Abstract
Mutations associated with hepatitis B virus (HBV) resistance to lamivudine have not been extensively addressed in human immunodeficiency virus (HIV)-HBV coinfection. We have studied the HBV polymerase sequences from nine coinfected patients who experienced HBV recurrence while under lamivudine treatment. In seven of these nine patients, Met550, belonging to the highly conserved YMDD motif, was mutated to Val and was associated with a substitution of Met for Leu526 in each case. In the two remaining patients, we found a Met550-to-Ile change that was associated in only one case with a Leu526-to-Met mutation. No mutation was observed in three control patients not receiving lamivudine. This study demonstrates the emergence of particular genetic profiles in HBV-HIV-coinfected patients experiencing a loss of control of HBV infection despite high doses of lamivudine.
Hepatitis B virus (HBV) and human immunodeficiency virus (HIV) share the same routes of transmission. Chronic infection with HBV affects about 5% of the population worldwide and as many as 20% of patients infected with HIV (14). The nucleoside analog lamivudine (2′,3′-dideoxy-3′-thiacytidine) has been shown to be very efficient in reducing both HIV and HBV replication by inhibiting the reverse transcriptase enzyme (5, 16). HIV resistance to lamivudine occurs rapidly within the first months of treatment and mostly involves mutations in the conserved YMDD motif of the reverse transcriptase of the virus (18, 19). Recent studies of patients treated with lamivudine for chronic hepatitis or post liver transplantation have suggested that mutations of the HBV polymerase could also confer resistance to this drug (1, 4, 6, 11, 13, 17). However, these HBV polymerase mutations have not been described in HIV-HBV-coinfected patients. These patients usually receive several antiviral drugs, and some of them may have impaired immune functions. To investigate the HBV mutations possibly associated with lamivudine resistance in HIV-HBV-coinfected patients, we studied the nucleotide sequence of the HBV polymerase gene from patients receiving lamivudine as part of their HIV infection treatment.
Patients.
Patients belonged to a cohort of 226 HIV-seropositive and hepatitis B surface antigen (HBsAg)-positive subjects monitored by the Liver Disease Department of our institution. All of these patients have had at least four visits per year for clinical examinations, biological tests, CD4 measurements, HBV serological marker analysis, serum HBV DNA evaluation, and HIV viral load measurements. Sixty-six of them received lamivudine, 150 mg twice daily, as part of their antiretroviral therapy. The recommended lamivudine dosage for HBV therapy is currently 100 mg/day; however, these patients received 300 mg/day, as recommended for HIV infection. Nine of these 66 HIV-HBV-coinfected patients were retrospectively selected on the basis of HBV replication recurrence during therapy and the availability of serum samples at the time of interest. Replication recurrence or breakthrough was defined as an initial drop in viral replication after initiation of therapy, leading to an undetectable HBV DNA level (threshold, 2.5 pg/ml), followed by a subsequent increase while therapy was maintained. Three patients who were not treated with lamivudine and whose serum had persistent high levels of HBV DNA (more than 2,000 pg/ml) served as controls. These 12 patients were homosexual men with a mean age of 36 (range, 31 to 47) years.
Biological markers.
CD4 lymphocytes were counted by dual-color flow cytometry. HIV antibodies were detected with both Sanofi Pasteur Diagnostics Genescreen HIV1/2 and Abbott HIV 1/2 Recombigen 3+. HIV loads were assessed by using a Roche Monitor reverse transcription-PCR kit (Roche, Neuilly, France). The detection threshold of the test is 200 RNA copies/ml. HBV serological markers were determined by using Axsym (HBsAg, core antibody, hepatitis B e antigen [HBeAg], and hepatitis B e antibody [HBeAb]) Abbott tests (Abbott, Les Ulis, France) in accordance with the manufacturer’s specifications. HBV DNA levels in serum were assessed by Digene Hybrid Capture test (Murex, Châtillon, France). The range of detection was 2.5 to 2,000 pg/ml.
Sequencing procedures.
For each patient, two serum samples were studied, one at the beginning of lamivudine treatment and one at breakthrough. For the three control patients, two sera were also studied at 9-month intervals. After viral DNA purification (QIAamp blood kit; Qiagen) starting from 200 μl of each serum, we amplified a 667-nucleotide fragment by using primers VT366 (5′-TGGTTATCGCTGGATGTGTC) and VT1015 (5′-CCCAAAAGACCCACAATTC). Sequencing of the purified PCR products was conducted on a Vistra DNA sequencer by using two inner primers labeled with Texas red at the 5′ end (VT462R [5′-TATGTTGCCCGTTTGTCCTC] and VT842R [5′-AGGGTTC/TAAATGTATACCC]). These two sets of conserved primers located in the polymerase gene have been designed from a consensus sequence generated from published HBV gene sequences and accessible from GenBank. Analysis of the sequences was accomplished with Geneworks software (Intelligenetics, Oxford, United Kingdom). There are a variety of amino acid numbering systems for HBV; we decided to use that of Pillay et al. (15).
Results.
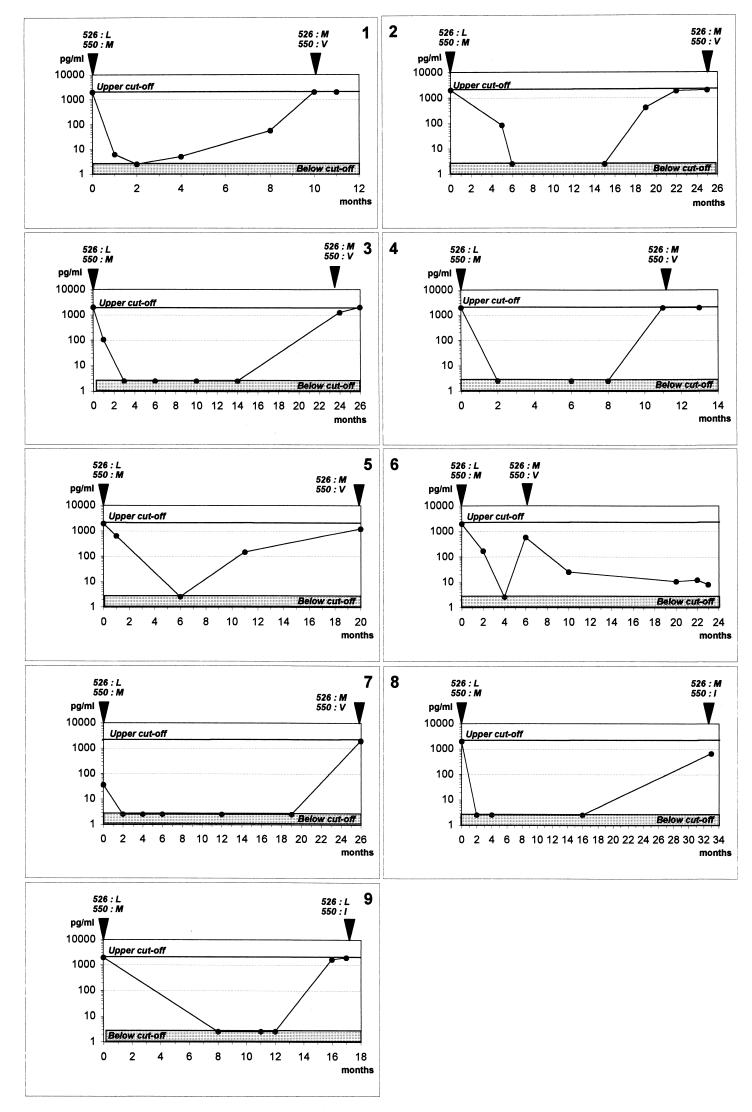
The nine selected patients undergoing lamivudine therapy had their treatment started in association with another nucleoside analog, either zidovudine (n = 5) or stavudine (n = 4). None of them was receiving an HIV protease inhibitor at the time of initiation of therapy. At breakthrough, six patients were receiving a protease inhibitor, either indinavir (n = 3), ritonavir (n = 2), or nelfinavir (n = 1), in association with lamivudine and another nucleoside analog, either zidovudine (n = 2) or stavudine (n = 4). Median HIV loads, alanine aminotransferase (ALT) levels in serum, CD4 lymphocyte counts, serological HBV markers, and HBV DNA levels in serum are summarized in Table 1. All patients but one were HBeAg positive and HBeAb negative throughout the study. Interestingly, patient 8, who had HBeAbs at the beginning of lamivudine treatment, was found to be HBeAg positive at breakthrough. In the sera of all patients but patient 2, HBV DNA was undetectable after 2 months of lamivudine therapy and HBV replication recurrence, suggesting the emergence of resistance, occurred after a mean delay of 17 (range, 4 to 33) months (Fig. 1). At the time of initiation of therapy, the sequences of the HBV polymerase gene were identical to those previously published for the wild-type virus (Fig. 1). When viral resistance emerged, HBV DNA sequence analysis revealed mutations mostly at position 550 of the polymerase, where a methionine was replaced with a valine (n = 7 of 9) or an isoleucine (n = 2 of 9) (Fig. 1). HBV 550M→V or 550M→I mutations were associated with an L→M substitution at position 526 in eight of nine cases. Only one patient (no. 9) had a unique mutation at position 550 (M→I) without any substitution at position 526. As expected, for the three patients not receiving lamivudine, sequences were similar to those of the wild-type virus throughout the study (data not shown).
TABLE 1.
Main characteristics of nine HIV-HBV-coinfected patients receiving lamivudine therapy and experiencing HBV relapse
| Parameter | Beginning of lamivudine therapy | Breakthrough |
|---|---|---|
| Median ALT level in serum (times above normal) | 2.6 (1–3)a | 1.8 (1.3–2) |
| Median no. of CD4 lymphocytes/μl | 90 (27–318) | 246 (218–469) |
| HIV type 1 load | ||
| No. of RNA copies/ml of plasmab | 61,674 (393–213,427) | 3,909 (1,851–7,299) |
| No. of patients with no detectable viral load | 2 | 3 |
| No. of untested patients | 2 | 1 |
| HBV load (pg of HBV DNA/ml)c | >2,000 (1,450–>2,000) | 680 (150–1,701) |
| HBV serological markers (no. of subjects positive) | ||
| HBsAg | 9 | 9 |
| HBsAb | 0 | 0 |
| HbeAg | 8 | 9 |
| HbeAb | 1 | 0 |
The values in parentheses are 95% confidence intervals, when applicable.
Measured by Monitor (Roche). Range of detection, 200 to 1,500,000 copies/ml.
Measured by Hybrid Capture (Murex). Range of detection, 2.5 to 2,000 pg/ml.
FIG. 1.
Course of HBV DNA during lamivudine treatment and resistance occurrence. HBV DNA from each of nine patients was measured at different time points and is represented by a specific dot. The time point of sequence analysis is represented by an arrowhead above each graph with the position of each residue.
As already described in healthy and immunocompromised transplant patients, HIV-HBV-coinfected patients may develop HBV resistance to lamivudine. In HIV-uninfected individuals, HBV mutations associated with lamivudine resistance have been found in patients receiving 100 mg of lamivudine per day (1, 4, 6, 9, 11). However, in HIV-HBV-coinfected patients, a higher daily dose of lamivudine (300 mg daily) did not appear to prevent the emergence of these mutations. In our study, HBV resistance to lamivudine occurred after a mean period of 17 (95% confidence interval, 9.8 to 24.2) months and was always associated with mutations in the YMDD motif. It is noteworthy that a more recent study in this laboratory of the whole cohort of HIV-HBV-infected patients (n = 226) indicates that after 2 years of treatment, 50% of patients develop lamivudine-resistant HBV strains (unpublished data). As previously described, HBV resistance to lamivudine develops much more slowly than that of HIV (16). The observed mutations are analogous to that found in lamivudine-resistant HIV strains, where M184 of the HIV reverse transcriptase is replaced with either Val or Ile. However, in contrast to the mutations of HIV reverse transcriptase associated with lamivudine resistance, which were described as single mutations, HBV 550M→V or 550M→I mutations are associated with an L→M substitution at position 526 in eight of nine cases. The 550M→V substitution associated with 526L→M has been commonly described. In our study, the mutation 550M→I was also detected; in one case it was a single mutation, and in the other it was associated with 526L→M. The mutation 550M→I could correspond, as it has been reported for HIV, to transient variants which will eventually evolve toward more fit species with the 550M→V-526L→M double mutation (3). These findings may also indicate the selection of progressive conformational rearrangements of the polymerase toward more replication-efficient resistant strains under drug pressure. In vitro studies assessing replication efficiency in the presence of 3TC are necessary to confirm this hypothesis. It is noteworthy that the replication level of YMDD-mutated strains usually increases slowly after breakthrough and only sometimes reaches the pretreatment level. However, many parameters may influence HBV replication, especially in coinfected patients. Although a larger study is needed to strengthen our findings, CD4 counts did not seem to correlate with the occurrence of breakthrough, nor did the HIV level of replication.
Other mutations, particularly at position 519, have been described in non-HIV-infected patients; however, none of these mutations was found in the nine patients studied here (1, 4, 8, 11, 15). Interestingly, although most of these HIV-infected patients were treated for several months with other nucleoside analogs, none showed particular mutations in the HBV polymerase catalytic site before lamivudine introduction. These findings agree with the absence of activity against HBV of the other nucleoside analogs and therefore confirm the absence of HBV resistance selection by these molecules. Although we studied only three patients not treated with lamivudine, it is highly unlikely that the mutations found were solely due to a high level of replication over time. Indeed, most of the patients have been chronic HBV carriers for several years and polymerase sequences before lamivudine therapy were found analogous to that of the wild type. Furthermore, several in vitro studies have clearly shown that mutations occurring within the polymerase B and C domains can confer resistance to lamivudine (2, 4, 7, 10, 12, 20).
It is of note that although underlying liver disease could not be precisely evaluated due to the lack of liver biopsies, the low level of ALT in serum at breakthrough indicates a low activity of HBV hepatitis in the context of lamivudine resistance and might suggest impaired replication of the virus.
In summary, in HIV-HBV-coinfected patients, treatment with high daily doses of lamivudine does not prevent the emergence of HBV-resistant strains carrying mutations within the polymerase B and C domains. These mutations also confer resistance to other nucleoside analogs (10). Hence, the selection of lamivudine-resistant HBV mutants raises concern about the future of hepatitis B management in both HIV-infected and non-HIV-infected persons.
Acknowledgments
We are sincerely grateful to Jack Gauldie for English revision of this article.
REFERENCES
- 1.Allen M I, Deslauriers M, Andrews C W, Tipples G A, Walters K A, Tyrrell D L, Brown N, Condreay L D. Identification and characterization of mutations in hepatitis B virus resistant to lamivudine. Lamivudine Clinical Investigation Group. Hepatology. 1998;27:1670–1677. doi: 10.1002/hep.510270628. [DOI] [PubMed] [Google Scholar]
- 2.Aye T T, Bartholomeusz A, Shaw T, Bowden S, Breschkin A, McMillan J, Angus P, Locarnini S. Hepatitis B virus polymerase mutations during antiviral therapy in a patient following liver transplantation. J Hepatol. 1997;26:1148–1153. doi: 10.1016/s0168-8278(97)80125-0. [DOI] [PubMed] [Google Scholar]
- 3.Back N K, Nijhuis M, Keulen W, Boucher C A, Oude Essink B O, van Kuilenburg A B, van Gennip A H, Berkhout B. Reduced replication of 3TC-resistant HIV-1 variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO J. 1996;15:4040–4049. [PMC free article] [PubMed] [Google Scholar]
- 4.Bartholomew M M, Jansen R W, Jeffers L J, Reddy K R, Johnson L C, Bunzendahl H, Condreay L D, Tzakis A G, Schiff E R, Brown N A. Hepatitis-B-virus resistance to lamivudine given for recurrent infection after orthotopic liver transplantation. Lancet. 1997;349:20–22. doi: 10.1016/S0140-6736(96)02266-0. [DOI] [PubMed] [Google Scholar]
- 5.Benhamou Y, Katlama C, Lunel F, Coutellier A, Dohin E, Hamm N, Tubiana R, Herson S, Poynard T, Opolon P. Effects of lamivudine on replication of hepatitis B virus in HIV-infected men. Ann Intern Med. 1996;125:705–712. doi: 10.7326/0003-4819-125-9-199611010-00001. [DOI] [PubMed] [Google Scholar]
- 6.Chayama K, Suzuki Y, Kobayashi M, Kobayashi M, Tsubota A, Hashimoto M, Miyano Y, Koike H, Kobayashi M, Koida I, Arase Y, Saitoh S, Murashima N, Ikeda K, Kumada H. Emergence and takeover of YMDD motif mutant hepatitis B virus during long-term lamivudine therapy and re-takeover by wild type after cessation of therapy. Hepatology. 1998;27:1711–1716. doi: 10.1002/hep.510270634. [DOI] [PubMed] [Google Scholar]
- 7.Fischer K P, Tyrrell D L J. Generation of duck hepatitis B virus polymerase mutants through site-directed mutagenesis which demonstrate resistance to lamivudine [(−)-β-l-2′,3′-dideoxy-3′-thiacytidine] in vitro. Antimicrob Agents Chemother. 1996;40:1957–1960. doi: 10.1128/aac.40.8.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu L, Cheng Y C. Role of additional mutations outside the YMDD motif of hepatitis B virus polymerase in L(−)SddC (3TC) resistance. Biochem Pharmacol. 1998;55:1567–1572. doi: 10.1016/s0006-2952(98)00050-1. [DOI] [PubMed] [Google Scholar]
- 9.Honkoop P, Niesters H G, de Man R A, Osterhaus A D, Schalm S W. Lamivudine resistance in immunocompetent chronic hepatitis B. Incidence and patterns. J Hepatol. 1997;26:1393–1395. doi: 10.1016/s0168-8278(97)80476-x. [DOI] [PubMed] [Google Scholar]
- 10.Ladner S, Miller T, Otto M, King R. The hepatitis B virus M539V polymerase variation responsible for 3TC resistance also confers cross-resistance to other nucleoside analogues. Antiviral Chem Chemother. 1998;9:65–72. [PubMed] [Google Scholar]
- 11.Ling R, Mutimer D, Ahmed M, Boxall E H, Elias E, Dusheiko G M, Harrison T J. Selection of mutations in the hepatitis B virus polymerase during therapy of transplant recipients with lamivudine. Hepatology. 1996;24:711–713. doi: 10.1002/hep.510240339. [DOI] [PubMed] [Google Scholar]
- 12.Melegari M, Scaglioni P P, Wands J R. Hepatitis B virus mutants associated with 3TC and famciclovir administration are replication defective. Hepatology. 1998;27:628–633. doi: 10.1002/hep.510270243. [DOI] [PubMed] [Google Scholar]
- 13.Niesters H G, Honkoop P, Haagsma E B, de Man R A, Schalm S W, Osterhaus A D. Identification of more than one mutation in the hepatitis B virus polymerase gene arising during prolonged lamivudine treatment. J Infect Dis. 1998;177:1382–1385. doi: 10.1086/517819. [DOI] [PubMed] [Google Scholar]
- 14.Ockenga J, Tillmann H L, Trautwein C, Stoll M, Manns M P, Schmidt R E. Hepatitis B and C in HIV-infected patients. Prevalence and prognostic value. J Hepatol. 1997;27:18–24. doi: 10.1016/s0168-8278(97)80274-7. [DOI] [PubMed] [Google Scholar]
- 15.Pillay D, Bartholomeusz A, Cane P, Mutimer D, Schinazi R F, Locarnini S. Mutations in the hepatitis B virus DNA polymerase associated with antiviral resistance. Int Antiviral News. 1998;6:167–169. [Google Scholar]
- 16.Schuurman R, Nijhuis M, van Leeuwen R, Schipper P, de Jong D, Collis P, Danner S A, Mulder J, Loveday C, Christopherson C, et al. Rapid changes in human immunodeficiency virus type 1 RNA load and appearance of drug-resistant virus populations in persons treated with lamivudine (3TC) J Infect Dis. 1995;171:1411–1419. doi: 10.1093/infdis/171.6.1411. [DOI] [PubMed] [Google Scholar]
- 17.Tipples G A, Ma M M, Fischer K P, Bain V G, Kneteman N M, Tyrrell D L. Mutation in HBV RNA-dependent DNA polymerase confers resistance to lamivudine in vivo. Hepatology. 1996;24:714–717. doi: 10.1002/hep.510240340. [DOI] [PubMed] [Google Scholar]
- 18.Tisdale M, Kemp S D, Parry N R, Larder B A. Rapid in vitro selection of human immunodeficiency virus type 1 resistant to 3′-thiacytidine inhibitors due to a mutation in the YMDD region of reverse transcriptase. Proc Natl Acad Sci USA. 1993;90:5653–5656. doi: 10.1073/pnas.90.12.5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wainberg M A, Salomon H, Gu Z, Montaner J S, Cooley T P, McCaffrey R, Ruedy J, Hirst H M, Cammack N, Cameron J, et al. Development of HIV-1 resistance to (−)2′-deoxy-3′-thiacytidine in patients with AIDS or advanced AIDS-related complex. AIDS. 1995;9:351–357. [PubMed] [Google Scholar]
- 20.Xiong X, Flores C, Yang H, Toole J J, Gibbs C S. Mutations in hepatitis B DNA polymerase associated with resistance to lamivudine do not confer resistance to adefovir in vitro. Hepatology. 1998;28:1669–1673. doi: 10.1002/hep.510280629. [DOI] [PubMed] [Google Scholar]