Abstract
Understanding the molecular basis for immune recognition of SARS-CoV-2 spike (S) glycoprotein antigenic sites will inform development of improved therapeutics. We determined the structures of two human monoclonal antibodies AZD8895 and AZD1061, which form the basis of the investigational antibody cocktail AZD7442, in complex with the receptor binding domain (RBD) of SARS-CoV-2, in order to define the genetic and structural basis of neutralization. AZD8895 forms an “aromatic cage” at the heavy/light chain interface using germline-encoded residues in complementarity determining regions (CDRs) 2 and 3 of the heavy chain and CDRs 1 and 3 of the light chain. These structural features explain why highly similar antibodies (public clonotypes) have been isolated from multiple individuals. AZD1061 has an unusually long LCDR1, and HCDR3 make interactions with the opposite face of the RBD from that of AZD8895. Using deep mutational scanning and neutralization escape selection experiments, we comprehensively mapped the crucial binding residues of both antibodies and identified positions of concern with regards to virus escape from antibody-mediated neutralization. Both AZD8895 and AZD1061 have strong neutralizing activity against SARS-CoV-2 and variants of concern with antigenic substitutions in the RBD. We conclude that germline-encoded antibody features enable recognition of the SARS-CoV-2 spike RBD and demonstrate the utility of the cocktail AZD7442 in neutralizing emerging variant viruses.
Keywords: Coronavirus, SARS-CoV-2, SARS-CoV, COVID-19, Antibodies, Monoclonal, Human, Adaptive Immunity
The current coronavirus disease 2019 (COVID-19) pandemic is caused by SARS-CoV-2, a clade B betacoronavirus (Sarbecovirus subgenus). The S glycoprotein mediates viral attachment via binding to the host receptor angiotensin converting enzyme 2 (ACE2) and possibly other host factors, enabling subsequent entry into cells after priming by the host transmembrane protease serine 2 (TMPRSS2)1–3. The trimeric S protein consists of two subunits, designated S1 and S2. The S1 subunit binds to ACE2 with its receptor binding domain (RBD), while the central trimeric S2 subunits function as a fusion apparatus after S protein sheds the S1 subunits4. The human humoral immune response to SARS-CoV-2 has been well documented5–7, and numerous groups have isolated monoclonal antibodies (mAbs) from B cells of previously infected patients that react to SARS-CoV-2 S protein. A subset of the human mAbs neutralize virus in vitro and protect against disease in animal models7–17. Studies of the human B cell response to SARS-CoV-2 have focused mostly on S protein so far, due to its critical functions in attachment and entry into host cells7–17. For these S-protein-targeting antibodies, the RBD of S protein is the dominant target of human neutralizing antibody responses7–17. This high frequency of molecular recognition may be related to the accessibility of the RBD to B cell receptors, stemming from a low number of obscuring glycans (only 2 glycosylation sites on the RBD versus 8 or 9 sites on the N-terminal domain [NTD] or S2 subunit, respectively)7. The RBD also occupies an apical position and exhibits exposure due to the “open-closed” dynamics of the S trimer observed in S protein cryo-EM structures18–20. Potently neutralizing mAbs predominantly target the RBD, since this region is directly involved in receptor binding.
In previous studies, we isolated a large panel of human mAbs that bind to the SARS-CoV-2 S protein from the B cells of patients previously infected with the virus21. A subset of these mAbs was shown to bind to recombinant RBD and S protein ectodomain and exhibit neutralization activity against SARS-CoV-2 by blocking S-protein-mediated binding to receptor17. Two noncompeting antibodies, designated COV2–2196 and COV2–2130 (later engineered to be long-acting IgG molecules designated as AZD8895 and AZD1061, respectively), synergistically neutralized SARS-CoV-2 in vitro and protected against SARS-CoV-2 infection in mouse models and a rhesus macaque model when used separately or in combination17,21. Several Phase III clinical trials are ongoing to study the antibody cocktail AZD7442, which incorporates AZD8895 and AZD1061, for post-exposure prophylaxis (ClinicalTrials.gov Identifier: NCT04625972), prevention (Identifier: NCT04625725), out-patient treatment (Identifier: NCT04723394 and NCT04518410) and in-patient treatment (NCT04501978) of COVID-19. Thus, it is important to define the binding sites of these two antibodies to understand how they interact with the RBD and their ability to neutralize new virus variants.
To understand the atomic details of the recognition of RBD by AZD8895 and AZD1061, we determined the crystal structures of the S protein RBD in complex with AZD8895 at 2.50 Å (Fig. 1, Supplementary Data Table 1) and in complex with both AZD8895 and AZD1061 at 3.00 Å (Fig. 2, Supplementary Data Table 1). The substructure of RBD/AZD8895 in the RBD/AZD8895/AZD1061 complex is superimposable with that in the structure of the RBD/AZD8895 complex (Extended Data Fig. 1). AZD8895 binds to the receptor-binding ridge of RBD, and AZD1061 binds to one side of the RBD edge around residue K444 and the saddle region of the receptor binding motif RBM), both partially overlapping the ACE2 binding site (Fig. 1a,b,c, 2a–b). These features explain the competition between the antibodies and ACE2 for RBD binding from our previous study, e.g., both AZD8895 and AZD1061 neutralize the virus by blocking RBD access to the human receptor ACE217. Aromatic residues from the AZD8895 heavy and light chains form a hydrophobic pocket that surrounds RBD residue F486 and adjacent residues (G485, N487) (Fig. 1a, 1d; Extended Data Fig. 2a–c). This mode of antibody-antigen interaction is unusual in that the formation of the antibody pocket is caused by wide spatial separation of the HCDR3 and LCDR3. Overlays of the substructure of RBD in complex with AZD1061 (Fig. 2c) and the structure of RBD in complex with both AZD8895 and AZD1061 (Fig. 2d) suggest that AZD1061 is able to bind RBD in both “up” and “down” conformations of the S trimer. We compared the RBD/AZD1061 crystal structure with the published cryo EM structures of the human mAbs C119 and C13513. AZD1061, C119, and C135 have overlapping but different epitopes, as R346 and K444 of the RBD are key residues for AZD1061 and C13522 binding but were not important for C119 binding (Extended Data Fig. 5e).
Fig. 1. Crystal structure of S protein RBD in complex with Fab AZD8895.
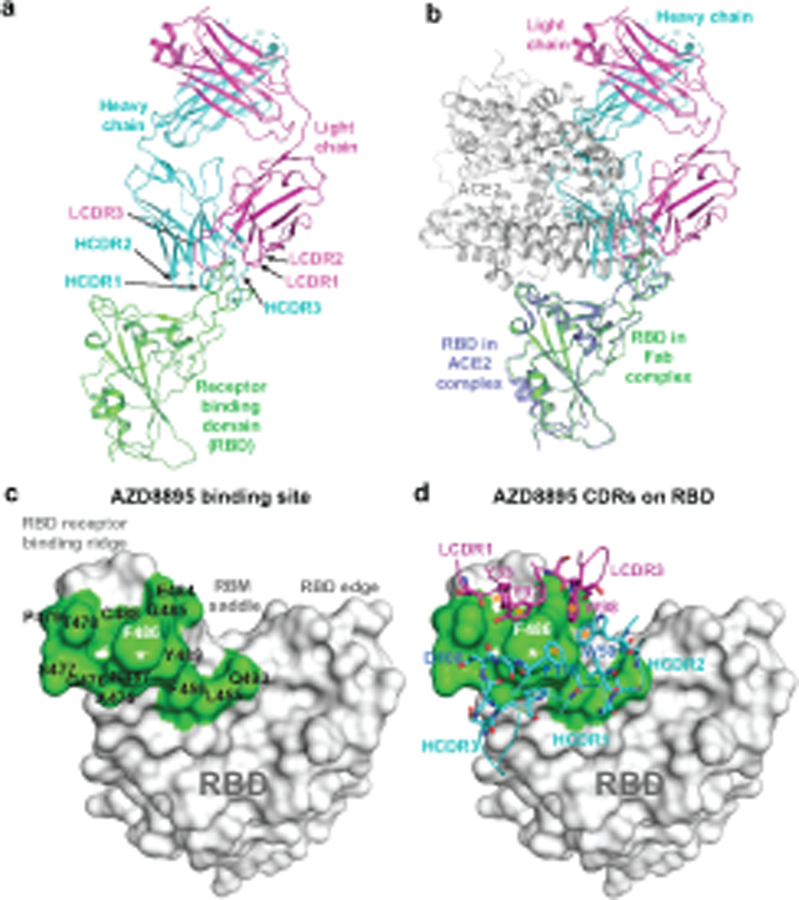
a. Cartoon representation of AZD8895 in complex with RBD. AZD8895 heavy chain is shown in cyan, light chain in magenta, and RBD in green.
b. Structure of AZD8895-RBD complex is superimposed onto the structure of RBD/human ACE2 complex (PDB ID: 6M0J), using the RBD structure as the reference. The color scheme of RBD/AZD8895 complex is the same as that in Fig. 1a. The RBD in the RBD/ACE2 complex is colored in blue, the human ACE2 peptidase domain in grey.
c. Structure of the RBD/AZD8895 complex is superimposed onto the structure of spike with single RBD in the “up” conformation (PDB ID: 6XM4), using the RBD in “up” conformation as the reference. The color scheme of the RBD/AZD8895 complex is the same as that in Fig. 1a. The three subunits of spike are colored in grey, yellow, or blue respectively (the subunit with its RBD in “up” conformation is yellow).
d. Surface representation of RBD epitope recognized by AZD8895. The epitope residues are colored in different shades of green and labeled in black with the critical contact residue F486 labled in white.
e. Antibody-antigen interactions between AZD8895 and RBD. RBD is shown in the same surface representation and orientation as that in Fig. 1d. AZD8895 paratope residues are shown in stick representation. The heavy chain is colored in cyan, and light chain is colored in magenta. Aromatic cage residues Y33, Y92, W98, F110, and W50 are all colored with darker shades of blue or purple, and labelled with an orange star.
Fig. 2. Crystal structure of S protein RBD in complex with both Fabs AZD8895 and AZD1061.
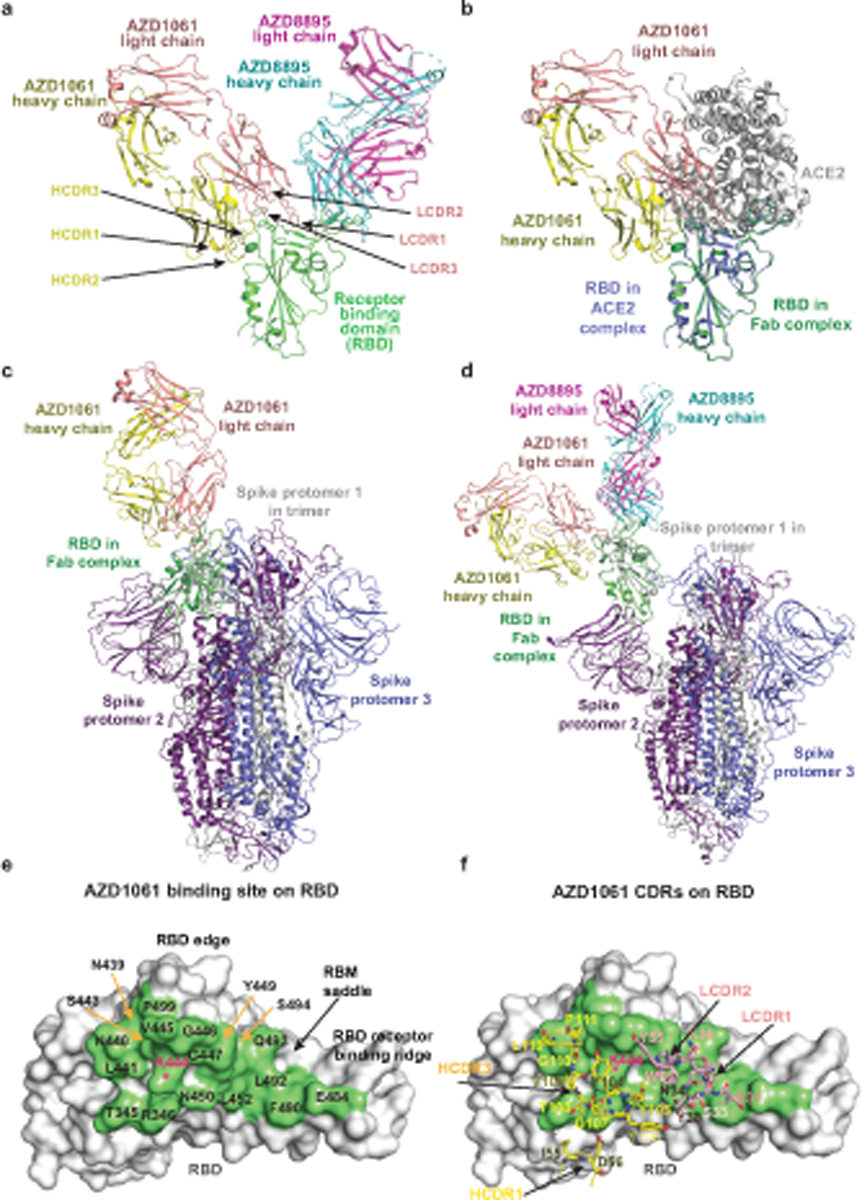
a. Cartoon representation of crystal structure of S protein RBD in complex with AZD8895 and AZD1061 Fabs. RBD is shown in green, AZD8895 heavy chain in cyan, AZD8895 light chain in magenta, AZD1061 heavy chain in yellow, and AZD1061 light chain in pink. CDRs of AZD1061 are labeled.
b. Structure of RBD/AZD1061 complex is superimposed onto the structure of the RBD/ACE2 complex (PDB ID: 6M0J), using the RBD structure as the reference. The color scheme of the RBD/AZD1061 complex is the same as that in Fig. 2a. The RBD in the RBD/ACE2 complex is colored in blue, the human ACE2 peptidase domain in grey.
c. Structure of RBD/AZD1061 complex is superimposed onto the structure of spike with all RBD in “down” conformation (PDB ID: 6ZOY), using the RBD in one protomer as the reference. The color scheme of RBD/AZD1061 complex is the same as that in Fig. 2a. The three protomers of spike are colored in grey, blue, or purple respectively.
d. Structure of the RBD/AZD8895/AZD1061 complex is superimposed onto the structure of spike with one RBD in “up” conformation (PDB ID: 7CAK), using the RBD in “up” conformation as the reference. The color scheme of RBD/AZD1061 complex is the same as that in Fig. 2a. The three protomers of spike are colored in grey, blue, or purple respectively.
e. Surface representation of RBD epitope recognized by AZD1061. The epitope residues are indicated in different colors and labeled in black; the key residue K444 is labelled in red.
f. Interactions of AZD1061 paratope residues with the epitope. RBD is shown in the same surface representation and orientation as those in Fig. 2e. The paratope residues are shown in stick representation. The heavy chain is colored in yellow, and the light chain in orange. The key residue K444 is labelled in red.
The AZD8895 interaction with the RBD F486 residue is distinctive. The RBD aromatic residue interacts extensively via a hydrophobic effect and van der Waals interactions with a hydrophobic pocket formed between AZD8895 heavy/light chains (residue P99 of heavy chain and an “aromatic cage” formed by 5 aromatic side chains) (Fig. 1d, Extended Data Fig. 2a–b). A hydrogen bond (H-bond) network, constructed with 4 direct antibody-RBD H-bonds and 16 water-mediated H-bonds, surround residue F486 and strengthen the antibody-RBD interaction (Extended Data Fig. 2c). Importantly, all residues that interact extensively with the epitope except one (residue P99 of the heavy chain) are encoded by germline sequences (IGHV1–58*01 and IGHJ3*02 for the heavy chain, IGKV3–20*01 and IGKJ1*01 for the light chain) (Fig. 3a) or only their backbone atoms are involved in the antibody-RBD interactions, such as heavy chain N107 and G99 and light chain S94. We noted another antibody in the literature, S2E12, that is encoded by the same IGHV/IGHJ and IGKV/IGKJ recombinations, with similar but most likely different IGHD genes to those of AZD8895 (IGHD2–15 vs IGHD2–2)23. A comparison of the cryo-EM structure of S2E12 in complex with S protein (PDB 7K4N) suggests that the mAb S2E12 likely uses nearly identical antibody-RBD interactions as those of AZD8895, although variations in conformations of interface residue side-chains can be seen (Extended Data Fig. 2a–b). For example, the phenyl rings of light chain residue Y92 are perpendicular to each other in the two structures. These analyses suggest that AZD8895 and S2E12 have similar modes of recognition of RBD.
Fig. 3. Characterization of important sequence features of the AZD8895 public clonotype.
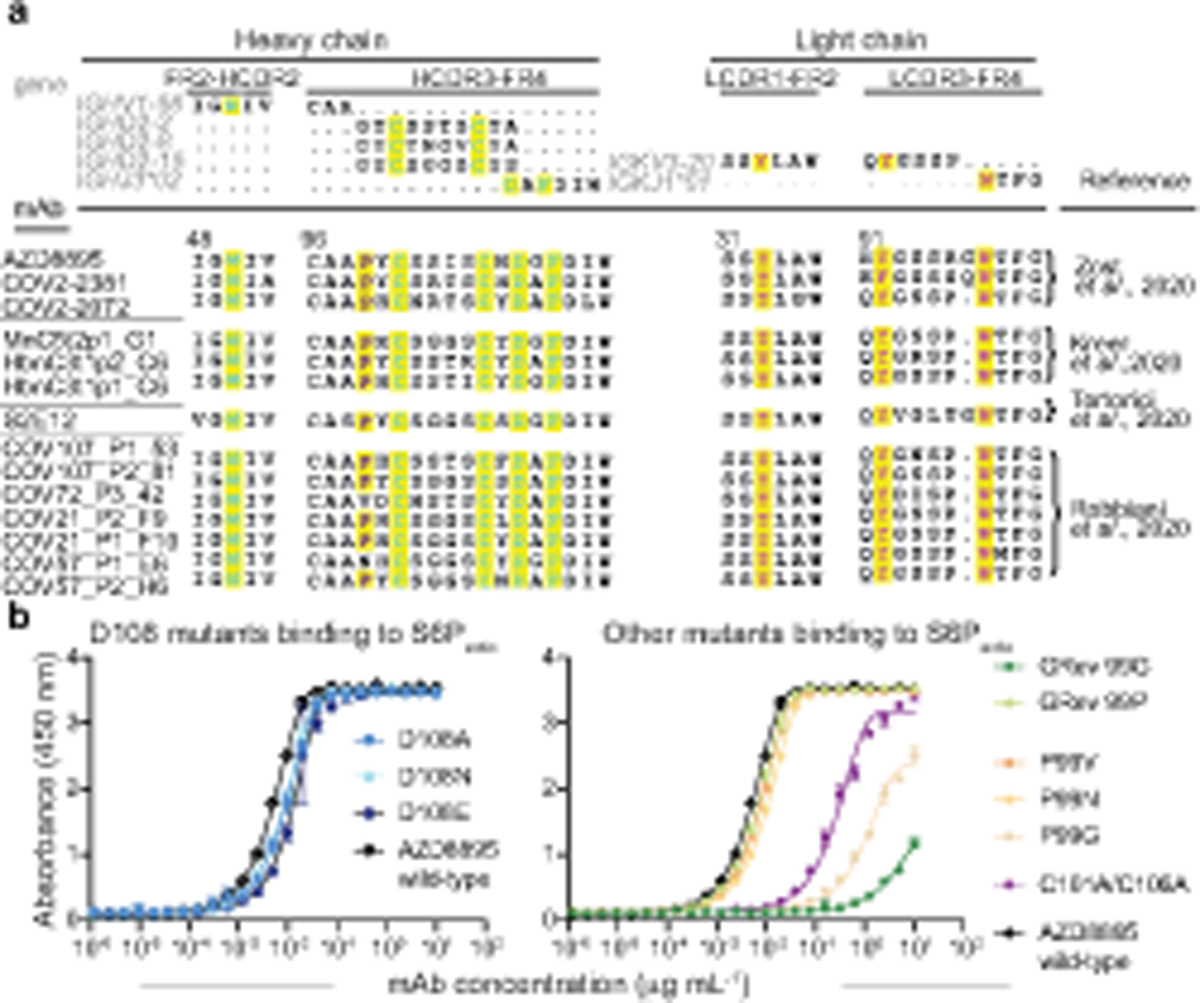
a. IMGT/DomainGapAlign results of AZD8895 heavy and light chains with germline V (IGHV1–58 and IGLV3–20), D (IGHD2–2, IGHD 2–8, or IGHD 2–15) or J (IGHJ3*02 and IGKJ1*01) gene segments and with representative variable gene sequences of mAbs in this public clonotype. Key interacting residues and their corresponding residues in germline genes are highlighted in yellow and colored in blue except for P99 in purple (heavy chain) or in red (light chain).
b. Binding curves of point mutants of AZD8895. Mutants of D108 residue are in blue, revertant mutation of inferred somatic mutations to germline sequence (GRev) are in green, P99 mutants are in orange, and C101A/C106A mutations removing the disulfide bond in HCDR3 is in purple. Data points show the mean ± SD for each tested antibody dilution. Experiments were performed in technical triplicate, with data shown from a single experiment repeated twice. Data for the AZD8895 wild-type binding curve shown in both panels are from the same experiment.
We searched genetic databases to determine if these structural features are present in additional SARS-CoV-2 mAbs isolated by others and found additional members of the clonotype (Fig. 3a). Two other studies reported the same or a similar clonotype of antibodies isolated from multiple COVID-19 convalescent patients13,23, and one study found three antibodies with the same IGHV1–58 and IGKV3–20 pairing, without providing information on D or J gene usage24. All of these antibodies are reported to bind SARS-CoV-2 RBD avidly and to neutralize virus with high potency13,17,23,24. So far, there are only two atomic resolution structures of antibodies encoded by these VH-DH-JH and VK-JK recombinations available, the structure for AZD8895 presented here and that for S2E1223. We performed homology modeling with the Structuropedia webserver25 for two additional antibodies of this clonotype from our own panel of anti-SARS-CoV-2 antibodies, designated COV2–2072 and COV2–2381, using the RBD/AZD8895 crystal structure as the template to construct the homology models. As expected, given that these antibodies are members of a shared genetic clonotype, the modeled structures of RBD/COV2–2072 and RBD/COV2–2381 complexes are virtually superimposable with those of RBD/AZD8895 and RBD/S2E12 at the RBD/antibody interfaces (Extended Data Fig. 3a–e). Additionally, COV2–2072 encodes an N-linked glycosylation sequon in the HCDR3, which we modeled using the GlyProt webserver26 (Extended Data Fig. 3d). This HCDR3 glycosylation is similar to that of the recently reported antibody 253H55L27 and represents an unusual feature for antibodies, given that glycosylation of CDRs might adversely affect antigen recognition. However, the AZD8895 structure shows that the disulfide-stapled HCDR3 in this clonotype is angled away from the binding site, explaining how this unusual HCDR3 glycosylation in COV2–2072 can be tolerated without compromising binding (Extended Data Fig. 3d). Inspecting the sequences carefully, we speculate that it is likely that the AZD8895 sequence derives from an independent rearrangement and that COV2–2072 and COV2–2381 derive from a common rearrangement, based on cDNA sequences. Differences in the HCDR3 are consistent with this interpretation, since differing nucleotide substitutions encoding the same amino acid are present in the two inferred lineages. However, it is impossible to be certain about this for technical reasons. In our original antibody discovery effort, two PBMC samples were pooled to expedite the efficiency of the large-scale screening effort21. Thus, these particular antibodies theoretically could have arisen from 2 different donors, or from 2 clones that occurred independently in one donor with similar genetic features, or from 1 clone in a single donor that diverged in a complex manner into diverse somatic variants.
We next determined whether we could identify potential precursors of this public clonotype in the antibody variable gene repertoires of circulating B cells from SARS-CoV-2-naïve individuals. We searched for the V-D-J and V-J genes in previously described comprehensive repertoire datasets originating from 3 healthy human donors without a history of SARS-CoV-2 infection and in datasets from cord blood collected prior to the COVID-19 pandemic28. A total of 386, 193, 47, or 7 heavy chain sequences for this SARS-CoV-2 reactive public clonotype was found in each donor or cord blood repertoire, respectively (Extended Data Fig. 4a). Additionally, we found 516,738 human antibody sequences with the same light chain V-J recombination (IGKV3–20-IGKJ1*01). A total of 103,534, 191,039, or 222,165 light chain sequences was found for this public clonotype in each donor respectively. Due to the large number of sequences, the top five abundant sequences were aligned from each donor. Multiple sequence alignments were generated for each donor’s sequences and logo plots were generated. The top 5 sequences with the same recombination event in each donor were identical, resulting in the same logo plots (Extended Data Fig. 4a,b).
We noted that 8 of the 9 common residues important for RBD binding in the antibody were encoded by germline gene sequences. Interestingly, these residues were present in all 14 members of the public clonotype that we or others have described (Fig. 3a)13,17,23,24. To validate the importance of these features, we expressed variant antibodies with point mutations in the HCDR3 of the paratope to determine the effect of variation at conserved residues on antibody binding to RBD (Fig. 3b).
We focused site-directed mutagenesis efforts on the P99 and D108 residues since these positions could be impacted at the stage of VDJ recombination. Altering the D108 residue to A, N, or E had little effect, but removing the disulfide bond in the HCDR3 through cysteine to alanine substitutions greatly reduced binding. While altering the P99 residue to V or N (observed in other mature antibodies) had little effect, a P99G substitution had a dramatic effect on binding. Additionally, we made two germline revertants of the AZD8895 antibody. The P99 residue is not templated by the V-gene IGHV1–58 nor by the D gene IGHD2–2. However, IGHD2–2 has a likely templated G at position 99. Therefore, two germline revertants were tested - one with P99 and the other with G99. As the P99 residue orients the HCDR3 loop away from the interaction site with antigen, the G99 germline revertant exhibited reduced binding, whereas the P99 germline revertant bound antigen equivalently to wt AZD8895 (Fig. 3b).
Unlike AZD8895, AZD1061 uses the HCDR3 for critical contacts. The HCDR3 comprises 22 amino acid residues, which is relatively long for human antibodies. The HCDR3 forms a long, structured loop that is stabilized by short-ranged hydrogen bonds and hydrophobic interactions/aromatic stackings within the HCDR3 and is further strengthened by its interactions (hydrogen bonds and aromatic stackings) with residues of the light chain (Extended Data Fig. 5a,b). The AZD1061 heavy or light chain is encoded by the germline gene IGHV3–15 or IGKV4–1, respectively, and the two genes encode the longest germline-encoded HCDR2 (10 residues) and LCDR1 (12 residues) loops. The heavy chain V-D-J recombination, HCDR3 mutations, and the pairing of heavy and light chains result in a binding cleft between the heavy and light chains, matching the shape of the RBD region centered at the S443 – Y449 loop (Fig. 2a, Extended Data Fig. 5c). Closely related to these structural features, only HCDR3, LCDR1, HCDR2, and LCDR2 are involved in formation of the paratope (Fig. 2e–f, Extended Data Fig. 2e–f). Inspection of the antibody-RBD interface reveals a region that likely drives much of the energy of interaction. The RBD residue K444 sidechain is surrounded by subloop Y104 – V109 of the HCDR3 loop, and the positive charge on the side chain nitrogen atom is neutralized by the HCDR3 residue D107 side chain, three mainchain carbonyl oxygen atoms from Y105, D107, and V109, and the electron-rich face of the Y104 phenyl ring (cation-π interaction) (Extended Data Fig. 2e). In addition, AZD1061 light chain LCDR1 and LCDR2 make extensive contacts with the RBD (Extended Data Fig. 2f). In the crystal structure of the RBD in complex with both AZD8895 and AZD1061, we noted that the closely spaced AZD8895 and AZD1061 Fabs may interact directly with each other when bound to RBD (Extended Data Fig. 6).
To better understand the RBD residues critical for binding of AZD8895 and AZD1061, we used a deep mutational scanning (DMS) approach to map all RBD mutations that escape antibody binding29; (Extended Data Fig. 7). Antibody escape in the DMS studies was quantified as an ‘escape fraction’ ranging from 0 (no cells with the mutation in the antibody-escape bin) to 1 (all cells with the mutation in the antibody-escape bin). For both antibodies, we identified several key positions, nearly all in the antibody binding site, where RBD mutations strongly disrupted binding (Fig. 4a–d). We leveraged our previous work quantifying the effects of RBD mutations on ACE2 binding30 to overlay the effect on ACE2 binding for mutations that abrogated antibody binding to the Wuhan-1 strain RBD (Fig. 4a,b). For AZD8895, many mutations to F486 and N487 had escape fractions approaching 1 (i.e., all cells expressing this RBD mutation fell into the antibody-escape bin), reinforcing the importance of the contributions of these two residues to antibody binding. Similarly, for AZD1061, mutation of residue K444 to any of the other 19 amino acids abrogated antibody binding, indicating that the lysine at this position is critical for the antibody-RBD interaction.
Fig. 4. Critical residues for AZD8895 and AZD1061 binding.
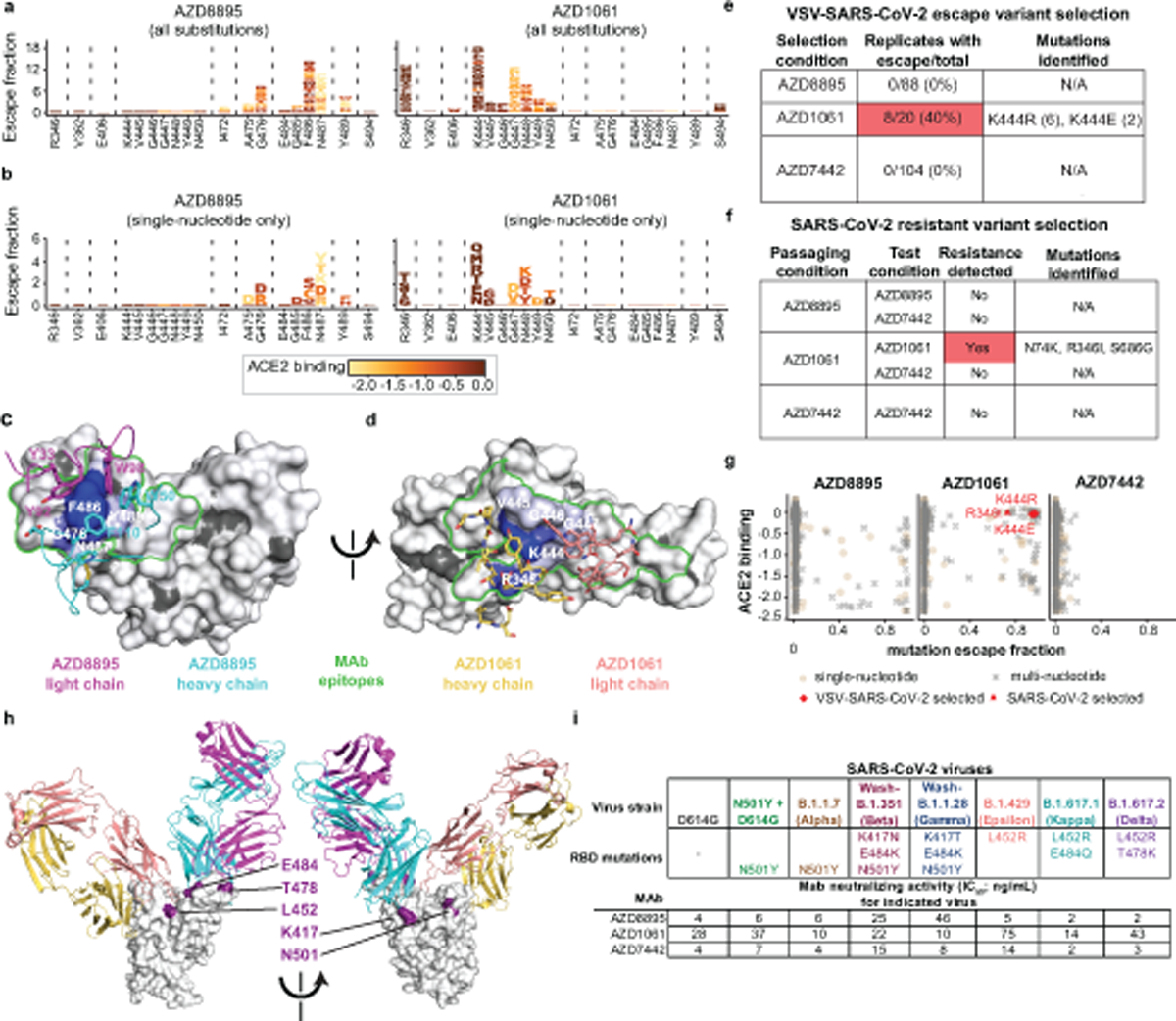
a. Logo plots of mutation escape fractions at RBD sites with strong escape for AZD8895 (left) or AZD1061 (right). Taller letters indicate greater escape. Mutations colored by degree to which they reduce RBD binding to hACE2. Data shown are the average of two independent escape experiments using two independent yeast libraries; correlations are shown in Extended Data Fig. 7b,c. Interactive plots - https://jbloomlab.github.io/SARS-CoV-2-RBD_MAP_AZ_Abs/.
b. Logo plots of mutation escape fractions for that are accessible by single nucleotide substitutions from the Wuhan-Hu-1 reference strain (e,f). Effect represented as in Fig. 4a.
c. Mapping DMS escape mutations for AZD8895 onto the RBD surface. Blue - RBD site with the greatest cumulative antibody escape; white - no escape detected. Grey - residues where deleterious effects on RBD expression prevented assessment. Heavy chain, cyan; light chain, magenta. Two replicates were performed with independent libraries, as described in (a). Green - structurally-defined AZD8895 footprint on RBD.
d. Mapping DMS escape mutations for AZD1061 onto the RBD surface in the RBD/AZD1061 structure. Mutations that abrogate AZD1061 binding are displayed on the RBD structure using a heatmap as in Fig. 4c. Heavy chain, yellow; light chain pink. Green - structurally-defined AZD1061 footprint on RBD.
e. Table showing the results of VSV-SARS-CoV-2 escape selection experiments.
f. Table showing the results of passage of SARS-CoV-2 in the presence of sub-neutralizing concentrations of mAbs.
g. Scatter plot showing DMS data from (a), with mutation escape fraction on the x-axis and effect on ACE2 binding on the y-axis. Crosses - mutations accessible only by multi-nucleotide substitutions; circles - mutations accessible by single-nucleotide substitution. Substitutions selected by AZD1061 in VSV-SARS-CoV-2 (K444R, K444E) or authentic SARS-CoV-2 (R346I) are denoted.
h. Locations of mutations in VOCs/VOIs mapped onto the RBD/AZD8895/AZD1061 crystal structure.
i. Neutralization (FRNT) against SARS-CoV-2 VOC/VOIs. Assays were performed in duplicate, repeated twice. Data are the mean of the two independent experiments. Wash-B.1.351 refers to a chimeric, recombinant virus with the WA1/2020 backbone expressing the B.1.351 (Beta) spike gene; Wash-B.1.1.28 refers to a similar virus expressing the B.1.1.28 [P1] (Gamma) spike gene.
Nevertheless, few antibody binding site residues were identified as sites where mutations greatly reduced binding. Several explanations are possible: 1) some binding site residues may not be critical for binding, 2) some RBD residues do not use their side chains to form interactions with the mAbs or 3) mutations at some sites may not be tolerated for RBD expression30. For instance, residues L455, F456, and Q493 are part of the structurally-defined binding site for AZD8895 (Fig. 1d), but mutations to these sites did not impact antibody binding (Fig. 4a), suggesting that these residues do not make critical binding contributions. Superimposition of the RBD/AZD8895 structure onto the RBD/S2E12 structure clearly demonstrates a flexible hinge region between the RBD ridge and the rest of the RBD that is maintained when antibody is bound (Extended Data Fig. 2d). This finding indicates that mutations at these three positions could be well-tolerated for antibody-RBD binding and supports the non-essential nature of these particular residues for AZD8895 or S2E12 binding.
Importantly, AZD8895 and AZD1061 do not compete with one another for binding to the RBD17, suggesting they could comprise an escape-resistant cocktail for prophylactic or therapeutic use. Indeed, the binding sites and escape variant maps for these two antibodies are non-overlapping. To test whether there were single mutations that could escape binding of both antibodies, we performed escape variant mapping experiments with AZD7442, but we did not detect any mutation that had an escape fraction of greater than 0.2, indicating only partial reduction in antibody binding, whereas the individual amino acid mutations with the largest effects for each of the single antibodies was approximately 1, indicating substantial loss of antibody binding due to that amino acid change (Extended Data Fig. 7d).
Although these experiments map all individual amino acid mutations that escape antibody binding to the RBD, we also sought to determine which mutations have the potential to arise during viral growth. To address this question, we first attempted to select escape mutations using a recombinant VSV expressing the SARS-CoV-2 S glycoprotein (VSV-SARS-CoV-2)31; (Fig 4e). We expected that the only amino acid mutations that would be selected during viral growth were those 1) arising by single-nucleotide RNA changes, 2) causing minimal deleterious effect on ACE2 binding and expression, and 3) substantially impacting antibody binding29,30. Indeed, we did not detect any AZD8895-induced mutations that were both single-nucleotide accessible and relatively well-tolerated with respect to effects on ACE2 binding (Fig. 4b), which may explain why escape mutants were not selected in any of the 88 independent replicates of recombinant VSV growth in the presence of antibody (Fig. 4e Extended Data Fig. 7g). For AZD1061, mutations to site K444, a site that is relatively tolerant to mutation30, demonstrated the most frequent escape from antibody binding in neutralization assays with the the VSV chimeric virus. K444R (selected in 6 out of 20 replicates) or K444E (selected in 2 out of 20 replicates) were identified in 40% of the replicates of recombinant VSV growth in the presence of AZD1061 (Fig. 4e, Extended Data Fig. 7g).
To explore resistance with authentic infectious virus, SARS-CoV-2 strain USA-WA1/2020 was passaged serially in Vero cell cultures with AZD8895, AZD1061 or AZD7442, at concentrations beginning at their respective IC50 values and increased step-wise to their IC90 value with each passage (Extended Data Fig. 8a). As a control, virus was passaged in the absence of antibody. Following the final passage, viruses were evaluated for susceptibility against the partner antibody at a final concentration of 10× the IC90 concentration by plaque assay. We did not detect any plaques resistant to neutralization by AZD8895 or the AZD7442 cocktail. Virus that was passaged serially in the presence of AZD1061 formed plaques to a titer of 1.2 × 107 PFU/mL after selection in 10× the IC90 value concentration of AZD1061, but plaques were not formed in the presence of AZD7442. Plaques (n=6) were selected randomly and the phenotype of the escaped viruses was characterized in virus neutralization tests (Extended Data Fig. 8b). The S gene form each plaque isolate was amplified and sequenced, revealing the same 3 amino acid changes in all 6 of the plaques: N74K, R346I and S686G (Fig. 4f). The S686G change in SARS-CoV-2 has been reported previously to be associated with serial passaging in Vero cells32 and identified in SARS-CoV-2 isolated from infected ferrets33 or NHPs34, and it is predicted to decrease furin activity32. The N74K residue is located in the N-terminal domain outside of the AZD1061 binding site and results in the loss of a glycan35. The R346I residue is located in the binding site of AZD1061 and may be associated with AZD1061-resistance. In a biolayer-interferometry-based binding test, AZD1061 retained binding to spike proteins with N74K and S686G to levels similar to the reference spike protein. (Extended Data Fig. 8c,d). However, there were dramatic reductions in AZD1061 binding to spike proteins containing either the R346I substitution or the K444R or K444E substitutions selected in the VSV-SARS-CoV-2 virus (Extended Data Fig. 8c,d). Importantly, AZD8895 binding was unaffected by these substitutions (Extended Data Fig. 8c,d). The data confirmed that N74K and S686G were mutations that accumulated during the passaging of SARS-CoV-2 in Vero cells, and that R346I is the primary amino acid change in RBD that contributed to escape from AZD1061.The K444R and K444E substitutions selected in the VSV-SARS-CoV-2 system and the R346I substitution selected by passage with authentic SARS-CoV-2 are accessible by single nucleotide substitution and preserve ACE2 binding activity (Fig. 4g), indicating that our DMS analysis using yeast predicted the mutations in SARS-CoV-2 selected in the presence of AZD1061 antibody. Taken together, these results comprehensively map the effects of all amino acid substitutions on the binding of AZD8895 and AZD1061 and identify sites of possible concern for viral evolution. That said, variants containing mutations at residues K444 and R346 are rare among all sequenced viruses present in the GISAID databases (all ≤ 0.08% when accessed on 11 May 2021).
Viral variants of concern (VOCs) with increased transmissibility and antigenic mutations have been reported in clinical isolates. Due to the potential of these VOCs to undermine the protective effects of immunity induced by infection, vaccination, or monoclonal antibody therapeutics, VOCs are the subject of intense study36–41. We next sought to determine whether some of the amino acid substitutions present in these emerging VOCs affect the activity of these potently neutralizing antibodies in focus reduction neutralization tests42. We tested authentic SARS-CoV-2 viruses that contained S proteins matching the B.1.1.7 (Alpha), B1.1.351 (Beta), B.1.1.28 (Gamma), B.1.617.1 (Kappa) or B.1.617.2 (Delta) VOCs or the B.1.429 (Epsilon) virus of interest (VOI). Substitutions at position E484 were of special interest, since this residue is located within 5 Å of each of the mAbs in the complex of Fabs and RBD, albeit at the very peripheral edge of the binding sites. E484K is present in emerging lineages B.1.35138 and B.1.1.2839, along with substitutions at K417, and has been demonstrated to alter the binding of some monoclonal antibodies40,43,44 as well as human polyclonal serum antibodies45. Variants containing E484K also have been shown to be neutralized less efficiently by convalescent serum and plasma from SARS-CoV-2 survivors46–48. An E484Q substitution is present in the B.1.617.1 (Kappa) VOC, which has also been shown to have reduced susceptibility to neutralization40. Additionally, a L452R substitution is present in the B.1.617.1 and B.1.617.240 VOCs and B.1.42941 VOI, which also abrogates the binding of some neutralizing antibodies. B.1.617.2 contains an additional T478K mutation in the RBD40. AZD8895, AZD1061, and AZD7442 all potently neutralized representatives of the B.1.1.7, B.1.351, B.1.1.28, B.1.617.1, and B.1.617.2 VOCs and B.1.429 VOI, with IC50 values comparable to those for the D614G strain, with at most a 6- to 11-fold reduction for AZD8895 in 2 cases (Fig. 4h,i). Recent reports from others also have described neutralization data for recombinant IgGs incorporating the Fv of COV2–2196 and COV2–2130, the predecessors of the engineered long-acting AZD8895 and AZD1061 antibodies, based on Fv sequences we previously published. Those studies also show that amino acid substitutions present in B.1.1.7, B.1.351, or P.1 (B.1.1.28) VOCs had little effect on neutralization of these mAbs42,49–52.
Discussion.
The process of B cell development, in which diverse variable gene segments are recombined, results in human naïve B cell repertoires containing an enormous amount of structural diversity in the complementarity determining regions (CDRs) of the antibodies (Abs) that they encode. Despite this extensive and diverse pool of naïve B cells, infection or vaccination with viral pathogens sometimes elicit antibodies in diverse individuals that share common structural features encoded by the same antibody variable genes. Examples of recurring variable gene usage have been described for antibody responses to human rotavirus16,53, human immunodeficiency virus54–57 influenza A virus58–61, and hepatitis C virus62,63, among others. The recognition of the use of common variable genes in antiviral responses has led to the general concept of B cell public clonotypes, or B cells with similar genetic features in their variable regions that encode for antibodies with similar patterns of specificity and function in different individuals. A number of recent reports have described the identification of public clonotypes in the Ab responses to SARS-CoV-28,13,64,65. Identifying and understanding the genetic and structural basis for selection of public clonotypes is valuable, as this information forms the central conceptual underpinning for many current rational structure-based vaccine design efforts66. Our structural analyses define the molecular basis for the frequent selection of a public clonotype of human antibodies sharing heavy chain V-D-J and light chain V-J recombinations that target the same region of the SARS-CoV-2 S RBD. Germline antibody gene-encoded residues in heavy and light chains play a vital role in antigen recognition, suggesting that few somatic mutations are required for antibody maturation of this clonotype. The existence of potenty neutralizing public clonotypes across multiple individuals may in part account for the remarkable efficacy of S protein-based vaccines that is being observed in the clinic. One might envision an opportunity to elicit serum neutralizing antibody titers with even higher neutralization potency using domain- or motif-based vaccine designs for this antigenic site to prime human immune responses to elicit this clonotype.
The recent emergence of variant virus lineages with increased transmissibility and altered sequences in known sites of neutralization is concerning for the capacity of SARS-CoV-2 to evade current antibody countermeasures in development and testing. Our comprehensive mapping of the effect of RBD mutations on the binding of AZD8895 and AZD1061 underscores their use as a rationally designed cocktail, given that they have different escape mutations. The data from out DMS experiments are also consistent with the binding sites determined by our antibody-RBD crystal structures, and the DMS results predict the mutations present in resistant variants selected by in vitro passaging experiments. We tested the activity of the individual antibodies or the cocktail against authentic viruses containing mutations from several important SARS-CoV-2 VOCs, and demonstrate that the individual antibodies or their combination are capable of potently neutralizing these emerging variants. Recent work from others also has demonstrated that some circulating VOCs exhibit substantial escape from neutralization of many human monoclonal antibodies in clinical development, but AZD8895 and AZD1061 still potently neutralized viruses that included the B.1.1.7, B.1.351, B.1.1.28, B.1.617.1 and B.1.617.2 VOCs and B.1.429 VOI. While other VOIs have been described contaning N439K67, S477N44, or F48668 substitutions with potential antigenic effects, our DMS experiments suggest that these mutations are not likely to have substantial effects on binding and neutralization of AZD8895, AD1061, or AZD7442. Taken together, this work defines the molecular basis for potent neutralization of SARS-CoV-2 by AZD8895 and AZD1061 and demonstrates that these antibodies efficiently neutralize emerging antigenic variants either separately or in combination, underscoring the promise of the AZD7442 investigational cocktail for use in the prevention and treatment of COVID-19.
Materials and Methods
Expression and purification of recombinant receptor binding domain (RBD) of SARS-CoV-2 spike protein
The DNA segments correspondent to the S protein RBD (residues 319 to 528) was sequence optimized for expression, synthesized, and cloned into the pTwist-CMV expression DNA plasmid downstream of the IL-2 signal peptide (MYRMQLLSCIALSLALVTNS) (Twist Bioscience). A three amino acid linker (GSG) and a His-tag were incorporated at the C-terminus of the expression constructs to facilitate protein purification. Expi293F cells were transfected transiently with the plasmid encoding RBD, and culture supernatants were harvested after 5 days. RBD was purified from the supernatants by nickel affinity chromatography with HisTrap Excel columns (GE Healthcare Life Sciences). For protein production used in crystallization trials, 5 μM kifunensine was included in the culture medium to produce RBD with high mannose glycans. The high mannose glycoproteins subsequently were treated with endoglycosidase F1 (Millipore) to obtain homogeneously deglycosylated RBD.
Expression and purification of recombinant AZD8895 and AZD1061 Fabs
The DNA fragments corresponding to the AZD8895 and AZD1061 heavy chain variable domains with human IgG1 CH1 domain and light chain variable domains with human kappa chain constant domain were synthesized and cloned into the pTwist vector (Twist Bioscience). This vector includes the heavy chain of each Fab, followed by a GGGGS linker, a furin cleavage site, a T2A ribosomal cleavage site, and the light chain of each Fab. Expression of the heavy and light chain are driven by the same CMV promoter. AZD8895 and AZD1061 Fabs were expressed in ExpiCHO cells by transient transfection with the expression plasmid. The recombinant Fab was purified from culture supernatant using an anti-CH1 CaptureSelect column (Thermo Fisher Scientific). For the RBD/AZD8895 complex, the wt sequence of AZD8895 was used for expression. For the RBD/AZD8895/AZD1061 complex, a modified version of AZD8895 Fab was used in which the first two amino acids of the variable region were mutated from QM to EV.
Crystallization and structural determination of antibody-antigen complexes
Purified AZD8895 Fab was mixed with deglycosylated RBD in a molar ratio of 1:1.5, and the mixture was purified further by size-exclusion chromatography with a Superdex-200 Increase column (GE Healthcare Life Sciences) to obtain the antibody-antigen complex. To obtain RBD/AZD8895/AZD1061 triple complex, purified and deglycosylated RBD was mixed with both AZD8895 and AZD1061 Fabs in a molar ratio of 1:1.5:1.5, and the triple complex was purified with a Superdex-200 Increase column. The complexes were concentrated to about 10 mg/mL and subjected to crystallization trials. The RBD/AZD8895 complex was crystallized in 16% - 18% PEG 3350, 0.2 Tris-HCl pH 8.0 – 8.5, and the RBD/AZD8895/AZD1061 complex was crystallized in 5% (w/v) PEG 1000, 100 mM sodium phosphate dibasic/citric acid pH 4.2, 40% (v/v) reagent alcohol. Cryo-protection solution was made by mixing crystallization solution with 100% glycerol in a volume ratio of 20:7 for crystals of both complexes. Protein crystals were flash-frozen in liquid nitrogen after a quick soaking in the cryo-protection solution. Diffraction data were collected at 100 K at the beamline 21-ID-F (wavelength: 0.97872 Å) for RBD/AZD8895 complex and 21-ID-G (wavelength: 0.97857 Å) for RBD/AZD8895/AZD1061 complex at the Advanced Photon Source. The diffraction data were processed with XDS69 and CCP4 suite70. The crystal structures were solved by molecular replacement using the structure of RBD in complex with Fab CC12.1 (PDB ID 6XC2) and Fab structure of MR78 (PDB ID 5JRP) with the program Phaser71. The structures were refined and rebuilt manually with Phenix72 or Coot73, respectively. The Ramachandran statistics for final structure of RBD/AZD8895 are: 95.82% favored, 4.18% allowed, and 0.00% disallowed, and the Ramachandran statistics for final structure of RBD/AZD8895/AZD1061: 95.34% favored, 4.37% allowed, and 0.00% disallowed. The models have been deposited into the Protein Data Bank. PyMOL software74 was used to make all of the structural figures.
AZD8895 mutant generation
Struturally-important residues in the AZD8895 heavy chain sequence were identified as D108, P99, and the disulfide bond in HCDR3. The D108 residue was mutated to alanine, asparagine, and glutamic acid. The P99 residue was mutanted to valine, asparagine, and glycine. The disulfide bond was removed by introducing C101A/C106A mutations. Additionally, the germline revertant (GRev) forms of AZD8895 were generated by aligning the sequence to identified germline sequences using IgBlast, and reverting back the residues that were not germline-encoded. DNA fragments corresponding to the AZD8895 mutant heavy chain variable domains with human IgG1 and light chain variable domain with human kappa chain constant domain were synthesized and cloned into the pTwist_mCis vector (Twist Bioscience) as previously described21. Constructs were transformed into E. coli, and DNA was purified. Antibodies then were produced by transient tranfection of ExpiCHO cells following the manufacturer’s protocol (Gibco). Supernatants were filter-sterilized using 0.45 µm pore size filters and samples were applied to HiTrap MabSelect Sure columns (Cytiva).
ELISA binding of AZD8895 mutants
Wells of 384-well microtiter plates were coated with purified recombinant SARS-CoV-2 S 6P protein at 4°C overnight. Plates were blocked with 2% non-fat dry milk and 2% normal goat serum in DPBS containing 0.05% Tween-20 (DPBS-T) for 1 h. Antibodies were diluted to 10 µg/mL and titrated two-fold 23 times in DPBS-T and added to the wells, followed by an incubation for 1 h at room temperature. After washing with DPBS-T, a goat anti-human IgG secondary antibody conjugated with horseradish peroxidase (Southern Biotech, Cat# 2040–05, 1:5,000 dilution) was added and incubated for one hour. Plates were then washed with DPBS-T and TMB substrate was added to each well (Thermo Fischer Scientific). Reactions were quenched with 1 M hydrochloric acid and absorbance was measured at 450 nm using a spectrophotometer (Biotek).
Mapping of all mutations that escape antibody binding
All mutations that escape antibody binding were mapped via a DMS approach29. We used previously described yeast-display RBD mutant libraries29,30. Briefly, duplicate mutant libraries were constructed in the spike receptor binding domain (RBD) from SARS-CoV-2 (isolate Wuhan-Hu-1, Genbank accession number MN908947, residues N331-T531) and contain 3,804 of the 3,819 possible amino-acid mutations, with >95% present as single mutants. Each RBD variant was linked to a unique 16-nucleotide barcode sequence to facilitate downstream sequencing. As previously described, libraries were sorted for RBD expression and ACE2 binding to eliminate RBD variants that are completely misfolded or non-functional (i.e., lacking modest ACE2 binding affinity29).
Antibody escape mapping experiments were performed in biological duplicate using two independent mutant RBD libraries, as previously described29, with minor modifications. Briefly, mutant yeast libraries induced to express RBD were washed and incubated with antibody at 400 ng/mL for 1 h at room temperature with gentle agitation. After the antibody incubations, the libraries were secondarily labeled with 1:100 FITC-conjugated anti-MYC antibody (Immunology Consultants Lab, CYMC-45F) to label for RBD expression and 1:200 PE-conjugated goat anti-human-IgG (Jackson ImmunoResearch 109–115-098) to label for bound antibody. Flow cytometric sorting was used to enrich for cells expressing RBD variants with reduced antibody binding via a selection gate drawn to capture unmutated SARS-CoV-2 cells labeled at 1% the antibody concentration of the library samples. For each sample, approximately 10 million RBD+ cells were processed on the cytometer. Antibody-escaped cells were grown overnight in SD-CAA (6.7 g/L Yeast Nitrogen Base, 5.0 g/L Casamino acids, 1.065 g/L MES acid, and 2% w/v dextrose) to expand cells prior to plasmid extraction.
Plasmid samples were prepared from pre-selection and overnight cultures of antibody-escaped cells (Zymoprep Yeast Plasmid Miniprep II) as previously described29. The 16-nucleotide barcode sequences identifying each RBD variant were amplified by PCR and sequenced on an Illumina HiSeq 2500 with 50 bp single-end reads as described29,30.
Escape fractions were computed as described29, with minor modifications as noted below. We used the dms_variants package (https://jbloomlab.github.io/dms_variants/, version 0.8.2) to process Illumina sequences into counts of each barcoded RBD variant in each pre-sort and antibody-escape population using the barcode/RBD look-up table previously described75.
For each antibody selection, we computed the “escape fraction” for each barcoded variant using the deep sequencing counts for each variant in the original and antibody-escape populations and the total fraction of the library that escaped antibody binding via a previously described formula29. These escape fractions represent the estimated fraction of cells expressing that specific variant that fall in the antibody escape bin, such that a value of 0 means the variant is always bound by serum and a value of 1 means that it always escapes antibody binding. We then applied a computational filter to remove variants with low sequencing counts or highly deleterious mutations that might cause antibody escape simply by leading to poor expression of properly folded RBD on the yeast cell surface29,30. Specifically, we removed variants that had (or contained mutations with) ACE2 binding scores < −2.35 or expression scores < −1, using the variant- and mutation-level deep mutational scanning scores as previously described30. Note that these filtering criteria are slightly more stringent than those previously used to map a panel of human antibodies29 but are identical to those used in recent studies defining RBD residues that impact the binding of mAbs75 and polyclonal serum45.
We next deconvolved variant-level escape scores into escape fraction estimates for single mutations using global epistasis models76 implemented in the dms_variants package, as detailed at (https://jbloomlab.github.io/dms_variants/dms_variants.globalepistasis.html) and described29. The reported escape fractions throughout the paper are the average across the libraries (correlations shown in Extended Data Fig. 7a,b); these scores are also in Supplementary Data Table 2. Sites of strong escape from each antibody for highlighting in logo plots were determined heuristically as sites whose summed mutational escape scores were at least 10 times the median sitewise sum of selection, and within 10-fold of the sitewise sum of the most strongly selected site. Full documentation of the computational analysis is at https://github.com/jbloomlab/SARS-CoV-2-RBD_MAP_AZ_Abs. These results are also available in an interactive form at https://jbloomlab.github.io/SARS-CoV-2-RBD_MAP_AZ_Abs/.
For plotting and analyses that required identifying RBD sites of “strong escape” (e.g., choosing which sites to show in logo plots in Fig 4a,b), we considered a site to mediate strong escape if the total escape (sum of mutation-level escape fractions) for that site exceeded the median across sites by > 5-fold, and was at least 5% of the maximum for any site. A markdown rendering of the identification of these sites of strong escape is at https://github.com/jbloomlab/SARS-CoV-2-RBD_MAP_AZ_Abs/blob/main/results/summary/call_strong_escape_sites.md.
Antibody escape selection experiments with VSV-SARS-CoV-2.
For escape selection experiments with AZD8895 and AZD1061, we used a replication competent recombinant VSV virus encoding the spike protein from SARS-CoV-2 with a 21 amino-acid C-terminal deletion31. The spike-expressing VSV virus was propagated in MA104 cells (African green monkey, ATCC CRL-2378.1) as described previously31, and viral stocks were titrated on Vero E6 cell monolayer cultures. Plaques were visualized using neutral red staining. To screen for escape mutations selected in the presence of AZD8895, AZD1061, or AZD7442, we used a real-time cell analysis assay (RTCA) and xCELLigence RTCA MP Analyzer (ACEA Biosciences Inc.) and a previously described escape selection scheme29. Briefly, 50 μL of cell culture medium (DMEM supplemented with 2% FBS) was added to each well of a 96-well E-plate to obtain a background reading. Eighteen thousand (18,000) Vero E6 cells in 50 μL of cell culture medium were seeded per well, and plates were placed on the analyzer. Measurements were taken automatically every 15 min and the sensograms were visualized using RTCA software version 2.1.0 (ACEA Biosciences Inc). VSV-SARS-CoV-2 virus (5,000 plaque forming units [PFU] per well, ∼0.3 MOI) was mixed with a saturating neutralizing concentration of AZD8895, AZD1061, or AZD7442 antibody (5 μg/mL total concentration of antibodies) in a total volume of 100 μL and incubated for 1 h at 37°C. At 16 to 20 h after seeding the cells, the virus-antibody mixtures were added to cell monolayers. Wells containing only virus in the absence of antibody and wells containing only Vero E6 cells in medium were included on each plate as controls. Plates were measured continuously (every 15 min) for 72 h. Escape mutations were identified by monitoring the cell index for a drop in cellular viability. To verify escape from antibody selection, wells where cytopathic effect was observed in the presence of AZD1061 were assessed in a subsequent RTCA experiment in the presence of 10 μg/mL of AZD1061 or AZD8895. After confirmation of resistance of selected viruses to neutralization by AZD1061, viral isolates were expanded on Vero E6 cells in the presence of 10 μg/mL of AZD1061. Viral RNA was isolated using a QiAmp Viral RNA extraction kit (QIAGEN) according to manufacturer protocol, and the SARS-CoV-2 spike gene was reverse-transcribed and amplified with a SuperScript IV One-Step RT-PCR kit (ThermoFisher Scientific) using primers flanking the S gene (forward primer: 5′-AGCTTCTGAACAATCCCCGG-3′, reverse primer: 5′-GAGGCCTCTTTGAGCATGGT-3′). The amplified PCR product was purified using SPRI magnetic beads (Beckman Coulter) at a 1:1 ratio and sequenced by the Sanger method, using primers giving forward and reverse reads of the RBD.
Serial passaging and testing of SARS-CoV-2 to select for mAb resistant mutations.
SARS-CoV-2 strain USA-WA1/2020 was passaged serially in Vero cell monolayer cultures with AZD8895, AZD1061 or AZD7442, at concentrations beginning at their respective IC50 values and increased step-wise to their IC90 value with each passage. As a control, virus was passaged in the absence of antibody. Following the final passage, viruses were evaluated for susceptibility to neutralization by AZD8895, AZD1061 or AZD7442, at a final concentration of 10× the IC90 concentration by plaque assay. Plaques (n=6) were selected randomly for AZD1061 cultures, and their virus spike-encoding gene was sequenced.
The escape phenotype of selected plaque progeny viruses was validated by testing each for the 6 plaque purified progency viruses for neutralization in a PRNT in comparison with the parent virus, SARS-CoV-2 (strain WA-1). All plaque-purified viruses resulted from the same monoclonal antibody passage.
The escape phenotype of the 6 selected plaque progeny viruses also was validated by demonstration of a loss-of-binding phenotype in biochemical tests with variant proteins incorporating selected mutations. Spike trimer variants were cloned into a HexaPro ectodomain soluble trimer construct77 with the D614G mutation and then expressed in FreeStyle™ 293-F Cells (Thermo Fisher Scientific). Expression supernatants were harvested five days post-transfection and buffer-exchanged into 1× PBS and then diluted 10-fold into assay buffer (1× HBS-EP+ buffer [Cytiva]). R2 values indicating goodness of the fit for AZD8895 or AZD1061 kinetic binding values to the spike trimer variants were calculated from binding traces. Relative fold-shifts in KD were calculated in comparison to binding to wild-type S protein.
Generation of authentic SARS-CoV-2 viruses, including viruses with variant residues.
The 2019n-CoV/USA_WA1/2020 isolate of SARS-CoV-2 was obtained from the US Centers for Disease Control (CDC) and passaged on Vero E6 cells. The B.1.429 isolate was generously provided by Raul Andino and Charles Chiu (University of California, San Francisco) and passaged once on Vero-TMPRRS2 cells. Benjamin Pinsky (Stanford) and Mehul Suthar (Emory) provided the B.1.617.1 variant, and Richard Webby (St. Jude Children’s Research Hospital) provided the B.1.617.2 variant. Individual point mutations in the spike gene (D614G and E484K/D614G) were introduced into an infectious cDNA clone of the 2019n-CoV/USA_WA1/2020 strain as described previously78. Chimeric viruses expressing the spoike genes of B.1.351 or B.1.1.28 were generated in the WA1/2020 backbone and have been described previously42. Nucleotide or spike gene substitutions were introduced into an existing WA1/2020 infectious cDNA clone and assembled by in vitro ligation of seven contiguous cDNA fragments following the previously described protocol79. In vitro transcription then was performed to synthesize full-length genomic RNA. To recover the mutant or chimeric viruses, the RNA transcripts were electroporated into Vero E6 cells. The viruses from the supernatant of cells were collected 40 h later and served as p0 stocks. All virus stocks were confirmed by next generation sequencing.
Focus reduction neutralization test.
Serial dilutions of mAbs or serum were incubated with 102 focus-forming units (FFU) of different strains or variants of SARS-CoV-2 for 1 h at 37°C. Antibody-virus complexes were added to Vero-hACE2-TMPRSS2 cell monolayer cultures in 96-well plates and incubated at 37°C for 1 h. Subsequently, cells were overlaid with 1% (w/v) methylcellulose in MEM supplemented with 2% FBS. Plates were harvested 20 h later by removing overlays and fixed with 4% PFA in PBS for 20 min at room temperature. Plates were washed and sequentially incubated with an oligoclonal pool of anti-S murine mAbs44 and HRP-conjugated goat anti-human IgG in PBS supplemented with 0.1% saponin and 0.1% bovine serum albumin. SARS-CoV-2-infected cell foci were visualized using TrueBlue peroxidase substrate (KPL) and quantitated on an ImmunoSpot microanalyzer (Cellular Technologies).
Multiple sequence alignments
We searched for antibody variable gene sequences originating with the same features as those encoding AZD8895 and retrieved the matching sequences from the repertoires of each individual examined. We searched for similar sequences in the publicly available large-scale antibody sequence repertoires for three healthy individuals and cord blood repertoires (deposited at SRP174305). The search parameters for the heavy chain were sequences with IGHV1–58 and IGHJ3 with the P99, D108, and F110 residues. Additionally, the search parameters for the light chain were sequences with Y92 and W98 residues. Sequences from a matching clonotype that belonged to each individual were aligned with either ClustalO80 (heavy chains) or with MUSCLE81 (light chains). Then, LOGOs plots of aligned sequences were generated using WebLogo82.
Extended Data
Extended Data Fig. 1.
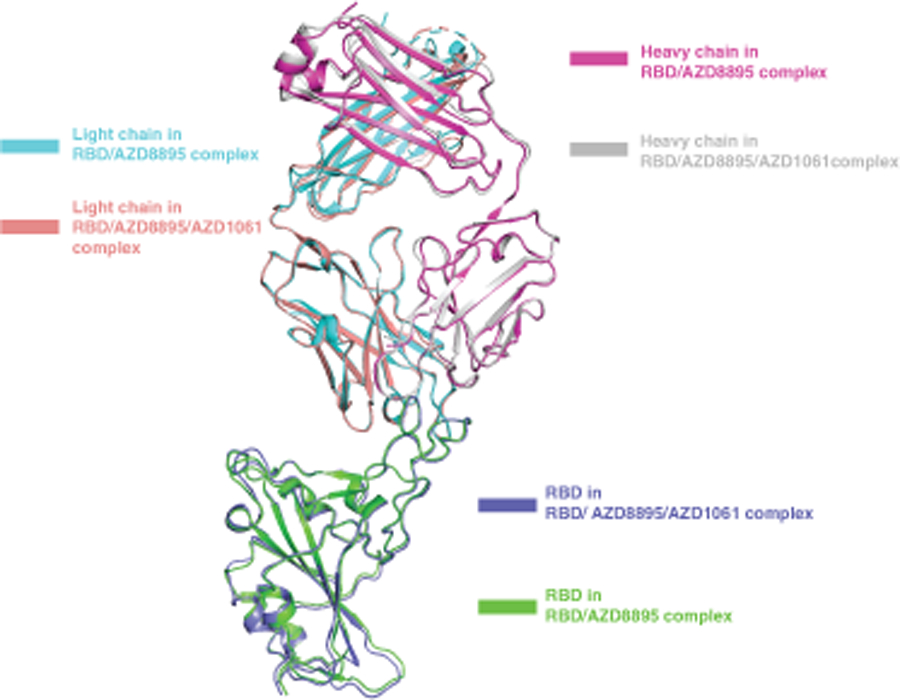
Overlay of substructure of RBD/AZD8895 in RBD/AZD8895/AZD1061 complex and RBD/AZD8895 crystal structure.
Extended Data Fig. 2. Similarities in structural details of interactions between RBD and AZD8895, AZD1061 and those seen in the spike/S2E12 complex.
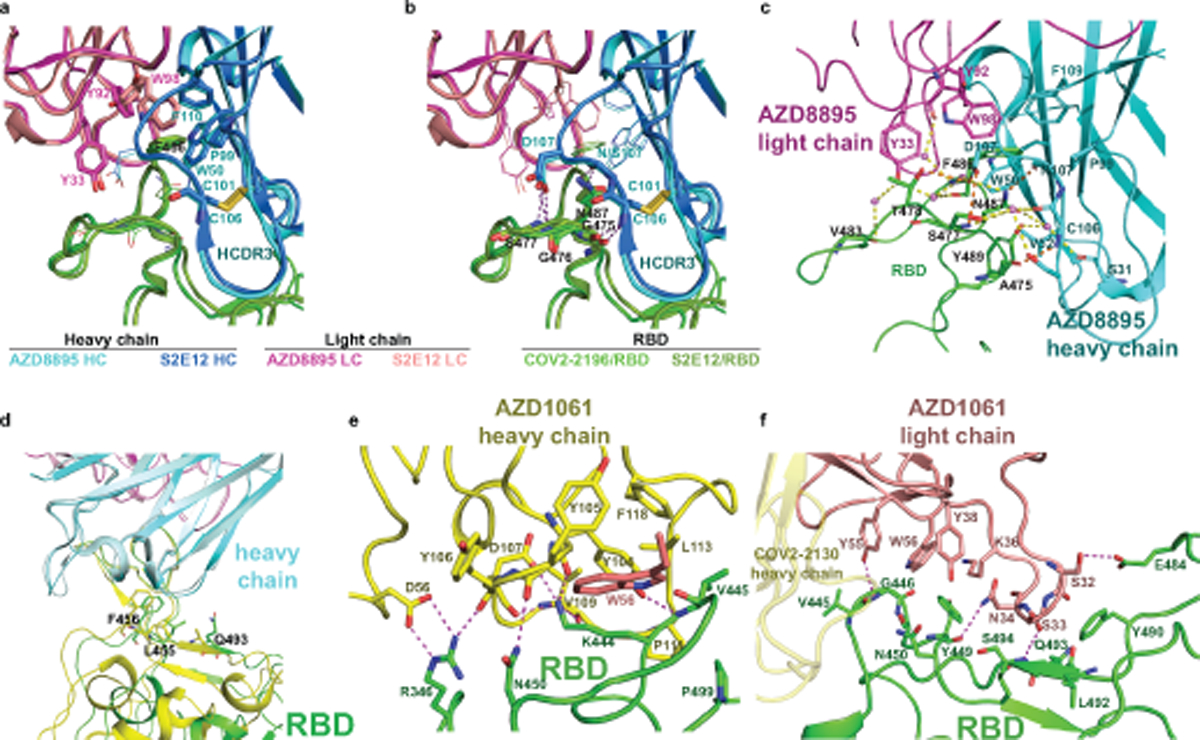
a. Similar aromatic stacking and hydrophobic interaction patterns at the RBD site F486 shared between RBD/AZD8895 and spike/S2E12 complexes.
b. Same hydrogen bonding pattern surrounding residue F486 in the structures of the RBD/AZD8895 and spike/S2E12 complexes .
c. Detailed interactions between AZD8895 and RBD. AZD8895 heavy chain is colored in cyan, the light chain is colored in magenta, and RBD is colored in green. Important interacting residues are shown in stick representation. Water molecules involved in antibody-RBD interaction are represented as pink spheres. Direct hydrogen bonds are shown as orange dashed lines, and water-mediated hydrogen bonds as yellow dashed lines.
d. Superimposition of RBD/S2E12 cryo-EM structure onto the RBD/AZD8895 crystal structure, with the variable domains of antibodies as references. AZD8895 heavy chain is in cyan, and its light chain in magenta; S2E12 heavy chain is in pale cyan, and its light chain in light pink. The two corresponding RBD structures are colored in green or yellow, respectively.
e. Detailed interactions between AZD1061 heavy chain and RBD. Paratope residues are shown in stick representation and colored in yellow, epitope residues in green sticks. Hydrogen-bonds or strong polar interactions are represented as dashed magenta lines.
f. Detailed interactions between AZD1061 light chain and RBD. Paratope residues are shown in stick representation and colored in orange, epitope residues in green sticks. Hydrogen-bonds are represented as dashed magenta lines.
Extended Data Fig. 3. A common clonotype of anti-RBD antibodies with the same binding mechanism.
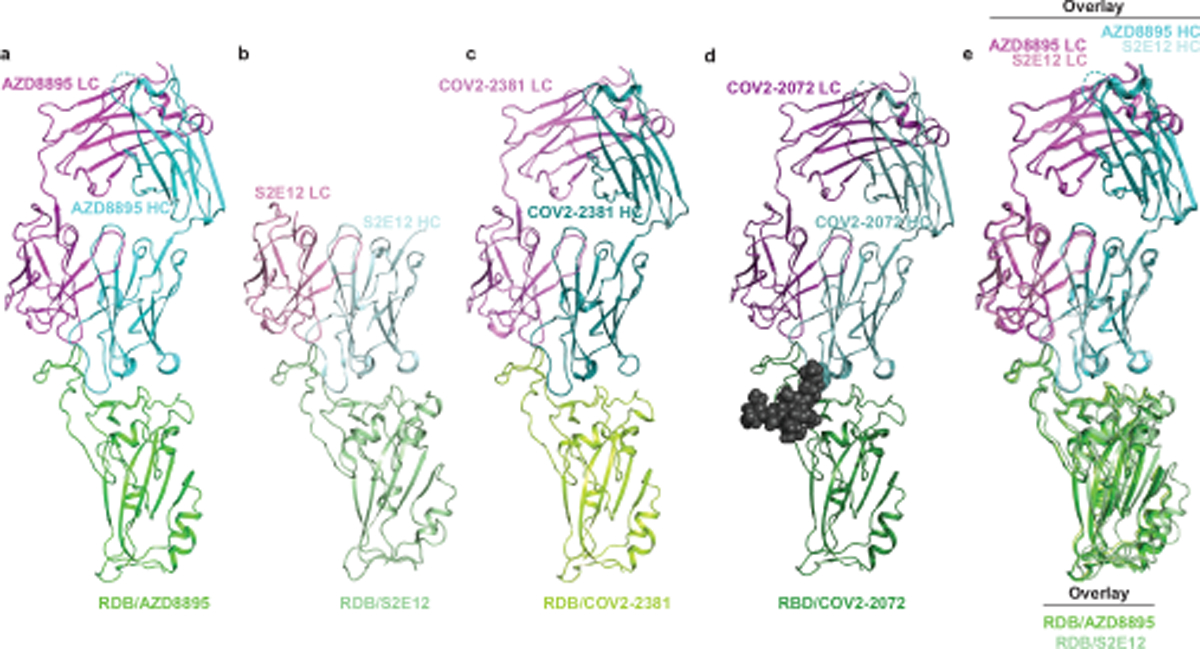
a. RBD/AZD8895 crystal structure.
b. RBD/S2E12 cryo-EM structure.
c. RBD/COV2–2381 homology model generated with Structuropedia25. COV2–2072 encodes an N-linked glycosylation sequon in the HCDR3, indicated by the gray spheres, which was modeled using the GlyProt webserver.
d. RBD/COV2–2072 homology model generated with Structuropedia25.
e. Overlay of the RBD/AZD8895 crystal structure (a) and RBD/S2E12 cryo-EM structure (b).
Extended Data Fig. 4. Identification of putative public clonotype members genetically similar to AZD8895 in the antibody variable gene repertoires of virus-naïve individuals.

Antibody variable gene sequences collected from healthy individuals (HIP1, 2, or 3) prior to the pandemic with the same sequence features as AZD8895 heavy chain and light chain are aligned.
a. WebLogo plots of heavy chain (top) and light chain (bottom) sequences from three different adult donors and cord blood samples with the features of the public clonotype. The sequence features and contact residues used in AZD8895 are highlighted in red boxes below each multiple sequence alignment.
b. Since the light chain plots in (a) showed restricted diversity, here we show amino acid alignments for the top five representative light chains that occurred most frequently in the three adult donors studied (HIP1, 2, or 3).
Extended Data Fig. 5. Details of AZD1061 interaction with SARS-CoV-2 S protein RBD.
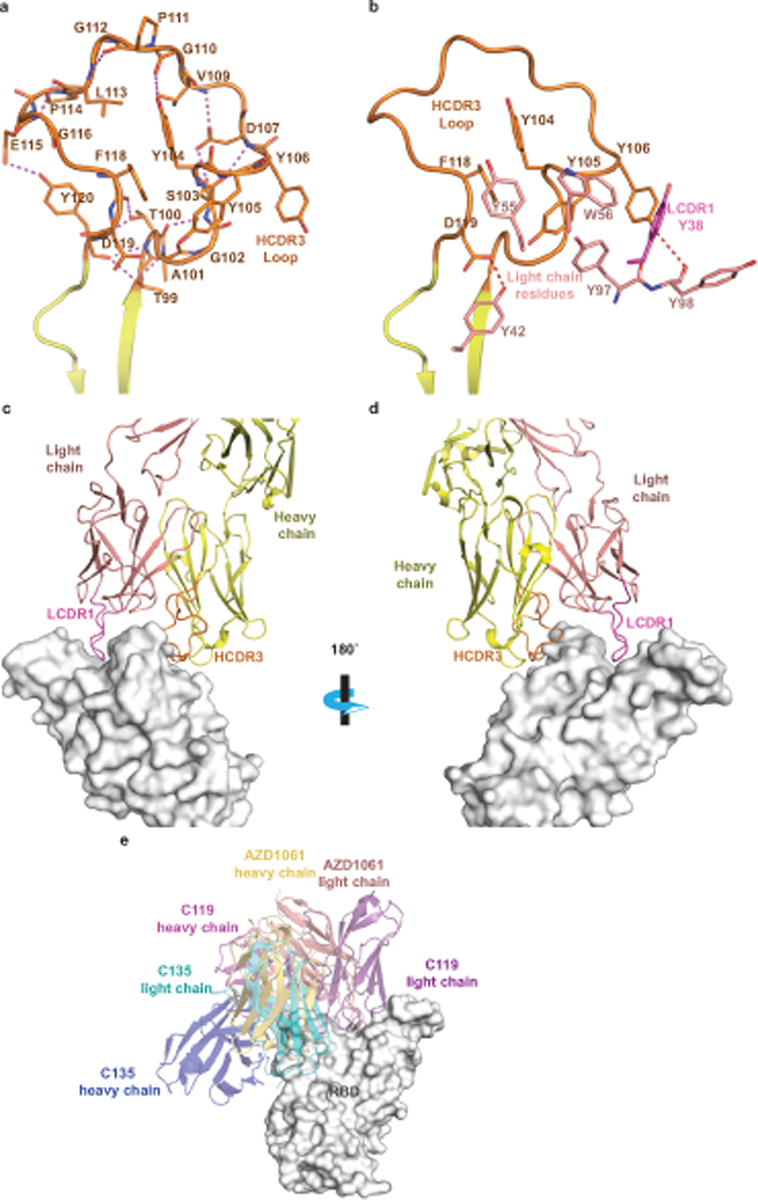
a. Detailed AZD1061 HCDR3 loop structure. Short-range hydrogen bonds, stabilizing the loop conformation, are shown as dashed magenta lines.
b.Residues of AZD1061 light chain form aromatic stacking interactions and hydrogen bonds with HCDR3 to further stabilize the HCDR3 loop. LCDR1 residue Y38 is colored in magenta to match the LCDR1 coloring in panels (c) and (d).
c. Long LCDR1, HCDR2, and HCDR3 form complementary binding surface to the RBD epitope. RBD is shown as surface representation in grey. AZD1061 heavy chain is colored in yellow with HCDR3 in orange, and the light chain in pink with LCDR1 in magenta.
d. 180° rotation view of panel c.
e. Comparison of AZD1061 binding with the previously published mAbs C119 and C135.
Extended Data Fig. 6. Interface between AZD8895 and AZD1061 in the RBD/AZD8895/AZD1061 crystal structure.
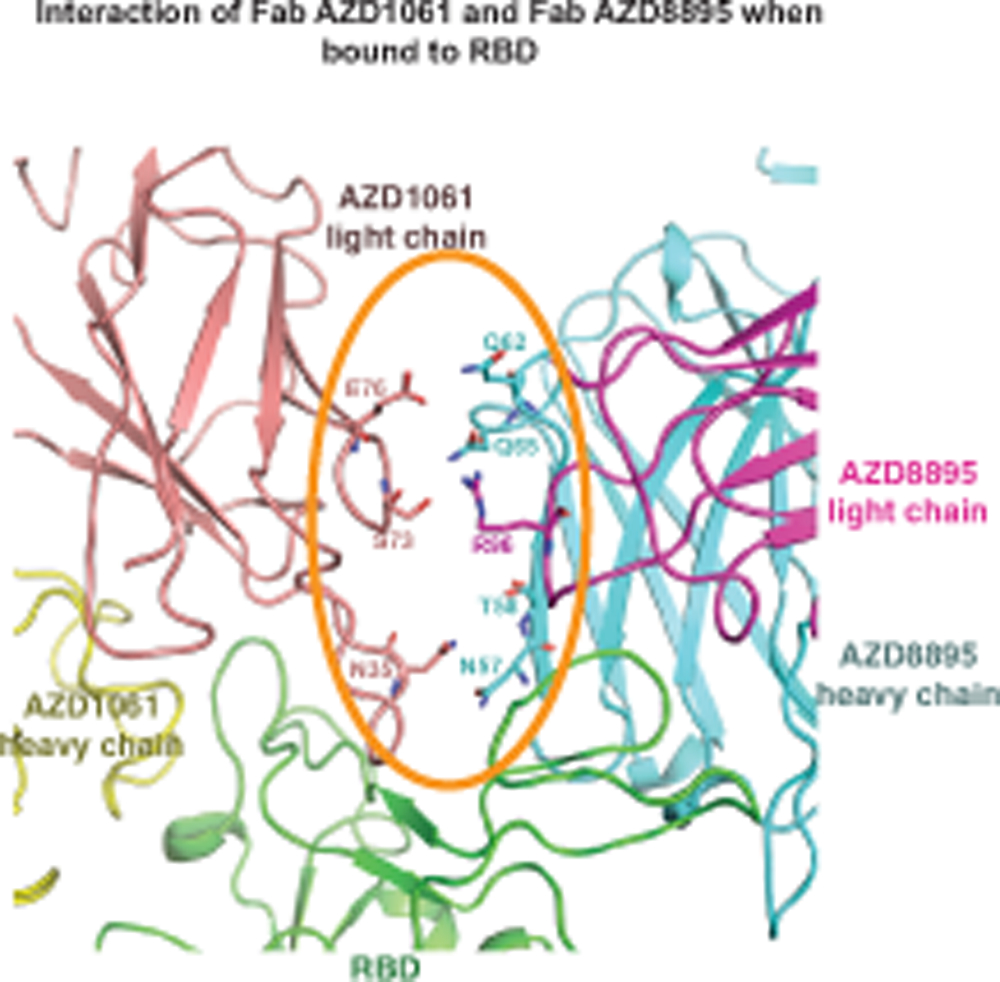
AZD8895 heavy or light chain are shown as cartoon representation in cyan or magenta, respectively, and AZD1061 heavy or light chains in yellow or pink, respectively. The RBD is colored in green. Interface residues are shown in stick representation.
Extended Data Fig. 7. Identification by deep mutational scanning of mutations affecting antibody binding and method of selection of antibody resistant mutants with VSV-SARS-CoV-2 virus.
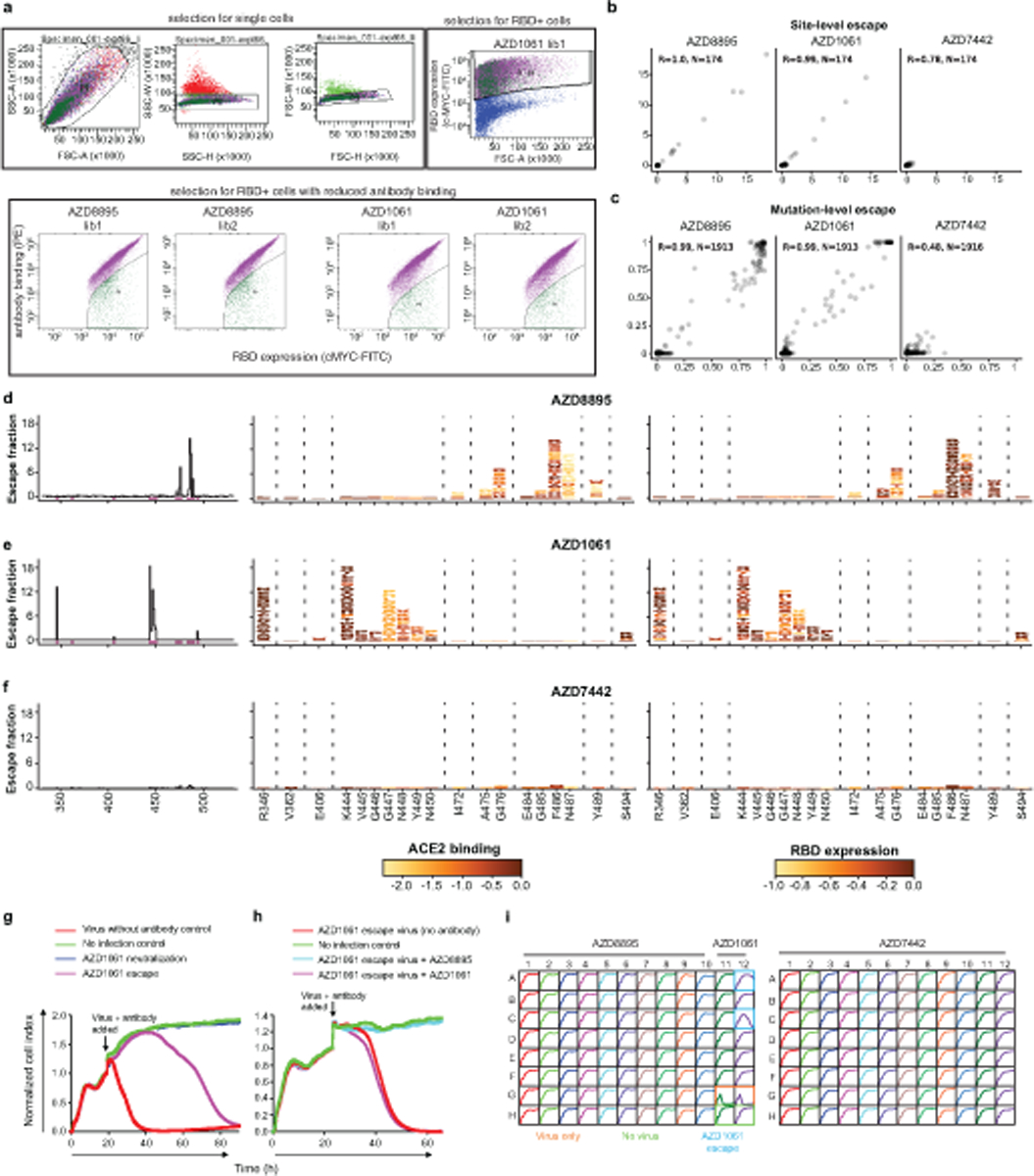
a. Top: Plots showing gating strategy for selection of single yeast cells using forward- and side-scatter (first three panels) or RBD expression (right panel). Each plot is derived from the preceding gate. Bottom: Plots showing gating for RBD+, antibody− yeast cells. Selection experiments are shown for AZD8895 or AZD1061 - two independent libraries each.
b. Correlation of sites of escape between yeast library selection experiments. The x-axes show cumulative escape fraction for each site for library 1, and the y-axes show cumulative escape fraction for each site for library 2. Correlation coefficient and n are denoted for each graph.
c. Correlation of observed mutations that escape antibody binding between yeast library selection experiments. The x-axes show each amino acid mutation’s escape fraction for library 1, and the y-axes show each amino acid mutation’s escape fraction for library 2. Correlation coefficient and n are denoted for each graph.
d–f. DMS results for AZD8895 (d), AZD1061 (e), or AZD7442 (f). Left panels: sites of escape across the entire RBD indicated by peaks that correspond to the logo plots in the middle/right panels. Middle panel: logo plot of cumulative escape mutation fractions of all RBD sites with strong escape mutations. Mutations are colored based on degree to which they abrogate RBD binding to hACE2. Right: logo plots show cumulative escape fractions, but colored based on degree to which mutations affect RBD expression.
g. RTCA sensograms showing neutralization escape. Cytopathic effect was monitored kinetically in cells inoculated with virus in the presence of 5 μg/mL AZD1061. Representative escape (magenta) or lack of escape (blue) are shown. Green - uninfected cells; red -cells inoculated with virus without antibody. Magenta/blue curves - a single representative well; red/green controls - mean of technical duplicates.
h. Representative RTCA sensograms validating that a virus selected by AZD1061 in (g) escaped AZD1061 (magenta) but not AZD8895 (light blue).
i. Example sensograms from wells of 96-well E-plate analysis for escape selection. Instances of escape from AZD1061 are noted, while escape was not detected in the presence of AZD8895 or AZD7442. Positive and negative controls are denoted on the first plate.
Extended Data Fig. 8. Antibody resistant mutants selected with VSV-SARS-CoV2 or authentic SARS-CoV-2 virus.
a. The method for assessing monoclonal antibody resistant spike protein variants is shown. SARS-CoV-2 was passaged serially in the presence of monoclonal antibodies at the increasing concentrations indicated in the figure or without antibody (no monoclonal antibody). Following passage at IC90 concentrations, samples were treated with 10× IC90 concentrations of monoclonal antibodies and any resultant resistant virus collected, and the genome was sequenced. Red viruses in the schematic represent selection of escape variants.
b. The escape phenotype of 6 independent plaques selected with AZD1061 was validated by demonstration of escape by testing in a PRNT. Antibody neutralization as measured by PRNT against the 6 plaque-purified, AZD1061-resistant SARS-CoV-2 viruses (blue) was compared to the parent virus WA-1 (orange) during treatment with AZD1061 or AZD7442. All plaque-purified viruses resulted from the same monoclonal antibody passage as detailed in (a). Data shown are from a single technical replicate for each of the six selected escape mutants.
c,d. The escape phenotype of independent plaques selected with AZD1061 also was validated by demonstration of loss of binding to proteins incorporating variant residues in the selected plaques using biolayer interferometry (BLI). Data shown are from a single experiment.
c. Binding traces of AZD8895 and AZD1061 to various spike trimers with kinetics curve fits. An inability to fit AZD1061 binding to the K444E S variant is due to a lack of detectable binding even at 5 µM for AZD1061.
d. Summary of AZD8895 and AZD1061 kinetic binding values to the S trimer variants from binding traces with R2 indicating goodness of the fit. Relative fold-change in KD is shown in comparison to wild-type. No detectable binding is indicated as NB.
Supplementary Material
Per-mutation escape fractions for deep mutational scanning experiments
Viral neutralization and binding kinetic data
ELISA binding source data
Acknowledgments:
At Fred Hutchinson Cancer Research Center, we thank Amin Addetia for experimental assistance, the Flow Cytometry and Genomics core facilities, and Scientific Computing, supported by ORIP grant S10OD028685. We thank Adrian Creanga and Barney Graham of the U.S. National Institutes of Health for the Vero-hACE2-TMPRSS2 cells. At AstraZeneca, we thank Paul Warrener, Christopher Morehouse and Dave Tabor for virus genome sequencing and spike variant analysis, and Kuishu Ren for generation of protein reagents and related binding data. We thank Benjamin Pinsky (Stanford) and Mehul Suthar (Emory) for providing the B.1.617.1 variant, and Richard Webby (St. Jude Children’s Research Hospital) for the B.1.617.2 variant. Some schematics were created with Biorender.com.
Funding:
This work was supported by Defense Advanced Research Projects Agency (DARPA) grants HR0011–18-2–0001 and HR0011–18-3–0001; U.S. N.I.H. contracts 75N93019C00074 and 75N93019C00062; N.I.H. grants AI150739, AI157155, AI141707, AI083203, AI095202, and UL1TR001439, the Dolly Parton COVID-19 Research Fund at Vanderbilt, a grant from Fast Grants, Mercatus Center, George Mason University, and funding from AstraZeneca. T.N.S. is a Washington Research Foundation Innovation Fellow at the University of Washington Institute for Protein Design and a Howard Hughes Medical Institute Fellow of the Damon Runyon Cancer Research Foundation (DRG-2381–19). J.E.C. is a recipient of the 2019 Future Insight Prize from Merck KGaA, which supported this work with a grant. J.D.B. is an Investigator of the Howard Hughes Medical Institute. P.-Y.S. was supported by awards from the Sealy & Smith Foundation, Kleberg Foundation, the John S. Dunn Foundation, the Amon G. Carter Foundation, the Gilson Longenbaugh Foundation, and the Summerfield Robert Foundation. J.B.C. is supported by a Helen Hay Whitney Foundation postdoctoral fellowship. X-ray diffraction data were collected at Beamline 21-ID-F and 21-ID-G at the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the Office of Science by Argonne National Laboratory under contract no. DE-AC02–06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri- Corridor (grant 085P1000817). Support for crystallography was provided from the Vanderbilt Center for Structural Biology. The content is solely the responsibility of the authors and does not necessarily represent the official views of the U.S. government or the other sponsors.
Footnotes
Materials availability: Requests for reagents may be directed to and be fulfilled by the Lead Contact: Dr. James E. Crowe, Jr. (james.crowe@vumc.org). Materials reported in this study will be made available but may require execution of a Materials Transfer Agreement.
Code availability: The computational pipeline for the deep mutational scanning analysis of antibody escape mutations is available on GitHub: (https://github.com/jbloomlab/SARS-CoV-2-RBD_MAP_AZ_Abs).
Competing interests: T.B., K.T., H.B., Y.M-L., P.M., and M.E. are employees of and may own stock in AstraZeneca. M.S.D. is a consultant for Inbios, Vir Biotechnology, Fortress Biotech, and Carnival Corporation and on the Scientific Advisory Boards of Moderna and Immunome. The Diamond laboratory has received funding support in sponsored research agreements from Moderna, Vir Biotechnology, and Emergent BioSolutions. J.E.C. has served as a consultant for Luna Biologics, is a member of the Scientific Advisory Boards of CompuVax and Meissa Vaccines and is Founder of IDBiologics. The Crowe laboratory at Vanderbilt University Medical Center has received sponsored research agreements from Takeda Vaccines, IDBiologics, and AstraZeneca. Vanderbilt University has applied for patents concerning antibodies that are related to this work; Vanderbilt University has licensed certain rights to antibodies described in this paper to Astra Zeneca. All other authors declare no competing interests.
Additional information
Supplementary information is available for this paper.
Data availability:
The crystal structures reported in this paper have been deposited to the Protein Data Bank (https://www.rcsb.org) under the accession numbers 7L7D (COV2–2196 + RBD) and 7L7E COV2–2196 and COV2–2130 + RBD). The following were obtained from the PDB and used for visualization or molecular replacement: PDB IDs: 7K4N, 6M0J, 6XM4, 7CAK, 6ZOY, 6XC2, 5JRP. The FASTQ files for deep mutational scanning are available on the NCBI Sequence Read Archive under BioSample SAMN17532001 as part of BioProject PRJNA639956. Per-mutation escape fractions are available on GitHub (https://github.com/jbloomlab/SARS-CoV-2-RBD_MAP_AZ_Abs/blob/main/results/supp_data/AZ_cocktail_raw_data.csv) and in Supplementary Data Table 2. All other data are available in the main text or the supplementary materials.
References
- 1.Hoffmann M et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–280 e278, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Letko M, Marzi A & Munster V Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol 5, 562–569, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wahba L et al. An extensive meta-metagenomic search identifies SARS-CoV-2-homologous sequences in pangolin lung viromes. mSphere 5, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walls AC et al. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc Natl Acad Sci U S A 114, 11157–11162, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Algaissi A et al. SARS-CoV-2 S1 and N-based serological assays reveal rapid seroconversion and induction of specific antibody response in COVID-19 patients. Sci Rep 10, 16561, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Long QX et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med 26, 845–848, (2020). [DOI] [PubMed] [Google Scholar]
- 7.Piccoli L et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell, (2020). [DOI] [PMC free article] [PubMed]
- 8.Brouwer PJM et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science 369, 643–650, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao Y et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell 182, 73–84 e16, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansen J et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science 369, 1010–1014, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ju B et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 584, 115–119, (2020). [DOI] [PubMed] [Google Scholar]
- 12.Liu L et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 584, 450–456, (2020). [DOI] [PubMed] [Google Scholar]
- 13.Robbiani DF et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 584, 437–442, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rogers TF et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science 369, 956–963, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi R et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 584, 120–124, (2020). [DOI] [PubMed] [Google Scholar]
- 16.Weitkamp JH et al. Infant and adult human B cell responses to rotavirus share common immunodominant variable gene repertoires. J Immunol 171, 4680–4688, (2003). [DOI] [PubMed] [Google Scholar]
- 17.Zost SJ et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature 584, 443–449, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benton DJ et al. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature, (2020). [DOI] [PMC free article] [PubMed]
- 19.Wrapp D et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wrobel AG et al. SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects. Nat Struct Mol Biol 27, 763–767, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zost SJ et al. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nat Med 26, 1422–1427, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greaney AJ et al. Mapping mutations to the SARS-CoV-2 RBD that escape binding by different classes of antibodies. Nat Commun 12, 4196, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tortorici MA et al. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science, (2020). [DOI] [PMC free article] [PubMed]
- 24.Kreer C et al. Longitudinal isolation of potent near-Germline SARS-CoV-2-neutralizing antibodies from COVID-19 patients. Cell 182, 843–854 e812, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farooq A http://www.farooqed.com/mod/, <http://www.farooqed.com/mod/> (2021).
- 26.Bohne-Lang A & von der Lieth CW GlyProt: in silico glycosylation of proteins. Nucleic Acids Res 33, W214–219, (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dejnirattisai W et al. The antigenic anatomy of SARS-CoV-2 receptor binding domain. Cell 184, 2183–2200 e2122, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soto C et al. High frequency of shared clonotypes in human B cell receptor repertoires. Nature 566, 398–402, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greaney AJ et al. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe, (2020). [DOI] [PMC free article] [PubMed]
- 30.Starr TN et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell 182, 1295–1310 e1220, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Case JB et al. Neutralizing antibody and soluble ACE2 inhibition of a replication-competent VSV-SARS-CoV-2 and a clinical isolate of SARS-CoV-2. Cell Host Microbe 28, 475–485 e475, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klimstra WB et al. SARS-CoV-2 growth, furin-cleavage-site adaptation and neutralization using serum from acutely infected hospitalized COVID-19 patients. J Gen Virol 101, 1156–1169, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sawatzki K et al. Host barriers to SARS-CoV-2 demonstrated by ferrets in a high-exposure domestic setting. Proc Natl Acad Sci U S A 118, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baum A et al. REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters. Science 370, 1110–1115, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Q et al. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell 182, 1284–1294 e1289, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galloway SE et al. Emergence of SARS-CoV-2 B.1.1.7 Lineage - United States, December 29, 2020-January 12, 2021. MMWR Morb Mortal Wkly Rep 70, 95–99, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leung K, Shum MH, Leung GM, Lam TT & Wu JT Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Euro Surveill 26, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tegally H et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature 592, 438–443, (2021). [DOI] [PubMed] [Google Scholar]
- 39.Voloch CM et al. Genomic characterization of a novel SARS-CoV-2 lineage from Rio de Janeiro, Brazil. J Virol, (2021). [DOI] [PMC free article] [PubMed]
- 40.Liu J et al. BNT162b2-elicited neutralization of B.1.617 and other SARS-CoV-2 variants. Nature, (2021). [DOI] [PubMed]
- 41.Deng X et al. Transmission, infectivity, and neutralization of a spike L452R SARS-CoV-2 variant. Cell, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen RE et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat Med 27, 717–726, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weisblum Y et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife 9, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Z et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe 29, 477–488 e474, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Greaney AJ et al. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe 29, 463–476 e466, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wibmer CK et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat Med 27, 622–625, (2021). [DOI] [PubMed] [Google Scholar]
- 47.Andreano E et al. SARS-CoV-2 escape in vitro from a highly neutralizing COVID-19 convalescent plasma. bioRxiv, 2020.2012.2028.424451, (2020). [DOI] [PMC free article] [PubMed]
- 48.Cele S et al. Escape of SARS-CoV-2 501Y.V2 from neutralization by convalescent plasma. Nature 593, 142–146, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang P et al. Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. Cell Host Microbe 29, 747–751 e744, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang P et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature 593, 130–135, (2021). [DOI] [PubMed] [Google Scholar]
- 51.Wang Z et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature, (2021). [DOI] [PMC free article] [PubMed]
- 52.Zhou P et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tian C et al. Immunodominance of the VH1–46 antibody gene segment in the primary repertoire of human rotavirus-specific B cells is reduced in the memory compartment through somatic mutation of nondominant clones. J Immunol 180, 3279–3288, (2008). [DOI] [PubMed] [Google Scholar]
- 54.Wu X et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 333, 1593–1602, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou T et al. Structural repertoire of HIV-1-neutralizing antibodies targeting the CD4 supersite in 14 donors. Cell 161, 1280–1292, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang CC et al. Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120. Proc Natl Acad Sci U S A 101, 2706–2711, (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Williams WB et al. Diversion of HIV-1 vaccine-induced immunity by gp41-microbiota cross-reactive antibodies. Science 349, aab1253, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Joyce MG et al. Vaccine-induced antibodies that neutralize rroup 1 and group 2 influenza A viruses. Cell 166, 609–623, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pappas L et al. Rapid development of broadly influenza neutralizing antibodies through redundant mutations. Nature 516, 418–422, (2014). [DOI] [PubMed] [Google Scholar]
- 60.Sui J et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol 16, 265–273, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wheatley AK et al. H5N1 vaccine-elicited memory B cells Are genetically constrained by the IGHV locus in the recognition of a neutralizing epitope in the hemagglutinin stem. J Immunol 195, 602–610, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bailey JR et al. Broadly neutralizing antibodies with few somatic mutations and hepatitis C virus clearance. JCI Insight 2, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Giang E et al. Human broadly neutralizing antibodies to the envelope glycoprotein complex of hepatitis C virus. Proc Natl Acad Sci U S A 109, 6205–6210, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yuan M et al. Structural basis of a shared antibody response to SARS-CoV-2. Science 369, 1119–1123, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nielsen SCA et al. Human B cell clonal expansion and convergent antibody responses to SARS-CoV-2. Cell Host Microbe 28, 516–525 e515, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rappuoli R, Bottomley MJ, D’Oro U, Finco O & De Gregorio E Reverse vaccinology 2.0: Human immunology instructs vaccine antigen design. J Exp Med 213, 469–481, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thomson EC et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell 184, 1171–1187 e1120, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Dorp L et al. Recurrent mutations in SARS-CoV-2 genomes isolated from mink point to rapid host-adaptation. bioRxiv, 2020.2011.2016.384743, (2020).
- 69.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr 66, 125–132, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Winn MD et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67, 235–242, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McCoy AJ et al. Phaser crystallographic software. J Appl Crystallogr 40, 658–674, (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Emsley P & Cowtan K Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–2132, (2004). [DOI] [PubMed] [Google Scholar]
- 74.Schrodinger LLC. The PyMOL Molecular Graphics System, Version 1.8 (2015).
- 75.Starr TN et al. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science 371, 850–854, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Otwinowski J, McCandlish DM & Plotkin JB Inferring the shape of global epistasis. Proc Natl Acad Sci U S A 115, E7550–E7558, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hsieh CL et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science 369, 1501–1505, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Plante JA et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature, (2020). [DOI] [PMC free article] [PubMed]
- 79.Xie X et al. An infectious cDNA clone of SARS-CoV-2. Cell Host Microbe 27, 841–848 e843, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sievers F et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7, 539, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Edgar RC MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113, (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Crooks GE, Hon G, Chandonia JM & Brenner SE WebLogo: a sequence logo generator. Genome Res 14, 1188–1190, (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Per-mutation escape fractions for deep mutational scanning experiments
Viral neutralization and binding kinetic data
ELISA binding source data
Data Availability Statement
The crystal structures reported in this paper have been deposited to the Protein Data Bank (https://www.rcsb.org) under the accession numbers 7L7D (COV2–2196 + RBD) and 7L7E COV2–2196 and COV2–2130 + RBD). The following were obtained from the PDB and used for visualization or molecular replacement: PDB IDs: 7K4N, 6M0J, 6XM4, 7CAK, 6ZOY, 6XC2, 5JRP. The FASTQ files for deep mutational scanning are available on the NCBI Sequence Read Archive under BioSample SAMN17532001 as part of BioProject PRJNA639956. Per-mutation escape fractions are available on GitHub (https://github.com/jbloomlab/SARS-CoV-2-RBD_MAP_AZ_Abs/blob/main/results/supp_data/AZ_cocktail_raw_data.csv) and in Supplementary Data Table 2. All other data are available in the main text or the supplementary materials.


