Abstract
During infection Legionella bacteria translocate a variety of effectors into host cells that modify host cell membrane trafficking for the benefit of the intracellular pathogen. Here we found a self-organizing system consisting of a bacterial phosphoinositide kinase and its opposing phosphatase that formed spatiotemporal patterns, including traveling waves, to remodel host cellular membranes. The Legionella effector MavQ, a phosphatidylinositol (PI) 3-kinase, was targeted to the endoplasmic reticulum (ER). MavQ and the Legionella PI 3-phosphatase SidP, even in the absence of other bacterial components, drove rapid PI 3-phosphate turnover on the ER, spontaneously forming traveling waves that spread along ER subdomains inducing vesicle/tubule budding. Thus, bacteria can exploit a self-organizing membrane-targeting mechanism to hijack host cellular structures for survival.
One Sentence Summary:
Legionella translocate a kinase–phosphatase duo into host cells that self-organizes on the endoplasmic reticulum for membrane remodeling.
Intracellular bacterial pathogens employ a variety of strategies for survival and proliferation inside the host cell, and many manipulate the host endomembrane system (1). Because phosphoinositides function as identifiers for membrane-bound organelles and play critical roles in vesicle trafficking, they are frequently targeted by these pathogens, including Legionella pneumophila, the causative agent of Legionnaires’ disease. Two hallmarks of host cell subversion by L. pneumophila are reprogramming of membrane trafficking via phosphoinositide conversion and hijacking the host endoplasmic reticulum (ER) membrane to establish a replicative niche known as the Legionella containing vacuole (LCV) (1). L. pneumophila translocates more than 300 effector proteins via the type 4 secretion system (T4SS) into the host cell to manipulate cellular machinery, and a few Legionella effectors acting as phosphoinositide kinases or phosphatases have been identified (2–5). However, none of these Legionella phosphoinositide kinases and phosphatases are known to target the ER.
Here we describe a two-component module consisting of the Legionella effector MavQ, an atypical PI 3-kinase, and the Legionella PI 3-phosphatase SidP that dynamically self-organize to remodel the host ER.
The Legionella effector MavQ is an atypical PI 3-kinase
We analyzed the Legionella T4SS effectors and identified MavQ (Lpg2975) as a distant homolog of the protein kinase superfamily (fig. S1A). To gain insight into the molecular function of MavQ, we solved the crystal structure of MavQ1–580 (residues 1–580), which resolved an atypical kinase domain (KD) followed by a four-helical bundle (4HB) (Fig. 1, A and B). The KD consists of a β-strand-rich N-lobe and an α-helical-rich C-lobe. The catalytic pocket of the KD adopts an active conformation with ADP/Mg2+ bound in the active site (Fig. 1C).
Fig. 1. The Legionella effector MavQ is an atypical PI 3-kinase.
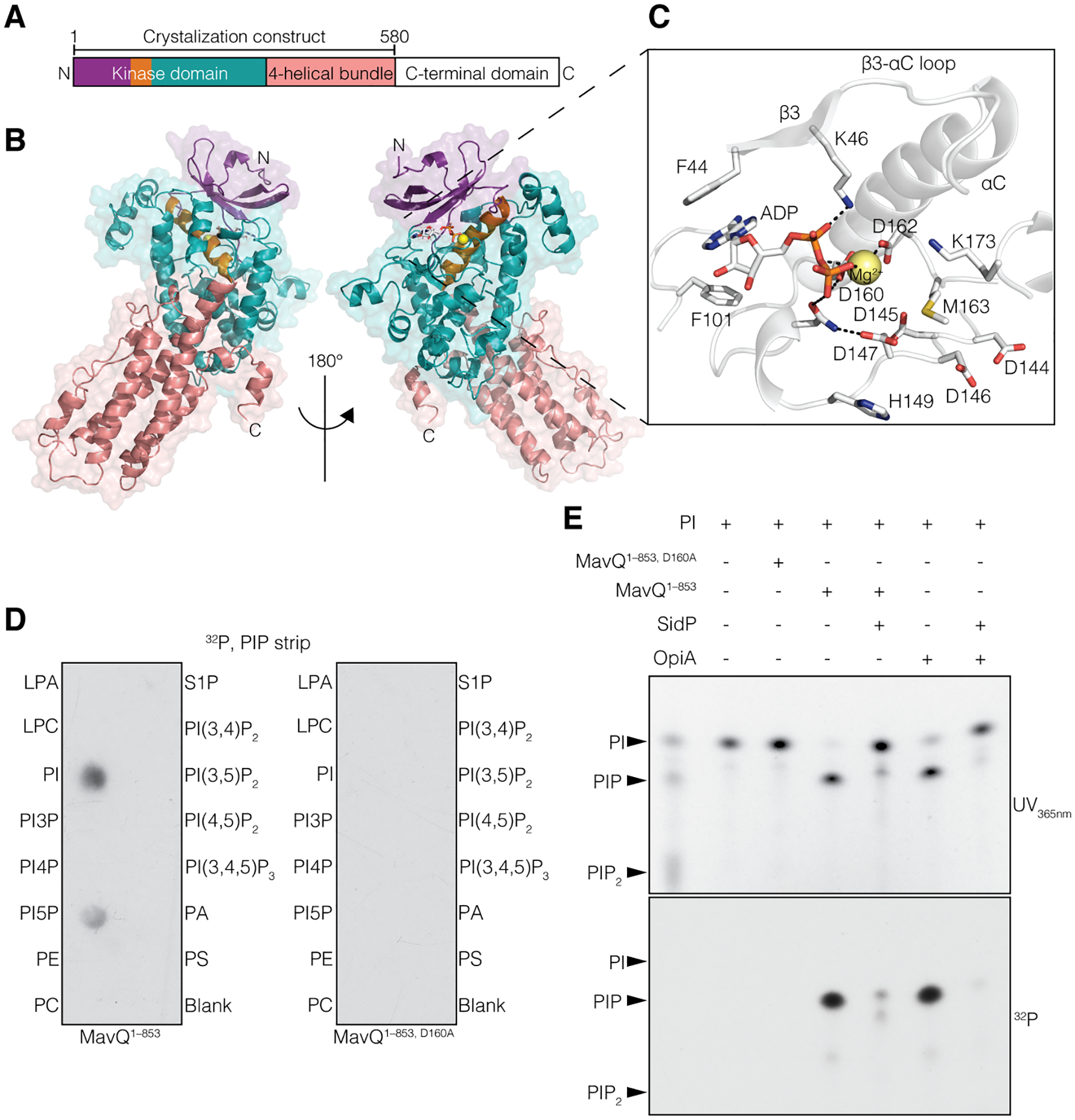
(A and B) Domain architecture of MavQ (A) and overall structure of MavQ1–580 (B) depicting the N-lobe (violet), the αC-helix (orange), the C-lobe (teal), the four-helical bundle (4HB; salmon) and the C-terminal domain (CTD; white). (C) Magnified view of MavQ1–580 kinase active site depicting the interactions (dashed lines) involved in nucleotide binding. The ADP is shown as sticks and the Mg2+ ion as a yellow sphere. (D) Autoradiograph depicting the transfer of γ−32P from [γ−32P]ATP onto PI and PI5P on a PIP strip by MavQ1–853 but not the predicted catalytically inactive mutant MavQ1–853, D160A. (E) Chromatogram depicting the incorporation of γ−32P from [γ−32P]ATP by MavQ1–853 or the inactive mutant using BODIPY-labeled PI as a substrate. Reactions were subsequently treated with or without the PI 3-phosphatase SidP. OpiA, a Francisella tularenesis PI 3-kinase (5), was used as a positive control. Reaction products were separated by thin layer chromatography and visualized by UV365nm fluorescence and autoradiography.
Searches using the DALI server (6) identified several phosphoinositide kinases as the closest structural homologs of MavQ (fig. S1B). A systems biology screen also suggests that MavQ may be a phosphoinositide kinase (7). Indeed, MavQ1–853, but not its predicted catalytically inactive mutant MavQ1–853, D160A, phosphorylated PI and PI 5-phosphate (PI5P) (Fig. 1D and fig. S1C). Ala substitutions of active site residues decreased MavQ’s kinase activity (fig. S1, D and E). MavQ1–853 produced mono- and di-phosphorylated PI species when PI and PI5P were used as substrates, respectively, and treatment with the Legionella PI 3-phosphatase SidP (3) removed the phosphates introduced by MavQ (Fig. 1E and fig. S1F). Furthermore, the MavQ1–853 reaction products with PI-containing liposomes as substrates were detected by the PI3P-specific probe PI3P Grip (fig. S1G). Thus, MavQ is a PI 3-kinase that phosphorylates PI and PI5P in vitro.
MavQ generates PI3P on the ER and atypical vesicular-tubular structures
Fluorescence and immunogold electron microscopy revealed that GFP-MavQ localized to the ER or atypical vesicular-tubular membrane structures in HeLa cells and RAW264.7 macrophages (Fig 2, A and B, fig. S2A–D and movie S1). MavQ, but not MavQD160A, redistributed the PI3P probes 2xFYVEHrs and 2xFYVEEEA1 from early endosomes, the major PI3P-positive membrane compartment, to the ER or vesicular-tubular structures (Fig. 2C and fig. S2E–G). Similar results were obtained using a doxycycline inducible expression system (fig. S2H) and when recombinant GFP-MavQ1–853 was microinjected into the cell (fig. S2I). Notably, MavQ-positive vesicular-tubular structures rarely colocalized with the early endosomal marker RAB5B (fig. S2G), indicating that these structures were unlikely to be of endosomal origin. Moreover, catalytically inactive MavQD160A was mostly cytosolic (Fig. 2C and fig. S2E), suggesting that MavQ’s membrane association depended on its kinase activity. The PI(3,5)P2 probe mCherry-2xML1NTRPML1 was not redistributed to MavQ-positive compartments (fig. S2J), likely because of the absence of PI5P on MavQ-targeted organelles or the accuracy of the probe (8).
Fig. 2. MavQ generates PI3P on the ER and atypical vesicular-tubular structures.
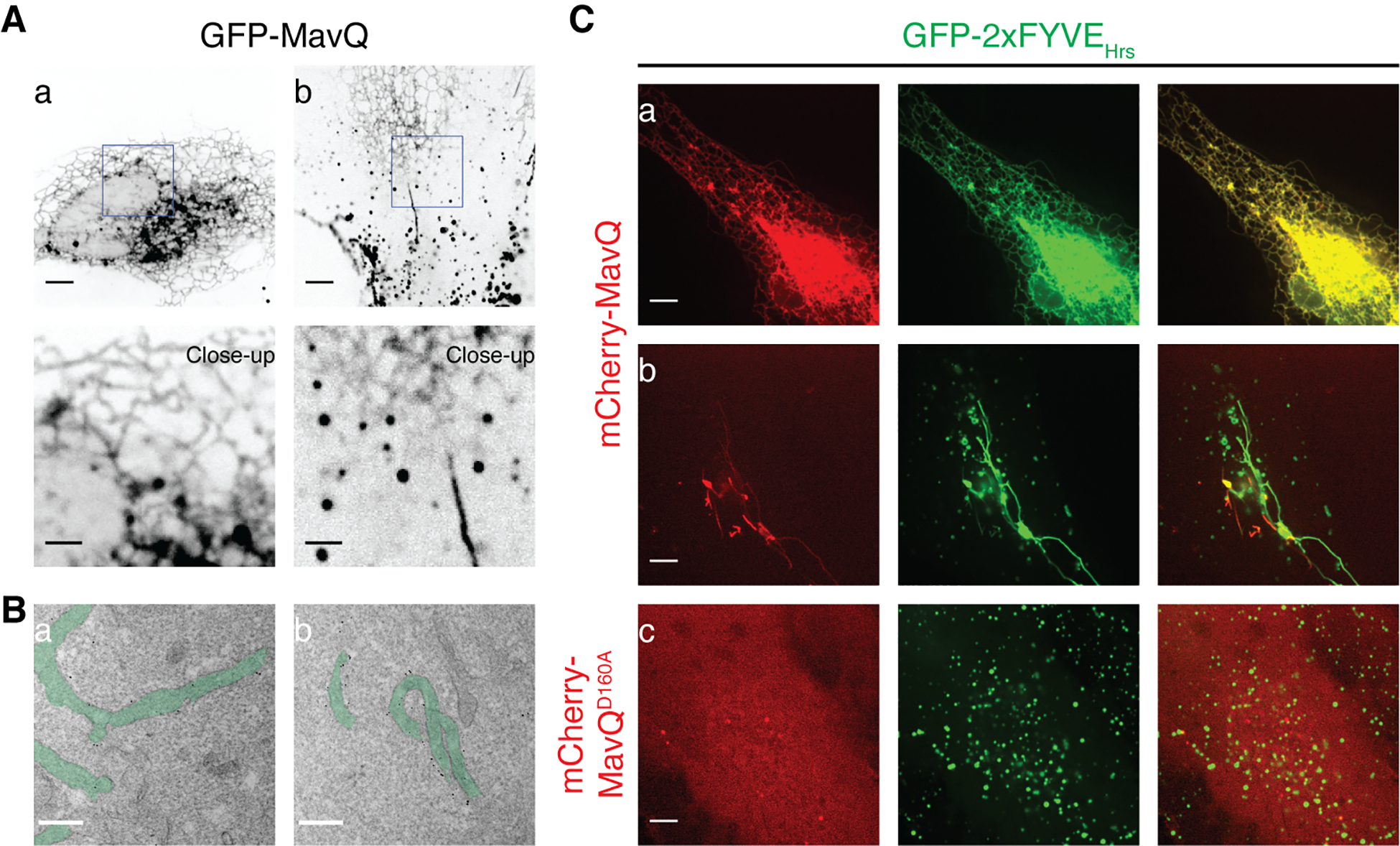
(A) Confocal images of HeLa cells expressing GFP-MavQ. In (a), GFP-MavQ localizes to an ER-like compartment. In (b), GFP-MavQ concentrates on vesicular-tubular structures, in addition to the weak ER-like localization. Bars: 5 and 2 (close-ups) μm. (B) Transmission electron micrographs depicting the ultrastructural localization of GFP-MavQ in HeLa cells. GFP-MavQ was visualized by immunogold labeling with anti-GFP antibody. Membrane structures positive for GFP-MavQ are highlighted in green. The micrographs were taken with ultrathin sections from a cell where GFP-MavQ localizes to an ER-like compartment (a) or from a cell where GFP-MavQ localizes to vesicular-tubular structures (b). See also fig. S2, A and B. Bars: 200 nm. (C) Confocal images of HeLa cells expressing the PI3P probe GFP-2xFYVEHrs, along with mCherry-MavQ (a and b) or mCherry-MavQD160A (c). In addition to its normal endosomal localization still visible in (b), GFP-2xFYVEHrs distributes to MavQ-positive ER (a) or vesicular-tubular structures (b). MavQD160A is mostly cytosolic and does not affect normal endosomal localization of GFP-2xFYVEHrs (c). Bars: 5 μm.
MavQ exhibits spatiotemporal oscillatory behavior and remodels the host ER membrane
The ER constitutes the major intracellular PI pool (9) and targeting of the host PI 3-kinase VPS34 to the ER produces PI3P to drive autophagosome formation (10). By analogy, we wondered whether MavQ could target the ER and drive the formation of MavQ-positive vesicular-tubular structures. Sequence analysis predicted that MavQ contains noncanonical, ER-targeting FFAT motifs (fig. S3A), and mutations in three of them decreased MavQ’s ER targeting (fig. S3B). We used live cell imaging to track the dynamics of MavQ and observed budding of MavQ-enriched vesicles/tubules from the ER (Fig. 3A, fig. S3C and movie S2). These MavQ-enriched vesicles/tubules were capable of fusing with the ER (Fig. 3B and movie S3). Remarkably, MavQ was able to oscillate back and forth between the ER and vesicles/tubules on the order of minutes and this oscillation propagated in a wave-like manner on the order of microns (Fig. 3C, fig. S3D, movies S4 and S5). The spatial switching of MavQ and the propagation of switching events as waves suggested a feedback mechanism and possibly cooperativity underlying its ER association.
Fig. 3. MavQ exhibits spatiotemporal oscillatory behavior and remodels the host ER membrane.
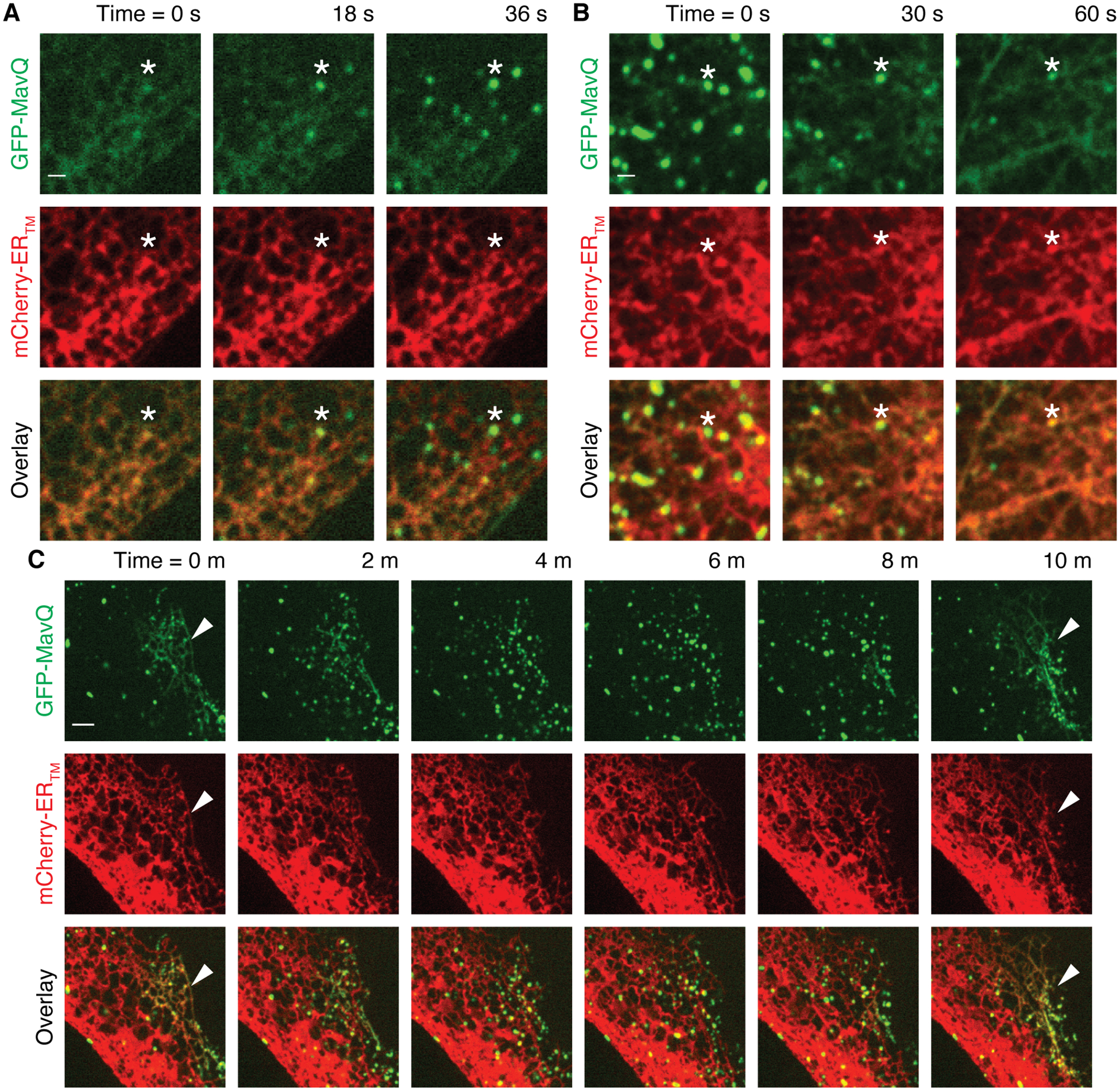
(A–C) Localization of GFP-MavQ relative to the ER membrane probe mCherry-ERTM in live HeLa cells by time-lapse confocal imaging. (A) GFP-MavQ distributes mostly on the ER initially, but several MavQ-enriched vesicles (an instance marked by asterisks) then emerge within 45 s; see also movie S2. Note the timestamps here are −54 s from the ones shown in movie S2. (B) GFP-MavQ distributes mostly on vesicles initially, but these MavQ-enriched vesicles (an instance marked by asterisks) then fuse with the ER within 75 s. Notice the increase in intensity of GFP-MavQ on the ER over time while MavQ-enriched vesicles vanish; see also movie S3. (C) At 0 m, MavQ localizes on the ER subdomain in the right corner (marked by arrowheads) and on vesicles in the rest of the cell. ER-localizing MavQ then dissipates while more MavQ-positive vesicles form in the region. At 10 m, MavQ redistributes to the ER subdomain in the right corner again (marked by arrowheads). See also movie S4. Bars: 1 (A and B) and 3 μm (C).
The C-terminal domain of MavQ recognizes MavQ-positive compartments, which leads to self-enhancement of MavQ’s membrane association
We hypothesized that MavQ may promote its own membrane association because positive feedback is generally involved in biological oscillators (11). While MavQD160A was mostly cytosolic (Fig. 2C, fig. S2E and S4A), expression of WT MavQ redistributed MavQD160A to the ER or vesicular-tubular structures (fig. S4A). Furthermore, the C-terminal domain of MavQ (MavQCTD, residues 581–871), but not the KD (MavQKD, residues 1–345), was also redistributed to the ER or vesicular-tubular structures by WT MavQ (fig. S4B). Thus, the MavQCTD recognizes MavQ-positive compartments, likely by interacting with membrane-bound MavQ or with its reaction product PI3P. Indeed, MavQ1–853 predominantly co-sedimented with liposomes containing PI3P or PI(3,5)P2, (Fig. 4A and fig. S4C), and the MavQCTD was necessary and sufficient for PI3P binding (fig. S4D). Thus, MavQ promotes its own membrane association in part by interaction with its reaction product PI3P.
Fig. 4. Positive and negative feedback loops facilitate MavQ protein dynamics and MavQ-induced membrane dynamics.
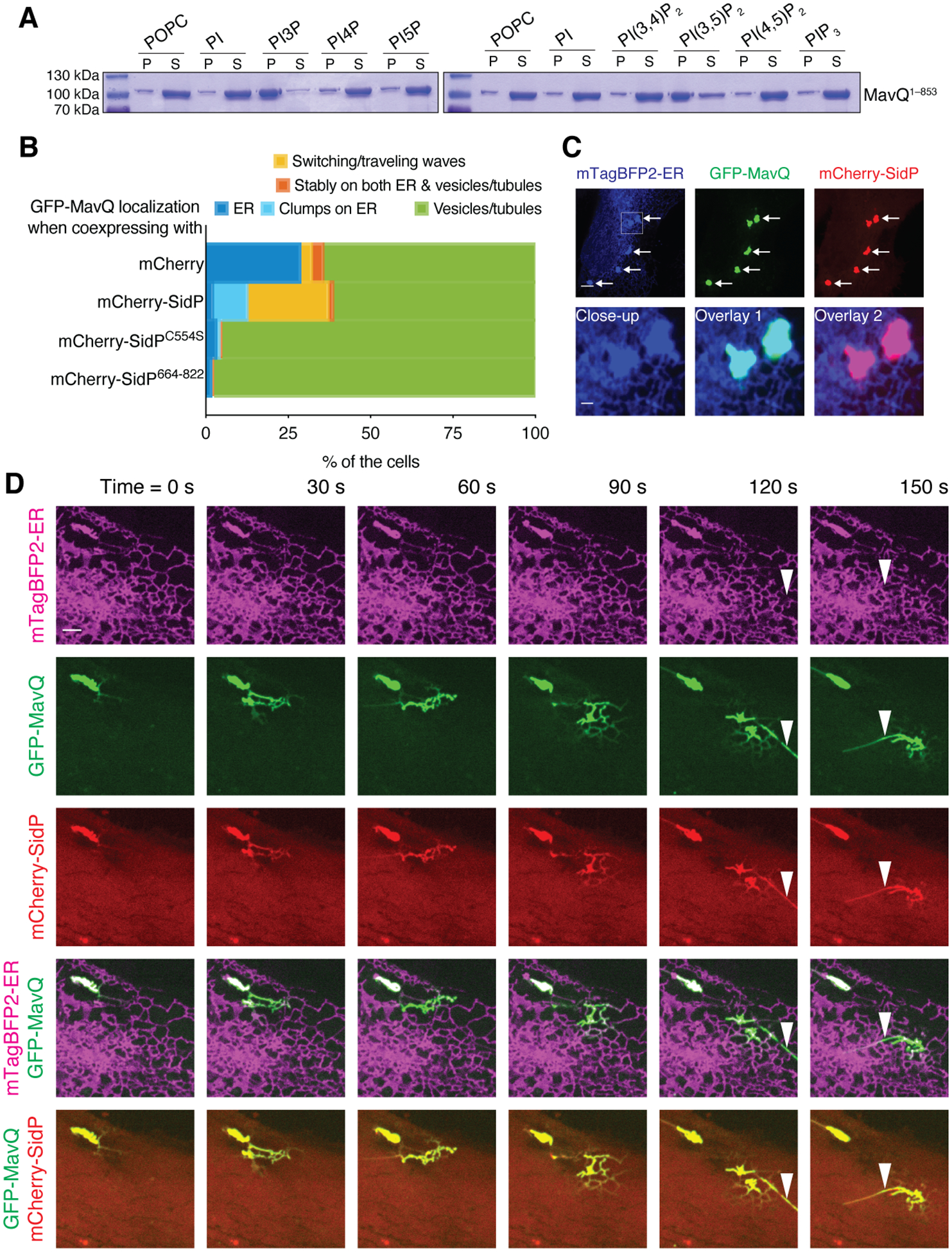
(A) Sedimentation of MavQ1–853 with POPC or phosphoinositide-containing liposomes. Pellet (P) and supernatant (S) fractions were resolved by SDS-PAGE and visualized by Coomassie staining. Liposome-bound proteins are present in the pellet. See fig. S4C for quantification.
(B) Frequency bar plot showing the proportions of cells with different types of MavQ localization when coexpressed with mCherry, mCherry-SidP, mCherry-SidPC554S or mCherry-SidP664–822 (200 cells/condition pooled from 4 independent experiments). See also fig. S5, B and C.
(C and D) Localization of GFP-MavQ and mCherry-SidP relative to the ER in live HeLa cells by confocal imaging. (C) mCherry-SidP sequesters GFP-MavQ from the main ER network into clumps that still retain ER identity, as indicated by arrows. The close-up shows two such clumps. Bars: 5 and 1 (close-up) μm. (D) Initially, GFP-MavQ and mCherry-SidP concentrate on an ER clump. A wave of ER-bound GFP-MavQ and mCherry-SidP then emits and propagates on the ER network. MavQ/SidP-positive tubules, which are negative for the ER luminal probe mTagBFP2-ER, form during the process (instances marked by arrowheads). See also movie S6. Bars: 3 μm.
The Legionella PI 3-phosphatase SidP facilitates spatiotemporal oscillatory behavior of MavQ
For a biological system to oscillate, negative feedback is required (11). We reasoned that a counteracting PI 3-phosphatase could be the negative regulator underlying the spatiotemporal oscillation of MavQ. Host PI 3-phosphatases may fulfill the role to some extent, as some type of oscillatory behavior occurred when only MavQ was expressed in the cell (Fig. 3C, fig. S3D, movies S4 and S5). However, the Legionella PI3-phosphatase SidP is functionally linked to MavQ (7). Cytosolic mCherry-SidP was redistributed to MavQ-positive compartments when coexpressed with GFP-MavQ (fig. S5A) and caused the sequestration of GFP-MavQ from the main ER network into clumps that retained ER identity (Fig. 4, B and C, and fig. S5B). Live-cell imaging revealed that SidP facilitated spatiotemporal oscillation of MavQ (Fig. 4B and fig. S5C). MavQ/SidP-enriched ER clumps or subdomains were able to generate MavQ/SidP protein waves, which propagated on the ER membrane and drove membrane remodeling during propagation (Fig. 4D, fig. S5D, movies S6 and S7). Such wave propagation correlated with PI3P turnover on the ER (fig. S5E and movie S8), suggesting that the concerted PI 3-kinase/phosphatase activities of MavQ/SidP constitute positive and negative feedback loops to drive MavQ protein dynamics and MavQ-induced membrane dynamics.
To further test the role of PI3P turnover in MavQ dynamics, we recruited the human PI3P phosphatase MTM1 to the ER using the inducible FKBP–FRB system (12). GFP-MavQ was rapidly released from the ER, and MavQ-positive vesicular-tubular structures formed upon rapamycin-induced ER recruitment of WT MTM1, but not the catalytically inactive MTM1C375S mutant (fig. S6, A and B, movies S9 and S10). Thus, PI3P is necessary to sustain the ER localization of MavQ, and PI3P depletion is sufficient to trigger the spatial switching of MavQ.
SidP contains an N-terminal phosphatase domain, an insertion domain and an α-helical C-terminal domain (fig. S6C) (3). We reasoned that SidP may modulate the MavQ–ER interaction not only through its phosphatase activity but also in another manner. Indeed, catalytically inactive SidPC554S as well as the SidPCTD (residues 664–822) colocalized with MavQ on vesicular-tubular structures (Fig. S6D) and both were sufficient to reduce the association of MavQ to the ER (Fig. 4B and fig. S5B). Thus, while the phosphatase activity of SidP was required for facilitating MavQ dynamics (Fig. 4B and fig. S5C), SidP also modulates MavQ’s ER association via its CTD. SidPC554S co-sedimented with MavQ1–853 when using PI3P-containing liposomes (fig. S6, E and F). Furthermore, both the MavQCTD and the SidPCTD were necessary for this interaction (fig. S6, E and F). Thus, SidP interacts with membrane-bound MavQ via its CTD. Collectively, PI3P-dependent MavQ–SidP interaction and SidP’s PI 3-phosphatase activity constitute the negative feedback to facilitate the spatiotemporal oscillation of MavQ.
MavQ exhibits spatiotemporal oscillatory behavior and remodels the host ER membrane during infection
We asked if any membrane remodeling occurs in association with PI3P turnover during Legionella infection and if MavQ is involved in this process. We observed dynamic PI3P-positive vesicular-tubular structures originating near the LCV (fig. S7A and movie S11) as well as sparks of the PI3P signal around the LCV (fig. S7B and movie S11) around 1 hr post infection (hpi). These PI3P-positive vesicular-tubular structures underwent fusion and fission and subsequently developed into vesicular-tubular networks (fig. S7A and movie S11). The T4SS was required for the formation of these PI3P-positive structures, and deletion of mavQ diminished their occurrence (fig. S7C).
We adopted the SunTag system (13) and developed a live-cell imaging approach to track the dynamics of translocated Legionella effectors in the host cell. As a proof-of-principle, we tracked the Legionella effector SidC, which binds to the LCV during infection (1). In uninfected cells, constitutively expressed anti-SunTag (single-chain variable fragment antibody against GCN4, fused to GFP, the solubility tag GB1, and a nuclear localization sequence) was sequestered in the nucleus (fig. S7D). When the cell was challenged with a Legionella strain expressing SunTag24x-SidC (SidC tagged with 24 copies of the GCN4 peptide), anti-SunTag redistributed to the polar regions of the LCV within 0.5 hpi (fig. S7E), indicating the onset of SunTag24x-SidC translocation at the bacterial poles. Notably, this redistribution was T4SS-dependent (fig. S7, E and F). Around 0.5–1 hpi, the entire membrane of some LCVs was decorated with SidC (fig. S7G). SidC remained associated with the LCV and did not redistribute to PI3P-positive vesicular-tubular structures or networks at later time points (fig. S7H).
When the cell was challenged with a Legionella strain expressing SunTag24x-MavQ, anti-SunTag decorated the polar regions of the LCV and sometimes the entire membrane starting around 1–1.5 hpi (fig. S8A). At this time point, the LCV had been associated with the ER or acquired ER-derived membranes (fig. S8A), and the majority of LCVs had lost PI3P that had accumulated during the early phase of LCV formation (fig. S8B). Remarkably, we observed budding of MavQ-enriched, PI3P-positive vesicular-tubular structures from the ER-associated LCV (Fig. 5A, fig. S8C, movies S12 and S13). These structures later developed into dynamic vesicular-tubular networks spreading throughout the cytoplasm (fig. S9 and movie S14). There were fast, “catastrophic” releases of MavQ together with the PI3P probe mTagBFP2–2xFYVEHrs from vesicular-tubular structures to the cytosol, as well as shrinkage of these structures (fig. S9 and movie S14). Starting around 3.5 hpi, MavQ decorated the whole ER network or ER subdomains in some cells (Fig. 5, B and C). Furthermore, MavQ was able to form protein waves in concert with PI3P turnover on the ER and redistribute from the ER to vesicular-tubular structures (Fig. 5C, fig. S10A, movies S15, S16 and S17). We also performed co-infection using a Legionella strain expressing SunTag24x-SidP and a Legionella strain expressing untagged MavQ. SidP localized on transient PI3P-positive vesicular-tubular structures in the cell co-infected by both strains (fig. S10B and movie S18), supporting the role of SidP in facilitating the PI3P turnover and PI3P-positive membrane dynamics. Thus, MavQ translocated from L. pneumophila exhibits spatiotemporal oscillatory behavior and remodels the host ER membrane in the context of infection.
Fig. 5. MavQ exhibits spatiotemporal oscillatory behavior and remodels the host ER membrane during infection.
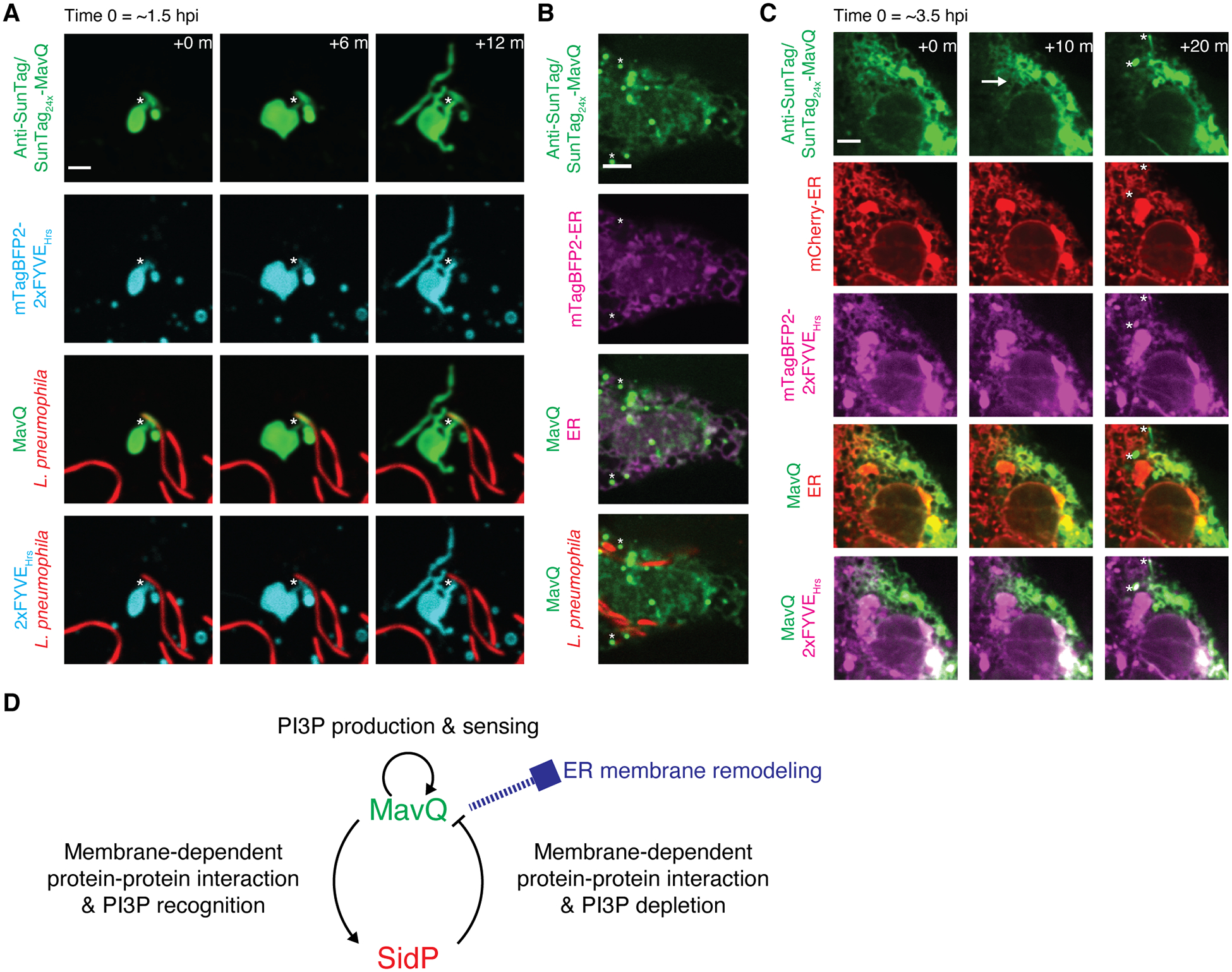
(A and B) Confocal images of live COS-7 cells expressing anti-SunTag together with the PI3P probe mTagBFP2-2xFYVEHrs (A) or the ER luminal probe mTagBFP2-ER (B) challenged with opsonized, mCherry-labeled L. pneumophila expressing SunTag24x-MavQ. (A) Around 1.5 hr post infection (hpi), MavQ-enriched, PI3P-positive vesicular-tubular structures bud from the LCV. Note that the LCV subdomain positive for MavQ is not PI3P-positive. The neck connecting the LCV and vesicular-tubular structures is marked by asterisks. See also movie S12. (B) Around 5 hpi, MavQ weakly decorates the ER in addition to vesicular-tubular structures (instances marked by asterisks). Bars: 3 (A), 5 μm (B).
(C) Time-lapse confocal images of a live COS-7 cell expressing anti-SunTag, mCherry-ER and mTagBFP2-2xFYVEHrs around 3.5 hpi with opsonized, unlabeled L. pneumophila expressing SunTag24x-MavQ. A wave of MavQ dissociation from the ER occurs (direction of propagation indicated by the arrow), and MavQ-enriched, PI3P-positive vesicles emerge during the process (instances marked by asterisks). Bar: 5 μm. See also movie S15 and movie S16.
(D) Model depicting interlinked positive and negative feedback loops between MavQ and SidP that drive spatiotemporal oscillation of MavQ to facilitate ER membrane remodeling.
To assess the functional significance of MavQ and SidP, we monitored replication of Legionella strains bearing a catalytically inactive mutation of mavQ (Lp02, mavQD160A) or the sidP deletion (Lp02, ΔsidP) in Acanthamoeba castellanii. The mutant strains displayed an intracellular growth defect that was complemented by expression of WT MavQ or WT SidP (fig. S11). Thus, both MavQ and SidP are required for full Legionella virulence in a eukaryotic host.
Discussion
ER manipulation is a hallmark of host cell subversion by L. pneumophila (1). Our results uncover that the atypical Legionella PI 3-kinase MavQ alters the phosphoinositide identity of the ER and is capable of ER membrane remodeling.
During Legionella infection, the nascent LCV is first enriched with PI3P and then matures into a PI 4-phosphate (PI4P)-enriched vacuole (1). Legionella effectors are not required for the initial PI3P acquisition of the LCV (14). Sequential action of the Legionella PI 4-kinase LepB and PI 3-phosphatase SidF may then mediate the PI3P-to-PI4P conversion in the LCV membrane (2, 4). A recent report suggests that MavQ acts in tandem with LepB and SidF to supply PI4P in the LCV membrane (15). While it is possible that MavQ indirectly contributes to PI4P generation in the LCV membrane and thus intracellular Legionella growth, we favor the idea that the membrane remodeling capabilities of MavQ play more important roles based on the following observations: 1) coevolution of MavQ and SidP during Legionella speciation (7) and 2) the requirement of biochemical synergy between MavQ and SidP for effective, dynamic membrane remodeling.
The positive and negative feedback loops between MavQ and SidP constitute a simple molecular circuit that displays oscillatory behavior and enables MavQ/SidP to form traveling waves on the membrane (Fig. 5D). We propose the following model based on Alan Turing’s discovery of reaction–diffusion pattern-forming systems (16): Initially, MavQ and SidP are freely diffusing in the cytosol, which would lead to a spatially homogeneous state of the MavQ/SidP system if there were no ATP for MavQ to consume. However, with energy from cytosolic ATP, any MavQ molecule transiently associated with the ER membrane has a chance to convert PI into PI3P, which promotes the recruitment of other MavQ molecules through MavQ’s PI3P-binding CTD. Local accumulation of membrane-bound MavQ then triggers the recruitment of SidP. Intuitively, membrane-bound SidP would cause MavQ and then SidP itself to dissociate from the membrane owing to SidP-catalyzed PI3P hydrolysis. Nevertheless, if SidP diffuses quicker on the membrane than MavQ does, it is possible that MavQ/SidP co-clusters will form on the membrane according to Turing’s model (16, 17). Such MavQ/SidP co-clusters may correspond to the MavQ/SidP-enriched ER clumps or subdomains we observed. Once local concentrations of membrane-bound MavQ/SidP reach certain thresholds, membrane remodeling can occur due to protein crowding, possible membrane insertion of the MavQCTD, or even PI3P accumulation (18), ultimately giving rise to MavQ/SidP-positive vesicles/tubules. These nascent ER-derived vesicles/tubules can fuse back with the ER or dissipate, thereby releasing MavQ/SidP back to the ER or the cytosol. This completes one cycle of spatiotemporal oscillation of MavQ/SidP. In addition, traveling waves can also occur in reaction–diffusion systems according to Turing’s theory (16, 17). Indeed, waves of MavQ and/or SidP form and propagate on the ER membrane.
The MavQ/SidP system resembles the exemplary minimal biochemical oscillator MinD/MinE. MinD/MinE forms traveling waves (19, 20) that can be explained by a Turing-like reaction–diffusion mechanism (20). This makes the MavQ/SidP system a strong candidate for a simple two-component Turing-like reaction–diffusion system. Striking spatiotemporal patterns of phosphoinositides have been recently reconstituted in vitro using a kinase–phosphatase competitive reaction (21), and a synthetic biology investigation also demonstrated that artificial molecular circuits containing competing kinases and phosphatases are capable of exhibiting a particular self-organizing behavior, i.e., cell polarization (22). Traveling waves of phosphoinositides occur at the plasma membrane of Dictyostelium discoideum and mast cells (23, 24) and can be induced at the trans-Golgi network upon inhibition of the PI 4-kinase PI4KIIIβ (25). Our results exemplify the importance of self-organizing behaviors due to chemically interacting kinases and phosphatases in complex cellular behaviors, and reveal a previously unappreciated mechanism utilized by intracellular bacterial pathogens to remodel host cellular membranes for survival.
Supplementary Material
Acknowledgements:
We thank members of the Tagliabracci and Liou laboratories for discussions. We thank A. Darehshouri for help with TEM and M. Gradowski for help with bioinformatics. Results shown in this report are derived from work performed at the Structural Biology Center, Advanced Photon Source, Argonne National Laboratory.
Funding:
This work was funded by NIH Grants DP2GM137419 (V.S.T.), R01GM113079 (J.L.), T32GM008203-29 (V.A.L), F30HL143859-01 (M.H.B.), Welch Foundation Grants I-1911 (V.S.T.), I-1789 (J.L.), CPRIT grant RP170674 (V.S.T) and Polish National Agency for Scientific Exchange scholarship PPN/BEK/2018/1/00431 (K.P.). Research reported in this publication was supported by the Office of the Director, NIH under Award Numbers S10OD021685 and S10OD025018. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. J.L. is a Sowell Family Scholar in Medical Research. V.S.T. is a Michael L. Rosenberg Scholar in Medical Research, a CPRIT Scholar (RR150033) and a Searle Scholar.
Footnotes
Data and materials availability:
All materials developed in this study will be made available upon request. The atomic coordinates have been deposited in the Protein Data Bank with accession code 7M7A.
References and Notes:
- 1.Steiner B, Weber S, Hilbi H, Formation of the Legionella-containing vacuole: phosphoinositide conversion, GTPase modulation and ER dynamics. Int. J. Med. Microbiol 308, 49–57 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Hsu F et al. , Structural basis for substrate recognition by a unique Legionella phosphoinositide phosphatase. Proc. Natl. Acad. Sci. USA 109, 13567–13572 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Toulabi L, Wu X, Cheng Y, Mao Y, Identification and structural characterization of a Legionella phosphoinositide phosphatase. J. Biol. Chem 288, 24518–24527 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong N et al. , Modulation of membrane phosphoinositide dynamics by the phosphatidylinositide 4-kinase activity of the Legionella LepB effector. Nat. Microbiol 2, 16236 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Ledvina HE et al. , A Phosphatidylinositol 3-Kinase Effector Alters Phagosomal Maturation to Promote Intracellular Growth of Francisella. Cell Host Microbe 24, 285–295 e288 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holm L, Laakso LM, Dali server update. Nucleic Acids Res. 44, W351–355 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Urbanus ML et al. , Diverse mechanisms of metaeffector activity in an intracellular bacterial pathogen, Legionella pneumophila. Mol. Syst. Biol 12, 893 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hammond GR, Takasuga S, Sasaki T, Balla T, The ML1Nx2 Phosphatidylinositol 3,5-Bisphosphate Probe Shows Poor Selectivity in Cells. PLoS One 10, e0139957 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim YJ, Guzman-Hernandez ML, Balla T, A highly dynamic ER-derived phosphatidylinositol-synthesizing organelle supplies phosphoinositides to cellular membranes. Dev. Cell 21, 813–824 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Axe EL et al. , Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol 182, 685–701 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsai TY et al. , Robust, tunable biological oscillations from interlinked positive and negative feedback loops. Science 321, 126–129 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rivera VM et al. , A humanized system for pharmacologic control of gene expression. Nat. Med 2, 1028–1032 (1996). [DOI] [PubMed] [Google Scholar]
- 13.Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD, A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159, 635–646 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weber S, Wagner M, Hilbi H, Live-cell imaging of phosphoinositide dynamics and membrane architecture during Legionella infection. mBio 5, e00839–00813 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li G, Liu H, Luo ZQ, Qiu J, Modulation of phagosome phosphoinositide dynamics by a Legionella phosphoinositide 3-kinase. EMBO Rep. 22, e51163 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turing AM, The chemical basis of morphogenesis. Philos. Trans. R. Soc. London, Ser. B 237, 37–72 (1952). [Google Scholar]
- 17.Halatek J, Brauns F, Frey E, Self-organization principles of intracellular pattern formation. Philos. Trans. R. Soc. London, Ser. B 373, 20170107 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Renard HF, Johannes L, Morsomme P, Increasing Diversity of Biological Membrane Fission Mechanisms. Trends Cell Biol. 28, 274–286 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Raskin DM, de Boer PA, Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc. Natl. Acad. Sci. USA 96, 4971–4976 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loose M, Fischer-Friedrich E, Ries J, Kruse K, Schwille P, Spatial regulators for bacterial cell division self-organize into surface waves in vitro. Science 320, 789–792 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Hansen SD et al. , Stochastic geometry sensing and polarization in a lipid kinase-phosphatase competitive reaction. Proc. Natl. Acad. Sci. USA 116, 15013–15022 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chau AH, Walter JM, Gerardin J, Tang C, Lim WA, Designing synthetic regulatory networks capable of self-organizing cell polarization. Cell 151, 320–332 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arai Y et al. , Self-organization of the phosphatidylinositol lipids signaling system for random cell migration. Proc. Natl. Acad. Sci. USA 107, 12399–12404 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiong D et al. , Frequency and amplitude control of cortical oscillations by phosphoinositide waves. Nat. Chem. Biol 12, 159–166 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Mesmin B et al. , Sterol transfer, PI4P consumption, and control of membrane lipid order by endogenous OSBP. EMBO J. 36, 3156–3174 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang CL et al. , Feedback regulation of receptor-induced Ca2+ signaling mediated by E-Syt1 and Nir2 at endoplasmic reticulum-plasma membrane junctions. Cell Rep. 5, 813–825 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Subach OM, Cranfill PJ, Davidson MW, Verkhusha VV, An enhanced monomeric blue fluorescent protein with the high chemical stability of the chromophore. PLoS One 6, e28674 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gillooly DJ et al. , Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 19, 4577–4588 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X et al. , Genetically encoded fluorescent probe to visualize intracellular phosphatidylinositol 3,5-bisphosphate localization and dynamics. Proc. Natl. Acad. Sci. USA 110, 21165–21170 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hammond GR, Machner MP, Balla T, A novel probe for phosphatidylinositol 4-phosphate reveals multiple pools beyond the Golgi. J. Cell Biol 205, 113–126 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sexton JA, Yeo HJ, Vogel JP, Genetic analysis of the Legionella pneumophila DotB ATPase reveals a role in type IV secretion system protein export. Mol. Microbiol 57, 70–84 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Kovach ME et al. , Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175–176 (1995). [DOI] [PubMed] [Google Scholar]
- 33.Jeong KC, Sexton JA, Vogel JP, Spatiotemporal regulation of a Legionella pneumophila T4SS substrate by the metaeffector SidJ. PLoS Pathogens 11, e1004695 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merriam JJ, Mathur R, Maxfield-Boumil R, Isberg RR, Analysis of the Legionella pneumophila fliI gene: intracellular growth of a defined mutant defective for flagellum biosynthesis. Infect. Immun 65, 2497–2501 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chatfield CH, Cianciotto NP, Culturing, media, and handling of Legionella. Methods Mol. Biol 954, 151–162 (2013). [DOI] [PubMed] [Google Scholar]
- 36.de Lorenzo V, Cases I, Herrero M, Timmis KN, Early and late responses of TOL promoters to pathway inducers: identification of postexponential promoters in Pseudomonas putida with lacZ-tet bicistronic reporters. J. Bacteriol 175, 6902–6907 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M, HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr 62, 859–866 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Borek D, Minor W, Otwinowski Z, Measurement errors and their consequences in protein crystallography. Acta Crystallogr. D Biol. Crystallogr 59, 2031–2038 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Otwinowski Z, Borek D, Majewski W, Minor W, Multiparametric scaling of diffraction intensities. Acta Crystallogr. A 59, 228–234 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Borek D, Cymborowski M, Machius M, Minor W, Otwinowski Z, Diffraction data analysis in the presence of radiation damage. Acta Crystallogr. D Biol. Crystallogr 66, 426–436 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Borek D, Dauter Z, Otwinowski Z, Identification of patterns in diffraction intensities affected by radiation exposure. J. Synchrotron Radiat 20, 37–48 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schneider TR, Sheldrick GM, Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr 58, 1772–1779 (2002). [DOI] [PubMed] [Google Scholar]
- 43.Otwinowski Z, in CCP4 Study Weekend, Wolf M, Evans PR, Leslie AGW, Eds. (Daresbury Laboratory: Science and Engineering Research Council, 1991), pp. 80–86. [Google Scholar]
- 44.Cowtan K, Recent developments in classical density modification. Acta Crystallogr. D Biol. Crystallogr 66, 470–478 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cowtan K, The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr 62, 1002–1011 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adams PD et al. , PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N, Computer control of microscopes using μManager. Curr. Protoc. Mol. Biol 92, 14.20.11–14.20.17 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reddick LE, Alto NM, Correlative light and electron microscopy (CLEM) as a tool to visualize microinjected molecules and their eukaryotic sub-cellular targets. J. Vis. Exp, e3650 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu D, Jaroszewski L, Li Z, Godzik A, FFAS-3D: improving fold recognition by including optimized structural features and template re-ranking. Bioinformatics 30, 660–667 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katoh K, Rozewicki J, Yamada KD, MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform 20, 1160–1166 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crooks GE, Hon G, Chandonia JM, Brenner SE, WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schindelin J et al. , Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rueden CT et al. , ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18, 529 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Murphy SE, Levine TP, VAP, a Versatile Access Point for the Endoplasmic Reticulum: Review and analysis of FFAT-like motifs in the VAPome. Biochim. Biophys. Acta 1861, 952–961 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Chen VB et al. , MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All materials developed in this study will be made available upon request. The atomic coordinates have been deposited in the Protein Data Bank with accession code 7M7A.


