Abstract
Background:
Despite increased interest in MSC-based cell therapies for the acute respiratory distress syndrome (ARDS), clinical investigations have not yet been successful and understanding of the potential in vivo mechanisms of MSC actions in ARDS remain limited. ARDS is driven by an acute severe innate immune dysregulation, often characterized by inflammation, coagulation, and cell injury. How this inflammatory microenvironment influences MSC functions remains to be determined.
Aim:
To comparatively assess how the inflammatory environment present in ARDS lungs vs. the lung environment present in healthy volunteers alters MSC behaviors.
Methods:
Clinical grade human bone marrow-derived MSCs (hMSCs) were exposed to bronchoalveolar lavage fluid (BALF) samples obtained from ARDS patients or from healthy volunteers. Following exposure, hMSCs and their conditioned media were evaluated for a broad panel of relevant properties including viability, levels of expression of inflammatory cytokines, gene expression, cell surface HLA expression, and activation of coagulation and complement pathways.
Results:
Pro-inflammatory, pro-coagulant, and major histocompatibility complex (self recognition) related gene expression was markedly up-regulated in hMSCs exposed ex vivo to BALF obtained from healthy volunteers. In contrast, these changes were less apparent and often opposite in hMSCs exposed to ARDS BALF samples.
Conclusion:
These data provide new insights into how hMSCs behave in healthy vs. inflamed lung environments strongly suggesting that the inflamed environment in ARDS induces hMSC responses potentially benefical for cell survival and actions. This further highlights the need to understand how different disease environments affect hMSC functions.
Keywords: Cell Therapy, Inflammation, Acute Respiratory Distress Syndrome, Mesenchymal Stromal Cells, Bronchoalveolar Lavage Fluid
INTRODUCTION
Mesenchymal stromal cells (MSCs) are being increasingly investigated as a cell-based therapy to suppress excessive inflammation in the acute respiratory distress syndrome (ARDS) [1, 2]. However, results of clinical investigations of MSCs in ARDS, while uniformly demonstrating safety, have not as yet demonstrated efficacy [3–5]. While a number of factors may be responsible for the lack of improved outcome, there remains a fundamental lack of knowledge as to the fate and actions of the administered MSCs in vivo in the diseased human lung microenvironment (reviewed in [6]). This raises the possibility that the inflammatory environment encountered may significantly alter potential MSC efficacy and potency.
A growing number of studies, including our own, have found that MSC functions, and thus potential therapeutic actions, differ depending on the inflammatory environment encountered [7–13]. Ex vivo exposure to bronchoalveolar lavage fluid (BALF) or serum samples from ARDS patients has significant impact on MSC functions including profile of secreted mediators and downstream effects on macrophage functions, often enhancing anti-inflammatory actions [8–12]. In one example, BALF from cystic fibrosis patients who have pulmonary Aspergillus infection is rapidly toxic to MSCs, in part related to the fungal product gliotoxin [13]. This raises the possibility that certain inflammatory lung environments may have deleterious effects on MSCs with according implications for potential therapeutic use. Moreover, contrary to previously held beliefs, systemically administered allogeneic MSCs rapidly undergo clearance and/or inactivation [14–17]. In part, this may be related to a phenomenon known as the instant blood mediated inflammatory reaction (IBMIR), an immediate inflammatory response to systemically administered allogeneic MSCs [18–20].
Thus, to investigate the effects of the ARDS inflammatory lung environment on MSC viability and function, clinical grade human bone marrow-derived MSCs obtained from healthy volunteers (hMSCs) were exposed ex vivo to individual BALF samples obtained from ARDS patients and from healthy volunteers (HC) for comparison. Unexpectedly, hMSCs exposed to HC BALF developed an inflammatory response and also increased gene and protein expression associated with self- vs non-self recognition, notably increased Class II HLA expression, and increased complement expression. These results suggest that an otherwise non-inflamed normal lung environment stimulates mechanisms for clearance of allogeneic hMSCs. In contrast, changes in gene and protein expression associated with self- vs non-self recognition were either mitigated, absent, or opposite in hMSCs exposed to BALF from ARDS patients. These findings provide evidence of the plasticity of hMSC responses in different clinically relevant lung environments and shed new light into the potential mechanisms of action of MSC-based cell therapy for ARDS.
METHODS
BALF samples
BALF samples from ARDS patients without sepsis were collected prospectively as part of an unrelated clinical investigation conducted by the National Heart Lung Blood Institute (NHLBI) ARDSNET (ClinicalTrials.gov NCT0011216) [21]. ARDS BALF samples were obtained by mini-BAL from a phase 2 NIH trial conducted by the ARDSNET (PETAL) (ref 21, published in 2008). The technique utilized was a standard 40 ml mini-BAL utilizing sterile saline in intubated ARDS patients. BALF samples were subsequently centrifuged and stored at −80°C. The healthy volunteers underwent standard fiberoptic bronchoscopy of the RML at Dartmouth under appropriate institutional IRB protocols. Twenty cc sterile saline was utilized and samples were comparably centrifuged and supernatants stored at −70°C. These samples are more recent, being obtained between January and July 2018. and stored at −70°C. Healthy volunteers were excluded if they had any history of cardiopulmonary disease, if they smoked or vaped regularly, or were taking any immunomodulatory medications.
Ex vivo exposure of hMSCs
Human MSCs (hMSCs) were obtained from the NHLBI’s PACT program and routinely cultured. The hMSCs utilized were obtained from a single volunteer (a second donor was also used in the complement and flow cytometry experiments) and were the same as those utilized in the recent START trial [3, 4]. Cells at passage 3–5 were used. hMSCs were exposed to individual ARDS or HC BALF samples diluted into serum free media (20% BALF as delineated in prior studies [11, 13]) for 24 hours. Serum free media only or with 20% PBS was added to control and unstimulated hMSCs, respectively. After 24 hours incubation, cells and conditioned media (CM) was collected.
RNA Sequencing Analysis
Total RNA was extracted using standard Trizol extraction followed by a cleaning step using RNeasy spin columns (Qiagen). RNA was quality assessed on a Fragment Analyzer instrument (Agilent) and quantified on a qubit fluorometer. RNA sequencing analyses were performed on RNA extracted from hMSCs exposed to PBS (n=4), ARDS BALF (n=5), and HC samples (n=5) and were aligned to human genes using salmon [22]. Transcript level information from salmon was imported into R using tximport [23], normalized in edgeR based on library size to create counts per million (CPM) for each gene and differential gene expression was assessed. Pathway analysis genes that differed significantly from unstimulated control samples were identified using IPA (www.ingenuity.com).
Statistical Analyses
Mann-Whitney test was used to assess differences between two groups. Kruskal-Wallis tests (Dunn’s post hoc test) or one-way ANOVA (Dunnett’s post hoc test) were used to assess differences between three or more groups. Statistical analyses were performed using GraphPad Prism software. P-values ≤0.05 were considered as significant, except in the case of RNA sequencing data analyzed in edgeR, where a multiple hypothesis corrected false discovery rates (FDR) less than 0.05 were considered significant. Spearman correlations were calculated in base R, using the t distribution to calculate p-values in those cases that included ties in rank. Additional detailed descriptions of these and other methods used in this study are provided in the online supplement.
RESULTS
ARDS BALF contains elevated inflammatory mediators compared to HC BALF
To initially determine if the BALF samples differed between the HC and ARDS patient lungs, inflammatory mediators in clinical BALF samples were assessed utilizing a human magnetic Luminex Assay kit (R&D Systems). The BALF samples utilized for each assay in the overall study are depicted in Supplementary Table 1. Although there were variations between the different clinical isolates, the levels of total protein, dsDNA, and a range of inflammatory mediators were significantly elevated in ARDS compared to HC BALF samples (Supplementary Table 2). There were no significant differences between ARDS and HC BALF in the levels of anti-inflammatory and Th2 mediators such as IL-10, IL-4, and IL-13.
BALF from both ARDS and HC patients is non-toxic to hMSCs
To determine if BALF samples from ARDS and HC lungs were associated with increased cell death, hMSCs were exposed ex vivo to individual clinical BALF samples. There was no significant difference in toxicity between hMSCs exposed to ARDS or HC BALF samples as determined by light microscopy (Figure 1A) and by LDH release (Figure 1B). To further determine toxicity, mitochondrial respiration in hMSCs exposed to different BALFs was assessed. Neither HC or ARDS BALF significantly altered hMSC basal respiration rate, maximal respiration rate, spare respiratory capacity, or other mitochondrial functions compared to PBS-exposed hMSCs (Figure 1C–E, Supplementary Figure 1). Interestingly, a significant reduction in spare respiratory capacity was similarly observed in ARDS and HC BALF-exposed hMSCs compared to control hMSCs (serum free media) (p=0.030 and p=0.034, respectively, Figure 1E). However, this reduction was also observed in PBS-exposed hMSCs compared to control hMSCs (p=0.059).
Figure 1. Exposure to BALF from ARDS patients and healthy controls are non-toxic to hMSCs.
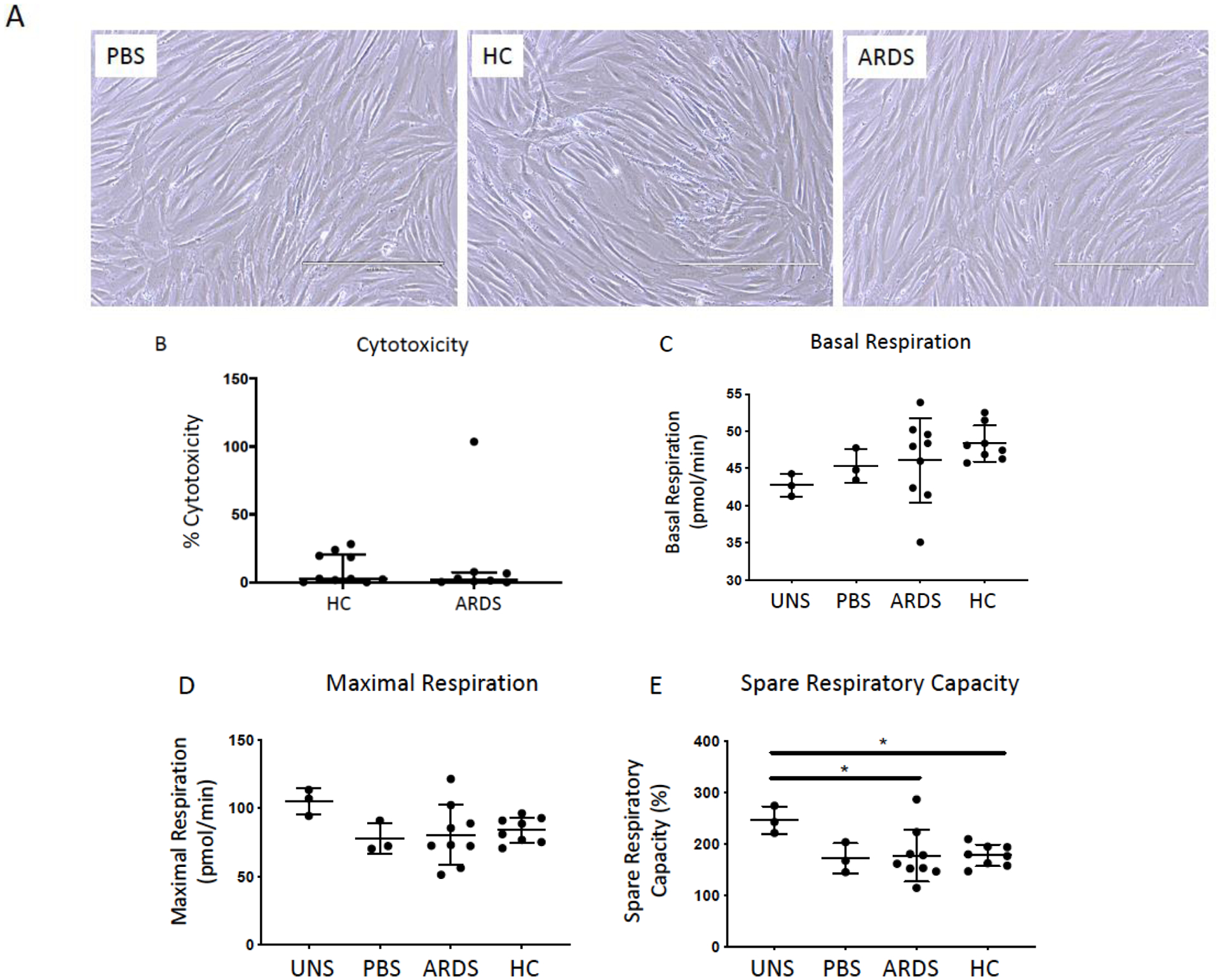
Representative phase contrast photomicrographs (10X) of hMSCs exposed for 24 hours to 20% BALF samples obtained from ARDS patients or from healthy control subjects (HC). PBS-exposed (20%) hMSCs were used as controls. Scale bar represents 400μm and photomicrographs have been brightness/contrast adjusted (A). Cytotoxicity was evaluated in conditioned medium utilizing a standard LDH assay following 24 hours exposure (ARDS: n= 8 and HC: n=10). Data are presented as median with interquartile range of % cytotoxicity (B). To assess the impact of ARDS (n=9) and healthy control BALF (n=8) samples on hMSC mitochondrial function, basal respiration (C), maximal respiration (D), and spare respiration capacity (E) was measured in hMSCs pre-exposed (24 hours) utilizing XF-96e Extracellular Flux Analyzer and compared to PBS-exposed (n=3) and unstimulated (serum free media only, n=3). Data are presented as means ± with SD and statistical analysis was performed by Shapiro-Wilk test, followed by a one-way ANOVA with Dunnett’s post hoc test. Abbreviations: HC, healthy control; ARDS, acute respiratory distress syndrome; LDH, lactate dehydrogenase; PBS, phosphate-buffered saline; uns, unstimulated (serum free media only); *, p≤0.05.
ARDS and HC BALF activate hMSCs to release a spectrum of mediators
Level of IL-6 (p=0.0034 and p=0.0257, respectively) and other pro-inflammatory mediators such as IL-8 (p=0.0008 and p=0.0084, respectively) and IL-18 (p=0.0591 and p=0.0157, respectively) were increased in hMSC-CM following exposure to either HC or ARDS BALF samples compared to PBS-exposed hMSCs (Figure 2A–C, Table 1). Moreover, significantly increased levels of CD44 (p=0.0041 and p=0.0342), respectively) and SP-D (p=0.0162 and p=0.0055, respectively) were comparably observed in hMSCs exposed to both ARDS and HC BALFs compared to PBS-exposed hMSCs (Figure 2D–E, Table 1). Interestingly, hMSCs exposed to ARDS but not HC BALF samples induced significantly higher levels of HGF (p=0.0180) and in particular MMP-3 (p=0.0041) compared to controls (Figure 2F–G). In contrast, hMSCs exposed to HC but not ARDS BALF induced significantly higher CCL2 levels (p=0.0208) compared to controls (Figure 2H). These data suggest that hMSCs can acquire both pro- and anti-inflammatory phenotype in response to specific mediators present or absent in the BALF.
Figure 2. ARDS BALF as well as healthy control BALF exposure activates hMSCs to release a spectrum of mostly pro- but some anti-inflammatory mediators.
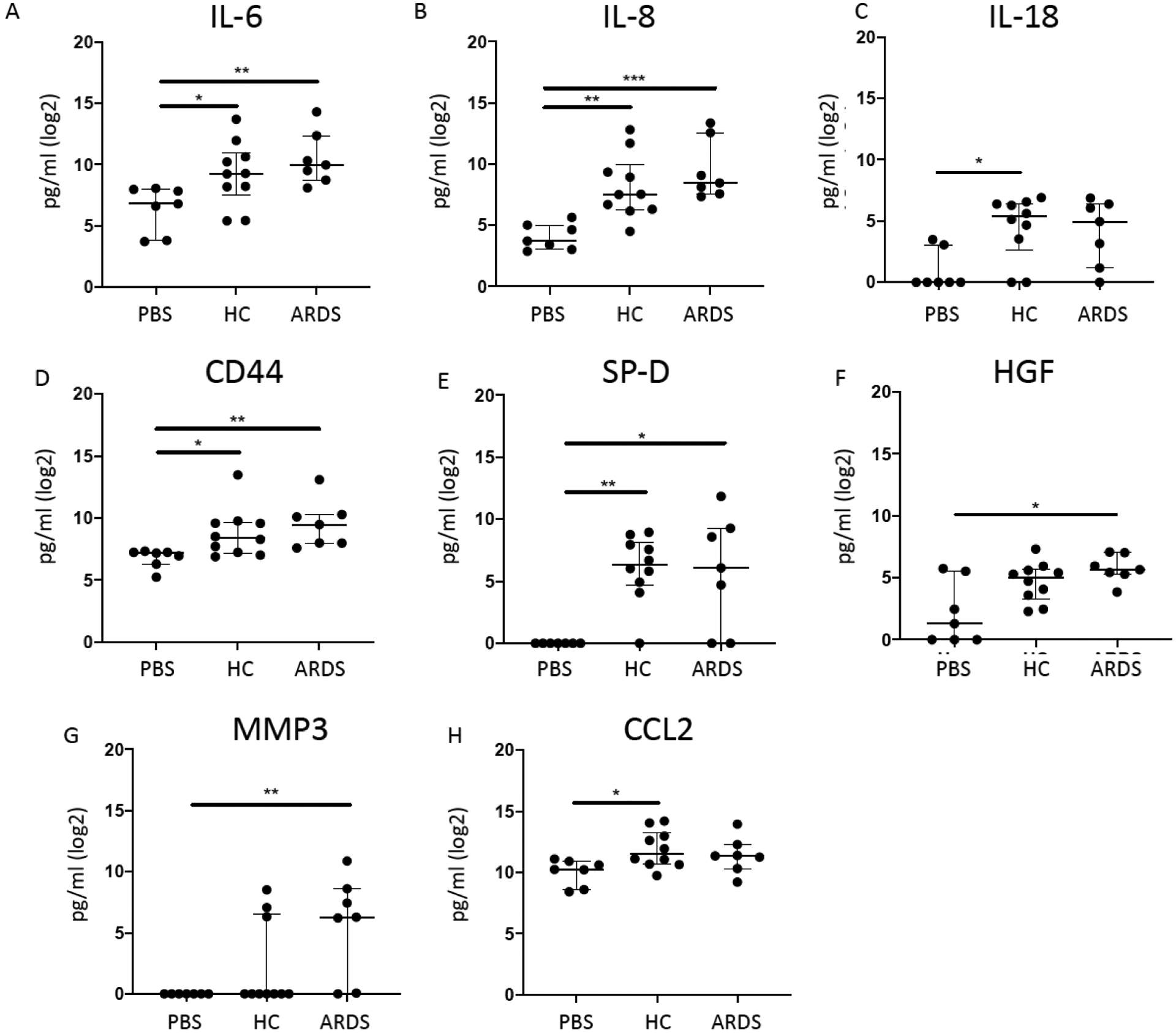
To assess if the secretome profiles of hMSCs exposed to ARDS BALF (n=7) samples differed from healthy control BALF-exposed hMSCs (n=10) and PBS-exposed hMSCs (n=7), conditioned media after BALF or PBS exposure was assessed for a range of inflammatory and other mediators including IL-6 (A), IL-8 (B), IL-18 (C), CD44 (D), SP-D (E), HGF (F), MMP3 (G), and CCL2 (H). Data are presented as median with interquartile range of log2 normalized values, and statistical analysis was performed by Shapiro-Wilk test, followed by Kruskal-Wallis followed by Dunn’s post hoc test by comparing to the unstimulated control group. Abbreviations: HC, healthy control; ARDS, acute respiratory distress syndrome; PBS, phosphate-buffered saline; IL-6, Interleukin 6; IL-8, Interleukin 8; IL-18, Interleukin 18; CD44, CD44 Molecule/Hyaluronate Receptor; SP-D, Surfactant protein D; HGF, Hepatocyte growth factor; MMP3, matrix metalloproteinase-3; CCL2, chemokine (C-C motif) ligand 2/ monocyte chemoattractant protein 1; *, p≤0.05; **, p≤0.01. 0.01.
Table 1.
Cytokines detected in CM from hMSCs exposed to ARDS and healthy volunteer BALF samples
| Cytokine | Cytokines in conditioned media (pg/ml) | Kruskal-Wallis with Dunn’s Compared to UNS | |||
|---|---|---|---|---|---|
| UNS | HC | ARDS | HC | ARDS | |
| Mean (SD) | Mean (SD) | Mean (SD) | P-value | P-value | |
| ADAMTS13 | 1.0 (0.0) | 1.0 (0.0) | 1.0 (0.0) | NA | NA |
| CXCL8/IL-8 | 20.9 (15.9) | 1227.8 (2307.8) | 2583.7 (4101.4) | 0.0084 (**) | 0.0008 (***) |
| Fas Ligand | 1.0 (0.0) | 1.3 (1.0) | 1.0 (0.0) | 0.3019 | >0.9999 |
| GM-CSF | 1.8 (1.0) | 2.5 (2.4) | 5.7 (9.3) | >0.9999 | 0.3280 |
| IL-10 | 1.0 (0.0) | 1.2 (0.6) | 1.0 (0.0) | 0.6403 | >0.9999 |
| IL-13 | 1.0 (0.0) | 1.0 (0.0) | 1.0 (0.0) | NA | NA |
| IL-2 | 1.0 (0.0) | 10.7 (22.5) | 23.7 (39.7) | 0.6189 | 0.2913 |
| IL-4 | 1.0 (0.0) | 6.2 (7.3) | 10.8 (10.6) | 0.2390 | 0.0457 (*) |
| Leptin | 1.0 (0.0) | 1.0 (0.0) | 1.0 (0.0) | NA | NA |
| MIF | 2056.0 (2742.6) | 20575.8 (30710.7) | 14619.7 (34703.7) | 0.2957 | 0.3239 |
| CCL4 | 1.0 (0.0) | 1.0 (0.0) | 96.6 (166.5) | >0.9999 | 0.1171 |
| Osteopontin | 7638.4 (937.2) | 9799.8 (3483.3) | 9289.5 (3196.5) | 0.4702 | 0.6699 |
| TNF-α | 1.0 (0.0) | 1.0 (0.0) | 1.0 (0.0) | NA | NA |
| CD44 | 121.7 (46.4) | 1508.0 (3486.9) | 1786.9 (3101.3) | 0.0342 (*) | 0.0041 (**) |
| Fas | 1.0 (0.0) | 47.3 (79.6) | 260.2 (493.0) | 0.1293 | 0.0477 (*) |
| G-CSF | 1.0 (0.0) | 1.0 (0.0) | 10.1 (24.0) | >0.9999 | 0.3809 |
| HGF | 15.8 (23.4) | 41.7 (45.7) | 68.0 (47.0) | 0.2356 | 0.0180 (*) |
| IL-1β | 1.0 (0.0) | 4.1 (9.8) | 32.0 (74.2) | >0.9999 | 0.2291 |
| IL-12 p70 | 11.9 (10.9) | 16.8 (29.4) | 8.7 (13.2) | >0.9999 | >0.9999 |
| IL-18 | 3.5 (4.4) | 49.9 (42.0) | 44.4 (45.5) | 0.0157 (*) | 0.0591 |
| IL-36β | 1.0 (0.0) | 1.3 (0.7) | 2.3 (2.3) | >0.9999 | 0.3667 |
| IL-6 | 140.0 (108.4) | 2202.3 (4080.5) | 4179.4 (7296.9) | 0.0257 (*) | 0.0034 (**) |
| CCL2 | 1268.4 (716.0) | 6290.1 (6588.2) | 4377.0 (5283.3) | 0.0208 (*) | 0.0903 |
| CCL3 | 1.0 (0.0) | 38.8 (67.3) | 47.6 (80.6) | 0.3145 | 0.3667 |
| MMP-3 | 1.0 (0.0) | 58.8 (117.8) | 372.4 (681.0) | 0.4444 | 0.0041 (**) |
| SP-D | 1.0 (0.0) | 163.8 (176.5) | 682.3 (13344.2) | 0.0055 (**) | 0.0162 (*) |
| IFN-γ # | 0.07 (0.14) | 1.5 (2.2) | 0 (0) | 0.9276 | >0.9999 |
Cytokines detected in conditioned media from hMSCs exposed to ARDS BALF samples (n=7), healthy control BALF samples (n=10), or PBS-exposed (n=7) using 27-plex Luminex assay. Samples were analyzed on two luminex plates. Mean with SD of extrapolated values are presented. Values out of range below were set to 1.0. IFN-γ was measured on a separate ELISA on conditioned media from hMSCs exposed to ARDS BALF samples (n=3), healthy control BALF samples (n=5), or PBS-exposed (n=4). Data are presented as mean with SD. Kruskal-Wallis tests (Dunn’s post hoc test) was used to assess differences between ARDS/HC exposed hMSCs and unstimulated hMSCs. CM, conditioned media; hMSCs, human mesenchymal stromal cells; BALF, bronchoalveolarlavage fluid; UNS, unstimulated; ARDS, acute respiratory distress syndrome; HC, healthy control subjects;
IFN-γ was measured on a separate ELISA; SD, standard deviation; NA, not available;
≤ 0.05;
≤ 0.01,
≤ 0.001.
BALF IL-1β predicts hMSC cytokine secretion
We next determined whether specific BALF cytokines correlated (Spearman correlations) with hMSC inflammatory mediator production. Notably, IL-1β in both ARDS or HC BALF samples was predictive of the presence of several inflammatory and apoptosis-inducing mediators in hMSC-CM including IL-6 (p=0.0173), IL-36 (p=0.0334), IL-2 (p=0.0340), MMP-3 (p=0.0034), FAS (p=0.0427), and IL-8 (p=0.0346) (Figure 3A, Supplementary Table 3, Supplementary Figure 2). As shown in Figure 3B, expression of IL-1β was not correlated with other cytokines measured in BALF samples. However, expression of cytokines detected in CM were frequently correlated with each other (Figure 3C). Interestingly, none of the cytokines measured in CM were co-expressed with FAS (Figure 3C). These data suggest that the presence of higher concentrations of IL-1β in BALF may be used to predict presence of pro-inflammatory mediators.
Figure 3. BALF IL-1β predicts hMSC pro-inflammatory cytokine secretion.
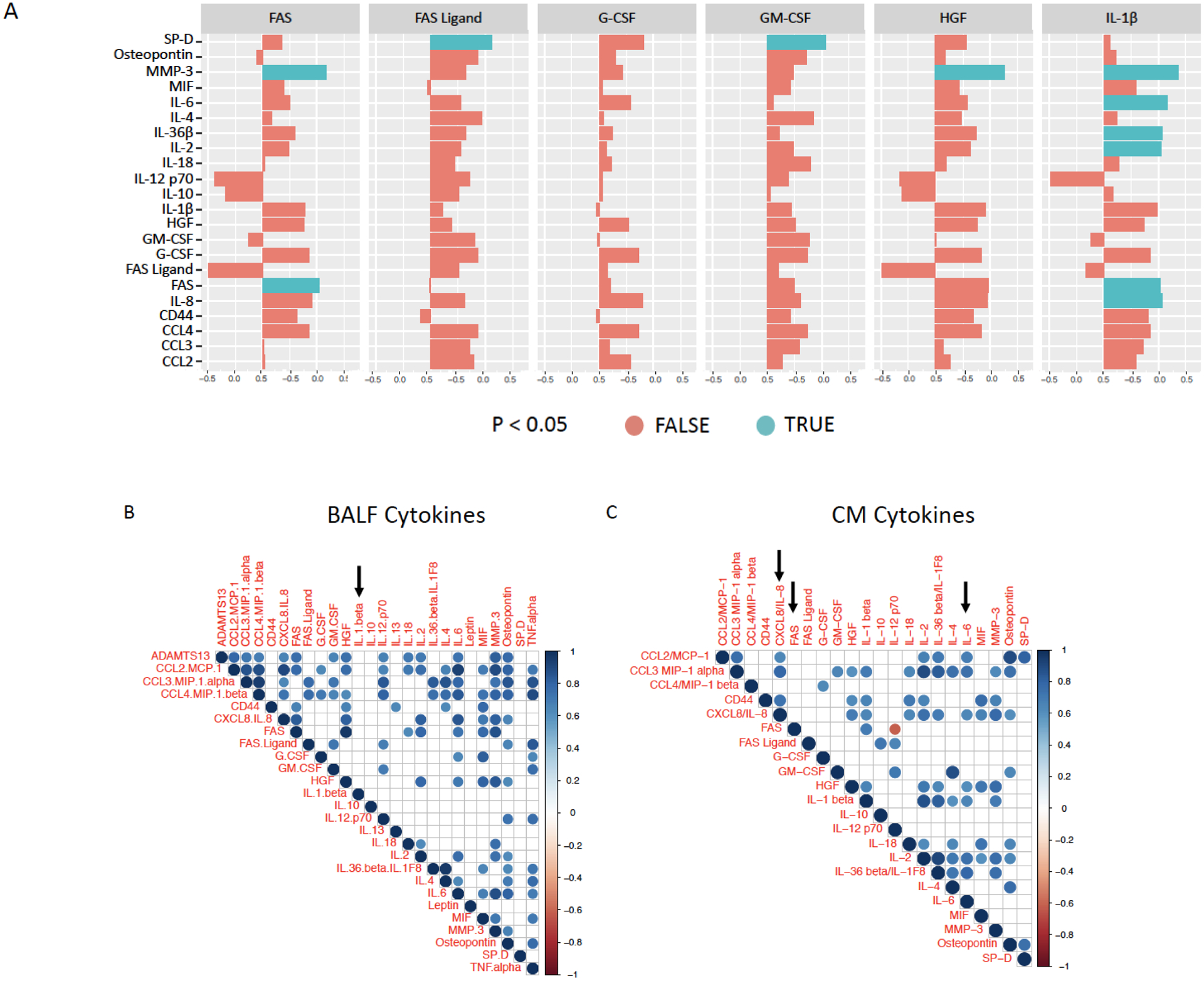
Cytokines measured in BALF samples (top) were correlated with cytokines detected in conditioned media (left) from ARDS (n=6) and HC (n=10) BALF-exposed hMSC cultures (A). Red color indicates no significant difference (p<0.05) and blue color indicates significant difference (p<0.05). Cytokines measured in BALF samples were correlated with each other (B) and cytokines measured in conditioned media from BALF-exposed hMSC cultures was correlated with each other (C). Red color indicates significantly decreased expression (p<0.01) and blue color indicates significantly increased expression (p<0.01). Arrows indicate cytokines of specific interest (IL-1β, IL-6, and FAS). Spearman correlation was calculated in R. P-values are estimated and not exact because there were ties in the data.
HC BALF-exposed hMSCs demonstrate increased overall gene expression compared to ARDS BALF-exposed hMSCs
To further probe BALF exposure effects on hMSC functions, BALF-exposed hMSCs were analyzed by RNA sequencing and compared to PBS-exposed hMSCs. A heat map demonstrates the cytokine profiles of the individual BALF samples utilized for hMSC exposures prior to RNA sequencing analyses (Figure 4A). These data demonstrate that both the HC and ARDS BALF samples utilized were representative of the full set of BALF samples analyzed (Supplementary Table 2). Despite high levels of inflammatory mediators in ARDS BALF samples, RNA sequencing demonstrated that HC BALF samples were more potent overall in inducing increased hMSC gene expression whereas ARDS BALF decreased gene expression compared to that induced by control PBS exposure (Figure 4B). Notably increased expression of many genes observed with HC BALF exposures were decreased with ARDS BALF exposure (Figure 4B). Interestingly, the COVID-19 binding and entry receptors ACE2 and TMPRSS2 were expressed in the hMSCs, however only at minimal levels with no differences observed between the BALF exposure groups (Supplementary Table 4).
Figure 4. Healthy control-exposed hMSCs demonstrate increased gene expression compared to ARDS- and PBS-exposed hMSCs.
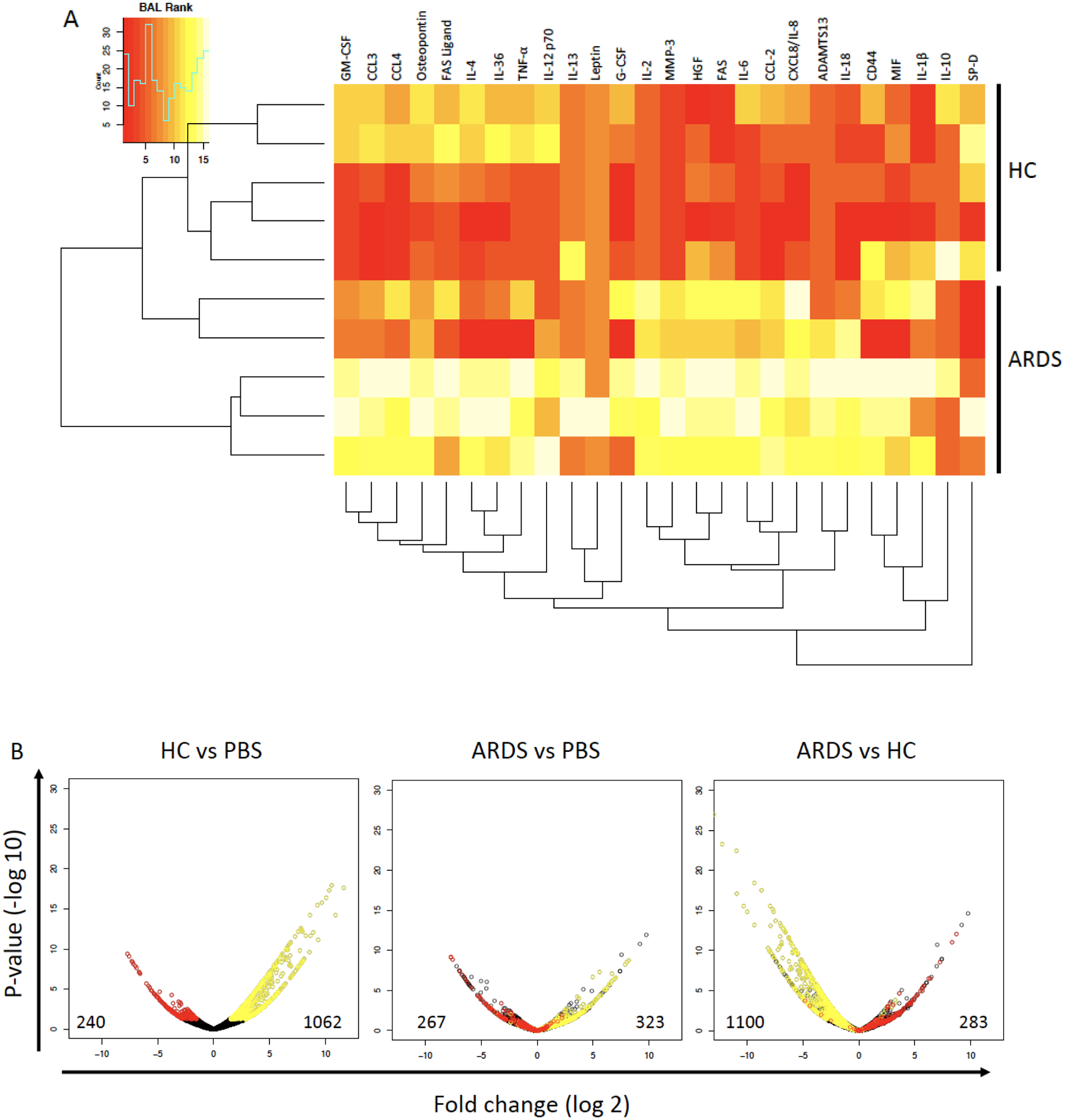
Heat map clustering of ranked cytokines measured in ARDS (n=5) and healthy control (n=5) BALF samples utilized in the RNA sequencing analysis. Red color indicates lower and yellow color higher concentrations of cytokines (A). Volcano plots of genes identified by RNA sequencing of ARDS BALF (n=5), healthy control BALF (n=5), or PBS-exposed hMSCs (n=4). Each volcano plot shows a different comparison. Y-axis shows −log10 p-value and X-axis is log2 fold change for each gene. Genes highlighted in red were significantly repressed in the comparison between HC BALF and PBS or ARDS vs PBS. Genes highlighted in yellow were significantly induced in the comparison between HC BALF and PBS or ARDS ve PBS. The number of genes that were significantly repressed (left) or induced (right) in each of the three comparisons is also shown. Abbreviations: HC, healthy control; ARDS, acute respiratory distress syndrome; PBS, phosphate-buffered saline.
Exposure to HC but not ARDS BALF increases pro-inflammatory cytokine genes
hMSCs exposed to HC BALF samples demonstrated an overall increased expression of genes involved in multiple immune-regulatory pathways as compared to PBS-exposed hMSCs, including TNF, ICAM-1, CXCL10, CCL2, CCL8, and IFN-β1 (Figure 5A). In striking contrast ARDS BALF-exposed hMSCs demonstrated expression of the majority of those genes at levels similar to those observed in PBS-exposed hMSCs. However, ARDS BALF-exposed hMSCs demonstrated an increased expression of cytokines known to be involved in neutrophil trafficking including CXCL1, CXCL2 (MIP-2α), CXCL3, CXCL8/IL-8, and IL-6 (Figure 5A). However, these genes were expressed at levels lower than those observed following exposure to HC BALF.
Figure 5. Exposure to healthy control but not ARDS BALF increases pro-inflammatory cytokine gene and interacting gene expression.
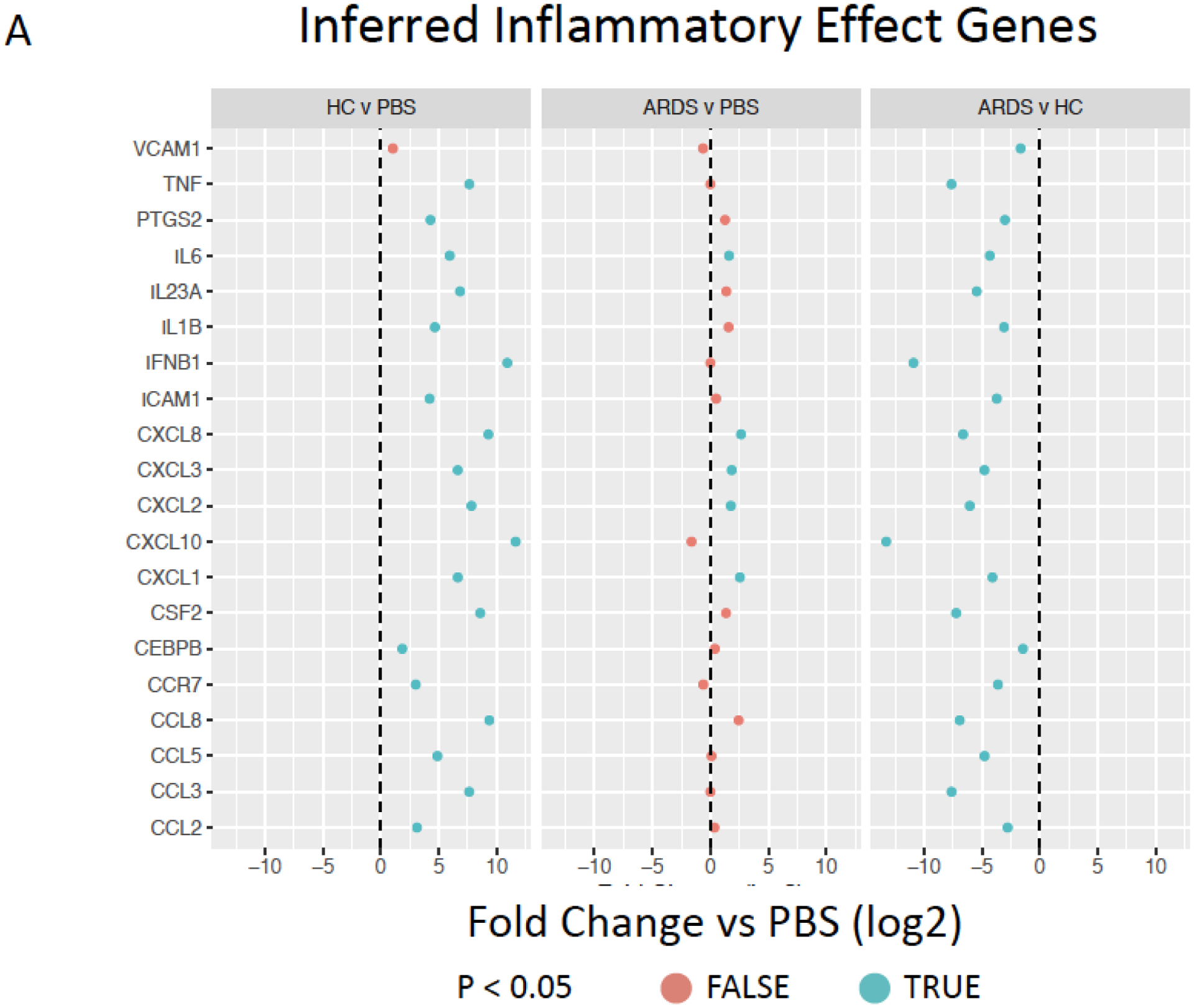
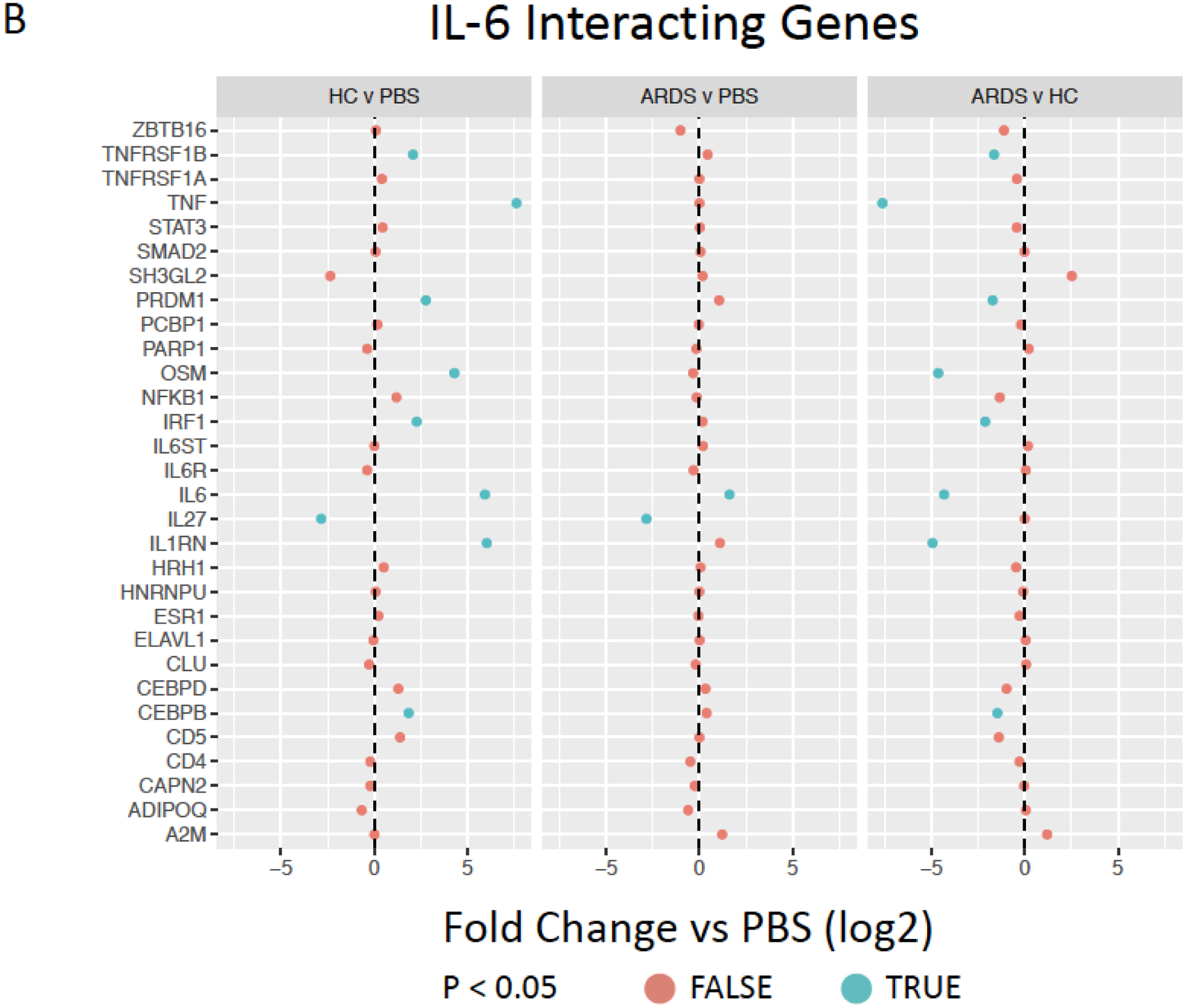
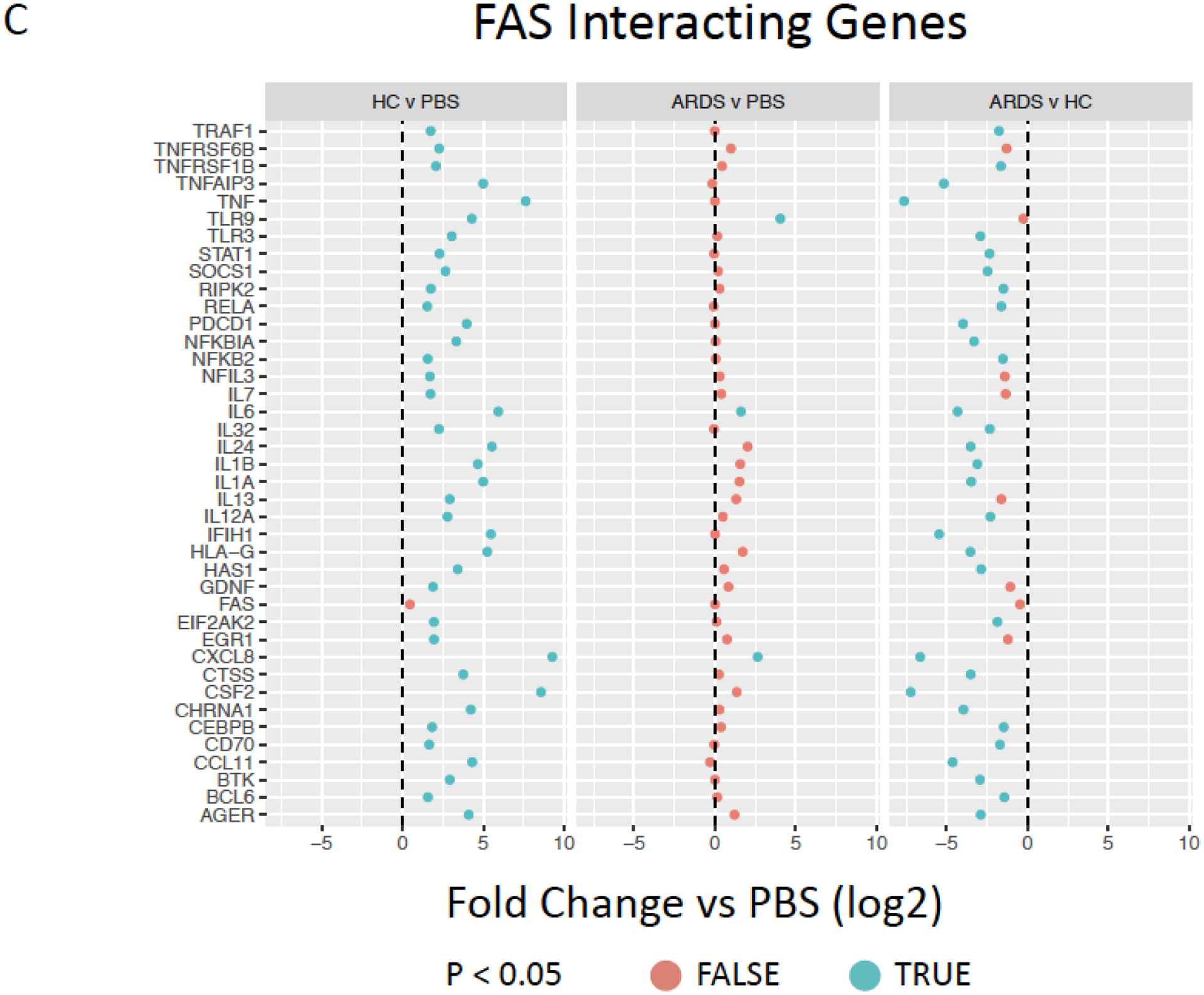
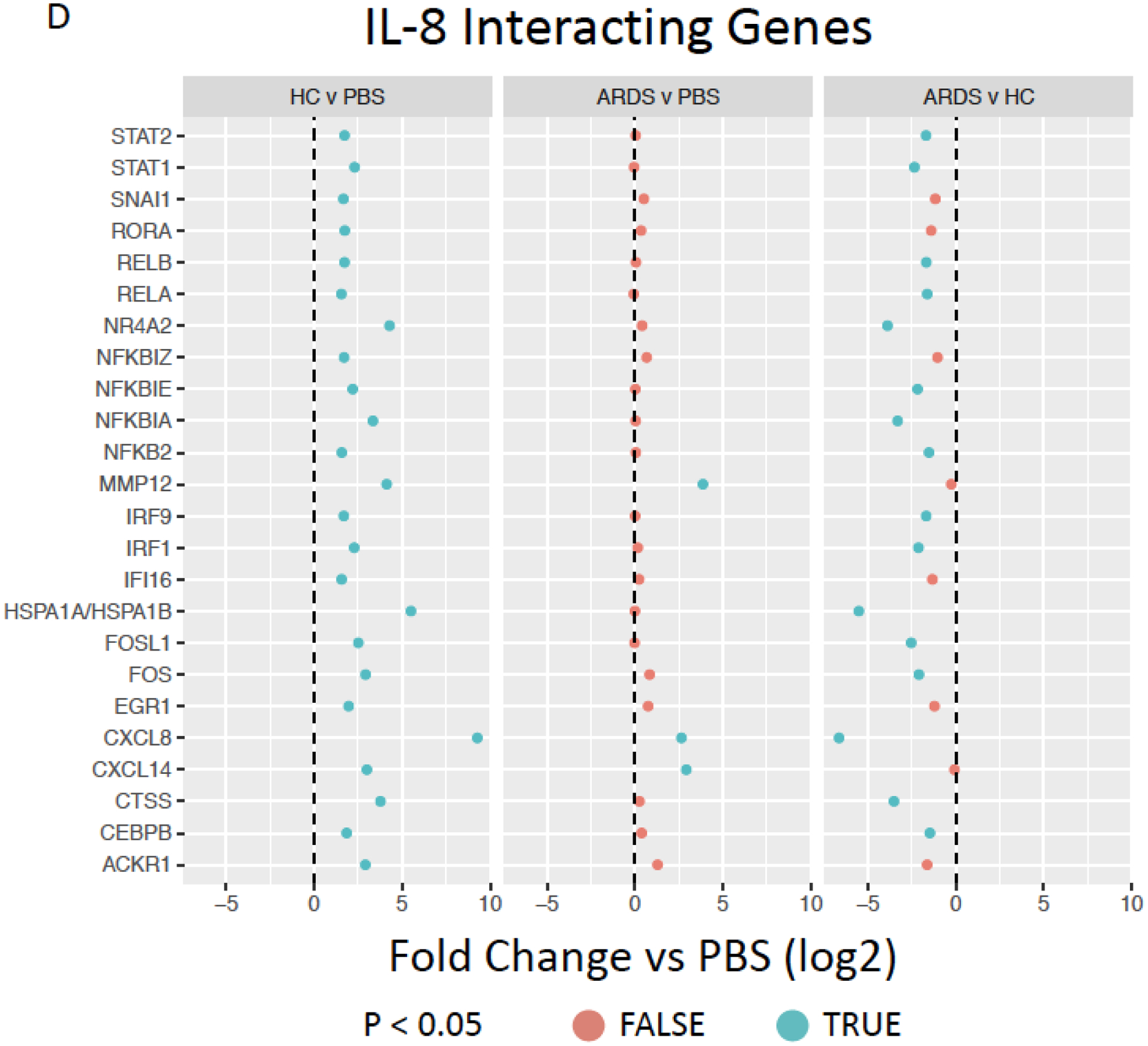
Inferred inflammatory genes resulting from Ingenuity software data analysis including (A), IL-6 (B), CXCL8/IL-8 (C), and FAS (D) interacting genes expressed by ARDS-exposed (left, n=5) and healthy control BALF (right, n=5) samples were compared to PBS-exposed hMSCs (n=4). Red color indicates no significant different (p>0.05) and blue color indicates significantly difference (p<0.05). Data are presented as mean of log2 fold change.
Further evaluations were undertaken to assess BALF exposure effect on hMSC expression of other inter-related genes associated with selected prominent inflammatory and other mediators for which both gene (Figures 3B, F) and protein levels (Figure 2A, Supplementary Figure 3) were increased following BALF exposures, specifically IL-6, IL-8, and FAS. Notably, HC BALF exposures significantly induced a range of IL-6 interacting genes whereas in contrast ARDS BALF exposures resulted in similar expression as observed in PBS-exposed hMSCs with only a significant increase in IL-6 gene expression itself. Interestingly, both HC and ARDS BALF exposure had an inhibitory effect on secretion of the anti-inflammatory cytokine IL-27 compare to PBS-exposed cells (Figure 5B). Similarly, a marked induction of a wide range of FAS interactive genes was only observed in hMSCs exposed to HC BALF samples (Figure 5C). A similar pattern was observed for the pro-inflammatory cytokine IL-8 in which hMSC gene but not protein expression was increased following BALF exposures. Increase in expression of a range of IL-8 interacting genes was increased following HC BALF exposure with ARDS BALF exposure increasing expression of only a few genes (Figure 5D). Taken together, these data suggest that HC BALF exposure induces increased expression of genes involved in multiple immune-regulatory pathways. However, this is not seen in ARDS BALF-exposed hMSCs except for a few genes, some of which are involved in neutrophil trafficking.
Exposure to HC but not ARDS BALF increases complement gene and protein expression but not tissue factor or other coagulation cascade gene expression
hMSCs exposed to HC BALF samples demonstrated increased gene expression of complements C3b and C4a as well as the C3A complement receptor (C3AR) compared to PBS-exposed hMSCs (Figure 6A). In contrast, ARDS BALF-exposed hMSCs demonstrated only increased C3b expression with expression of other complement cascade genes at levels similar to those observed in PBS-exposed hMSCs (Figure 6A). Direct comparison of ARDS vs. HC BALF-exposed hMSCs demonstrated respective decrease in C4a and C3AR as well as C2a and BFb gene expression. Assessment of the CM from BALF and PBS exposed to hMSCs obtained from two different donors demonstrated no detectable complement (C3) production by either HC BALF or PBS-exposed hMSCs (Figure 6B–C). However, low but detectable levels of complement were seen in some of the conditioned media from hMSCs exposed to ARDS BALF (Figure 6B–C).
Figure 6. Exposure to healthy control but not ARDS BALF increases complement gene and protein but not coagulation cascade gene expression.
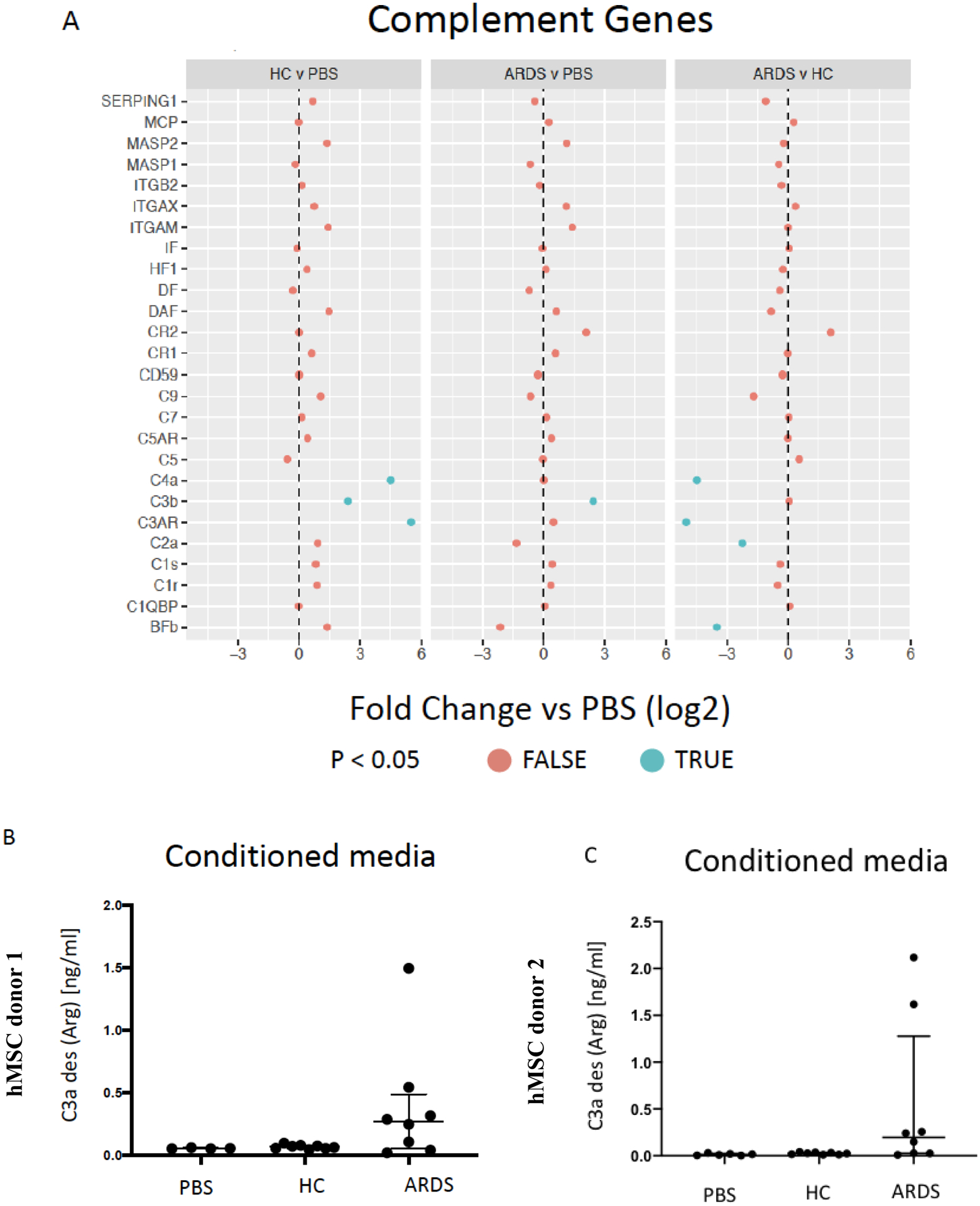
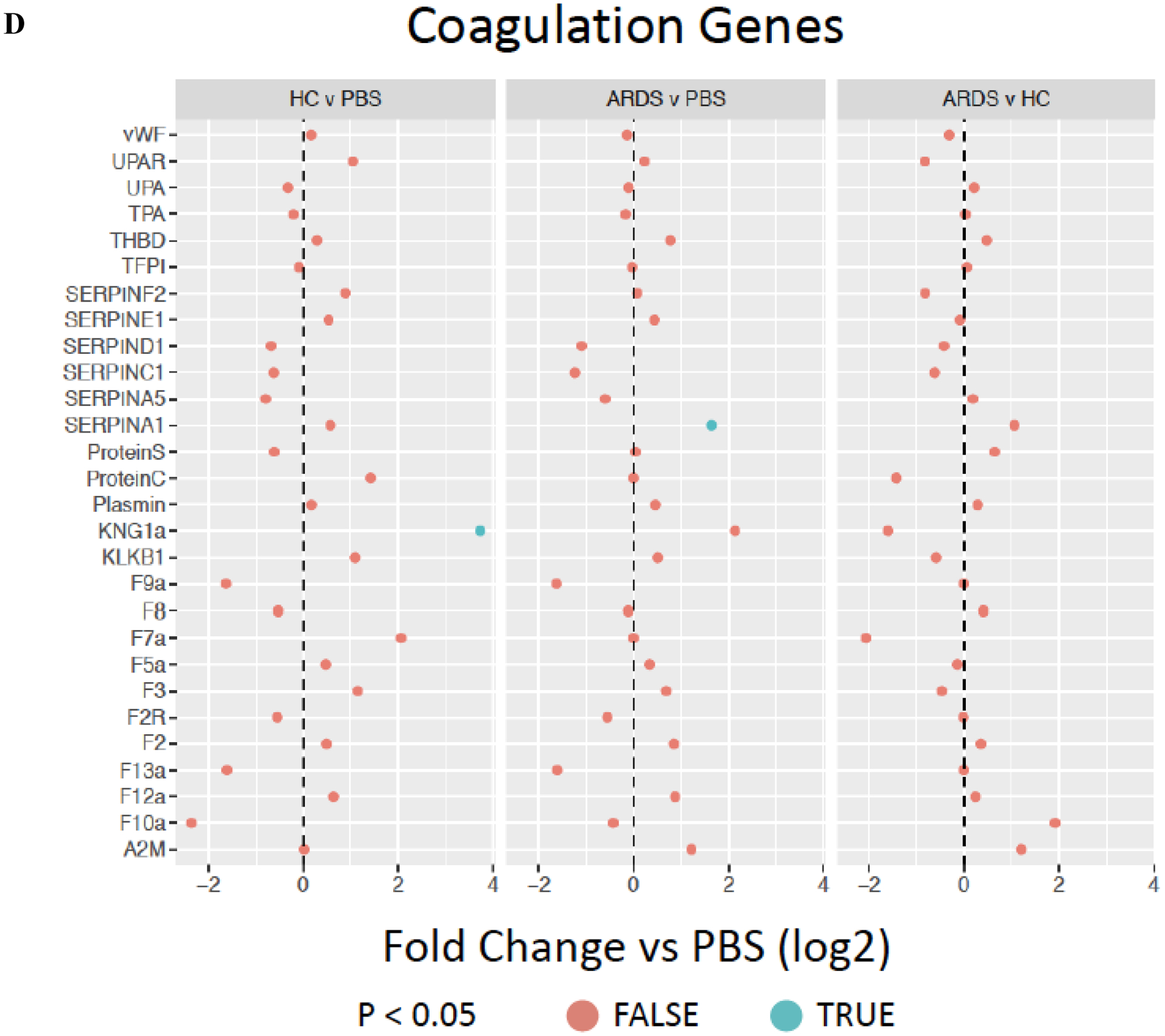
Complement genes (A) expressed by ARDS-exposed (n=5) and healthy control BALF (n=5) samples were compared to PBS-exposed hMSCs (n=4). Levels of complement secreted into the conditioned media by HC or ARDS BALF compared to PBS-exposed hMSCs are depicted in B-C. Two different hMSC donors were used. Abbreviations: HC, healthy control; ARDS, acute respiratory distress syndrome; PBS, phosphate-buffered saline; TFP1, tissue factor. Coagulation cascade genes (D) expressed by ARDS-exposed (n=5) and healthy control BALF (n=5) samples were compared to PBS-exposed hMSCs (n=4). Gene expression data are presented as mean of log2 fold change. Complement (C3a) levels are depicted media with interquartile range.
Parallel assessment of coagulation-associated gene expression demonstrated no changes in tissue factor (TFP1) expression in hMSCs exposed to either HC or ARDS BALF as compared to PBS-exposed cells (Figure 6D). Isolated increase in gene expression of KNG1a, part of the intrinsic coagulation cascade was observed in HC-exposed hMSCs whereas isolated increase in SERPIN A1 (plasminogen activator-1) gene expression was observed in ARDS BALF-exposed hMSCs (Figure 6D). These results suggest that although complement-related gene and protein expression is increased following BALF exposure, unlike IBMIR, there is no increase tissue factor and only isolated changes in other coagulation cascade gene expression.
Exposure to HC but not ARDS BALF increases hMSC HLA gene and cell surface protein expression
We next assessed BALF exposure effects on expression of genes and proteins that might result in recognition of the MSCs by the host immune system, notably HLA expression. HC BALF exposure resulted in significantly increased expression of a number of HLA Class I and II genes and pseudogenes when compared to PBS-exposed hMSCs. These included classical (A, B, C) and non-classical (E, F, G) HLA Class I and also HLA Class II including DRA and DRB1, DMA, DMB, and DPA1 (major Class I and II genes depicted in Figure 7A with all HLA genes evaluated depicted in Supplementary Figure 4). In contrast, exposure to ARDS BALF resulted in significant increase only in C and F and significant decrease in G, with no changes in A, B, or E compared to PBS-exposed hMSCs (Figure 7A, Supplementary Figure 4). Comparably, exposure to ARDS BALF resulted in significant increase only in DMA and significant decrease in DRA, with no changes in any of the other Class II genes. Strikingly, a significant decrease in expression of genes encoding for HLA class I genes A, B, and G, and HLA class II DRB1 was observed when comparing ARDS BALF to HC BALF-exposed hMSCs (Figure 7A).
Figure 7. Exposure to healthy control but not ARDS BALF increases Class II hMSC HLA gene and protein expression.
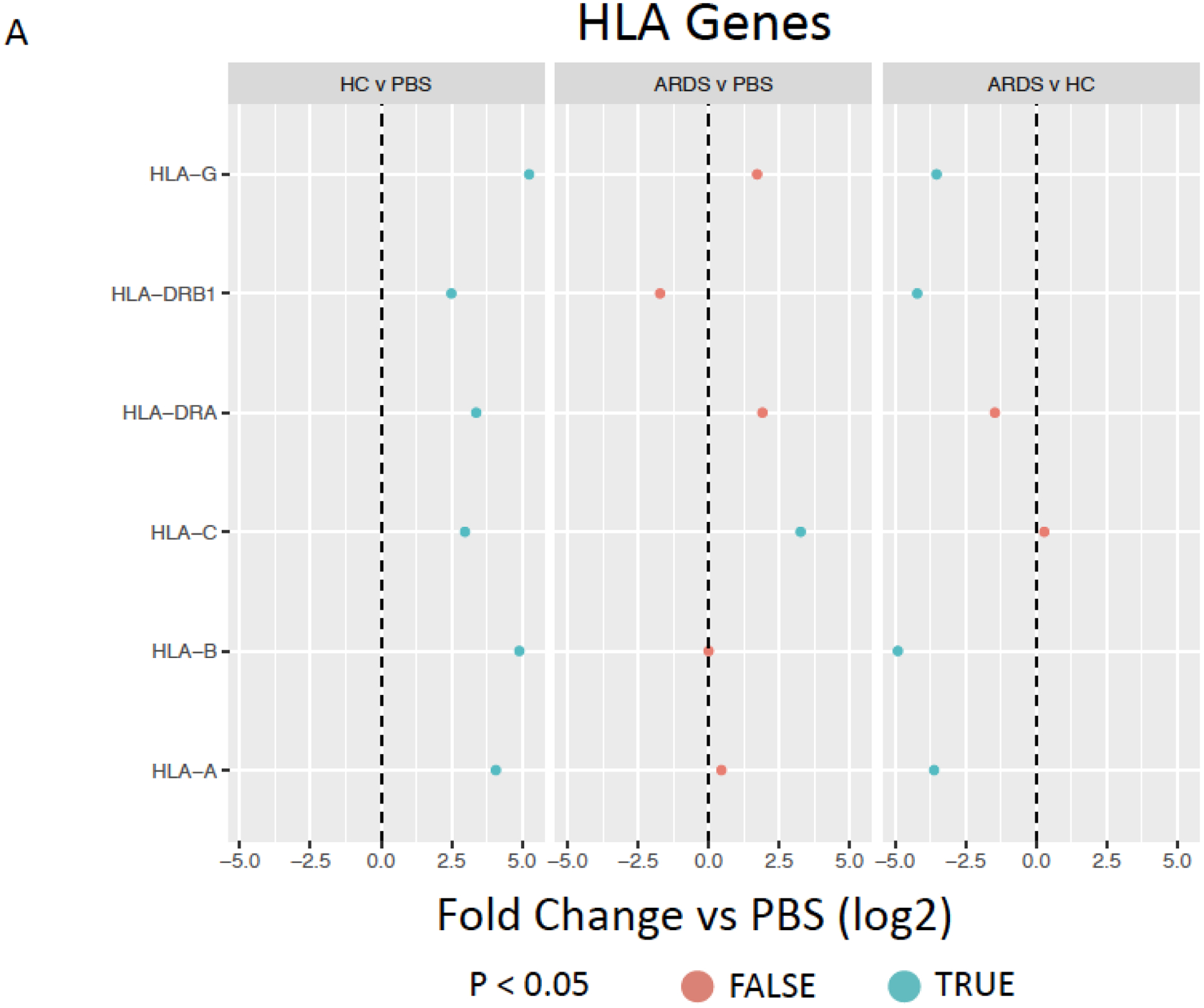
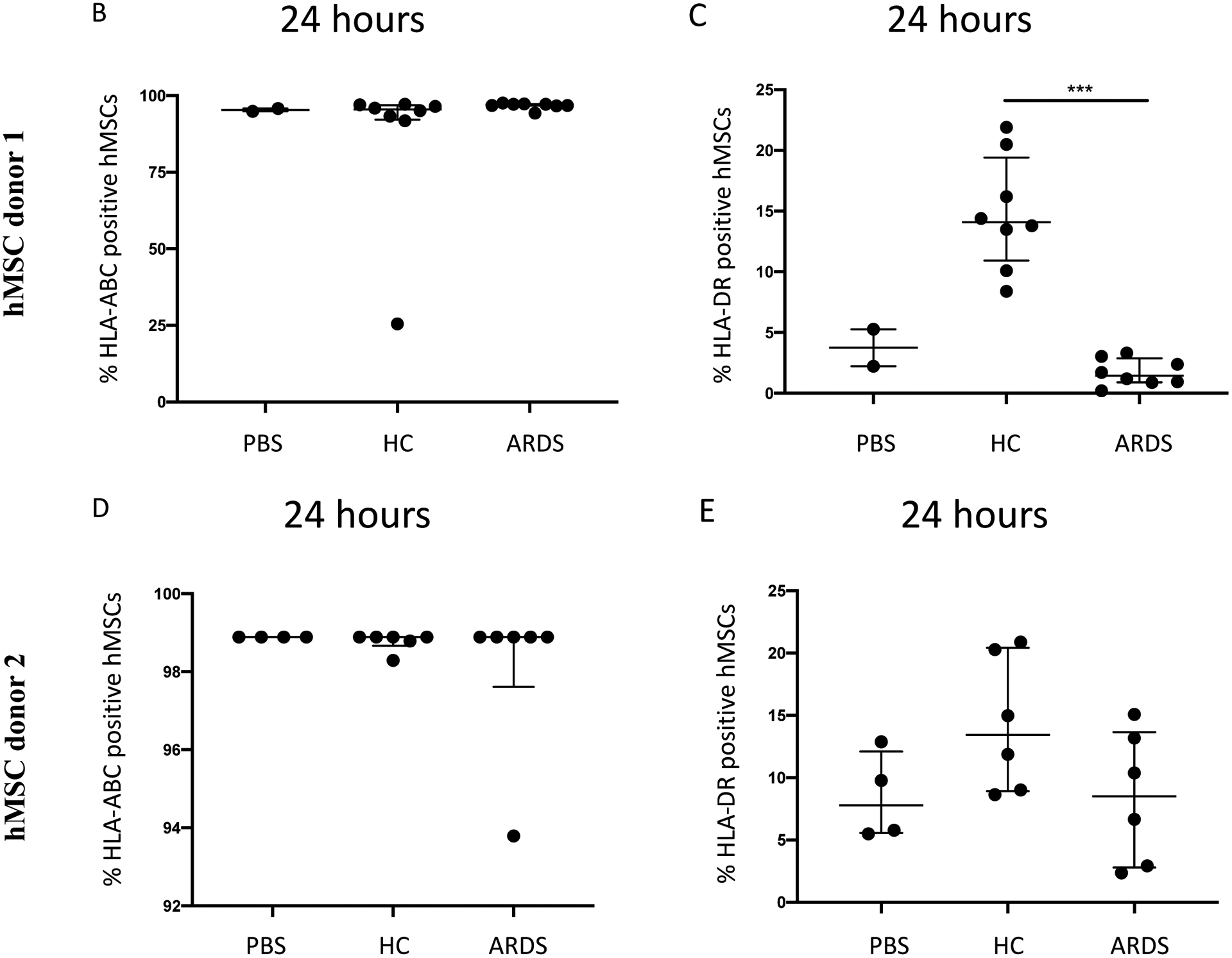
HLA genes expressed by ARDS-exposed (n=5) and healthy control BALF (n=5) samples were compared to PBS-exposed hMSCs (n=4) (A). Data are presented as mean of log2 fold change. HLA-ABC (B, D) and HLA-DR (C, E) surface marker expression was measured with flow cytometry on hMSCs exposed to PBS, HC BALF samples, and ARDS BALF samples for 24 hours. Two different hMSC donors were used. Data are presented as median with interquartile range of % positive cells – % positive cells on DAPI only stainings. Statistical analysis was performed by Kruskal-Wallis followed by Dunn’s post hoc test. Abbreviations: HC, healthy control; ARDS, acute respiratory distress syndrome; PBS, phosphate-buffered saline; HLA, human leukocyte antigen.
To further validate these observations, hMSC HLA-ABC and DR protein expression was evaluated by flow cytometry following exposure to HC vs. ARDS BALF using two different hMSC donors. These results demonstrated an upregulation of HLA-DR expression in HC BALF-exposed hMSCs (Figure 7C, E). In contrast, exposure to ARDS BALF or to PBS resulted in no upregulation of HLA-DR surface marker expression (Figure 7C, E). No change in HLA-ABC expression compared to control was observed after exposure to either BALF samples or PBS (Figure 7B, D). Scattergrams from these studies are shown in Supplementary Figure 4. These results demonstrate that exposure of allogeneic hMSCs to a normal lung environment up-regulates expression of a non-self antigen recognized by the host immune system that will provoke clearance of the hMSCs. In contrast, hMSCs exposed to inflammatory ARDS BALF appear to be protected in this regard.
DISCUSSION
There remains a fundamental lack of knowledge as to the fate and actions of MSCs in clinical lung disease inflammatory environments. Here we found that hMSCs exposed to ARDS BALF behaved quite differently from the same hMSCs exposed to HC BALF. Not only was the pro-inflammatory gene response observed with HC BALF exposure blunted with ARDS BALF exposure but also the increased expression of genes and proteins associated with self- vs non-self recognition following HC BALF exposure. These are novel observations that provide insight into the potential mechanisms of action of MSC-based cell therapies in ARDS.
Our data support the tantalizing hypothesis that a mechanism similar to IBMIR may be responsible for the aggressive removal of hMSCs in healthy individual lungs. Similar to the IBMIR literature centered on blood exposure, when stimulated with HC BALF, hMSCs responded by engaging in behaviors that will provoke an acute innate immune response including marked increase in pro-inflammatory, complement, and Class II HLA gene and protein expression [19, 20]. In striking contrast, stimulation of the same hMSCs with BALF from ARDS patients either failed or comparatively resulted in decreased expression of these self antigen-encoding genes and proteins. However, in contrast to IBMIR, exposure of hMSCs to either HC or ARDS BALF did not increase tissue factor gene expression, an important observation supporting the safety of these cells in clinical practice. Notably, a recent study found that allogeneic MSC exposure to blood of trauma patients was less potent in inducing tissue factor expression than was exposure to healthy volunteer blood [24]. This suggests that IBMIR can be affected by the patient’s inflammatory status but there is little other data in this regards. hMSCs are not well recognized as producing complement factors although they do produce anti-bacterial peptides such as LL-37 [25]. Paradoxically, some of the ARDS but not HC BALF samples stimulated detectable levels of complement (C3a) in hMSC-CM. At present, the significane of these findings remains unknown.
Human MSCs generally express low levels of HLA class I molecules and no constitutive expression of Class II molecules [18, 26, 27], attributes that have long been thought to minimize recognition of systemically administered allogeneic hMSCs by host immune surveillance mechanisms [28]. However, HLA-ABC and HLA-DR expression can be induced by exposure to several factors, including IFN-γ, which contributes to increased immune recognition and clearance of hMSCs [29, 30]. Thus, the observed results demonstrating increased expression of several Class II molecules following exposure to HC but not ARDS BALF further suggests that an intact normal allogeneic lung environment can activate hMSCs to participate in immune surveillance activities that may result in their own inactivation and clearance. Importantly, similar results were observed using hMSCs from two different donors, which strengthens the hypothesis that this is a general hMSC response. These observations parallel data from the literature showing hMSCs may persist for longer time in inflamed vs non-inflamed lungs presumably allowing for more opportunity to exert effects on inflammatory pathways [24, 31, 32]. However, although IFN-γ was detected at low levels in both groups there was no significant difference. As such, even though exposure to HC BALF samples triggers increased Class II HLA expression, this does not appear to be correlated with BALF IFN-γ. One possible explanation is that low doses of IFN-γ have been demonstrated to have less effects on MSC immunosuppressive potency compared to higher doses [33]. An increased expression of the IFN-γ-dependent HLA class II transactivator C2Ta was observed, however the significance of this finding remains unknown [34].
Notably, BALF IL-1β was significantly predictive of hMSC production of a range of important pro-inflammatory mediators. This suggests that IL-1β, commonly elevated in ARDS lungs, can drive hMSC behaviors and that blocking IL-1β may potentially alter hMSC actions. However, the overall picture is complex, as demonstrated in Figure 3 and Supplemental Figure 2. This suggests that healthy lung environments, are capable of provoking inflammatory behaviors in allogeneic bone marrow-derived hMSCs.
This study has several strengths. First, the hMSCs utilized are clinically relevant, having also been utilized in the recent START trial of systemic hMSC administration in ARDS patients [3, 4]. Second, the BALF samples were assessed individually rather than as pooled samples. Importantly, the current findings are robust and reproducible across multiple individual healthy or ARDS BALF samples. Further, the effects on complement and HLA protein expression were observed with two different bone marrow-derived MSC isolates obtained from different HC. However, one caveat is that the BALF samples utilized came from a range of participating institutions and were obtained by different operators. As such, it is not possible to fully demonstrate uniformity in either bronchoscopy procedures utilized or in any potential sample dilutions. Further, the BALF samples utilized were obtained using different isolation protocols, were stored differently both at the originating institutions −80°C (ARDS) and −70°C (HC) and also in our laboratory at −20°C (ARDS) and −70°C (HC) respectively. As such, although we feel these difference unlikely to account for the current observations, further prospective studies will attempt to better control for these variable Further, given HIPAA limitations, no clinical data, including microbiologic data, on the underlying etiology of the ARDS patients is available except that for the ARDSNET clinical study under which these samples were obtained, patients with sepsis/septic shock were excluded [21]. Similarly, limited clinical information is available for the healthy volunteers. One additional significant caveat to this study is that the BALF samples utilized were cell-free supernatants. As such, we speculate that the absence of direct toxicity following overnight hMSC exposure to the BALF samples likely reflects the absence of immune effector cells that would then clear the hMSCs through efferocytosis and possibly other means of clearance and/or inactivation (15). Further investigations, for example with mixed lymphocyte studies utilizing BALF-exposed hMSCs or otherwise adding immune effector cells to the BALF samples, will further provide important information on the lung inflammatory environment effects on hMSC behaviors.
In summary, hMSC exposure to healthy lung environments induces expression of a range of genes encoding for inflammation and also for recognition as foreign to the host immune system. These changes are not observed or in some cases are opposite following hMSC exposure to inflammatory ARDS lung environments. Nonetheless, both environments provoke often comparable production and/or release of both pro- and anti-inflammatory-related mediators by the hMSCs. Further, selected components in the BALF are correlated with and in some cases predictive of hMSC mediator release. These observations provide a growing understanding of the complex interplay of inflammatory and other pathways involved in hMSC actions in the lung and provide important information towards developing more effective MSC-based cell therapies for ARDS and other lung diseases.
Supplementary Material
Take home message:
MSCs exposed to a healthy lung environment induces an inflammatory response with increased gene/protein expression associated with self vs non-self-recognition. These changes were either absent or opposite in MSCs exposed to an inflamed ARDS environment.
ACKNOWLEDGEMENTS
The authors gratefully acknowledge Drs. Jennifer L. Ather and Matthew E. Poynter for assistance with the cytokine measurements, and Evan Hoffman for excellent overall technical assistance. The RNA sequencing was carried out by Dr. Fred W. Kolling at the Geisel School of Medicine at Dartmouth in the Genomics Shared Resources, which was established by equipment grants from the NIH and NSF and is supported by a Cancer Centre Core Grant (P30CA023108) from the National Cancer Institute. The authors gratefully thanks the European Union (EU) and European Respiratory Society (ERS) for financial support.
All sources of support
A.A is supported by R01HL122372 (NIH/NHLBI), ASHARE15A0 (CFF), and the Translational Research Core (STANTO19R0 to BAS and P30DK117469 to Dean Madden). T.H.H and B.A.S are supported by grants from the NIH (P30-DK117469 and R01 HL151385) and the Cystic Fibrosis Foundation (STANTO19R0, STANTO19GO and STANTO02PO). S.R.E is supported by a Marie Curie Postdoctoral Research Fellowship (RESPIRE3) from the European Respiratory Society and the European Union’s H2020 research and innovation programme (Marie Sklodowska-Curie grant agreement No. 713406). D.J.W is supported by the National Heart Lung Blood Institute (HL127144, EB024329), Department of Defense, Cystic Fibrosis Foundation, and the University of Vermont. D.H.M MSC manufacturing was funded through the NIH Production Assistance for Cellular Therapies (PACT) contract with the University of Minnesota, Molecular and Cellular Therapeutics (HHSN268201600014I; PI: DHM). K.E is supported by an Irish Reserch Council Laureate award IRCLA/2017/288. A.K is supported by UK Medical Research Council Research Awards (MRC MR/R025096/1 and MR/S009426/1). P.R.M.R is supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico e Fundação Carlos Chagas Filho de Amparo a Pesquisa do Estado do Rio de Janeiro. M.A.M is supported by HL134828 and HL140026.
Publisher's Disclaimer: This manuscript has recently been accepted for publication in the European Respiratory Journal. It is published here in its accepted form prior to copyediting and typesetting by our production team. After these production processes are complete and the authors have approved the resulting proofs, the article will move to the latest issue of the ERJ online.
This article has an online data supplement.
REFERENCES
- [1].Matthay MA, Zemans RL, Zimmerman GA, Arabi YM, Beitler JR, Mercat A, Herridge M, Randolph AG, Calfee CS, Acute respiratory distress syndrome, Nat Rev Dis Primers 5(1) (2019) 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Khoury M, Cuenca J, Cruz FF, Figueroa FE, Rocco PRM, Weiss DJ, Current Status of Cell-Based Therapies for Respiratory Virus Infections: Applicability to COVID-19, Eur Respir J (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wilson JG, Liu KD, Zhuo H, Caballero L, McMillan M, Fang X, Cosgrove K, Vojnik R, Calfee CS, Lee JW, Rogers AJ, Levitt J, Wiener-Kronish J, Bajwa EK, Leavitt A, McKenna D, Thompson BT, Matthay MA, Mesenchymal stem (stromal) cells for treatment of ARDS: a phase 1 clinical trial, Lancet Respir Med 3(1) (2015) 24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Matthay MA, Calfee CS, Zhuo H, Thompson BT, Wilson JG, Levitt JE, Rogers AJ, Gotts JE, Wiener-Kronish JP, Bajwa EK, Donahoe MP, McVerry BJ, Ortiz LA, Exline M, Christman JW, Abbott J, Delucchi KL, Caballero L, McMillan M, McKenna DH, Liu KD, Treatment with allogeneic mesenchymal stromal cells for moderate to severe acute respiratory distress syndrome (START study): a randomised phase 2a safety trial, Lancet Respir Med 7(2) (2019) 154–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Simonson OE, Mougiakakos D, Heldring N, Bassi G, Johansson HJ, Dalen M, Jitschin R, Rodin S, Corbascio M, El Andaloussi S, Wiklander OP, Nordin JZ, Skog J, Romain C, Koestler T, Hellgren-Johansson L, Schiller P, Joachimsson PO, Hagglund H, Mattsson M, Lehtio J, Faridani OR, Sandberg R, Korsgren O, Krampera M, Weiss DJ, Grinnemo KH, Le Blanc K, In Vivo Effects of Mesenchymal Stromal Cells in Two Patients With Severe Acute Respiratory Distress Syndrome, Stem Cells Transl Med 4(10) (2015) 1199–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sara Rolandsson Enes GW-T, Comparison of the Regenerative Potential for Lung Tissue of Mesenchymal Stromal Cells from Different Sources/Locations Within the Body, in: Janette IHH Burgess K (Ed.), Stem Cell-Based Therapy for Lung Disease, Springer; 2019. [Google Scholar]
- [7].Kusuma GD, Carthew J, Lim R, Frith JE, Effect of the Microenvironment on Mesenchymal Stem Cell Paracrine Signaling: Opportunities to Engineer the Therapeutic Effect, Stem Cells Dev 26(9) (2017) 617–631. [DOI] [PubMed] [Google Scholar]
- [8].Xu AL, Rodriguez LA 2nd, Walker KP 3rd, Mohammadipoor A, Kamucheka RM, Cancio LC, Batchinsky AI, Antebi B, Mesenchymal Stem Cells Reconditioned in Their Own Serum Exhibit Augmented Therapeutic Properties in the Setting of Acute Respiratory Distress Syndrome, Stem Cells Transl Med 8(10) (2019) 1092–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Islam D, Huang Y, Fanelli V, Delsedime L, Wu S, Khang J, Han B, Grassi A, Li M, Xu Y, Luo A, Wu J, Liu X, McKillop M, Medin J, Qiu H, Zhong N, Liu M, Laffey J, Li Y, Zhang H, Identification and Modulation of Microenvironment Is Crucial for Effective Mesenchymal Stromal Cell Therapy in Acute Lung Injury, Am J Respir Crit Care Med 199(10) (2019) 1214–1224. [DOI] [PubMed] [Google Scholar]
- [10].Bustos ML, Huleihel L, Meyer EM, Donnenberg AD, Donnenberg VS, Sciurba JD, Mroz L, McVerry BJ, Ellis BM, Kaminski N, Rojas M, Activation of human mesenchymal stem cells impacts their therapeutic abilities in lung injury by increasing interleukin (IL)-10 and IL-1RN levels, Stem Cells Transl Med 2(11) (2013) 884–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Abreu SC, Enes SR, Dearborn J, Goodwin M, Coffey A, Borg ZD, Dos Santos CC, Wargo MJ, Cruz FF, Loi R, DeSarno M, Ashikaga T, Antunes MA, Rocco PRM, Liu KD, Lee JW, Matthay MA, McKenna DH, Weiss DJ, Lung Inflammatory Environments Differentially Alter Mesenchymal Stromal Cell Behavior, Am J Physiol Lung Cell Mol Physiol (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Abreu SC, Xisto DG, de Oliveira TB, Blanco NG, de Castro LL, Kitoko JZ, Olsen PC, Lopes-Pacheco M, Morales MM, Weiss DJ, Rocco PRM, Serum from Asthmatic Mice Potentiates the Therapeutic Effects of Mesenchymal Stromal Cells in Experimental Allergic Asthma, Stem Cells Transl Med 8(3) (2019) 301–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Soraia THH Abreu C, Hoffman Evan, Dearborn Jacob, Ashare Alix, Singh Sidhu Karatatiwant, Matthews Dwight E., McKenna David H., Amiel Eyal, Barua Jayita, Krasnodembskaya Anna, English Karen, Mahon Bernard, dos Santos Claudia, Cruz Fernanda F., Chambers Daniel C., Liu Kathleen D., Matthay Michael A., Cramer Robert A., Stanton Bruce A., Rocco Patricia R. M., Wargo Matthew J., Weiss Daniel J., Rolandsson Enes Sara, Differential effects of the cystic fibrosis lung inflammatory environment on mesenchymal stromal cells, American Journal of Physiology-Lung Cellular and Molecular Physiology In Press (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Galleu A, Riffo-Vasquez Y, Trento C, Lomas C, Dolcetti L, Cheung TS, von Bonin M, Barbieri L, Halai K, Ward S, Weng L, Chakraverty R, Lombardi G, Watt FM, Orchard K, Marks DI, Apperley J, Bornhauser M, Walczak H, Bennett C, Dazzi F, Apoptosis in mesenchymal stromal cells induces in vivo recipient-mediated immunomodulation, Sci Transl Med 9(416) (2017). [DOI] [PubMed] [Google Scholar]
- [15].Weiss DJ, English K, Krasnodembskaya A, Isaza-Correa JM, Hawthorne IJ, Mahon BP, The Necrobiology of Mesenchymal Stromal Cells Affects Therapeutic Efficacy, Front Immunol 10 (2019) 1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Weiss ARR, Dahlke MH, Immunomodulation by Mesenchymal Stem Cells (MSCs): Mechanisms of Action of Living, Apoptotic, and Dead MSCs, Front Immunol 10 (2019) 1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Galipeau J, Sensebe L, Mesenchymal Stromal Cells: Clinical Challenges and Therapeutic Opportunities, Cell Stem Cell 22(6) (2018) 824–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Moll G, Hult A, von Bahr L, Alm JJ, Heldring N, Hamad OA, Stenbeck-Funke L, Larsson S, Teramura Y, Roelofs H, Nilsson B, Fibbe WE, Olsson ML, Le Blanc K, Do ABO blood group antigens hamper the therapeutic efficacy of mesenchymal stromal cells?, PLoS One 9(1) (2014) e85040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hoogduijn MJ, Roemeling-van Rhijn M, Engela AU, Korevaar SS, Mensah FK, Franquesa M, de Bruin RW, Betjes MG, Weimar W, Baan CC, Mesenchymal stem cells induce an inflammatory response after intravenous infusion, Stem Cells Dev 22(21) (2013) 2825–35. [DOI] [PubMed] [Google Scholar]
- [20].Moll G, Jitschin R, von Bahr L, Rasmusson-Duprez I, Sundberg B, Lonnies L, Elgue G, Nilsson-Ekdahl K, Mougiakakos D, Lambris JD, Ringden O, Le Blanc K, Nilsson B, Mesenchymal stromal cells engage complement and complement receptor bearing innate effector cells to modulate immune responses, PLoS One 6(7) (2011) e21703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Liu KD, Levitt J, Zhuo H, Kallet RH, Brady S, Steingrub J, Tidswell M, Siegel MD, Soto G, Peterson MW, Chesnutt MS, Phillips C, Weinacker A, Thompson BT, Eisner MD, Matthay MA, Randomized clinical trial of activated protein C for the treatment of acute lung injury, Am J Respir Crit Care Med 178(6) (2008) 618–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C, Salmon provides fast and bias-aware quantification of transcript expression, Nat Methods 14(4) (2017) 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Soneson C, Love MI, Robinson MD, Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences, F1000Res 4 (2015) 1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].George MJ, Prabhakara K, Toledano-Furman NE, Gill BS, Wade CE, Cotton BA, Cap AP, Olson SD, Cox CS Jr., Procoagulant in vitro effects of clinical cellular therapeutics in a severely injured trauma population, Stem Cells Transl Med 9(4) (2020) 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Krasnodembskaya A, Song Y, Fang X, Gupta N, Serikov V, Lee JW, Matthay MA, Antibacterial effect of human mesenchymal stem cells is mediated in part from secretion of the antimicrobial peptide LL-37, Stem Cells 28(12) (2010) 2229–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wang Y, Huang J, Gong L, Yu D, An C, Bunpetch V, Dai J, Huang H, Zou X, Ouyang H, Liu H, The Plasticity of Mesenchymal Stem Cells in Regulating Surface HLA-I, iScience 15 (2019) 66–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kot M, Baj-Krzyworzeka M, Szatanek R, Musial-Wysocka A, Suda-Szczurek M, Majka M, The Importance of HLA Assessment in “Off-the-Shelf” Allogeneic Mesenchymal Stem Cells Based-Therapies, Int J Mol Sci 20(22) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ryan JM, Barry FP, Murphy JM, Mahon BP, Mesenchymal stem cells avoid allogeneic rejection, J Inflamm (Lond) 2 (2005) 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Chinnadurai R, Copland IB, Garcia MA, Petersen CT, Lewis CN, Waller EK, Kirk AD, Galipeau J, Cryopreserved Mesenchymal Stromal Cells Are Susceptible to T-Cell Mediated Apoptosis Which Is Partly Rescued by IFNgamma Licensing, Stem Cells 34(9) (2016) 2429–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Stagg J, Pommey S, Eliopoulos N, Galipeau J, Interferon-gamma-stimulated marrow stromal cells: a new type of nonhematopoietic antigen-presenting cell, Blood 107(6) (2006) 2570–7. [DOI] [PubMed] [Google Scholar]
- [31].Moll G, Drzeniek N, Kamhieh-Milz J, Geissler S, Volk HD, Reinke P, MSC Therapies for COVID-19: Importance of Patient Coagulopathy, Thromboprophylaxis, Cell Product Quality and Mode of Delivery for Treatment Safety and Efficacy, Front Immunol 11 (2020) 1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Reilly JP, Calfee CS, Christie JD, Acute Respiratory Distress Syndrome Phenotypes, Semin Respir Crit Care Med 40(1) (2019) 19–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Romieu-Mourez R, Francois M, Boivin MN, Stagg J, Galipeau J, Regulation of MHC class II expression and antigen processing in murine and human mesenchymal stromal cells by IFN-gamma, TGF-beta, and cell density, J Immunol 179(3) (2007) 1549–58. [DOI] [PubMed] [Google Scholar]
- [34].Tang KC, Trzaska KA, Smirnov SV, Kotenko SV, Schwander SK, Ellner JJ, Rameshwar P, Down-regulation of MHC II in mesenchymal stem cells at high IFN-gamma can be partly explained by cytoplasmic retention of CIITA, J Immunol 180(3) (2008) 1826–33. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


