Abstract
Direct amplification and sequencing of the hemagglutinin (HA) genes of equine influenza virus subtype H3N8 was undertaken in order to characterize strains of this virus circulating in Sweden. The majority of viruses from outbreaks during 1997 analyzed belonged to the American lineage of H3 equine influenza, and one strain was shown to belong to the European lineage. Furthermore, it was shown that recent American-lineage strains are mutated at amino acid position 190 of the HA during serial passage in embryonated hens’ eggs. Host cell adaptation of these viruses thus takes place at antigenic region B of the HA.
Equine influenza virus is a common respiratory pathogen of horses, which causes severe acute disease and also predisposes the host to sequelae, such as chronic obstructive pulmonary disease and bacterial superinfection (4). Two different subtypes of equine influenza virus, H7N7 and H3N8, have been associated with disease in the horse. H7N7 has not been isolated from horses for over 20 years and is presumed not to circulate at the present time (9). H3N8, on the other hand, has been shown to circulate in large parts of the world excluding Australia, New Zealand, and Iceland.
Recent studies of the H3N8 subtype of equine influenza viruses have demonstrated that these strains have diverged into two distinct evolutionary lineages (1, 11). Based on the geographic origins of virus strains comprising these two lineages, the lineages have been designated as European and American. Virus isolated in the United States has, however, been shown to belong to the European lineage, and vice versa. The functional significance of this phylogenetic dichotomy has been examined in two studies of virus antigenicity, one employing polyclonal sera (1) and the other employing monoclonal antibodies (11) to study the hemagglutinin (HA) proteins of the viruses. Both studies concluded that differences in antigens characteristic of members of each lineage could be seen and that these differences could be sufficient to compromise cross-lineage protection after vaccination or infection.
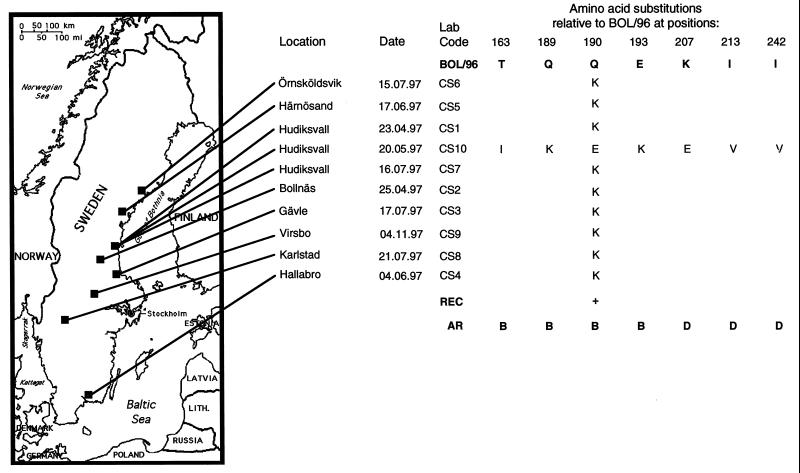
To determine which types of virus are circulating in the Swedish horse population, we chose to amplify and sequence the HA gene directly from clinical samples obtained from our routine diagnostics laboratory. Nasal swabs collected from horses displaying clinical signs of influenza were assayed by nested reverse transcription-PCR (RT-PCR) as described by Oxburgh and Hagström (13), and amplicons were subsequently nucleotide sequenced. Sequences were analyzed with the University of Wisconsin Genetics Computer Group software package (2). The primers used for partial amplification of HA in this study generate a 522-bp fragment (nucleotides 322 to 842) encoding amino acids 84 to 256. This region of HA was chosen because it contains all amino acids which have been shown to make up the receptor binding pocket of the molecule, and it also contains all amino acids composing antigenic regions A, B, and D. These regions have been demonstrated in previous studies to be extremely variable among equine influenza virus subtype H3N8 strains (1, 12), and they are presumably under strong immune system-mediated selective pressure in the horse. A comparison of the deduced amino acid sequences generated in this study with those of the most recently isolated Swedish virus (BOL/96 [11]) (Fig. 1) revealed complete homology of samples CS1 to CS9 with BOL/96 with the exception of position 190. Position 190 forms the membrane-distal limitation of the receptor binding site in the human H3N2 influenza virus strain (14) and is thus significant in determining both the affinity and the specificity of receptor binding of this virus. To exclude the possibility that the CS1 to CS9 sequences originated from the same virus due to laboratory contamination, we determined the entire sequences of the HA proteins of CS1, CS5, and CS9. The complete sequence of CS10 HA was also determined. All sequences showed heterogeneity: CS1 and CS5 differed at positions in the HA1 region, CS9 differed from CS1 only in the HA2 region (alanine to leucine change at position 42), and CS10 differed from CS1 at a number of positions. Amino acid differences in HA1 are shown in Fig. 2A. On the basis of this sequence heterogeneity we conclude that samples CS1 to CS10 are of different origins. CS10 shows extreme divergence from the BOL/96 strain, with substantial differences at antigenic regions B and C and also at the receptor-binding site. Comparisons with sequences of previously reported isolates of equine influenza virus show that CS10 has more likeness to European-lineage equine influenza virus than to American-lineage virus, to which BOL/96 belongs. A sequence alignment of CS10 HA1 with HA1 of the most recently isolated Swedish strain of the European lineage (AVE/93) (Fig. 2B) shows the close relatedness of these two strains at the amino acid level.
FIG. 1.
Alignment of deduced amino acid sequences of HA from 10 circulating strains of the H3N8 subtype of equine influenza virus (CS1 through CS10) and the sequence of the most recently identified Swedish virus strain (BOL/96). Differences at amino acid positions comprising the receptor binding site of HA (REC), and at positions defined as antigenic regions of the protein (AR), are specified below the alignment. The geographic origins of samples used in the study are shown on the map to the left.
FIG. 2.
(A) Amino acid sequence alignment of HA1 regions of three field strains of the H3N8 subtype of equine influenza virus with the sequence of BOL/96, the most recently isolated Swedish strain. The sequence of CS9 differs from that of CS1 only at amino acid 42 of HA2 (alanine to leucine change) and has therefore not been included in the alignment. (B) Amino acid sequence alignment of HA1 of CS10 with HA1 of AVE/93, the most recently isolated Swedish representative of the European lineage of equine influenza virus subtype H3N8. Where present, amino acid differences in field strains are denoted below the BOL/96 and AVE/93 sequences. Possible N-linked glycosylation sites are boxed. The positions of amino acids involved in the receptor activity of HA are shown above the sequence alignment in the row named REC. v denotes an amino acid which makes up the receptor binding pocket structure, x denotes an amino acid involved in stabilizing the pocket, a dot denotes an amino acid which can interact with the receptor, ∗ denotes an amino acid found to vary in epidemic strains of human influenza virus subtype H3N2, and ° denotes an amino acid found to vary in laboratory-selected escape mutants. The positions of amino acids which have been identified as components of antigenic regions, and their corresponding antigenic regions, are shown below the sequence alignment on the row designated AR.
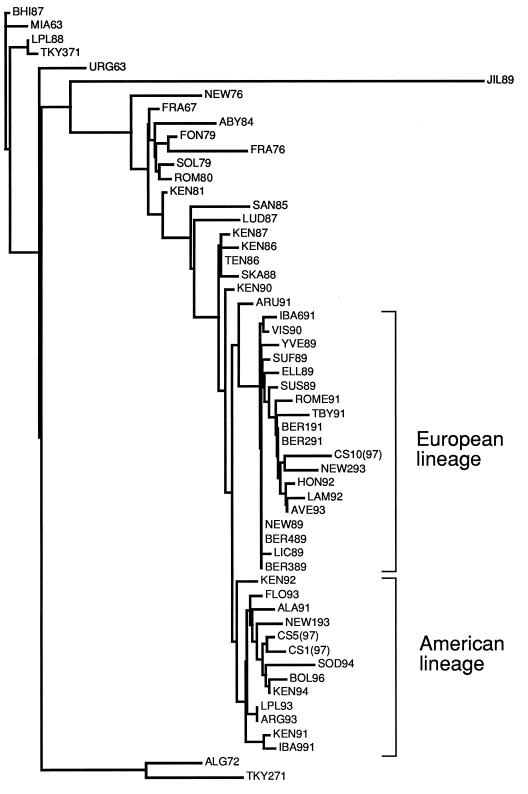
In order to properly ascertain the phylogenetic relationships of recently circulating strains of equine influenza virus in Sweden we performed a phylogenetic analysis based on the sequence of the entire HA1 subunit of HA. Since this is the globular domain of HA and is subjected to the most intense immune system-related selective pressure, it was chosen as the most revealing sequence for phylogenetic comparison. Figure 3 shows the results of the phylogenetic analysis performed by using the PHYLIP software package (3). It is apparent that CS1 and CS5, which were chosen as representatives of the CS1 to CS9 group, belong to the American lineage of H3 equine influenza virus and are closely related to BOL/96 and SOD/94, the strains most recently found to circulate in Sweden. CS10, however, is more closely related to NEW/2/93, one of the most recently isolated members of the European lineage of virus. This is surprising, since no virus belonging to the European lineage has been isolated in this country since 1993.
FIG. 3.
Phylogenetic analysis of HA1 amino acid sequences from CS1, CS5, and CS10 samples. A distance matrix was calculated and an unrooted tree was fitted by using the Fitch program of the PHYLIP software package (3).
Given the importance of position 190 in receptor binding (8), the fact that the only consensus amino acid difference seen in the sequences of CS1, CS5, and CS9 was at position 190, and the fact that all directly amplified strains are divergent from egg-adapted strains at this position, the question of whether this change could be due to adaptation of BOL/96 to the culture system during passaging was raised. In order to answer this, we assayed four passages of two individual virus strains by hemagglutination assay and partial sequencing of the HA protein. Nasal swab samples which had previously been found positive by immunofluorescence testing using an anti-A/equi 2/Visingsö/90 rabbit antiserum (5) were inoculated into the allantoic cavities of 10-day embryonated hens’ eggs after the addition of antibiotics and antimycotics. The eggs were harvested after 2 days, and the allantoic fluid was tested for the presence of virus by hemagglutination assay with 1% chick erythrocytes in phosphate-buffered saline. An aliquot of allantoic fluid was then inoculated into another embryonated hen’s egg, and virus was passaged in this way five times until HA titers ranging from 1:128 to 1:256 were attained. Table 1 shows the result of our analyses. The hemagglutination assay titration showed an increase in titer first between passages 2 and 3 and subsequently between passages 3 and 4. Viral RNA became detectable at passage 3, and it became more abundant at passages 4 and 5. The deduced amino acid sequences of RT-PCR products from each of the passages shows that the amino acid at position 190 changed from lysine to glutamine between passages 3 and 4, at the same time as the second titer increase. Based on this observation we propose that the variability seen at position 190 between directly amplified and egg-adapted strains is due to adaptation of the virus to the culture system. This data apparently contradicts that presented in a recent paper by Ilobi and colleagues, who showed that some amino acid changes could be expected upon culture of virus in embryonated hens’ eggs but that the positions of these changes were not consistent (6). However, this study was performed with European-lineage virus, which, like the CS10 virus analyzed in this study, showed no consistent evidence of adaptation to the culture system. Comparison of CS10 with sequences of closely related strains indicates that this adaptation is necessary only for strains with lysine at position 190 (i.e., CS1 to CS9), since the glutamic acid of CS10 is also seen at this position in the most closely related egg-adapted strains (Fig. 2B) (11).
TABLE 1.
Analysis of serial passages of two strains of H3N8 by hemagglutination assay and RT-PCR/sequencing
| Strain | Passage no. | Hemagglutination assay titera | Amino acid at position 190b |
|---|---|---|---|
| BOL/96 | 2 | <4 | −c |
| 3 | 8 | K | |
| 4 | 64 | Q | |
| 5 | 128 | Q | |
| ALV/96 | 2 | <4 | −c |
| 3 | 16 | K | |
| 4 | 126 | Q | |
| 5 | 256 | Q |
Hemagglutinating assay titer is expressed as the reciprocal of the highest dilution of allantoic fluid agglutinating an equal volume of 1% chick erythrocytes.
Deduced amino acid at position 190 of the virus HA, determined by sequencing of amplification products.
−, no amplification product detected.
Two major conclusions can be drawn based on the data presented in this paper. First, representatives of both phylogenetic lineages of the H3N8 subtype of equine influenza virus are circulating in the Swedish horse population. Because of the antigenic differences that have previously been demonstrated between representatives of the two lineages, incorporation of virus from both lineages in vaccines should be considered. Second, care must be taken in the isolation and propagation of equine influenza viruses of the H3N8 subtype. The change seen at position 190 of HA is presumably of importance in eliciting a neutralizing immune response because of the central location of this residue in antigenic region B. When virus of the American lineage is cultured in embryonated hens’ eggs, adaptation seems to occur through substitution of this amino acid. To obtain cultured virus with the wild-type genotype we are faced with the prospect of either finding an alternative culture system or cultivating large batches of allantoic fluid at very low titer from eggs inoculated with an early passage of the virus. An alternative to both of these approaches might be to use a DNA vaccination approach, i.e., inoculating horses with HA genes cloned directly from clinical strains into eukaryotic expression vectors. Recent research exploring the possible applications of this technique for equine influenza virus has yielded promising results (7, 10). Further studies will reveal whether DNA vaccination elicits an adequate response in the horse.
Acknowledgments
Many thanks are due to Åsa Hagström for skillful technical assistance.
This work was supported by a grant from the Swedish Horse Racing Association (ATG).
REFERENCES
- 1.Daly J M, Lai A C, Binns M M, Chambers T M, Barrandeguy M, Mumford J A. Antigenic and genetic evolution of equine H3N8 influenza A viruses. J Gen Virol. 1996;77:661–671. doi: 10.1099/0022-1317-77-4-661. [DOI] [PubMed] [Google Scholar]
- 2.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.5c. Seattle: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 4.Gerber H. Second International Conference of Equine Infectious Diseases. Paris, France: Karger; 1969. Clinical features, sequelae and epidemiology of equine influenza; pp. 63–80. [Google Scholar]
- 5.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 6.Ilobi C P, Nicolson C, Taylor J, Mumford J A, Wood J M, Robertson J S. Direct sequencing of the HA gene of clinical equine H3N8 influenza virus and comparison with laboratory derived viruses. Arch Virol. 1998;143:891–901. doi: 10.1007/s007050050340. [DOI] [PubMed] [Google Scholar]
- 7.Larsen D L, Dybdahl-Sissoko N, McGregor M W, Drape R, Neumann V, Swain W F, Lunn D P, Olsen C W. Coadministration of DNA encoding interleukin-6 and hemagglutinin confers protection from influenza virus challenge in mice. J Virol. 1998;72:1704–1708. doi: 10.1128/jvi.72.2.1704-1708.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin J, Wharton S, Lin Y P, Takemoto D, Skehel J J, Wiley D, Steinhauer D A. Studies of the binding properties of influenza hemagglutinin receptor-site mutants. Virology. 1998;241:101–111. doi: 10.1006/viro.1997.8958. [DOI] [PubMed] [Google Scholar]
- 9.Mumford J, Wood J. WHO/OIE meeting: consultation on newly emerging strains of equine influenza. Vaccine. 1993;11:1172–1175. doi: 10.1016/0264-410x(93)90092-c. [DOI] [PubMed] [Google Scholar]
- 10.Olsen C W, McGregor M W, Dybdahl-Sissoko N, Schram B R, Nelson K M, Lunn D P, Macklin M D, Swain W F, Hinshaw V S. Immunogenicity and efficacy of baculovirus-expressed and DNA-based equine influenza virus hemagglutinin vaccines in mice. Vaccine. 1997;15:1149–1156. doi: 10.1016/s0264-410x(96)00309-x. [DOI] [PubMed] [Google Scholar]
- 11.Oxburgh L, Akerblom L, Fridberger T, Klingeborn B, Linne T. Identification of two antigenically and genetically distinct lineages of H3N8 equine influenza virus in Sweden. Epidemiol Infect. 1998;120:61–70. doi: 10.1017/s0950268897008315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oxburgh L, Berg M, Klingeborn B, Emmoth E, Linne T. Evolution of H3N8 equine influenza virus from 1963 to 1991. Virus Res. 1994;34:153–165. doi: 10.1016/0168-1702(94)90097-3. [DOI] [PubMed] [Google Scholar]
- 13.Oxburgh L, Hagström Å. A PCR based method for the identification of equine influenza virus from clinical samples. Vet Microbiol. 1999;67:161–174. doi: 10.1016/s0378-1135(99)00041-3. [DOI] [PubMed] [Google Scholar]
- 14.Weis W, Brown J H, Cusack S, Paulson J C, Skehel J J, Wiley D C. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature. 1988;333:426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]