Abstract
Purpose
The putative presence of SARS-CoV-2 in ocular specimen puts healthcare workers at risk. We thoroughly examined conjunctival swabs and tear fluid in a large cohort of COVID‐19 patients.
Methods
A total of 243 symptomatic laboratory-confirmed COVID-19 patients were included in this observational multicenter study. Conjunctival swabs were analyzed by reverse transcription polymerase chain reaction for detection of SARS-CoV-2 RNA. Next-generation sequencing and phylogenetic analysis were performed to identify viral strains and to determine tissue tropism. Schirmer tear samples from 43 hospitalized COVID-19 patients and 25 healthy controls were analyzed by multiplex cytokine immunoassays.
Results
Viral SARS-CoV-2 RNA was detected in conjunctival swabs from 17 (7.0%) of 243 COVID-19 patients. Conjunctival samples were positive for viral SARS-CoV-2 RNA as long as 12 days after disease onset. Cycle threshold (Ct) values for conjunctival swabs (mean 34.5 ± 5.1) were significantly higher than nasopharyngeal swabs (mean 16.7 ± 3.6). No correlation between Ct values of conjunctival and nasopharyngeal swabs was observed. The majority of positive conjunctival samples were detected only once and primarily during the first visit. Next-generation sequencing analysis revealed that the virus strain found in the conjunctiva was most often identical to the one found in the nasopharynx. Tear cytokine levels IL-1β and IL-6 were elevated in COVID-19 patients compared to healthy controls.
Conclusions
Conjunctival samples that were positive for SARS-CoV-2 RNA contained the same viral strain as the nasopharynx.
Translational Relevance
The presence of SARS-CoV-2 viral RNA and elevated cytokines in tear fluid confirm the involvement of the ocular surface in COVID-19 disease.
Keywords: COVID-19, SARS-CoV-2, sequencing, tear fluid, conjunctival swab
Introduction
Coronavirus disease 2019 (COVID‐19), caused by the severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2), has rapidly become a global health issue since it originated in Wuhan, China, in December 2019.1 On March 11, 2020, the World Health Organization (WHO) declared COVID-19 a pandemic. The clinical presentation of a SARS-CoV-2 infection can range from asymptomatic infection to severe disease.1 The main clinical features of COVID‐19 are fever, cough, fatigue, anorexia, dyspnea, and myalgia.1,2 Conjunctivitis has been occasionally reported among COVID-19 symptoms.3 There are cases reported in which conjunctivitis was the only sign of COVID-19,4 whereas other reports showed that patients developed conjunctivitis later in their course of disease after hospitalization.5 The estimated prevalence of conjunctival congestion is 0.8%, as stated by the WHO report of the China Joint Mission on COVID‐196 (summarizing 55.924 laboratory‐confirmed cases) and by Guan et al.2 in a large cohort of 1099 patients.
Human-to-human transmission occurs mainly through respiratory droplets, but other routes are under investigation because SARS-CoV-2 has been detected in several body fluids, including blood, stool, and saliva.7 SARS-CoV-2 has also been found in tear fluid, conjunctival secretions, and corneal tissue8,9 of COVID-19 patients. A recent meta-analysis including 252 COVID-19 patients identified 11 patients (4.4%) with a positive tear–polymerase chain reaction (PCR).10 The prevalence of positive tear-PCR cases ranges from 3%11 to 57.1%,12 whereas other studies fail to detect viral RNA in tear samples.13–16 This considerable inconsistency may be attributed to low sample size and variability in sample collection, handling, and testing methods. The main purpose of this study was to investigate the prevalence of RNA of SARS‐CoV‐2 in conjunctival swabs in a large cohort (n = 243) of COVID‐19 patients, during the second wave of the pandemic, in the Netherlands. In addition, we characterized the phylogenetic relationship between conjunctival and nasopharyngeal samples and determined tear fluid cytokine concentrations.
Methods
Study Subjects
A total of 244 laboratory‐confirmed COVID‐19 patients were enrolled in this study. Patients were recruited from the Maastricht University Medical Center, Maastricht,17 and the Zuyderland Medical Center, Heerlen, the Netherlands. Healthcare workers were recruited from the Corona employee outpatient testing facility at the Maastricht University Medical Center. Ethical approval was obtained from the medical ethics committee of the Maastricht University Medical Center. The study followed the tenets of the Declaration of Helsinki 2013. Written informed consent was obtained from all subjects. Inclusion criterion was age ≥18 years; pregnant women were excluded from participation. Samples from one intensive care unit (ICU)–hospitalized patient were omitted because of unreliable sample handling. All subjects were symptomatic for COVID-19. Most patients were hospitalized for COVID-19, although some patients were hospitalized originally for other reasons (e.g., an urgent surgical intervention) and became infected during their stay in the hospital.
Specimen Sampling
Respiratory samples were collected from the nasopharynx with disposable sterile polyamide-tipped swabs.18 Conjunctival samples were collected by the tear/conjunctival swab technique.19 Eyelids were everted, and samples were obtained by gently sweeping the inferior conjunctival fornices of both eyes with the same disposable sampling swab without topical anesthesia. Samples were placed into 3.5 mL viral transport medium and temporally stored at −30°C until analysis followed by storage at −80°C.
RT-PCR Analysis
Viral RNA was extracted from 90 µL viral transport medium with the MagNA Pure 96 DNA and Viral NA Small Volume Kit using the Pathogen Universal 200 system protocol (MagNA Pure 96 system; Roche Diagnostics, Basel, Switzerland), eluted in 50 µL elution buffer and diluted with 50 µL water. Reverse transcription (RT) PCR was carried out on a Quantstudio 5 system (Applied Biosystems, Foster City, CA, USA) using a validated multiplex in-house developed assay targeting the envelop (E) and nucleocapsid (N1) gene.20,21 The experimental set-up was validated to detect SARS-CoV-2 in conjunctival specimen. A sample process control (SPC) was spiked into all samples before extraction to verify correct sample handling. A positive test control and negative test control were processed throughout the process for each run to evaluate run validity. Final reaction volume contained 5 µL 4x Taqpath 1-step RT-qPCR MasterMix (Applied Biosystems), 5 µL primer/probe mix, and 10 µL sample. Cycling conditions consisted of uracil-N-glycosylase incubation at 25°C for two minutes, RT incubation at 50°C for 30 minutes, enzyme activation at 95°C for two minutes, and 42 cycles of denaturation at 94°C for three seconds and annealing/extension at 60°C for 30 seconds. Cycle threshold (Ct) values ≤ 42.0 are interpreted as positive for SARS-CoV-2 RNA. Negative results are reported as undetermined. Samples found positive by the in-house PCR were repeated with the commercially available GeneXpert assay (Xpert Xpress SARS-CoV-2 [Cepheid, Sunnyvale, CA, USA]) targeting the E and N2 gene. Detection of both target genes or the N2 gene alone within 45 cycles is interpreted positive. Detection of the E gene alone within 45 cycles is interpreted presumptive positive. We considered a conjunctival sample positive when at least one target gene by both in-house and commercially available GeneXpert assay was detectable. Conjunctival samples with positive signals by in-house RT-PCR, but not confirmed by GeneXpert assay, are listed in Supplementary Table S1. Standards for COVID-19 PCR testing can differ between countries and continents. The Dutch National Control of Infectious Diseases guidelines were followed at the time of testing.
Next-generation Sequencing With Phylogenetic Analysis
The PCR tiling of COVID-19 virus with Native Barcoding Expansion 96 (EXP-NBD-196) protocol (version PTCN_9103_v109_revG_13Jul2020, last update 22-01-2021) from Oxford Nanopore Technologies was followed with slight modifications to increase the amount of purified sample for sequencing, as written below. Primers for 89 overlapping amplicons spanning the entire genome of SARS-CoV-2 were used to generate 500 base-pair amplicons with a 75-base-pair overlap,22 and PCR consisted of 37 cycles. Samples were multiplexed using Native Barcoding Expansion 96 kit (EXP-NBD-196; Oxford Nanopore Technologies, Oxford, UK). One hundred nanograms of amplicons were end-repaired and dA-tailed at room temperature for 10 min, followed by a purification step. Barcoded libraries were prepared using Ligation Sequencing Kit (SQK-LSK-109; Oxford Nanopore Technologies). Ligation of native barcodes was performed with 2 µL of end-prepped DNA, after which the ligation of native barcodes was performed for 40 minutes at room temperature. Barcoded libraries were again purified, after which barcoded libraries were pooled and purified 1:1 using AMPure XP beads. The first wash step was performed with 80% EtOH, followed by adapter ligation using 65 µL of pooled barcoded sample, 5 µL adapter mix, 20 µL ligation reaction buffer, and 10 µL of DNA ligase. Final clean-up of the library was carried out with 80 µL AMPure beads and 250 µL Short Fragment Buffer. A total amount of 15 ng prepared barcoded library was loaded onto a MinION R9.4.1 Flow Cell (Oxford Nanopore Technologies) and sequenced with MinION Mk1B device (Oxford Nanopore Technologies) for approximately 24 hours. The resulting reads were base called with an in-house pipeline, based on the ARTIC pipeline, using Guppy v4.2.2 and aligned to the SARS-CoV-2 reference genome (GenBank accession numbers MN908947 and LR757998). Genome quality control was performed using Nextclade, phylogenetic analysis was carried out utilizing Nextstrain pipelines, phylogenetic tree visualization was generated in Auspice, and Pangolin (Phylogenetic Assignment of Named Global Outbreak Lineages) lineages were assigned using the Pangolin COVID-19 Lineage Assigner web application.
Tear Cytokine Analysis
Tear samples for cytokine analysis were collected from 43 hospitalized COVID-19 patients and 25 healthy controls (collected in 2018–2019 during the screening visit of the Ocular Coil and Drug Delivery and Comfort Trial).23 Tear samples were collected from the left eye using Schirmer tear strips (TEAR strips; Contacare Ophthalmics & Diagnostics, Panjiva, India) without topical anesthesia. Immediately after sampling, samples were stored at −80°C. Tear proteins were extracted in 150 µL PBS + 25X Complete protease inhibitor cocktail (Roche Diagnostics). Tear cytokine (interferon (IFN)-γ, interleukin (IL)-1β, IL-2, IL-4, IL-6, IL-8, IL-10, IL-12p70, IL-13, and tumor necrosis factor (TNF)-α) concentrations were measured in duplicate using the proinflammatory panel I 10-plex assay (Meso Scale Discovery; Meso Scale Diagnostics, Rockville, MD, USA). Values below the detection limit were excluded.
Statistical Analysis
Statistical analyses were performed using GraphPad Prism version 9 (GraphPad, San Diego, CA, USA). The dot plot was created using Microsoft Excel 2016. Mean Ct values were calculated using the lowest Ct value of both target genes by in-house PCR. The difference between conjunctival and nasopharyngeal Ct values was analyzed using paired t-testing, and correlations were assessed using Pearson's correlation analysis. The difference in tear cytokine levels between groups was tested using the Mann-Whitney test, and correlations were tested using Spearman's correlation analysis.
Results
Patient Demographics
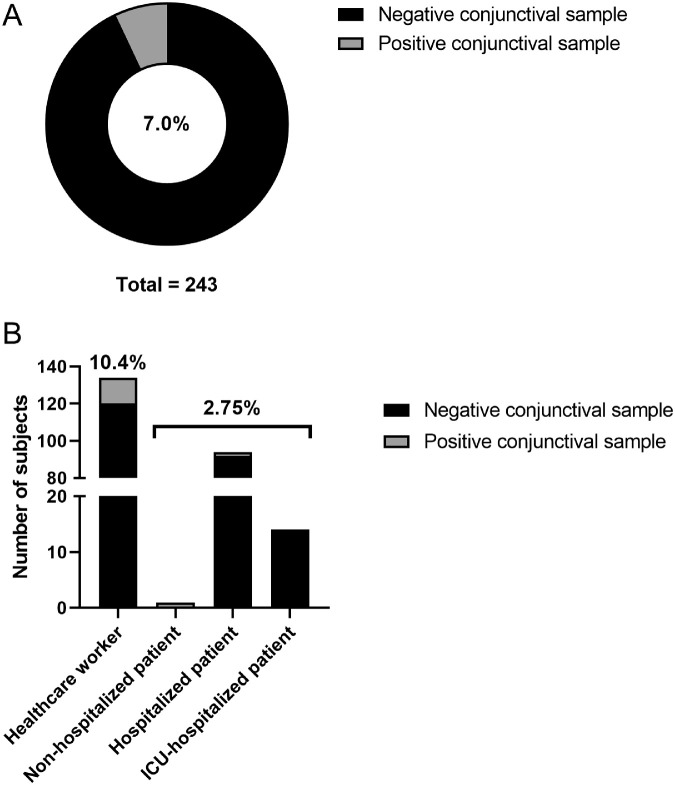
Between August and November 2020, a total of 487 samples were collected from 243 subjects with laboratory-confirmed SARS-CoV-2 infection. The mean age of all subjects was 50.9 ± 18.3 years (range, 20–93) and 44% were male. The clinical spectrum of SARS-CoV-2 infection ranged from mild symptomatic infection to critical illness. The majority of subjects (n = 134 [55%]) were healthcare workers, 108 (44%) patients required hospitalization, of whom 14 patients were admitted to the ICU. Eleven patients (4.5%) died. The prevalence of positive conjunctival samples among subjects was 7.0% (17/243) (Fig. 1A). Subjects with a positive conjunctival sample included 14 healthcare workers (10.4% of all healthcare workers) and three patients (2.7% of all patients), specifically one non-hospitalized patient and two hospitalized patients (Fig. 1B, Table 1). One patient (case 12) was hospitalized for COVID-19 disease and died on hospitalization day 8. Another patient (case 14) was originally hospitalized for an urgent surgical intervention but became infected during his stay in the hospital. One healthcare worker was admitted to the hospital nine days after the first positive nasopharyngeal and conjunctival sample for persistent symptoms of fever and cough. None of the subjects with a positive conjunctival sample reported ocular symptoms (such as pain or conjunctival secretions) among COVID-19 symptoms, except for case 15 who described tearing eyes two days before conjunctival sampling. Although most subjects did not report any comorbidities, three patients had an underlying respiratory disease.
Figure 1.
Prevalence (%) of positive conjunctival samples in (A) all subjects and (B) per subgroup.
Table 1.
Laboratory Findings and Clinical Characteristics of Patients With a Positive Conjunctival Sample (Confirmed By Two PCR Assays)
| Case 1 | Case 2 | Case 3 | Case 4 | Case 5 | Case 6 | Case 7 | Case 8 | Case 9 | Case 10 | Case 11 | Case 12 | Case 13 | Case 14 | Case 15 | Case 16 | Case 17 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Demographics | |||||||||||||||||
| Age | 54 | 50 | 34 | 28 | 50 | 42 | 26 | 55 | 61 | 28 | 28 | 80 | 30 | 74 | 51 | 49 | 53 |
| Sex | F | F | F | F | F | F | M | F | M | F | F | M | F | M | F | F | F |
| Group | Non-hospitalized patient | Health care worker | Health care worker | Health care worker | Health care worker | Health care worker | Health care worker | Health care worker | Health care worker | Health care worker | Health care worker | Hospitalized patient | Health care worker | Hospitalized patient | Health care worker | Health care worker | Health care worker |
| Hospitalization days | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8a | 0 | 36 | 0 | 0 | 5b |
| Hospitalization days at ICU | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
| Laboratory specimen results | |||||||||||||||||
| Nasopharynx Ct values (in-house PCR) | |||||||||||||||||
| E gene | 14.9 | 19.4 | 14.0 | 13.8 | 16.6 | 17.3 | 15.8 | 19.2 | 24.3 | 20.0 | 24.3 | 13.8 | 18.1 | 12.6 | 18.7 | 13.7 | 12.6 |
| N1 gene | 14.7 | 18.6 | 13.4 | 13.8 | 16.6 | 17.2 | 15.5 | 19.0 | 24.5 | 19.6 | 23.6 | 13.6 | 18.2 | 12.6 | 18.2 | 13.1 | 12.1 |
| Conjunctival Ct values (in-house PCR) | |||||||||||||||||
| E gene | 33.1 | 36.7 | 30.1 | 36.3 | 31.5 | 33.1 | UD | 36.0 | 32.9 | 40.1 | 29.6 | 22.6 | 31.4 | UD | UD | 35.8 | 42.0 |
| N1 gene | 33.7 | 42.0 | 30.3 | 37.4 | 31.1 | 33.3 | 38.1 | 42.0 | 32.7 | UD | 28.6 | 23.0 | 32.0 | 39.6 | 42.0 | 33.0 | UD |
| Conjunctival Ct values (GeneXpert PCR) | |||||||||||||||||
| E gene | 35.0 | 44.4 | 36.6 | 35.4 | 33.1 | 33.1 | 38.7 | 37.5 | 30.7 | 42.3 | 29.6 | 22.7 | 31.6 | UD | UD | 35.0 | UD |
| N2 gene | 37.5 | 44.6 | 39.2 | 38.9 | 34.6 | 35.9 | 42.7 | 40.2 | 31.1 | UD | 30.9 | 24.4 | 34.1 | 40.3 | 42.8 | 36.3 | 42.0 |
Table 1.
Continued
| Case 1 | Case 2 | Case 3 | Case 4 | Case 5 | Case 6 | Case 7 | Case 8 | Case 9 | Case 10 | Case 11 | Case 12 | Case 13 | Case 14 | Case 15 | Case 16 | Case 17 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| COVID symptoms | Fever, fatigue, anorexia, dyspnea, myalgia, gastrointestinal symptoms | Cough, cold, sore throat, headache, myalgia, feeling ill | Fever, loss of smell and taste, mouth ulcers | Sore throat, headache, feeling ill | Cough, cold, sore throat, headache | Cough, fever, cold, headache | Cough, cold, headache, loss of smell | Cold, loss of smell and taste, myalgia, mouth ulcers, back pain | Cough, fever, cold, sore throat, headache, feeling ill | Cough, cold, headache, dyspnea, feeling ill, extreme fatigue | Cough, cold, sore throat, loss of smell and taste, myalgia, feeling ill, extreme fatigue | Cough, fever, dyspnea | Cough, cold, sore throat, flu feeling | Fatigue, decreased sense of taste, gastrointestinal symptoms, ground-glass opacity bilateral lungs | Cough, fever, cold, sore throat, headache, tearing eyes | Cough, cold, headache, loss of smell and taste, feeling ill, extreme fatigue, vomit | Cough, cold, headache, feeling ill |
| Medical history | Chronic obstructive pulmonary disease | Kidney disease | / | / | Kidney disease | / | / | / | Cardiovascular disease | Asthma | / | Heart condition, lung emphysema | / | Hospitalized for splenectomy and pancreatic tail resection for suspected malignancy, melanoma, chronic pancreatitis | / | / | / |
F, female; M, male; UD, undetectable.
Died on hospitalization day 8.
Admitted to the hospital 9 days after positive conjunctival swab.
Prevalence of SARS-CoV-2 in Conjunctival Samples
We identified 17 conjunctival samples that were positive for SARS-CoV-2 RNA (Table 1). The mean age of patients with positive conjunctival samples was 46.7 ± 16.2 (range, 26–80) years. Females were overrepresented (13/17, 76%). The vast majority (88%) were not hospitalized at the time of conjunctival sample positivity.
Ct Values in Conjunctival Samples
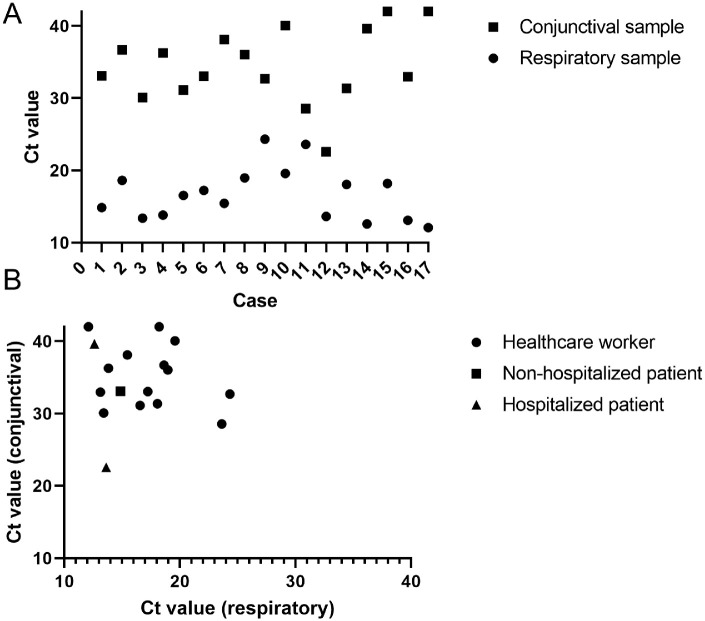
Figure 2A presents the Ct values per case. In all cases, Ct values of the conjunctival swabs (mean 34.5 ± 5.1; range, 22.6–42.0) were significantly higher (indicating lower viral load) than nasopharyngeal swabs (mean 16.7 ± 3.6; range, 12.1–24.3) (P < 0.001) (Fig. 2A). Although the lowest conjunctival Ct value (22.6) was detected in a patient (case 12) with a low nasopharyngeal Ct value (13.1), no correlation (r = −0.10, P = 0.70) of Ct values between conjunctival and nasopharyngeal swabs was observed (Fig. 2B).
Figure 2.
Ct values of patients with a positive conjunctival sample. (A) Ct value pattern per case. (B) Correlation between Ct value of conjunctival and nasopharyngeal swab.
Relationship Between Positive Conjunctival Sample and Course of the Disease
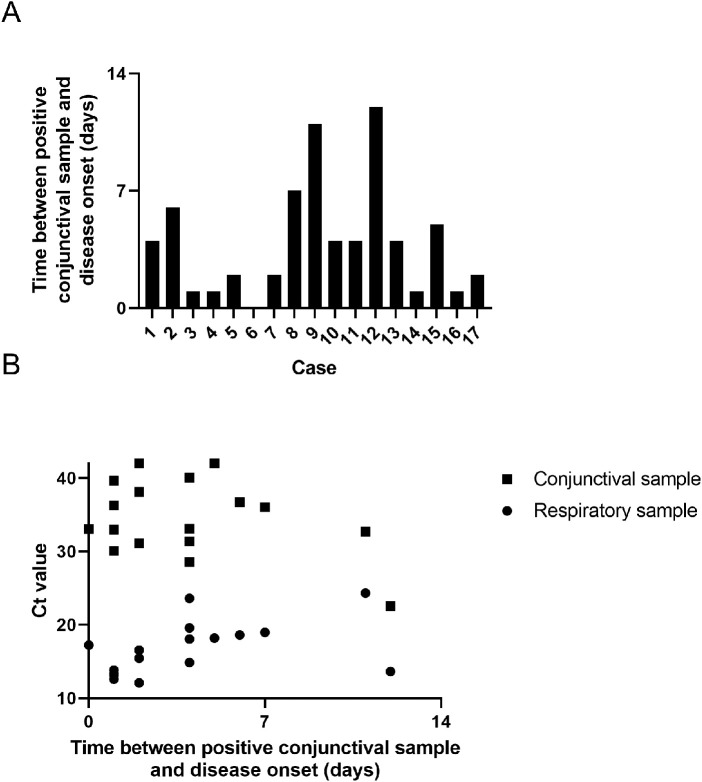
Conjunctival sample positivity was observed early in the course of the disease. In approximately half of the subjects (8/17), the positive conjunctival sample was detected within the first two days of the disease. The vast majority (15/17) of positive conjunctival samples was identified during the first week, whereas two cases (case 9 and 12) only displayed positivity on day 11 and 12, respectively (Fig. 3A). In contrast to the moderate positive correlation (r = 0.46, P = 0.06) between nasopharyngeal Ct value and days since disease onset, a negative correlation (r = −0.37, P = 0.17) was observed for conjunctival Ct values (Fig. 3B).
Figure 3.
Relationship between positive conjunctival sample and course of the disease. (A) Time between positive conjunctival sample disease onset per case. (B) Correlation between conjunctival and nasopharyngeal Ct values and time between positive conjunctival sample disease onset.
Consecutive Analyses of Subjects With a Positive Conjunctival Sample
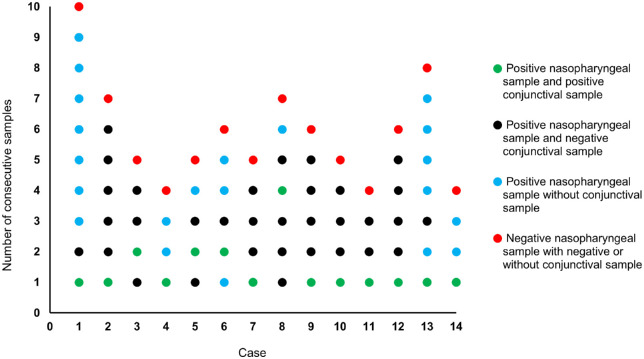
Healthcare workers with a positive nasopharyngeal sample were re-tested approximately every three days until a negative test result (Fig. 4, red dots). During each follow-up visit, they could voluntarily participate in this study. The visits where they did not participate are colored in blue (Fig. 4). Consecutive samples were available from 14 healthcare workers with a positive conjunctival sample. Figure 4 shows that most positive conjunctival samples (green dots) were found during the first visit. In three cases (cases 3, 5, and 6) the positive conjunctival sample occurred in the second follow-up visit. One case (case 8) showed a positive conjunctival sample only in the fourth follow-up visit. None of the cases displayed more than one visit with a positive conjunctival sample.
Figure 4.
Consecutive analysis of 13 healthcare workers with a positive conjunctival sample. The number of dots represents the number of follow-up visits per case. Green dots indicate a follow-up visit with a positive nasopharyngeal sample and positive conjunctival sample, black dots indicate a follow-up visit with a positive nasopharyngeal sample and negative conjunctival sample, blue dots represent a follow-up visit with a positive nasopharyngeal sample but without conjunctival sample, and red dots present a follow-up visit with a negative nasopharyngeal sample and with negative or without conjunctival sample.
Phylogenetic Analysis of Conjunctival and Nasopharyngeal Samples
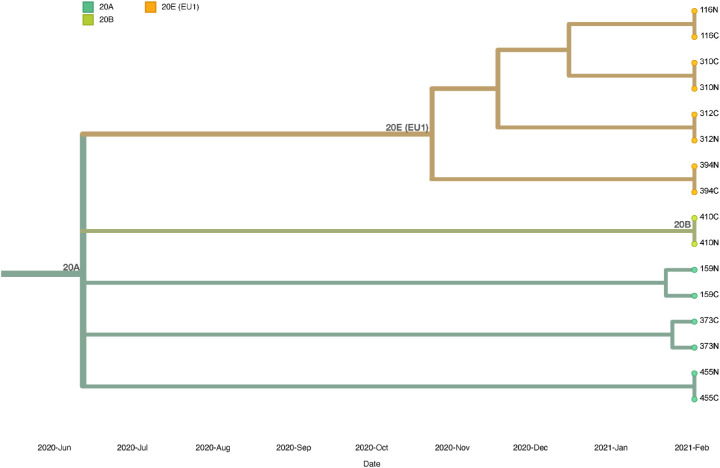
Figure 5 shows the genetic relationship between matched conjunctival and nasopharyngeal samples of eight individuals with Ct values ≤ 33. All samples were grouped in different subclusters of clade 20A, 20B and 20E (EU1). All lineages that were identified belonged to lineage B.1, of these 37 (42%) belonged to lineage B.1.221, 10 (11.3%) to lineage B.1.177.50, and nine (10.2%) to lineage B.1.160.
Figure 5.
Phylogenetic analysis of conjunctival samples (C) and nasopharyngeal samples (N) from eight matched individuals.
Conjunctival and nasopharyngeal samples of subjects 116, 310, 312, and 394 were grouped in clade 20E (EU1) and carried no mutations between matched samples. Identified lineages from these samples belonged to lineages B.1.177, B.1.177.50, W.2 (alias B.1.177.53.2), and B.1.177.36, respectively. Conjunctival and nasopharyngeal samples of subject 410 were grouped in clade 20B without mutations, lineage AP.1 (alias B.1.1.70.1) Both samples of subject 159 were placed in clade 20A; the conjunctival sample carried no mutations, whereas the nasopharyngeal sample carried nucleotide mutations (A1629T, T7767C, G11557T) and amino acid mutations in ORF1a, also known as the replicase/transcriptase gene (Q455L, I2501T, E3764D). The nasopharyngeal sample of subject 159 was included in lineage B.1.258, whereas the analysis for the conjunctival sample failed. The nasopharyngeal sample of subject 373, placed in clade 20A, carried no differences in nucleotide and amino acid mutations, whereas the conjunctival sample carried two nucleotide mutations (C19017T, A25255G), both samples were allied to lineage B.1.160. Samples from subject 455 were also placed in clade 20A, without mutations between matched samples, and could be affiliated to lineage B.1.221.
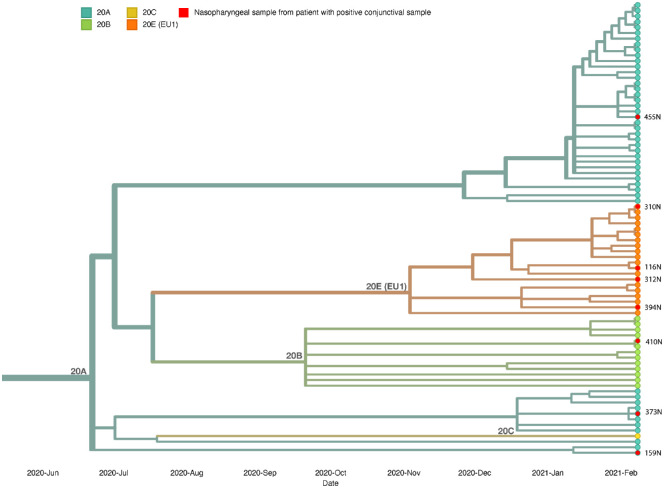
In Figure 6 the phylogenetic tree of 83 nasopharyngeal samples from eight patients with a positive conjunctival sample and 75 patients with a negative conjunctival sample is shown. No major differences were observed between variants in nasopharyngeal samples All samples were grouped in clades 20A, 20B, 20C, and 20E (EU1), lineage B.1, in different subclusters. From the eight nasopharyngeal samples from subjects with a positive conjunctival swab, three samples (37.5%) were classified in clade 20A, one sample (12.5%) in clade 20B, and four samples (50.0%) in clade 20E (EU1). The four samples that were classified in clade 20E can be traced back to lineage B.1.177. In the group with nasopharyngeal samples from subjects with negative conjunctival swabs, 44 (60.2%) samples were grouped in clades 20A, 12 (16.4%) in clade 20B, 1 (0.01%) in clade 20C, and 16 (21.9%) in clade 20E (EU1). The most common lineages from these samples correspond to lineages B.1.221 (49.3%), B.1.177 (19.2%), and B.1.160 (9.6%). These results are consistent with the circulating SARS-CoV-2 variants at the time of sample collection.
Figure 6.
Phylogenetic analysis of nasopharyngeal samples from subjects positive in conjunctival swabs versus subjects negative in conjunctival swabs. Nasopharyngeal samples from subjects 158 and 458 were excluded because of too many missing nucleotides.
Tear Cytokine Analysis in COVID-19 Patients
The concentration of ten proinflammatory cytokines in tear fluid from COVID-19 patients was compared to those from healthy controls (Table 2). Tear concentrations of interleukins IL1b and IL-6 were significantly higher in COVID-19 patients (P = 0.026 and P < 0.0001, respectively), whereas IL-13 and IL-8 concentrations were higher in healthy controls (P = 0.0115 and P < 0.0001, respectively) (Fig. 7). Concentrations of IL-12p70 and IL-4 were undetectable. In this subset of hospitalized COVID-19 patients, one case displayed a positive conjunctival sample by both PCR assays and one other case by a single PCR assay. However, their individual cytokine values were not elevated but rather below the mean values.
Table 2.
Tear Cytokine Concentrations (pg/mL) in COVID-19 Patients and Healthy Controls
| IFN-γ | IL10 | IL13 | IL1β | IL2 | IL6 | IL8 | TNFα | |
|---|---|---|---|---|---|---|---|---|
| COVID-19 patients | ||||||||
| Mean ± SD | 3.56 ± 3.63 | 0.17 ± 0.21 | 3.57 ± 2.18 | 8.40 ± 17.86 | 0.26 ± 0.19 | 16.93 ± 88.10 | 587.98 ± 1820.26 | 3.63 ± 9.44 |
| Range (min-max) | 0.80–7.68 | 0.08–0.76 | 2.11–13.11 | 0.58–89.35 | 0.13–0.78 | 0.20–530.50 | 23.99–11806.71 | 0.29–28.81 |
| Healthy controls | ||||||||
| Mean ± SD | 2.98 ± 2.81 | 0.16 ± 0.10 | 4.85 ± 2.31 | 3.78 ± 6.07 | 0.18 ± 0.04 | 7.21 ± 7.92 | 827.13 ± 939.91 | 0.63 ± 0.51 |
| Range (min-max) | 0.74–6.88 | 0.06–0.43 | 2.11–12.04 | 0.64–25.07 | 0.14–0.26 | 0.83–35.17 | 119.84–4930.47 | 0.18–2.27 |
| P value | 0.257 | 0.265 | 0.0115 | 0.026 | 0.656 | <0.0001 | <0.0001 | 0.809 |
Figure 7.
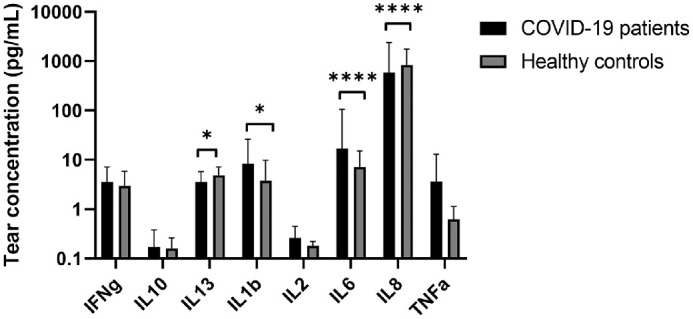
Tear cytokine analysis of COVID-19 patients and healthy controls.
Discussion
This observational cohort study characterized the presence of SARS‐CoV‐2 RNA in conjunctival swabs in a large cohort of COVID‐19 patients. Before the start of the study, the experimental set-up was validated to detect SARS-CoV-2 in conjunctival specimen.24 Samples found positive by the in-house PCR were repeated with the commercially available GeneXpert assay. Within our dataset, six samples were found positive by the in-house PCR and could not be confirmed by the GeneXpert assay. These samples were, however, not considered positive samples. Another sample tested positive for the E gene but not the N1 gene with both the in-house PCR (Ct 40.1) and the GeneXpert assay (Ct 42.3). However, the nasopharyngeal sample of this patient was positive for both genes (E gene Ct 20.0; N1 gene Ct 19.6). Based on that result we conclude that conjunctival sample positivity of that sample was most likely due to SARS-CoV-2 and not another coronavirus.
Our study population covered a broad range of COVID-19 patients, from mild symptomatic disease to severely affected and from home illness to hospitalized at the ICU. It did not include asymptomatic laboratory-confirmed individuals. We observed a prevalence of 7.0% of SARS‐CoV‐2 RNA in conjunctival swabs that was slightly higher than the range of 3% to 5% reported in a systematic review of 11 small cohort studies10 and a meta-analysis of seven studies.25 Remarkably, in the current study population, positivity did not relate to disease severity. Firstly, because most subjects with a positive conjunctival sample in our cohort were healthcare workers with mild COVID-19 symptoms not requiring hospitalization. Secondly, because 13 out of 17 subjects with a positive conjunctival sample were female, while male patients usually present a more severe form of the disease26,27 (as demonstrated by our group of hospitalized patients, 69 males versus 39 females). Among the four male subjects with positive conjunctival sample, two of them were hospitalized, of whom one died after hospitalization day 8. Finally, because the majority of conjunctival-positive subjects (10/17) did not carry underlying comorbidities.
Our results show significantly higher Ct values (corresponding to lower viral loads) in conjunctival samples compared to nasopharyngeal samples. The same trend has been observed in other nonrespiratory body samples, such as feces or blood.7 We postulate that this is partly contributed to removal of the virus from the ocular and conjunctival space by tearing, whereas the viral load in the pharynx typically grows during the first week after the onset of symptoms and remains high for another week.28,29 The viral load of the conjunctival samples was relatively low, as indicated by high Ct values and Ct values equal to or close to the detection limit. For this reason, a few conjunctival samples showed positivity for SARS-CoV-2 RNA in one target gene, but not in the other gene. A second RT-PCR assay was used to confirm these samples. We found no correlation between Ct values of conjunctival and respiratory samples for individual subjects indicating that a high viral load in the conjunctiva does not always co-occur with a high respiratory viral load. The majority of positive conjunctival samples were detected early (<7 days) in the course of the disease. There were no significant correlations between the time of disease onset in individual patients and Ct values. Nevertheless, although nasopharyngeal samples display higher Ct values (and thus lower viral loads) at the end of the disease (positive correlation), as shown in our study cohort and other reports,30 the opposite trend (negative correlation) for conjunctival samples suggests that Ct values decrease (and thus the viral load increases) later on in the course of the disease. Two cases displayed conjunctival sample positivity only at day 11 and 12 after disease onset. The majority (15/18) of patients with a positive conjunctival sample were healthcare workers. This may be due to the fact that more healthcare workers than patients were included (134 vs. 110), and because healthcare workers were tested earlier in the disease course because they had easy access to the employee testing facility. The majority of positive conjunctival samples were detected during the first visit, which is the closest to disease onset. However, four subjects tested negative in the conjunctival sample during the first visit (with positive nasopharyngeal sample) and only tested positive during the second or even fourth visit. Therefore it cannot be concluded that when subjects test negative during the first visit, they will remain negative during the course of their disease. All subjects with available follow-up samples tested positive in the conjunctival samples only once, whereas they tested up to nine times positive in the nasopharynx. These results are consistent with an animal (rhesus macaque) study that could detect the virus in ocular swabs only once (at day 1) after ocular conjunctival inoculation.31 However, Colavita and co-workers32 were able to detect viral RNA in six subsequent ocular samples from a single patient with severe bilateral conjunctivitis.
Phylogenetic analysis showed no variability in virus variants between matched conjunctival and nasopharyngeal samples in individual subjects. Intra-host variation was observed in one subject with amino acid mutations in ORF1a, the replicase/transcriptase gene in which mutations are detected at high frequency.33–35 Although nucleotide mutations were observed in one conjunctival sample, further investigation is required to determine whether active viral replication accounts for this result. Hui et al.36 observed that SARS-CoV-2 replicates better than other human coronaviruses in human conjunctival explant cultures. Indications of tissue tropism could not be identified in nasopharyngeal samples from subjects with positive conjunctival swabs versus subjects with negative conjunctival swabs. These findings suggest there is no specific SARS-CoV-2 variant responsible for conjunctival infections and support the theory that patients infect themselves through, for example, hand-eye contact.
We observed significantly higher tear cytokine levels (IL-1β and IL-6) in hospitalized COVID-19 patients compared to healthy controls. These cytokines, which are mediators of the acute phase of inflammation, have been shown to be elevated in serum samples of severe cases of COVID-19.37–39 Moreover, IL-1 and IL-6 have been proposed as anti-cytokine therapy for mitigating against the hyperinflammation.40 A previous study identified other tear cytokines (IL-9, IL-15, granulocyte colony-stimulating factor (G-CSF), granulocyte-macrophage colony-stimulating factor (GM-CSF), IFN-γ, platelet derived growth factor (PDGF), and vascular endothelial growth factor (VEGF)) that were elevated in COVID-19 patients.41 These results suggest that the hyperinflammation of the body is also reflected in peripheral parts of the body such as the ocular surface. Our results also show that a positive conjunctival sample (by RT-PCR) is not always associated with elevated tear cytokines. Similarly, COVID-19 patients with remarkably high tear cytokine concentrations do not always have detectable levels of viral RNA. The (long-term) effect of COVID-19–associated elevated tear cytokines on the ocular surface awaits clarification.
The occurrence of viral RNA in conjunctival swabs raises questions about its origin. SARS-CoV-2 is primarily spread through respiratory smaller particles (aerosols) and larger particles (droplets) that are released when an infected person coughs, sneezes, or speaks.42 Those infectious particles could land on the patient's own ocular surface and subsequently result in a positive conjunctival swab.43 Second, contact transmission involving direct or indirect contact with a contaminated object or surface may also result in a positive conjunctival swab. Hand-eye contact typically occurs during eye rubbing, (emotional) crying, eye drop administration, contact lenses insertion, or when putting on make-up. Thirdly, Azzolini et al.12 speculate that the virus may diffuse in tear fluid from the lacrimal glands because of systemic viremia. Although these explanations are speculative, direct contagion from airborne droplets seems to be the most likely theory.12 Despite viral RNA positivity in tear fluid and induced infection by conjunctival inoculation in rhesus macaques,31 the conjunctiva does not seem a common route of transmission.44–46
Conjunctivitis has been occasionally reported among COVID-19 symptoms. A recent meta-analysis showed that the overall rate of conjunctivitis was 3% in severe and 0.7% and non-severe COVID‐19 patients.47 Initially, many positive tear-PCR cases in the literature were accompanied with conjunctivitis,8 suggesting that positivity likely resulted from the virus in the exudation of conjunctivitis.10,48 However, more recent reports showed that conjunctivitis does not seem to be a prerequisite for a positive conjunctival swab,45 because these studies found no difference in number of positive swabs between patients with or without conjunctivitis49 or identified a positive tear swab in a severely affected patient without conjunctivitis.11 This may suggest that viral RNA is also present in the tear film in absence of conjunctival secretions (typically composed of degenerating epithelial cells and proteinaceous secretions from conjunctival glands).
One limitation of this study was the lack of asymptomatic patients with laboratory-confirmed SARS-CoV-2 infection. Knowledge of the prevalence of positive conjunctival swabs in these individuals is needed because these people will present themselves at the general practitioner or hospital, whereas the visit or surgery of symptomatic COVID-19 patients will generally be postponed.
Supplementary Material
Acknowledgments
The authors thank Roel Oliver and Rosalien Hoedemaker from the Corona employee outpatient testing facility of the Maastricht University Medical Center, Maastricht, the Netherlands. We thank Marjo vande Waarenburg from the Department of Internal Medicine of the Maastricht University for her help with the MesoScale Discovery platform. Finally, we greatly thank Brian van der Veer and Erik Beuken for their assistance with the next-generation sequencing analysis.
Disclosure: M. Gijs, None; J.M.J. Veugen, None; P.F.G. Wolffs, None; P.H.M. Savelkoul, None; J. Tas, None; B.C.T. van Bussel, None; M.D. de Kruif, None; R.M.A. Henry, None; C.A.B. Webers, None; M.M. Dickman, None; R.M.M.A. Nuijts, None
References
- 1. Organization WH. Clinical management of COVID-19: interim guidance, 27 May 2020. Geneva: World Health Organization; 2020. [Google Scholar]
- 2. Guan WJ, Ni ZY, Hu Y, et al.. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020; 382: 1708–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cheema M, Aghazadeh H, Nazarali S, et al.. Keratoconjunctivitis as the initial medical presentation of the novel coronavirus disease 2019 (COVID-19). Can J Ophthalmol. 2020; 55(4): e125–e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Scalinci SZ, Trovato Battagliola E.. Conjunctivitis can be the only presenting sign and symptom of COVID-19. IDCases. 2020; 20: e00774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Danthuluri V, Grant MB.. Update and recommendations for ocular manifestations of COVID-19 in adults and children: a narrative review. Ophthalmol Ther. 2020; 9: 853–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gomes C. Report of the WHO-China Joint Mission on Coronavirus Disease 2019 (COVID-19). Brazil J Implantol Health Sci. 2020; 2(3). [Google Scholar]
- 7. Wang W, Xu Y, Gao R, et al.. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020; 323: 1843–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yan Y, Diao B, Liu Y, Zhang W, Wang G, Chen X.. Severe acute respiratory syndrome coronavirus 2 nucleocapsid protein in the ocular tissues of a patient previously infected with coronavirus disease 2019. JAMA Ophthalmol. 2021; 138: 1201–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sawant OB, Singh S, Wright RE 3rd, et al.. Prevalence of SARS-CoV-2 in human post-mortem ocular tissues. Ocul Surf. 2021; 19: 322–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Aiello F, Gallo Afflitto G, Mancino R, et al.. Coronavirus disease 2019 (SARS-CoV-2) and colonization of ocular tissues and secretions: a systematic review. Eye (Lond). 2020; 34: 1206–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liang L, Wu P.. There may be virus in conjunctival secretion of patients with COVID-19. Acta Ophthalmol. 2020; 98(3): 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Azzolini C, Donati S, Premi E, et al.. SARS-CoV-2 on Ocular Surfaces in a Cohort of Patients With COVID-19 From the Lombardy Region, Italy. JAMA Ophthalmol. 2021; 139: 956–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pirraglia MP, Ceccarelli G, Cerini A, et al.. Ocular findings and retinal involvement in COVID-19 pneumonia patients: A cross-sectional study in an Italian referral centre [published online ahead of print October 15, 2020]. Research Square, 10.21203/rs.3.rs-48240/v2. [DOI] [Google Scholar]
- 14. Seah IYJ, Anderson DE, Kang AEZ, et al.. Assessing viral shedding and infectivity of tears in coronavirus disease 2019 (COVID-19) patients. Ophthalmology. 2020; 127: 977–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Meduri A, Oliverio GW, Mancuso G, et al.. Ocular surface manifestation of COVID-19 and tear film analysis. Sci Rep. 2020; 10(1): 20178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kuo IC, Mostafa HH.. Detection of SARS-CoV-2 RNA in the corneal epithelium of a patient after recovery from COVID-19. Am J Ophthalmol Case Rep. 2021; 22: 101074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tas J, van Gassel RJJ, Heines SJH, et al.. Serial measurements in COVID-19-induced acute respiratory disease to unravel heterogeneity of the disease course: design of the Maastricht Intensive Care COVID cohort (MaastrICCht). BMJ Open. 2020; 10(9): e040175–e040175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Garnett L, Bello A, Tran KN, et al.. Comparison analysis of different swabs and transport mediums suitable for SARS-CoV-2 testing following shortages. J Virol Methods. 2020; 285: 113947–113947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Arora R, Goel R, Kumar S, et al.. Evaluation of SARS-CoV-2 in Tears of Patients with Moderate to Severe COVID-19. Ophthalmology. 2021; 128: 494–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Corman VM, Landt O, Kaiser M, et al.. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020; 25(3): 2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lu X, Wang L, Sakthivel SK, et al.. US CDC real-time reverse transcription PCR panel for detection of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis. 2020; 26(8): 1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Munnink BBO, Nieuwenhuijse DF, Stein M, et al.. Rapid SARS-CoV-2 whole-genome sequencing and analysis for informed public health decision-making in the Netherlands. Nat Med. 2020; 26: 1405–1410. [DOI] [PubMed] [Google Scholar]
- 23. Bertens CJ, Dunker SL, Dias AJ, van den Biggelaar FJ, Nuijts RM, Gijs M.. Safety and comfort of an innovative drug delivery device in healthy subjects. Transl Vis Sci Technol. 2020; 9: 35–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kuo IC. A Rashomon moment? Ocular involvement and COVID-19. Ophthalmology. 2020; 127: 984–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ulhaq ZS, Soraya GV.. The prevalence of ophthalmic manifestations in COVID-19 and the diagnostic value of ocular tissue/fluid. Graefes Arch Clin Exp Ophthalmol. 2020; 258: 1351–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jin J-M, Bai P, He W, et al.. Gender differences in patients with COVID-19: focus on severity and mortality. Front Public Health. 2020; 8: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schiffer VM, Janssen EB, van Bussel BC, et al.. The “sex gap” in COVID-19 trials: a scoping review. EClinicalMedicine. 2020: 100652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Walsh KA, Jordan K, Clyne B, et al.. SARS-CoV-2 detection, viral load and infectivity over the course of an infection. J Infection. 2020; 81: 357–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wölfel R, Corman VM, Guggemos W, et al.. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020; 581(7809): 465–469. [DOI] [PubMed] [Google Scholar]
- 30. He X, Lau EHY, Wu P, et al.. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020; 26: 672–675. [DOI] [PubMed] [Google Scholar]
- 31. Deng W, Bao L, Gao H, et al.. Ocular conjunctival inoculation of SARS-CoV-2 can cause mild COVID-19 in Rhesus macaques. Nat Commun. 2020; 11(1): 4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Colavita F, Lapa D, Carletti F, et al.. SARS-CoV-2 isolation from ocular secretions of a patient with COVID-19 in Italy with prolonged viral RNA detection. Ann Intern Med. 2020; 173: 242–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Armero A, Berthet N, Avarre J-C.. Intra-host diversity of SARS-Cov-2 should not be neglected: case of the state of Victoria, Australia. Viruses. 2021; 13: 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Al Khatib HA, Benslimane FM, Elbashir IE, et al.. Within-host diversity of SARS-CoV-2 in COVID-19 patients with variable disease severities. Frontiers Cell Infect Microbiol. 2020; 10: 575613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang Y, Wang D, Zhang L, et al.. Intra-host variation and evolutionary dynamics of SARS-CoV-2 populations in COVID-19 patients. Genome Med. 2021; 13: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hui KPY, Cheung MC, Perera R, et al.. Tropism, replication competence, and innate immune responses of the coronavirus SARS-CoV-2 in human respiratory tract and conjunctiva: an analysis in ex-vivo and in-vitro cultures. Lancet Respir Med. 2020; 8: 687–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chi Y, Ge Y, Wu B, et al.. Serum cytokine and chemokine profile in relation to the severity of coronavirus disease 2019 in China. J Infect Dis. 2020; 222: 746–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kox M, Waalders NJ, Kooistra EJ, Gerretsen J, Pickkers P.. Cytokine levels in critically ill patients with COVID-19 and other conditions. JAMA. 2020; 324: 1565–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huang C, Wang Y, Li X, et al.. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020; 395(10223): 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McGonagle D, Sharif K, O'Regan A, Bridgewood C.. The role of cytokines including interleukin-6 in COVID-19 induced pneumonia and macrophage activation syndrome-like disease. Autoimmunity Rev. 2020; 19(6): 102537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Burgos-Blasco B, Guemes-Villahoz N, Santiago JL, et al.. Hypercytokinemia in COVID-19: tear cytokine profile in hospitalized COVID-19 patients. Exp Eye Res. 2020; 200: 108253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gandhi RT, Lynch JB, Del Rio C.. Mild or moderate Covid-19. N Engl J Med. 2020; 383(18): 1757–1766. [DOI] [PubMed] [Google Scholar]
- 43. World Health Organization. Transmission of SARS-CoV-2: implications for infection prevention precautions. Geneva: World Health Organization; 2020. [Google Scholar]
- 44. Zeng W, Wang X, Li J, et al.. Association of daily wear of eyeglasses with susceptibility to coronavirus disease 2019 infection. JAMA Ophthalmol. 2020; 138: 1196–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu Z, Sun C-B.. Conjunctiva is not a preferred gateway of entry for SARS-CoV-2 to infect respiratory tract. J Med Virol. 2020; 92: 1410–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Guo D, Xia J, Shen Y, Tong J.. SARS-CoV-2 may be related to conjunctivitis but not necessarily spread through the conjunctiva SARS-CoV-2 and conjunctiva. J Med Virol. 2020; 92: 1757–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Loffredo L, Pacella F, Pacella E, Tiscione G, Oliva A, Violi F.. Conjunctivitis and COVID-19: a meta-analysis. J Med Virol. 2020; 92: 1413–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Peng Y, Zhou YH.. Is novel coronavirus disease (COVID-19) transmitted through conjunctiva? J Med Virol. 2020; 92: 1408–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Guemes-Villahoz N, Burgos-Blasco B, Arribi-Vilela A, et al.. Detecting SARS-CoV-2 RNA in conjunctival secretions: is it a valuable diagnostic method of COVID-19? J Med Virol. 2021; 93: 383–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.