Concerning the article “With or Without W? Molecular and Cytogenetic Markers Are Not Sufficient for Identification of Environmentally Induced Sex Reversal in the Bearded Dragon” [Sexual Development, this issue, DOI: 10.1159/000514195] by Ehl et al., the following additions should be observed.
The article describes the sexing protocol used in Quinn et al. [2010] as equivalent in the region amplified to that used in Holleley et al. [2015] and Castelli et al. [2020]. The protocol by Quinn et al. [2010] is based on the amplification of a fragment by the primer pair F4/F1, while the one used by Holleley et al. [2015] and Castelli et al. [2020] is by the primer pair H2/F, both published in Quinn et al. [2010]. Our claim of the equivalence of these tests was based on the information that both fragments are a part of the same short sequence assigned as “contig C” as depicted by Quinn et al. [2010] and also reported like this by Holleley et al. [2015]. However, after the online publication of our paper, Prof. Arthur Georges and members of his team informed us that the fragments amplified by these two primer pairs are not a part of the same short continuous sequence (contig), which was not possible to determine from published resources. Instead, according to their information, the amplified fragments are parts of different paralogs of a repetitive sequence previously assigned as “contig C”. Therefore, these two protocols are not equivalent.
Sentence 3 of the Materials and Methods section should then read:
During a genetic screening of captive-bred central bearded dragons and the previous experiment with sex-reversed individuals [Ehl et al., 2017], we uncovered a male with a mismatch between the phenotypic sex and the genotype: it possessed a female-specific allele in the locus assigned as “contig C” by Quinn et al. [2010], serving as a sex-specific PCR marker previously used for detection of sex-reversed individuals in this species [Quinn et al., 2010; Ehl et al., 2017].
Consequently, the second part of the paragraph 3 of the Discussion (since the fifth sentence) should read:
In the recent study [Castelli et al., 2020], the mismatch between phenotypic and genotypic sex was assigned by the molecular marker used by Holleley et al. [2015] and was found in 5% out of 534 examined individuals of P. vitticeps covering the whole species range. Notably, all 28 animals with the mismatch were phenotypic females and they were recorded only in the south-western part of the species range. While this clustered distribution is consistent with non-random occurrence of sex reversals in certain environmental conditions at the edge of the species distribution as interpreted by Castelli et al. [2020], the spatial clustering can also reflect the geographic spread of a mutation, recombination, or other rearrangement concerning the region of the W chromosome containing the otherwise female-specific marker used for identification of the individuals with the mismatch. The current study discloses that the causes of the mismatch between the phenotypic and genotypic sex should be investigated more rigorously. Holleley et al. [2015] tested the reliability of their molecular marker by a congruence with cytogenetics, but our current work demonstrates that the presence of the accumulation of AAGG repeats is not a fully reliable marker as well.
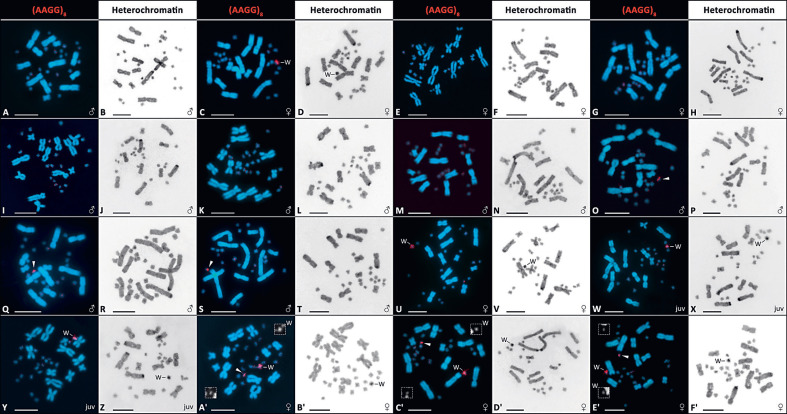
The Editors also pointed out that the labelling of the metaphases in Figure 1 and supplementary Figure S2 could be misleading for readers and the figures should be relabelled. We state in the figure legends that “Wm in the caption in males reflects the presence of the W-specific marker in PCR”, not the presence of the W chromosome. We hoped that it would prevent a misunderstanding that we claim that the male with the W-specific molecular fragment possesses the W chromosome (our interpretation of all the data is just the opposite). However, we agree that readers could be confused by our labelling. Therefore, we change the figures and their legends to the version below (Fig. 1, suppl. Fig. S2).
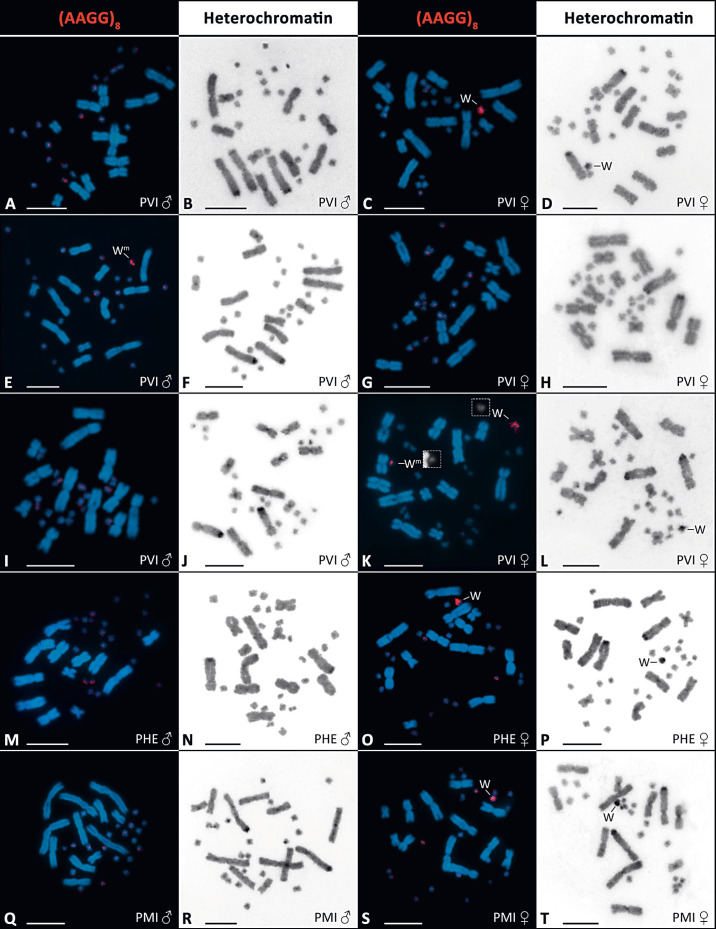
Fig. 1.
Visualization of the accumulation of AAGG repeats and heterochromatin in selected individuals of P. vitticeps (PVI; A-L), P. henrylawsoni (PHE; M-P), and P. minor (PMI; Q-T). In P. vitticeps, the figure represents successively a standard male ZZ karyotype with no accumulations (A, B), standard female ZW karyotype with AAGG (C) and heterochromatin accumulation (D), karyotype of a male with the W-specific molecular marker amplified by primers F4/F1 and with AAGG accumulation (E) but without heterochromatinization (F), karyotype of a sex-reversed ZZ female with no accumulations (G, H), karyotype of a male positive for the molecular marker and no accumulations (I, J), and karyotype of a female with 2 AAGG accumulations (K) and unpaired heterochromatic block (L). In P. henrylawsoni and P. minor, male karyotypes with no accumulations (M, N, Q, R) and female karyotypes with AAGG (O, S) and heterochromatin accumulations (P, T) are shown. Boxes in K show the W chromosome and the chromosome with the AAGG accumulation inherited from the father in separated blue channel mode to present their size difference. Scale bars, 10 ìm.
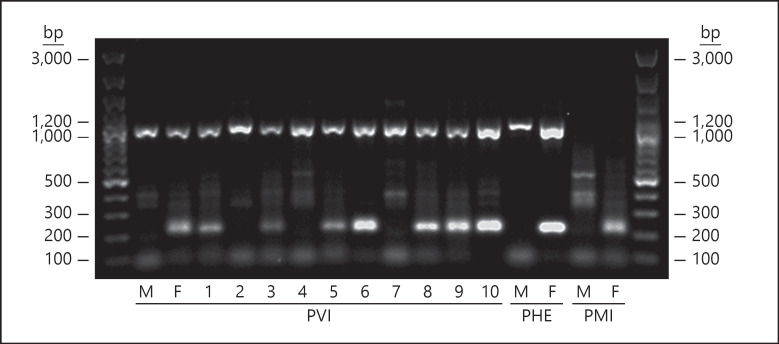
Suppl. Fig. S2.
Results of duplex PCR with E/C primers used for positive control and F4/F1 (W-specific fragment) primers in Pogona vitticeps (PVI), P. henrylawsoni (PHE), and P. minor (PMI) following the protocol of Quinn et al. (2010). All animals show the specific control product (around 1,000 bp) except for both PMI individuals. Standard females (F) of PVI, PHE and PMI display the W-specific fragment (224 bp), whereas in standard males (M) this fragment is not amplified. PVI individuals 1-10 include: (1) male with the W-specific PCR fragment and AAGG accumulation, used for the crosses, (2) sex-reversed ZZ female, (3, 4) male offspring of sex-reversed females and the male positive for the W-specific PCR fragment with AAGG accumulation, (5) offspring of unknown sex from the same cross, (6) standard ZW female used in the cross with this male, i.e. the mother of the offspring 7-10 depicted here (7, 8) male offspring of standard female and the male positive for the Wspecific PCR fragment with AAGG accumulation, and (9, 10) female offspring of standard ZW female and the male positive for the W-specific PCR fragment with AAGG accumulation.
Figure 2 in the article presents the results for the W-specific fragment only, for clarity. The original gel produced as described in the methods including positive controls, and used for analysis, is included as supplementary Figure S3 below.
Suppl. Fig. S3.
Results of duplex PCR with E/C primers used for positive control and F4/F1 (W-specific fragment) primers in Pogona vitticeps (PVI), P. henrylawsoni (PHE), and P. minor (PMI) following the protocol of Quinn et al. (2010). All animals show the specific control product (around 1,000 bp) except for both PMI individuals. Standard females (F) of PVI, PHE and PMI display the W-specific fragment (224 bp), whereas in standard males (M) this fragment is not amplified. PVI individuals 1-10 include: (1) male with the W-specific PCR fragment and AAGG accumulation, used for the crosses, (2) sex-reversed ZZ female, (3, 4) male offspring of sex-reversed females and the male positive for the W-specific PCR fragment with AAGG accumulation, (5) offspring of unknown sex from the same cross, (6) standard ZW female used in the cross with this male, i.e. the mother of the offspring 7-10 depicted here (7, 8) male offspring of standard female and the male positive for the Wspecific PCR fragment with AAGG accumulation, and (9, 10) female offspring of standard ZW female and the male positive for the W-specific PCR fragment with AAGG accumulation.