Abstract
The Pseudomonas aeruginosa orphan sensor SagS (PA2824) was initially reported as one of three orphan sensor kinases capable of activating HptB, a component of the HptB signaling pathway that intersects with the Gac/Rsm signaling pathway and fine-tunes P. aeruginosa motility and pathogenesis. Since then, this orphan sensor has been reported to be involved in other, unorthodox signaling pathways serving additional functions. The present review is aimed at summarizing the various functions of SagS, with an emphasis on its toggle or dual switch functions, and highlighting the role of SagS as a hub at which the various signaling pathways intersect, to regulate the transition from the planktonic to the sessile mode of growth, as well as the transition of surface-associated cells to a drug tolerant state.
1. Introduction
Pseudomonas aeruginosa is a Gram-negative, facultatively aerobic, rod-shaped bacterium. P. aeruginosa may not be a typical pseudomonad in natural environments given its optimum temperature for growth is 37 °C but this bacterium is ubiquitous in moist environments, having been isolated from surface water, soil and vegetation, including vegetables and salads [1,2]. P. aeruginosa has simple nutritional needs but can tolerate/is resistant to high concentrations of salts and dyes, weak antiseptics, and many commonly used antibiotics. These properties of P. aeruginosa are critical factors for its ecological success, which also help explain the ubiquitous nature of the organism and its prominence as an opportunistic pathogen, capable of causing disease in plants and animals, including humans.
In these various niches, P. aeruginosa exists in two main modes of growth, planktonic and as biofilm. During planktonic growth, bacteria appear as single, independent free-floating cells (planktonic), while during the biofilm mode of growth, bacteria are organized in aggregates and surface attached sessile communities encased in a hydrated polymeric matrix of their own synthesis [[3], [4], [5], [6], [7]]. The ability to form biofilms has also been recognized as the dominant mode of bacterial growth in nature and a common trait of various microorganisms, including lower-order eukaryotes [3,4,7,8], likely because the biofilm mode of growth provides protection from environmental challenges and outside threats including antimicrobial compounds and host immune responses [6,9,10]. The mode of growth has additional consequences on the behaviour of the bacteria. Key characteristics relating to the sessile mode of growth include loss of flagella gene expression, production of biofilm matrix components and increased levels of some virulence determinants [[11], [12], [13]]. Moreover, biofilms demonstrate increased innate resistance to host immune defenses and tolerance to stresses, including starvation, dehydration and antimicrobials. Biofilms have been reported to be 10–1000-times more tolerant to various antibiotics compared to their planktonic counterparts [[14], [15], [16]]. Various factors contribute to the observed tolerance of biofilms including restricted penetration of antimicrobial agents, starvation-induced growth arrest, reduced cellular metabolic and divisional rates, and the distinct biofilm phenotype associated with increased expression of several multidrug efflux pumps and ABC transporters [[17], [18], [19], [20], [21], [22], [23], [24]]. These biofilm characteristics pose severe consequences in industrial and medical settings, with biofilms being refractory to antimicrobial treatment, enabled by low-grade inflammation, and causing chronic state infections [25]. In general, chronic biofilm infections are observed in patients with implants [26,27], patients being immunocompromised or otherwise have a predisposing condition such as cystic fibrosis (CF) causing viscous mucus in their lungs or diabetics or edema impairing the vascularization of the lower limps resulting in chronic wounds [28,29]. Biofilm infections, such as chronic pneumonia in CF patients and ventilator-associated pneumonia, affect 17 million Americans each year, causing an estimated 268,000 deaths 550,000 patients in the USA alone, with over $18 billion in direct costs spent on the treatment of these infections [30].
Although it is now widely accepted that most if not all bacterial species form biofilms in a cyclic process, the first biofilm developmental life cycle has been reported for P. aeruginosa [31,32]. Biofilm formation of P. aeruginosa has been described as a stage-specific process that is initiated by free-living, planktonic bacteria attaching to a surface [31,32]. Once attached, they multiply and produce biofilm matrix components to form mushroom-like, differentiated, structures called microcolonies. The biofilm developmental cycle comes full circle when cells disperse from the mature biofilm to restart a planktonic mode of growth [[33], [34], [35]]. It has been reported that P. aeruginosa exhibits distinct pattern of protein phosphorylation, one of the well-characterized bacterial regulatory signals, at each stage of biofilm development [36]. Biofilm formation is governed by a sophisticated regulatory network that coordinates the temporal alterations in gene expression in response to inter- and intracellular signaling molecules and environmental stimuli as well as contribute to observable phenotypic changes such as differential productions of appendages (flagella and type IV pili) and biofilm polymeric matrix [32,[36], [37], [38], [39], [40], [41], [42], [43], [44], [45], [46]].
In P. aeruginosa, several factors have been reported to contribute to the formation of biofilms [[47], [48], [49]]. For example, transition from a planktonic to a surface-associated lifestyle, specifically the transition to the irreversible attachment stage, is regulated by two membrane-bound proteins, the diguanylate cyclase SadC (PA4332) and the phosphodiesterase BifA (PA4367), that inversely regulate the levels of second messenger cyclic-di-GMP (c-di-GMP) [50,51]. The status of this SadC- and BifA-modulated c-di-GMP pool in turn acts as a signal to the downstream constituents, reciprocally impacting Pel polysaccharide production and flagellar functions. Additional factors contributing to the progression toward irreversible attachment include the sadB, pelA, and motAB genes, and components of the CheIV chemotaxis-like cluster [[52], [53], [54], [55]]. Additionally, surface behavior is regulated by the Wsp (wrinkly spreader) chemosensory system, that is homologous to the enteric chemotaxis signal transduction system (Che) [56,57]. The proteins involved in the pathway are a methyl-accepting protein (WspA), CheW homologues (WspB and WspD), a CheA homologue (WspE), a diguanylate cyclase response regulator (WspR), a methylesterase response regulator (WspF) and a methyltransferase (WspC), encoded by the wspABCDEFR operon (PA3708-3702). The Wsp system responds to mechanical pressure associated with surface growth [58]. Association with surfaces leads to the activation of WspA which in turn promotes the autophosphorylation of WspE. WspE phosphorylates its two response regulators, WspR and WspF. Phosphorylated WspR catalyzes the synthesis of c-di-GMP that in turn, induces the production of the exopolysaccharide Pel and Psl, and thus, enhances attachment. In contrast, phosphorylated WspF acts to reset the system by removing methyl groups from WspA, reducing its ability to activate WspE. The methylesterase activity of WspF is opposed by the constitutive methyltransferase activity of WspC [56]. Another regulatory layer is provided by components of the two-component regulatory systems (TCSs) [48,59], SagS, BfiRS, BfmRS, and MifRS. These TCSs work in concert to coordinate the sequential progression of biofilm formation by P. aeruginosa, with SagS and BfiRS regulating initial attachment, BfmRS biofilm maturation, and MifRS microcolony formation [36,[60], [61], [62], [63]].
Classical TCSs, where a sensor kinase recognizes a signal by modifying the phosphorylation state of a cognate response regulator, are generally encoded in operons that include both the sensor kinase and response regulator genes [64]. This supports the co-expression of the corresponding proteins and the formation of an exclusively one-to-one phosphotransfer pair. The genome of P. aeruginosa encodes 127 TCS genes, of which 107 are adjacent or closely located each other, while the remainder genes are not adjacent to another TCS gene are referred to as “orphan” TCSs [65]. While BfiRS, BfmRS, and MifRS are classical TCSs, SagS (Surface-attached growth Sensor, PA2824) is an orphan TCS. An interesting aspect of orphan TCSs is that they have been reported to be involved in unorthodox or branched pathways involving more than one partner via multistep phosphorelays (also referred to as “one-to-many” and “many-to-one” phosphotransfer relationships) [64] and as a result, they participate in cross-communication to modify gene expression and physiological change [65,66].
The findings indicate that SagS is only one of few orphan sensors by P. aeruginosa Moreover, SagS also contributes to more than one biofilm-related pathway. Indeed, SagS has been shown to not only contribute to biofilm formation, with inactivation arresting biofilm formation at an early attachment stage [36,61], but also to contribute to two distinct pools of c-di-GMP present in biofilms [46,67], biofilms being rendered tolerant to antimicrobial agents [68], and biofilm formation and drug tolerance in in vivo models of infection [69].
Given the orphan sensor kinase SagS’ unusual and multifaceted contributions to the P. aeruginosa biofilm phenotype, this review will focus on its involvement in unorthodox signaling pathways to enable the transition from the planktonic to the biofilm mode of growth and the switch by biofilm cells to a highly tolerant state. Specifically, we will focus on the phosphorelay by SagS under planktonic conditions with the Gac (global activator of antibiotic and cyanide synthesis) and HptB (histidine phosphotransfer protein B) pathways, the phosphorelay with the TCS BfiSR to modulate c-di-GMP and the small regulatory RNA (sRNA) RsmZ post-attachment to enable biofilm formation, as well as its phosphotransfer-independent role in activating the c-di-GMP responsive transcriptional regulator BrlR (biofilm resistance locus regulator, PA4878) to enable biofilm drug tolerance. Overall, we aim at discussing signal transduction mechanisms by which SagS acts as a regulatory hub for both biofilm development and biofilm tolerance.
1.1. SagS and the Gac system network
The orphan sensor SagS (PA2824) was first described by Hsu et al. [70] as a component of the Gac system network. The major components of the Gac system are GacA and GacS. GacS (LemA, lesion manifestation) was initially identified in Pseudomonas syringae in 1992 [71], while GacA (global antibiotic and cyanide control) was identified in P. fluorescens CHAO in the same year [72]. Rich et al. [73] provided the first evidence that the orphans GacS (LemA) and GacA form a TCS in Pseudomonas syringae, by demonstrating that gacA encodes the cognate response regulator for the lemA (gacS) sensor. The system has since then gained special notoriety in the genus Pseudomonas due to its involvement in bacterial virulence, by regulating the reversible transition from acute to chronic infections by the opportunistic pathogen P. aeruginosa [74,75]. Acute virulence is associated with twitching and swimming motility, expression of a type III secretion system (T3SS), and the absence of alginate, Psl, or Pel polysaccharide production. Traits associated with chronic infection include growth as a biofilm, reduced motility, and expression of a type VI secretion system (T6SS). In P. aeruginosa, the Gac system consists at its core of a transmembrane sensor kinase, GacS (LemA [PA0928]), and its cognate regulator, GacA (PA2586) [73] (Fig. 1). Upon receiving a signal, GacS phosphorylates its cognate response regulator GacA which in turn upregulates the expression of the sRNAs RsmZ (PA3621.1) and RsmY (PA0527.1). Two additional sRNAs have been reported as RsmV and RsmW of which expression is Gac-independent [76,77]. RsmY, RsmZ, RsmV and RsmW sRNAs bind to sRNA-binding proteins of the CsrA family, namely RsmA and RsmF, with sRNA binding post-transcriptionally regulating and fine-tune gene RsmA/RsmF-dependent expression [78] (Fig. 1). For example, at low sRNA concentrations, RsmA and RsmF bind to specific mRNA targets and alter their stability by preventing translation of mRNA transcripts required for the establishment of chronic infections and blocking accessibility to the ribosome-binding site, thus enhancing the expression of genes encoding several acute virulence factors and those involved in bacterial motility while repressing the production of virulence factors associated with chronic infections [79,80]. At high sRNA concentrations, sRNAs bind to sRNA-binding proteins, by functioning as decoys to sequester CsrA-family proteins from target mRNAs, and thus, release the repression of virulence factors associated with chronic infections but repress the production of acute infection-associated factors. Therefore, sequestered RsmA/RsmF has been linked to the expression of genes involved in biofilm formation but the repression of genes involved in acute virulence and motility [[79], [80], [81]].
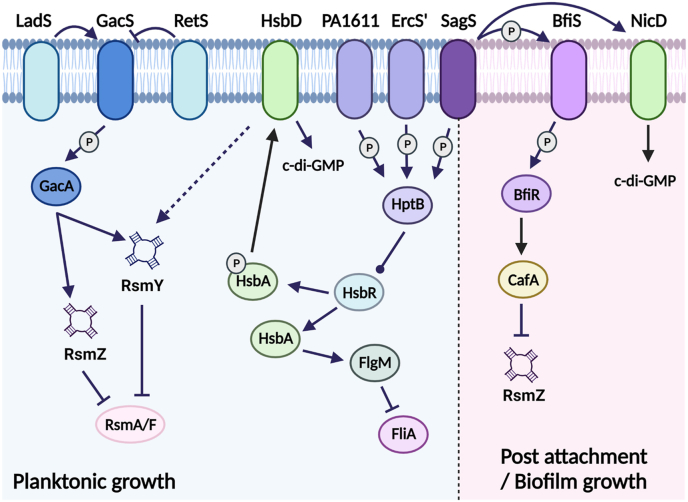
Fig. 1.
Schematic representation of the signal transduction system in which SagS participates to modulate biofilm development of P. aeruginosa. Under planktonic conditions, SagS regulates small regulatory RNA (sRNA) levels in an HptB-dependent manner. HptB and GacAS contribute to the regulation of the sRNAs, RsmY and both RsmY and RsmZ, respectively [70,86]. These sRNAs sequester the sRNA-binding proteins RsmA and RsmF from target mRNAs [78,79]. Upon transition to surface associated growth (post-attachment and biofilm growth), SagS promotes biofilm formation via hierarchical phosphotransfer to BfiSR [61], which in turn contributes to the suppression of the sRNA RsmZ level [60]. Arrows and blunt-ended lines indicate stimulatory interactions and inhibitory interactions, respectively. Bulb-ended lines show interactions that can differ depending on conditions. P, protein phosphorylation or phosphotransfer reaction. The image was created using biorender.com.
Two additional transmembrane sensor kinases, LadS (PA3974) and RetS (PA4856), reciprocally modulate gene expression via GacA [82] (Fig. 1). LadS (lost adherence sensor) works in parallel with GacS in response to calcium [38,75,83], while RetS (regulator of exopolysaccharide and type III secretion) hinders GacS/GacA signaling by controlling GacA in an opposite manner to GacS and LadS, thus blocking RsmY and RsmZ production and thus, promoting acute infection and repressing the expression of genes associated with biofilm production [82] (Fig. 1). The role of RetS is supported by the finding of a P. aeruginosa retS mutant being unable to establish an infection in a mouse model of acute pneumonia relative to its parental strain [74] (Fig. 1).
An additional component of the Gac-Rsm system is the HptB pathway (Fig. 1). The pathway directly modulates flagellar gene expression as well as fine-tunes sRNA levels, by intersecting with the Gac-Rsm system. The pathway is composed of the histidine phosphotransfer protein HptB (PA3345), the response regulator HsbR (PA3346), the anti-anti sigma factor HsbA, and the diguanylate cyclase HsbD. When HptB is phosphorylated, it activates the phosphatase domain of HsbR, which in turn dephosphorylates the anti-anti-sigma factor HsbA [70,84]. Dephosphorylated HsbA sequesters the anti-sigma factor FlgM from the sigma factor FliA, with free FliA inducing flagellar gene expression and thereby motility. In contrast, when HptB is not phosphorylated, HsbR functions as a kinase, phosphorylates HsbA, which in turn interacts with the diguanylate cyclase HsbD [85]. The switch in HsbA interaction partners from FlgM to HsbD coincides with increased levels of c-di-GMP, decreased swarming motility and increased of biofilm formation. In addition, phosphorylated HptB antagonizes GacA in a manner similar to RetS, likely via phosphorylated HsbA. However, while RetS affects the expression of both RsmY and RsmZ, HptB exclusively regulates RsmY expression [86] (Fig. 1). Input by HptB thus enables finetuning of sRNA levels and thus, biofilm formation, twitching, swimming, swarming motility and chemotaxis in P. aeruginosa in response to environmental cues. These inputs are provided by three orphan sensor kinase hybrids, namely PA1611, PA1976 (ErcS), and SagS, capable of activating HptB [70,87] (Fig. 1). Therefore, SagS indirectly contributes to the reversible transition from acute to chronic infections and the switch from the planktonic to the sessile lifestyle, by finetuning RsmY and RsmZ levels and the modulation of c-di-GMP levels in P. aeruginosa.
1.2. SagS is part of a dedicated signaling pathway to regulate biofilm development in a stage-specific manner
Biofilms by P. aeruginosa display multiple phenotypes with distinct physiological characteristics (e.g. 3D-biofilm structure, cell-cell communication, and metabolic changes) that can be correlated to distinct episodes or stages of biofilm development [31]. These stages were referred to as reversible and irreversible attachment, maturation, and dispersion (Fig. 2). Each biofilm developmental stage corresponds to unique patterns of protein production and gene expression [31,32]. Several regulators have been identified to affect and contribute to biofilm formation [50,51,56,57]. However, only one regulatory signaling network has been identified that links the regulation of biofilm formation to committed biofilm developmental steps by P. aeruginosa. This signaling network is composed of several TCSs named BfiSR, BfmRS, and MifRS that are sequentially activated/phosphorylated (BfiSR<BfmSR<MifSR) over the course of biofilm formation [36]. This is supported by the finding that inactivation of either TCSs arrested biofilm formation at distinct developmental stages. For example, ΔbfiS biofilms have been demonstrated to be arrested at the irreversible attachment stage, while biofilms formed by ΔbfmR and ΔmifR were found to be arrested at the maturation-1 and -2 stages of biofilm development, respectively [36,[60], [61], [62], [63]]. An additional component of this signaling network is the orphan sensor SagS. Moreover, similar to ΔbfiS, ΔsagS biofilms have been demonstrated to be arrested at the irreversible attachment stage [61]. Several lines of evidence suggest that SagS is functioning prior to BfiSR to enable biofilm formation (Fig. 2). For one, t SagS activates BfiS [61]. Secondly, multicopy expression of bfiR restored biofilm formation by ΔsagS to wild-type levels, but not vice versa [61]. Additionally, despite P. aeruginosa mutant strains ΔsagS and ΔbfiS forming impaired biofilms [36,60,61], phosphoproteome analysis indicated ΔsagS biofilms to be arrested prior to ΔbfiS biofilms, at the transition from the planktonic to the initial attachment stage [61]. And lastly, biofilms by ΔsagS and ΔbfiS differed in c-di-GMP levels. Biofilms formed by ΔbfiS harbored wild-type levels of c-di-GMP, approximately 75–78 pmol/mg c-di-GMP, while biofilms formed by ΔsagS exhibited significantly reduced c-di-GMP levels when compared to wild-type biofilms. On average, ΔsagS biofilms only harbored 33 ± 2 pmol/mg c-di-GMP [88,89]. Despite these low levels, ΔsagS biofilms harbor 2–3 times higher c-di-GMP levels relative to planktonic cells [88,89]. Recent findings suggest SagS to likely affect two diguanylate cyclases, PA3177 and NicD (Nutrient-induced cyclase D). SagS interacts with the membrane bound diguanylate cyclase NicD(45) (Fig. 2). Although evidence is lacking that NicD functions in a SagS-dependent manner, it is likely that NicD activity contributes to the c-di-GMP levels present in biofilms. However, inactivation of nicD has been shown to coincide with biofilms being similar in architecture to ΔsagS [90]. PA3177 will be discussed below.
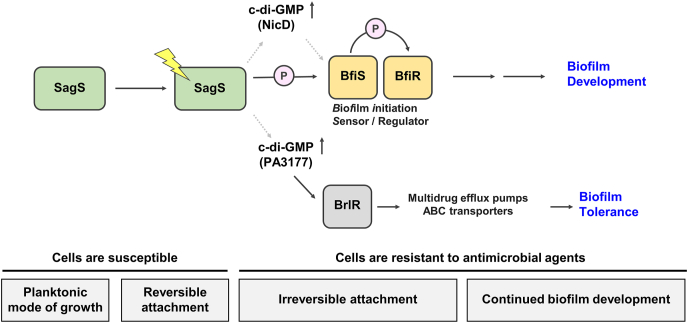
Fig. 2.
Contribution of SagS to the motile-sessile and susceptible-resistance switch in P. aeruginosa. Transition to the irreversible attachment stage, in which SagS directly interacts with and phosphorylates the TCS BfiSR [60,61], marks the timing when surface-associated cells start to be resistant to antimicrobial agents [68]. SagS regulates the switch from an antimicrobial susceptible to a highly tolerant state via regulation of c-di-GMP levels [88], which are generated by the diguanylate cyclase PA3177 [67], and subsequent activation of BrlR in a phosphotransfer-independent manner [67,68,95]. BrlR, in turn, modulates the expression of multidrug efflux pumps and ABC transporter [12,22]. Gray arrows with dashed lines indicate unidentified mechanism(s).
It is thus likely that SagS contributes to the cellular c-di-GMP pool of biofilm cells in two different ways, upon the transition to the surface via the HptB pathway (Fig. 1), and post-attachment via NicD (Fig. 2) and PA3177. Additionally, the findings further support SagS to be part of a genetic pathway that regulates the stage-specific biofilm developmental transitions in a hierarchical ordered manner in response to environmental cues, with SagS functioning as an early switch between the planktonic and sessile mode of growth.
1.3. SagS affects RsmZ levels in biofilms via BfiSR in a manner independent of HptB
Given that SagS affects the cellular level of c-di-GMP upon surface associated growth, it is not surprising that SagS not only contributes to biofilm formation but also the maintenance of the mature biofilm architecture, as loss of sagS expression in mature P. aeruginosa biofilms coincided with the collapse of the biofilm architecture [46,60,61,91]. But c-di-GMP is not the only reason for SagS contributing to biofilm formation, as SagS also affects biofilm formation in a manner dependent to sRNAs. In planktonic cells, SagS suppresses RsmY and RsmZ sRNAs accumulation and sequestration of RsmA in an HptB-dependent manner (Fig. 1), with inactivation of sagS resulting in an HptB-dependent increase in RsmY and RsmZ expression, increased Psl polysaccharide production, and increased virulence in planktonic cells. Under biofilm growth conditions, however, SagS suppresses sRNA accumulation in an HptB-dependent manner [61] (Fig. 1). Instead, RsmZ levels were found to be dependent on the SagS interaction partners BfiSR (biofilm initiation sensor/regulator) [61]. Post-attachment, SagS phosphorylates the transmembrane sensory protein BfiS which in turn phosphorylates its cognate response regulator BfiR (Fig. 1, Fig. 2). Phosphorylated BfiR then activates the expression of cafA encoding ribonuclease G [60] capable of selectively degrading RsmZ. Activation of BfiR thus effectively results in the reduction of RsmZ levels, without affecting RsmY, under biofilm growth conditions (Fig. 1).
The findings suggested SagS to indirectly affect RsmZ and RsmY levels via the HptB signaling pathway under planktonic growth conditions, while upon surface associated growth, SagS was found to contribute to the modulation of sRNA levels in a manner independent of HptB, but dependent on BfiSR (Fig. 1). Thus, SagS acts as a switch by linking the GacA-dependent sensory system under planktonic conditions to the suppression of sRNAs, particularly RsmZ, post-attachment.
1.4. Antimicrobial tolerance requires SagS
Biofilms have been reported to be difficult to eradicate by conventional antibiotic treatment [3,15,92] This biofilm drug tolerance has been reported to be distinct from mechanisms commonly associated with planktonic cells, such as the acquisition of resistance via uptake of plasmid-borne resistance markers or random mutation and be multifactorial as requiring a combination of several mechanisms [15,19,93]. Instead, biofilm drug tolerance has been reported to be multifactorial, and based on starvation-induced growth arrest, reduced metabolic rates, increased stress tolerance, reduced diffusion rates, and the presence of extracellular polymeric matrices [[17], [18], [19], [20], [21], [22], [23], [24]].
In an effort to understand the timing of induction of this biofilm drug tolerance, Gupta et al. [68] determined when over the course of their development P. aeruginosa biofilms gain their tolerance to antimicrobial agents. The study made use of wild-type cells grown to the reversible, irreversible, early and late maturation biofilm developmental stages under flowing conditions. Additionally, the study made use of mutant strains (ΔsagS, ΔbfiS and ΔbfmR) that had previously been demonstrated to be arrested at distinct stages of biofilm development. Specifically, ΔsagS has been reported to be arrested at the transition from the reversible to the irreversible attachment stage, ΔbfiS at the irreversible attachment stage, and ΔbfmR at the biofilm maturation [36,[60], [61], [62], [63]]. Relative to wild-type biofilms, ΔsagS biofilms were significantly more susceptible to various antimicrobial agents when exposed for 1 h compared to wild-type biofilms. Moreover, biofilms by ΔsagS were fully eradicated when exposed for 24 h to tobramycin concentrations exceeding 100 ug/ml while biofilms by P. aeruginosa wild-type and ΔbfiS remained viable at tobramycin concentrations exceeding 400 ug/ml [46,68,88]. The findings suggested that in P. aeruginosa, transition to the irreversible attachment stage, regulated by the orphan sensor SagS, marks the timing when biofilms switch to the high-level tolerance phenotype [68]. The findings furthermore suggested that additional factors than biofilm biomass accumulation contribute to biofilm drug tolerance. This was supported by the findings of biofilms formed by ΔsagS and ΔbfiS differing in drug tolerance despite featuring similar biofilm architectural features [68]. One of such factors is SagS. Several lines of evidence suggest SagS-dependent biofilm drug tolerance is linked to c-di-GMP and the c-di-GMP responsive transcriptional regulator BrlR (Fig. 2). BrlR has been found to contribute to the high-level antimicrobial tolerance of P. aeruginosa biofilms by activating the expression of the multidrug efflux pump operons, mexAB-oprM and mexEF-oprN, several ABC transport systems including PA1874-77, and by repressing the expression of the oprH-phoPQ operon [12,22,[94], [95], [96]]. For one, expression of brlR is SagS-dependent [68]. Little is known how brlR expression is regulated. However, inactivation of sagS coincided with decreased levels of BrlR production, with BrlR derived from ΔsagS biofilm cells being impaired in binding to the brlR promoter and promoters of the multidrug efflux pump operons, mexAB-oprM and mexEF-oprN [68]. Restoring c-di-GMP to wild-type biofilm-like levels in ΔsagS biofilms restored brlR expression, DNA binding by BrlR, and recalcitrance to killing by antimicrobial agents of ΔsagS biofilm cells [88]. In agreement with c-di-GMP levels contributing to drug tolerance, Gupta et al. [88] demonstrated that reducing cellular c-di-GMP levels of biofilm cells to ≤40 pmol/mg correlated with increased susceptibility and reduced brlR expression. Conversely, the authors demonstrated that planktonic cells are rendered significantly more resistant to antimicrobial agents upon increasing c-di-GMP levels present in planktonic cells to biofilm-like levels (≥55 pmol/mg), with increased resistance correlating with increased brlR, mexA, and mexE expression and BrlR production. It is of interest to note that ΔsagS biofilms harbored on average 33 ± 2 pmol/mg c-di-GMP [88,89].
Together, these data led to suggest that SagS directly or indirectly modulates c-di-GMP levels to activate BrlR and thus, biofilm drug tolerance (Fig. 2). Moreover, the findings suggested the presence of a SagS-dependent c-di-GMP pool that contributes to the resistance of P. aeruginosa biofilms. The c-di-GMP modulating enzyme to provide the necessary pool of c-di-GMP was identified as PA3177 encoding an active diguanylate cyclase (Fig. 2). PA3177 was found to contribute to the cellular c-di-GMP levels in biofilms cells, but not planktonic cells [67]. Moreover, inactivation of PA3177 did not affect biofilm formation but rendered ΔPA3177 mutant biofilms susceptible to antimicrobial agents such as tobramycin and hydrogen peroxide. Similar to ΔsagS biofilms, biofilms formed by ΔPA3177 demonstrated reduced BrlR abundance, with ΔPA3177 biofilm cell-derived BrlR unable to bind its target promoters [67]. The findings suggested the presence of a local pool of PA3177-generated c-di-GMP necessary to modulate BrlR activity and in turn, biofilm drug tolerance. Although P. aeruginosa biofilm tolerance is linked to PA3177-generated c-di-GMP and BrlR activity, the mechanism(s) of how SagS regulates PA3177 activity still remains to be elucidated. However, the findings strongly suggest SagS to induce the switch of attached cells to transition from a susceptible to a tolerant phenotype.
Given the link between SagS and BrlR, induction of brlR gene expression has been used to determine the timing of SagS switch function. Using a brlR promoter reporter, Chambers et al. [95] demonstrated brlR expression to be detectable under flowing conditions as early as 6 h post initial attachment. Activation of brlR expression as early as 6 h post attachment is in agreement with SagS functioning as a switch to regulate the transition to the irreversible attachment stage under flowing conditions. These data suggest that SagS has dual switch functions, regulating the transition from the planktonic to the sessile mode of growth as well as the transition to a heightened drug tolerant phenotype (Fig. 2).
1.5. Role of distinct amino acid residues and domains in the dual switch functions of SagS
SagS regulates both, (i) the transition to biofilm formation and (ii) the transition to cells gaining their enhanced tolerance to antimicrobials. The two functions are associated with distinct pathways or interactions, including the HptB pathway, the interactions with BfiSR, and the modulation with BrlR (Fig. 1, Fig. 2). Much research has focused on elucidating the role of each of the SagS domains in order to determine how SagS carries out its multiple functions [46,91]. SagS harbors multiple domains [46,91] (Fig. 3). Its N-terminus is composed of a periplasmic HmsP sensory domain, flanked by transmembrane helices, followed by a cytoplasmic portion composed of a HAMP (Histidine kinases, Adenylyl cyclases, Methyl binding proteins, Phosphatases) domain, a histidine kinase (HisKA) domain, and a C-terminal receiver (Rec) domain [46,91] (Fig. 3). SagS activation leads to auto-phosphorylation at the histidine residue H315 at the HisKA domain, and subsequent phosphoryl-group transfer to the aspartate residue D713 located in the Rec domain (Fig. 3). The HmsP domain of SagS is homologous to the N-terminal domains of the c-di-GMP phosphodiesterases BifA and HmsP in P. aeruginosa and Yersinia pestis, respectively [50,97,98]. As sensory domains, HmsP is a likely control point for SagS switch functions. Indeed, a mutant analysis revealed 32 residues located within the HmsP domain that only contribute to blocking one of the two SagS switch functions [91]. Specifically, the study revealed amino acid residue mutations affecting only attachment and subsequent biofilm formation but not biofilm tolerance. The same residues impaired histidine kinase signaling via BfiS. In turn, phosphorylated BfiS activates its cognate response regulator BfiR to transcribe CafA to modulate RsmZ levels [46,61]. These included residues F131 and L154 [91] (Fig. 3). In contrast, residues affecting biofilm drug tolerance, but not attachment and subsequent biofilm formation, negatively impacted BrlR transcription factor levels. These included residues D105, R116 and L126 [91] (Fig. 3). Structural prediction suggested the two sets of residues affecting sensory functions to be located in distinct areas that were previously described to be involved in ligand binding interactions [91].
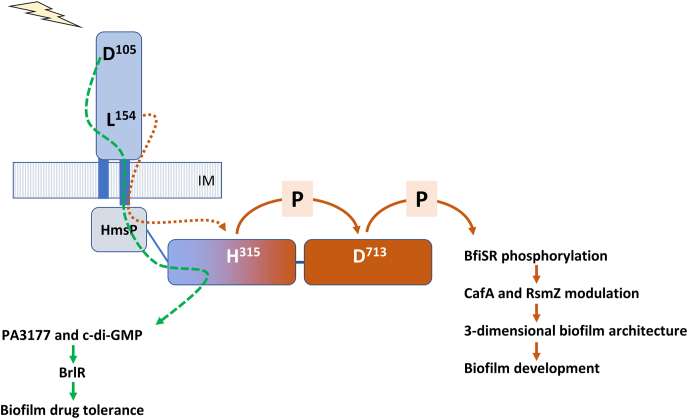
Fig. 3.
Domain-dependent modular functions of SagS. Upon sensing signal or cue such as glucose-6-phosphate [45], a signal is transduced from the periplasmic HmsP sensory domain of SagS to its HisKA domain via HAMP domain, followed by phosphotransfer between the histidine residue H315 in the HisKA domain and the aspartate residue D713 in the Rec domain, resulting in subsequent phosphorylation of BfiSR and biofilm development [46]. Residue L154 of HmsP domain contributes to biofilm development via signal cascade from SagS to BfiSR [91]. In contrast, HmsP-HisKA domain of SagS, in which residue D105 of HmsP domain is involved, contributes to biofilm tolerance via regulation of c-di-GMP levels and activation of BrlR [46,91]. Arrows indicate modulatory effect of domains. P, protein phosphorylation or phosphotransfer reaction.
The role of two select amino acid resides, L154 and D105, was analyzed using a mouse models of chronic pneumonia [69]. Relative to wild-type P. aeruginosa, ΔsagS mutants demonstrated significantly reduced bacterial colonization and increased susceptibility to the antibiotic tobramycin. Recombinant P. aeruginosa ΔsagS strains chromosomally expressing the SagS variant SagS_L154A showed reduced bacterial loads in the chronic pneumonia model but were as susceptible as wild-type biofilms. In contrast, interference with the D105 residue had no effect on bacterial colonization but enhanced the susceptibility of P. aeruginosa biofilms during tobramycin treatment [69]. The findings supported the notion that interference with the distinct regulatory circuits of SagS affected P. aeruginosa pathogenicity and drug tolerance independently in chronic infections in a manner similar to in vitro conditions. Additionally, the findings indicated SagS to affect two distinct regulatory pathways, with division of signal relay occurring via the HmsP domain.
Overall, the findings suggested that the periplasmic HmsP domain of SagS to be a control point in the regulation of biofilm formation and biofilm cells transitioning to a drug-tolerant state. However, if the HmsP domain is central, the distinct regulatory pathways that are activated by SagS, are likely enabled via the modular composition of SagS [46] (Fig. 3). Using SagS constructs lacking one or more domains, Petrova et al. [46] demonstrated that activation of the biofilm formation pathway via BfiSR only requires the HisKA-Rec domain and its His315-Asp713-phosphorelay. Moreover, the two domains were sufficient to restore c-di-GMP levels and biofilm formation by ΔsagS to wild-type levels [46] (Fig. 3). In contrast, regulation of the antimicrobial tolerant state was independent of the HisKA-Rec mediated His315-Asp713-phosphorelay. Instead, only the HmsP-HisKA domains were found to be necessary to restore drug tolerance by ΔsagS biofilms [46] (Fig. 3). Expression of the HmsP-HisKA domains coincided with restoration of brlR expression, BrlR activation, and c-di-GMP to wild-type levels [46]. Interestingly, however, while expression of both SagS domain constructs, HisKA-Rec and HmsP-HisKA, restored c-di-GMP to wild-type levels, only HmsP-HisKA and this variant-derived c-di-GMP rescued the drug tolerance phenotype of ΔsagS biofilms. It is likely that the HmsP-HisKA-derived pool of c-di-GMP is generated by the diguanylate cyclase PA3177 [67] (Fig. 2, Fig. 3). In addition to pointing at the presence of distinct pools of c-di-GMP that serve distinct functions, e.g. to support biofilm formation or enable biofilm drug tolerance, the findings also strongly suggest that biofilm drug tolerance is not a function of biofilm biomass accumulation but instead, that biofilm formation and biofilm drug tolerance are regulated independently (Fig. 2, Fig. 3).
1.6. Signals and SagS activation
The periplasmic sensory HmsP domain of SagS has been described as the control point of the dual functions of SagS [91]. In addition, several essential amino acid residues contributing to SagS functions were predicted to be located in an areas of the SagS HmsP domain that were previously suggested to be involved in ligand binding interactions [91]. The findings suggested HmsP to also contributes to signal perception (Fig. 3). Using a modified Biolog phenotypic microarray assays, glucose-6-phosphate, one of many host-derive hexose phosphates, has recently been identified as an extracellular signal of SagS [45]. Exposure to glucose-6-phosphate stimulated attachment and biofilm formation in a manner dependent SagS. The increase in surface attached biomass accumulation in response to glucose-6-phosphate coincided with enhanced intracellular c-di-GMP levels by biofilms formed by P. aeruginosa wild type and a complemented ΔsagS strain. However, no difference in c-di-GMP levels was noted in biofilms formed by ΔsagS in the absence or presence of glucose-6-phosphate. The findings indicated SagS modulates the c-di-GMP pool in response to glucose-6-phosphate under biofilm growth conditions. Specifically, SagS was found to likely modulates cellular c-di-GMP levels in a manner dependent on the diguanylate cyclase NicD [45]. While NicD has been previously reported to directly perceive sugars and amino acids such as glutamate, with cue perception resulting in NicD dephosphorylation and increased diguanylate cyclase activity [90], modulation of c-di-GMP levels in response to glucose-6-phosphate was independent of NicD sensing [45]. Instead, NicD activity appeared to be dependent on its interaction with SagS [45]. While these findings indicate that SagS-mediated biofilm formation and c-di-GMP regulation are stimulated by glucose-6-phosphate, it remains to be elucidated whether glucose-6-phosphate directly binds to the sensory domain HmsP of SagS, whether other signals and cues are perceived by SagS, and how SagS modulates NicD (or other diguanylate cyclases) upon signal perception to affect the cellular levels of c-di-GMP present in biofilms.
2. Conclusion and future work
The transition from a planktonic to a sessile lifestyle is regulated by multiple overlapping signaling system. A key player in this transition is SagS. In this review, we highlighted the roles of SagS in three distinct pathways under planktonic and surface associated growth conditions. A summary of these functions with SagS functioning as a regulatory hub connecting these multiple overlapping signaling system is shown in Fig. 4. Specifically, we focused on the role of SagS under planktonic conditions in which SagS functions as part of the Gac-Rsm and HptB pathways to modulate RsmY and RsmZ sRNAs, c-di-GMP and thus, modulation of virulence motility, polysaccharide production. Thus, in addition to regulates expression of genes encoding several acute virulence factors and those involved in bacterial motility, the system prepares P. aeruginosa for the transition from the planktonic to the surface associated mode of growth (Fig. 4). Transition to the surface associated lifestyle induces a switch in SagS, with SagS ceasing to function within the Gac-Rsm and HptB pathways but instead interacting with the TCS BfiSR. This switch leads to SagS partly countering the Gac-Rsm/HptB-dependent sRNA modulation, by suppressing the sRNA RsmZ post-attachment to enable continued biofilm development post-attachment (Fig. 4). SagS-mediated RsmZ degradation and subsequent biofilm formation depends on the phosphotransfer functions of SagS as well as c-di-GMP regulation [46,60,61]. Additionally, the transition coincides with a switch from acute virulence factors to the production of virulence factors associated with chronic infections.
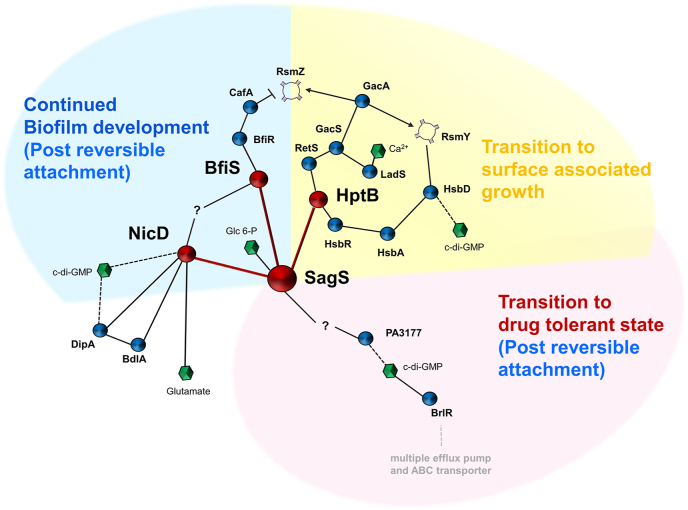
Fig. 4.
SagS is a hub protein, connecting multiple overlapping signaling system, to enable the transition from a planktonic to a sessile, drug tolerant mode of growth.?, unknown, Arrows and blunt-ended lines indicate stimulatory interactions and inhibitory interactions, respectively. Dashes lines indicate modulation of c-di-GMP levels. Confirmed SagS interaction partners are shown in red. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
But this is not the only switch that occurs post-attachment. SagS furthermore contributes to the transition of surface-associated cells to a drug tolerant state (Fig. 4). Activation of biofilm drug tolerance is independent of the phosphotransfer functions of SagS and involves the coordination of a distinct pool of c-di-GMP, likely derived by the diguanylate cyclase PA3177, and the c-di-GMP responsive transcriptional regulator BrlR [67,68,88] (Fig. 4). Activation of biofilm-associated antimicrobial tolerance by SagS contributes to the drug tolerance of chronic P. aeruginosa infections [46,69,91]. While the modulation of c-di-GMP levels upon the transition to the surface-associated growth are likely derived by HptB and other c-di-GMP modulating pathways, subsequently increased c-di-GMP levels detected in P. aeruginosa cells post attachment are dependent on SagS. In fact, SagS contributes to two distinct pools of c-di-GMP, one required for biofilm formation, and what appears to be a local PA3177-derived c-di-GMP pool necessary for the activation of biofilm drug tolerance.
SagS contributing to distinct but overlapping signaling system to overall affect distinct phenotypes has several implications. For one, SagS functions as a toggle switch to enable the transition from the planktonic to the surface associated mode of growth. The transition coincides with SagS switching from enabling RsmY and RsmZ sRNAs accumulation to the suppression of sRNA RsmZ, and a shift from an acute to a chronic virulence phenotype (Fig. 4). Moreover, SagS is modular in its function, capable of separately regulating biofilm development and biofilm drug tolerance post-attachment (Fig. 4). As interference with the biofilm or tolerance regulatory circuits of SagS affects P. aeruginosa pathogenicity in chronic models of infections, the findings furthermore reveal SagS to be a promising new target to treat P. aeruginosa biofilm infections.
However, certain gaps in our understanding of SagS remain. For examples, it is not known how SagS switches from interaction with the Gac/Rsm/HptB pathway to initiate a phosphor-signaling relay with BfiSR. It is likely that cues or signals perceived by SagS contribute to this switch. However, so far, only glucose-6-phosphate has been identified as a signal capable of activating SagS [45]. It is of interest to note that the same study also identified several other compounds that appeared to suppress SagS and biofilm formation [45]. While the study did not elucidate whether these compounds shift SagS from interacting with BfiSR to affecting the Gac/Rsm/HptB pathway, it is thus likely that an as of yet unknown compound may contribute to SagS toggling between its pre- and post-attachment functions (Fig. 4). Likewise, the specific mechanism of how SagS modulates the diguanylate cyclases PA3177 and NicD to stimulate distinct c-di-GMP pools is not known. As NicD activity has previously been shown to be modulated upon phosphorylation, it is possible that SagS affects the c-di-GMP pool via protein-protein interactions and subsequent phosphotransfer. However, direct evidence is lacking. Likewise, it is unclear whether additional proteins interact with SagS to regulate its hub functions and/or the two distinct biofilm phenotypic switches.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Karin Sauer reports financial support was provided by National Institutes of Health. Soyoung Park reports financial support was provided by National Institutes of Health.
Acknowledgments
This work was supported by a grant from the National Institutes of Health (2R01 AI080710).
References
- 1.Moradali M.F., Ghods S., Rehm B.H. Pseudomonas aeruginosa lifestyle: a paradigm for adaptation, survival, and persistence. Front Cell Infect Microbiol. 2017;7:39. doi: 10.3389/fcimb.2017.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neves P.R., McCulloch J.A., Mamizuka E.M., Lincopan N. In: Encyclopedia of food microbiology. second ed. Batt C.A., Tortorello M.L., editors. Academic Press; Oxford: 2014. PSEUDOMONAS | Pseudomonas aeruginosa, p; pp. 253–260. [Google Scholar]
- 3.Costerton J.W., Stewart P.S., Greenberg E.P. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 4.Flemming H.-C., Wingender J., Szewzyk U., Steinberg P., Rice S.A., Kjelleberg S. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol. 2016;14:563. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 5.Flemming H.-C. EPS—then and now. Microorganisms. 2016;4:41. doi: 10.3390/microorganisms4040041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flemming H.C., Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 7.Geesey G.G., Richardson W.T., Yeomans H.G., Irvin R.T., Costerton J.W. Microscopic examination of natural sessile bacterial populations from an alpine stream. Can J Microbiol. 1977;23:1733–1736. doi: 10.1139/m77-249. [DOI] [PubMed] [Google Scholar]
- 8.Costerton J.W., Cheng K.J., Geesey G.G., Ladd T., Nickel J.C., Dasgupta M., Marrie J.T. Bacterial biofilms in nature and disease. Annu Rev Microbiol. 1987;41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- 9.Costerton J.W., Lewandowski Z., Caldwell D.E., Korber D.R., Lappin-Scott H.M. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 10.Petrova O.E., Sauer K. Escaping the biofilm in more than one way: desorption, detachment or dispersion. Curr Opin Microbiol. 2016;30:67–78. doi: 10.1016/j.mib.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heacock-Kang Y., Sun Z., Zarzycki-Siek J., McMillan I.A., Norris M.H., Bluhm A.P., Cabanas D., Fogen D., Vo H., Donachie S.P., Borlee B.R., Sibley C.D., Lewenza S., Schurr M.J., Schweizer H.P., Hoang T.T. Spatial transcriptomes within the Pseudomonas aeruginosa biofilm architecture. Mol Microbiol. 2017;106:976–985. doi: 10.1111/mmi.13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liao J., Schurr M.J., Sauer K. The MerR-like regulator BrlR confers biofilm tolerance by activating multidrug efflux pumps in Pseudomonas aeruginosa biofilms. J Bacteriol. 2013;195:3352–3363. doi: 10.1128/JB.00318-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williamson K.S., Richards L.A., Perez-Osorio A.C., Pitts B., McInnerney K., Stewart P.S., Franklin M.J. Heterogeneity in Pseudomonas aeruginosa biofilms includes expression of ribosome hibernation factors in the antibiotic-tolerant subpopulation and hypoxia-induced stress response in the metabolically active population. J Bacteriol. 2012;194:2062–2073. doi: 10.1128/JB.00022-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies D. Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov. 2003;2:114. doi: 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]
- 15.Lewis K. Multidrug tolerance of biofilms and persister cells. Curr Top Microbiol Immunol. 2008;322:107–131. doi: 10.1007/978-3-540-75418-3_6. [DOI] [PubMed] [Google Scholar]
- 16.Luppens S.B., Rombouts F.M., Abee T. The effect of the growth phase of Staphylococcus aureus on resistance to disinfectants in a suspension test. J Food Protect. 2002;65:124–129. doi: 10.4315/0362-028x-65.1.124. [DOI] [PubMed] [Google Scholar]
- 17.Anwar H., Strap J.L., Chen K., Costerton J.W. Dynamic interactions of biofilms of mucoid Pseudomonas aeruginosa with tobramycin and piperacillin. Antimicrob Agents Chemother. 1992;36:1208–1214. doi: 10.1128/aac.36.6.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall C.W., Mah T.F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol Rev. 2017;41:276–301. doi: 10.1093/femsre/fux010. [DOI] [PubMed] [Google Scholar]
- 19.Mah T.F., O'Toole G.A. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001;9:34–39. doi: 10.1016/s0966-842x(00)01913-2. [DOI] [PubMed] [Google Scholar]
- 20.Mah T.F., Pitts B., Pellock B., Walker G.C., Stewart P.S., O’Toole G.A. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature. 2003;426:306–310. doi: 10.1038/nature02122. [DOI] [PubMed] [Google Scholar]
- 21.Nguyen D., Joshi-Datar A., Lepine F., Bauerle E., Olakanmi O., Beer K., McKay G., Siehnel R., Schafhauser J., Wang Y., Britigan B.E., Singh P.K. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science. 2011;334:982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poudyal B., Sauer K. The ABC of biofilm drug tolerance: the MerR-like regulator BrlR is an activator of ABC transport systems, with PA1874-77 contributing to the tolerance of Pseudomonas aeruginosa biofilms to tobramycin. Antimicrob Agents Chemother. 2018;62 doi: 10.1128/AAC.01981-17. e01981-01917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sternberg C., Christensen B.B., Johansen T., Toftgaard Nielsen A., Andersen J.B., Givskov M., Molin S. Distribution of bacterial growth activity in flow-chamber biofilms. Appl Environ Microbiol. 1999;65:4108–4117. doi: 10.1128/aem.65.9.4108-4117.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yasuda H., Ajiki Y., Aoyama J., Yokota T. Interaction between human polymorphonuclear leucocytes and bacteria released from in-vitro bacterial biofilm models. J Med Microbiol. 1994;41:359–367. doi: 10.1099/00222615-41-5-359. [DOI] [PubMed] [Google Scholar]
- 25.Hoiby N., Bjarnsholt T., Moser C., Bassi G.L., Coenye T., Donelli G., Hall-Stoodley L., Hola V., Imbert C., Kirketerp-Moller K., Lebeaux D., Oliver A., Ullmann A.J., Williams C. ESCMID guideline for the diagnosis and treatment of biofilm infections 2014. Clin Microbiol Infect. 2015;21(Suppl 1):S1–25. doi: 10.1016/j.cmi.2014.10.024. [DOI] [PubMed] [Google Scholar]
- 26.del Pozo J.L., Patel R. Clinical practice. Infection associated with prosthetic joints. N Engl J Med. 2009;361:787–794. doi: 10.1056/NEJMcp0905029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kapadia B.H., Berg R.A., Daley J.A., Fritz J., Bhave A., Mont M.A. Periprosthetic joint infection. Lancet. 2016;387:386–394. doi: 10.1016/S0140-6736(14)61798-0. [DOI] [PubMed] [Google Scholar]
- 28.González J.F., Hahn M.M., Gunn J.S. Chronic biofilm-based infections: skewing of the immune response. Pathog Dis. 2018;76 doi: 10.1093/femspd/fty023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moser C., Jensen P.Ø., Thomsen K., Kolpen M., Rybtke M., Lauland A.S., Trøstrup H., Tolker-Nielsen T. Immune responses to Pseudomonas aeruginosa biofilm infections. Front Immunol. 2021;12:237. doi: 10.3389/fimmu.2021.625597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Omar A., Wright J.B., Schultz G., Burrell R., Nadworny P. Microbial biofilms and chronic wounds. Microorganisms. 2017;5:9. doi: 10.3390/microorganisms5010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sauer K., Camper A.K., Ehrlich G.D., Costerton J.W., Davies D.G. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol. 2002;184:1140–1154. doi: 10.1128/jb.184.4.1140-1154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stoodley P., Sauer K., Davies D.G., Costerton J.W. Biofilms as complex differentiated communities. Annu Rev Microbiol. 2002;56:187–209. doi: 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- 33.Davies D.G. Biofilm highlights. Springer; Berlin: 2011. Biofilm dispersion; pp. 1–28. [Google Scholar]
- 34.Marques C.N., Davies D.G., Sauer K. Control of biofilms with the fatty acid signaling molecule cis-2-decenoic acid. Pharmaceuticals. 2015;8:816–835. doi: 10.3390/ph8040816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rumbaugh K.P., Sauer K. Biofilm dispersion. Nat Rev Microbiol. 2020;18:571–586. doi: 10.1038/s41579-020-0385-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petrova O.E., Sauer K. A novel signaling network essential for regulating Pseudomonas aeruginosa biofilm development. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brencic A., McFarland K.A., McManus H.R., Castang S., Mogno I., Dove S.L. The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol Microbiol. 2009;73:434–445. doi: 10.1111/j.1365-2958.2009.06782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Broder U.N., Jaeger T., Jenal U. LadS is a calcium-responsive kinase that induces acute-to-chronic virulence switch in Pseudomonas aeruginosa. Nat Microbiol. 2016;2:16184. doi: 10.1038/nmicrobiol.2016.184. [DOI] [PubMed] [Google Scholar]
- 39.Davey M.E., O'Toole G.A. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev. 2000;64:847–867. doi: 10.1128/mmbr.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghafoor A., Hay I.D., Rehm B.H. Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture. Appl Environ Microbiol. 2011;77:5238–5246. doi: 10.1128/AEM.00637-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klausen M., Heydorn A., Ragas P., Lambertsen L., Aaes-Jorgensen A., Molin S. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol Microbiol. 2003;48:1511–1524. doi: 10.1046/j.1365-2958.2003.03525.x. [DOI] [PubMed] [Google Scholar]
- 42.Lee J.H., Lee J. Indole as an intercellular signal in microbial communities. FEMS Microbiol Rev. 2010;34:426–444. doi: 10.1111/j.1574-6976.2009.00204.x. [DOI] [PubMed] [Google Scholar]
- 43.O’Toole G.A., Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 44.O'Toole G.A., Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 45.Park S., Dingemans J., Gowett M., Sauer K. Glucose-6-Phosphate acts as an extracellular signal of SagS to modulate Pseudomonas aeruginosa c-di-GMP levels, attachment, and biofilm formation. mSphere. 2021;6:e01231–1220. doi: 10.1128/mSphere.01231-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petrova O.E., Gupta K., Liao J., Goodwine J.S., Sauer K. Divide and conquer: the Pseudomonas aeruginosa two-component hybrid SagS enables biofilm formation and recalcitrance of biofilm cells to antimicrobial agents via distinct regulatory circuits. Environ Microbiol. 2017;19:2005–2024. doi: 10.1111/1462-2920.13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fazli M., Almblad H., Rybtke M.L., Givskov M., Eberl L., Tolker‐Nielsen T. Regulation of biofilm formation in Pseudomonas and Burkholderia species. Environ Microbiol. 2014;16:1961–1981. doi: 10.1111/1462-2920.12448. [DOI] [PubMed] [Google Scholar]
- 48.Mikkelsen H., Sivaneson M., Filloux A. Key two-component regulatory systems that control biofilm formation in Pseudomonas aeruginosa. Environ Microbiol. 2011;13:1666–1681. doi: 10.1111/j.1462-2920.2011.02495.x. [DOI] [PubMed] [Google Scholar]
- 49.Xavier J.B., Foster K.R. Cooperation and conflict in microbial biofilms. Proc Natl Acad Sci Unit States Am. 2007;104:876–881. doi: 10.1073/pnas.0607651104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuchma S.L., Brothers K.M., Merritt J.H., Liberati N.T., Ausubel F.M., O’Toole G.A. BifA, a cyclic-Di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol. 2007;189:8165–8178. doi: 10.1128/JB.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Merritt J.H., Brothers K.M., Kuchma S.L., O'Toole G.A. SadC reciprocally influences biofilm formation and swarming motility via modulation of exopolysaccharide production and flagellar function. J Bacteriol. 2007;189:8154–8164. doi: 10.1128/JB.00585-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Caiazza N.C., O'Toole G.A. SadB is required for the transition from reversible to irreversible attachment during biofilm formation by Pseudomonas aeruginosa PA14. J Bacteriol. 2004;186:4476–4485. doi: 10.1128/JB.186.14.4476-4485.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Caiazza N.C., Merritt J.H., Brothers K.M., O'Toole G.A. Inverse regulation of biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol. 2007;189:3603–3612. doi: 10.1128/JB.01685-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O Toole G.A. How Pseudomonas aeruginosa regulates surface behaviors. Microbe-Am Soc Microbiol. 2008;3:65. [Google Scholar]
- 55.Toutain C.M., Caizza N.C., Zegans M.E., O'Toole G.A. Roles for flagellar stators in biofilm formation by Pseudomonas aeruginosa. Res Microbiol. 2007;158:471–477. doi: 10.1016/j.resmic.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 56.Hickman J.W., Tifrea D.F., Harwood C.S. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. P Natl Acad Sci USA. 2005;102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guvener Z.T., Harwood C.S. Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol Microbiol. 2007;66:1459–1473. doi: 10.1111/j.1365-2958.2007.06008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Francis V.I., Stevenson E.C., Porter S.L. Two-component systems required for virulence in Pseudomonas aeruginosa. FEMS (Fed Eur Microbiol Soc) Microbiol Lett. 2017;364 doi: 10.1093/femsle/fnx104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu C., Sun D., Zhu J., Liu W. Two-component signal transduction systems: a major strategy for connecting input stimuli to biofilm formation. Front Microbiol. 2018;9:3279. doi: 10.3389/fmicb.2018.03279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Petrova O.E., Sauer K. The novel two-component regulatory system BfiSR regulates biofilm development by controlling the small RNA rsmZ through CafA. J Bacteriol. 2010;192:5275–5288. doi: 10.1128/JB.00387-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Petrova O.E., Sauer K. SagS contributes to the motile-sessile switch and acts in concert with BfiSR to enable Pseudomonas aeruginosa biofilm formation. J Bacteriol. 2011;193:6614–6628. doi: 10.1128/JB.00305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Petrova O.E., Schurr J.R., Schurr M.J., Sauer K. The novel Pseudomonas aeruginosa two-component regulator BfmR controls bacteriophage-mediated lysis and DNA release during biofilm development through PhdA. Mol Microbiol. 2011;81:767–783. doi: 10.1111/j.1365-2958.2011.07733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Petrova O.E., Schurr J.R., Schurr M.J., Sauer K. Microcolony formation by the opportunistic pathogen Pseudomonas aeruginosa requires pyruvate and pyruvate fermentation. Mol Microbiol. 2012;86:819–835. doi: 10.1111/mmi.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Laub M.T., Goulian M. Specificity in two-component signal transduction pathways. Annu Rev Genet. 2007;41:121–145. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- 65.Rodrigue A., Quentin Y., Lazdunski A., Mejean V., Foglino M. Two-component systems in Pseudomonas aeruginosa: why so many? Trends Microbiol. 2000;8:498–504. doi: 10.1016/s0966-842x(00)01833-3. [DOI] [PubMed] [Google Scholar]
- 66.Raghavan V., Groisman E.A. Orphan and hybrid two-component system proteins in health and disease. Curr Opin Microbiol. 2010;13:226–231. doi: 10.1016/j.mib.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Poudyal B., Sauer K. The PA3177 gene encodes an active diguanylate cyclase that contributes to biofilm antimicrobial tolerance but not biofilm formation by Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2018;62:e01049–18. doi: 10.1128/AAC.01049-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gupta K., Marques C.N.H., Petrova O.E., Sauer K. Antimicrobial tolerance of Pseudomonas aeruginosa biofilms is activated during an early developmental stage and requires the two-component hybrid SagS. J Bacteriol. 2013;195:4975–4987. doi: 10.1128/JB.00732-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dingemans J., Al-Feghali R.E., Lau G.W., Sauer K. Controlling chronic Pseudomonas aeruginosa infections by strategically interfering with the sensory function of SagS. Mol Microbiol. 2019;111:1211–1228. doi: 10.1111/mmi.14215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hsu J.L., Chen H.C., Peng H.L., Chang H.Y. Characterization of the histidine-containing phosphotransfer protein B-mediated multistep phosphorelay system in Pseudomonas aeruginosa PAO1. J Biol Chem. 2008;283:9933–9944. doi: 10.1074/jbc.M708836200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hrabak E., Willis D.K. The lemA gene required for pathogenicity of Pseudomonas syringae pv. syringae on bean is a member of a family of two-component regulators. J Bacteriol. 1992;174:3011–3020. doi: 10.1128/jb.174.9.3011-3020.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Laville J., Voisard C., Keel C., Maurhofer M., Défago G., Haas D. Global control in Pseudomonas fluorescens mediating antibiotic synthesis and suppression of black root rot of tobacco. Proc Natl Acad Sci Unit States Am. 1992;89:1562–1566. doi: 10.1073/pnas.89.5.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rich J.J., Kinscherf T.G., Kitten T., Willis D.K. Genetic evidence that the gacA gene encodes the cognate response regulator for the lemA sensor in Pseudomonas syringae. J Bacteriol. 1994;176:7468–7475. doi: 10.1128/jb.176.24.7468-7475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Goodman A.L., Kulasekara B., Rietsch A., Boyd D., Smith R.S., Lory S. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev Cell. 2004;7:745–754. doi: 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 75.Goodman A.L., Merighi M., Hyodo M., Ventre I., Filloux A., Lory S. Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev. 2009;23:249–259. doi: 10.1101/gad.1739009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Janssen K.H., Diaz M.R., Gode C.J., Wolfgang M.C., Yahr T.L. RsmV, a small noncoding regulatory RNA in Pseudomonas aeruginosa that sequesters RsmA and RsmF from target mRNAs. J Bacteriol. 2018;200 doi: 10.1128/JB.00277-18. e00277-00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Miller C.L., Romero M., Karna S.R., Chen T., Heeb S., Leung K.P. RsmW, Pseudomonas aeruginosa small non-coding RsmA-binding RNA upregulated in biofilm versus planktonic growth conditions. BMC Microbiol. 2016;16:1–16. doi: 10.1186/s12866-016-0771-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marden J.N., Diaz M.R., Walton W.G., Gode C.J., Betts L., Urbanowski M.L. An unusual CsrA family member operates in series with RsmA to amplify posttranscriptional responses in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2013;110:15055–15060. doi: 10.1073/pnas.1307217110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brencic A., Lory S. Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol Microbiol. 2009;72:612–632. doi: 10.1111/j.1365-2958.2009.06670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Romero M., Silistre H., Lovelock L., Wright V.J., Chan K.G., Hong K.W. Genome-wide mapping of the RNA targets of the Pseudomonas aeruginosa riboregulatory protein RsmN. Nucleic Acids Res. 2018;46:6823–6840. doi: 10.1093/nar/gky324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gebhardt M.J., Kambara T.K., Ramsey K.M., Dove S.L. Widespread targeting of nascent transcripts by RsmA in Pseudomonas aeruginosa. Proc Natl Acad Sci Unit States Am. 2020;117:10520–10529. doi: 10.1073/pnas.1917587117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ventre I., Goodman A.L., Vallet-Gely I., Vasseur P., Soscia C., Molin S. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc Natl Acad Sci U S A. 2006;103:171–176. doi: 10.1073/pnas.0507407103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chambonnier G., Roux L., Redelberger D., Fadel F., Filloux A., Sivaneson M. The hybrid histidine kinase LadS forms a multicomponent signal transduction system with the GacS/GacA two-component system in Pseudomonas aeruginosa. PLoS Genet. 2016;12 doi: 10.1371/journal.pgen.1006032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bhuwan M., Lee H.J., Peng H.L., Chang H.Y. Histidine-containing phosphotransfer protein-B (HptB) regulates swarming motility through partner-switching system in Pseudomonas aeruginosa PAO1 strain. J Biol Chem. 2012;287:1903–1914. doi: 10.1074/jbc.M111.256586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Valentini M., Laventie B.J., Moscoso J., Jenal U., Filloux A. The diguanylate cyclase HsbD intersects with the HptB regulatory cascade to control Pseudomonas aeruginosa biofilm and motility. PLoS Genet. 2016;12 doi: 10.1371/journal.pgen.1006354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bordi C., Lamy M.C., Ventre I., Termine E., Hachani A., Fillet S. Regulatory RNAs and the HptB/RetS signalling pathways fine-tune Pseudomonas aeruginosa pathogenesis. Mol Microbiol. 2010;76:1427–1443. doi: 10.1111/j.1365-2958.2010.07146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Laskowski M.A., Kazmierczak B.I. Mutational analysis of RetS, an unusual sensor kinase-response regulator hybrid required for Pseudomonas aeruginosa virulence. Infect Immun. 2006;74:4462–4473. doi: 10.1128/IAI.00575-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gupta K., Liao J., Petrova O.E., Cherny K.E., Sauer K. Elevated levels of the second messenger c-di-GMP contribute to antimicrobial resistance of Pseudomonas aeruginosa. Mol Microbiol. 2014;92:488–506. doi: 10.1111/mmi.12587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Roy A.B., Petrova O.E., Sauer K. The phosphodiesterase DipA (PA5017) is essential for Pseudomonas aeruginosa biofilm dispersion. J Bacteriol. 2012;194:2904–2915. doi: 10.1128/JB.05346-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Basu Roy A., Sauer K. Diguanylate cyclase NicD-based signalling mechanism of nutrient-induced dispersion by Pseudomonas aeruginosa. Mol Microbiol. 2014;94:771–793. doi: 10.1111/mmi.12802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dingemans J., Poudyal B., Sondermann H., Sauer K. The Yin and Yang of SagS: distinct residues in the HmsP domain of SagS independently regulate biofilm formation and biofilm drug tolerance. mSphere. 2018;3:e00442–19. doi: 10.1128/mSphere.00442-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Spoering A.L., Lewis K. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J Bacteriol. 2001;183:6746–6751. doi: 10.1128/JB.183.23.6746-6751.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ciofu O., Tolker-Nielsen T. Tolerance and resistance of Pseudomonas aeruginosa biofilms to antimicrobial agents-how P. aeruginosa can escape antibiotics. Front Microbiol. 2019;10:913. doi: 10.3389/fmicb.2019.00913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chambers J.R., Sauer K. The MerR-like regulator BrlR impairs Pseudomonas aeruginosa biofilm tolerance to colistin by repressing PhoPQ. J Bacteriol. 2013;195:4678–4688. doi: 10.1128/JB.00834-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chambers J.R., Liao J., Schurr M.J., Sauer K. BrlR from Pseudomonas aeruginosa is a c-di-GMP-responsive transcription factor. Mol Microbiol. 2014;92:471–487. doi: 10.1111/mmi.12562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liao J.L., Sauer K. The MerR-like transcriptional regulator BrlR contributes to Pseudomonas aeruginosa biofilm tolerance. J Bacteriol. 2012;194:4823–4836. doi: 10.1128/JB.00765-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bobrov A.G., Kirillina O., Perry R.D. The phosphodiesterase activity of the HmsP EAL domain is required for negative regulation of biofilm formation in Yersinia pestis. FEMS Microbiol Lett. 2005;247:123–130. doi: 10.1016/j.femsle.2005.04.036. [DOI] [PubMed] [Google Scholar]
- 98.Kirillina O., Fetherston J.D., Bobrov A.G., Abney J., Perry R.D. HmsP, a putative phosphodiesterase, and HmsT, a putative diguanylate cyclase, control Hms-dependent biofilm formation in Yersinia pestis. Mol Microbiol. 2004;54:75–88. doi: 10.1111/j.1365-2958.2004.04253.x. [DOI] [PubMed] [Google Scholar]