Abstract
Background and Objectives:
Along with the rapid improvement of imaging technology, convex probe endobronchial ultrasound (CP-EBUS) sonographic features play an increasingly important role in the diagnosis of intrathoracic lymph nodes (LNs). Conventional qualitative and quantitative methods for EBUS multimodal imaging are time-consuming and rely heavily on the experience of endoscopists. With the development of deep-learning (DL) models, there is great promise in the diagnostic field of medical imaging.
Materials and Methods:
We developed DL models to retrospectively analyze CP-EBUS images of 294 LNs from 267 patients collected between July 2018 and May 2019. The DL models were trained on 245 LNs to differentiate benign and malignant LNs using both unimodal and multimodal CP-EBUS images and independently evaluated on the remaining 49 LNs to validate their diagnostic efficiency. The human comparator group consisting of three experts and three trainees reviewed the same test set as the DL models.
Results:
The multimodal DL framework achieves an accuracy of 88.57% (95% confidence interval [CI] [86.91%–90.24%]) and area under the curve (AUC) of 0.9547 (95% CI [0.9451–0.9643]) using the three modes of CP-EBUS imaging in comparison to the accuracy of 80.82% (95% CI [77.42%–84.21%]) and AUC of 0.8696 (95% CI [0.8369–0.9023]) by experts. Statistical comparison of their average receiver operating curves shows a statistically significant difference (P < 0.001). Moreover, the multimodal DL framework is more consistent than experts (kappa values 0.7605 vs. 0.5800).
Conclusions:
The DL models based on CP-EBUS imaging demonstrated an accurate automated tool for diagnosis of the intrathoracic LNs with higher diagnostic efficiency and consistency compared with experts.
Keywords: convex probe endobronchial ultrasound, deep learning, lymph nodes, multimodal imaging
INTRODUCTION
Convex probe endobronchial ultrasound-guided transbronchial needle aspiration (CP-EBUS-TBNA) is a minimally invasive diagnostic tool for intrathoracic lymph nodes (LNs).[1,2] However, the samples obtained by puncture needles cannot reflect the entire LN, especially for the diagnosis of various benign diseases and LNs with tumor micrometastasis. Although a 90% average diagnosis rate for lung cancer can be reached, a 20% false negative is still possible.[3] With the improvement of ultrasound imaging, CP-EBUS sonographic features can be used to predict the diagnosis of LNs during EBUS-TBNA.[4] Bronchoscopists can use ultrasonographic features to judge the benign and malignant LNs during the process of EBUS-TBNA just as ultrasound doctors can distinguish the benign and malignant lesions by ultrasound images. EBUS imaging can guide the selection of LNs for biopsy as well as internal puncture site within the LN and improve biopsy efficiency. In addition, for patients with negative puncture results or insufficient tissue volume, EBUS imaging can supplement the diagnostic evaluation of EBUS-TBNA.[5]
CP-EBUS multimodal imaging includes lymph gray scale, blood flow Doppler, and elastography (simplified as G, F, and E, respectively).[6] Grayscale image reflects morphological features such as the shape, size, and heterogeneity of LNs.[7] The blood flow Doppler mode clearly shows the contours and distribution of blood vessels as well as the direction and volume of blood flow within the node.[5,8] Elastography measures the degree of tissue deformation in grayscale mode and quantifies the tissue elasticity which is displayed in different colors.[9] In addition, it also distinguishes the features inside LNs and the boundary from the surrounding tissues. Tumor tissue has harder textures than normal tissue, as it has more cells and blood vessels.[10] In general, elastography has better diagnostic efficiency than a single grayscale feature.[11] However, fibrosis within benign LNs and central necrosis within malignant LNs may degrade the accuracy of elastic evaluation since the hardness of tissue is not equivalent to benign and malignant.[12] Thus, the diagnostic accuracy of any single mode is inherently limited. Prior studies demonstrated that the combination of gray scale and blood flow Doppler can achieve better diagnostic accuracy than a single feature,[5,13] and the combination of gray scale and elastography is also better than a single mode.[14] The combination of all the three ultrasound modes has not been evaluated but holds the promise to improve the noninvasive diagnostic efficacy of CP-EBUS.
Qualitative and quantitative methods for CP-EBUS sonographic features are conventionally used to aid the diagnosis of LNs in clinical operation. However, these qualitative methods are inherently subjective and based on the experience level of the bronchoscopist. Quantitative methods have had limited clinical value. For example, elastography has many indicators, each with no uniform cutoff value.[15,16] The normal analysis of ultrasound multimodal imaging is part of the specialty of ultrasound doctors and the operator is afforded the time to review the images on the video to make conclusions: It is obviously difficult for the bronchoscopist to make accurate judgments in real-time during a procedure.
In recent years, deep learning (DL) has proved to be powerful in image recognition and classification in numerous medical fields.[17] Artificial intelligence (AI) is able to extract features that are difficult to be recognized by human eyes at different levels and may achieve diagnostic efficiency similar to that of experts.[18] Besides, the inference of the DL model is fast and the deployment of the DL model is cost-efficient. It can diagnose diseases by deeply analyzing images, which is convenient for many clinical applications. For example, convolutional neural networks (CNNs) have been used in radio probe EBUS images to differentiate benign and malignant lesions for early detection of lung cancer.[19] To date, DL models have not been employed to diagnose CP-EBUS images, especially using the combination of multimodal images.
In our study, DL models were developed to automatically discriminate benign and malignant LNs with CP-EBUS images. Unimodal and multimodal DL frameworks were constructed and then validated. To test them further, the models were compared with analysis from a human observation group.
MATERIALS AND METHODS
Study population
We performed a retrospective analysis on prospective patients meeting the following inclusion and exclusion criteria between July 2018 and May 2019 at Shanghai Chest Hospital. Inclusion criteria were chest computed tomography (CT) shows at least one enlarged intrathoracic LNs (short diameter >1 cm) or positron-emission tomography/CT shows patients with increased fluorodeoxyglucose uptake (standard uptake value 2.5) in intrathoracic LNs; determination by doctors that EBUS-TBNA should be performed on LNs for diagnosis or preoperative staging of lung cancer; and patients agree to undergo EBUS-TBNA, sign informed consent, and have no contraindications. Patients having contraindications to EBUS-TBNA were excluded from the study. This study was approved by the Local Ethics Committee of Shanghai Chest Hospital (No. KS1947).
Dataset
LNs in this study were classified based on the international staging system.[20] Before undergoing EBUS-TBNA, the target LN and peripheral vessels were examined using an ultrasound processor (EU-ME2, Olympus or Hi-vision Avius, Hitachi) equipped with elastography and Doppler functions and ultrasound bronchoscope (BF-UC260FW, Olympus or EB1970UK, Pentax). First, the target LN was examined with the grayscale mode and a 10-s video was recorded for analysis. Then, the flow characteristics (H-flow or Fine Flow) were observed by switching to Doppler mode, and a 20-s video was recorded after the flow image was stable. Finally, ultrasound elastography mode was used to examine the target and make sure the frame would include the target LN and surrounding tissues as much as possible. When the CP-EBUS probe touches the airway wall, the fluctuation of thoracic blood vessels and the patient’s respiratory movement can exert pressure to form elastography images. If the images were not ideal, the operator pressed the spiral part of the handle of the ultrasonic bronchoscope at a frequency of 3–5 times per second to pressurize the airway wall to achieve elastography and recorded two 20-s videos after the elastic image became stable. The final diagnosis of LNs was determined by EBUS-TBNA pathological results, thoracotomy, thoracoscopy, microbiological examinations, or clinical follow-up for at least 6 months. Blinded to the final results of LNs, three representative images were selected from videos of each CP-EBUS mode by two experts for analysis. During selection, grayscale images containing the largest profile of the LN were used to best reflect its characteristics, such as density and the hilum. Blood flow Doppler images reflected the overall blood flow without artifacts, and elastography images had good repeatability and were as complete as possible. With preprocess procedures that remove redundant information from the picture, a dataset of 2646 images was used to establish the basis of this study, and each mode had 882 images. We randomly divided the dataset into six parts (the images of the same LN were categorized into the same part) where the ratio of benign and malignant LNs was the same. The first five parts were used to perform fivefold cross-validation, while the sixth part was used as an independent test set. Region of interest (ROI) is determined by an expert and labeled by the open-source software “labelme” (https://github.com/wkentaro/labelme).
Preprocessing of convex probe endobronchial ultrasound images and region of interest detection
All the images were cropped to the minimum size covering ROI, which causes different image sizes due to the varying sizes of effective areas in videos. However, DL models expect input images with invariant size. To minimize the deformation of objects, we resized images to 224 along the short dimension proportionally and cropped 224 × 224 pixels in the center of resized images.[21]
CP-EBUS images of LNs usually contain other lung or mediastinal tissues, which may affect the diagnosis of DL models. In clinical practice, experienced doctors can ignore surrounding tissues and focus on the target LN for image feature analysis. It is essential for machine learning to accurately identify ROI from the whole image for automatic diagnosis. In this study, DL models were applied to detect automatically the LN area on our dataset. The mainstream methods for medical image segmentation were evaluated on CP-EBUS images and U-Net was selected for this study. Detailed process and segmentation results can be found in online Supplementary File [Supplementary Table 1 and Supplementary Figure 1 (923.6KB, tif) ].
Supplementary Table 1.
The results of region of interest segmentation on three endobronchial ultrasound modes
| Modes | Methods | AUC | Accuracy (%) | DC (%) |
|---|---|---|---|---|
| G | U-Net | 0.9820±0.0042 | 93.35±0.58 | 85.46±2.51 |
| Attention U-Net | 0.9823±0.0035 | 93.49±0.39 | 83.68±1.94 | |
| R2U-Net | 0.8971±0.0398 | 80.91±1.62 | 44.01±11.83 | |
| Attention R2U-Net | 0.8776±0.0602 | 82.81±3.16 | 51.33±11.73 | |
| F | U-Net | 0.9662±0.0027 | 88.81±0.67 | 88.66±1.10 |
| Attention U-Net | 0.9672±0.0047 | 88.84±0.29 | 88.21±1.84 | |
| R2U-Net | 0.9136±0.0391 | 80.88±3.50 | 77.49±5.68 | |
| Attention R2U-Net | 0.9004±0.0475 | 79.99±3.22 | 77.48±4.70 | |
| E | U-Net | 0.9701±0.0027 | 89.79±0.49 | 90.40±1.68 |
| Attention U-Net | 0.9679±0.0018 | 89.42±0.24 | 90.13±0.72 | |
| R2U-Net | 0.9128±0.0290 | 80.35±3.06 | 83.13±4.23 | |
| Attention R2U-Net | 0.9097±0.0114 | 79.48±1.46 | 83.17±2.34 |
Bold indicates values with the best performance for each statistical indicator in each mode. E: Elastography; G: Gray scale; F: Blood flow Doppler; AUC: Area under the curve; DC: Dice
Experiments were conducted to analyze the impact of segmented ROI on diagnosis using DL models. Cropped images and corresponding ROI images for the same training examples were fed into the same DL model to generate diagnostic results, respectively. The two results were obtained under the same settings (e.g., hyperparameters, random seed, and training scheme) to ensure that only the input data differed.
Development of the deep-learning architecture for convex probe endobronchial ultrasound images
Figure 1 depicts a DL model to diagnose benign and malignant LNs using arbitrary one of the three modes of CP-EBUS imaging. Design principles are shown in the Supplementary File. Since elastography has the best diagnostic efficiency, we named the DL model as ENet. Then, we compared ENet with mainstream DL architectures to validate the effectiveness of ENet using elastography images. For each LN, one image is randomly selected from the three images for each epoch during the training process to keep samples independent. All architectures share the same training scheme. The details of the training strategy are presented in the Supplementary File.
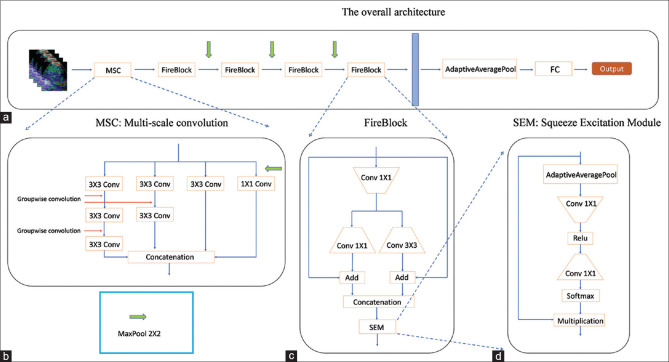
Figure 1.
Illustration of ENet architecture. (a) The overall architecture. The input size is 3 × 224 × 224. This architecture consists of a multiscale convolution module (illustrated in [b]), four FireBlocks (illustrated in [c]), and full connected layer. The green arrow is max-pooling operation of 2 × 2, and the blue block is the feature of size 512 × 14 × 14. (b) Multiscale convolution. This module consists of four branches. The stride of the first convolution operation in the first three branches is 2 to reduce the feature size, and the fourth branch adds a 2 × 2 pooling operation before the 1 × 1 convolution to keep the same feature size as other branches. Other convolution operations are group-wise convolution used to expand the receptive field. (c) FireBlock module. This module is borrowed from SqueezeNet. There are two modifications. One is that we add two skip connections before the concatenation operation. The other is that we apply a Squeeze excitation module at last. (d) Squeeze excitation module. This module is borrowed from SENet, which applies a channel attention mechanism to extracted features
Automated diagnosis with multimodal deep learning
To further improve the diagnostic accuracy, we proposed a multimodal DL framework named EBUSNet to jointly exploit all three modal images. Figure 2 illustrates our framework that substitutes the weighted sum operation with weight concatenation in CentralNet[22] to accommodate the multimodal CP-EBUS images. CentralNet is a multimodal fusion method that recursively weights the features from different modes at the current layer and the fused feature from the preceding layer. However, the weighted sum of features from different modes may degrade the performance in CP-EBUS multimodality, as features extracted from the CP-EBUS images of three modes are not aligned in one feature space due to their implications for diagnostic factors in quite different respects. Thus, weighted concatenation can consider the diagnostic factors derived from all the modes. The detailed training scheme for our framework is presented in the Supplementary File.
Figure 2.
The architecture of EBUSNet. (a) The overall architecture of EBUSNet. The stride of the initial convolution operations is 2. (b) The architecture of the initial CentralBlock. The FireBlocks in (b) and (c) are identical to those in Figure 2. The weights of the weighted concatenation are learnable and initialized as 0.33. (c) The architecture of other CentralBlocks. The weights of the weighted concatenation are learnable and initialized as 0.33. The weights of the weighted summarization are learnable and initialized as 0.5
Comparison between the human group and the deep-learning model
For evaluations in a clinical application, the DL models were compared with the human group using the same test set. The human group consists of three experts (experience of CP-EBUS image observation >300 LNs) and three trainees (experience of CP-EBUS image observation <30 LNs). Experts first educated the trainees according to the traditional assessment using the three modes.[5,7,23] Then, the six doctors independently diagnosed with unimodal and multimodal images using conventional methods blind to the final diagnosis of LNs. The test process performed by humans is consistent with the scenario of deep model testing, i.e., randomly selecting one from three images of each LN at a time, and then, repeating the test five times. For further comparison, doctors were asked to assign a diagnostic confidence level, i.e., a score of 1, 2, 4, or 5 indicating benign, benign tendency, malignant tendency, malignant, respectively, to each LN. Figure 3 illustrates the whole framework of this study.
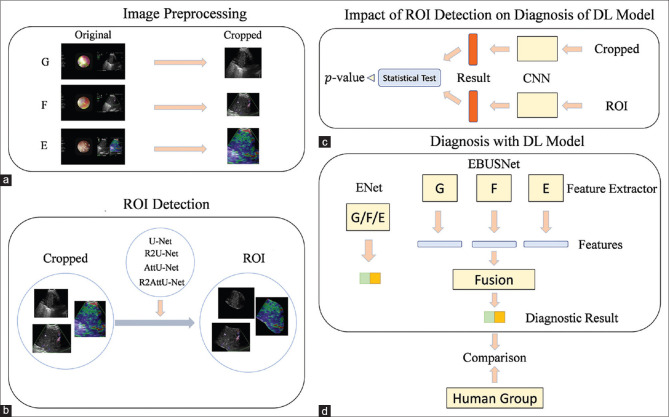
Figure 3.
The whole framework of this study. (a) The collected CP-EBUS images are preprocessed to remove redundant information initially. (b) Various deep-learning models that automatically detect the LN area are applied to CP-EBUS images. (c) The impact of ROI detection on diagnostic performance. (d) A multimodal framework named EBUSNet is designed, and the comparison between multimodal and unimodal is conducted. G: gray scale; F: blood flow Doppler; E: elastography; ROI: region of interest; CP-EBUS: convex probe endobronchial ultrasound
Statistical analysis
Statistical analysis is performed using R-studio 1.2.5033 with R 3.6.1. and SPSS version 20.0 (IBM, New York, NY, USA). The method proposed[24] was used to evaluate the difference of the average receiver operating characteristic (ROC) curves between various methods. P < 0.05 is regarded as significant. Consistency between the DL models and experts are calculated using Cohen’s kappa value.[25] The implementation of DL models is based on Pytorch 1.3.1 and Python 3.6.9. The implementation of mainstream CNN architectures refers to https://github.com/Cadene/pretrained-models.pytorch.
RESULTS
Clinical characteristic of patients
There are 294 LNs from 267 patients in our study. The location and final diagnosis of LNs are displayed in Table 1.
Table 1.
Characteristics of patients and lymph nodes included in the study
| Characteristic | Cases (%) |
|---|---|
| Number of patients | 267 |
| Sex | |
| Female | 99 (37.08) |
| Male | 168 (62.92) |
| Location | |
| 2R | 2 (0.68) |
| 4L | 20 (6.80) |
| 4R | 99 (33.67) |
| 7 | 100 (34.01) |
| 10L | 6 (2.04) |
| 10R | 6 (2.04) |
| 11L | 33 (11.22) |
| 11Ri | 14 (4.76) |
| 11Rs | 14 (4.76) |
| Diagnosis (malignant) | 169 (57.5) |
| Adenocarcinoma | 68 (23.13) |
| Squamous carcinoma | 31 (10.5) |
| Adenosquamous carcinoma | 1 (0.3) |
| NSCLC-NOS | 7 (2.4) |
| Small cell carcinoma | 41 (13.9) |
| Large cell neuroendocrine carcinoma | 1 (0.3) |
| NET-NOS | 6 (2.0) |
| Unknown type of lung cancer | 8 (2.7) |
| Metastatic tumors (nonlung primary malignancy) | 4 (1.4) |
| Diagnosis (benign) | 125 (42.5) |
| Inflammation | 81 (27.6) |
| Sarcoidosis | 30 (10.2) |
| Tuberculosis | 13 (4.4) |
| Nontuberculous mycobacterium infection | 1 (0.3) |
NSCLC-NOS: Nonsmall cell lung cancer not otherwise specified; NET-NOS: Neuroendocrine tumor not otherwise specified
Role of region of interest detection
Table 2 shows that the DL models trained with ROI have significantly higher performance in some fold in fivefold cross-validation than those trained with the cropped images but show no statistical difference on the whole. That means predefined ROI is not very important for DL models.
Table 2.
Impact of region of interest detection on diagnostic efficiency of convex probe endobronchial ultrasound multimodal images
| Modes | Five-fold cross-validation | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| 1 | 2 | 3 | 4 | 5 | Average | |||||||
|
| ||||||||||||
| AUC | P | AUC | P | AUC | P | AUC | P | AUC | P | Average AUC 95% CI | P | |
| G w/o ROI | 0.6725 | 0.7843 | 0.6850 | 0.0847 | 0.5661 | 0.1206 | 0.6705 | 0.3635 | 0.6011 | 0.0938 | 0.6390 (0.5740-0.7041) | 0.91984 |
| G w/ROI | 0.6814 | 0.6181 | 0.6297 | 0.6212 | 0.6586 | 0.6418 (0.6079-0.6757) | ||||||
| F w/o ROI | 0.6363 | 0.3089 | 0.6541 | 0.2958 | 0.6574 | 0.9650 | 0.6866 | 0.6029 | 0.5201 | 0.0253 | 0.6309 (0.5508-0.7110) | 0.62530 |
| F w/ROI | 0.5841 | 0.7053 | 0.6552 | 0.6603 | 0.6386 | 0.6487 (0.5944-0.7030) | ||||||
| E w/o ROI | 0.9548 | 0.0625 | 0.9488 | 0.4774 | 0.9456 | 0.2956 | 0.9579 | 0.3211 | 0.9447 | 0.0195 | 0.9503 (0.9432-0.9575) | 0.13900 |
| E w/ROI | 0.9806 | 0.9348 | 0.9626 | 0.9727 | 0.9792 | 0.9660 (0.9426-0.9894) | ||||||
Bold indicates P<0.05. The first row shows the performance of the DL model which is trained and tested with cropped images, while the second row corresponds to the models trained and tested with ROI. For each fold, AUCs are calculated for each mode with and without ROI, and P value is obtained using the Delong test between the two ROCs. For average AUCs of five-folds, P value in the last column is obtained with paired-samples t-test. G: Gray scale; F: Blood flow Doppler; E: Elastography; DL: Deep learning; ROI: Region of interest; w/o ROI: Without ROI; w/ROI: With ROI; AUCs: Area under the curves; CI: Confidence interval
Diagnostic performance of ENet on single-modal convex probe endobronchial ultrasound images
The evaluation is repeated for five times to guarantee the stability under the random sampling. These settings were kept throughout the study. Table 3 shows that ENet achieves a higher area under curve (AUC) on E mode (0.9504 [0.9458–0.9549]) than on G mode (0.6390 [0.5980–0.6801]) and F mode (0.6309 [0.5803–0.6815]) with a statistical significance (E-G: P < 0.001, E-F: P < 0.001). Figure 4b and c illustrate the univariate AUCs of features extracted by ENet from elastography and their distribution, respectively. The results show that 63.9% features achieve an AUC exceeding 0.8, and 28.5% features exceed 0.9, which implies that ENet can find the features maximally distinguishing benign and malignant LNs on elastography. Thus, we focused on E mode in the context of unimodal evaluation. To validate the effectiveness of our design, we compared ENet with the state-of-the-art mainstream architectures for natural images, including VGG11,[26] ResNet,[27] InceptionNet-v4,[28] and SeResNet50[29] (pretrained on ImageNet[30]), and lightweight architectures designed for mobile devices such as ShuffleNet,[31] MobileNetV2,[32] SqueezeNet,[33] and NasMobile.[34] Table 4 demonstrates that ENet significantly outperforms most mainstream CNNs in the elastography diagnosis task.
Table 3.
Comparison of deep-learning models and human group on endobronchial ultrasound images
| Modes | 95% CI | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| AUC | F1-score | Accuracy, % | Sensitivity, % | Specificity, % | PPV, % | NPV, % | |
| AI group | |||||||
| G | 0.6390 (0.5980-0.6801) | 0.6873 (0.6558-0.7189) | 62.04 (58.81-65.27) | 71.17 (63.09-79.26) | 48.80 (35.39-62.21) | 67.60 (63.05-72.16) | 53.89 (48.54-59.23) |
| F | 0.6309 (0.5803-0.6815) | 0.7171 (0.6882-0.7460) | 62.37 (57.50-67.24) | 80.41 (74.79-86.04) | 36.20 (21.15-51.25) | 65.23 (60.46-69.99) | 53.75 (44.99-62.50) |
| E | 0.9504 (0.9458-0.9549) | 0.8854 (0.8759-0.8950) | 86.20 (85.04-87.36) | 90.20 (85.76-94.65) | 80.40 (72.99-87.81) | 87.42 (83.55-91.29) | 85.78 (81.49-90.07) |
| Expert group | |||||||
| G | 0.6559 (0.6335-0.6783) | 75.79 (74.53-77.04) | 66.8 (65.06-68.54) | 87.82 (84.61-91.02) | 36.33 (30.39-42.27) | 66.71 (65.13-68.30) | 67.55 (63.75-71.34) |
| F | 0.734 (0.7156-0.7523) | 79.03 (76.82-81.24) | 71.02 (67.57-74.47) | 92.18 (88.51-95.86) | 40.33 (31.22-49.45) | 69.25 (66.34-72.17) | 78.42 (71.94-84.90) |
| E | 0.9058 (0.8702-0.9414) | 0.8999 (0.8702-0.9296) | 87.89 (84.11-91.67) | 91.49 (88.92-94.07) | 83.00 (74.28-91.05) | 88.66 (83.50-93.82) | 87.05 (83.23-90.87) |
| Trainee group | |||||||
| G | 0.4946 (0.4458-0.5434) | 0.5512 (0.4780-0.6244) | 50.34 (45.63-55.05) | 52.41 (41.54-63.28) | 47.33 (35.85-58.81) | 58.98 (54.64-63.33) | 40.58 (36.07-45.10) |
| F | 0.4443 (0.3481-0.5404) | 0.5116 (0.3893-0.6339) | 46.26 (37.12-55.39) | 49.19 (33.55-64.84) | 42.00 (32.22-51.78) | 54.24 (46.49-61.99) | 36.96 (27.99-45.94) |
| E | 0.7750 (0.5606-0.9895) | 0.8167 (0.7155-0.9179) | 75.65 (60.60-90.69) | 88.50 (82.72-94.28) | 57.00 (27.91-86.09) | 76.22 (62.79-89.66) | 72.73 (50.46-95.01) |
Bold indicates values with the best performance for each statistical indicator in each group. G: Gray scale; F: Blood flow Doppler; E: Elastography; AUC: Area under the curve; PPV: Positive predictive value; NPV: Negative predictive value; AI: Artificial intelligence
Figure 4.
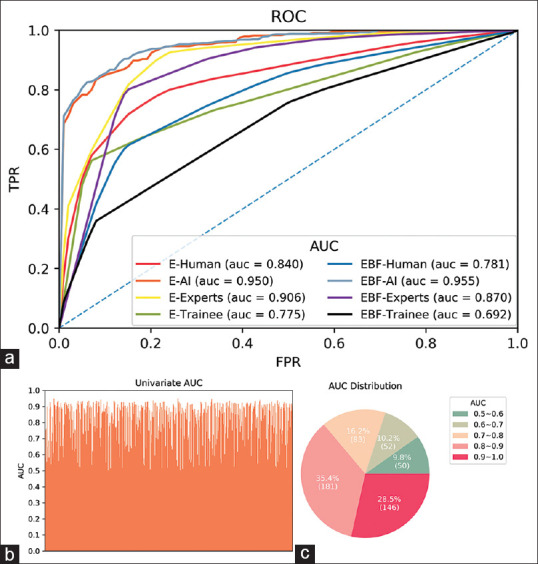
Comparison of diagnostic performance between DL models and human group. (a) The average ROC of the AI, experts, trainees, and the whole human group on elastography and multimodal image. (b) The univariate AUC of features extracted by ENet on elastography in the last layer. (c) The statistical of AUC in (b). DL: deep learning; ROC: receiver operating characteristic; AUC: area under the curve; AI: artificial intelligence
Table 4.
Comparison of ENet and mainstream architectures on elastography
| Modes | 95% CI | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| AUC | F1-score | Accuracy, % | Sensitivity, % | Specificity, % | PPV, % | NPV, % | |
| Squeezenet | 0.9313 (0.9209-0.9418) | 0.8561 (0.8345-0.8777) | 82.45 (78.56-86.34) | 87.03 (81.54-92.53) | 75.80 (59.60-92.00) | 85.62 (77.28-93.95) | 81.13 (77.73-84.54) |
| Mobilenet | 0.9331 (0.9199-0.9463) | 0.8702 (0.8558-0.8845) | 84.73 (83.35-86.12) | 86.76 (82.15-91.37) | 81.80 (75.57-88.03) | 87.69 (84.53-90.85) | 81.69 (77.15-86.23) |
| VGG11 | 0.5730 (0.5448-0.6012) | 0.6576 (0.6092-0.7059) | 59.02 (54.75-63.29) | 67.45 (58.49-76.40) | 46.80 (38.37-55.23) | 64.87 (61.52-68.21) | 50.81 (44.36-57.26) |
| Inception-v4 | 0.9177 (0.8760-0.9593) | 0.8634 (0.8243-0.9025) | 84.08 (79.11-89.05) | 84.55 (80.55-88.55) | 83.40 (73.92-92.88) | 88.50 (82.82-94.17) | 78.79 (73.17-84.40) |
| NasMobile | 0.9370 (0.9176-0.9564) | 0.8762 (0.8598-0.8926) | 84.98 (82.47-87.49) | 89.10 (85.70-92.51) | 79.00 (68.84-89.16) | 86.75 (80.82-92.68) | 83.85 (80.65-87.06) |
| SeResNet | 0.9445 (0.9365-0.9525) | 0.8747 (0.8606-0.8888) | 85.22 (83.57-86.88) | 87.17 (83.46-90.89) | 82.40 (76.06-88.74) | 88.08 (84.74-91.42) | 82.07 (78.16-85.98) |
| ResNet18 | 0.9445 (0.9364-0.9526) | 0.8606 (0.8390-0.8821) | 83.10 (80.00-86.21) | 88.00 (80.25-95.75) | 76.00 (60.45-91.55) | 86.02 (77.69-94.36) | 83.42 (77.44-89.41) |
| ENet | 0.9504 (0.9458-0.9549) | 0.8854 (0.8759-0.8950) | 86.20 (85.04-87.36) | 90.20 (85.76-94.65) | 80.40 (72.99-87.81) | 87.42 (83.55-91.29) | 85.78 (81.49-90.07) |
Bold indicates values with the best performance for each statistical indicator of ENet and mainstream architectures. AUC: Area under the curve; PPV: Positive predictive value; NPV: Negative predictive value; CI: Confidence interval
ENet was further compared with the human group. For both the expert group and trainee group, E mode outperforms G and F modes significantly (E-G: P < 0.001, E-F: P < 0.001). We then compared ENet with the human group on E mode. Table 3 shows that ENet has higher indicators than that of the trainee group and outperforms the trainee group in terms of average ROC significantly (P < 0.001). The expert group achieves better performance on all measurements except for AUC compared with ENet, but ENet also outperforms the expert group significantly (P = 0.004). Figure 4a plots the average ROC curves of the diagnostic results by the DL models, human group, expert group, and trainee group. ENet is better at distinguishing benign and malignant LNs than both experts and trainees.
Diagnostic performance of multimodal framework on convex probe endobronchial ultrasound images
To demonstrate the diagnostic efficiency of the DL models on multimodal images, EBUSNet was evaluated on different combinations of modes and achieved AUCs of 0.6543 (0.6177–0.6909), 0.9506 (0.9337–0.9674), 0.9512 (0.9440–0.9584), and 0.9547 (0.9451–0.9643) on G + F, G + E, F + E, and G + F + E, respectively [Table 5]. Here, we use “A + B” for the combination of modes A and B fed into the corresponding multimodal DL models. Note that G + F + E achieves the best performance among all combinations and is superior to elastography in terms of AUC, F-score, and accuracy.
Table 5.
Comparison of ENet, EBUSNet, and human group on multimodal imaging
| Modes | 95% CI | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| AUC | F1-score | Accuracy, % | Sensitivity, % | Specificity, % | PPV, % | NPV, % | |
| AI group | |||||||
| E | 0.9504 (0.9458-0.9549) | 0.8854 (0.8759-0.8950) | 86.20 (85.04-87.36) | 90.20 (85.76-94.65) | 80.40 (72.99-87.81) | 87.42 (83.55-91.29) | 85.78 (81.49-90.07) |
| G+F | 0.6543 (0.6177-0.6909) | 0.7312 (0.6955-0.7668) | 65.14 (61.82-68.47) | 80.97 (72.19-89.74) | 42.20 (30.82-53.58) | 67.26 (64.49-70.03) | 63.30 (55.58-71.02) |
| G+E | 0.9506 (0.9337-0.9674) | 0.8936 (0.8660-0.9213) | 86.53 (82.23-90.83) | 93.65 (91.31-95.99) | 76.20 (62.40-90.00) | 86.03 (79.25-92.81) | 89.80 (87.49-92.11) |
| F+E | 0.9512 (0.9440-0.9584) | 0.8972 (0.8933-0.9012) | 87.84 (87.36-88.31) | 89.79 (86.45-93.13) | 85.00 (79.65-90.35) | 89.94 (86.92-92.97) | 85.62 (82.16-89.07) |
| G+F+E | 0.9547 (0.9451-0.9643) | 0.9056 (0.8929-0.9183) | 88.57 (86.91-90.24) | 92.41 (91.64-93.18) | 83.00 (79.12-86.88) | 88.82 (86.53-91.11) | 88.29 (87.13-89.44) |
| Human group | |||||||
| E | 0.8404 (0.7198-0.9610) | 0.8583 (0.7959-0.9207) | 81.77 (72.60-90.94) | 90.00 (86.62-93.38) | 69.83 (51.54-88.12) | 82.44 (73.69-91.19) | 79.89 (67.22-92.56) |
| G+F+E | 0.7809 (0.6821-0.8797) | 0.7984 (0.7359-0.8608) | 75.03 (67.26-82.81) | 83.68 (75.43-91.93) | 62.50 (49.77-75.23) | 77.03 (69.45-84.60) | 73.90 (61.08-86.72) |
| Expert group | |||||||
| E | 0.9058 (0.8702-0.9414) | 0.8999 (0.8702-0.9296) | 87.89 (84.11-91.67) | 91.49 (88.92-94.07) | 82.67 (74.28-91.05) | 88.66 (83.50-93.82) | 87.05 (83.23-90.87) |
| G+F+E | 0.8696 (0.8369-0.9023) | 0.8505 (0.8199-0.8810) | 80.82 (77.42-84.21) | 92.64 (86.36-98.93) | 63.67 (58.25-69.08) | 78.75 (76.58-80.93) | 86.78 (77.33-96.23) |
| Trainee group | |||||||
| E | 0.7750 (0.5606-0.9895) | 0.8167 (0.7155-0.9179) | 75.65 (60.60-90.69) | 88.50 (82.72-94.28) | 57.00 (27.91-86.09) | 76.22 (62.79-89.66) | 72.73 (50.46-95.01) |
| G+F+E | 0.6922 (0.5587-0.8257) | 0.7463 (0.6584-0.8342) | 69.25 (57.23-81.28) | 74.71 (69.53-79.90) | 61.33 (36.52-86.15) | 75.30 (60.57-90.03) | 61.02 (49.06-72.98) |
Bold indicates values with the best performance for each statistical indicator in each group. G: Gray scale; F: Blood flow Doppler; E: Elastography; AUC: Area under the curve; PPV: Positive predictive value; NPV: Negative predictive value; CI: Confidence interval; AI: Artificial intelligence
We then compared EBUSNet models with our human group, and G + F + E outperforms both expert group and trainee group. The comparison of average ROC curves shows that G + F + E outperforms both expert group and trainee group significantly (P < 0.001). However, when we compare the diagnostic performance between E and G + F + E of the human group, there are decreases in AUC and accuracy in human, expert, and trainee groups. Consistency of E mode and G + F + E mode were compared for DL models and the expert group, and DL models achieve kappa values of 0.6834 and 0.7605 while experts achieve 0.6837 and 0.5800 for E and G + F + E mode respectively. The results show that EBUSNet is more stable compared with experts.
DISCUSSION
AI has been widely used in the field of digestive endoscopy but rarely in the field of CP-EBUS. As far as we know, this is the first study to combine DL with CP-EBUS imaging to differentiate benign and malignant LNs. Traditional diagnosis of LNs using qualitative and quantitative methods is time-consuming and relies heavily on the personal experience of the bronchoscopist. In contrast, DL models are more efficient and stable than experts, which is convenient for clinical application, especially for areas where medical resources are scarce.
In classification settings, it is not necessary for the DL models to delineate the ROI from the entire image. CNNs are employed to introduce the potential ability to focus on ROI and partially compensates for the loss of prior information caused by lacking ROI annotation. Thus, the entire CP-EBUS images are directly fed into the DL models in this study. ENet has significantly higher diagnostic ability than the expert group on elastography (P = 0.004). The univariate AUCs of 63.9% and 28.5% of its features exceed 0.8 and 0.9, respectively. It is hard for experts to discover these features, but DL models exploit these features to improve the diagnostic performance on CP-EBUS images.
We further develop EBUSNet to improve the diagnostic performance over unimodal features like elastography. Compared with ENet, EBUSNet enhances all metrics, especially sensitivity and specificity. The higher sensitivity reduces false-negative results, which will make the treatment more timely and would encourage more sampling of the node in question or consider more invasive staging prior to concluding the LN is benign. The higher specificity reduces false-positive results, which will effectively save medical resources and would lead to careful reconsiderations before concluding what stage a patient truly is. Interestingly, for the total human group, including experts and trainees, multimodal imaging had worse diagnostic efficiency than elastography alone, as G and F modes may have kinds of features disturbing the final diagnosis of doctors. The study related to EBUS sonographic features found that elastography has better diagnostic performance compared with G and F modes.[23] The AI models in this study also show that the E mode achieves a better diagnostic efficiency than the G and F modes. Thus, G and F modes may have less diagnostic value for malignant and benign diagnosis of LNs compared with E mode. Moreover, elastography can reflect the relative stiffness of tissues with different colors, which is intuitive and easy to quantify. The qualitative five score method that defines scores 1–3 as benign and 4–5 as malignant used by the human group in this study is convenient and easy to learn. However, G and F modes have a variety of sonographic characteristics, and there still lack unified qualitative diagnostic criteria. Therefore, the characteristics of G and F modes may interfere with the diagnosis of E mode. Some features that can be recognized by AI rather than human on G and F modes may lead to better performance of the model including G, F, and E modes than ENet. Notably, since that AI models were trained according to the final diagnosis of LNs and were not involved in human experience, the findings that G and F modes influenced the overall diagnosis in the human group had no effect in determining the application of an AI algorithm, which was also the advantage of AI models for the diagnosis of intrathoracic LNs.
EBUSNet has better generalization ability than the unimodal framework because of the more even distribution of data. In the setting of machine learning, due to limited data or uneven data distribution, the optimal decision threshold of a model may vary for different datasets. The larger difference in data distribution implies the higher deviation of the optimal threshold. In practical applications, the threshold of a model is often calibrated using the validation set, as it is hard to obtain the optimal threshold for data distribution. Thus, the larger difference between the distribution of actual data and the validation set would lead to the larger gap between the actual optimal threshold and the threshold calibrated on the validation set. This large gap may affect the accuracy, while the AUC does not change. By solving this problem, EBUSNet models achieve gains in metrics in addition to AUC based on the more even distribution of multimodal data (compared with the unimodal data). Table 6 lists the thresholds on the validation and test sets using G + F + E and E where the last column is the sum of the absolute values of the differences between the optimal thresholds on the validation and test sets. We find that the summarization of multimodality is significantly larger than that of elastography, which suggests a more even distribution of multimodal data. From another perspective, multimodal data represent the information of the LN in more aspects and lead to a smaller difference in data distribution than arbitrary unimodal.
Table 6.
Optimal decision thresholds for fivefold cross-validation on the validation set and test set
| Modes | Dataset | Optimal decision thresholds for five-fold cross-validation | Summarization | ||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| 1 | 2 | 3 | 4 | 5 | |||
| G+F+E | Validation | 0.6250 | 0.5714 | 0.5263 | 0.4173 | 0.5222 | 0.1224 |
| Test | 0.5849 | 0.5852 | 0.5358 | 0.4677 | 0.5135 | ||
| E | Validation | 0.5040 | 0.4492 | 0.4724 | 0.4989 | 0.4650 | 0.4170 |
| Test | 0.4850 | 0.4025 | 0.5391 | 0.3410 | 0.5969 | ||
Optimal thresholds are obtained by exhaustion. Summarization is the sum of the absolute values of the differences between the best threshold of the validation set and the best threshold of the test set. G: Gray scale; F: Blood flow Doppler; E: Elastography
There are limitations to our study. First, although two experts are employed to choose representative images, there may still be subjective factors that can affect results, which suggests that automatic selection by DL models from the videos may yield better results, and this is the next research direction to realize real-time diagnosis of EBUS videos in real-time examinations. Second, the number of LNs may have been insufficient and more CP-EBUS images may acquire better diagnostic efficiency for the AI that usually needs large datasets.[35] However, our results are robust enough to lessen this concern, and the model will continue to improve as our training set grows. Third, the training and test datasets of DL models came from the same hospital. Since the types and distribution of diseases in different hospitals are different, the diagnostic model may not be well applied to other hospitals. Thus, it is necessary to carry out multicenter research. Finally, our model can only intelligently identify LNs as benign and malignant, not specific disease types. Larger data sizes may ultimately realize this function as well.
CONCLUSIONS
EBUSNet, the multimodal framework, showed great potential in the diagnosis of intrathoracic LNs with the higher diagnostic efficiency and consistency compared with human experts, which indicates a significant application value in clinical practice.
Supplementary Materials
Supplementary information is linked to the online version of the paper on the Endoscopic Ultrasound website.
Financial support and sponsorship
This work was supported by the National Natural Science Foundation of China (grant number 81870078, 61720106001, 61971285, and 61932022); Shanghai Municipal Health and Medical Talents Training Program (grant number 2018BR09); Shanghai Municipal Education Commission-Gaofeng Clinical Medicine Grant Support (grant number 20181815); and the Program of Shanghai Academic Research Leader (grant 17XD1401900).
Conflicts of interest
There are no conflicts of interest.
Segmentation results of different methods. Original image is images after preprocessing; Ground truth indicates region of interest delinated by experts. Yellow area indicates region of interest predicted by deep-learning models
SUPPLEMENTARY FILE
Supplementary Materials and Methods
REGION OF INTEREST DETECTION
U-Net[1] is widely used in medical image segmentation and achieves excellent performance in a variety of segmentation tasks, and much effort[2,3,4] has been made for further improvement. We evaluate the segmentation performance by various U-Net based methods on our dataset to determine the proper architecture for EBUS images. The main idea behind U-Net is adding skip connections in the encoder–decoder structure. Thus, the loss of information caused by downsampling can be averted. Attention U-Net[3] adds the attention gate (AG) to skip connections in U-Net. Instead of concatenating features directly, Attention U-Net employs an attention mechanism on the feature fusion step to keep the most informative features. Inspired by the ResNet[5] and RCNN,[6] Alom et al. proposed R2UNet[4] in which the residual module and recurrent convolution module are incorporated into the convolution operations in UNet. Attention R2U-Net simply combines the AG, residual module, and recurrent convolution into U-Net since they are not conflicted in implementation. Although Attention U-Net, R2UNet, and Attention UNet declare to outperform U-Net, a well-trained U-Net is still supposed to be state-of-the-art (SOTA).[7] Thus, an evaluation of these methods still makes sense.
To find the segmentation model that best fits EBUS images, we trained U-Net, Attention U-Net, R2U-Net, and Attention R2U-Net on our dataset under the same settings. For training, the total number of epochs was set to 200 and the batch size is 8. Adam with a weight decay of 5e-4 and an initial learning rate of 0.001 was applied to update parameters. We adopted the warm-up scheme to increase the learning rate to 0.01 linearly in the first 10 epochs and then decreased it by a factor of 0.4 on the 40th, 80th, 150th, and 180th epochs. Dice loss was used as the loss function and its formula is elaborated in the following section. Fivefold cross-validation was leveraged to find the models achieving the highest accuracy on the validation set for evaluation on the test set.
The segmentation model outputs a mask with the same size as the input image, in which the value of each pixel indicates the probability that the pixel at the same position in the input image belongs to ROI. To evaluate the performance of a model, we divide pixels in each mask into true positive (TP), true negative (TN), false positive (FP), and false negative (FN) by comparing them with the corresponding ground truth (given by human experts). Consequently, we calculate accuracy (AC), the area under the curve (AUC), and Dice (DC) for each mask and average them on the entire test set. DC is two times the ratio of the intersection of the mask and the ground truth to the number of pixels belonging to ROI in the mask or the ground truth, as elaborated in the following section. We obtain AC, AUC, and DC for each validation, and their means and standard deviations in the five-fold cross-validation are used to evaluate the performance of methods.
BINARY CROSS ENTROPY-DICE LOSS
Binary cross-entropy (BCE) loss is widely used in binary classification tasks, which is defined as:
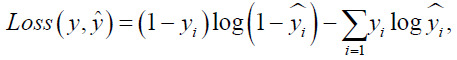
Where y is the score yˆ predicted by the model and is the actual label. However, BCE loss cannot handle the tasks of semantic segmentation when the target area is too small for the whole image. Dice is one of the widely adopted evaluation metrics for the tasks of semantic segmentation. It is defined as:

Where Y is the output mask predicted by the model and Ŷ is the ground truth. When the target area is too small to the whole image, Dice loss changes sharply with little changes in prediction and makes the training unstable.
To address this problem, a weighted BCE-Dice loss is adopted as the loss function to improve both training stability and evaluation performance. The weighted BCE-Dice loss is defined as:
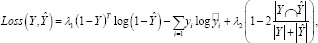
Where and can be adjusted to meet the demand.
Supplementary Table 1 shows the segmentation performance by different methods on the three modes of CP-EBUS images. U-Net and attention U-Net obviously outperform the other methods. This problem is probably caused by the recurrent module. Supplementary Figure 1 (923.6KB, tif) further compares these methods on the three modes of EBUS images. R2U-Net and Attention R2U-Net suffer from serious distortion in certain cases, and their edges are rough even discontinuous. Attention U-Net and U-net can present approximate contour of the lymph node, but we find distortion in edges and lost subtle structures for Attention U-Net.
THE DESIGN PRINCIPLES OF ENET
LNs are characterized by a wide variety of features, such as margin, shape, echogenicity, and central hilar structure.[8] It is difficult to extract these features of different shapes and sizes with convolution kernels of one single size.[9] Thus, we adopted the inception module to employ convolution kernels of different sizes to extract features of various scales. Compared to the natural image, the pattern of CP-EBUS image is simpler, we employed modules from computation efficient CNN architectures to suppress the overfitting caused by excessive parameters as well as instant inference. Lightweight architectures lead to structural risk due to fewer parameters. Skip connection[5] between the squeeze and expand operation was introduced into the fire block[10] to relieve the vanishing and exploding gradients in deep neural networks and the Squeeze-and-Excitation module[11] was employed after the expansion operation to select informative features from channels under the attention mechanism.
TRAINING SCHEMES OF ENET AND EBUSNET
To train ENet, we adopted Adam optimizer with a weight decay of 5e-4 and an initialize learning rate of 0.005. The learning rate decreased to 4e-5 from the 20th epoch to the 130th epoch by decaying every 5 epochs and was maintained until the 150th epoch. The batch size was 32, and the cross-entropy function weighted by [1, 0.66] was used as the loss function. For EBUSNet, we used the same optimizer as the ENet with an initial learning rate of 0.004. The learning rate decreased to 4e-5 from the 20th epoch to the 430th epoch by decaying every 5 epochs and was maintained until the 450th epoch. The batch size was 32, and the cross-entropy weighted by [1, 0.66] was used as the loss function for each branch of the multimodal framework. We applied fivefold cross-validation in model evaluation. The model with the best performance on the validation set was selected in each fold and evaluated on the test set.
SUPPLEMENTARY REFERENCES
- 1.Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. Paper Presented at: International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI) 2015 [Google Scholar]
- 2.Wang B, Qiu S, He H. Dual Encoding U-Net for Retinal Vessel Segmentation. Paper Presented at: International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI) 2019 [Google Scholar]
- 3.Oktay O, Schlemper J, Folgoc LL, et al. Attention U-Net: Learning where to look for the pancreas. arXiv preprint arXiv: 1804.03999. 2018 [Google Scholar]
- 4.Alom MZ, Hasan M, Yakopcic C, et al. Recurrent Residual Convolutional Neural Network based on U-Net (r2u-net) for Medical Image Segmentation. arXiv preprint arXiv: 1802.06955. 2018 [Google Scholar]
- 5.He K, Zhang X, Ren S, et al. Deep Residual Learning for Image Recognition. Paper Presented at: 2016 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 2016 [Google Scholar]
- 6.Liang M, Hu X. Recurrent Convolutional Neural Network for Object Recognition. Paper Presented at: 2015 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 2015 [Google Scholar]
- 7.Isensee F, Kickingereder P, Wick W, et al. No new-net. Paper Presented at: International MICCAI Brainlesion Workshop. 2018 [Google Scholar]
- 8.Fujiwara T, Yasufuku K, Nakajima T, et al. The utility of sonographic features during endobronchial ultrasound-guided transbronchial needle aspiration for lymph node staging in patients with lung cancer: a standard endobronchial ultrasound image classification system. Chest. 2010;138:641–7. doi: 10.1378/chest.09-2006. [DOI] [PubMed] [Google Scholar]
- 9.Qi X, Zhang L, Chen Y, et al. Automated diagnosis of breast ultrasonography images using deep neural networks. Med Image Anal. 2019;52:185–98. doi: 10.1016/j.media.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 10.Iandola FN, Han S, Moskewicz MW, et al. SqueezeNet: AlexNet-Level Accuracy with 50x Fewer Parameters and < 0.5 MB Model Size. arXiv preprint arXiv: 1602.07360. 2016 [Google Scholar]
- 11.Hu J, Shen L, Sun G. Squeeze-and-Excitation Networks. Paper Presented at: 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 2018 [Google Scholar]
REFERENCES
- 1.Yasufuku K, Chiyo M, Sekine Y, et al. Real-time endobronchial ultrasound-guided transbronchial needle aspiration of mediastinal and hilar lymph nodes. Chest. 2004;126:122–8. doi: 10.1378/chest.126.1.122. [DOI] [PubMed] [Google Scholar]
- 2.Erer OF, Erol S, Anar C, et al. Diagnostic yield of EBUS-TBNA for lymphoma and review of the literature. Endosc Ultrasound. 2017;6:317–22. doi: 10.4103/2303-9027.180762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anantham D, Koh MS, Ernst A. Endobronchial ultrasound. Respir Med. 2009;103:1406–14. doi: 10.1016/j.rmed.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 4.Wahidi MM, Herth F, Yasufuku K, et al. Technical aspects of endobronchial ultrasound-guided transbronchial needle aspiration: CHEST Guideline and Expert Panel Report. Chest. 2016;149:816–35. doi: 10.1378/chest.15-1216. [DOI] [PubMed] [Google Scholar]
- 5.Wang L, Wu W, Hu Y, et al. Sonographic features of endobronchial ultrasonography predict intrathoracic lymph node metastasis in lung cancer patients. Ann Thorac Surg. 2015;100:1203–9. doi: 10.1016/j.athoracsur.2015.04.143. [DOI] [PubMed] [Google Scholar]
- 6.Zhi X, Chen J, Xie F, et al. Diagnostic value of endobronchial ultrasound image features: A specialized review. Endosc Ultrasound. 2020 doi: 10.4103/eus.eus_43_20. doi: 10.4103/eus.eus_43_20. https://www.eusjournal.com/preprintarticle.asp?id=290305;type=0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujiwara T, Yasufuku K, Nakajima T, et al. The utility of sonographic features during endobronchial ultrasound-guided transbronchial needle aspiration for lymph node staging in patients with lung cancer: A standard endobronchial ultrasound image classification system. Chest. 2010;138:641–7. doi: 10.1378/chest.09-2006. [DOI] [PubMed] [Google Scholar]
- 8.Nakajima T, Anayama T, Shingyoji M, et al. Vascular image patterns of lymph nodes for the prediction of metastatic disease during EBUS-TBNA for mediastinal staging of lung cancer. J Thorac Oncol. 2012;7:1009–14. doi: 10.1097/JTO.0b013e31824cbafa. [DOI] [PubMed] [Google Scholar]
- 9.Dietrich CF, Jenssen C, Herth FJ. Endobronchial ultrasound elastography. Endosc Ultrasound. 2016;5:233–8. doi: 10.4103/2303-9027.187866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krouskop TA, Wheeler TM, Kallel F, et al. Elastic moduli of breast and prostate tissues under compression. Ultrason Imaging. 1998;20:260–74. doi: 10.1177/016173469802000403. [DOI] [PubMed] [Google Scholar]
- 11.Rozman A, Malovrh MM, Adamic K, et al. Endobronchial ultrasound elastography strain ratio for mediastinal lymph node diagnosis. Radiol Oncol. 2015;49:334–40. doi: 10.1515/raon-2015-0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin CK, Yu KL, Chang LY, et al. Differentiating malignant and benign lymph nodes using endobronchial ultrasound elastography. J Formos Med Assoc. 2019;118:436–43. doi: 10.1016/j.jfma.2018.06.021. [DOI] [PubMed] [Google Scholar]
- 13.Wang L, Wu W, Teng J, et al. Sonographic features of endobronchial ultrasound in differentiation of benign lymph nodes. Ultrasound Med Biol. 2016;42:2785–93. doi: 10.1016/j.ultrasmedbio.2016.07.019. [DOI] [PubMed] [Google Scholar]
- 14.Fujiwara T, Nakajima T, Inage T, et al. The combination of endobronchial elastography and sonographic findings during endobronchial ultrasound-guided transbronchial needle aspiration for predicting nodal metastasis. Thorac Cancer. 2019;10:2000–5. doi: 10.1111/1759-7714.13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hernández Roca M, Pérez Pallarés J, Prieto Merino D, et al. Diagnostic value of elastography and endobronchial ultrasound in the study of hilar and mediastinal lymph nodes. J Bronchology Interv Pulmonol. 2019;26:184–92. doi: 10.1097/LBR.0000000000000550. [DOI] [PubMed] [Google Scholar]
- 16.He HY, Huang M, Zhu J, et al. Endobronchial ultrasound elastography for diagnosing mediastinal and hilar lymph nodes. Chin Med J (Engl) 2015;128:2720–5. doi: 10.4103/0366-6999.167296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bi WL, Hosny A, Schabath MB, et al. Artificial intelligence in cancer imaging: Clinical challenges and applications. CA Cancer J Clin. 2019;69:127–57. doi: 10.3322/caac.21552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Misawa M, Kudo SE, Mori Y, et al. Accuracy of computer-aided diagnosis based on narrow-band imaging endocytoscopy for diagnosing colorectal lesions: Comparison with experts. Int J Comput Assist Radiol Surg. 2017;12:757–66. doi: 10.1007/s11548-017-1542-4. [DOI] [PubMed] [Google Scholar]
- 19.Chen CH, Lee YW, Huang YS, et al. Computer-aided diagnosis of endobronchial ultrasound images using convolutional neural network. Comput Methods Programs Biomed. 2019;177:175–82. doi: 10.1016/j.cmpb.2019.05.020. [DOI] [PubMed] [Google Scholar]
- 20.Rusch VW, Asamura H, Watanabe H, et al. The IASLC lung cancer staging project: A proposal for a new international lymph node map in the forthcoming seventh edition of the TNM classification for lung cancer. J Thorac Oncol. 2009;4:568–77. doi: 10.1097/JTO.0b013e3181a0d82e. [DOI] [PubMed] [Google Scholar]
- 21.Qi X, Zhang L, Chen Y, et al. Automated diagnosis of breast ultrasonography images using deep neural networks. Med Image Anal. 2019;52:185–98. doi: 10.1016/j.media.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 22.Vielzeuf V, Lechervy A, Pateux S, et al. CentralNet: A Multilayer Approach for Multimodal Fusion. Paper Presented at: 2018 European Conference on Computer Vision (ECCV) 2018 [Google Scholar]
- 23.Sun J, Zheng X, Mao X, et al. Endobronchial ultrasound elastography for evaluation of intrathoracic lymph nodes: A pilot study. Respiration. 2017;93:327–38. doi: 10.1159/000464253. [DOI] [PubMed] [Google Scholar]
- 24.Lee MT, Rosner BA. The average area under correlated receiver operating characteristic curves: A nonparametric approach based on generalized two-sample Wilcoxon statistics. J R Stat Soc. 2001;50:337–44. [Google Scholar]
- 25.Kundel HL, Polansky M. Measurement of observer agreement. Radiology. 2003;228:303–8. doi: 10.1148/radiol.2282011860. [DOI] [PubMed] [Google Scholar]
- 26.Simonyan K, Zisserman A. Very Deep Convolutional Networks for Large-Scale Image Recognition. Paper Presented at: 3rd International Conference on Learning Representations (ICLR) 2015 [Google Scholar]
- 27.He K, Zhang X, Ren S, et al. Deep Residual Learning for Image Recognition. Paper Presented at: 2016 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 2016 [Google Scholar]
- 28.Szegedy C, Ioffe S, Vanhoucke V, et al. Inception-v4, Inception-Resnet and the Impact of Residual Connections on Learning. Paper Presented at: 31rd AAAI Conference on Artificial Intelligence (AAAI) 2017 [Google Scholar]
- 29.Hu J, Shen L, Sun G. Squeeze-and-Excitation Networks. Paper Presented at: 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 2018 [Google Scholar]
- 30.Deng J, Dong W, Socher R, et al. ImageNet: A Large-Scale Hierarchical Image Database. Paper Presented at: 2009 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 2009 [Google Scholar]
- 31.Zhang X, Zhou X, Lin M, et al. ShuffleNet: An Extremely Efficient Convolutional Neural Network for Mobile Devices. Paper Presented at: 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 2018 [Google Scholar]
- 32.Sandler M, Howard A, Zhu M, et al. MobileNetv2: Inverted Residuals and Linear Bottlenecks. Paper Presented at: 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 2018 [Google Scholar]
- 33.Iandola FN, Han S, Moskewicz MW, et al. SqueezeNet: AlexNet-Level Accuracy with 50x Fewer Parameters and <0.5 MB Model Size. arXiv: 1602.07360v4 [Preprint] 2016. [[Last cited 2021 Jan 14]]. p. 13. Available from: https://arxiv.org/abs/1602.07360 .
- 34.Zoph B, Vasudevan V, Shlens J, et al. Learning Transferable Architectures for Scalable Image Recognition. Paper Presented at: 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 2018 [Google Scholar]
- 35.Neumann H, Bisschops R. Artificial intelligence and the future of endoscopy. Dig Endosc. 2019;31:389–90. doi: 10.1111/den.13391. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Segmentation results of different methods. Original image is images after preprocessing; Ground truth indicates region of interest delinated by experts. Yellow area indicates region of interest predicted by deep-learning models