging and grading) might have contributed to differences in patient characteristics. The archived tissue specimens were obtained from single centers and were not from a randomized trial; this also represents a limitation of the present study. Grading, T stage, and lymph node involvement represent robust prognostic parameters in ER-positive patients, and these parameters are less significant in TNBC cancer. Kashiwagi et al. [26] showed that in TNBC clinical parameters such as grading and T stage lost their statistical significance in multivariate OS analyses. The statistical significance for lymph node status was, however, retained. In the presented patient populations, the lymph node status (N0 vs. N+) was comparable.
In the anthracycline-treated patient population, a higher percentage of grade 3 was observed compared to the untreated population. However, it has been previously demonstrated in ER-positive patients that the value of PITX2 DNA methylation for prediction of clinical outcomes is independent of tumor grading [4, 5]. In addition, it needs to be mentioned that TNBC comprises a disease with substantial heterogeneity on a morphological level as well as on a molecular level [27, 28, 29, 30]. Unfortunately, molecular profiling regarding these subtypes was not available for the cohorts; therefore, the potential impact of subtypes on outcomes was not investigated. Evidence has been published demonstrating a high recurrence and OS event rates for TNBC within the first 3–5 years [31, 32]. In the present study the OS event rates seemed to be lower, as expected, (12% in the untreated cohort vs. 14% in the treated cohort) and increased in the untreated population to 20 and 42% at 5 and 10 years, respectively. The increase in the treated population was less significant, with 14% at 5 years and 18% at 10 years, which is most probably due to the treatment effect. No explanation could be found to explain why the effect in the first 3 years was not as prominent as reported by others.
In addition, the evaluation of DFS had major limitations based on the incomparability of imaging technologies used for both patient cohorts. In the late 1980s high-resolution ultrasound machines became available; multidetector CT were only developed in the late 1990s and became part of routine imaging follow-up in oncology in the early 2000s, i.e., these modern technologies were not available for imaging follow-up of cohort 1 (untreated patient cohort). Therefore, the DFS results in the untreated subgroup can be regarded as less precise and the observed DFS events in this cohort were mostly death. Nevertheless, the OS results suggest that PITX2 DNA methylation is rather of predictive value in anthracycline-treated TNBC patients for 2 reasons. First, we could not identify a clinically relevant cutoff value for PITX2 in the untreated patient cohort, whereas in the anthracycline-treated patient cohort the cutoff value for PITX2 already at PMR 1 identified patients with poor versus good outcomes for OS with a high statistical significance. Second, while the risk for death increased strongly with rising PITX2 DNA methylation in the anthracycline-treated patient population but not in the untreated population at 5-years of follow-up, only a slight increase in the untreated population at 10-years could be observed.
The data of Nimmrich et al. [16] showed clinical relevance of PITX2 in untreated ER-positive breast cancer patients, indicating a prognostic value. Therefore, a slight prognostic value in TNBC cannot be ruled out and should be further investigated. In comparison to the publication of Absmaier et al. [6] our results showed an inverse clinical outcome, associating hypermethylated PITX2 values >2 with a poor OS compared to hypomethylation as shown by Absmaier et al. [6]. This may be due to noncomparable TNBC collectives investigated with regard to possibly different TNBC subtypes analyzed, heterogeneous chemotherapy regimens including cotherapies with CMF in the study of Absmaier et al. [6], and use of a nonvalidated PITX2 assay with differences in qPCR assay set-up and detection threshold settings, which may have led to different sensitivities of the qPCR assay.
PITX2 shows a strong correlation with specific drug responses (e.g., doxorubicin and letrozole) in preclinical cancer models mediated by ABC type drug transporters, importers, and other pathways [22, 23, 24], which supports the role of PITX2 as a drug-specific response marker. This is also supported by our results, showing a HR of 5.71 in the subgroup treated with anthracycline without taxanes, compared to a HR of 3.96 in the overall anthracycline-treated group (including or not including taxanes in the therapy regimen).
Based on the discussed differences in the patient characteristics, the different time intervals of sampling, and the fact that no specimen was available from a randomized prospective clinical trial, the study results should be interpreted with caution and considered as hypothesis generating only. In order to unequivocally prove that PITX2 DNA methylation is predictive of anthracycline treatment in TNBC, additional studies are required to investigate the effect in patients treated with non-anthracycline versus anthracycline-containing regimens according to current treatment guidelines. Furthermore, the effect of taxanes (anthracycline-taxane vs. anthracycline-non-taxane) regimens may merit further investigation. The proposed study should utilize tissue specimens preferably from a randomized trial comparing both treatment modalities and by applying a formal statistical test of the treatment-by-biomarker interaction which should be significant as described by Ballman [7].
In conclusion, in this hypothesis-generating study DNA methylation of PITX2 identified anthracycline-treated patients with a poor outcome with a high statistical significance. The risk of death (OS) increased with increasing PITX2 DNA methylation with a high statistical significance in the anthracycline-treated patient cohort but not in the untreated patient cohort. These results suggest that PITX2 DNA methylation has only a minor prognostic value (for 10-year OS only) but a significant predictive value for anthracycline-based treatment in TNBC patients. To conclusively confirm the predictive value of PITX2 DNA methylation, the results warrant a confirmatory study with tumor specimens from a prospective trial with a predefined follow-up period in anthracycline-treated patients versus anthracycline-free treated patients and distinction regarding taxane cotreatment.
Statement of Ethics
This study was conducted in accordance with the Declaration of Helsinki (1964). All of the subjects included in this study provided their written informed consent (archived at each site). This study was approved by the Ethical Commission of the TUM (reference No. 101/16s).
Disclosure Statement
O.G.W., G.S., R.N., and M.A. are employees of Therawis Diagnostics. J.P. is an employee of QIAGEN. O.G.W., R.N., M.A., V.M., M.K., and G.S. are equity owners of Therawis Diagnostics GmbH. W.W. is a member of the following advisory boards: AstraZeneca, Novartis, BMS, MSD, Roche, Pfizer, and Takeda. This work was in part performed under a research agreement supported by Therawis Diagnostics. W.W. has attended advisory boards and served as speaker for Roche, MSD, BMS, AstraZeneca, Pfizer, Merck, Lilly, Boehringer, Novartis, Takeda, Amgen, and Astellas. W.W. receives research funding from Roche, MSD, BMS, and Bruker. M.K. received remuneration for advisory boards, travel costs, and lectures from Celgene, Astra Zeneca, Myriad Genetics, and TEVA. M.H., G.A., K.U., and A.W. report no conflict of interests.
Funding Sources
None declared.
Author Contributions
All of the authors participated in the conception and design of this study. They also read and approved it. O.G.W., M.A., G.S., R.N., and M.H. set up the experimental concept and designed this study. G.A., M.K., W.W., and A.W. provided tumor samples and histopathological data from a tumor bank and gave pathological approval of the tissues. V.M. coordinated probe sampling and provision of the data base of patients. J.P. and R.N. were responsible for assay design and development and quality control. K.U. verified the analytical methods and the statistical project set-up and performed statistical analyses of the results obtained.
Acknowledgment
We thank Elisabeth Schüren for her technical assistance. We would also like to thank 2 independent Therawis reviewers for their insights and support in preparing the graphs and tables for this publication. This work has been partly supported by Therawis Diagnostics GmbH. The funding had no impact on or role in the study design, data collection, analysis, and interpretation, or the writing of this report.
References
- 1.Aubele M, Schmitt M, Napieralski R, Paepke S, Ettl J, Absmaier M, et al. The Predictive Value of PITX2 DNA Methylation for High-Risk Breast Cancer Therapy: Current Guidelines, Medical Needs, and Challenges. Dis Markers. 2017;2017:4934608. doi: 10.1155/2017/4934608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baylin SB. DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol. 2005 Dec;2((S1 Suppl 1)):S4–11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- 3.Basu M, Roy SS. Wnt/β-catenin pathway is regulated by PITX2 homeodomain protein and thus contributes to the proliferation of human ovarian adenocarcinoma cell, SKOV-3. J Biol Chem. 2013 Feb;288((6)):4355–67. doi: 10.1074/jbc.M112.409102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hartmann O, Spyratos F, Harbeck N, Dietrich D, Fassbender A, Schmitt M, et al. DNA methylation markers predict outcome in node-positive, estrogen receptor-positive breast cancer with adjuvant anthracycline-based chemotherapy. Clin Cancer Res. 2009 Jan;15((1)):315–23. doi: 10.1158/1078-0432.CCR-08-0166. [DOI] [PubMed] [Google Scholar]
- 5.Schmitt M, Wilhelm OG, Noske A, Schricker G, Napieralski R, Vetter M, et al. Clinical Validation of PITX2 DNA Methylation to Predict Outcome in High-Risk Breast Cancer Patients Treated with Anthracycline-Based Chemotherapy. Breast Care (Basel) 2018 Dec;13((6)):425–33. doi: 10.1159/000493016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Absmaier M, Napieralski R, Schuster T, Aubele M, Walch A, Magdolen V, et al. PITX2 DNA-methylation predicts response to anthracycline-based adjuvant chemotherapy in triple-negative breast cancer patients. Int J Oncol. 2018 Mar;52((3)):755–67. doi: 10.3892/ijo.2018.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ballman KV. Biomarker: predictive or Prognostic? J Clin Oncol. 2015 Nov;33((33)):3968–71. doi: 10.1200/JCO.2015.63.3651. [DOI] [PubMed] [Google Scholar]
- 8.Napieralski R, Schricker G, Piednoir E, Manner O, Bona A, Segalas S, et al. therascreen PITX2 RGQ PCR assay for the assessment of PITX2 DNA-methylation status to investigate the role of the transcription factor PITX2 and the regulation of the Wnt/ß-catenin pathway in pathophysiological processes. In: Perkins J, Noske A, Magdolen V, Weichert W, Kiechle M, Wilhelm O, Schmitt M, editors. Protocol Exchange. 2018. [Google Scholar]
- 9.Napieralski R, Schricker G, Schueren E, Perkins J, Magdolen V, Weichert W, Kiechle M, Wilhelm O. Establishment of a RESEARCH USE ONLY Condensed-Efficient-Fast (CEF) PITX2 workflow for analysis of PITX2 DNA methylation in small tumor tissue samples. Protocol Exchange. 2020 [Google Scholar]
- 10.QIAGEN therascreen PITX2 RGQ PCR Kit Handbook. QIAGEN GmbH. 2017 [Google Scholar]
- 11.Maier S, Nimmrich I, Koenig T, Eppenberger-Castori S, Bohlmann I, Paradiso A, et al. European Organisation for Research and Treatment of Cancer (EORTC) PathoBiology group DNA-methylation of the homeodomain transcription factor PITX2 reliably predicts risk of distant disease recurrence in tamoxifen-treated, node-negative breast cancer patients—technical and clinical validation in a multi-centre setting in collaboration with the European Organisation for Research and Treatment of Cancer (EORTC) PathoBiology group. Eur J Cancer. 2007 Jul;43((11)):1679–86. doi: 10.1016/j.ejca.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 12.Hothorn T. Maxstat: maximally selected rank statistics. In: R package version 0.7-14. 2002 [Google Scholar]
- 13.Cox DR, Oakes D. Analysis of survival data. London, United Kingdom: Chapman and Hall; 1984. [Google Scholar]
- 14.Harrell FE. Regression modeling strategies: with applications to linear models, logistic regression, and survival analysis. New York, NY. Cham: Springer; 2001. [Google Scholar]
- 15.Schricker G, Napieralski R, Noske A, Piednoir E, Manner O, Schüren E, et al. Clinical performance of an analytically validated assay in comparison to microarray technology to assess PITX2 DNA-methylation in breast cancer. Sci Rep. 2018 Nov;8((1)):16861. doi: 10.1038/s41598-018-34919-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nimmrich I, Sieuwerts AM, Meijer-van Gelder ME, Schwope I, Bolt-de Vries J, Harbeck N, et al. DNA hypermethylation of PITX2 is a marker of poor prognosis in untreated lymph node-negative hormone receptor-positive breast cancer patients. Breast Cancer Res Treat. 2008 Oct;111((3)):429–37. doi: 10.1007/s10549-007-9800-8. [DOI] [PubMed] [Google Scholar]
- 17.Harbeck N, Nimmrich I, Hartmann A, Ross JS, Cufer T, Grützmann R, et al. Multicenter study using paraffin-embedded tumor tissue testing PITX2 DNA methylation as a marker for outcome prediction in tamoxifen-treated, node-negative breast cancer patients. J Clin Oncol. 2008 Nov;26((31)):5036–42. doi: 10.1200/JCO.2007.14.1697. [DOI] [PubMed] [Google Scholar]
- 18.Fung FK, Chan DW, Liu VW, Leung TH, Cheung AN, Ngan HY. Increased expression of PITX2 transcription factor contributes to ovarian cancer progression. PLoS One. 2012;7((5)):e37076. doi: 10.1371/journal.pone.0037076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pillai SG, Dasgupta N, Siddappa CM, Watson MA, Fleming T, Trinkaus K, et al. Paired-like Homeodomain Transcription factor 2 expression by breast cancer bone marrow disseminated tumor cells is associated with early recurrent disease development. Breast Cancer Res Treat. 2015 Oct;153((3)):507–17. doi: 10.1007/s10549-015-3576-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang Y, Guigon CJ, Fan J, Cheng SY, Zhu GZ. Pituitary homeobox 2 (PITX2) promotes thyroid carcinogenesis by activation of cyclin D2. Cell Cycle. 2010 Apr;9((7)):1333–41. doi: 10.4161/cc.9.7.11126. [DOI] [PubMed] [Google Scholar]
- 21.Vela I, Morrissey C, Zhang X, Chen S, Corey E, Strutton GM, et al. PITX2 and non-canonical Wnt pathway interaction in metastatic prostate cancer. Clin Exp Metastasis. 2014 Feb;31((2)):199–211. doi: 10.1007/s10585-013-9620-7. [DOI] [PubMed] [Google Scholar]
- 22.Lee WK, Chakraborty PK, Thévenod F. Pituitary homeobox 2 (PITX2) protects renal cancer cell lines against doxorubicin toxicity by transcriptional activation of the multidrug transporter ABCB1. Int J Cancer. 2013 Aug;133((3)):556–67. doi: 10.1002/ijc.28060. [DOI] [PubMed] [Google Scholar]
- 23.Lee WK, Thévenod F. Oncogenic PITX2 facilitates tumor cell drug resistance by inverse regulation of hOCT3/SLC22A3 and ABC drug transporters in colon and kidney cancers. Cancer Lett. 2019 May;449:237–51. doi: 10.1016/j.canlet.2019.01.044. [DOI] [PubMed] [Google Scholar]
- 24.Xu YY, Yu HR, Sun JY, Zhao Z, Li S, Zhang XF, Liao ZX, Cui MK, Li J, Li C, Zhang Q. Upregulation of PITX2 Promotes Letrozole Resistance Via Transcriptional Activation of IFITM1 Signaling in Breast Cancer Cells. Cancer research and treatment: official journal of Korean Cancer Association. 2019;51:576–592. doi: 10.4143/crt.2018.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang JX, Tong ZT, Yang L, Wang F, Chai HP, Zhang F, et al. PITX2: a promising predictive biomarker of patients' prognosis and chemoradioresistance in esophageal squamous cell carcinoma. Int J Cancer. 2013 Jun;132((11)):2567–77. doi: 10.1002/ijc.27930. [DOI] [PubMed] [Google Scholar]
- 26.Kashiwagi S, Yashiro M, Takashima T, Aomatsu N, Ikeda K, Ogawa Y, et al. Advantages of adjuvant chemotherapy for patients with triple-negative breast cancer at Stage II: usefulness of prognostic markers E-cadherin and Ki67. Breast Cancer Res. 2011;13((6)):R122. doi: 10.1186/bcr3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elsawaf Z, Sinn HP. Triple-Negative Breast Cancer: Clinical and Histological Correlations. Breast Care (Basel) 2011;6((4)):273–8. doi: 10.1159/000331643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pareja F, Geyer FC, Marchiò C, Burke KA, Weigelt B, Reis-Filho JS. Triple-negative breast cancer: the importance of molecular and histologic subtyping, and recognition of low-grade variants. NPJ Breast Cancer. 2016 Nov;2((1)):16036. doi: 10.1038/npjbcancer.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen X, Li J, Gray WH, Lehmann BD, Bauer JA, Shyr Y, et al. TNBCtype: A Subtyping Tool for Triple-Negative Breast Cancer. Cancer Inform. 2012;11:147–56. doi: 10.4137/CIN.S9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lehmann BD, Jovanović B, Chen X, Estrada MV, Johnson KN, Shyr Y, et al. Refinement of Triple-Negative Breast Cancer Molecular Subtypes: Implications for Neoadjuvant Chemotherapy Selection. PLoS One. 2016 Jun;11((6)):e0157368. doi: 10.1371/journal.pone.0157368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007 Aug;13((15 Pt 1)):4429–34. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- 32.Bagegni NA, Tao Y, Ademuyiwa FO. Clinical outcomes with neoadjuvant versus adjuvant chemotherapy for triple negative breast cancer: A report from the National Cancer Database. PLoS One. 2019 Sep;14((9)):e0222358. doi: 10.1371/journal.pone.0222358. [DOI] [PMC free article] [PubMed] [Google Scholar]