Abstract
Background
Diabetes mellitus (DM) is a complex metabolic disease that is caused by a complex interplay between genetic and environmental factors. This research aimed to investigate the association of genetic polymorphisms in PDX1 and MC4R with T2DM risk.
Methods
The genotypes of 10 selected SNPs in PDX1 and MC4R were identified using the Agena MassARRAY platform. We utilized odds ratio (OR) and 95% confidence intervals (CIs) to assess the correlation between genetic polymorphisms and T2DM risk.
Results
We found that PDX1-rs9581943 decreased susceptibility to T2DM among in a Chinese Han population (OR = 0.76, p = 0.045). We also found that selected genetic polymorphisms in PDX1 and MC4R could modify the risk of T2DM, which might also be influenced by age, sex, BMI, smoking status, and drinking status (p < 0.05).
Conclusions
We concluded that PDX1 and MC4R genetic variants were significantly associated with T2DM risk in a Chinese Han population. These single polymorphic markers may be considered to be new targets in the assessment and prevention of T2DM among Chinese Han people.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12920-021-01037-3.
Keywords: Type 2 diabetes mellitus, PDX1, MC4R, Polymorphism, Susceptibility
Background
Diabetes mellitus (DM) is a metabolic disease characterized by the presence of chronic hyperglycemia, which results from either weakened insulin secretion or insulin action or both [1]. The global prevalence of diabetes reached 9.3% (463 million) in 2019, and it is expected to increase to 10.9% (700 million) by 2045 [2]. China has the highest number of adults with diabetes, approximatedly116 million, ranking first in diabetes prevalence worldwide [2]. Type 2 diabetes mellitus (T2DM) accounts for nearly 90% of the total diabetes patients. There are multiple reasons for the incidence of T2DM including aging, sedentary lifestyles and genetic factors [3]. It has been reported that subjects withT2DM-affected siblings have a two- to three fold increased risk of developing T2DM compared with the general population [4]. Having one parent with diabetes increases the risk of developing T2DM by 30–40%, and having two parents with diabetes increases the risk to 70% [5]. Furthermore, some research reported that genetic polymorphisms in candidate genes could influence the formation and course of T2DM [6, 7].
Pancreatic and duodenal homeobox-1 (PDX1) modulates pancreas development and β-cell function. The PDX1 gene encodes a protein of 283 amino acids in humans. It also regulates many genes, such as those encoding insulin and glucokinase (GK), involved in maintaining the function of β-cells. In adults, PDX1 is highly expressed in β-cells, where it is required for efficient insulin gene transcription [8]. Indeed, PDX1 has been proposed to be an oncogene, since its overexpression increased pancreatic cancer cell proliferation, invasion, and growth in humans [9]. Gurevich et al. also illustrated that PDX1 was upregulated in neuroendocrine tumors, including pancreatic ductal and acinar cell tumors and gastric signet ring cell carcinomas [10]. It has previously been noted that PDX1 deficiency inhibits the development of pancreatic buds, leading to extreme hyperglycemia [11]. These findings demonstrated that PDX1 plays a pivotal role in the development of pancreas-related disease. However, no literature supports the effect of PDX1 polymorphisms on T2DM.
Melanocortin receptor 4 (MC4R) belongs to class A of G protein-coupled receptors and is a member of the melanocortin receptor family [12]. MC4R can control energy homeostasis, sympathetic nervous system activity, and blood pressure in rodents and humans [13]. For instance, MC4R knockdown mice were severely obese and the loss of one MC4R allele resulted in an intermediate obesity phenotype [14]. Greenfield et al. demonstrated reduction in blood pressure and circulating catecholamine levels in humans with MC4R deficiency [15]. In addition, previous research has established that MC4R deletion or mutation results in obesity, hyperphagia, and insulin resistance [16]. These observations highlight a potential role for MC4R in obesity-related diseases. In addition, obesity is believed to be an independent risk factor for T2DM [17]. Based on the above information, we hypothesized that MC4R may be involved in the occurrence of T2DM.
Therefore, we mainly examined the role of PDX1 and MC4R genetic polymorphisms in T2DM development in a Chinese population. We identified four polymorphisms in PDX1 (rs11619319, rs2293941, rs9581943 and rs7981781) and six polymorphisms in MC4R (rs6567160, rs663129, rs17782313, rs12969709, rs11663816, and rs12970134) to investigate the correlations between genetic polymorphisms and T2DM susceptibility. The current study will provide new targets for the early assessment and prevention of T2DM.
Methods
Study population
A total of 500 T2DM patients and 501 healthy controls were enrolled from the First Affiliated Hospital of Xi’an Jiaotong University in the present study. All patients were diagnosed with T2DM based on fasting plasma glucose ≥ 7.0 mmol/L or postprandial plasma glucose ≥ 11.1 mmol/L or HbA1c ≥ 6.5% [18]. Patients with type 1 diabetes mellitus; gestational diabetes; acute or chronic diseases of the liver, kidney, or heart; other endocrine diseases; inflammatory diseases; or malignant tumors were excluded. The inclusion criteria for controls were no history of diabetes, metabolic disorders or severe diseases. The demographic and clinical characteristics of all subjects, including age, sex, smoking status, drinking status, complications, and body mass index (BMI), were collected from medical records and questionnaires.
This research received approval from the Ethics Committee of the First Affiliated Hospital of Xi’an Jiaotong University, and conformed to the Declaration of Helsinki. Informed consent was acquired from each participant at recruitment after fully describing our research to them.
SNP genotyping
We selected four SNPs in PDX1 and six SNPs in MC4R and all SNPs had minor allele frequencies (MAFs) ≥ 5% in the 1000 Genomes Chinese Han Beijing population. Peripheral blood samples (5 mL) were collected from each subject, and genomic DNA was extracted using the GoldMag whole-blood DNA purification kit (GoldMag Co.Ltd., Xi’an, China) following the manufacturer’s protocol. Genotyping of PDX1 and MC4R polymorphisms was performed by the Agena MassARRAY platform (Agena Bioscience, San Diego, CA, USA). Moreover, Agena Typer 4.0 software was used to analyze and manage data.
Gene expression analysis
We performed PDX1 and MC4R mRNA expression analysis with blood samples from 100 unrelated Chinese Han individuals. Total RNA was isolated from peripheral blood using a Qiagen kit (Qiagen) according to the manufacturer’s instructions. RNA was reverse transcribed to synthesize first-strand cDNA using the PrimeScript-1st strand cDNA Synthesis Kit (Takara Bio, Shiga, Japan), as described by the manufacturers. The mRNA expression of the PDX1 and MC4R genes and the internal control GAPDH were assessed using quantitative real-time PCR (ABI PRISM 7500 Real-Time PCR System; Applied Biosystems). The relative mRNA expression was calculated by the 2−Δ(ΔCt) comparative method and normalized to GAPDH expression.
The primer sequences for the mRNA expression of PDX1, MC4R and GAPDH are shown in Additional file 1: Table S1. Amplification was performed in a reaction mixture containing 10 pM each primer, 10 μl SYBR Green/High ROX (Amplicon), 7 μl nuclease-free water, and 2 μl cDNA solution. Experiments were performed in triplicate.
Statistical analysis
Statistical differences in demographic characteristics of the participants were assessed using the χ2 test and Student′s t-test. Hardy–Weinberg equilibrium (HWE) of each SNP among controls was evaluated using the χ2 test. The association of the selected SNPs with T2DM susceptibility was examined by odds ratio (ORs) and 95% confidence intervals (CIs) by logistic regression analysis in multiple inheritance models and different subgroups (age, sex, smoking, drinking and BMI). The potential functions of the selected SNPs were forecasted using HaploReg v4.1 (https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php). Haploview software and PLINK software were used for Haploview analysis and linkage disequilibrium [19, 20]. The mRNA expression was analyzed using Student’s t-test in the case and control groups. The effects of the polymorphisms on mRNA expression were examined by one-way analysis of variance (ANOVA). A p value < 0.05 was considered statistically significant.
Results
Characteristics of the study population
As presented in Table 1, there were 500 T2DM patients (358 men and 142 women) and 501 healthy controls (358 men and 143 women) in this study. The average ages were 59.87 ± 12.87 years for cases and 59.85 ± 9.34 years for controls. There were no significant differences in age (p = 0.973) or sex (p = 0.960) between the case and control groups. In addition, significant differences were observed in total cholesterol (p < 0.001), low-density lipoprotein cholesterol (LDL-C, p = 0.012), high-density lipoprotein cholesterol (HDL-C, p = 0.024), fasting blood glucose (p < 0.001) and urea (p < 0.001) between the two groups.
Table 1.
Characteristics of the study population
| Characteristics | Cases (n = 500) | Controls (n = 501) | p |
|---|---|---|---|
| Age, years | |||
| Mean ± SD (years) | 59.87 ± 12.87 | 59.85 ± 9.34 | 0.973a |
| > 60 | 240 (48%) | 268 (54%) | |
| ≤ 60 | 260 (52%) | 233 (46%) | |
| Sex | 0.960b | ||
| Male | 358 (72%) | 358 (71%) | |
| Female | 142 (28%) | 143 (29%) | |
| Smoking | |||
| Yes | 219 (44%) | 98 (20%) | |
| No | 280 (56%) | 164 (33%) | |
| Absence | 1 | 239 (47%) | |
| Drinking | |||
| Yes | 109 (22%) | 103 (21%) | |
| No | 385 (77%) | 140 (28%) | |
| Absence | 6 (1%) | 258 (51%) | |
| BMI | |||
| ≤ 24 | 203 (41%) | 130 (26%) | |
| > 24 | 239 (48%) | 188 (38%) | |
| Absence | 58 (11%) | 183 (36%) | |
| Complication | |||
| One | 107 (21%) | ||
| Multiple | 337 (67%) | ||
| Absence | 56 (12%) | ||
| Total cholesterol (mmol/L) | 4.19 ± 2.01 | 4.93 ± 4.00 | < 0.001a |
| LDL-C (mmol/L) | 2.45 ± 0.90 | 2.62 ± 0.76 | 0.012a |
| HDL-C (mmol/L) | 1.05 ± 0.72 | 1.15 ± 0.55 | 0.024a |
| Fasting blood glucose | 7.35 ± 3.40 | 6.05 ± 1.60 | < 0.001a |
| Triglyceride | 1.91 ± 1.91 | 1.74 ± 0.10 | 0.088 |
| GFR(ml/min) | 96.62 ± 22.22 | 96.01 ± 19.78 | 0.710 |
| Urea | 6.52 ± 3.26 | 5.42 ± 2.78 | < 0.001a |
| Creatinine | 71.20 ± 52.66 | 68.74 ± 12.87 | 0.371 |
Bold indicates a statistically significant (p < 0.05).
SD standard deviation, BMI body mass index, HDL-C high-density lipoprotein cholesterol, LDL-C low-density lipoprotein cholesterol
pa value obtained from an independent sample t-test
pb value obtained from Pearson's χ2 test
T2DM risk assessment
Four candidate SNPs in PDX1 (rs11619319, rs2293941, rs9581943, and rs7981781) and six SNPs in MC4R (rs6567160, rs663129, rs17782313, rs12969709, rs11663816, and rs12970134) were successfully genotyped, as shown in Additional file 1: Table S2. Deviation from HWE was assessed in controls and all candidate SNPs reached the expected p values (p > 0.05). There were no significant associations between allele frequencies of any SNP and susceptibility to T2DM (p > 0.05).
Additionally, we investigated the correlation of PDX1 and MC4R polymorphisms with T2DM risk in multiple inheritance models by logistic regression analyses (Table 2). The results revealed that the AG genotype of PDX1-rs9581943 decreased susceptibility to T2DM in the study subjects (OR = 0.76, 95% CI = 0.58–0.99, p = 0.045).
Table 2.
Relationships of polymorphisms in PDX1 and MC4R and T2DM risk
| Gene | SNP | Model | Genotype | Without adjustment | With adjustment | ||
|---|---|---|---|---|---|---|---|
| OR (95% CI) | pa | OR (95% CI) | pb | ||||
| PDX1 | rs11619319 | Codominant | AA | 1.00 | 1.00 | ||
| GG | 1.09 (0.76–1.56) | 0.629 | 1.09 (0.76–1.56) | 0.629 | |||
| GA | 0.90 (0.68–1.20) | 0.471 | 0.90 (0.68–1.20) | 0.471 | |||
| Dominant | AA | 1.00 | 1.00 | ||||
| GG-GA | 0.95 (0.73–1.25) | 0.715 | 0.95 (0.73–1.25) | 0.717 | |||
| Recessive | GA-AA | 1.00 | 1.00 | ||||
| GG | 1.17 (0.85–1.59) | 0.333 | 1.17 (0.85–1.59) | 0.334 | |||
| Additive | – | 1.03 (0.86–1.23) | 0.755 | 1.03 (0.86–1.23) | 0.756 | ||
| PDX1 | rs2293941 | Codominant | GG | 1.00 | 1.00 | ||
| AA | 1.09 (0.76–1.56) | 0.646 | 1.09 (0.76–1.56) | 0.646 | |||
| AG | 0.89 (0.67–1.18) | 0.425 | 0.89 (0.67–1.18) | 0.426 | |||
| Dominant | GG | 1.00 | 1.00 | ||||
| AA-AG | 0.94 (0.72–1.23) | 0.666 | 0.94 (0.72–1.23) | 0.667 | |||
| Recessive | AG-GG | 1.00 | 1.00 | ||||
| AA | 1.17 (0.85–1.60) | 0.332 | 1.17 (0.85–1.60) | 0.333 | |||
| Additive | – | 1.02 (0.86–1.22) | 0.791 | 1.02 (0.86–1.22) | 0.792 | ||
| PDX1 | rs9581943 | Codominant | GG | 1.00 | 1.00 | ||
| AA | 0.96 (0.65–1.42) | 0.844 | 0.96 (0.65–1.41) | 0.842 | |||
| AG | 0.76 (0.58–0.99) | 0.046 | 0.76 (0.58–0.99) | 0.045 | |||
| Dominant | GG | 1.00 | 1.00 | ||||
| AA-AG | 0.80 (0.63–1.04) | 0.090 | 0.80 (0.62–1.04) | 0.090 | |||
| Recessive | AG-GG | 1.00 | 1.00 | ||||
| AA | 1.11 (0.77–1.59) | 0.574 | 1.11 (0.77–1.59) | 0.574 | |||
| Additive | – | 0.92 (0.77–1.10) | 0.353 | 0.92 (0.77–1.10) | 0.354 | ||
| PDX1 | rs7981781 | Codominant | GG | 1.00 | 1.00 | ||
| AA | 1.08 (0.75–1.54) | 0.681 | 1.08 (0.75–1.54) | 0.681 | |||
| AG | 0.85 (0.64–1.13) | 0.263 | 0.85 (0.64–1.13) | 0.263 | |||
| Dominant | GG | 1.00 | 1.00 | ||||
| AA-AG | 0.91 (0.70–1.19) | 0.486 | 0.91 (0.70–1.19) | 0.487 | |||
| Recessive | AG-GG | 1.00 | 1.00 | ||||
| AA | 1.19 (0.87–1.63) | 0.289 | 1.19 (0.86–1.63) | 0.290 | |||
| Additive | – | 1.01 (0.85–1.21) | 0.898 | 1.01 (0.85–1.21) | 0.899 | ||
| MC4R | rs6567160 | Codominant | TT | 1.00 | 1.00 | ||
| CC | 0.88 (0.53–1.47) | 0.627 | 0.88 (0.53–1.47) | 0.626 | |||
| CT | 1.10 (0.84–1.44) | 0.475 | 1.10 (0.84–1.44) | 0.475 | |||
| Dominant | TT | 1.00 | 1.00 | ||||
| CC-CT | 1.06 (0.83–1.37) | 0.634 | 1.06 (0.83–1.37) | 0.635 | |||
| Recessive | CT-TT | 1.00 | 1.00 | ||||
| CC | 0.85 (0.51–1.41) | 0.527 | 0.85 (0.51–1.41) | 0.526 | |||
| Additive | – | 1.01 (0.83–1.24) | 0.899 | 1.01 (0.83–1.24) | 0.900 | ||
| MC4R | rs663129 | Codominant | GG | 1.00 | 1.00 | ||
| AA | 0.89 (0.53–1.48) | 0.646 | 0.89 (0.53–1.48) | 0.645 | |||
| AG | 1.12 (0.86–1.47) | 0.395 | 1.12 (0.86–1.47) | 0.396 | |||
| Dominant | GG | 1.00 | 1.00 | ||||
| AA-AG | 1.08 (0.84–1.39) | 0.545 | 1.08 (0.84–1.39) | 0.546 | |||
| Recessive | AG-GG | 1.00 | 1.00 | ||||
| AA | 0.85 (0.51–1.41) | 0.527 | 0.85 (0.51–1.41) | 0.526 | |||
| Additive | – | 1.02 (0.84–1.25) | 0.818 | 1.02 (0.84–1.25) | 0.820 | ||
| MC4R | rs17782313 | Codominant | TT | 1.00 | 1.00 | ||
| CC | 0.89 (0.53–1.49) | 0.664 | 0.89 (0.53–1.49) | 0.663 | |||
| CT | 1.14 (0.88–1.49) | 0.324 | 1.14 (0.88–1.49) | 0.324 | |||
| Dominant | TT | 1.00 | 1.00 | ||||
| CC-CT | 1.10 (0.85–1.42) | 0.463 | 1.10 (0.85–1.41) | 0.464 | |||
| Recessive | CT-TT | 1.00 | 1.00 | ||||
| CC | 0.85 (0.51–1.41) | 0.527 | 0.85 (0.51–1.41) | 0.526 | |||
| Additive | – | 1.04 (0.85–1.27) | 0.740 | 1.04 (0.85–1.27) | 0.741 | ||
| MC4R | rs12969709 | Codominant | CC | 1.00 | 1.00 | ||
| AA | 0.70 (0.40–1.22) | 0.203 | 0.70 (0.40–1.21) | 0.202 | |||
| AC | 1.06 (0.81–1.38) | 0.694 | 1.06 (0.81–1.38) | 0.695 | |||
| Dominant | CC | 1.00 | 1.00 | ||||
| AA-AC | 0.99 (0.77–1.28) | 0.969 | 0.99 (0.77–1.28) | 0.967 | |||
| Recessive | AC-CC | 1.00 | 1.00 | ||||
| AA | 0.68 (0.40–1.18) | 0.174 | 0.68 (0.40–1.18) | 0.173 | |||
| Additive | – | 0.94 (0.77–1.16) | 0.578 | 0.94 (0.77–1.16) | 0.577 | ||
| MC4R | rs11663816 | Codominant | TT | 1.00 | 1.00 | ||
| CC | 0.88 (0.50–1.55) | 0.659 | 0.88 (0.50–1.55) | 0.657 | |||
| CT | 0.98 (0.75–1.27) | 0.854 | 0.98 (0.75–1.27) | 0.852 | |||
| Dominant | TT | 1.00 | 1.00 | ||||
| CC-CT | 0.96 (0.75–1.24) | 0.766 | 0.96 (0.75–1.24) | 0.764 | |||
| Recessive | CT-TT | 1.00 | 1.00 | ||||
| CC | 0.89 (0.51–1.55) | 0.678 | 0.89 (0.51–1.55) | 0.676 | |||
| Additive | – | 0.96 (0.78–1.18 | 0.688 | 0.96 (0.78–1.18) | 0.686 | ||
| MC4R | rs12970134 | Codominant | GG | 1.00 | 1.00 | ||
| AA | 0.83 (0.46–1.50) | 0.543 | 0.83 (0.46–1.50) | 0.542 | |||
| AG | 0.97 (0.74–1.26) | 0.801 | 0.97 (0.74–1.26) | 0.800 | |||
| Dominant | GG | 1.00 | 1.00 | ||||
| AA-AG | 0.95 (0.73–1.23) | 0.683 | 0.95 (0.73–1.23) | 0.682 | |||
| Recessive | AG-GG | 1.00 | 1.00 | ||||
| AA | 0.84 (0.47–1.51) | 0.564 | 0.84 (0.47–1.51) | 0.564 | |||
| Additive | – | 0.94 (0.76–1.17) | 0.582 | 0.94 (0.76–1.17) | 0.581 | ||
SNP single nucleotide polymorphism, OR odds ratio, 95% CI 95% confidence interval
pa values were calculated by logistic regression analysis with the comparison between diabetes patients and healthy controls
pb values were calculated by logistic regression analysis with adjustment for age and gender
Bold values indicate statistical significance (p < 0.05)
Stratified analysis
Stratification analysis was carried out by age, sex, smoking, drinking and BMI. The results of stratification by age and sex are shown in Table 3. We found that PDX1-rs9581943 significantly decreased the risk of T2DM among patients aged ≤ 60 years in the codominant (OR = 0.66, 95% CI = 0.45–0.98, p = 0.039) and dominant models (OR = 0.69, 95% CI = 0.48–1.00, p = 0.049). Rs6567160, rs663129, rs17782313, rs12969709 and rs11663816 in MC4R reduced the susceptibility to T2DM among individuals aged ≤ 60 years under the codominant (rs6567160: OR = 0.33, 95% CI = 0.13–0.81, p = 0.015; rs663129: OR = 0.33, 95% CI = 0.13–0.82, p = 0.017; rs17782313: OR = 0.34, 95% CI = 0.14–0.83, p = 0.018; rs12969709: OR = 0.27, 95% CI = 0.10–0.75, p = 0.012; rs11663816: OR = 0.31, 95% CI = 0.11–0.88, p = 0.027) and recessive (rs6567160: OR = 0.33, 95% CI = 0.14–0.81, p = 0.016; rs663129: OR = 0.33, 95% CI = 0.14–0.81, p = 0.016; rs17782313: OR = 0.33, 95% CI = 0.14–0.81, p = 0.016; rs12969709: OR = 0.27, 95% CI = 0.10–0.75, p = 0.012; and rs11663816: OR = 0.32, 95% CI = 0.11–0.91, p = 0.032) models. After stratifying by sex, rs9581943 (OR = 0.73, 95% CI = 0.5–1.00, p = 0.049) and rs7981781 (OR = 0.70, 95% CI = 0.56–0.97, p = 0.033) were found to be associated with a decreased risk of T2DM in males under the codominant model.
Table 3.
Relationships of PDX1 and MC4R polymorphisms with T2DM risk stratified by age and sex
| Gene SIP | Model | Genotype | > 60 | ≤ 60 | Male | Female | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | p | OR (95% CI) | p | OR (95% CI) | p | OR (95% CI) | p | |||
|
PDX1 rs9581943 |
Allele | G | 1.00 | 1.00 | 1.00 | 1.00 | ||||
| A | 0.99 (0.76–1.28) | 0.919 | 0.84 (0.65–1.09) | 0.180 | 0.94 (0.75–1.16) | 0.544 | 0.87 (0.62–1.22) | 0.424 | ||
| Codominant | GG | 1.00 | 1.00 | 1.00 | 1.00 | |||||
| AA | 1.05 (0.60–1.84) | 0.852 | 0.79 (0.45–1.38) | 0.409 | 1.05 (0.67–1.66) | 0.829 | 0.77 (0.37–1.60) | 0.481 | ||
| AG | 0.86 (0.58–1.27) | 0.439 | 0.66 (0.45–0.98) | 0.039 | 0.73 (0.5–1.00) | 0.049 | 0.85 (0.51–1.40) | 0.516 | ||
| Dominant | GG | 1.00 | 1.00 | 1.00 | 1.00 | |||||
| AA-AG | 0.90 (0.63–1.30) | 0.580 | 0.69 (0.48–1.00) | 0.049 | 0.80 (0.59–1.07) | 0.130 | 0.83 (0.51–1.34) | 0.439 | ||
| Recessive | AG-GG | 1.00 | 1.00 | 1.00 | 1.00 | |||||
| AA | 1.14 (0.67–1.92) | 0.634 | 0.99 (0.59–1.67) | 0.982 | 1.24 (0.81–1.89) | 0.330 | 0.84 (0.43–1.66) | 0.620 | ||
| Additive | – | 0.98 (0.76–1.27) | 0.873 | 0.83 (0.63–1.08) | 0.156 | 0.94 (0.76–1.16) | 0.554 | 0.87 (0.62–1.22) | 0.421 | |
|
PDX1 rs7981781 |
Allele | G | 1.00 | 1.00 | 1.00 | 1.00 | ||||
| A | 0.96 (0.75–1.23) | 0.753 | 1.08 (0.84–1.40) | 0.542 | 0.94 (0.76–1.16) | 0.558 | 1.22 (0.88–1.70) | 0.241 | ||
| Codominant | GG | 1.00 | 1.00 | 1.00 | 1.00 | |||||
| AA | 1.00 (0.60–1.67) | 0.999 | 1.23 (0.72–2.08) | 0.449 | 0.96 (0.64–1.46) | 0.856 | 1.46 (0.72–2.96) | 0.300 | ||
| AG | 0.89 (0.59–1.35) | 0.584 | 0.91 (0.61–1.35) | 0.628 | 0.70 (0.50–0.97) | 0.033 | 1.41 (0.83–2.39) | 0.203 | ||
| Dominant | GG | 1.00 | 1.00 | 1.00 | 1.00 | |||||
| AA-AG | 0.92 (0.62–1.36) | 0.683 | 0.98 (0.67–1.43) | 0.925 | 0.77 (0.56–1.05) | 0.096 | 1.42 (0.86–2.35) | 0.172 | ||
| Recessive | AG-GG | 1.00 | 1.00 | 1.00 | 1.00 | |||||
| AA | 1.07 (0.69–1.68) | 0.757 | 1.30 (0.81–2.09) | 0.281 | 1.19 (0.83–1.72) | 0.352 | 1.18 (0.63–2.20) | 0.615 | ||
| Additive | – | 0.99 (0.77–1.27) | 0.928 | 1.07 (0.83–1.38) | 0.603 | 0.94 (0.77–1.16) | 0.567 | 1.24 (0.88–1.75) | 0.225 | |
|
MC4R rs6567160 |
Allele | T | 1.00 | 1.00 | 1.00 | 1.00 | ||||
| C | 1.32 (0.99–1.75) | 0.060 | 0.77 (0.57–1.04) | 0.091 | 0.96 (0.7–1.23) | 0.756 | 1.16 (0.79–1.71) | 0.460 | ||
| Codominant | TT | 1.00 | 1.00 | 1.00 | 1.00 | |||||
| CC | 1.81 (0.91–3.58) | 0.091 | 0.33 (0.13–0.81) | 0.015 | 0.70 (0.37–1.32) | 0.271 | 1.41 (0.57–3.54) | 0.459 | ||
| CT | 1.22 (0.82–1.80) | 0.322 | 0.96 (0.65–1.40) | 0.815 | 1.11 (0.81–1.52) | 0.522 | 1.08 (0.65–1.81) | 0.758 | ||
| Dominant | TT | 1.00 | 1.00 | 1.00 | 1.00 | |||||
| CC-CT | 1.31 (0.91–1.89) | 0.144 | 0.84 (0.58–1.21) | 0.340 | 1.04 (0.77–1.39) | 0.820 | 1.14 (0.70–1.84) | 0.596 | ||
| Recessive | CT-TT | 1.00 | 1.00 | 1.00 | 1.00 | |||||
| CC | 1.68 (0.86–3.28) | 0.129 | 0.33 (0.14–0.81) | 0.016 | 0.68 (0.36–1.26) | 0.215 | 1.37 (0.56–3.37) | 0.489 | ||
| Additive | – | 1.29 (0.97–1.71) | 0.077 | 0.77 (0.57–1.04) | 0.085 | 0.96 (0.76–1.23) | 0.760 | 1.14 (0.79–1.66) | 0.484 | |
|
MC4R rs663129 |
Allele | G | 1.00 | 1.00 | 1.00 | 1.00 | ||||
| A | 1.32 (0.99–1.75) | 0.060 | 0.79 (0.59–1.07) | 0.125 | 0.98 (0.77–1.25) | 0.852 | 1.16 (0.79–1.71) | 0.460 | ||
| Codominant | GG | 1.00 | 1.00 | 1.00 | 1.00 | |||||
| AA | 1.81 (0.91–3.58) | 0.091 | 0.33 (0.13–0.82) | 0.017 | 0.71 (0.38–1.33) | 0.284 | 1.41 (0.57–3.54) | 0.459 | ||
| AG | 1.22 (0.82–1.80) | 0.322 | 0.99 (0.68–1.45) | 0.966 | 1.14 (0.83–1.55) | 0.424 | 1.08 (0.65–1.81) | 0.758 | ||
| Dominant | GG | 1.00 | 1.00 | 1.00 | 1.00 | |||||
| AA-AG | 1.31 (0.91–1.89) | 0.144 | 0.87 (0.60–1.25) | 0.441 | 1.06 (0.79–1.43) | 0.704 | 1.14 (0.70–1.84) | 0.596 | ||
| Recessive | AG-GG | 1.00 | 1.00 | 1.00 | 1.00 | |||||
| AA | 1.68 (0.86–3.28) | 0.129 | 0.33 (0.14–0.81) | 0.016 | 0.68 (0.36–1.26) | 0.215 | 1.37 (0.56–3.37) | 0.489 | ||
| Additive | – | 1.29 (0.97–1.71) | 0.077 | 0.78 (0.58–1.06) | 0.116 | 0.98 (0.77–1.24) | 0.854 | 1.14 (0.79–1.66) | 0.484 | |
|
MC4R rs17782313 |
Allele | T | 1.00 | 1.00 | 1.00 | 1.00 | ||||
| C | 1.32 (0.99–1.75) | 0.060 | 0.81 (0.60–1.09) | 0.167 | 0.98 (0.77–1.26) | 0.901 | 1.18 (0.80–1.74) | 0.403 | ||
| Codominant | TT | 1.00 | 1.00 | 1.00 | 1.00 | |||||
| CC | 1.81 (0.91–3.58) | 0.091 | 0.34 (0.14–0.83) | 0.018 | 0.71 (0.38–1.34) | 0.291 | 1.43 (0.57–3.58) | 0.443 | ||
| CT | 1.22 (0.82–1.80) | 0.322 | 1.03 (0.71–1.51) | 0.867 | 1.15 (0.84–1.58) | 0.378 | 1.12 (0.67–1.87) | 0.660 | ||
| Dominant | TT | 1.00 | 1.00 | 1.00 | 1.00 | |||||
| CC-CT | 1.31 (0.91–1.89) | 0.144 | 0.90 (0.62–1.30) | 0.569 | 1.07 (0.80–1.44) | 0.648 | 1.17 (0.73–1.90) | 0.515 | ||
| Recessive | CT-TT | 1.00 | 1.00 | 1.00 | 1.00 | |||||
| CC | 1.68 (0.86–3.28) | 0.129 | 0.33 (0.14–0.81) | 0.016 | 0.68 (0.36–1.26) | 0.215 | 1.37 (0.56–3.37) | 0.489 | ||
| Additive | – | 1.29 (0.97–1.71) | 0.077 | 0.80 (0.59–1.09) | 0.159 | 0.99 (0.77–1.25) | 0.903 | 1.16 (0.80–1.69) | 0.426 | |
|
MC4R rs12969709 |
Allele | C | 1.00 | 1.00 | 1.00 | 1.00 | ||||
| A | 1.13 (0.85–1.51) | 0.406 | 0.78 (0.57–1.06) | 0.111 | 0.91 (0.71–1.16) | 0.449 | 1.03 (0.69–1.53) | 0.884 | ||
| Codominant | CC | 1.00 | 1.00 | 1.00 | 1.00 | |||||
| AA | 1.37 (0.67–2.80) | 0.396 | 0.27 (0.10–0.75) | 0.012 | 0.58 (0.29–1.15) | 0.117 | 1.02 (0.39–2.70) | 0.965 | ||
| AC | 1.12 (0.76–1.65) | 0.583 | 0.98 (0.67–1.43) | 0.900 | 1.06 (0.77–1.45) | 0.726 | 1.05 (0.63–1.75) | 0.867 | ||
| Dominant | CC | 1.00 | 1.00 | 1.00 | 1.00 | |||||
| AA-AC | 1.16 (0.80–1.67) | 0.442 | 0.85 (0.59–1.23) | 0.397 | 0.98 (0.72–1.32) | 0.879 | 1.04 (0.64–1.69) | 0.871 | ||
| Recessive | AC-CC | 1.00 | 1.00 | 1.00 | 1.00 | |||||
| AA | 1.31 (0.65–2.66) | 0.449 | 0.27 (0.10–0.75) | 0.012 | 0.57 (0.29–1.11) | 0.099 | 1.01 (0.39–2.62) | 0.989 | ||
| Additive | – | 1.14 (0.86–1.53) | 0.361 | 0.77 (0.56–1.05) | 0.098 | 0.91 (0.71–1.16) | 0.451 | 1.03 (0.70–1.51) | 0.893 | |
|
MC4R rs11663816 |
Allele | T | 1.00 | 1.00 | 1.00 | 1.00 | ||||
| C | 1.21 (0.90–1.61) | 0.203 | 0.75 (0.55–1.02) | 0.067 | 0.92 (0.72–1.17) | 0.488 | 1.07 (0.72–1.60) | 0.727 | ||
| Codominant | TT | 1.00 | 1.00 | 1.00 | 1.00 | |||||
| CC | 1.78 (0.85–3.73) | 0.127 | 0.31 (0.11–0.88) | 0.027 | 0.70 (0.35–1.39) | 0.304 | 1.44 (0.53–3.96) | 0.476 | ||
| CT | 1.07 (0.73–1.58) | 0.726 | 0.88 (0.60–1.28) | 0.504 | 0.98 (0.72–1.34) | 0.923 | 0.94 (0.57–1.57) | 0.823 | ||
| Dominant | TT | 1.00 | 1.00 | 1.00 | 1.00 | |||||
| CC-CT | 1.17 (0.81–1.68) | 0.412 | 0.80 (0.55–1.15) | 0.223 | 0.94 (0.70–1.27) | 0.703 | 1.01 (0.62–1.64) | 0.968 | ||
| Recessive | CT-TT | 1.00 | 1.00 | 1.00 | 1.00 | |||||
| CC | 1.74 (0.84–3.59) | 0.137 | 0.32 (0.11–0.91) | 0.032 | 0.70 (0.35–1.38) | 0.305 | 1.47 (0.54–3.98) | 0.447 | ||
| Additive | – | 1.21 (0.90–1.61) | 0.210 | 0.75 (0.55–1.03) | 0.072 | 0.92 (0.71–1.17) | 0.486 | 1.07 (0.73–1.57) | 0.739 | |
SNP single nucleotide polymorphism, OR odds ratio, 95% CI 95% confidence interval
p values were calculated by logistic regression analysis with adjustment for age and gender
Bold values indicate statistical significance (p < 0.05)
In addition, as shown in Table 4, PDX1-rs7981781 reduced the susceptibility to T2DM among smokers under the codominant (OR = 0.50, 95% CI = 0.29–0.89, p = 0.018) and dominant (OR = 0.55, 95% CI = 0.32–0.95, p = 0.030) models. However, MC4R-rs6567160 could increase the occurrence of T2DM among nonsmokers under the codominant (OR = 1.60, 95% CI = 1.04–2.45, p = 0.032) and dominant (OR = 1.56, 95% CI = 1.04–2.34, p = 0.031) models. MC4R-rs663129 induced a significantly higher susceptibility to T2DM among individuals who were nonsmokers in the codominant (OR = 1.64, 95% CI = 1.07–2.52, p = 0.023), dominant (OR = 1.60, 95% CI = 1.07–2.40, p = 0.023) and additive (OR = 1.40, 95% CI = 1.00–1.95, p = 0.049) models. Moreover, rs17782313 in MC4R was related to a higher risk of T2DM among nonsmokers under the allelic (OR = 1.43, 95% CI = 1.00–1.95, p = 0.036), codominant (OR = 1.72, 95% CI = 1.12–2.64, p = 0.014), dominant (OR = 1.66, 95% CI = 1.11–2.50, p = 0.014) and additive (OR = 1.44, 95% CI = 1.03–2.01, p = 0.034) models.
Table 4.
The associations between PDX1 and MC4R polymorphisms and the risk of T2DM stratified by smoking, drinking status
| Gene SIP | Model | Genotype | Smoking | Non-smoking | Drinking | Non-drinking | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | p | OR (95% CI) | p | OR (95% CI) | p | OR (95% CI) | p | |||
|
PDX1 rs11619319 |
Allele | A | 1.00 | 1.00 | 1.00 | 1.00 | ||||
| G | 0.82 (0.58–1.15) | 0.246 | 0.92 (0.70–1.21) | 0.535 | 0.80 (0.55–1.18) | 0.263 | 0.93 (0.71–1.22) | 0.608 | ||
| Codominant | AA | 1.00 | 1.00 | 1.00 | 1.00 | |||||
| GG | 0.75 (0.37–1.50) | 0.410 | 0.84 (0.48–1.49) | 0.558 | 0.67 (0.30–1.47) | 0.313 | 0.89 (0.50–1.56) | 0.676 | ||
| GA | 0.61 (0.34–1.09) | 0.098 | 0.77 (0.48–1.23) | 0.274 | 0.51 (0.27–0.97) | 0.039 | 0.81 (0.51–1.29) | 0.381 | ||
| Dominant | AA | 1.00 | 1.00 | 1.00 | 1.00 | |||||
| GG-GA | 0.65 (0.38–1.13) | 0.125 | 0.79 (0.51–1.23) | 0.299 | 0.55 (0.30–1.01) | 0.054 | 0.83 (0.54–1.29) | 0.418 | ||
| Recessive | GA-AA | 1.00 | 1.00 | 1.00 | 1.00 | |||||
| GG | 1.02 (0.57–1.83) | 0.943 | 1.00 (0.62–1.62) | 0.998 | 1.01 (0.63–1.63) | 0.957 | 1.01 (0.63–1.63) | 0.957 | ||
| Additive | – | 0.85 (0.61–1.19) | 0.346 | 0.91 (0.69–1.20) | 0.501 | 0.93 (0.71–1.23) | 0.623 | 0.93 (0.71–1.23) | 0.623 | |
|
PDX1 rs2293941 |
Allele | G | 1.00 | 1.00 | 1.00 | 1.00 | ||||
| A | 0.83 (0.59–1.16) | 0.274 | 0.91 (0.69–1.20) | 0.515 | 0.80 (0.55–1.18) | 0.264 | 0.94 (0.71–1.23) | 0.647 | ||
| Codominant | GG | 1.00 | 1.00 | 1.00 | ||||||
| AA | 0.77 (0.38–1.54) | 0.454 | 0.85 (0.48–1.50) | 0.568 | 0.68 (0.31–1.48) | 0.326 | 0.90 (0.51–1.57) | 0.703 | ||
| AG | 0.64 (0.36–1.13) | 0.124 | 0.80 (0.50–1.26) | 0.331 | 0.51 (0.27–0.97) | 0.040 | 0.85 (0.54–1.34) | 0.477 | ||
| Dominant | GG | 1.00 | 1.00 | 1.00 | 1.00 | |||||
| AA-AG | 0.67 (0.39–1.16) | 0.156 | 0.81 (0.52–1.25) | 0.345 | 0.56 (0.30–1.02) | 0.056 | 0.86 (0.56–1.33) | 0.499 | ||
| Recessive | AG-GG | 1.00 | 1.00 | 1.00 | 1.00 | |||||
| AA | 1.02 (0.57–1.83) | 0.943 | 0.98 (0.61–1.59) | 0.939 | 1.03 (0.53–2.00) | 0.935 | 1.00 (0.62–1.61) | 0.986 | ||
| Additive | – | 0.86 (0.62–1.21) | 0.384 | 0.91 (0.69–1.20) | 0.511 | 0.79 (0.54–1.16) | 0.231 | 0.94 (0.71–1.24) | 0.654 | |
|
PDX1 rs7981781 |
Allele | G | 1.00 | 1.00 | 1.00 | 1.00 | ||||
| A | 0.76 (0.54–1.07) | 0.117 | 0.95 (0.72–1.25) | 0.726 | 0.73 (0.50–1.08) | 0.111 | 0.96 (0.73–1.27) | 0.786 | ||
| Codominant | GG | 1.00 | 1.00 | 1.00 | 1.00 | |||||
| AA | 0.68 (0.34–1.36) | 0.277 | 0.92 (0.53–1.61) | 0.775 | 0.57 (0.26–1.26) | 0.167 | 0.94 (0.54–1.64) | 0.834 | ||
| AG | 0.50 (0.29–0.89) | 0.018 | 0.89 (0.57–1.41) | 0.628 | 0.47 (0.25–0.88) | 0.019 | 0.92 (0.59–1.44) | 0.716 | ||
| Dominant | GG | 1.00 | 1.00 | 1.00 | 1.00 | |||||
| AA-AG | 0.55 (0.32–0.95) | 0.030 | 0.90 (0.59–1.38) | 0.636 | 0.49 (0.27–0.90) | 0.022 | 0.93 (0.61–1.41) | 0.724 | ||
| Recessive | AG-GG | 1.00 | 1.00 | 1.00 | 1.00 | |||||
| AA | 1.03 (0.57–1.86) | 0.919 | 0.99 (0.61–1.60) | 0.962 | 0.92 (0.47–1.81) | 0.811 | 0.99 (0.61–1.60) | 0.972 | ||
| Additive | – | 0.80 (0.57–1.11) | 0.175 | 0.95 (0.72–1.26) | 0.737 | 0.72 (0.49–1.07) | 0.100 | 0.97 (0.74–1.27) | 0.803 | |
|
MC4R rs6567160 |
Allele | T | 1.00 | 1.00 | 1.00 | 1.00 | ||||
| C | 0.92 (0.61–1.37) | 0.665 | 1.36 (0.98–1.90) | 0.068 | 0.90 (0.56–1.46) | 0.682 | 1.14 (0.83–1.58) | 0.416 | ||
| Codominant | TT | 1.00 | 1.00 | 1.00 | 1.00 | |||||
| CC | 0.99 (0.33–2.93) | 0.980 | 1.37 (0.58–3.20) | 0.472 | 1.13 (0.29–4.41) | 0.857 | 0.99 (0.45–2.18) | 0.988 | ||
| CT | 0.86 (0.52–1.44) | 0.569 | 1.60 (1.04–2.45) | 0.032 | 0.80 (0.44–1.45) | 0.460 | 1.34 (0.87–2.04) | 0.180 | ||
| Dominant | TT | 1.00 | 1.00 | 1.00 | 1.00 | |||||
| CC-CT | 0.88 (0.54–1.43) | 0.601 | 1.56 (1.04–2.34) | 0.031 | 0.83 (0.47–1.47) | 0.532 | 1.27 (0.85–1.90) | 0.237 | ||
| Recessive | CT-TT | 1.00 | 1.00 | 1.00 | 1.00 | |||||
| CC | 1.04 (0.36–3.04) | 0.942 | 1.16 (0.50–2.69) | 0.725 | 1.22 (0.32–4.67) | 0.776 | 0.89 (0.41–1.92) | 0.772 | ||
| Additive | – | 0.92 (0.62–1.37) | 0.688 | 1.37 (0.99–1.92) | 0.061 | 0.90 (0.56–1.45) | 0.673 | 1.15 (0.83–1.58) | 0.407 | |
|
MC4R rs663129 |
Allele | G | 1.00 | 1.00 | 1.00 | 1.00 | ||||
| A | 0.93 (0.62–1.38) | 0.713 | 1.39 (0.99–1.94) | 0.053 | 0.96 (0.59–1.55) | 0.869 | 1.14 (0.83–1.58) | 0.416 | ||
| Codominant | GG | 1.00 | 1.00 | 1.00 | 1.00 | |||||
| AA | 0.99 (0.33–2.95) | 0.990 | 1.38 (0.59–3.23) | 0.459 | 1.17 (0.30–4.54) | 0.825 | 0.99 (0.45–2.18) | 0.988 | ||
| AG | 0.88 (0.53–1.47) | 0.624 | 1.64 (1.07–2.52) | 0.023 | 0.88 (0.48–1.58) | 0.660 | 1.34 (0.87–2.04) | 0.180 | ||
| Dominant | GG | 1.00 | 1.00 | 1.00 | 1.00 | |||||
| AA-AG | 0.89 (0.55–1.46) | 0.655 | 1.60 (1.07–2.40) | 0.023 | 0.91 (0.51–1.60) | 0.735 | 1.27 (0.85–1.90) | 0.237 | ||
| Recessive | AG-GG | 1.00 | 1.00 | 1.00 | 1.00 | |||||
| AA | 1.04 (0.36–3.04) | 0.942 | 1.16 (0.50–2.69) | 0.725 | 1.22 (0.32–4.67) | 0.776 | 0.89 (0.41–1.92) | 0.772 | ||
| Additive | – | 0.93 (0.63–1.39) | 0.734 | 1.40 (1.00–1.95) | 0.049 | 0.96 (0.60–1.54) | 0.855 | 1.15 (0.83–1.58) | 0.407 | |
|
MC4R rs17782313 |
Allele | T | 1.00 | 1.00 | 1.00 | 1.00 | ||||
| C | 0.93 (0.62–1.38) | 0.713 | 1.43 (1.02–2.00) | 0.036 | 0.96 (0.59–1.55) | 0.869 | 1.18 (0.85–1.63) | 0.329 | ||
| Codominant | TT | 1.00 | 1.00 | 1.00 | 1.00 | |||||
| CC | 0.99 (0.33–2.95) | 0.990 | 1.40 (0.60–3.28) | 0.439 | 1.17 (0.30–4.54) | 0.825 | 1.01 (0.46–2.21) | 0.977 | ||
| CT | 0.88 (0.53–1.47) | 0.624 | 1.72 (1.12–2.64) | 0.014 | 0.88 (0.48–1.58) | 0.660 | 1.40 (0.92–2.15) | 0.118 | ||
| Dominant | TT | 1.00 | 1.00 | 1.00 | 1.00 | |||||
| CC-CT | 0.89 (0.55–1.46) | 0.655 | 1.66 (1.11–2.50) | 0.014 | 0.91 (0.51–1.60) | 0.735 | 1.33 (0.89–1.99) | 0.163 | ||
| Recessive | CT-TT | 1.00 | 1.00 | 1.00 | 1.00 | |||||
| CC | 1.04 (0.36–3.04) | 0.942 | 1.16 (0.50–2.69) | 0.725 | 1.22 (0.32–4.67) | 0.776 | 0.89 (0.41–1.92) | 0.772 | ||
| Additive | – | 0.93 (0.63–1.39) | 0.734 | 1.44 (1.03–2.01) | 0.034 | 0.96 (0.60–1.54) | 0.855 | 1.18 (0.85–1.63) | 0.317 | |
SNP single nucleotide polymorphism, OR odds ratio, 95% CI 95% confidence interval
p values were calculated by logistic regression analysis with adjustment for age and gender
Bold values indicate statistical significance (p < 0.05)
Additionally, PDX1-rs11619319 (OR = 0.51, 95% CI = 0.27–20.97, p = 0.039) and rs2293941 (OR = 0.51, 95% CI = 0.27–0.97, p = 0.040) were predominantly related to a reduced risk of T2DM among drinkers under the codominant model. Rs7981781 was correlated with a lower risk of T2DM among drinkers under the codominant (OR = 0.47, 95% CI = 0.25–0.88, p = 0.019) and dominant (OR = 0.49, 95% CI = 0.27–0.90, p = 0.022) models.
When stratified by BMI (Table 5), PDX1-rs7981781 was correlated with a lower risk of T2DM among subjects with BMI > 24 kg/m2 under the codominant model (OR = 0.64, 95% CI = 0.41–1.00, p = 0.049).
Table 5.
The association between PDX1 polymorphisms and the risk of T2DM stratified by BMI
| Gene SIP | Model | Genotype | ≤ 24 | > 24 | ||
|---|---|---|---|---|---|---|
| OR (95% CI) | p | OR (95% CI) | p | |||
|
PDX1 rs7981781 |
Allele | G | 1.00 | 1.00 | ||
| A | 1.07 (0.78–1.46) | 0.675 | 0.94 (0.72–1.24) | 0.682 | ||
| Codominant | GG | 1.00 | 1.00 | |||
| AA | 1.10 (0.59–2.06) | 0.766 | 0.97 (0.54–1.72) | 0.910 | ||
| AG | 0.98 (0.58–1.64) | 0.928 | 0.64 (0.41–1.00) | 0.049 | ||
| Dominant | GG | 1.00 | 1.00 | |||
| AA-AG | 1.01 (0.62–1.65) | 0.956 | 0.72 (0.47–1.09) | 0.117 | ||
| Recessive | AG-GG | 1.00 | 1.00 | |||
| AA | 1.12 (0.65–1.92) | 0.690 | 1.26 (0.75–2.10) | 0.383 | ||
| Additive | – | 1.04 (0.76–1.43) | 0.790 | 0.92 (0.70–1.22) | 0.559 | |
SNP single nucleotide polymorphism, OR odds ratio, 95% CI 95% confidence interval
p values were calculated by logistic regression analysis with adjustment for age and gender
Bold values indicate statistical significance (p < 0.05)
Haplotype analysis
We next conducted linkage disequilibrium (LD) analysis for the polymorphisms in MC4R1 and PDX1. Our results indicated two blocks (block1: rs11619319 and rs2293941; block2: rs9581943 and rs7981781) in PDX1 (Fig. 1) and two blocks (block1: rs6567160, rs663129, and rs17782313; block2: rs11663816 and rs12970134) in MC4R (Fig. 2). Besides, Table 6 shows that there was no association between haplotype frequency and T2DM risk (p > 0.05).
Fig. 1.
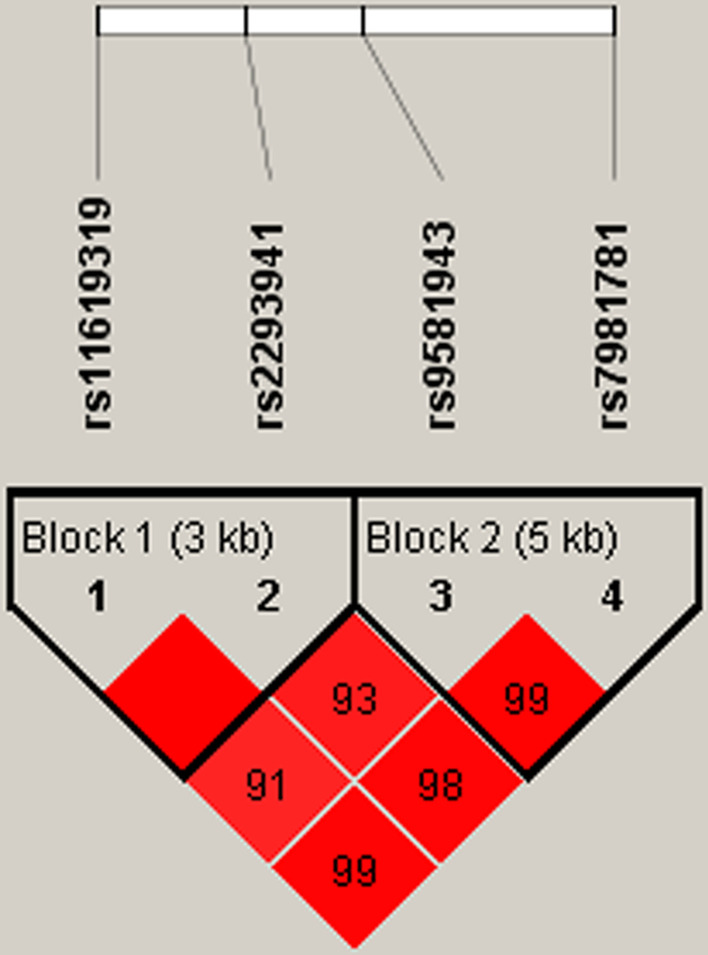
Haplotype block map for SNPs in PDX1. Block 1 includes rs11619319 and rs2293941. Block 2 includes rs9581943 and rs7981781. The numbers inside the diamonds indicate the D’ for pairwise analyses
Fig. 2.
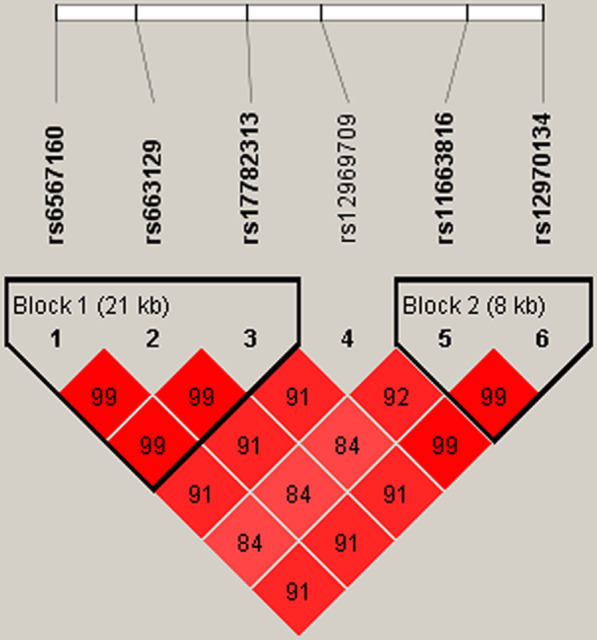
Haplotype block map for SNPs in MC4R.Block 1 includes rs6567160, rs663129 and rs17782313. Block 2 includes rs11663816 and rs12970134. The numbers inside the diamonds indicate the D’ for pairwise analyses
Table 6.
Haplotype analysis of PDX1 and MC4R SNPs with T2DM risk
| Gene | SNP | Haplotype | Frequency in cases | Frequency in controls | With adjustment | Without adjustment | ||
|---|---|---|---|---|---|---|---|---|
| OR (95%CI) | p | OR (95%CI) | p | |||||
| PDX1 | rs11619319|rs2293941 | GA | 0.446 | 0.440 | 1.02 (0.86–1.22) | 0.791 | 1.02 (0.86–1.22) | 0.792 |
| PDX1 | rs11619319|rs2293941 | AG | 0.451 | 0.444 | 1.03 (0.86–1.23) | 0.755 | 1.03 (0.86–1.23) | 0.756 |
| PDX1 | rs9581943|rs7981781 | GA | 0.432 | 0.431 | 1.00 (0.84–1.20) | 0.969 | 1.00 (0.84–1.20) | 0.970 |
| PDX1 | rs9581943|rs7981781 | AG | 0.350 | 0.372 | 0.91 (0.72–1.09) | 0.307 | 0.91 (0.76–1.09) | 0.308 |
| PDX1 | rs9581943|rs7981781 | GG | 0.216 | 0.197 | 1.13 (0.91–1.40) | 0.283 | 1.13 (0.91–1.40) | 0.284 |
| MC4R | rs6567160|rs663129|rs17782313 | CAC | 0.237 | 0.233 | 1.02 (0.84–1.25) | 0.819 | 1.02 (0.84–1.25) | 0.820 |
| MC4R | rs6567160|rs663129|rs17782313 | TGT | 0.239 | 0.235 | 1.02 (0.84–1.25) | 0.818 | 1.02 (0.84–1.25) | 0.819 |
| MC4R | rs11663816|rs12970134 | CA | 0.204 | 0.217 | 0.93 (0.75–1.15) | 0.510 | 0.93 (0.75–1.15) | 0.509 |
| MC4R | rs11663816|rs12970134 | CG | 0.015 | 0.011 | 1.38 (0.63–3.04) | 0.423 | 1.38 (0.63–3.04) | 0.423 |
| MC4R | rs11663816|rs12970134 | TG | 0.221 | 0.228 | 0.97 (0.78–1.19) | 0.746 | 0.97 (0.78–1.19) | 0.744 |
SNP single nucleotide polymorphism, OR odd ratios, CI confidence interval
The relative mRNA expression of PDX1 and MC4R
The MC4R mRNA expression levels in T2DM case subjects decreased compared with those in their nondiabetic counterparts (p = 0.040, Fig. 3a). In addition, although no significant differences were observed in the expression levels of PDX1 mRNA between the two groups, we did observe a decreased pattern of PDX1 expression in individual samples between the cases and controls (p = 0.054, Fig. 3b).
Fig. 3.
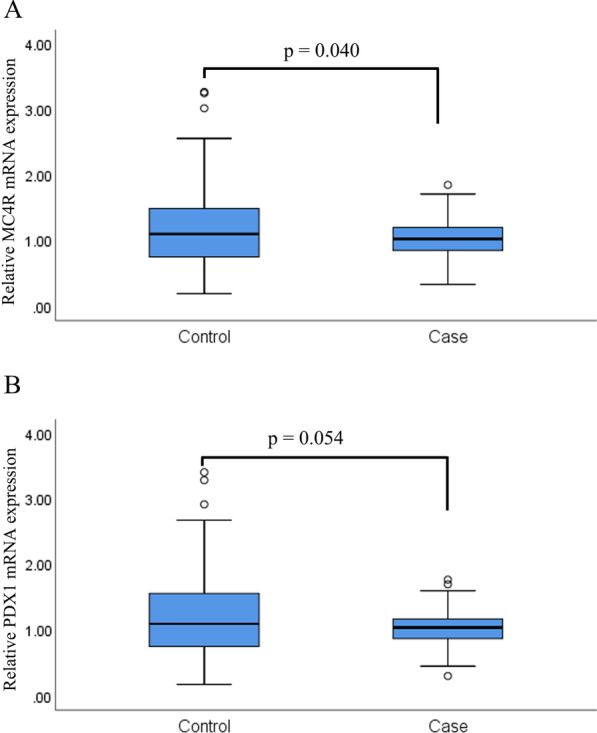
The relative mRNA expression of the MC4R and PDX1 genes in T2DM patients and controls. T2DM, type 2 diabetes mellitus
The association of relative mRNA expression and PDX1 and MC4R polymorphisms
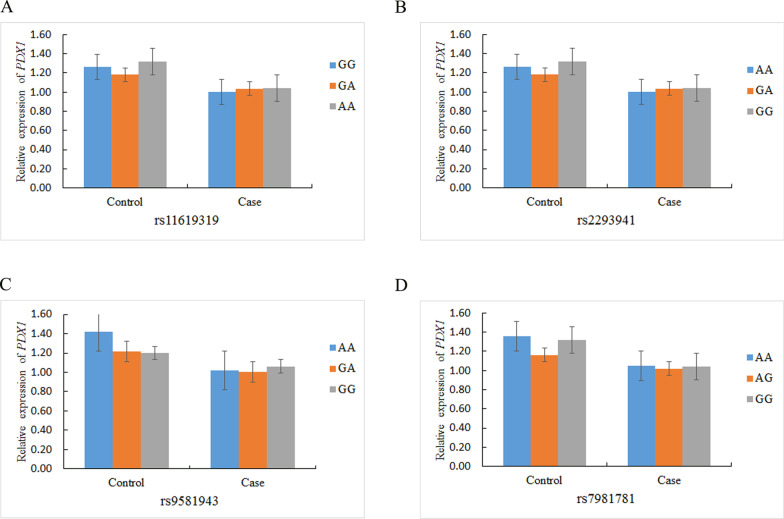
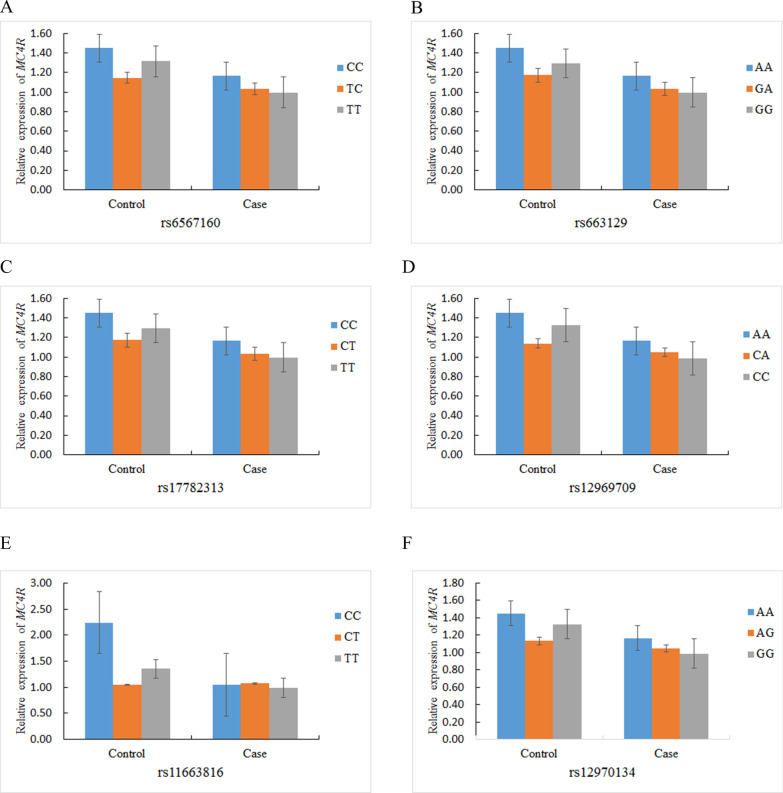
The PDX1 and MC4R polymorphisms were not associated with the relative PDX1 and MC4R mRNA expression in the T2DM patients and controls (Figs. 4, 5).
Fig. 4.
The association of relative PDX1 mRNA expression and genetic polymorphisms in T2DM patients and healthy controls
Fig. 5.
The association of relative MC4R mRNA expression and genetic polymorphisms in T2DM patients and healthy controls
Discussion
This research focused on the association of PDX1 and MC4R polymorphisms with susceptibility to T2DM in Chinese Han people. We found that PDX1-rs9581943 was correlated with a decreased risk of T2DM among the study subjects. In addition, the effects of PDX1 and MC4R polymorphisms on T2DM susceptibility were dependent on age, sex, smoking status, drinking status and BMI. These findings suggest that genetic polymorphisms in PDX1 and MC4R may play a crucial role in the development of T2DM.
In humans, the PDX1 gene is located on chromosome 13q12.1. It is a key transcription factor involved in pancreatic development, islet hormone and insulin expression. Data from several studies suggested that deletion and mutation in PDX1 caused overt diabetes and maturity-onset diabetes of the young [21, 22]. Additionally, Steinthorsdottir et al. found that rare frameshift variants in PDX1 were associated with a higher risk of T2DM in Icelanders [6]. Recently, a homozygous mutation in PDX1 was detected in a 65-day-old Iranian patient with neonatal diabetes [23]. However, there are few studies on rs11619319, rs2293941, rs9581943, and rs7981781. In the present study, we found that only rs9581943 decreased the incidence of T2DM among the study subjects. Moreover, we found that the relative mRNA expression of the PDX1 gene was lower in T2DM patients than in controls, but the difference was insignificant. Interestingly, stratified analysis results revealed that rs9581943, rs11619319, rs2293941, and rs7981781were associated with susceptibility to T2DM in different subgroups. Manning et al. [24] illustrated that rs2293941 was associated with fasting glucose levels in individuals of European ancestry. However, this correlation was not observed among participants in the Chinese Han population in the present study (not shown). The inconsistencies in these reports may result from subjects of different ethnicities and different environments. Taken together, these results demonstrated that the PDX1 polymorphism is important in the development and risk assessment of T2DM.
MC4R is a G-protein-coupled receptor that is highly expressed in the hypothalamus, where it regulates appetite, energy expenditure and body weight [25]. It is located on chromosome 18q21 in humans. Disruption of the MC4R gene leads to the obesity phenotype, which is related to T2DM [26]. Vaisse et al. claimed that rare heterozygous MC4R variants have been identified in obese children and adults in many populations [27]. Obesity is an important risk factor for the progression of T2DM [17].
Herein, we explored whether MC4R polymorphisms could contribute to T2DM risk in a Chinese Han population. In this study, we found that the mRNA level of MC4R was decreased in T2DM patients compared to healthy controls. However, the overall analysis revealed that the association between MC4R polymorphisms and T2DM risk was insignificant. Subsequently, we examined the correlation of MC4R polymorphisms and T2DM risk by stratification analysis. We found that rs17782313 in MC4R obviously reduced the susceptibility toT2DM among individuals younger than 60 years old. It has previously been demonstrated that the MC4R-rs17782313 polymorphism is strongly related to obesity in adults and children of European descent [28]. Moreover, Hardy et al. also demonstrated that rs17782313 was associated with weight and BMI. The association of this polymorphism with weight strengthened during childhood and adolescence, and weakened during adulthood [29]. This result suggested that the effect of MC4R-rs17782313 on disease risk was dependent on age. In addition, a study showed that rs12970134 increased the risk of T2DM among individuals of European descent [30], although this effect was not found in our study. In our analysis, rs6567160 reduced the susceptibility to T2DM among individuals ≤ 60 years old but was not associated with the clinical characteristics. However, Carvalho et al. suggested that rs6567160 was associated with a greater postpartum increase in HbA1c in women who had experienced gestational diabetes mellitus than in those who had not [31]. Additionally, rs663129 decreased the risk of T2DM among Han Chinese people. This finding was inconsistent with the discovery of Nikpay et al., which indicated that allele A of rs663129 increased the risk of both coronary artery disease and obesity in individuals of European ancestry [32]. The reason for these inconsistent results may be that the occurrence and development of T2DM are related to a variety of factors, including population, sample size, and environment. Together, these data highlighted the important role of MC4R polymorphisms in the occurrence of T2DM.
Moreover, these selected SNPs in the PDX1 and MC4R genes can affect promoter histone marks, enhancer histone marks, DNAse, proteins bound, motifs changed, NHGRI/EBI GWAS hits, and GRASP QTL hits. Therefore, we presumed that these functions could modify the risk of T2DM by influencing gene expression. The specific mechanisms underlying these effects require further investigation.
There were several limitations in this study. First, this research was performed based on a Chinese Han population. Therefore, further research with subjects of different genetic backgrounds should be conducted to validate our results. Second, selection bias was an unavoidable problem in our research.
Conclusions
In conclusion, our findings demonstrated that the variants in the PDX1 and MC4R genes were related to susceptibility to T2DM in the Chinese Han population. These single polymorphic markers are considered to be new targets in the assessment and prevention of T2DM among Chinese Han people.
Supplementary Information
Additional file 1. Table S1. Primer sequences of PDX1 and MC4R for PCR. Table S2. Basic information of candidate SNPs in the study.
Acknowledgements
We sincerely thank all participators in this study.
Abbreviations
- DM
diabetes mellitus
- T2DM
type 2 diabetes mellitus
- PDX1
pancreatic and duodenal homeobox-1
- MC4R
melanocortin receptor 4
- HWEL
Hardy–Weinberg equilibrium
- ORs
odds ratio
- CI
95% confidence intervals
Authors' contributions
NW, RT and WC designed this study protocol and drafted the manuscript; JX and JC performed the DNA extraction and genotyping; YT and JP performed the data analysis; HC, YP, SF and SY performed the sample collection and information recording. LW and JY polished the manuscript, WC conceived and supervised the study. All authors read and approved the final manuscript.
Funding
This study was supported by the Bethune-Merck Diabetes Research Foundation (G-X-2019-056), the Clinical Research Project of the First Affiliated Hospital of Xi’an Jiaotong University (XJTU1AF-CRF-2019-007) and the Social Development Science and Technology Project of Shaanxi Province (2016sf-314).
Availability of data and material
The datasets generated and/or analyzed during the current study are not publicly available due patient privacy but are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
This research received approval from the Ethics committee of the Frist Affiliated Hospital of Xi’an Jiaotong University, and conformed to the Declaration of Helsinki. Informed consent was acquired from each participant at recruitment after fully describing our research to them.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflict of interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Co-first author: Ning Wang and Rui Tong
References
- 1.Chatterjee S, Davies MJ. Accurate diagnosis of diabetes mellitus and new paradigms of classification. Nat Rev Endocrinol. 2018;14:386–387. doi: 10.1038/s41574-018-0025-1. [DOI] [PubMed] [Google Scholar]
- 2.Saeedi P, Petersohn I, Salpea P, Malanda B, Karuranga S, Unwin N, et al. Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: results from the International Diabetes Federation Diabetes Atlas. Diabetes Res Clin Pract. 2019;157:107843. doi: 10.1016/j.diabres.2019.107843. [DOI] [PubMed] [Google Scholar]
- 3.Cho NH, Shaw JE, Karuranga S, Huang Y, da Rocha Fernandes JD, Ohlrogge AW, et al. IDF diabetes Atlas: global estimates of diabetes prevalence for 2017 and projections for 2045. Diabetes Res Clin Pract. 2018;138:271–281. doi: 10.1016/j.diabres.2018.02.023. [DOI] [PubMed] [Google Scholar]
- 4.Hemminki K, Li X, Sundquist K, Sundquist J. Familial risks for type 2 diabetes in Sweden. Diabetes Care. 2010;33:293–297. doi: 10.2337/dc09-0947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meigs JB, Cupples LA, Wilson PW. Parental transmission of type 2 diabetes: the Framingham Offspring Study. Diabetes. 2000;49:2201–2207. doi: 10.2337/diabetes.49.12.2201. [DOI] [PubMed] [Google Scholar]
- 6.Steinthorsdottir V, Thorleifsson G, Sulem P, Helgason H, Grarup N, Sigurdsson A, et al. Identification of low-frequency and rare sequence variants associated with elevated or reduced risk of type 2 diabetes. Nat Genet. 2014;46:294–298. doi: 10.1038/ng.2882. [DOI] [PubMed] [Google Scholar]
- 7.Scott RA, Scott LJ, Mägi R, Marullo L, Gaulton KJ, Kaakinen M, et al. An expanded genome-wide association study of type 2 diabetes in Europeans. Diabetes. 2017;66:2888–2902. doi: 10.2337/db16-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu Y, Liu Q, Zhou Z, Ikeda Y. PDX1, Neurogenin-3, and MAFA: critical transcription regulators for beta cell development and regeneration. Stem Cell Res Ther. 2017;8:240. doi: 10.1186/s13287-017-0694-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu S, Ballian N, Belaguli NS, Patel S, Li M, Templeton NS, et al. PDX-1 acts as a potential molecular target for treatment of human pancreatic cancer. Pancreas. 2008;37:210–220. doi: 10.1097/MPA.0b013e31816a4a33. [DOI] [PubMed] [Google Scholar]
- 10.Gurevich LE, Proshchina AE, Voronkova IA, Ashevskaya VE, Korosteleva PA. Dolzhansky OV [Differential diagnostic value of the expression of the transcription factor PDX-1 in neuroendocrine and non-neuroendocrine tumors of the pancreas and other organs] Arkh Patol. 2019;81:11–21. doi: 10.17116/patol20198105111. [DOI] [PubMed] [Google Scholar]
- 11.Ahlgren U, Jonsson J, Edlund H. The morphogenesis of the pancreatic mesenchyme is uncoupled from that of the pancreatic epithelium in IPF1/PDX1-deficient mice. Development. 1996;122:1409–1416. doi: 10.1242/dev.122.5.1409. [DOI] [PubMed] [Google Scholar]
- 12.Cai M, Hruby VJ. The melanocortin receptor system: a target for multiple degenerative diseases. Curr Protein Pept Sci. 2016;17:488–496. doi: 10.2174/1389203717666160226145330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kishi T, Aschkenasi CJ, Lee CE, Mountjoy KG, Saper CB, Elmquist JK. Expression of melanocortin 4 receptor mRNA in the central nervous system of the rat. J Comp Neurol. 2003;457:213–235. doi: 10.1002/cne.10454. [DOI] [PubMed] [Google Scholar]
- 14.Huszar D, Lynch CA, Fairchild-Huntress V, Dunmore JH, Fang Q, Berkemeier LR, et al. Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell. 1997;88:131–141. doi: 10.1016/S0092-8674(00)81865-6. [DOI] [PubMed] [Google Scholar]
- 15.Greenfield JR, Miller JW, Keogh JM, Henning E, Satterwhite JH, Cameron GS, et al. Modulation of blood pressure by central melanocortinergic pathways. N Engl J Med. 2009;360:44–52. doi: 10.1056/NEJMoa0803085. [DOI] [PubMed] [Google Scholar]
- 16.Srisai D, Gillum MP, Panaro BL, Zhang XM, Kotchabhakdi N, Shulman GI, et al. Characterization of the hyperphagic response to dietary fat in the MC4R knockout mouse. Endocrinology. 2011;152:890–902. doi: 10.1210/en.2010-0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malone JI, Hansen BC. Does obesity cause type 2 diabetes mellitus (T2DM)? Or is it the opposite? Pediatr Diabetes. 2019;20:5–9. doi: 10.1111/pedi.12787. [DOI] [PubMed] [Google Scholar]
- 18.American Diabetes Association. Standards of medical care in diabetes—2010. Diabetes Care 2010;33(Suppl 1):S11–61. [DOI] [PMC free article] [PubMed]
- 19.Du J, Jin T, Cao Y, Chen J, Guo Y, Sun M, et al. Association between genetic polymorphisms of MMP8 and the risk of steroid-induced osteonecrosis of the femoral head in the population of northern China. Medicine (Baltimore) 2016;95:e4794. doi: 10.1097/MD.0000000000004794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao Q, Liao S, Wei H, Liu D, Li J, Zhang X, et al. CDKN2BAS polymorphisms are associated with coronary heart disease risk a Han Chinese population. Oncotarget. 2016;7:82046–82054. doi: 10.18632/oncotarget.12575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weng J, Macfarlane WM, Lehto M, Gu HF, Shepherd LM, Ivarsson SA, et al. Functional consequences of mutations in the MODY4 gene (IPF1) and coexistence with MODY3 mutations. Diabetologia. 2001;44:249–258. doi: 10.1007/s001250051608. [DOI] [PubMed] [Google Scholar]
- 22.Gao T, McKenna B, Li C, Reichert M, Nguyen J, Singh T, et al. Pdx1 maintains beta cell identity and function by repressing an alpha cell program. Cell Metab. 2014;19:259–271. doi: 10.1016/j.cmet.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sahebi L, Niknafs N, Dalili H, Amini E, Esmaeilnia T, Amoli M, et al. Iranian neonatal diabetes mellitus due to mutation in PDX1 gene: a case report. J Med Case Rep. 2019;13:258. doi: 10.1186/s13256-019-2149-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manning AK, Hivert MF, Scott RA, Grimsby JL, Bouatia-Naji N, Chen H, et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat Genet. 2012;44:659–669. doi: 10.1038/ng.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mountjoy KG, Mortrud MT, Low MJ, Simerly RB, Cone RD. Localization of the melanocortin-4 receptor (MC4-R) in neuroendocrine and autonomic control circuits in the brain. Mol Endocrinol. 1994;8:1298–1308. doi: 10.1210/mend.8.10.7854347. [DOI] [PubMed] [Google Scholar]
- 26.Heymsfield SB, Wadden TA. Mechanisms, pathophysiology, and management of obesity. N Engl J Med. 2017;376:254–266. doi: 10.1056/NEJMra1514009. [DOI] [PubMed] [Google Scholar]
- 27.Vaisse C, Clement K, Guy-Grand B, Froguel P. A frameshift mutation in human MC4R is associated with a dominant form of obesity. Nat Genet. 1998;20:113–114. doi: 10.1038/2407. [DOI] [PubMed] [Google Scholar]
- 28.Loos RJ, Lindgren CM, Li S, Wheeler E, Zhao JH, Prokopenko I, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40:768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hardy R, Wills AK, Wong A, Elks CE, Wareham NJ, Loos RJ, et al. Life course variations in the associations between FTO and MC4R gene variants and body size. Hum Mol Genet. 2010;19:545–552. doi: 10.1093/hmg/ddp504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morris AP, Voight BF, Teslovich TM, Ferreira T, Segrè AV, Steinthorsdottir V, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Carvalho AM, Shao P, Liu H, Cheng H-L, Zheng Y, Leng J, et al. The MC4R genotype is associated with postpartum weight reduction and glycemic changes among women with prior gestational diabetes: longitudinal analysis. Sci Rep. 2017;7:9654. doi: 10.1038/s41598-017-10101-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nikpay M, Turner AW, McPherson R. Partitioning the pleiotropy between coronary artery disease and body mass index reveals the importance of low frequency variants and central nervous system-specific functional elements. Circ Genom Precis Med. 2018;11:e002050. doi: 10.1161/CIRCGEN.117.002050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Table S1. Primer sequences of PDX1 and MC4R for PCR. Table S2. Basic information of candidate SNPs in the study.
Data Availability Statement
The datasets generated and/or analyzed during the current study are not publicly available due patient privacy but are available from the corresponding author on reasonable request.