Abstract
Resistance to anti-cancer treatments is a critical and widespread health issue that has brought serious impacts on lives, the economy and public policies. Mounting research has suggested that a selected spectrum of patients with advanced colorectal cancer (CRC) tend to respond poorly to both chemotherapeutic and targeted therapeutic regimens. Drug resistance in tumours can occur in an intrinsic or acquired manner, rendering cancer cells insensitive to the treatment of anti-cancer therapies. Multiple factors have been associated with drug resistance. The most well-established factors are the emergence of cancer stem cell-like properties and overexpression of ABC transporters that mediate drug efflux. Besides, there is emerging evidence that signalling pathways that modulate cell survival and drug metabolism play major roles in the maintenance of multidrug resistance in CRC. This article reviews drug resistance in CRC as a result of alterations in the MAPK, PI3K/PKB, Wnt/β-catenin and Notch pathways.
Keywords: Colorectal cancer, Drug resistance, Chemotherapy, Targeted therapy, Signalling pathways, Mitogen-activated protein kinases, Protein kinase B, Notch receptor
Introduction
Colorectal cancer (CRC) is ranked as the most prevalent malignancy globally, after cancers of the lungs and the breast. In 2020 alone, two million new cases of CRC have been estimated worldwide, whereby 940,000 CRC cases potentially result in mortalities (International Agency for Research on Cancer, 2020). Risk factors of CRC are primarily genetic predisposition and environmental influences. As such, the development of CRC epitomizes gene-environment interaction, and multiple aetiologies that have been ascribed to CRC include genetic disorders (familial adenomatous polyposis and Lynch syndrome), family history of sporadic CRC, as well as unhealthy lifestyle (tobacco smoking, physical inactivity and heavy alcohol consumption) (Stigliano et al., 2014; Macrae, 2016; Yurgelun et al., 2017).
The genetic model of CRC carcinogenesis theorizes CRC as an accumulation of a set of driver mutations occurring in genes essential for the growth and differentiation of intestinal epithelium (Fearon & Vogelstein, 1990). Such mutations dysregulate cell signalling events in the intestinal epithelium, leading to CRC progression. Activation of oncogenes (e.g., KRAS or BRAF) and deletions of tumour suppressor genes (e.g., APC and p53) are known to disrupt cell development which results in uncontrolled cell division and cancer metastasis (Baker et al., 1989; Sansom et al., 2004; Aoki et al., 2007; Raskov et al., 2020).
Individuals diagnosed with CRC are subsequently treated according to the severity of CRC. The treatment options are summarised in Table 1. Surgery is employed to resect early-stage cancer to prevent the metastasis of CRC and its accompanying complications (Rentsch et al., 2016; Dekker et al., 2019). Meanwhile, radiotherapy is often applied as a supplemental treatment before CRC surgery and is targeted towards locally advanced CRC in order to reduce the size of tumour prior to surgery. This renders the surgical procedure less radical and may reduce local relapse (Häfner & Debus, 2016; Ma et al., 2017). On the other hand, systemic treatments of CRC are prescribed when patients suffer from metastatic CRC (mCRC). Systemic treatments can be divided into combination chemotherapy and targeted therapy. In combination chemotherapy, cancer drugs are combined in synergistically to produce more effective cytotoxic effects on the cancer cells (Carethers, 2008; Wolpin & Mayer, 2008). FOLFOX is a standard adjuvant chemotherapy for treating advanced CRC that is made up of folinic acid (leucovorin), oxaliplatin (L-OHP) and 5-fluorouracil (5-FU) (De Gramont et al., 2000). While FOLFOX has been proven effective for treating stage III and IV CRC, FOLFOX might not be suitable for treating high-risk stage II CRC harbouring BRAF V660E mutation with or without microsatellite stable (MSS) status due to a higher chance of tumour relapse after treatment (Seppälä et al., 2015). Clinical trial data also suggests that a 6-month FOLFOX regimen results in significant neurotoxicity for high-risk stage II CRC patients, suggesting the need to reduce the duration of adjuvant chemotherapy for better treatment outcomes (Iveson et al., 2021). XELOX (also known as CAPOX) is an alternative first-line or second-line treatment for high-risk stage II CRC which comprises capecitabine and oxaliplatin. It has been reported that a 3-month XELOX regimen exhibits a similar curative effect to a 6-month FOLFOX regimen. However, treatment benefits vary according to the patients’ medical condition and side effects of the treatment (Guo et al., 2016; Petrelli et al., 2020; Iveson et al., 2021). Targeted therapy on the other hand involves the use of small-molecule drugs or antibodies to specifically target genes or proteins that drive cancer survival and cancer metastasis (Xie, Chen & Fang, 2020a). In most cases, combination chemotherapy and targeted therapy are employed to treat mCRC after surgical removal of tumours (Kuo et al., 2005; Townsley et al., 2006; Berlin et al., 2007).
Table 1. List of treatments available for CRC patients.
| Treatment method | Characteristic | Reference |
|---|---|---|
| Complete mesocolic excision (CME) | Surgery that involves removal of the affected colon and its lateral lymphatic supply by cutting the mesentry. | Dimitriou & Griniatsos (2015) |
| Single incision laparoscopic surgery (SILS) | Surgery involving the use of one umbilical port. | Greaves & Nicholson (2011) |
| Natural orifice transluminal endoscopic surgery (NOTES) | Surgery that involves entering the peritoneal cavity via the gastrointestinal tract using a natural orifice. | Kalloo (2007) |
| Robotics laparoscopic surgery | Minimally invasive bowel resections performed by robotic system. | Giulianotti et al. (2003) |
| Short-course radiotherapy | Procedure regarding patients being exposed to radiation for 1 week with a clinical dosage of 25 Gray in 5 fractions followed by surgery 1 week later. | Påhlman (1997) |
| Long-course chemoradiotherapy | Procedure that covers radiation exposure for 5 weeks with a clinical dosage of 45–50 Gray in 25–28 fractions together with 5 concurrent fluoropyrimidine-based regimens as radiation sensitiser. This is followed by surgery 4–8 weeks later. | Bosset et al. (2006) |
| FOLFOX | Chemotherapeutic regimen involving the use of folinic acid, 5-fluorouracil (5-FU) and oxaliplatin to promote DNA cross-linking, hence inhibiting DNA synthesis and eventually induces cell death. | Gramont et al. (2000), Parker & Cheng (1990), Goldberg et al. (2004), Raymond et al. (1998) |
| FOLFIRI | Chemotherapeutic regimen involving the use of folinic acid, 5-FU and irinotecan to interfere with DNA uncoiling during DNA replication which ultimately induces cell death. | Klein et al. (2002), Douillard et al. (2000), Saltz et al. (2000) |
| Growth factor receptor inhibitors (e.g., Bevacizumab, Cetuximab) | Therapeutic agents designed to target specific pathways supporting cancer proliferation and formation of new blood vessels that allow the spread of mCRC. | Sherwood, Parris & Folkman (1971), Venook (2005) |
| Tyrosine kinase inhibitors (e.g., Gefitinib, Erlotinib, Sorafenib) | Therapeutic agents designed to target tyrosine kinases that mediate downstream signalling events of mCRC. | Huang et al. (2004), Townsley et al. (2006), Wilhelm et al. (2006) |
Drug resistance in CRC
Despite advances in the diagnostics and treatments, the global age-standardised mortality rates of CRC remain high (8.9 per 100,000 population in both sexes) (Siegel et al., 2017; Rawla, Sunkara & Barsouk, 2019). This is mainly due to the development of resistance to the standard chemotherapeutic regimens (5-FU and L-OHP) or combinational treatments (FOLFOX and XELOX) (Swanton, 2012; Weeks et al., 2012; Pai et al., 2017). In such cases, targeted therapeutic agents such as growth factor receptor inhibitors and protein kinase inhibitors are combined with the standard treatments to improve drug efficacy and patients’ response rates (Cunningham et al., 2004; Heinemann et al., 2014). Nevertheless, CRC patients have also been reported to develop resistance against these targeted therapeutic agents (Chen et al., 2018a; Negri et al., 2019; Vitiello et al., 2019; Rimassa et al., 2019). Hence, cancer drug resistance represents a significant obstacle to the successful treatment of CRC patients.
Drug resistance occurs when a tumour has become insensitive to the prescribed drugs, leading to the emergence of drug-tolerant cancer persister cells which support the growth of cancer cells under treatment pressure (Ramirez et al., 2016; Russo et al., 2019). While intrinsic or primary drug resistance occurs before drug treatment, acquired or secondary drug resistance manifests itself as a gradual reduction in drug efficacy against CRC (Lippert, Ruoff & Volm, 2008). Among the factors culminating in drug resistance, overexpression of ATP-binding cassette (ABC) transporters has been identified as the main driver. ABC transporters function to mediate the efflux of drugs from the tumours, leading to reduced drug concentration and drug efficacy (Giacomini et al., 2010; Hu et al., 2016). On top of that, the development of drug resistance has been attributed to genetic and epigenetic alterations, such as the (i) overexpression and gain-of-function of oncogenes (e.g., epidermal growth factor receptor (EGFR), Kirsten rat sarcoma virus (KRAS)) (Lièvre et al., 2006; Misale et al., 2014; Wang et al., 2019; Wang, Zhang & Chen, 2019), (ii) loss-of-function of tumour suppressor genes (e.g., p53, phosphatase and tensin homolog (PTEN)) (Boyer et al., 2004; Frattini et al., 2007; Sartore-Bianchi et al., 2009), (iii) under-expression of cell signalling regulator (e.g., thymidine phosphorylase) (Meropol et al., 2006), and (iv) the change in the binding site of drug target (e.g., topoisomerase I) (Gongora et al., 2011). In addition, the evolution of CRC subclones further complicates CRC treatment due to the limited ability of the cancer therapeutics to counteract the diverse drug resistance mechanisms present in the heterogeneous cancer subpopulations (Molinari et al., 2018; Wang et al., 2019; Wang, Zhang & Chen, 2019).
It has increasingly been acknowledged that various molecular mechanisms contribute to cancer drug resistance, among which the dysregulation of signalling pathways has been shown to play critical roles (Nisar et al., 2020). As such, the study of cell signalling pathways can provide valuable insights into the cancer biology of drug-resistant CRC and improve the treatment strategies (Wan et al., 2020). Previous studies have attributed four major signalling pathways (MAPK, PI3K/PKB, Wnt/β-catenin and Notch) to the development of resistance against CRC treatment (Li et al., 2011; Corcoran et al., 2012; Xu et al., 2017; He et al., 2018). There are significant efforts focusing on delineating tumour evolution and the underlying molecular mechanisms of drug resistance linked to these signalling pathways. Numerous studies have revealed that genetic mutations and/or epigenetic alterations of these pathways contribute to drug resistance (Normanno et al., 2015; Jeantet et al., 2016; Yamada et al., 2020). Apart from that, recent evidence also indicates that resistance of the tumour cells involves highly complex and tightly controlled crosstalk between different signal transduction pathways (Duong et al., 2018). Additionally, emerging findings suggest that signalling related to tumour microenvironment (TME), metabolic reprogramming and gut microbiome are also associated with the development of drug resistance (Lotti et al., 2013; Endo et al., 2020). A summary of the molecular alterations and clinical implications associated with the treatment of CRC is provided in Table 2.
Table 2. Molecular alterations and clinical implications associated with the dysregulation of targeted signalling pathways in multidrug-resistant CRC.
| Therapeutic agent | Targeted signalling pathway | CRC mutational status | Molecular alteration | Clinical implication | Reference |
|---|---|---|---|---|---|
| Anti-EGFR antibodies (cetuximab and panitumumab) alone or in combination with chemotherapy | MAPK pathway | Wild-type KRAS | KRAS, NRAS, BRAF and PI3KCA mutations | •Poor prognosis for overall survival •Low response rate to anti-EGFR therapy |
De Roock et al. (2010), Diaz et al. (2012) |
| Anti-EGFR antibodies (cetuximab, panitumumab, SYM004, MM151, trastuzumab, pertuzumab and duligotuzumab) alone or in combination with chemotherapy | MAPK pathway | Wild-type KRAS, NRAS, BRAF and PI3KCA | •HER2 gene amplification and activating mutations •Sustained signalling of PI3K/PKB and MAPK pathways |
•Poor therapeutic response •Oncogenic transformation of colon epithelial cells |
Kavuri et al. (2015), Belli et al. (2019) |
| RAF inhibitor (vemurafenib) | MAPK pathway | BRAF(V600E) | Feedback activation of EGFR | Treatment failure | Prahallad et al. (2012) |
| Combined RAF inhibitors (vemurafenib and cetuximab or vemurafenib and selumetinib) | MAPK pathway | BRAF(V600E) | Reactivation of MAPK pathway | Tumour relapses | Ahronian et al. (2015) |
| RAF inhibitors (GDC-0879 and vemurafenib) | MAPK pathway | BRAF(V600E) | RAF dimerization and MEK/ERK phosphorylation | Enhanced tumour growth | Hatzivassiliou et al. (2010) |
| Vemurafenib | MAPK pathway | KRAS(G13D) | Activation of ERK leads to the activation of Hippo and Rho pathways | Cancer metastasis | Kubiniok et al. (2017) |
| Chemotherapeutic drug (oxaliplatin) | MAPK pathway | Not applicable | miRNA-625-3p-mediated downregulation of MAP2K6 | Cancer progression due to reduced apoptosis. | Rasmussen et al. (2016) |
| Combinational chemotherapeutic drugs (FOLFOX and FOLFIRI) | PI3K/PKB pathway | Not applicable | PIK3CA mutations (E545K, E542K and E545D on exon 9; H1047R and H1047L on exon 20) | LGR5+ CRC stem cells survival and proliferation | Wang et al. (2018) |
| Cetuximab | PI3K/PKB pathway | Wild type KRAS and BRAF | PIK3CA mutations on exon 19 (K944N, V955I, F930S, V955G and K966E) | Decrease in progression-free survival | Xu et al. (2017) |
| Cetuximab and panitumumab | •MAPK pathway •PI3K/PKB pathway |
Wild type KRAS | BRAF, NRAS, PTEN and PIK3CA mutations | •Poor prognosis for overall survival •Cancer metastasis |
Sartore-Bianchi et al. (2009), Laurent-Puig et al. (2009), De Roock et al. (2010) |
| NVP-BEZ235 (dual PI3K/MTOR inhibitor) | •MAPK pathway •PI3K/PKB pathway |
Not applicable | KRAS and PIK3CA mutations leads to additive activation of PI3K/PKB pathway | Suppression of BIM-induced apoptosis.which leads to cancer survival | Kim et al. (2013) |
| Chemotherapeutic drug (paclitaxel) | PI3K/PKB pathway | Not applicable | miR-29a-mediated PTEN inhibition | Reduction in drug sensitivity suppress apoptosis which supports cancer growth | Yuan et al. (2018) |
| Chemotherapeutic drug (doxorubicin) | PI3K/PKB pathway | Not applicable | miR-29a-mediated P-gp inhibition and upregulation of PTEN | Enhanced drug sensitivity which thwart cancer growth | Shi et al. (2020) |
| Chemotherapeutic drug (5-FU) | PI3K/PKB pathway | Not applicable | miR-543-mediated PTEN inhibition | Reduced drug sensitivity which supports cancer growth | Liu, Zhou & Dong (2019) |
| Chemotherapeutic drug (vincristine) | Wnt/β-catenin pathway | Not applicable | Overexpression of Dvl1-3 leads to β-catenin/TCF-induced transcription of ABC transporters (P-gp, MRP2 and BCRP)‘and anti-apoptotic proteins (Survivin and Bcl-2) | CRC is protected from Vincristine-induced apoptosis which drives cancer growth | Zhang et al. (2017b) |
| 5-FU and oxaliplatin | Wnt/β-catenin pathway | Not applicable | Overexpression of LINC00152 inhibits CK1α-dependant β-catenin phosphorylation | •Cancer metastasis •Expression of EMT markers |
Yue et al. (2016), Yue et al. (2018), Bian et al. (2017) |
| 5-FU | Wnt/β-catenin pathway | Not applicable | miR-30-5p-mediated inhibition of USP22 and Wnt target genes (Axin2 and c-Myc) | Suppression of cancer stemness and chemoresistance | Ning et al. (2014), Jiang et al. (2019) |
| 5-FU and oxaliplatin | Wnt/β-catenin pathway | Not applicable | LncRNA CRNDE-mediated repression of miR-181a-5p promotes β-catenin/TCF transcriptional activity | CRC cell proliferation and chemoresistance | Han et al. (2017) |
| Small-molecule multi kinase inhibitor (regorafenib) | Notch pathway | Not applicable | Upregulation of Notch-1 and the target genes (HES1 and HEY1) | CRC cell proliferation due to reduced sensitivity to Regorafenib | Mirone et al. (2016) |
| Anti-VEGF antibody (bevacizumab) | Notch pathway | Not applicable | Upregulation of NICD | Cancer stemness | Negri & Ardizzoni (2015), Negri et al. (2019) |
| 5-FU | Notch pathway | Not applicable | HES1-mediated overexpression of ABC transporters (ABCC1, ABCC2 and P-gp1) with depressed E-cadherins and elevated N-cadherins | Tumour relapses | Gao et al. (2014), Sun et al. (2017) |
| Chemotherapeutic drug (methotrexate) | •Notch pathway •Wnt/β-catenin pathway |
Not applicable | Dvl-3-related Wnt and Notch crosstalk. | Cancer stemness | Zhao et al. (2020) |
| 5-FU and Irinotecan | •Notch pathway •KRAS/Erk/ADAM pathway |
KRAS(G12D, G12A, G13D, Q61L) | Aberrant Jagged1 processing leads to sustained Jag1-ICDs-mediated intrinsic reverse signalling. | Cancer progression and chemoresistance | Van Schaeybroeck et al. (2011), Pelullo et al. (2019) |
| 5-FU | Notch pathway | Not applicable | miR-139-5p-mediated inhibition of Notch-1 and downstream multidrug-resistant genes (MRP-1 and BCL-2) | Increased sensitivity to 5-FU | Liu et al. (2016) |
| 5-FU | Notch pathway | Not applicable | miR-195-5p-mediated inhibition of Notch-2 and RBPJ | Inhibition of cancer stemness and 5-FU resistance | Jin et al. (2018) |
| 5-FU | Notch pathway | Not applicable | miR-34a-mediated ABCG2 inhibition | Enhanced chemosensitivity to 5-FU | Xie, Chen & Fang, (2020a), Xie et al. (2020b) |
In this review article, we attempt to summarize the gap in knowledge in understanding the link between modulation of the signalling mechanisms due to diverse exogenous and endogenous factors with drug resistance in CRC. We aim to provide current updates related to the dysregulation of the four selected signal transduction pathways and their roles in conferring drug resistance in CRC. In addition, future perspectives pertinent to the involvement of other signalling pathways and resistance mechanisms due to TME, metabolic reprogramming and gut microbiome are also discussed.
Survey methodology
To ensure a thorough and unbiased coverage of the literature, we searched fthe PubMed database for published articles written in English from 1990 until present. The search strings include “colorectal cancer AND (crosstalk OR communication) AND (signalling OR pathway) AND therapy resistance”, “colorectal cancer AND (monotherapy OR combinational therapy) AND drug resistance AND MAPK pathway”, “colorectal cancer AND (monotherapy OR combinational therapy) AND drug resistance AND PI3K pathway”, “colorectal cancer AND (monotherapy OR combinational therapy) AND drug resistance AND Wnt pathway”, “colorectal cancer AND drug resistance AND Notch”, and “colorectal cancer AND (MAPK OR PI3K OR Wnt OR Notch) AND drug resistance”. The searches were performed by two authors independently of each other. The abstracts of these articles were then assessed to exclude papers that were not relevant to the scope of this review. This review paper is intended for scientist who work in the same scientific area as well as other readers in the field of molecular and cancer biology in general.
Regulation of signalling pathways associated with drug resistance in CRC
Mitogen-activated protein kinase (MAPK) pathway
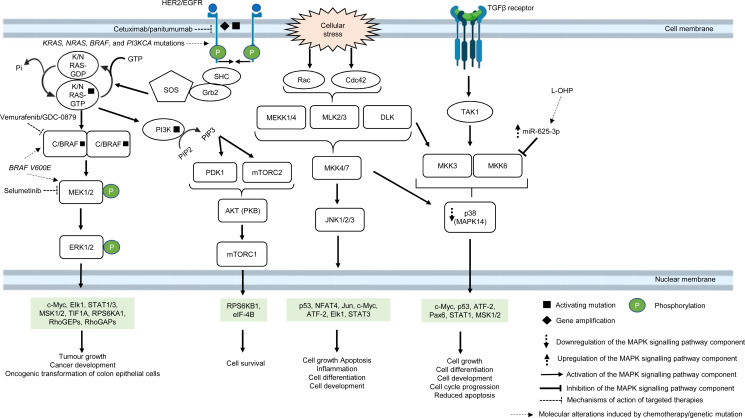
The MAPK pathway is mediated by mitogen-activated protein kinases (MAPKs), which comprise a family of serine/threonine-specific protein kinases that regulate a variety of cellular processes and play crucial roles in the pathogenesis of many diseases such as cancer, infection, inflammatory and autoimmune diseases (Kim & Choi, 2010). The MAPK family is divided into several subgroups. Conventional MAPKs are (i) extracellular signal-regulated kinase 1 and 2 (ERK1/2), (ii) c-Jun N-terminal kinases (JNKs), (iii) p38 (also known as MAPK14) and (iv) ERK5, all are summarised in Fig. 1 (Morrison, 2012). Atypical MAPKs comprise ERK3/4, ERK7/8 and Nemo-like kinase (NLK) (Cargnello & Roux, 2011). In the canonical ERK/MAPK signalling pathway, extracellular signals (e.g., growth factors, stress, mitogens) bind to the receptors, most of which are receptor tyrosine kinases (RTKs) at the surface of the cell membrane, leading to auto-phosphorylation of growth factors receptors and recruitment of adaptor proteins (e.g., growth factor receptor-bound protein 2 (GRB2) and son of sevenless 1 (SOS1)) (Morrison, 2012). This in turn results in the switching of the inactivated form of Ras-family GTPase (Ras in GDP bound form, Ras-GDP) to the active form of Ras-family GTPase (Ras in GTP bound form, Ras-GTP). The external signal is then transmitted via Ras-GTP to other downstream phosphorylation targets within the cytoplasm where the signalling cascade converges at the activation of a series of MAPKs, starting from MAPK kinase kinase (MAPKKK, e.g., Raf1) followed by MAPK kinase (MAPKK, e.g., MEK1/2) and MAP kinase (MAPK, e.g., ERK1/2). Finally, the MAP kinase translocates to the nucleus to phosphorylate transcription factors (e.g., c-Jun, STAT1, c-Myc) that regulate transcription of genes for different cellular processes (Wei & Liu, 2002; Fang & Richardson, 2005; Morrison, 2012) (Fig. 1).
Figure 1. Deregulation of the canonical MAPK signalling pathway during CRC treatment.
Region highlighted green represents substrates for MAPKs that regulate biological processes in CRC cells treated with drug therapy.
In multidrug-resistant CRC, MAPK pathway is often reprogrammed, usually by the overexpression of RTKs, Ras and Raf; or gain-of-function mutations of Ras and Raf, which sustain the activity of MAPK signalling pathway upon treatment with MAPK and RTK inhibitors, 5-FU and oxaliplatin (Wan et al., 2004; Kavuri et al., 2015; Martinelli et al., 2017; Ressa et al., 2018). EGFR has been a favourable target for the treatment of mCRC since the last decade, mainly because they are highly expressed in most human tumours, including CRC (Yarden & Pines, 2012). In particular, monoclonal antibody targeting EGFR, such as cetuximab and panitumumab are widely used to treat mCRC patients due to their initial benefit of improving patients’ survival (Jonker et al., 2007; Vermorken et al., 2008; Pirker et al., 2009). However, some studies have shown that anti-EGFR based therapy may not be effective in treating mCRC, indicating that a subset of CRC is resistant to the anti-EGFR treatment (Bokemeyer et al., 2009; Van Cutsem et al., 2009). Common molecular mechanisms associated with the resistance are KRAS, NRAS, BRAF and PI3KCA mutations (De Roock et al., 2010; Diaz et al., 2012). In CRC which is quadruple wild-type for KRAS, NRAS, BRAF and PI3KCA genes, HER2 gene amplification and activating mutations at the phosphorylation sites of the catalytic domain have been shown to bypass EGFR blockade by activating a compensatory signalling mechanism for cell survival (Kavuri et al., 2015; Belli et al., 2019). It has been reported that HER2 could form heterodimers with either EGFR or ERBB3 with consequent activation of ERK and Akt signalling respectively, in which the latter has been shown to promote anti-EGFR resistance (Zhang et al., 2014a; Zhang et al., 2014b). Besides, it has been found that aberrant ERBB2 activation could result in the stimulation of ERK 1/2 signalling that mediates cetuximab resistance (Yonesaka et al., 2011).
B-Raf is a MAPKKK that mediates cell growth and differentiation via the ERK/MAPK subfamily of MAPK pathway, in response to growth factors and mitogens (Morrison, 2012). B-Raf mutations, more often BRAF V600E, occur in approximately 8% of CRC and are associated with poor prognosis (Davies et al., 2002; Richman et al., 2009). BRAF V600E mutation leads to conformational changes at the catalytic domain which renders B-Raf constitutively active, independent of Ras-GTP activation and dimerization with Raf-1 (also known as C-Raf) (Durrant & Morrison, 2018). This results in prolonged phosphorylation and activation of MEK1/2 and ERK1/2 kinases which, in turn, activates downstream substrates that mediate cell growth and survival (Fang & Richardson, 2005). It has been reported that targeting BRAF V600E using mono-therapeutic agents, such as vemurafenib (a B-Raf inhibitor, also known as PLX4032) which binds to the ATP-binding site of BRAF V600E to inhibit its activity, shows limited therapeutic response in CRC (Prahallad et al., 2012). This is because targeting BRAF V600E results in feedback activation of EGFR characterised by enhanced phosphorylation of RAS and CRAF upstream of MAPK pathway and downstream activation of RAF, MEK and ERK (Corcoran et al., 2012). In order to circumvent resistance to B-Raf inhibition, B-Raf inhibitor is used in combination with EGFR inhibitor or MEK inhibitor or both which were initially shown to offer therapeutic benefit of at least 12% response rate to the drugs and improve the suppression of ERK/MAPK pathway (Bendell et al., 2014; Corcoran et al., 2014; Tabernero et al., 2014). Despite the initial success in suppressing B-Raf resistance using the multi-target approach, there is compelling evidence that BRAF V600E mutant CRC patients could also develop resistance to the new treatment (Oddo et al., 2016). Ahronian et al. (2015) has shown that the reactivation of the MAPK pathway confers cross-resistance to the combined RAF/EGFR or RAF/MEK inhibition in BRAF-mutant CRC and further demonstrated that the use of ERK inhibitor could overcome the resistance by suppressing the MAPK signalling.
On the other hand, it has been reported that treatment using ATP-competitive inhibitors produces opposing mechanisms of action that is dependent on the cellular context and genotype of the tumour. It was found that RAF inhibitors effectively block MAPK pathway in BRAF V600E cells but activate the MAPK pathway in wild-type BRAF tumours by inducing RAF dimerization and MEK/ERK phosphorylation leading to enhanced tumour growth, suggesting that other strategies to block RAF activation are needed to improve the treatment efficacy (Hatzivassiliou et al., 2010). Furthermore, RAS mutant tumours are also known to exhibit poor response to RAF inhibitors. A time-course phosphoproteomic analysis of vemurafenib-treated RAS mutant CRC cell lines has found potential cross-talk between ERK signalling with Hippo and Rho pathways and revealed novel functional targets downstream of ERK (Kubiniok et al., 2017).
Apart from post-translational regulation of proteins as a regulatory checkpoint for cellular signalling, microRNAs (miRNAs) also exhibit a functional role in the regulation of MAPK pathway in CRC drug resistance (Rasmussen et al., 2016; Angius et al., 2019). A previous miRNA profiling study has uncovered the link between high expression of miRNA-625-3p and poor clinical response towards oxaliplatin-based therapy (Arango et al., 2004; Rasmussen et al., 2013). Mechanistically, it has been demonstrated that miRNA-625-3p mediates oxaliplatin resistance by targeting MAPK kinase MAP2K6 and abrogates MAPK14 signalling, leading to increased cell cycle progression and reduced apoptosis (Rasmussen et al., 2016).
As one of the most frequently altered signalling pathways and its important roles in CRC drug resistance, MAPK pathway represents a promising target for cancer therapy. Significant progress has been made on the development of therapeutics targeting MAPK kinases with considerable clinical success (Bendell et al., 2014). Notably, the combination of BRAF and MEK inhibitors has been shown to improve response rates and may offer potential therapeutic benefit in BRAF-mutated CRC (Corcoran et al., 2014). Nevertheless, it is clear that we are still far from any complete understanding of the MAPK pathway. Moreover, the emergence of new resistance mechanisms prompts more further research to provide a deeper understanding on the complex regulation and interconnectivity of the underlying biological processes to overcome resistance and increase therapeutic efficacy.
Phosphoinositide 3-kinase (PI3K)/protein kinase B (PKB, also known as AKT) pathway
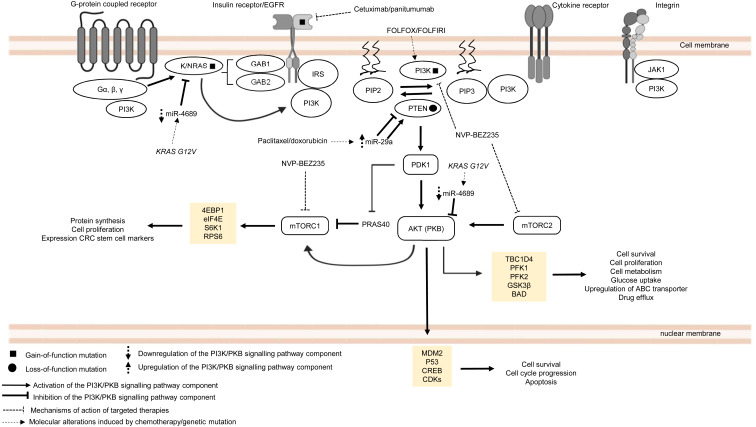
The PI3K/PKB pathway regulates cell metabolism, cell growth and cell survival. In normal condition, PI3K/PKB pathway is activated by four major sensors upstream of the pathway, namely (i) receptor tyrosine kinases (RTKs) which bind to growth factors (Hemmings, 1997), (ii) cytokine receptors (Chang et al., 2003), (iii) G protein-coupled receptors (GPCRs) that are activated by various biological molecules (Murga et al., 1998), and (iv) integrins which detect cell–cell or cell–matrix communication (Su et al., 2007). Upon ligand binding, these receptors, together with their cofactors, will activate PI3K family proteins. There are three classes of PI3K family proteins, among which only class I PI3Ks and the signalling networks they regulate are covered in this review (Fig. 2). Information about Class II and Class III PI3Ks and their roles in cellular signalling are covered in other reviews or journal articles (Falasca & Maffucci, 2012; Okkenhaug, 2013; Backer, 2016; Hawkins & Stephens, 2016). Within the class I PI3K subfamily itself, there are four catalytic isoforms (p110α, p110β, p110γ and p110δ encoded by PIK3CA, PIK3CB, PIK3CG, and PIK3CD respectively) which catalyse the phosphorylation of phosphatidylinositol-4,5-bisphosphate (PI(4,5)P2) to phosphatidylinositol (3,4,5)-trisphosphate (PI(3,4,5)P3) or PIP3. p110α and p110β are expressed ubiquitously while p110γ and p110δ are expressed in immune cells. Each catalytic isoform forms a dimer with a regulatory subunit that controls the activity and subcellular localisation of the PI3K complex (Fig. 2). In response to specific external stimuli, PIP3, which acts as a secondary messenger, will recruit cytoplasmic proteins with PIP3 binding domains (typical examples of which are phosphoinositide-dependent kinase-1 (PDK1) and PKB) to specific cell membrane locations. Shortly after the transmission of the signal to downstream effectors, PIP3 is then metabolised by phosphatase and tensin homolog (PTEN), which is a tumour suppressor that negatively regulates the PI3K signal by removing 3′-phosphate from PIP3 (Danielsen et al., 2015; Fruman et al., 2017). Activation of PKB is a two-step process, whereby PDK1 phosphorylates PKB on threonine-308 to partially activate PKB (Alessi et al., 1997), followed by phosphorylation of PKB by mTORC2 on serine-473 to fully activate PKB (Sarbassov et al., 2005). The activated PKB will subsequently regulate the phosphorylation of the target substrates (the most well-known examples are glycogen synthase kinase 3 beta (GSK3β), BCL2-antagonist of death (BAD), mouse double minute homolog 2 (MDM2), forkhead box O (FOXO) and mechanistic target of rapamycin complex (mTORC1)) which mediate important cellular functions, such as glucose uptake, protein synthesis, cell survival and cell cycle progression (Manning & Cantley, 2007). The activity of PKB is also negatively regulated by protein phosphatase 2A (PP2A), PH domain leucine-rich repeat protein phosphatase 1/2 (PHLPP1/2) and carboxyl-terminal modulator protein (CTMP) (Hemmings & Restuccia, 2012) (Fig. 2). In multidrug-resistant CRC, PI3K signalling is prolonged by PIK3CA mutations, null mutation of PTEN, and RAS mutations upon treatment with standard chemotherapeutic drugs and targeted therapeutic agents (Laurent-Puig et al., 2009; De Roock et al., 2010; Kim et al., 2013; Hamada, Nowak & Ogino, 2017).
Figure 2. Deregulation of the canonical PI3K signalling pathway during CRC treatment.
Region highlighted yellow represents substrates for PKB/AKT that regulate biological processes in CRC cells treated with drug therapy.
PIK3CA is regarded as one of the most frequently mutated genes in CRC, which accounts for approximately 10–30% of all CRC cases (Samuels et al., 2004; Velho et al., 2005; Hamada, Nowak & Ogino, 2017). It is reported that non-random somatic mutations occurring in the coding region, mainly exon 9 (helical domain) and exon 20 (catalytic domain), have heightened basal PI3K and PKB activities which then promote cancer progression (Kang, Bader & Vogt, 2005; Ikenoue et al., 2005). Furthermore, it has been shown that PIK3CA mutations (E545K, E542K and E545D on exon 9; H1047R and H1047L on exon 20) could mediate resistance to standard chemotherapy (FOLFOX and FOLFORI) by inducing phosphorylation of PKB and expression CRC stem cell markers (LGR5) via sustained PI3K signalling, thereby promoting cancer cell survival and proliferation (Wang et al., 2018). Interestingly, PIK3CA mutations have also been found to mediate acquired cetuximab resistance in mCRC complementary to the previously reported RAS mutations. A recent circulating tumour DNA sequencing analysis from mCRC patients has revealed five novel mutations on exon 19 of PIK3CA (K944N, V955I, F930S, V955G and K966E) that may potentially drive resistance to cetuximab via EGFR-mediated activation of the PI3K/PKB signalling pathway, suggesting that combined regimens of PI3K/mTOR inhibitors (PP242 and NVP-BEZ235) with anti-EGFR therapy may be beneficial to overcome the resistance (Xu et al., 2017).
Apart from PIK3CA mutations, there is increasing evidence of the emergence of mutations involving components of MAPK and PI3K/PKB signalling pathways when treated with targeted therapeutic agents (De Roock et al., 2010; Vitiello et al., 2019). Although anti-EGFR based therapy is commonly prescribed for KRAS wild-type CRC patients, clinical evidence has indicated that the KRAS mutation status alone is insufficient to predict therapeutic response to the therapy (Allegra et al., 2009). Retrospective cohort studies of CRC cases have identified several key players in the EGFR signalling pathway (which is a signalling network shared by both MAPK and PI3K/PKB pathways) that hinder the effectiveness of anti-EGFR monoclonal antibodies in KRAS wild-type CRC patients (Sartore-Bianchi et al., 2009; Laurent-Puig et al., 2009; De Roock et al., 2010). It has been previously reported that BRAF, NRAS, PTEN and PIK3CA mutations are associated with the efficacy and clinical outcome of EGFR-targeted therapy but their exact roles in driving the resistance are still unclear (Laurent-Puig et al., 2009; De Roock et al., 2010). Surprisingly, KRAS mutations also negatively affect the outcome of treatment against PIK3CA mutant CRC. Kim et al. (2013) have shown that KRAS and PIK3CA mutations attenuate sensitivity to treatment with a dual inhibitor of PI3K and mTOR by suppressing BIM-induced apoptosis via activation of PI3K/MTOR pathway, leading to cell survival.
More recently, miRNAs have also been reported to control CRC pathogenesis via the modulation of PI3K/PKB pathway (Soleimani et al., 2019). Based on previous studies, it has been shown that miR-29a could induce or suppress tumour progression in drug-resistant cancer cells (Zhong et al., 2013; Liu et al., 2018). Yuan et al. (2018) showed that higher level of miR-29a is expressed in CRC cell lines resistant to paclitaxel which resulted in downregulation of PTEN and upregulation of phosphorylated AKT. This suggests that miR-29a has a regulatory function to PI3K/PKB pathway via inhibition of PTEN which reduces drug sensitivity and supports cancer growth. In contrast, miR-29a could potentially reduce P-gp-mediated chemoresistance via modulation of PTEN and P-gp expression in doxorubicin-resistant CRC cell lines. This means that miR-29a exerts tumour suppressive function by inhibiting membrane transporter activity through PI3K/PKB pathway (Shi et al., 2020). These seemingly contradictory roles of miR-29a have also been observed in other malignancies. In the context of regulation of drug resistance, miR-29a has been reported to increase sensitisation to gemcitabine in pancreatic cancer cells (Kwon et al., 2016) as well as sensitization to tamoxifen in breast cancer cells (Muluhngwi et al., 2017). On the other hand, miR-29a was shown to play a role in mediating adriamycin resistance in breast cancer cells via inhibiting the PTEN/AKT/GSK3β pathway (Shen et al., 2016). It is unclear whether the opposing results are due to heterogeneity in the cancer cells or that miR-29a could exhibit multifaceted functions in a context dependent manner. Nevertheless, these paradoxical findings demand the need for further work to clarify and elucidate the function of miR-29a more comprehensively. Similar to miR-29a, miR-543 has recently been identified as the mediator of chemoresistance in CRC cells, also by suppressing the expression of PTEN which activates PI3K/PKB pathway (Liu, Zhou & Dong, 2019). Interestingly, miR-4689 which has been identified as a negative regulator of KRAS and AKT is downregulated in KRAS mutant CRC cells and confers resistance to molecular-targeted therapy, suggesting that miR-4689 could be a promising therapeutic agent to control multidrug resistance in CRC via modulation of PI3K/PKB pathway and MAPK pathway (Hiraki et al., 2015).
Taken together, current evidence clearly shows that mutations of the MAPK/PI3K/PKB signalling pathway components are frequently observed in cancer drug resistance. However, the signalling mechanisms associated with these mutations are still not well elucidated which necessitate further investigation. Importantly, distinct molecular events that regulate drug resistance such as in KRAS wild-type and mutant CRC suggests the importance of identifying relevant drug targets in CRC with different mutational status. Moreover, future research should also focus on dissecting the link of miRNAs with cancer drug resistance and their underlying molecular mechanisms.
Wnt/β-catenin pathway
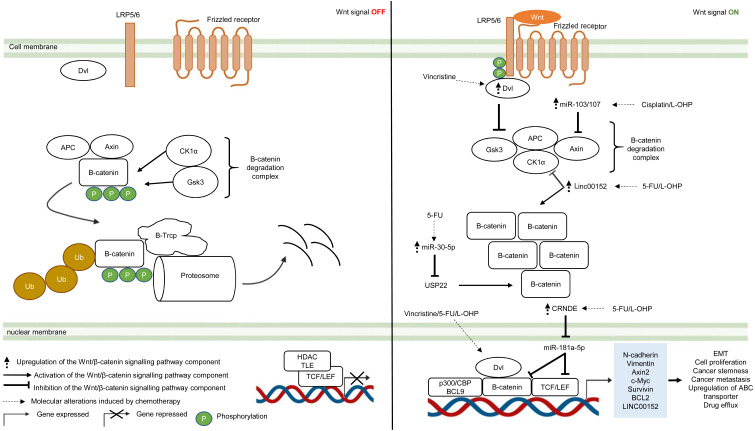
The Wnt/β-catenin pathway is an evolutionarily conserved system that regulates cell development, cell differentiation, cell proliferation and cell migration. The Wnt/β-catenin pathway can be grouped into β-catenin-dependent Wnt pathway (canonical Wnt pathway) and β-catenin-independent Wnt pathway (non-canonical Wnt pathway) which are further divided into the planar cell polarity Wnt pathway and the Wnt/Ca2+ pathway (Komiya & Habas, 2008). The canonical Wnt/β-catenin pathway is made up of the membrane proteins, degradation complex and β-catenin protein. In the absence of Wnt ligands, the degradation complex which comprises of adenomatous polyposis coli (APC), Axin, GSK3 and CK1α is formed through phosphorylation of Axin and APC by GSK3 and casein kinase 1α (CK1α). As a result, β-catenin is ubiquitinated by E3-ligase protein βTrCP (β-transducin repeats-containing proteins) through phosphorylation and targeted for proteasomal degradation (Van Kappel & Maurice, 2017) (Fig. 3). In the presence of Wnt ligands, lipoprotein receptor-related protein 5/6 (LRP5/6) co-receptor and Frizzled (Fzd) receptor are activated which leads to phosphorylation of LRP5/6 co-receptors and binding of adaptor protein disheveled (Dvl) to the phosphorylated LRP5/6. This is followed by the recruitment of the remaining degradation complex components to the Fzd-LRP5/6 complex to inactivate the degradation complex (Janda et al., 2012) (Fig. 3). The molecular mechanism for Wnt-mediated degradation complex inactivation is still heavily disputed due to conflicting findings on the inhibition of GSK3 in the presence of Wnt signal. Several models have been proposed: (1) blockade of GSK3 catalytic site by binding to the phosphorylation motif of LRP5/6 (Wu et al., 2009). (2) Wnt-mediated dissociation of APC from GSK3 (Valvezan et al., 2012). (3) sequestration of GSK3 in endosomal vesicles through endocytosis of Fzd-LRP5/6 complex (Taelman et al., 2010). Disruption of the degradation complex integrity in the presence of Wnt signal promotes stabilisation of β-catenin, leading to accumulation of newly synthesised β-catenin in the cytoplasm and their subsequent translocation to the nucleus. Interestingly, it is also known that, in the presence of Wnt signal, the degradation complex could remain intact to target β-catenin for degradation via phosphorylation, but ubiquitination of β-catenin is impaired which inhibits its degradation by the degradation complex (Gerlach et al., 2014). Within the nucleus, β-catenin binds to T-cell factor/lymphoid enhancing factor (TCF/LEF) which are the transcription factors that activate Wnt-responsive genes required for cell growth and survival (e.g., c-Myc, Cyclin D1). In addition, β-catenin interacts with TCF/LEF to recruit transcriptional co-activators (p300/CBP and BCL9) to the transcription factors to activate gene expression (Kretzschmar & Clevers, 2017; Taciak et al., 2018) (Fig. 3). In multidrug-resistant CRC, Wnt/β-catenin pathway is reprogrammed by overexpression of Dvl protein and non-coding RNAs that interfere with the activities of downstream signalling mediators (Yue et al., 2016; Han et al., 2017; Bian et al., 2017; Zhang et al., 2017b; Jiang et al., 2019).
Figure 3. Deregulation of the canonical Wnt/β-catenin signalling pathway during CRC treatment.
Left-hand-side of the figure describes signalling events in the absence of the Wnt signal (OFF). Right-hand-side of the figure describes signalling events in the presence of the Wnt signal (ON). Region highlighted blue represents Wnt target genes that regulate the biological processes of CRC cells treated with drug therapy.
Dysregulation in the key Wnt/β-catenin pathway components such as upstream regulator (Dvl protein), β-catenin degradation complex and its downstream targets (β-catenin and TCF/LEF) instigate tumour progression in many types of cancer (Van Kappel & Maurice, 2017). Moreover, recent studies indicate that aberrant Wnt/β-catenin signalling could trigger anti-cancer drug resistance (Zhang et al., 2017a). Zhang et al. (2017b) reported that DVL1-3 proteins are overexpressed in CRC resistant to vincristine (a chemotherapeutic drug that interferes with microtubule synthesis leading to cell cycle arrest) which results in overexpression of ABC transporters (P-glycoprotein (P-gp), MRP2 and BCRP) and anti-apoptotic proteins (Survivin and Bcl-2). Contrary to previous findings which suggest that DVL promotes β-catenin accumulation and subsequent translocation to the nucleus, it was found that DVL1-3 translocate to the nucleus and bind to β-catenin to form a transcriptional complex, independent of β-catenin accumulation and nuclear translocation (Gao & Chen, 2010; Shang, Hua & Hu, 2017). Moreover, the study also showed that silencing DVL1-3 could re-sensitise CRC cells to vincristine, 5-FU and oxaliplatin, suggesting that DVL could be a potential therapeutic target in multidrug-resistant CRC.
It has been increasingly recognised that long non-coding RNA (lncRNA, a non-coding regulatory RNA with greater than 200 nucleotides in length) regulates Wnt/β-catenin pathway in multidrug-resistant CRC (Ma et al., 2016; Lu et al., 2017). Recent studies have shown that lncRNA cytoskeleton regulator RNA (CYTOR, also known as LINC00152) is overexpressed in CRC which confers resistance to oxaliplatin-induced apoptosis (Yue et al., 2016). In addition, elevated expression of LINC00152 is also observed in CRC that gives rise to 5-FU resistance and cancer metastasis (Bian et al., 2017). However, the regulatory mechanism of LINC00152 in the Wnt/β-catenin pathway of mCRC is still unknown (Yue et al., 2016). In a recent study, Yue et al. (2018) demonstrated that LINC00152 competitively binds to β-catenin to prevent CK1α from phosphorylating β-catenin. As a result, β-catenin accumulates and translocates to the nucleus to activate the expression of epithelial-mesenchymal transition (EMT) markers (N-cadherin and Vimentin) which are hallmarks of metastatic cancer. Reciprocally, β-catenin/TCF4 transcriptional complex promotes the expression of LINC00152 to sustain Wnt/β-catenin signalling in mCRC.
Likewise, miRNA is also known to influence cancer growth and the sensitivity of cancer cells towards anti-cancer drugs (Jansson & Lund, 2012; Piletič & Kunej, 2016). Jiang et al. (2019) have demonstrated that overexpression of miR-30-5p negatively regulates the expression of Wnt/β-catenin pathway target genes (Axin2 and c-Myc) and inhibits chemoresistance in CRC cells by targeting ubiquitin-specific peptidase 22 (USP22). Mechanistically, it has been reported that USP22 induces β-catenin nuclear localisation and upregulates FoxM1 expression to promote G1/S cell cycle transition and cell proliferation (Ning et al., 2014). In another study conducted by Chen et al. (2019), miR-103/107 has been shown to repress the activity of Axin2 leading to sustained activation of Wnt/β-catenin signalling that potentiates cancer stemness and chemotherapeutic resistance. Nevertheless, despite that non-coding RNAs such as lncRNA and miRNA are known to promote resistance to therapeutic agents in CRC, the interaction network between LncRNA and miRNA is not well defined (Gao et al., 2019). Han et al. (2017) has reported that lncRNA Colorectal Neoplasia Differentially Expressed (CRNDE) binds to miR-181a-5p to repress its expression, resulting in increased levels of its downstream targets β-catenin and transcription factor TCF4 in the Wnt/β-catenin signaling pathway. It was also demonstrated that CRNDE knockdown and miR-181a-5p overexpression inhibit Wnt/β-catenin signaling and could reduce chemoresistance and attenuate cell proliferation in CRC cells, suggesting that it could be a novel cancer therapeutic strategy.
Over the years, substantial efforts have been directed to studying the molecular mechanisms and functional effects of Wnt signalling pathway. Accumulating studies confirm the critical role Wnt signalling in drug resistance and convey important insights into its underlying mechanisms that confer resistance to different therapies. Notably, such knowledge could be potentially harnessed to facilitate the development of specific inhibitors or drug combinations to improve anticancer efficacy. Nevertheless, owing to the complexity of Wnt signalling, there are still numerous details remain be uncovered with regard to its connections to therapy resistance that warrant further investigation.
Notch pathway
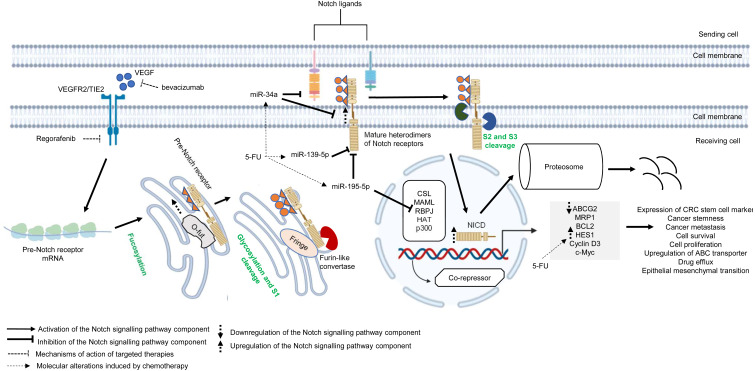
Similar to other signalling pathways (Wnt/β-catenin, Hedgehog (Hh), and transforming growth factor-beta (TGF-β)/bone morphogenic protein (BMP)), the Notch pathway is highly conserved across species and is known to control cell development, apoptosis, cell differentiation and proliferation (Artavanis-Tsakonas, Rand & Lake, 1999). Notch receptors (Notch 1–4) are synthesised as precursors from mRNAs (known as pre-Notch receptor) which then undergo fucosylation (a type of glycosylation) in the endoplasmic reticulum. In the Golgi apparatus, Notch receptors are further modified by enzymes (one typical example is Fringe) and cleaved at site 1 (S1) by furin-like convertase to induce heterodimerisation of Notch receptors (Siebel & Lendahl, 2017) (Fig. 4). At the cell surface, Notch receptors bind to Notch ligands (e.g., Jagged1, Jagged2, Delta-like ligand 1 (Dll1), Delta-like ligand 3 (Dll3) and Delta-like ligand 4 (Dll4)) of neighbouring cells. This initiates subsequent cleavages of Notch receptors by ADAM10/17 metalloproteases and presenilin–γ-secretase enzyme complex at the outer side (site 2 (S2)) and inner side (site 3 (S3)) of the cell membrane (Bray, 2006) (Fig. 4). The end product of the proteolytic cleavages of Notch receptors known as Notch intracellular cleaved domain (NICD, an active form of the molecules which acts as transcriptional activators), travels to the nucleus to displace co-repressors (e.g., recombining binding protein J-kappa (RBPJκ) or CSL) and interacts with transcriptional co-activators (e.g., mastermind-like (MAML), histone acetyltransferase (HAT), p300), in order to activate the transcription of Notch target genes (e.g., Hes and Hey family proteins, cyclin D3, c-Myc). Notch signalling is terminated when the intracellular domain of Notch (ICN or NICD) is targeted for proteasomal degradation through a ubiquitin pathway (Vinson et al., 2016; Siebel & Lendahl, 2017) (Fig. 4). In multidrug-resistant CRC, cross-regulation of signalling pathways, post-transcriptional regulation and overexpression of genes in the Notch signalling pathway are among the common mechanisms underlying the development of resistance towards targeted or chemotherapeutic regimens (Rodilla et al., 2009; Majidinia et al., 2018; Negri et al., 2019).
Figure 4. Deregulation of the canonical Notch signalling pathway during CRC treatment.
Region highlighted grey represents Notch target genes that regulate the biological processes of CRC cells treated with drug therapy.
There is growing evidence that upregulation of Notch receptors, Notch ligands and Notch target genes could lead to the maintenance of CRC stem cell populations and the acquisition of metastatic phenotype which are strongly related to poor survival of CRC patients and drug resistance (Mohamed et al., 2019; Weber et al., 2019; Shaik et al., 2020). It has been previously reported that Notch-1 signalling pathway induces EMT in non-small cell lung cancer resistant cells with acquired resistance to EGFR tyrosine kinase inhibitor, suggesting that Notch signalling may contribute to cancer drug resistance (Xie et al., 2013). In another study, Mirone et al. (2016) has reported that CRC cells that are resistant to regorafenib (a small-molecule multi kinase inhibitor) showed significant upregulation of Notch-1 and the target genes (HES1 and HEY1). The study demonstrated that knockdown of Notch-1 could partially restore the sensitivity to regorafenib and inhibit cell growth, indicating that Notch-1 may play a role in tumour resistance. In mCRC patients treated with bevacizumab-based therapy, high NICD expression was found to be associated with poorer response whereas no correlation was observed between Dll-4 expression and clinical response (Negri & Ardizzoni, 2015). In a follow-up study, Negri et al. (2019) has shown that high expression levels of NICD and CD44 are linked to cancer stemness in patients with advanced CRC treated with bevacizumab. Notably, the study demonstrated that NICDs (the functional components of Notch signalling pathway) instead of Dll-4 induce resistance to anti-angiogenic therapy in CRC via activation of Notch-induced regulation of colon cancer stem cells. Nevertheless, the functional roles of NICDs and CD44s in the CRC microenvironment during anti-angiogenic treatment are still unclear (Negri et al., 2019). HES1, which is a downstream target of Notch pathway and one of the important markers of CRC stem cells, is known to contribute to tumour relapses in CRC patients after 5-FU based chemotherapy, but the role of HES1 in chemoresistant CRC has not yet been elucidated (Gao et al., 2014). A recent study by Sun et al. (2017) has shown that HES1 modulates gene expression related to drug metabolism and EMT, notable overexpression of ABC transporters (ABCC1, ABCC2 and P-gp1) with depressed E-cadherins and elevated N-cadherins in CRC cell lines treated with 5-FU, supporting the crucial role of HES1 in promoting chemoresistance.
miRNAs are known to regulate Notch signalling pathway which results in various tumour pathology, such as metastasis, tumour relapses, cancer stemness and low survival rate (Majidinia et al., 2018; Khan et al., 2019). Recently, accumulating evidence also suggests that cross-regulation between miRNAs and Notch signalling pathway plays a critical role in cancer drug resistance. miR-139-5p is a tumour suppressor that has been found to be frequently downregulated in CRC (Shen et al., 2014). It has been reported that miR-139-5p targets Notch-1 and regulates its signal transduction to exert tumour suppressive effect in CRC (Zhang et al., 2014b). Furthermore, miR-139-5p/Notch-1 signalling has also been correlated with drug resistance in CRC. Liu et al. (2016) has shown that miR-139-5p sensitises CRC cells to 5-FU by inhibiting Notch-1 and its downstream multidrug-resistant genes (MRP-1 and BCL-2). Similarly, miR-195-5p is known to suppress cancer growth by inhibiting cell cycle progression, cell proliferation and cell migration (Luo et al., 2014). A recent study by Jin et al. (2018) has revealed that miR-195-5p could inhibit CRC cell stemness and 5-FU resistance by targeting the Notch signalling proteins Notch-2 and RBPJ, suggesting that miR-195-5p could be a potential therapeutic target in chemoresistance. On the other hand, a recent study by Xie et al. (2020b) reported that miR-34a could negatively regulate multidrug resistance protein ABCG2 via DLL1-mediated Notch signalling pathway and demonstrated that overexpression of miR-34a could overcome 5-FU resistance in CRC cells.
Overall, these findings suggest the critical role of Notch signalling pathway in cancer drug resistance. Notably, recent evidence indicates that Notch contribute to the maintenance of CRC stem cells and resistance to therapeutic agents, hence targeting Notch pathway may hold a promising prospect for cancer therapy. Thus, further studies are needed to elucidate the underlying mechanisms and the crosstalk between Notch and other signalling pathways to facilitate the design of better therapeutic approach.
Crosstalk of signalling pathways
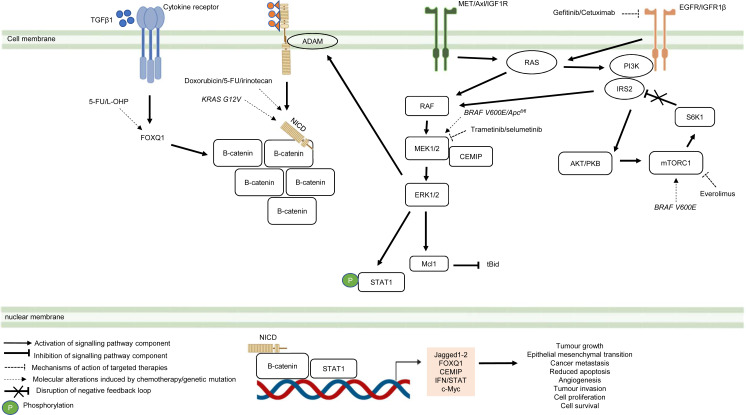
Besides deregulated signalling events mediated by single pathway during CRC treatment, the pathological link between the crosstalk of signalling pathways and the acquisition of drug resistance in CRC has also been documented in numerous studies (Fig. 5) (Hiraki et al., 2015; Ahmed et al., 2015; Zou et al., 2017; Mesange et al., 2018). It has been shown that MAPK-mediated pathway interacts with other signalling pathways to induce drug resistance in CRC (Watanabe et al., 2011). Tyrosine kinase inhibitors such as gefitinib (EGFR inhibitor) desensitises CRC cells to the antitumour effect of the drugs by promoting the heterodimerisation of EGFR and IGF1Rβ, leading to cross-regulation of the IGFR1β and MAPK signalling pathways (Yang et al., 2011). The EGFR signalling pathway has also been reported to cooperate with the MAPK signalling pathways mediated by RTKs (MET, Axl, and IGF1R) to promote resistance against EGFR inhibitors such as cetuximab (Hu et al., 2016). In addition to MAPK-based signalling involving RTKs, KRAS-mediated activation of the MAPK signalling pathway in CRC has also been demonstrated to confer resistance to MEK inhibition by instigating STAT1 phosphorylation and the activation of IFN/STAT signalling (Sakahara et al., 2019). Additionally, research findings also suggest that BRAF V600E regulates the crosstalk of the KRAS-mediated MAPK signalling pathway with other signalling pathways in multidrug-resistant CRC (He et al., 2018; Duong et al., 2018). BRAF V600E promotes the expression of endosomal protein CEMIP via a β-catenin-dependant pathway that sustains ERK1/2 activation after MEK1 inhibition. The crosstalk between the Wnt/β-catenin and MAPK signalling pathways that involves CEMIP enhances the expression of c-Myc to promote cell survival (Duong et al., 2018). Inhibition of mTORC1 in BRAF V600E CRC has also been shown to disrupt the S6K1-IRS-2/PI3K negative feedback loop, leading to ERK-dependant Mcl-1 stabilisation which blocks apoptosis (He et al., 2018).
Figure 5. Crosstalk of signalling pathways during CRC treatment.
Region highlighted orange represents substrates that regulate the biological processes of CRC cells treated with drug therapy.
On the hand, several reports have shown that there is crosstalk between Notch and other signalling pathways that are involved in the development of chemoresistance. It has been demonstrated that Notch-1 signalling pathway could activate Wnt/β-catenin pathway by NICD-1-mediated translocation of β-catenin to the nucleus upon binding of Notch-1 to its ligands (Ishiguro et al., 2017). On the other hand, it has been reported that the activation of Notch signalling pathway in CRC cell lines is mediated by β-catenin through up-regulation of Jagged1 (Rodilla et al., 2009). Furthermore, it has been shown that β-catenin promotes the expression of Jagged2 in CRC and contributes to tumour resistance to chemotherapy through modulation of p21 (Vaish, Kim & Shim, 2017). Besides, it has been reported that Notch and Wnt pathways were both upregulated and associated with the development of chemoresistance in CRC cells by upregulating HES1 expression (Kukcinaviciute et al., 2018). Interestingly, a recent study has revealed that Dvl scaffold protein acts as a key regulator of the Wnt and Notch crosstalk (Zhao et al., 2020). Findings from the study highlighted that Dvl-3 might have a functional role in the acquisition of Methotrexate (MTX, an inhibitor of the dihydrofolate reductase (DHFR) enzyme) resistance and stem cell-like properties in CRC cell lines, although the mechanistic details of the resistance still remain unknown. Also, it has been found that Notch pathway could mediate chemoresistance via crosstalk with KRAS pathway. It has been previously reported that KRAS mutations regulate growth factor shedding following chemotherapy treatment via the MEK/Erk/ADAM17 signalling axis and contribute to drug resistance in CRC tumours (Van Schaeybroeck et al., 2011). In a recent study conducted by Pelullo et al. (2019), it has been demonstrated that Jagged-1-ICDs (Jag1-ICDs) are produced by aberrant Jagged1 processing via KRAS/Erk/ADAM pathway in CRC tumours with mutant KRAS. The study highlighted a novel role of Jag1-ICD beyond the canonical Notch signalling in mediating the oncogenic KRAS pathway, which promotes malignant behaviour and confers chemoresistance to CRC cells.
Besides the crosstalk between the Wnt/β-catenin and Notch signalling pathways, research evidence also suggests that TGF β1 induces the expression of FOXQ1 to promote the nuclear translocation of β-catenin. FOXQ1-mediated crosstalk of the Wnt/β-catenin and TGFβ1 signalling pathways results in resistance to chemotherapy drug-induced apoptosis, EMT and tumour invasion (Peng et al., 2015). In view of the importance of the crosstalk of signalling pathways in CRC drug resistance, future research therefore should aim to identify key regulators that mediate the such interaction to provide a holistic view of the resistance mechanism.
Challenges and future perspectives
A variety of signalling pathways have also been demonstrated to induce drug resistance in CRC, other than the four signal transduction pathways discussed above (Lazzari et al., 2019; Kadioglu et al., 2021). For example, the activation of Hedgehog (Hh)-GLI pathway has been found to mediate the acquisition of chemoresistance via GLI-induced upregulation of ABC transporters in CRC (Po et al., 2020). It has also been reported that bone morphogenetic protein-2 (BMP-2) signalling activates STAT3 and promotes EMT and colon cancer stemness in CRC, which contribute to drug resistance (Kim et al., 2015). Likewise, studies have also suggested that Smad3/4 and IFN play important role in regulating multidrug-resistant CRC via STAT signalling (Moon et al., 2015; Sakahara et al., 2019). Some studies also feature other signalling events such as Hedgehog signalling in CRC during chemotherapy or molecular-targeted therapy, implying the underlying complexity of signalling mechanisms in multidrug-resistant CRC (Tang et al., 2018; Park et al., 2019).
On top of non-coding RNAs-based regulation of therapeutic resistance in CRC, epigenetic changes that involve DNA methylation and histone modifications have also increasingly been reported (Shen et al., 2018; Mahalakshmi, Husayn Ahmed & Mahadevan, 2018; Porcellini et al., 2018; Rezapour et al., 2019). For instances, hypermethylation of genes such as MEIS2, SLFN11 and B4GALT1 are associated with cancer progression and resistance to chemotherapy (cisplatin or oxaliplatin-based therapy) and anti-EGFR therapy (Baharudin et al., 2017; He et al., 2017; Picardo et al., 2019; Wang et al., 2019). Mechanistically, DNA methylation regulates the expression of miRNAs which influence the activity of signalling proteins in CRC with MSI-H to initiate anti-cancer drug resistance and cancer development (Shi et al., 2018). In addition, the regulatory function of histone methylation in multidrug-resistant CRC is also documented in studies that report the relationship between H3K27me3 level and oxaliplatin-induced apoptosis (Wang et al., 2020b), as well as the cancer-driving nature of histone methyltransferase SETDB1 in CRC resistant to cetuximab (Hou et al., 2020).
Besides cell-autonomous mechanisms of drug resistance, the association between tumour microenvironment (TME)-driven CRC pathogenesis and therapeutic failure has also been detailed in various studies (Hu et al., 2020; Ren et al., 2018; Hu et al., 2019; Jackstadt et al., 2019). Cancer-associated fibroblasts (CAFs), which constitute a main cellular component of the TME, have been identified as a key mediator of drug resistance in CRC by transferring exosomes to CRC cells (Kahlert & Kalluri, 2013; Herrera et al., 2018). Research evidence suggests that the transportation of exosomal miR-92a-3p from CAFs to CRC activates the Wnt/β-catenin pathway and inhibits mitochondrial apoptosis by targeting FBXW7 and MOAP1, contributing to cancer progression and chemotherapy resistance (Hu et al., 2019). The crosstalk between CAFs and CRC also involves the transfer of exosomal lncRNA H19 from CAFs to CRC cells (Ren et al., 2018). H19 activates the Wnt/β-catenin pathway by acting as a RNA sponge for miR-141, which inhibits the stemness of CRC cells (Ren et al., 2018). Correspondingly, exosomes derived from CRC cells also contain factors essential for reprogramming normal colonic fibroblast into CAFs which may in turn lead to chemoresistance in CRC (Rai et al., 2019). Potential strategies for CAFs-induced drug resistance in CRC include (i) suppressing the transformation of CAFs using small-molecule MSI-N1014 (Yadav et al., 2020), (ii) blocking tumoral IL1β-mediated signalling in normal colonic fibroblasts to thwart inflammatory CAF activation (Díaz-Maroto et al., 2021), and (iii) selective targeting of CAFs by engineered nanoparticles loaded with pro-apoptotic drug (Sitia et al., 2021). However, the failure to identify CRC subclones that mediate functional reprogramming of CAFs remains a big therapeutic hurdle for treating drug resistance in CRC that needs to be addressed. Furthermore, the components within the TME have also been reported to interact via multiple signal transduction pathways to confer drug resistance in CRC (Margolin et al., 2011). Paracrine signalling initiated by IL-17 derived from TH17 cells involves the crosstalk of the ERK pathway and NF-κB pathway to induce G-CSF expression, leading to the recruitment of immature myeloid cells to the TME and tumour resistance to anti-angiogenic therapy (Chung et al., 2013). Noncanonical TGFβ pathway interacts with PI3K/PKB pathway to sustain fibroblast activation during molecular-targeted treatment that inhibits IL-1β/TGF β signalling (Díaz-Maroto et al., 2019).
In recent decades, in addition to the conventional chemotherapy and targeted therapy, immunotherapy such as immune checkpoint blockade (ICB) has emerged as a promising strategy for cancer therapeutic. ICB works by inhibiting immune checkpoints to facilitate the activation of cytotoxic T cells and enhance anti-tumour immune response (Dosset et al., 2018; Woolston et al., 2019). The acquisition of resistance against ICB therapy, including those targeting cytotoxic T lymphocyte-associated protein 4 (CTLA-4), programmed death 1 (PD-1) and programmed death-ligand 1 (PD-L1), is well documented in numerous studies but the underlying mechanisms are still not well characterized (Gurjao et al., 2019; Liao et al., 2019). Molecular events associated with resistance against anti-PD-1 therapy include (i) suppressing IFN-γ-Stat1-Irf1 signalling in CRC and reducing cytotoxic tumour-infiltrating CD8+T cells by m6A methyltransferases-mediated downregulation of STAT1 (Wang et al., 2020a), (ii) the reduction of tumour suppressive myeloid cells via intracellular signalling initiated by myeloid receptor TREM2 (Molgora et al., 2020) and (iii) activating oncogenic myeloid-derived suppressor cells and regulatory T cells by enhanced transcription of the gene that encodes lactate transporter which induces lactate secretion (Li et al., 2020a; Li et al., 2020b). Emerging studies have also highlighted that several oncogenic signalling pathways that are involved in regulating immune response that renders resistance to ICB. Activation of the Wnt/β-catenin signalling pathway has been reported to be associated with a lack of T-cell infiltration within the tumour microenvironment in cancer patients (Spranger, Bao & Gajewski, 2015). This immunological defect was found to be mediated by decreased production of chemokine CCL4 that suppresses the recruitment of CD103+ dendritic cells, resulting in resistance to the immunotherapy. The interferon (IFN) signalling is another signalling pathway that has been implicated in resistance to ICB therapy. Patients that did not respond to anti-CTLA-4 antibody ipilimumab therapy was reported to harbour mutations in the interferon gamma (IFN-γ) pathway genes leading to the ability of the tumour cells to escape from T cells, which was identified as a primary resistance to anti-CTLA-4 therapy (Gao et al., 2016). Nevertheless, the roles of other signalling pathways that are involved in modulating the sensitivity and resistance to ICB are still largely unknown and require further studies. The current strategy to tackle anti-PD1 resistance involves reprogramming immunosuppressive myeloid cells to promote the expansion of tumour suppressive M1 macrophages (Lu et al., 2021).
In addition to CAFs and immune cells as the regulators of drug resistance in CRC, the emerging role of gut microbiota in the regulation of innate immune signalling and autophagy, which leads to chemoresistance in CRC, has also been reported. This suggests that crosstalk between different components of the TME has functional relevance in CRC development and clinical outcome, which prompts further investigation (Yu et al., 2017).
Tumour heterogeneity which is the cornerstone for the maintenance of cancer cell populations has been strongly correlated with therapy resistance (Schumacher et al., 2019). Mounting evidence has demonstrated the existence of various forms of tumour heterogeneity with the most frequently observed type being the genetic heterogeneity (Di et al., 2019; Loeb et al., 2019). Intra-tumour or inter-tumour genetic status was shown to influence the prognostic outcome and drug response and are determinants of resistance to anti-cancer therapy (Russo et al., 2016; De Angelis et al., 2016; Galofré et al., 2020; Bruun et al., 2020). Other types of tumour heterogeneity include cell type heterogeneity which is found between CRC subpopulations in the TME, as reported by Yoon et al. (2019) which showed that T-cell densities is highly variable in DNA mismatch repair-deficient tumour as compared to DNA mismatch repair-proficient tumour.
Tumour heterogeneity can be further classified into metabolic heterogeneity due to metabolic reprogramming in cells that contributes to disease development (Katoh, 2017). Metabolic reprogramming or alterations in the cellular metabolism is an important cancer hallmark to meet the increased energy and nutrient demand of malignant cells to promote tumour development. Notably, emerging evidence suggests that metabolic reprogramming could also contribute to resistance to antitumor drugs (Teng et al., 2017; Yu et al., 2021). The underlying mechanisms of metabolic adaptation during the development of drug resistance are still unclear, but available data implies that activation of oncogenic pathways are involved in the regulation of metabolic reprogramming implicated in resistance. Vellinga and colleagues (Vellinga et al., 2015) demonstrated that tumour metabolism was shifted from glycolysis towards oxidative phosphorylation in colon cancer cells that were exposed to chemotherapy to support tumour survival during treatment. It was discovered that the enhanced oxidative metabolism was mediated by histone deacetylase sirtuin-1 (SIRT1) and its substrate, the transcriptional coactivator PGC1α [239]. The study further showed that knockdown of SIRT1 or PGC1α sensitized the tumour cells to the drug treatment, suggesting that the SIRT1/PGC1α is a novel pathway of drug resistance that may be targeted for therapy. More recently, Barisciano et al. (2020) reported the role for miR-27a as a key regulator of metabolic reprogramming and enhancing drug resistance in CRC cells. The study revealed that miR-27a modulates several tumour-associated pathways that link metabolic rewiring with chemoresistance in CRC. It was found that miR-27a negatively regulates AMPK and positively regulates mTOR pathway to force anaerobic glycolytic metabolism supporting tumour growth and chemoresistance (Barisciano et al., 2020). Interestingly, a recent study has shown the colorectal tumour-derived exosomes could activate hepatic stellate cells in the liver to enhance lactate metabolism of tumour cells via the IL-6/STAT3 pathway to confer the resistance of SN38 (active metabolite of irinotecan) (Li et al., 2020a; Li et al., 2020b). Hence, this indicates a novel mechanism in which the tumour-derived exosomes are involved in regulating the metabolic reprogramming between the tumour cells and the microenvironment to promote drug resistance (Li et al., 2020a; Li et al., 2020b). Previous studies suggest that hypoxia-induced metabolic reprogramming of CRC can be reversed by targeting valine catabolism and the inhibition of PTEN/AKT/HIF1α signalling pathway to interfere with energy production in CRC (Wang et al., 2018; Shan et al., 2019). Alternative strategies to tackle metabolic heterogeneity in CRC warrant further investigation, given that the signalling mechanisms that contribute to metabolic reprogramming in CRC are complex.
Given the increasing complexity of molecular networks in multidrug-resistant CRC, multi-omics approaches encompassing genomics, epigenomics, transcriptomics, proteomics and metabolomics are applied to facilitate cancer biomarker discovery and guide cancer treatment strategies (Tong et al., 2016; Satoh et al., 2017; Ressa et al., 2018; Ishaque et al., 2018). Integrated multi-omics analyses of CRC cell lines have reported genetic and epigenetic alterations in the molecular landscape for CRC carcinogenesis such as (i) the identification of BRCA1-centred gene-miRNA-protein regulatory network as the main driver for liver metastasis of CRC and chemoresistance (Gerovska et al., 2020), (ii) co-occurrence of genetic alteration events in CRC that affects drug response (Zhou et al., 2020), (iii) heterogeneous Wnt/β-catenin activity that supports Runt-related transcription factor 2 (RUNX2)-based epigenetic regulation of EMT as the molecular implication for poor survival of CRC patients and failure of anti-cancer therapy (Yi et al., 2020), and (iv) the characterisation of CRC patients’ drug response patterns based on differential DNA methylation profiles in CRC stem cell populations (Visone et al., 2019). To better understand the relationship between immune landscape in the TME and drug efficacy against CRC, computational methods such as (i) the development of artificial intelligence platform to predict immunological responses to ICB therapy in MSI-H tumour (Cao et al., 2020) and (ii) the reconstruction of the intercellular network according to consensus molecular subtypes of CRC (Lee et al., 2020), have enabled the selection of better treatment options for individuals resistant to immunotherapy. Comprehensive analysis of multi-omics data such as metagenomic and metabolomic has also reported the role of gut microbiota in CRC progression and their influences on the DNA methylome of CRC, implying that the metabolic output of gut microbiota and the host’s epigenetic signatures could be important diagnostic targets for CRC management (Yachida et al., 2019; Sobhani et al., 2019; Zouggar, Haebe & Benoit, 2020).
Cancer biopsy-based bulk analysis of CRC is gradually replaced by patient-derived cancer organoid (PDCO) to study therapy resistance in vitro. This is because PDCO can recapitulate tumour heterogeneity in the patient tumour which potentiates the study of CRC at the single-cell level (Jeppesen et al., 2017; Chen et al., 2018c; Pasch et al., 2019; Demmers et al., 2020). However, limitations such as the time and cost to grow the organoids as well as limited amount of organoids available prompts the shift of focus to cancer tissue-originated spheroid (CTOS) as an alternative method for measuring chemotherapeutic heterogeneity and high-throughput drug screening for CRC patients (Jeppesen et al., 2017; Kondo et al., 2019). Technical advancement to improve the study of tumour heterogeneity in the spatial context is exemplified by RNA-based in situ hybridisation which complements the current genetic method to ease the detection of rare subclones in CRC (Baker et al., 2017). Non-invasive methods such as comprehensive genotyping of circulating tumour DNA (ctDNA) and integrated multi-omics data analyses for gene alteration events relating to drug responses, are also useful in identifying druggable mutational targets for personalising cancer medicine (Cao et al., 2019; Zhou et al., 2020). Besides improving on experimental methods to better understand tumour heterogeneity in CRC, the application of modern bioinformatic practice (e.g., reference component analysis of single-cell transcriptomes) and machine learning algorithm (e.g., deep learning for the prediction of treatment efficacy on CRC patients) are equally important to the management of increasingly complex cancer research data which influences cancer treatment policy (Li et al., 2017; Skrede et al., 2020; Vera-Yunca et al., 2020).
Conclusions
Therapy resistance in CRC remains a major obstacle to CRC management due to alterations in the molecular landscape that drive the survival of cancer cells. In particular, dysregulation of MAPK pathway, PI3K/PKB pathway, Wnt/β-catenin pathway and Notch pathway are frequently reported to induce resistance to anti-cancer drugs targeting CRC cells. On the other hand, other signal transduction pathways such as TGFβ/Smad, BMP and Hedgehog pathways have also been implicated in the development of therapeutic resistance in CRC but are not well studied yet. The complexity of drug resistance mechanisms is further widened by pre-existing genetic heterogeneity in CRC and cellular components of the TME (e.g., stromal cells, immune cells and gut bacteria) which results in the evolution of drug-resistant tumour subclones. To address this problem, integrated multi-omics data analysis using modern computational methods, three-dimensional cell culture model and other robust experimental methods are needed to identify new cancer biomarkers and drug targets for CRC treatment. Despite the effort in combating multidrug-resistant CRC, further studies are warranted to generate quality results for better cancer care delivery.
Disclaimer
Most experimental findings discussed this review are derived from studies using laboratory-based cancer cell lines. Hence, the results should be interpreted with caution and be further validated in animal models and human clinical studies.
Funding Statement
This work is supported by the Fundamental Research Grant Scheme (FRGS/1/2019/SKK08/ UKM/03/5) awarded by the Ministry of Higher Education, Malaysia. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Yeelon Yeoh performed the experiments, analyzed the data, prepared figures and/or tables, and approved the final draft.
Teck Yew Low and Nadiah Abu conceived and designed the experiments, authored or reviewed drafts of the paper, and approved the final draft.
Pey Yee Lee conceived and designed the experiments, performed the experiments, analyzed the data, authored or reviewed drafts of the paper, and approved the final draft.
Data Availability
The following information was supplied regarding data availability:
There are no raw data
References
- Ahmed et al. (2015).Ahmed M, Hussain AR, Siraj AK, Uddin S, Al-Sanea N, Al-Dayel F, Al-Assiri M, Beg S, Al-Kuraya KS. Co-targeting of Cyclooxygenase-2 and FoxM1 is a viable strategy in inducing anticancer effects in colorectal cancer cells. Molecular Cancer. 2015;14:1–14. doi: 10.1186/s12943-015-0406-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahronian et al. (2015).Ahronian LG, Sennott EM, Van Allen EM, Wagle N, Kwak EL, Faris JE, Godfrey JT, Nishimura K, Lynch KD, Mermel CH, Lockerman EL, Kalsy A, Gurski JM, Bahl S, Anderka K, Green LM, Lennon NJ, Huynh TG, Mino-Kenudson M, Getz G, Dias-Santagata D, Iafrate AJ, Engelman JA, Garraway LA, Corcoran RB. Clinical acquired resistance to RAF inhibitor combinations in BRAF-mutant colorectal cancer through MAPK pathway alterations. Cancer Discovery. 2015;5:358–367. doi: 10.1158/2159-8290.CD-14-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alessi et al. (1997).Alessi DR, James SR, Downes CP, Holmes AB, Gaffney PRJ, Reese CB, Cohen P. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Bα. Current Biology. 1997;7:261–269. doi: 10.1016/s0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- Allegra et al. (2009).Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, Hayes DF, McAllister PK, Morton RF, Schilsky RL. American society of clinical oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. Journal of Clinical Oncology. 2009;27:2091–2096. doi: 10.1200/JCO.2009.21.9170. [DOI] [PubMed] [Google Scholar]
- Andrew et al. (2018).Andrew AS, Parker S, Anderson JC, Rees JR, Robinson C, Riddle B, Butterly LF. Risk factors for diagnosis of colorectal cancer at a late stage: a population-based study. Journal of General Internal Medicine. 2018;33:2100–2105. doi: 10.1007/s11606-018-4648-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angius et al. (2019).Angius A, Pira G, Scanu AM, Uva P, Sotgiu G, Saderi L, Manca A, Serra C, Uleri E, Piu C, Caocci M, Ibba G, Zinellu A, Cesaraccio MR, Sanges F, Muroni MR, Dolei A, Cossu-Rocca P, De Miglio MR. Microrna-425-5p expression affects BRAF/RAS/MAPK pathways in colorectal cancers. International Journal of Medical Sciences. 2019;16:1480–1491. doi: 10.7150/ijms.35269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki et al. (2007).Aoki K, Aoki M, Sugai M, Harada N, Miyoshi H, Tsukamoto T, Mizoshita T, Tatematsu M, Seno H, Chiba T, Oshima M, Hsieh CL, Taketo MM. Chromosomal instability byβ-catenin/TCF transcription in APC orβ-catenin mutant cells. Oncogene. 2007;26:3511–3520. doi: 10.1038/sj.onc.1210141. [DOI] [PubMed] [Google Scholar]
- Arango et al. (2004).Arango D, Wilson AJ, Shi Q, Corner GA, Arañes MJ, Nicholas C, Lesser M, Mariadason JM, Augenlicht LH. Molecular mechanisms of action and prediction of response to oxaliplatin in colorectal cancer cells. British Journal of Cancer. 2004;91:1931–1946. doi: 10.1038/sj.bjc.6602215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artavanis-Tsakonas, Rand & Lake (1999).Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Backer (2016).Backer JM. The intricate regulation and complex functions of the Class III phosphoinositide 3-kinase Vps34. Biochemical Journal. 2016;473:2251–2271. doi: 10.1042/BCJ20160170. [DOI] [PubMed] [Google Scholar]
- Baharudin et al. (2017).Baharudin R, Mutalib NSAb, Othman SN, Sagap I, Rose IM, Mokhtar NM, Jamal R. Identification of predictive DNA methylation biomarkers for chemotherapy response in colorectal cancer. Frontiers in Pharmacology. 2017;8:47. doi: 10.3389/fphar.2017.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker et al. (1989).Baker SJ, Fearon ER, Nigro JM, Hamilton SR, Preisinger AC, Jessup JM, Vantuinen P, Ledbetter DH, Barker DF, Nakamura Y, White R, Vogelstein B. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science. 1989;244:217–221. doi: 10.1126/science.2649981. [DOI] [PubMed] [Google Scholar]
- Baker et al. (2017).Baker AM, Huang W, Wang XMM, Jansen M, Ma XJ, Kim J, Anderson CM, Wu X, Pan L, Su N, Luo Y, Domingo E, Heide T, Sottoriva A, Lewis A, Beggs AD, Wright NA, Rodriguez-Justo M, Park E, Tomlinson I, Graham TA. Robust RNA-based in situ mutation detection delineates colorectal cancer subclonal evolution. Nature Communications. 2017;8:1998. doi: 10.1038/s41467-017-02295-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baretti et al. (2018).Baretti M, Personeni N, Destro A, Santoro A, Rimassa L. Emergence of KRAS-mutation in liver metastases after an anti-EGFR treatment in patient with colorectal cancer: are we aware of the therapeutic impact of intratumor heterogeneity? Cancer Biology and Therapy. 2018;19:659–663. doi: 10.1080/15384047.2018.1450117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barisciano et al. (2020).Barisciano G, Colangelo T, Rosato V, Muccillo L, Taddei M, Ippolito L, Chiarugi P, Galgani M, Bruzzaniti S, Matarese G, Fassan M, Agostini M, Bergamo F, Pucciarelli S, Carbone A, Mazzoccoli G, Colantuoni V, Bianchi F, Sabatino L. miR-27a is a master regulator of metabolic reprogramming and chemoresistance in colorectal cancer. British Journal of Cancer. 2020;122:1354–1366. doi: 10.1038/s41416-020-0773-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belli et al. (2019).Belli V, Matrone N, Napolitano S, Migliardi G, Cottino F, Bertotti A, Trusolino L, Martinelli E, Morgillo F, Ciardiello D, De Falco V, Giunta EF, Bracale U, Ciardiello F, Troiani T. Combined blockade of MEK and PI3KCA as an effective antitumor strategy in HER2 gene amplified human colorectal cancer models. Journal of Experimental and Clinical Cancer Research. 2019;38:236. doi: 10.1186/s13046-019-1230-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendell et al. (2014).Bendell JC, Atreya CE, André T, Tabernero J, Gordon MS, Bernards R, Van Cutsem E, Tejpar S, Sidhu R, Go WY, Allred A, Motwani M, Suttle BB, Wu Y, Hoos A, Orford KW, Corcoran RB, Schellens JH. Efficacy and tolerability in an open-label phase I/II study of MEK inhibitor trametinib (T), BRAF inhibitor dabrafenib (D), and anti-EGFR antibody panitumumab (P) in combination in patients (pts) with BRAF V600E mutated colorectal cancer (CRC) Journal of Clinical Oncology. 2014;32:3515–3515. doi: 10.1200/jco.2014.32.15_suppl.3515. [DOI] [Google Scholar]
- Benson et al. (2018).Benson AB, Venook AP, Al-Hawary MM, Cederquist L, Chen Y-J, Ciombor KK, Cohen S, Cooper HS, Deming D, Engstrom PF, Garrido-Laguna I, Grem JL, Grothey A, Hochster HS, Hoffe S, Hu S, Freedman-Cass DA. NCCN guidelines insights: colon cancer, version 2.2018. Journal of the National Comprehensive Cancer Network. 2018;16:359–369. doi: 10.6004/jnccn.2018.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin et al. (2007).Berlin J, Posey J, Tchekmedyian S, Hu E, Chan D, Malik I, Yang L, Amado RG, Randolph Hecht J. Panitumumab with irinotecan/leucovorin/5-fluorouracil for first-line treatment of metastatic colorectal cancer. Clinical Colorectal Cancer. 2007;6:427–432. doi: 10.3816/CCC.2007.n.011. [DOI] [PubMed] [Google Scholar]
- Bian et al. (2017).Bian Z, Zhang J, Li M, Feng Y, Yao S, Song M, Qi X, Fei B, Yin Y, Hua D, Huang Z. Long non-coding RNA LINC00152 promotes cell proliferation, metastasis, and confers 5-FU resistance in colorectal cancer by inhibiting miR-139-5p. Oncogenesis. 2017;6:395. doi: 10.1038/s41389-017-0008-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokemeyer et al. (2009).Bokemeyer C, Bondarenko I, Makhson A, Hartmann JT, Aparicio J, De Braud F, Donea S, Ludwig H, Schuch G, Stroh C, Loos AH, Zubel A, Koralewski P. Fluorouracil, leucovorin, and oxaliplatin with and without cetuximab in the first-line treatment of metastatic colorectal cancer. Journal of Clinical Oncology. 2009;27:663–671. doi: 10.1200/JCO.2008.20.8397. [DOI] [PubMed] [Google Scholar]
- Bosset et al. (2006).Bosset JF, Collette L, Calais G, Mineur L, Maingon P, Radosevic-Jelic L, Daban A, Bardet E, Beny A, Ollier JC. Chemotherapy with preoperative radiotherapy in rectal cancer. New England Journal of Medicine. 2006;355:1114–1123. doi: 10.1056/NEJMoa060829. [DOI] [PubMed] [Google Scholar]
- Boyer et al. (2004).Boyer J, Mclean EG, Aroori S, Wilson P, Mcculla A, Carey PD, Longley DB, Johnston PG, Carey PDeclan, Longley DB, Johnston PG. Characterization of p53 Wild-type and null isogenic colorectal cancer cell lines resistant to 5-Fluorouracil, Oxaliplatin, and Irinotecan. Clinical Cancer Research. 2004;10:2158–2167. doi: 10.1158/1078-0432.CCR-03-0362. [DOI] [PubMed] [Google Scholar]
- Bray (2006).Bray SJ. Notch signalling: a simple pathway becomes complex. Nature Reviews Molecular Cell Biology. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- Bruun et al. (2020).Bruun J, Kryeziu K, Eide PW, Moosavi SH, Eilertsen IA, Langerud J, Røsok B, Totland MZ, Brunsell TH, Pellinen T, Saarela J, Bergsland CH, Palmer HG, Brudvik KW, Guren T, Dienstmann R, Guren MG, Nesbakken A, Bjørnbeth BA, Sveen A, Lothe RA. Patient-derived organoids from multiple colorectal cancer liver metastases reveal moderate intra-patient pharmacotranscriptomic heterogeneity. Clinical Cancer Research. 2020;26:4107–4119. doi: 10.1158/1078-0432.CCR-19-3637. [DOI] [PubMed] [Google Scholar]
- Cancer Research UK (2019).Cancer Research UK CT colonography — Tests and scans — Cancer Research UK. 2019. [22 May 2020]. https://www.cancerresearchuk.org/about-cancer/cancer-in-general/tests/ct-colonography https://www.cancerresearchuk.org/about-cancer/cancer-in-general/tests/ct-colonography
- Cao et al. (2019).Cao W, Xu Y, Chang L, Gong Y, Li L, Mo X, Zhang X, Lin G, Zhou J, Liu D, Yi Y, Dai P, Zhu C, Liu T, Chu Y, Guan Y, Chen Y, Wang J, Xia X, Yang L, Yi X, Cheng Y. Genotyping of circulating tumor DNA reveals the clinically actionable mutation landscape of advanced colorectal cancer. Molecular Cancer Therapeutics. 2019;18:1158–1167. doi: 10.1158/1535-7163.MCT-18-1247. [DOI] [PubMed] [Google Scholar]
- Cao et al. (2020).Cao R, Yang F, Ma SC, Liu L, Zhao Y, Li Y, Wu DH, Wang T, Lu WJ, Cai WJ, Zhu HB, Guo XJ, Lu YW, Kuang JJ, Huan WJ, Tang WM, Huang K, Huang J, Yao J, Dong ZY. Development and interpretation of a pathomics-based model for the prediction of microsatellite instability in Colorectal Cancer. Theranostics. 2020;10:11080–11091. doi: 10.7150/thno.49864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carethers (2008).Carethers JM. Review: systemic treatment of advanced colorectal cancer: tailoring therapy to the tumor. Therapeutic Advances in Gastroenterology. 2008;1:33–42. doi: 10.1177/1756283X08093607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cargnello & Roux (2011).Cargnello M, Roux PP. Activation and Function of the MAPKs and Their Substrates, the MAPK-Activated Protein Kinases. Microbiology and Molecular Biology Reviews. 2011;75:50–83. doi: 10.1128/mmbr.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Piedras et al. (2018).Castro-Piedras I, Sharma M, Den Bakker M, Molehin D, Martinez EG, Vartak D, Pruitt WM, Deitrick J, Almodovar S, Pruitt K. DVL1 and DVL3 differentially localize to CYP19A1 promoters and regulate aromatase mRNA in breast cancer cells. Oncotarget. 2018;9:35639–35654. doi: 10.18632/oncotarget.26257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang et al. (2003).Chang F, Lee JT, Navolanic PM, Steelman LS, Shelton JG, Blalock WL, Franklin RA, McCubrey JA. Involvement of PI3K/Akt pathway in cell cycle progression, apoptosis, and neoplastic transformation: a target for cancer chemotherapy. Leukemia. 2003;17:590–603. doi: 10.1038/sj.leu.2402824. [DOI] [PubMed] [Google Scholar]
- Chen et al. (2018a).Chen G, Gao C, Gao X, Zhang DH, Kuan SF, Burns TF, Hu J. Wnt/β-catenin pathway activation mediates adaptive resistance to BRAF inhibition in colorectal cancer. Molecular Cancer Therapeutics. 2018a;17:806–813. doi: 10.1158/1535-7163.MCT-17-0561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen et al. (2018b).Chen XQ, Jiang J, Wang XT, Zhang CL, Ji AY, Chen XJ. Role and mechanism of Dv13 in the esophageal squamous cell carcinoma. European Review for Medical and Pharmacological Sciences. 2018b;22:7716–7725. doi: 10.26355/eurrev-201811-16393. [DOI] [PubMed] [Google Scholar]
- Chen et al. (2019).Chen HY, Lang YD, Lin HN, Liu YR, Liao CC, Nana AW, Yen Y, Chen RH. miR-103/107 prolong Wnt/β-catenin signaling and colorectal cancer stemness by targeting Axin2. Scientific Reports. 2019;9:9687. doi: 10.1038/s41598-019-41053-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen et al. (2018c).Chen KY, Srinivasan T, Lin C, Tung KL, Gao Z, Hsu DS, Lipkin SM, Shen X. Single-cell transcriptomics reveals heterogeneity and drug response of human colorectal cancer organoids. Proceedings of the annual international conference of the IEEE engineering in medicine and biology society, EMBS 2018-July; 2018c. pp. 2378–2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung et al. (2013).Chung AS, Wu X, Zhuang G, Ngu H, Kasman I, Zhang J, Vernes JM, Jiang Z, Meng YG, Peale FV, Ouyang W, Ferrara N. An interleukin-17-mediated paracrine network promotes tumor resistance to anti-angiogenic therapy. Nature Medicine. 2013;19:1114–1123. doi: 10.1038/nm.3291. [DOI] [PubMed] [Google Scholar]
- Corcoran et al. (2014).Corcoran RB, Atreya CE, Falchook GS, Infante JR, Hamid O, Messersmith WA, Daud A, Kwak EL, Ryan D, Kurzrock R, Sun P, Cunningham EA, Orford KW, Motwani M, Bai Y, Patel K, Venook AP, Kopetz S. Phase 1-2 trial of the BRAF inhibitor dabrafenib (D) plus MEK inhibitor trametinib (T) in BRAF V600 mutant colorectal cancer (CRC): updated efficacy and biomarker analysis. Journal of Clinical Oncology. 2014;32:3517–3517. doi: 10.1200/jco.2014.32.15_suppl.3517. [DOI] [Google Scholar]
- Corcoran et al. (2012).Corcoran RB, Ebi H, Turke AB, Coffee EM, Nishino M, Cogdill AP, Brown RD, Pelle P Della, Dias-Santagata D, Hung KE, Flaherty KT, Piris A, Wargo JA, Settleman J, Mino-Kenudson M, Engelman JA. EGFR-mediated reactivation of MAPK signaling contributes to insensitivity of BRAF-mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discovery. 2012;2:227–235. doi: 10.1158/2159-8290.CD-11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham et al. (2004).Cunningham D, Humblet Y, Siena S, Khayat D, Bleiberg H, Santoro A, Bets D, Mueser M, Harstrick A, Verslype C, Chau I, Van Cutsem E. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan- refractory metastatic colorectal cancer. New England Journal of Medicine. 2004;351:337–345. doi: 10.1056/NEJMoa033025. [DOI] [PubMed] [Google Scholar]
- Danielsen et al. (2015).Danielsen SA, Eide PW, Nesbakken A, Guren T, Leithe E, Lothe RA. Portrait of the PI3K/AKT pathway in colorectal cancer. Biochimica et Biophysica Acta - Reviews on Cancer. 2015;1855:104–121. doi: 10.1016/j.bbcan.2014.09.008. [DOI] [PubMed] [Google Scholar]
- Davies et al. (2002).Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JWC, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Wooster R, Stratton MR, Futreal PA. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- De Angelis et al. (2016).De Angelis ML, Zeuner A, Policicchio E, Russo G, Bruselles A, Signore M, Vitale S, De Luca G, Pilozzi E, Boe A, Stassi G, Ricci-Vitiani L, Amoreo CA, Pagliuca A, Francescangeli F, Tartaglia M, De Maria R, Baiocchi M. Cancer stem cell-based models of colorectal cancer reveal molecular determinants of therapy resistance. Stem Cells Translational Medicine. 2016;5:511–523. doi: 10.5966/sctm.2015-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gramont et al. (2000).De Gramont A, Figer A, Seymour M, Homerin M, Hmissi A, Cassidy J, Boni C, Cortes-Funes H, Cervantes A, Freyer G, Papamichael D, Le Bail N, Louvet C, Hendler D, De Braud F, Wilson C, Morvan F, Bonetti A. Leucovorin and fluorouracil with or without oxaliplatin as first-line treatment in advanced colorectal cancer. Journal of Clinical Oncology. 2000;18:2938–2947. doi: 10.1200/JCO.2000.18.16.2938. [DOI] [PubMed] [Google Scholar]
- De Roock et al. (2010).De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, Kalogeras KT, Kotoula V, Papamichael D, Laurent-Puig P, Penault-Llorca F, Rougier P, Vincenzi B, Santini D, Tonini G, Cappuzzo F, Frattini M, Molinari F, Saletti P, De Dosso S, Martini M, Bardelli A, Siena S, Sartore-Bianchi A, Tabernero J, Macarulla T, Di Fiore F, Gangloff AO, Ciardiello F, Pfeiffer P, Qvortrup C, Hansen TP, Van Cutsem E, Piessevaux H, Lambrechts D, Delorenzi M, Tejpar S. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. The Lancet Oncology. 2010;11:753–762. doi: 10.1016/S1470. [DOI] [PubMed] [Google Scholar]
- Dekker et al. (2019).Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. The Lancet. 2019;394:1467–1480. doi: 10.1016/S0140-6736(19)32319-0. [DOI] [PubMed] [Google Scholar]
- Demmers et al. (2020).Demmers LC, Kretzschmar K, Van Hoeck A, Bar-Epraïm YE, van den Toorn HWP, Koomen M, Van Son G, Van Gorp J, Pronk A, Smakman N, Cuppen E, Clevers H, Heck AJR, Wu W. Single-cell derived tumor organoids display diversity in HLA class I peptide presentation. Nature Communications. 2020;11:5338. doi: 10.1038/s41467-020-19142-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di et al. (2019).Di J, Yang H, Wang Z, Yang J, Gao P, Jiang B, Su X. Clonality and heterogeneity of metachronous colorectal cancer. Molecular Carcinogenesis. 2019;58:447–457. doi: 10.1002/mc.22947. [DOI] [PubMed] [Google Scholar]
- Diaz et al. (2012).Diaz LA, Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, Allen B, Bozic I, Reiter JG, Nowak MA, Kinzler KW, Oliner KS, Vogelstein B. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537–540. doi: 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Maroto et al. (2021).Díaz-Maroto NG, Garcia-Vicién G, Polcaro G, Bañuls M, Albert N, Villanueva A, Molleví DG. The blockade of tumoral il1β-mediated signaling in normal colonic fibroblasts sensitizes tumor cells to chemotherapy and prevents inflammatory caf activation. International Journal of Molecular Sciences. 2021;22:4960. doi: 10.3390/ijms22094960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Maroto et al. (2019).Díaz-Maroto NG, Sanz-Pamplona R, Berdiel-Acer M, Cimas FJ, García E, Gonçalves-Ribeiro S, Albert N, Garcia-Vicien G, Capella G, Moreno V, Salazar R, Villanueva A, Mollev DG. Noncanonical TGFb pathway relieves the blockade of IL1B/TGFb-mediated crosstalk between tumor and stroma: TGFBR1 and TAK1 inhibition in colorectal cancer. Clinical Cancer Research. 2019;25:4466–4479. doi: 10.1158/1078-0432.CCR-18-3957. [DOI] [PubMed] [Google Scholar]
- Dimitriou & Griniatsos (2015).Dimitriou N, Griniatsos J. Complete mesocolic excision: techniques and outcomes. World Journal of Gastrointestinal Oncology. 2015;7:383–388. doi: 10.4251/wjgo.v7.i12.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosset et al. (2018).Dosset M, Vargas TR, Lagrange A, Boidot R, Végran F, Roussey A, Chalmin F, Dondaine L, Paul C, Marie-Joseph EL, Martin F, Ryffel B, Borg C, Adotévi O, Ghiringhelli F, Apetoh L. PD-1/PD-L1 pathway: an adaptive immune resistance mechanism to immunogenic chemotherapy in colorectal cancer. OncoImmunology. 2018;7:e1433981. doi: 10.1080/2162402X.2018.1433981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douillard et al. (2000).Douillard JY, Cunningham D, Roth AD, Navarro M, James RD, Karasek P, Jandik P, Iveson T, Carmichael J, Alakl M, Gruia G, Awad L, Rougier P. Irinotecan combined with fluorouracil compared with fluorouracil alone. as first-line treatment for metastatic colorectal cancer: a multicentre randomised trial. The Lancet. 2000;355:1041–1047. doi: 10.1016/S0140-6736(00)02034-1. [DOI] [PubMed] [Google Scholar]
- Duong et al. (2018).Duong HQ, Nemazanyy I, Rambow F, Tang SC, Delaunay S, Tharun L, Florin A, Buttner R, Vandaele D, Close P, Marine JC, Shostak K, Chariot A. The endosomal protein CEMIP links WNT signaling to MEK1ERK1/2 activation in selumetinib-resistant intestinal organoids. Cancer Research. 2018;78:4533–4548. doi: 10.1158/0008-5472.CAN-17-3149. [DOI] [PubMed] [Google Scholar]
- Durrant & Morrison (2018).Durrant DE, Morrison DK. Targeting the Raf kinases in human cancer: the Raf dimer dilemma. British Journal of Cancer. 2018;118:3–8. doi: 10.1038/bjc.2017.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo et al. (2020).Endo E, Okayama H, Saito K, Nakajima S, Yamada L, Ujiie D, Kase K, Fujita S, Endo H, Sakamoto W, Saito M, Saze Z, Momma T, Ohki S, Mimura K, Kono K. A TGFb-dependent stromal subset underlies immune checkpoint inhibitor efficacy in DNA mismatch repair-deficient/microsatellite instability-high colorectal cancer. Molecular Cancer Research. 2020;18:1402–1413. doi: 10.1158/1541-7786.MCR-20-0308. [DOI] [PubMed] [Google Scholar]
- Falasca & Maffucci (2012).Falasca M, Maffucci T. Regulation and cellular functions of class II phosphoinositide 3-kinases. Biochemical Journal. 2012;443:587–601. doi: 10.1042/BJ20120008. [DOI] [PubMed] [Google Scholar]
- Fang & Richardson (2005).Fang JY, Richardson BC. The MAPK signalling pathways and colorectal cancer. The Lancet Oncology. 2005;6:322–327. doi: 10.1016/S1470-2045(05)70168-6. [DOI] [PubMed] [Google Scholar]
- Fearon & Vogelstein (1990).Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-I. [DOI] [PubMed] [Google Scholar]
- Frattini et al. (2007).Frattini M, Saletti P, Romagnani E, Martin V, Molinari F, Ghisletta M, Camponovo A, Etienne LL, Cavalli F, Mazzucchelli L. PTEN loss of expression predicts cetuximab efficacy in metastatic colorectal cancer patients. British Journal of Cancer. 2007;97:1139–1145. doi: 10.1038/sj.bjc.6604009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruman et al. (2017).Fruman DA, Chiu H, Hopkins BD, Bagrodia S, Cantley LC, Abraham RT. The PI3K pathway in human disease. Cell. 2017;170:605–635. doi: 10.1016/j.cell.2017.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galofré et al. (2020).Galofré C, Geyik ÖG, Asensio E, Wangsa D, Hirsch D, Parra C, Saez J, Mollà M, Yüce Z, Castells A, Ried T, Camps J. Tetraploidy-associated genetic heterogeneity confers chemo-radiotherapy resistance to colorectal cancer cells. Cancer. 2020;12:1118. doi: 10.3390/cancers12051118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao & Chen (2010).Gao C, Chen YG. Dishevelled: the hub of Wnt signaling. Cellular Signalling. 2010;22:717–727. doi: 10.1016/j.cellsig.2009.11.021. [DOI] [PubMed] [Google Scholar]
- Gao et al. (2019).Gao Z, Fu P, Yu Z, Zhen F, Gu Y. Comprehensive analysis of lncRNA-miRNA- mRNA network ascertains prognostic factors in patients with colon cancer. Technology in Cancer Research & Treatment. 2019;18:1533033819853237. doi: 10.1177/1533033819853237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao et al. (2011).Gao J, Liu J, Fan D, Xu H, Xiong Y, Wang Y, Xu W, Wang Y, Cheng Y, Zheng G. Up-regulated expression of Notch1 and Jagged1 in human colon adenocarcinoma. Pathologie Biologie. 2011;59:298–302. doi: 10.1016/j.patbio.2010.11.001. [DOI] [PubMed] [Google Scholar]
- Gao et al. (2016).Gao J, Shi L, Zhao H, Chen J, Xiong L, He Q, Chen T, Roszik J, Bernatchez C, Woodman S, Chen P, Hwu P, Allison J, Futreal A, Wargo J, Sharma P. Loss of IFN-γ pathway genes in tumor cells as a mechanism of resistance to Anti-CTLA-4 therapy. Cell. 2016;167:397–404. doi: 10.1016/j.cell.2016.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao et al. (2014).Gao F, Zhang Y, Wang S, Liu Y, Zheng L, Yang J, Huang W, Ye Y, Luo W, Xiao D. Hes1 is involved in the self-renewal and tumourigenicity of stem-like cancer cells in colon cancer. Scientific Reports. 2014;4:1–9. doi: 10.1038/srep03963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach et al. (2014).Gerlach JP, Emmink BL, Nojima H, Kranenburg O, Maurice MM. Wnt signalling induces accumulation of phosphorylatedβ-catenin in two distinct cytosolic complexes. Open Biology. 2014;4:140120. doi: 10.1098/rsob.140120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerovska et al. (2020).Gerovska D, Larrinaga G, Solano-Iturria JD, Márquez J, Gallastegi PG, Khatib AM, Poschmann G, Stühler K, Armesto M, Lawrie CH, Badiola I, Araúzo-Bravo MJ. An integrative omics approach reveals involvement of BRCA1 in hepatic metastatic progression of colorectal cancer. Cancer. 2020;12:1–24. doi: 10.3390/cancers12092380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomini et al. (2010).Giacomini KM, Huang SM, Tweedie DJ, Benet LZ, Brouwer KLR, Chu X, Dahlin A, Evers R, Fischer V, Hillgren KM, Hoffmaster KA, Ishikawa T, Keppler D, Kim RB, Lee CA, Niemi M, Polli JW, Sugiyama Y, Swaan PW, Ware JA, Wright SH, Wah Yee S, Zamek-Gliszczynski MJ, Zhang L. Membrane transporters in drug development. Nature Reviews Drug Discovery. 2010;9:215–236. doi: 10.1038/nrd3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giulianotti et al. (2003).Giulianotti PC, Coratti A, Angelini M, Sbrana F, Cecconi S, Balestracci T, Caravaglios G. Robotics in general surgery: personal experience in a large community hospital. Archives of Surgery. 2003;138:777–784. doi: 10.1001/archsurg.138.7.777. [DOI] [PubMed] [Google Scholar]
- Goldberg et al. (2004).Goldberg RM, Sargent DJ, Morton RF, Fuchs CS, Ramanathan RK, Williamson SK, Findlay BP, Pitot HC, Alberts SR. A randomized controlled trial of fluorouracil plus leucovorin, irinotecan, and oxaliplatin combinations in patients with previously untreated metastatic colorectal cancer. Journal of Clinical Oncology. 2004;22:23–30. doi: 10.1200/JCO.2004.09.046. [DOI] [PubMed] [Google Scholar]
- Gongora et al. (2011).Gongora C, Vezzio-Vie N, Tuduri S, Denis V, Causse A, Auzanneau C, Collod-Beroud G, Coquelle A, Pasero P, Pourquier P, Martineau P, Del Rio M. New Topoisomerase I mutations are associated with resistance to camptothecin. Molecular Cancer. 2011;10:1–13. doi: 10.1186/1476-4598-10-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves & Nicholson (2011).Greaves N, Nicholson J. Single incision laparoscopic surgery in general surgery: a review. Annals of the Royal College of Surgeons of England. 2011;93:437–440. doi: 10.1308/003588411X590358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo et al. (2016).Guo Y, Xiong BH, Zhang T, Cheng Y, Ma L. XELOX vs. FOLFOX in metastatic colorectal cancer: An updated meta-analysis. Cancer Investigation. 2016;34:94–104. doi: 10.3109/07357907.2015.1104689. [DOI] [PubMed] [Google Scholar]
- Gupta, Brenner & Turgeon (2008).Gupta AK, Brenner DE, Turgeon DK. Early detection of colon cancer: new tests on the horizon. Molecular Diagnosis and Therapy. 2008;12:77–85. doi: 10.1007/BF03256273. [DOI] [PubMed] [Google Scholar]
- Gurjao et al. (2019).Gurjao C, Liu D, Hofree M, AlDubayan SH, Wakiro I, Su MJ, Felt K, Gjini E, Brais LK, Rotem A, Rosenthal MH, Rozenblatt-Rosen O, Rodig S, Ng K, Van Allen EM, Corsello SM, Ogino S, Regev A, Nowak JA, Giannakis M. Intrinsic resistance to immune checkpoint blockade in a mismatch repair–deficient colorectal cancer. Cancer Immunology Research. 2019;7:1230–1236. doi: 10.1158/2326-6066.CIR-18-0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häfner & Debus (2016).Häfner MF, Debus J. Radiotherapy for colorectal cancer: current standards and future perspectives. Visceral Medicine. 2016;32:172–177. doi: 10.1159/000446486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada, Nowak & Ogino (2017).Hamada T, Nowak JA, Ogino S. PIK3CA mutation and colorectal cancer precision medicine. Oncotarget. 2017;8:22305–22306. doi: 10.18632/oncotarget.15724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han et al. (2017).Han P, Li JW, Zhang BM, Lv JC, Li YM, Gu XY, Yu ZW, Jia YH, Bai XF, Li L, Liu YL, Cui BB. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/β-catenin signaling. Molecular Cancer. 2017;16:9. doi: 10.1186/s12943-017-0583-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzivassiliou et al. (2010).Hatzivassiliou G, Song K, Yen I, Brandhuber BJ, Anderson DJ, Alvarado R, Ludlam MJC, Stokoe D, Gloor SL, Vigers G, Morales T, Aliagas I, Liu B, Sideris S, Hoeflich KP, Jaiswal BS, Seshagiri S, Koeppen H, Belvin M, Friedman LS, Malek S. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464:431–435. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- Hawkins & Stephens (2016).Hawkins PT, Stephens LR. Emerging evidence of signalling roles for PI(3,4)P2 in Class I and II PI3K-regulated pathways. Biochemical Society Transactions. 2016;44:307–314. doi: 10.1042/BST20150248. [DOI] [PubMed] [Google Scholar]
- He et al. (2017).He T, Zhang M, Zheng R, Zheng S, Linghu E, Herman JG, Guo M. Methylation of SLFN11 is a marker of poor prognosis and cisplatin resistance in colorectal cancer. Epigenomics. 2017;9:849–862. doi: 10.2217/epi-2017-0019. [DOI] [PubMed] [Google Scholar]
- He et al. (2018).He L, Zhu H, Zhou S, Wu T, Wu H, Yang H, Mao H, SekharKathera C, Janardhan A, Edick AM, Zhang A, Hu Z, Pan F, Guo Z. Wnt pathway is involved in 5-FU drug resistance of colorectal cancer cells. Experimental and Molecular Medicine. 2018;50:1–12. doi: 10.1038/s12276-018-0128-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Health (2020).Health NI. Colonoscopy — MedlinePlus. 2020. https://medlineplus.gov/colonoscopy.html. [22 May 2020]. https://medlineplus.gov/colonoscopy.html
- Heinemann et al. (2014).Heinemann V, Weikersthal LF von, Decker T, Kiani A, Vehling-Kaiser U, Al-Batran S-E, Heintges T, Lerchenmüller C, Kahl C, Seipelt G, Kullmann F, Stauch M, Scheithauer W, Hielscher J, Scholz M, Müller S, Link H, Niederle N, Rost A, Höffkes H-G, Moehler M, Lindig RU, Modest DP, Rossius L, Kirchner T, Jung A, Stintzing S. FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab as first-line treatment for patients with metastatic colorectal cancer (FIRE-3): A randomised, open-label, phase 3 trial. The Lancet Oncology. 2014;15:1065–1075. doi: 10.1016/S1470-2045(14)70330-4. [DOI] [PubMed] [Google Scholar]
- Hemmings (1997).Hemmings BA. Akt signaling: linking membrane events to life and death decisions. Science. 1997;275:628–630. doi: 10.1126/science.275.5300.628. [DOI] [PubMed] [Google Scholar]
- Hemmings & Restuccia (2012).Hemmings BA, Restuccia DF. PI3K-PKB/Akt pathway. Cold Spring Harbor Perspectives in Biology. 2012;4:a011189. doi: 10.1101/cshperspect.a011189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera et al. (2018).Herrera M, Llorens C, Rodríguez M, Herrera A, Ramos R, Gil B, Candia A, Larriba MJ, Garre P, Earl J, Rodríguez-Garrote M, Caldés T, Bonilla F, Carrato A, García-Barberán V, Peña C. Differential distribution and enrichment of non-coding RNAs in exosomes from normal and Cancer-associated fibroblasts in colorectal cancer. Molecular Cancer. 2018;17:114. doi: 10.1186/s12943-018-0863-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraki et al. (2015).Hiraki M, Nishimura J, Takahashi H, Wu X, Takahashi Y, Miyo M, Nishida N, Uemura M, Hata T, Takemasa I, Mizushima T, Soh JW, Doki Y, Mori M, Yamamoto H. Concurrent targeting of KRAS and AKT by MiR-4689 is a novel treatment against mutant KRAS colorectal cancer. Molecular Therapy - Nucleic Acids. 2015;4:e231. doi: 10.1038/mtna.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou et al. (2020).Hou Z, Sun L, Xu F, Hu F, Lan J, Song D, Feng Y, Wang J, Luo X, Hu J, Wang G. Blocking histone methyltransferase SETDB1 inhibits tumorigenesis and enhances cetuximab sensitivity in colorectal cancer. Cancer Letters. 2020;487:63–73. doi: 10.1016/j.canlet.2020.05.029. [DOI] [PubMed] [Google Scholar]
- Hu et al. (2020).Hu PS, Li T, Lin JF, Qiu MZ, Wang DS, Liu ZX, Chen ZH, Yang LP, Zhang XL, Zhao Q, Chen YX, Lu YX, Wu QN, Pu HY, Zeng ZL, Xie D, Ju HQ, Luo HY, Xu RH. VDR–SOX2 signaling promotes colorectal cancer stemness and malignancy in an acidic microenvironment. Signal Transduction and Targeted Therapy. 2020;5:183. doi: 10.1038/s41392-020-00230-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu et al. (2016).Hu T, Li Z, Gao CY, Cho CH. Mechanisms of drug resistance in colon cancer and its therapeutic strategies. World Journal of Gastroenterology. 2016;22:6876–6889. doi: 10.3748/wjg.v22.i30.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu et al. (2019).Hu JL, Wang W, Lan XL, Zeng ZC, Liang YS, Yan YR, Song FY, Wang FF, Zhu XH, Liao WJ, Liao WT, Ding YQ, Liang L. CAFs secreted exosomes promote metastasis and chemotherapy resistance by enhancing cell stemness and epithelial-mesenchymal transition in colorectal cancer. Molecular Cancer. 2019;18:91. doi: 10.1186/s12943-019-1019-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huan et al. (2020).Huan L, Guo T, Wu Y, Xu L, Huang S, Xu Y, Liang L, He X. Hypoxia induced LUCAT1/PTBP1 axis modulates cancer cell viability and chemotherapy response. Molecular Cancer. 2020;19:11. doi: 10.1186/s12943-019-1122-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang et al. (2004).Huang S, Armstrong EA, Benavente S, Chinnaiyan P, Harari PM. Dual-agent molecular targeting of the epidermal growth factor receptor (EGFR): combining anti-EGFR antibody with tyrosine kinase inhibitor. Cancer Research. 2004;64:5355–5362. doi: 10.1158/0008-5472.CAN-04-0562. [DOI] [PubMed] [Google Scholar]
- Ikenoue et al. (2005).Ikenoue T, Kanai F, Hikiba Y, Obata T, Tanaka Y, Imamura J, Ohta M, Jazag A, Guleng B, Tateishi K, Asaoka Y, Matsumura M, Kawabe T, Omata M. Functional analysis of PIK3CA gene mutations in human colorectal cancer. Cancer Research. 2005;65:4562–4567. doi: 10.1158/0008-5472.CAN-04-4114. [DOI] [PubMed] [Google Scholar]
- International Agency for Research on Cancer (2020).International Agency for Research on Cancer Cancer Today. 2020. [24 February 2021]. https://gco.iarc.fr/today/home https://gco.iarc.fr/today/home
- Ishaque et al. (2018).Ishaque N, Abba ML, Hauser C, Patil N, Paramasivam N, Huebschmann D, Leupold JH, Balasubramanian GP, Kleinheinz K, Toprak UH, Hutter B, Benner A, Shavinskaya A, Zhou C, Gu Z, Kerssemakers J, Marx A, Moniuszko M, Kozlowski M, Reszec J, Niklinski J, Eils J, Schlesner M, Eils R, Brors B, Allgayer H. Whole genome sequencing puts forward hypotheses on metastasis evolution and therapy in colorectal cancer. Nature Communications. 2018;9:4782. doi: 10.1038/s41467-018-07041-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro et al. (2017).Ishiguro H, Okubo T, Kuwabara Y, Kimura M, Mitsui A, Sugito N, Ogawa R, Katada T, Tanaka T, Shiozaki M, Mizoguchi K, Samoto Y, Matsuo Y, Takahashi H, Takiguchi S. NOTCH1 activates the Wnt/β-catenin signaling pathway in colon cancer. Oncotarget. 2017;8:60378–60389. doi: 10.18632/oncotarget.19534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iveson et al. (2021).Iveson TJ, Sobrero AF, Yoshino T, Souglakos I, Ou FS, Meyers JP, Shi Q, Grothey A, Saunders MP, Labianca R, Yamanaka T, Boukovinas I, Hollander NH, Galli F, Yamazaki K, Georgoulias V, Kerr R, Oki E, Lonardi S, Harkin A, Rosati G, Paul J. Duration of Adjuvant Doublet Chemotherapy (3 or 6 months) in Patients With High-Risk Stage II Colorectal Cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2021;39:631–641. doi: 10.1200/JCO.20.01330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackstadt et al. (2019).Jackstadt R, Van Hooff SR, Leach JD, Cortes-Lavaud X, Lohuis JO, Ridgway RA, Wouters VM, Roper J, Kendall TJ, Roxburgh CS, Horgan PG, Nixon C, Nourse C, Gunzer M, Clark W, Hedley A, Yilmaz OH, Rashid M, Bailey P, Biankin AV, Campbell AD, Adams DJ, Barry ST, Steele CW, Medema JP, Sansom OJ. Epithelial NOTCH signaling rewires the tumor microenvironment of colorectal cancer to drive poor-prognosis subtypes and metastasis. Cancer Cell. 2019;36:319–336e7. doi: 10.1016/j.ccell.2019.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda et al. (2012).Janda CY, Waghray D, Levin AM, Thomas C, Garcia KC. Structural basis of Wnt recognition by frizzled. Science. 2012;336:59–64. doi: 10.1126/science.1222879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson & Lund (2012).Jansson MD, Lund AH. MicroRNA and cancer. Molecular Oncology. 2012;6:590–610. doi: 10.1016/j.molonc.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeantet et al. (2016).Jeantet M, Tougeron D, Tachon G, Cortes U, Archambaut C, Fromont G, Karayan-Tapon L. High intra-and inter-tumoral heterogeneity of RAS mutations in colorectal cancer. International Journal of Molecular Sciences. 2016;17 doi: 10.3390/ijms17122015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen et al. (2017).Jeppesen M, Hagel G, Glenthoj A, Vainer B, Ibsen P, Harling H, Thastrup O, Jørgensen LN, Thastrup J. Short-term spheroid culture of primary colorectal cancer cells as an in vitro model for personalizing cancer medicine. PLOS ONE. 2017;12:e0183074. doi: 10.1371/journal.pone.0183074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang et al. (2019).Jiang S, Miao D, Wang M, Lv J, Wang Y, Tong J. MiR-30-5p suppresses cell chemoresistance and stemness in colorectal cancer through USP22/Wnt/β-catenin signaling axis. Journal of Cellular and Molecular Medicine. 2019;23:630–640. doi: 10.1111/jcmm.13968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin et al. (2018).Jin Y, Wang M, Hu H, Huang Q, Chen Y, Wang G. Overcoming stemness and chemoresistance in colorectal cancer through miR-195-5p-modulated inhibition of notch signaling. International Journal of Biological Macromolecules. 2018;117:445–453. doi: 10.1016/j.ijbiomac.2018.05.151. [DOI] [PubMed] [Google Scholar]
- Jonker et al. (2007).Jonker DJ, O’Callaghan CJ, Karapetis CS, Zalcberg JR, Tu D, Au HJ, Berry SR, Krahn M, Price T, Simes RJ, Tebbutt NC, Van Hazel G, Wierzbicki R, Langer C, Moore MJ. Cetuximab for the treatment of colorectal cancer. New England Journal of Medicine. 2007;357:2040–2048. doi: 10.1056/NEJMoa071834. [DOI] [PubMed] [Google Scholar]
- Kadioglu et al. (2021).Kadioglu O, Saeed M, Mahmoud N, Azawi S, Mrasek K, Liehr T, Efferth T. Identification of potential novel drug resistance mechanisms by genomic and transcriptomic profiling of colon cancer cells with p53 deletion. Archives of Toxicology. 2021;95 doi: 10.1007/s00204-021-02979-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafka et al. (2017).Kafka A, Tomas D, Lechpammer M, Gabud T, Pažanin L, Pećina-Šlaus N. Expression levels and localizations of DVL3 and sFRP3 in glioblastoma. Disease Markers. 2017;2017:9253495. doi: 10.1155/2017/9253495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlert & Kalluri (2013).Kahlert C, Kalluri R. Exosomes in tumor microenvironment influence cancer progression and metastasis. Journal of Molecular Medicine. 2013;91:431–437. doi: 10.1007/s00109-013-1020-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalloo (2007).Kalloo AN. Natural orifice transluminal endoscopic surgery (NOTES) Gastroenterology & Hepatology. 2007;3:183–184. [PMC free article] [PubMed] [Google Scholar]
- Kang, Bader & Vogt (2005).Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:802–807. doi: 10.1073/pnas.0408864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh (2017).Katoh M. Canonical and non-canonical WNT signaling in cancer stem cells and their niches: cellular heterogeneity, omics reprogramming, targeted therapy and tumor plasticity (Review) International Journal of Oncology. 2017;51:1357–1369. doi: 10.3892/ijo.2017.4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavuri et al. (2015).Kavuri SM, Jain N, Galimi F, Cottino F, Leto SM, Migliardi G, Searleman AC, Shen W, Monsey J, Trusolino L, Jacobs SA, Bertotti A, Bose R. HER2 activating mutations are targets for colorectal cancer treatment. Cancer Discovery. 2015;5:832–841. doi: 10.1158/2159-8290.CD-14-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan et al. (2019).Khan A, Ahmed E, Elareer N, Junejo K, Steinhoff M, Uddin S. Role of miRNA-Regulated cancer stem cells in the pathogenesis of human malignancies. Cell. 2019;8:840. doi: 10.3390/cells8080840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim & Choi (2010).Kim EK, Choi EJ. Pathological roles of MAPK signaling pathways in human diseases. Biochimica et Biophysica Acta. 2010;1802:396–405. doi: 10.1016/j.bbadis.2009.12.009. [DOI] [PubMed] [Google Scholar]
- Kim et al. (2013).Kim A, Lee JE, Lee SS, Kim C, Lee SJ, Jang WS, Park S. Coexistent mutations of KRAS and PIK3CA affect the efficacy of NVP-BEZ235, a dual PI3K/MTOR inhibitor, in regulating the PI3K/MTOR pathway in colorectal cancer. International Journal of Cancer. 2013;133:984–996. doi: 10.1002/ijc.28073. [DOI] [PubMed] [Google Scholar]
- Kim et al. (2015).Kim BR, Oh SC, Lee DH, Kim JL, Lee SY, Kang MH, Lee S Il, Kang S, Joung SY, Min BW. BMP-2 induces motility and invasiveness by promoting colon cancer stemness through STAT3 activation. Tumor Biology. 2015;36:9475–9486. doi: 10.1007/s13277-015-3681-y. [DOI] [PubMed] [Google Scholar]
- Klein et al. (2002).Klein CE, Gupta E, Reid JM, Atherton PJ, Sloan JA, Pitot HC, Ratain MJ, Kastrissios H. Population pharmacokinetic model for irinotecan and two of its metabolites, SN-38 and SN-38 glucuronide. Clinical Pharmacology and Therapeutics. 2002;72:638–647. doi: 10.1067/mcp.2002.129502. [DOI] [PubMed] [Google Scholar]
- Komiya & Habas (2008).Komiya Y, Habas R. Wnt signal transduction pathways. Organogenesis. 2008;4:68–75. doi: 10.4161/org.4.2.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo et al. (2019).Kondo J, Ekawa T, Endo H, Yamazaki K, Tanaka N, Kukita Y, Okuyama H, Okami J, Imamura F, Ohue M, Kato K, Nomura T, Kohara A, Mori S, Dan S, Inoue M. High-throughput screening in colorectal cancer tissue-originated spheroids. Cancer Science. 2019;110:345–355. doi: 10.1111/cas.13843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar & Clevers (2017).Kretzschmar K, Clevers H. Wnt/β-catenin signaling in adult mammalian epithelial stem cells. Developmental Biology. 2017;428:273–282. doi: 10.1016/j.ydbio.2017.05.015. [DOI] [PubMed] [Google Scholar]
- Kubiniok et al. (2017).Kubiniok P, Lavoie H, Therrien M, Thibault P. Time-resolved phosphoproteome analysis of paradoxical RAF activation reveals novel targets of ERK. Molecular and Cellular Proteomics. 2017;16:663–679. doi: 10.1074/mcp.M116.065128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukcinaviciute et al. (2018).Kukcinaviciute E, Jonusiene V, Sasnauskiene A, Dabkeviciene D, Eidenaite E, Laurinavicius A. Significance of Notch and Wnt signaling for chemoresistance of colorectal cancer cells HCT116. Journal of Cellular Biochemistry. 2018;119:5913–5920. doi: 10.1002/jcb.26783. [DOI] [PubMed] [Google Scholar]
- Kuo et al. (2005).Kuo T, Cho CD, Halsey J, Wakelee HA, Advani RH, Ford JM, Fisher GA, Sikic BI. Phase II study of gefitinib, fluorouracil, leucovorin, and oxaliplatin therapy in previously treated patients with metastatic colorectal cancer. Journal of Clinical Oncology. 2005;23:5613–5619. doi: 10.1200/JCO.2005.08.359. [DOI] [PubMed] [Google Scholar]
- Kwon et al. (2016).Kwon JJ, Willy JA, Quirin KA, Wek RC, Korc M, Yin XM, Kota J. Novel role of miR-29a in pancreatic cancer autophagy and its therapeutic potential. Oncotarget. 2016;7:71635–71650. doi: 10.18632/oncotarget.11928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labianca et al. (2013).Labianca R, Nordlinger B, Beretta GD, Mosconi S, Mandalà M, Cervantes A, Arnold D, Group EGW. Early colon cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Annals of Oncology. 2013;24:164–172. doi: 10.1093/annonc/mdt354. [DOI] [PubMed] [Google Scholar]
- Laurent-Puig et al. (2009).Laurent-Puig P, Cayre A, Manceau G, Buc E, Bachet JB, Lecomte T, Rougier P, Lievre A, Landi B, Boige V, Ducreux M, Ychou M, Bibeau F, Bouché O, Reid J, Stone S, Penault-Llorca F. Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer. Journal of Clinical Oncology. 2009;27:5924–5930. doi: 10.1200/JCO.2008.21.6796. [DOI] [PubMed] [Google Scholar]
- Lazzari et al. (2019).Lazzari L, Corti G, Picco G, Isella C, Montone M, Arcela P, Durinikova E, Zanella ER, Novara L, Barbosa F, Cassingena A, Cancelliere C, Medico E, Sartore-Bianchi A, Siena S, Garnett MJ, Bertotti A, Trusolino L, Di Nicolantonio F, Linnebacher M, Bardelli A, Arena S. Patient-derived xenografts and matched cell lines identify pharmacogenomic vulnerabilities in colorectal cancer. Clinical Cancer Research. 2019;25:6243–6259. doi: 10.1158/1078-0432.CCR-18-3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee et al. (2020).Lee HO, Hong Y, Etlioglu HE, Cho YB, Pomella V, VandenBosch B, Vanhecke J, Verbandt S, Hong H, Min JW, Kim N, Eum HH, Qian J, Boeckx B, Lambrechts D, Tsantoulis P, De Hertogh G, Chung W, Lee T, An M, Shin HT, Joung JG, Jung MH, Ko G, Wirapati P, Kim SH, Kim HC, Yun SH, Tan IBH, Ranjan B, Lee WY, Kim TY, Choi JK, Kim YJ, Prabhakar S, Tejpar S, Park WY. Lineage-dependent gene expression programs influence the immune landscape of colorectal cancer. Nature Genetics. 2020;52:594–603. doi: 10.1038/s41588-020-0636-z. [DOI] [PubMed] [Google Scholar]
- Li et al. (2017).Li H, Courtois ET, Sengupta D, Tan Y, Chen KH, Goh JJL, Kong SL, Chua C, Hon LK, Tan WS, Wong M, Choi PJ, Wee LJK, Hillmer AM, Tan IB, Robson P, Prabhakar S. Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nature Genetics. 2017;49:708–718. doi: 10.1038/ng.3818. [DOI] [PubMed] [Google Scholar]
- Li et al. (2020b).Li N, Kang Y, Wang L, Huff S, Tang R, Hui H, Agrawal K, Gonzalez GM, Wang Y, Patel SP, Rana TM. ALKBH5 regulates anti–PD-1 therapy response by modulating lactate and suppressive immune cell accumulation in tumor microenvironment. Proceedings of the National Academy of Sciences of the United States of America. 2020b;117:20159–20170. doi: 10.1073/PNAS.1918986117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al. (2011).Li JL, Sainson RCA, Oon CE, Turley H, Leek R, Sheldon H, Bridges E, Shi W, Snell C, Bowden ET, Wu H, Chowdhury PS, Russell AJ, Montgomery CP, Poulsom R, Harris AL. DLL4-Notch signaling mediates tumor resistance to anti-VEGF therapy in vivo. Cancer Research. 2011;71:6073–6083. doi: 10.1158/0008-5472.CAN-11-1704. [DOI] [PubMed] [Google Scholar]
- Li et al. (2020a).Li F, Zhan L, Dong Q, Wang Q, Wang Y, Li X, Zhang Y, Zhang J. Tumor-derived exosome-educated hepatic stellate cells regulate lactate metabolism of hypoxic colorectal tumor cells via the IL-6/STAT3 pathway to confer drug resistance. OncoTargets and Therapy. 2020a;13:7851–7864. doi: 10.2147/OTT.S253485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao et al. (2019).Liao W, Overman MJ, Boutin AT, Shang X, Zhao D, Dey P, Li J, Wang G, Lan Z, Li J, Tang M, Jiang S, Ma X, Chen P, Katkhuda R, Korphaisarn K, Chakravarti D, Chang A, Spring DJ, Chang Q, Zhang J, Maru DM, Maeda DY, Zebala JA, Kopetz S, Wang YA, De Pinho RA. KRAS-IRF2 axis drives immune suppression and immune therapy resistance in colorectal cancer. Cancer Cell. 2019;35:559–572. doi: 10.1016/j.ccell.2019.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lièvre et al. (2006).Lièvre A, Bachet JB, Le Corre D, Boige V, Landi B, Emile JF, Côté JF, Tomasic G, Penna C, Ducreux M, Rougier P, Penault-Llorca F, Laurent-Puig P. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Research. 2006;66:3992–3995. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- Lippert, Ruoff & Volm (2008).Lippert TH, Ruoff HJ, Volm M. Intrinsic and acquired drug resistance in malignant tumors: the main reason for therapeutic failure. Arzneimittel-Forschung/Drug Research. 2008;58:261–264. doi: 10.1055/s-0031-1296504. [DOI] [PubMed] [Google Scholar]
- Liu et al. (2018).Liu X, Lv X, Yang Q, Jin H, Zhou W, Fan Q. MicroRNA-29a functions as a tumor suppressor and increases cisplatin sensitivity by targeting NRAS in lung cancer. Technology in Cancer Research and Treatment. 2018;17:153303381875890. doi: 10.1177/1533033818758905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu et al. (2016).Liu H, Yin Y, Hu Y, Feng Y, Bian Z, Yao S, Li M, You Q, Huang Z. miR-139-5p sensitizes colorectal cancer cells to 5-fluorouracil by targeting NOTCH-1. Pathology Research and Practice. 2016;212:643–649. doi: 10.1016/j.prp.2016.04.011. [DOI] [PubMed] [Google Scholar]
- Liu, Zhou & Dong (2019).Liu G, Zhou JP, Dong M. Down-regulation of miR-543 expression increases the sensitivity of colorectal cancer cells to 5-Fluorouracil through the PTEN/PI3K/AKT pathway. Bioscience Reports. 2019;39:BSR20190249. doi: 10.1042/BSR20190249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb et al. (2019).Loeb LA, Kohrn BF, Loubet-Senear KJ, Dunn YJ, Ahn EH, O’Sullivan JN, Salk JJ, Bronner MP, Beckman RA. Extensive subclonal mutational diversity in human colorectal cancer and its significance. Proceedings of the National Academy of Sciences of the United States of America. 2019;116:26863–26872. doi: 10.1073/pnas.1910301116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotti et al. (2013).Lotti F, Jarrar AM, Pai RK, Hitomi M, Lathia J, Mace A, Gantt GA, Sukhdeo K, DeVecchio J, Vasanji A, Leahy P, Hjelmeland AB, Kalady MF, Rich JN. Chemotherapy activates cancer-associated fibroblasts to maintain colorectal cancer-initiating cells by IL-17A. Journal of Experimental Medicine. 2013;210:2851–2872. doi: 10.1084/jem.20131195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu et al. (2021).Lu W, Yu W, He J, Liu W, Yang J, Lin X, Zhang Y, Wang X, Jiang W, Luo J, Zhang Q, Yang H, Peng S, Yi Z, Ren S, Chen J, Siwko S, Nussinov R, Cheng F, Zhang H, Liu M. Reprogramming immunosuppressive myeloid cells facilitates immunotherapy for colorectal cancer. EMBO Molecular Medicine. 2021;13:e12798. doi: 10.15252/emmm.202012798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu et al. (2017).Lu Y, Zhao X, Liu Q, Li C, Graves-Deal R, Cao Z, Singh B, Franklin JL, Wang J, Hu H, Wei T, Yang M, Yeatman TJ, Lee E, Saito-Diaz K, Hinger S, Patton JG, Chung CH, Emmrich S, Klusmann JH, Fan D, Coffey RJ. LncRNA MIR100HG-derived miR-100 and miR-125b mediate cetuximab resistance via Wnt/β-catenin signaling. Nature Medicine. 2017;23:1331–1341. doi: 10.1038/nm.4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo et al. (2014).Luo Q, Wei C, Li X, Li J, Chen L, Huang Y, Song H, Li D, Fang L. MicroRNA-195-5p is a potential diagnostic and therapeutic target for breast cancer. Oncology Reports. 2014;31:1096–1102. doi: 10.3892/or.2014.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma et al. (2017).Ma B, Gao P, Wang H, Xu Q, Song Y, Huang X, Sun J, Zhao J, Luo J, Sun Y, Wang Z. What has preoperative radio(chemo)therapy brought to localized rectal cancer patients in terms of perioperative and long-term outcomes over the past decades? A systematic review and meta-analysis based on 41, 121 patients. International Journal of Cancer. 2017;141:1052–1065. doi: 10.1002/ijc.30805. [DOI] [PubMed] [Google Scholar]
- Ma et al. (2016).Ma Y, Yang Y, Wang F, Moyer MP, Wei Q, Zhang P, Yang Z, Liu W, Zhang H, Chen N, Wang H, Qin H. Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wnt/β-catenin signalling pathway via suppression of activator protein 2α. Gut. 2016;65:1494–1504. doi: 10.1136/gutjnl-2014-308392. [DOI] [PubMed] [Google Scholar]
- Macrae (2016).Macrae FA. Colorectal cancer: epidemiology, risk factors, and protective factors - UpToDate. 2016. https://www.uptodate.com/contents/colorectal-cancer-epidemiology-risk-factors-and-protective-factors. [30 April 2020]. https://www.uptodate.com/contents/colorectal-cancer-epidemiology-risk-factors-and-protective-factors
- Mahalakshmi, Husayn Ahmed & Mahadevan (2018).Mahalakshmi R, Husayn Ahmed P, Mahadevan V. HDAC inhibitors show differential epigenetic regulation and cell survival strategies on p53 mutant colon cancer cells. Journal of Biomolecular Structure and Dynamics. 2018;36:938–955. doi: 10.1080/07391102.2017.1302820. [DOI] [PubMed] [Google Scholar]
- Majidinia et al. (2018).Majidinia M, Darband SG, Kaviani M, Nabavi SM, Jahanban-Esfahlan R, Yousefi B. Cross-regulation between Notch signaling pathway and miRNA machinery in cancer. DNA Repair. 2018;66–67:30–41. doi: 10.1016/j.dnarep.2018.04.002. [DOI] [PubMed] [Google Scholar]
- Manning & Cantley (2007).Manning BD, Cantley LC. AKT/PKB signaling: navigating Downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin et al. (2011).Margolin DA, Silinsky J, Grimes C, Spencer N, Aycock M, Green H, Cordova J, Davis NK, Driscoll T, Li L. Lymph node stromal cells enhance drug-resistant colon cancer cell tumor formation through SDF-1α/CXCR4 paracrine signaling. Neoplasia. 2011;13:874–886. doi: 10.1593/neo.11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli et al. (2017).Martinelli E, Morgillo F, Troiani T, Ciardiello F. Cancer resistance to therapies against the EGFR-RAS-RAF pathway: the role of MEK. Cancer Treatment Reviews. 2017;53:61–69. doi: 10.1016/j.ctrv.2016.12.001. [DOI] [PubMed] [Google Scholar]
- Meropol et al. (2006).Meropol NJ, Gold PJ, Diasio RB, Andria M, Dhami M, Godfrey T, Kovatich AJ, Lund KA, Mitchell E, Schwarting R. Thymidine phosphorylase expression is associated with response to capecitabine plus irinotecan in patients with metastatic colorectal cancer. Journal of Clinical Oncology. 2006;24:4069–4077. doi: 10.1200/JCO.2005.05.2084. [DOI] [PubMed] [Google Scholar]
- Mesange et al. (2018).Mesange P, Bouygues A, Ferrand N, Sabbah M, Escargueil AE, Savina A, Chibaudel B, Tournigand C, Andre T, De Gramont A, Larsen AK. Combinations of bevacizumab and erlotinib show activity in colorectal cancer independent of ras status. Clinical Cancer Research. 2018;24:2548–2558. doi: 10.1158/1078-0432.CCR-17-3187. [DOI] [PubMed] [Google Scholar]
- Mirone et al. (2016).Mirone G, Perna S, Shukla A, Marfe G. Involvement of Notch-1 in resistance to regorafenib in colon cancer cells. Journal of Cellular Physiology. 2016;231:1097–1105. doi: 10.1002/jcp.25206. [DOI] [PubMed] [Google Scholar]
- Misale et al. (2014).Misale S, Di Nicolantonio F, Sartore-Bianchi A, Siena S, Bardelli A. Resistance to Anti-EGFR therapy in colorectal cancer: From heterogeneity to convergent evolution. Cancer Discovery. 2014;4:1269–1280. doi: 10.1158/2159-8290.CD-14-0462. [DOI] [PubMed] [Google Scholar]
- Mohamed et al. (2019).Mohamed SY, Kaf RM, Ahmed MM, Elwan A, Ashour HR, Ibrahim A. The prognostic value of cancer stem cell markers (Notch1, ALDH1, and CD44) in primary colorectal carcinoma. Journal of Gastrointestinal Cancer. 2019;50:824–837. doi: 10.1007/s12029-018-0156-6. [DOI] [PubMed] [Google Scholar]
- Molgora et al. (2020).Molgora M, Esaulova E, Vermi W, Hou J, Chen Y, Luo J, Brioschi S, Bugatti M, Omodei AS, Ricci B, Fronick C, Panda SK, Takeuchi Y, Gubin MM, Faccio R, Cella M, Gilfillan S, Unanue ER, Artyomov MN, Schreiber RD, Colonna M. TREM2 modulation remodels the tumor myeloid landscape enhancing Anti-PD-1 immunotherapy. Cell. 2020;182:886–900. doi: 10.1016/j.cell.2020.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinari et al. (2018).Molinari C, Marisi G, Passardi A, Matteucci L, De Maio G, Ulivi P. Heterogeneity in colorectal cancer: a challenge for personalized medicine? International Journal of Molecular Sciences. 2018;19:3733–3750. doi: 10.3390/ijms19123733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon et al. (2015).Moon SU, Kang MH, Sung JH, Kim JW, Lee JO, Kim YJ, Lee KW, Bang SM, Lee JS, Kim JH. Effect of Smad3/4 on chemotherapeutic drug sensitivity in colorectal cancer cells. Oncology Reports. 2015;33:185–192. doi: 10.3892/or.2014.3582. [DOI] [PubMed] [Google Scholar]
- Morrison (2012).Morrison DK. MAP kinase pathways. Cold Spring Harbor Perspectives in Biology. 2012;4:a011254. doi: 10.1101/cshperspect.a011254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muluhngwi et al. (2017).Muluhngwi P, Alizadeh-Rad N, Vittitow SL, Kalbfleisch TS, Klinge CM. The miR-29 transcriptome in endocrine-sensitive and resistant breast cancer cells. Scientific Reports. 2017;7:5205. doi: 10.1038/s41598-017-05727-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murga et al. (1998).Murga C, Laguinge L, Wetzker R, Cuadrado A, Gutkind JS. Activation of Akt/protein kinase B by G protein-coupled receptors: a role for α and βγ subunits of heterotrimeric G proteins acting through phosphatidylinositol-3-OH kinase γ. Journal of Biological Chemistry. 1998;273:19080–19085. doi: 10.1074/jbc.273.30.19080. [DOI] [PubMed] [Google Scholar]
- Negri et al. (2019).Negri F, Bozzetti C, Pedrazzi G, Azzoni C, Bottarelli L, Squadrilli A, Lagrasta C, Tamagnini I, Bisagni A, Ragazzi M, Porzio R, Tomasello G, Mori D, Leonardi F, Gnetti L, Crafa P, Sala R, Cascinu S. High levels of Notch intracellular cleaved domain are associated with stemness and reduced bevacizumab efficacy in patients with advanced colon cancer. Oncology Reports. 2019;42:2750–2758. doi: 10.3892/or.2019.7349. [DOI] [PubMed] [Google Scholar]
- Negri et al. (2015).Negri FV, Crafa P, Pedrazzi G, Bozzetti C, Lagrasta C, Gardini G, Tamagnini I, Bisagni A, Azzoni C, Bottarelli L, Graiani G, Romano I, Porzio R, Bacchini GP, Paties C, Tomasello G, Marchetti G, Fanello S, Pinto C, Sala R, Ardizzoni A. Strong Notch activation hinders bevacizumab efficacy in advanced colorectal cancer. Future Oncology. 2015;11:3167–3174. doi: 10.2217/fon.15.218. [DOI] [PubMed] [Google Scholar]
- Ning et al. (2014).Ning Z, Wang A, Liang J, Xie Y, Liu J, Feng L, Yan Q, Wang Z. USP22 promotes the G1/S phase transition by upregulating FoxM1 expression via β-catenin nuclear localization and is associated with poor prognosis in stage II pancreatic ductal adenocarcinoma. International Journal of Oncology. 2014;45:1594–1608. doi: 10.3892/ijo.2014.2531. [DOI] [PubMed] [Google Scholar]
- Nisar et al. (2020).Nisar S, Hashem S, Macha MA, Yadav SK, Muralitharan S, Therachiyil L, Sageena G, Al-Naemi H, Haris M, Bhat AA. Exploring dysregulated signaling pathways in cancer. Current Pharmaceutical Design. 2020;26:429–445. doi: 10.2174/1381612826666200115095937. [DOI] [PubMed] [Google Scholar]
- Normanno et al. (2015).Normanno N, Rachiglio AM, Lambiase M, Martinelli E, Fenizia F, Esposito C, Roma C, Troiani T, Rizzi D, Tatangelo F, Botti G, Maiello E, Colucci G, Ciardiello F, Giuliani F, Simone G, Febbraro A, Tommaselli E, Cinieri S, Criscuolo M, Rinaldi A, Bordonaro R, Manusia M, Romito S, Bufo P, Carten G, Biglietto M, Nappi O, Montesarchio E, Micheli P, Nasti G, Chicchinelli N, Iannaccone A, Russo A, Cabibi D, Barone C, Rindi G, Tonini G, Onetti Muda A, Perrone G, Latiano T, Graziano P, Pisconti S, Sebastio A. Heterogeneity of KRAS, NRAS, BRAF and PIK3CA mutations in metastatic colorectal cancer and potential effects on therapy in the CAPRI GOIM trial. Annals of Oncology. 2015;26:1710–1714. doi: 10.1093/annonc/mdv176. [DOI] [PubMed] [Google Scholar]
- Oddo et al. (2016).Oddo D, Sennott EM, Barault L, Valtorta E, Arena S, Cassingena A, Filiciotto G, Marzolla G, Elez E, Van Geel RMJM, Bartolini A, Crisafulli G, Boscaro V, Godfrey JT, Buscarino M, Cancelliere C, Linnebacher M, Corti G, Truini M, Siravegna G, Grasselli J, Gallicchio M, Bernards R, Schellens JHM, Tabernero J, Engelman JA, Sartore-Bianchi A, Bardelli A, Siena S, Corcoran RB, Di Nicolantonio F. Molecular landscape of acquired resistance to targeted therapy combinations in BRAF-mutant colorectal cancer. Cancer Research. 2016;76:4504–4515. doi: 10.1158/0008-5472.CAN-16-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkenhaug (2013).Okkenhaug K. Signaling by the phosphoinositide 3-kinase family in immune cells. Annual Review of Immunology. 2013;31:675–704. doi: 10.1146/annurev-immunol-032712-095946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Påhlman (1997).Påhlman L. Improved survival with preoperative radiotherapy in resectable rectal cancer. New England Journal of Medicine. 1997;336:980–987. doi: 10.1056/NEJM199704033361402. [DOI] [PubMed] [Google Scholar]
- Pai et al. (2017).Pai SG, Carneiro BA, Chae YK, Costa RL, Kalyan A, Shah HA, Helenowski I, Rademaker AW, Mahalingam D, Giles FJ. Correlation of tumor mutational burden and treatment outcomes in patients with colorectal cancer. Journal of Gastrointestinal Oncology. 2017;8:858–866. doi: 10.21037/jgo.2017.06.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park et al. (2019).Park SH, Jo MJ, Kim BR, Jeong YA, Na YJ, Kim JL, Jeong S, Yun HK, Kim DY, Kim BG, Kang SH, Oh SC, Lee DH. Sonic hedgehog pathway activation is associated with cetuximab resistance and EPHB3 receptor induction in colorectal cancer. Theranostics. 2019;9:2235–2251. doi: 10.7150/thno.30678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker & Cheng (1990).Parker WB, Cheng YC. Metabolism and mechanism of action of 5-fluorouracil. Pharmacology and Therapeutics. 1990;48:381–395. doi: 10.1016/0163-7258(90)90056-8. [DOI] [PubMed] [Google Scholar]
- Pasch et al. (2019).Pasch CA, Favreau PF, Yueh AE, Babiarz CP, Gillette AA, Sharick JT, Karim MR, Nickel KP, De Zeeuw AK, Sprackling CM, Emmerich PB, De Stefanis RA, Pitera RT, Payne SN, Korkos DP, Clipson L, Walsh CM, Miller D, Carchman EH, Burkard ME, Lemmon KK, Matkowskyj KA, Newton MA, Ong IM, Bassetti MF, Kimple RJ, Skala MC, Deming DA. Patient-derived cancer organoid cultures to predict sensitivity to chemotherapy and radiation. Clinical Cancer Research. 2019;25:5376–5387. doi: 10.1158/1078-0432.CCR-18-3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelullo et al. (2019).Pelullo M, Nardozza F, Zema S, Quaranta R, Nicoletti C, Besharat ZM, Felli MP, Cerbelli B, D’Amati G, Palermo R, Capalbo C, Talora C, Marcotullio L Di, Giannini G, Checquolo S, Screpanti I, Bellavia D. Kras/ADAM17-dependent Jag1-ICD reverse signaling sustains colorectal cancer progression and chemoresistance. Cancer Research. 2019;79:5575–5586. doi: 10.1158/0008-5472.CAN-19-0145. [DOI] [PubMed] [Google Scholar]
- Peng et al. (2015).Peng X, Luo Z, Kang Q, Deng D, Wang Q, Peng H, Wang S, Wei Z. FOXQ1 mediates the crosstalk between TGF- and Wnt signaling pathways in the progression of colorectal cancer. Cancer Biology and Therapy. 2015;16:1099–1109. doi: 10.1080/15384047.2015.1047568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrelli et al. (2020).Petrelli F, Labianca R, Zaniboni A, Lonardi S, Galli F, Rulli E, Rosati G, Corallo S, Ronzoni M, Cardellino GGG, Mattioli R, Mambrini A, Ciuffreda L, Banzi M, Pusceddu V, Maiello E, Zampino M, Zagonel V, Marchetti P, Corsi D, Rimassa L, Cinieri S, Sobrero A. Assessment of Duration and Effects of 3 vs 6 Months of Adjuvant Chemotherapy in High-Risk Stage II Colorectal Cancer: A Subgroup Analysis of the TOSCA Randomized Clinical Trial. JAMA Oncology. 2020;6:547–551. doi: 10.1001/jamaoncol.2019.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picardo et al. (2019).Picardo F, Romanelli A, Muinelo-Romay L, Mazza T, Fusilli C, Parrella P, Barbazán J, Lopez-López R, Barbano R, De Robertis M, Taffon C, Bordoni V, Agrati C, Costantini M, Ricci F, Graziano P, Maiello E, Muscarella LA, Fazio VM, Poeta ML. Diagnostic and prognostic value of B4GALT1 hypermethylation and its clinical significance as a novel circulating cell-free DNA biomarker in colorectal cancer. Cancer. 2019;11:1598. doi: 10.3390/cancers11101598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piletič & Kunej (2016).Piletič K, Kunej T. MicroRNA epigenetic signatures in human disease. Archives of Toxicology. 2016;90:2405–2419. doi: 10.1007/s00204-016-1815-7. [DOI] [PubMed] [Google Scholar]
- Pirker et al. (2009).Pirker R, Pereira JR, Szczesna A, Pawel Jvon, Krzakowski M, Ramlau R, Vynnychenko I, Park K, Yu CT, Ganul V, Roh JK, Bajetta E, O’Byrne K, De Marinis F, Eberhardt W, Goddemeier T, Emig M, Gatzemeier U. Cetuximab plus chemotherapy in patients with advanced non-small-cell lung cancer (FLEX): an open-label randomised phase III trial. The Lancet. 2009;373:1525–1531. doi: 10.1016/S0140-6736(09)60569-9. [DOI] [PubMed] [Google Scholar]
- Po et al. (2020).Po A, Citarella A, Catanzaro G, Besharat ZM, Trocchianesi S, Gianno F, Sabato C, Moretti M, De Smaele E, Vacca A, Fiori ME, Ferretti E. Hedgehog-GLI signalling promotes chemoresistance through the regulation of ABC transporters in colorectal cancer cells. Scientific Reports. 2020;10:13988. doi: 10.1038/s41598-020-70871-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcellini et al. (2018).Porcellini E, Laprovitera N, Riefolo M, Ravaioli M, Garajova I, Ferracin M. Epigenetic and epitranscriptomic changes in colorectal cancer: diagnostic, prognostic, and treatment implications. Cancer Letters. 2018;419:84–95. doi: 10.1016/j.canlet.2018.01.049. [DOI] [PubMed] [Google Scholar]
- Prahallad et al. (2012).Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, Beijersbergen RL, Bardelli A, Bernards R. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–104. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- Rai et al. (2019).Rai A, Greening DW, Chen M, Xu R, Ji H, Simpson RJ. Exosomes Derived from Human Primary and Metastatic Colorectal Cancer Cells Contribute to Functional Heterogeneity of Activated Fibroblasts by Reprogramming Their Proteome. Proteomics. 2019;19:e1800148. doi: 10.1002/pmic.201800148. [DOI] [PubMed] [Google Scholar]
- Ramirez et al. (2016).Ramirez M, Rajaram S, Steininger RJ, Osipchuk D, Roth MA, Morinishi LS, Evans L, Ji W, Hsu C-H, Thurley K, Wei S, Zhou A, Koduru PR, Posner BA, Wu LF, Altschuler SJ. Diverse drug-resistance mechanisms can emerge from drug-tolerant cancer persister cells. Nature Communications. 2016;7:10690. doi: 10.1038/ncomms10690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskov et al. (2020).Raskov H, Søby JH, Troelsen J, Bojesen RD, Gögenur I. Driver gene mutations and epigenetics in colorectal cancer. Annals of Surgery. 2020;271:75–85. doi: 10.1097/SLA.0000000000003393. [DOI] [PubMed] [Google Scholar]
- Rasmussen et al. (2013).Rasmussen MH, Jensen NF, Tarpgaard LS, Qvortrup C, Rømer MU, Stenvang J, Hansen TP, Christensen LL, Lindebjerg J, Hansen F, Jensen BV, Hansen TF, Pfeiffer P, Brünner N, Ørntoft TF, Andersen CL. High expression of microRNA-625-3p is associated with poor response to first-line oxaliplatin based treatment of metastatic colorectal cancer. Molecular Oncology. 2013;7:637–646. doi: 10.1016/j.molonc.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen et al. (2016).Rasmussen MH, Lyskjær I, Jersie-Christensen RR, Tarpgaard LS, Primdal-Bengtson B, Nielsen MM, Pedersen JS, Hansen TP, Hansen F, Olsen JV, Pfeiffer P, Ørntoft TF, Andersen CL. MiR-625-3p regulates oxaliplatin resistance by targeting MAP2K6-p38 signalling in human colorectal adenocarcinoma cells. Nature Communications. 2016;7:12436–12436. doi: 10.1038/ncomms12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawla, Sunkara & Barsouk (2019).Rawla P, Sunkara T, Barsouk A. Epidemiology of colorectal cancer: incidence, mortality, survival, and risk factors. Prz Gastroenterol. 2019;14:89–103. doi: 10.5114/pg.2018.81072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond et al. (1998).Raymond E, Faivre S, Woynarowski JM, Chaney SG. Oxaliplatin: mechanism of action and antineoplastic activity. Seminars in Oncology. 1998;2:4–12. [PubMed] [Google Scholar]
- Ren et al. (2018).Ren J, Ding L, Zhang D, Shi G, Xu Q, Shen S, Wang Y, Wang T, Hou Y. Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics. 2018;8:3932–3948. doi: 10.7150/thno.25541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentsch et al. (2016).Rentsch M, Schiergens T, Khandoga A, Werner J. Surgery for colorectal cancer—Trends, developments, and future perspectives. Visceral Medicine. 2016;32:184–191. doi: 10.1159/000446490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressa et al. (2018).Ressa A, Bosdriesz E, De Ligt J, Mainardi S, Maddalo G, Prahallad A, Jager M, De La Fonteijne L, Fitzpatrick M, Groten S, Altelaar AFM, Bernards R, Cuppen E, Wessels L, Heck AJR. A system-wide approach to monitor responses to synergistic BRAF and EGFR inhibition in colorectal cancer cells. Molecular and Cellular Proteomics. 2018;17:1892–1908. doi: 10.1074/mcp.RA117.000486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezapour et al. (2019).Rezapour S, Hosseinzadeh E, Marofi F, Hassanzadeh A. Epigenetic-based therapy for colorectal cancer: Prospect and involved mechanisms. Journal of Cellular Physiology. 2019;234:19366–19383. doi: 10.1002/jcp.28658. [DOI] [PubMed] [Google Scholar]
- Richman et al. (2009).Richman SD, Seymour MT, Chambers P, Elliott F, Daly CL, Meade AM, Taylor G, Barrett JH, Quirke P. KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan: results from the MRC FOCUS trial. Journal of Clinical Oncology. 2009;27:5931–5937. doi: 10.1200/JCO.2009.22.4295. [DOI] [PubMed] [Google Scholar]
- Rimassa et al. (2019).Rimassa L, Bozzarelli S, Pietrantonio F, Cordio S, Lonardi S, Toppo L, Zaniboni A, Bordonaro R, Di Bartolomeo M, Tomasello G, Dadduzio V, Tronconi MC, Piombo C, Giordano L, Gloghini A, Di Tommaso L, Santoro A. Phase II study of tivantinib and cetuximab in patients with KRAS wild-type metastatic colorectal cancer with acquired resistance to EGFR inhibitors and emergence of MET overexpression: lesson learned for future trials with EGFR/MET dual inhibition. Clinical Colorectal Cancer. 2019;18:125–132. doi: 10.1016/j.clcc.2019.02.004. [DOI] [PubMed] [Google Scholar]
- Rodilla et al. (2009).Rodilla V, Villanueva A, Obrador-Hevia A, Robert-Moreno À, Fernández-Majada V, Grilli A, López-Bigas N, Bellora N, Albà MM, Torres F, Duñach M, Sanjuan X, Gonzalez S, Gridley T, Capella G, Bigas A, Espinosa L. Jagged1 is the pathological link between Wnt and Notch pathways in colorectal cancer. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:6315–6320. doi: 10.1073/pnas.0813221106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo et al. (2019).Russo M, Crisafulli G, Sogari A, Reilly NM, Arena S, Lamba S, Bartolini A, Amodio V, Magrì A, Novara L, Sarotto I, Nagel ZD, Piett CG, Amatu A, Sartore-Bianchi A, Siena S, Bertotti A, Trusolino L, Corigliano M, Gherardi M, Lagomarsino MC, Nicolantonio F Di, Bardelli A. Adaptive mutability of colorectal cancers in response to targeted therapies. Science. 2019;366:1473–1480. doi: 10.1126/science.aav4474. [DOI] [PubMed] [Google Scholar]
- Russo et al. (2016).Russo M, Siravegna G, Blaszkowsky LS, Corti G, Crisafulli G, Ahronian LG, Mussolin B, Kwak EL, Buscarino M, Lazzari L, Valtorta E, Truini M, Jessop NA, Robinson HE, Hong TS, Mino-Kenudson M, Di Nicolantonio F, Thabet A, Sartore-Bianchi A, Siena S, Iafrate J, Bardelli A, Corcoran RB. Tumor heterogeneity and Lesion-Specific response to targeted therapy in colorectal cancer. Cancer Discovery. 2016;6:147–153. doi: 10.1158/2159-8290.CD-15-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakahara et al. (2019).Sakahara M, Okamoto T, Oyanagi J, Takano H, Natsume Y, Yamanaka H, Kusama D, Fusejima M, Tanaka N, Mori S, Kawachi H, Ueno M, Sakai Y, Noda T, Nagayama S, Yao R. IFN/STAT signaling controls tumorigenesis and the drug response in colorectal cancer. Cancer Science. 2019;110:1293–1305. doi: 10.1111/cas.13964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltz et al. (2000).Saltz LB, Cox JV, Blanke C, Rosen LS, Fehrenbacher L, Moore MJ, Maroun JA, Ackland SP, Locker PK, Pirotta N, Elfring GL, Miller LL. Irinotecan plus fluorouracil and leucovorin for metastatic colorectal cancer. New England Journal of Medicine. 2000;343:905–914. doi: 10.1056/NEJM200009283431302. [DOI] [PubMed] [Google Scholar]
- Samuels et al. (2004).Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, Willson JKV, Markowitz S, Kinzler KW, Vogelstein B, Velculescu VE. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- Sansom et al. (2004).Sansom OJ, Reed KR, Hayes AJ, Ireland H, Brinkmann H, Newton IP, Batlle E, Simon-Assmann P, Clevers H, Nathke IS, Clarke AR, Winton DJ. Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes and Development. 2004;18:1385–1390. doi: 10.1101/gad.287404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov et al. (2005).Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- Sartore-Bianchi et al. (2009).Sartore-Bianchi A, Di Nicolantonio F, Nichelatti M, Molinari F, De Dosso S, Saletti P, Martini M, Cipani T, Marrapese G, Mazzucchelli L, Lamba S, Veronese S, Frattini M, Bardelli A, Siena S. Multi-determinants analysis of molecular alterations for predicting clinical benefit to EGFR-targeted monoclonal antibodies in colorectal cancer. PLOS ONE. 2009;4:e7287. doi: 10.1371/journal.pone.0007287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh et al. (2017).Satoh K, Yachida S, Sugimoto M, Oshima M, Nakagawa T, Akamoto S, Tabata S, Saitoh K, Kato K, Sato S, Igarashi K, Aizawa Y, Kajino-Sakamoto R, Kojima Y, Fujishita T, Enomoto A, Hirayama A, Ishikawa T, Taketo MM, Kushida Y, Haba R, Okano K, Tomita M, Suzuki Y, Fukuda S, Aoki M, Soga T. Global metabolic reprogramming of colorectal cancer occurs at adenoma stage and is induced by MYC. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:E7697–E7706. doi: 10.1073/pnas.1710366114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher et al. (2019).Schumacher D, Andrieux G, Boehnke K, Keil M, Silvestri A, Silvestrov M, Keilholz U, Haybaeck J, Erdmann G, Sachse C, Templin M, Hoffmann J, Boerries M, Schäfer R, Regenbrecht CRA. Heterogeneous pathway activation and drug response modelled in colorectal-tumor-derived 3D cultures. PLOS Genetics. 2019;15:e1008076. doi: 10.1371/journal.pgen.1008076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppälä et al. (2015).Seppälä TT, Böhm JP, Friman M, Lahtinen L, Väyrynen VMJ, Liipo TKE, Ristimäki AP, Kairaluoma MVJ, Kellokumpu IH, Kuopio THI, Mecklin JP. Combination of microsatellite instability and BRAF mutation status for subtyping colorectal cancer. British Journal of Cancer. 2015;112:1966–1975. doi: 10.1038/bjc.2015.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaik et al. (2020).Shaik JP, Alanazi IO, Pathan AAK, Parine NR, Almadi MA, Azzam NA, Aljebreen AM, Alharbi O, Alanazi MS, Khan Z. Frequent activation of notch signaling pathway in colorectal cancers and its implication in patient survival outcome. Journal of Oncology. 2020;2020:6768942. doi: 10.1155/2020/6768942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan et al. (2019).Shan Y, Gao Y, Jin W, Fan M, Wang Y, Gu Y, Shan C, Sun L, Li X, Yu B, Luo Q, Xu Q. Targeting HIBCH to reprogram valine metabolism for the treatment of colorectal cancer. Cell Death and Disease. 2019;10:618. doi: 10.1038/s41419-019-1832-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang, Hua & Hu (2017).Shang S, Hua F, Hu ZW. The regulation of β-catenin activity and function in cancer: therapeutic opportunities. Oncotarget. 2017;8:33972–33989. doi: 10.18632/oncotarget.15687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen et al. (2016).Shen H, Li L, Yang S, Wang D, Zhong S, Zhao J, Tang J. MicroRNA-29a contributes to drug-resistance of breast cancer cells to adriamycin through PTEN/AKT/GSK3β signaling pathway. Gene. 2016;593:84–90. doi: 10.1016/j.gene.2016.08.016. [DOI] [PubMed] [Google Scholar]
- Shen et al. (2014).Shen K, Mao R, Ma L, Li Y, Qiu Y, Cui D, Le V, Yin P, Ni L, Liu J. Post-transcriptional regulation of the tumor suppressor miR-139-5p and a network of miR-139-5p-mediated mRNA interactions in colorectal cancer. FEBS Journal. 2014;281:3609–3624. doi: 10.1111/febs.12880. [DOI] [PubMed] [Google Scholar]
- Shen et al. (2018).Shen Y, Tong M, Liang Q, Guo Y, Sun HQ, Zheng W, Ao L, Guo Z, She F. Epigenomics alternations and dynamic transcriptional changes in responses to 5-fluorouracil stimulation reveal mechanisms of acquired drug resistance of colorectal cancer cells. Pharmacogenomics Journal. 2018;18:23–28. doi: 10.1038/tpj.2016.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood, Parris & Folkman (1971).Sherwood LM, Parris EE, Folkman J. Tumor Angiogenesis: therapeutic Implications. New England Journal of Medicine. 1971;285:1182–1186. doi: 10.1056/NEJM197111182852108. [DOI] [PubMed] [Google Scholar]
- Shi et al. (2018).Shi L, Li X, Wu Z, Li X, Nie J, Guo M, Mei Q, Han W. DNA methylation-mediated repression of miR-181a/135a/302c expression promotes the microsatellite-unstable colorectal cancer development and 5-FU resistance via targeting PLAG1. Journal of Genetics and Genomics. 2018;45:205–214. doi: 10.1016/j.jgg.2018.04.003. [DOI] [PubMed] [Google Scholar]
- Shi et al. (2020).Shi X, Valizadeh A, Mir SM, Asemi Z, Karimian A, Majidina M, Safa A, Yosefi B. miRNA-29a reverses P-glycoprotein-mediated drug resistance and inhibits proliferation via up-regulation of PTEN in colon cancer cells. European Journal of Pharmacology. 2020;880:173138. doi: 10.1016/j.ejphar.2020.173138. [DOI] [PubMed] [Google Scholar]
- Siebel & Lendahl (2017).Siebel C, Lendahl U. Notch signaling in development, tissue homeostasis, and disease. Physiological Reviews. 2017;97:1235–1294. doi: 10.1152/physrev.00005.2017. [DOI] [PubMed] [Google Scholar]
- Siegel et al. (2017).Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A, Jemal A. Colorectal cancer statistics. 2017. CA: A Cancer Journal for Clinicians. 2017;67:177–193. doi: 10.3322/caac.21395. [DOI] [PubMed] [Google Scholar]
- Sitia et al. (2021).Sitia L, Bonizzi A, Mazzucchelli S, Negri S, Sottani C, Grignani E, Rizzuto MA, Prosperi D, Sorrentino L, Morasso C, Allevi R, Sevieri M, Silva F, Truffi M, Corsi F. Selective targeting of cancer-associated fibroblasts by engineered h-ferritin nanocages loaded with navitoclax. Cell. 2021;10:328. doi: 10.3390/cells10020328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrede et al. (2020).Skrede OJ, De Raedt S, Kleppe A, Hveem TS, Liestøl K, Maddison J, Askautrud HA, Pradhan M, Nesheim JA, Albregtsen F, Farstad IN, Domingo E, Church DN, Nesbakken A, Shepherd NA, Tomlinson I, Kerr R, Novelli M, Kerr DJ, Danielsen HE. Deep learning for prediction of colorectal cancer outcome: a discovery and validation study. The Lancet. 2020;395:350–360. doi: 10.1016/S0140-6736(19)32998-8. [DOI] [PubMed] [Google Scholar]
- Sobhani et al. (2019).Sobhani I, Bergsten E, Couffin S, Amiot A, Nebbad B, Barau C, De’Angelis N, Rabot S, Canoui-Poitrine F, Mestivier D, Pédron T, Khazaie K, Sansonetti PJ. Colorectal cancer-associated microbiota contributes to oncogenic epigenetic signatures. Proceedings of the National Academy of Sciences of the United States of America. 2019;116:24285–24295. doi: 10.1073/pnas.1912129116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soleimani et al. (2019).Soleimani A, Rahmani F, Ferns GA, Ryzhikov M, Avan A, Hassanian SM. Role of regulatory oncogenic or tumor suppressor miRNAs of PI3K/AKT signaling axis in the pathogenesis of colorectal cancer. Current Pharmaceutical Design. 2019;24:4605–4610. doi: 10.2174/1381612825666190110151957. [DOI] [PubMed] [Google Scholar]
- Spranger, Bao & Gajewski (2015).Spranger S, Bao R, Gajewski T. Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature. 2015;523:231–235. doi: 10.1038/nature14404. [DOI] [PubMed] [Google Scholar]
- Stigliano et al. (2014).Stigliano V, Sanchez-Mete L, Martayan A, Anti M. Early-onset colorectal cancer: a sporadic or inherited disease? World Journal of Gastroenterology. 2014;20:12420–12430. doi: 10.3748/wjg.v20.i35.12420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su et al. (2007).Su C-C, Lin Y-P, Cheng Y-J, Huang J-Y, Chuang W-J, Shan Y-S, Yang B-C. Phosphatidylinositol 3-Kinase/Akt activation by integrin-tumor matrix interaction suppresses fas-mediated apoptosis in T Cells. The Journal of Immunology. 2007;179:4589–4597. doi: 10.4049/jimmunol.179.7.4589. [DOI] [PubMed] [Google Scholar]
- Sun et al. (2017).Sun L, Ke J, He Z, Chen Z, Huang Q, Ai W, Wang G, Wei Y, Zou X, Zhang S, Lan P, Hong C. HES1 promotes colorectal cancer cell resistance to 5-Fu by inducing of EMT and ABC transporter proteins. Journal of Cancer. 2017;8:2802–2808. doi: 10.7150/jca.19142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanton (2012).Swanton C. Intratumor heterogeneity: evolution through space and time. Cancer Research. 2012;72:4875–4882. doi: 10.1158/0008-5472.CAN-12-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabernero et al. (2014).Tabernero J, Chan E, Baselga J, Blay J-Y, Chau I, Hyman DM, Raje NS, Wolf J, Sirzen F, Veronese ML, Mitchell L, Hidalgo M. VE-BASKET, a Simon 2-stage adaptive design, phase II, histology-independent study in nonmelanoma solid tumors harboring BRAF V600 mutations (V600m): activity of vemurafenib (VEM) with or without cetuximab (CTX) in colorectal cancer (CRC) Journal of Clinical Oncology. 2014;32:3518–3518. doi: 10.1200/jco.2014.32.15_suppl.3518. [DOI] [Google Scholar]
- Taciak et al. (2018).Taciak B, Pruszynska I, Kiraga L, Bialasek M, Krol M. Wnt signaling pathway in development and cancer. Journal of Physiology and Pharmacology. 2018;69:185–196. doi: 10.26402/jpp.2018.2.07. [DOI] [PubMed] [Google Scholar]
- Taelman et al. (2010).Taelman VF, Dobrowolski R, Plouhinec JL, Fuentealba LC, Vorwald PP, Gumper I, Sabatini DD, De Robertis EM. Wnt signaling requires sequestration of Glycogen Synthase Kinase 3 inside multivesicular endosomes. Cell. 2010;143:1136–1148. doi: 10.1016/j.cell.2010.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang et al. (2018).Tang YA, Chen YF, Bao Y, Mahara S, Yatim SMJM, Oguz G, Lee PL, Feng M, Cai Y, Tan EY, Fong SS, Yang ZH, Lan P, Wu XJ, Yu Q. Hypoxic tumor microenvironment activates GLI2 via HIF-1α and TGF-β2 to promote chemoresistance in colorectal cancer. Proceedings of the National Academy of Sciences of the United States of America. 2018;115:E5990–E5999. doi: 10.1073/pnas.1801348115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng et al. (2017).Teng W, Kuan N, Lu TX, Hua D. Elevated expression of TrpC5 and GLUT1 is associated with chemoresistance in colorectal cancer. Oncology Reports. 2017;37:1059–1065. doi: 10.3892/or.2016.5322. [DOI] [PubMed] [Google Scholar]
- Tong et al. (2016).Tong M, Zheng W, Li H, Li X, Ao L, Shen Y, Liang Q, Li J, Hong G, Yan H, Cai H, Li M, Guan Q, Guo Z. Multi-omics landscapes of colorectal cancer subtypes discriminated by an individualized prognostic signature for 5-fluorouracil-based chemotherapy. Oncogenesis. 2016;5:e242–e242. doi: 10.1038/oncsis.2016.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsley et al. (2006).Townsley CA, Major P, Siu LL, Dancey J, Chen E, Pond GR, Nicklee T, Ho J, Hedley D, Tsao M, Moore MJ, Oza AM. Phase II study of erlotinib (OSI-774) in patients with metastatic colorectal cancer. British Journal of Cancer. 2006;94:1136–1143. doi: 10.1038/sj.bjc.6603055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaish, Kim & Shim (2017).Vaish V, Kim J, Shim M. Jagged-2 (JAG2) enhances tumorigenicity and chemoresistance of colorectal cancer cells. Oncotarget. 2017;8:53262–53275. doi: 10.18632/oncotarget.18391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvezan et al. (2012).Valvezan AJ, Zhang F, Diehl JA, Klein PS. Adenomatous Polyposis Coli (APC) regulates multiple signaling pathways by enhancing glycogen synthase kinase-3 (GSK-3) activity. Journal of Biological Chemistry. 2012;287:3823–3832. doi: 10.1074/jbc.M111.323337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Cutsem et al. (2009).Van Cutsem E, Köhne CH, Hitre E, Zaluski J, Chien CRC, Makhson A, D’Haens G, Pintér T, Lim R, Bodoky G, Roh JK, Folprecht G, Ruff P, Stroh C, Tejpar S, Schlichting M, Nippgen J, Rougier P. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. New England Journal of Medicine. 2009;360:1408–1417. doi: 10.1056/NEJMoa0805019. [DOI] [PubMed] [Google Scholar]
- Van Kappel & Maurice (2017).Van Kappel EC, Maurice MM. Molecular regulation and pharmacological targeting of the β-catenin destruction complex. British Journal of Pharmacology. 2017;174:4575–4588. doi: 10.1111/bph.13922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Schaeybroeck et al. (2011).Van Schaeybroeck S, Kyula JN, Fenton A, Fenning CS, Sasazuki T, Shirasawa S, Longley DB, Johnston PG. Oncogenic Kras promotes chemotherapy-induced growth factor shedding via ADAM17. Cancer Research. 2011;71:1071–1080. doi: 10.1158/0008-5472.CAN-10-0714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velho et al. (2005).Velho S, Oliveira C, Ferreira A, Ferreira AC, Suriano G, Schwartz S, Duval A, Carneiro F, Machado JC, Hamelin R, Seruca R. The prevalence of PIK3CA mutations in gastric and colon cancer. European Journal of Cancer. 2005;41:1649–1654. doi: 10.1016/j.ejca.2005.04.022. [DOI] [PubMed] [Google Scholar]
- Vellinga et al. (2015).Vellinga T, Borovski T, De Boer V, Fatrai S, Van Schelven S, Trumpi K, Verheem A, Snoeren N, Emmink B, Koster J, Rinkes I, Kranenburg O. SIRT1/PGC1α-Dependent Increase in Oxidative Phosphorylation Supports Chemotherapy Resistance of Colon Cancer. Clinical Cancer Research. 2015;21:2870–2879. doi: 10.1158/1078-0432.CCR-14-2290. [DOI] [PubMed] [Google Scholar]
- Venook (2005).Venook AP. Epidermal growth factor receptor-targeted treatment for advanced colorectal carcinoma. Cancer. 2005;103:2435–2446. doi: 10.1002/cncr.21123. [DOI] [PubMed] [Google Scholar]
- Vera-Yunca et al. (2020).Vera-Yunca D, Girard P, Parra-Guillen ZP, Munafo A, Trocóniz IF, Terranova N. Machine learning analysis of individual tumor lesions in four metastatic colorectal cancer clinical studies: linking tumor heterogeneity to overall survival. AAPS Journal. 2020;22:58. doi: 10.1208/s12248-020-0434-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermorken et al. (2008).Vermorken JB, Mesia R, Rivera F, Remenar E, Kawecki A, Rottey S, Erfan J, Zabolotnyy D, Kienzer HR, Cupissol D, Peyrade F, Benasso M, Vynnychenko I, De Raucourt D, Bokemeyer C, Schueler A, Amellal N, Hitt R. Platinum-based chemotherapy plus cetuximab in head and neck cancer. New England Journal of Medicine. 2008;359:1116–1127. doi: 10.1056/NEJMoa0802656. [DOI] [PubMed] [Google Scholar]
- Visone et al. (2019).Visone R, Bacalini MG, Franco S Di, Ferracin M, Colorito ML, Pagotto S, Laprovitera N, Licastro D, Marco M Di, Scavo E, Bassi C, Saccenti E, Nicotra A, Grzes M, Garagnani P, Laurenzi V De, Valeri N, Mariani-Costantini R, Negrini M, Stassi G, Veronese A. DNA methylation of shelf, shore and open sea CpG positions distinguish high microsatellite instability from low or stable microsatellite status colon cancer stem cells. Epigenomics. 2019;11:587–604. doi: 10.2217/epi-2018-0153. [DOI] [PubMed] [Google Scholar]
- Vinson et al. (2016).Vinson KE, George DC, Fender AW, Bertrand FE, Sigounas G. The Notch pathway in colorectal cancer. International Journal of Cancer. 2016;138:1835–1842. doi: 10.1002/ijc.29800. [DOI] [PubMed] [Google Scholar]
- Vitiello et al. (2019).Vitiello PP, Cardone C, Martini G, Ciardiello D, Belli V, Matrone N, Barra G, Napolitano S, Corte CDella, Turano M, Furia M, Troiani T, Morgillo F, De Vita F, Ciardiello F, Martinelli E. Receptor tyrosine kinase-dependent PI3K activation is an escape mechanism to vertical suppression of the EGFR/RAS/MAPK pathway in KRAS-mutated human colorectal cancer cell lines. Journal of Experimental and Clinical Cancer Research. 2019;38:41. doi: 10.1186/s13046-019-1035-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan et al. (2004).Wan PTC, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, Jones CM, Marshall CJ, Springer CJ, Barford D, Marais R, Project CG, Jones CM, Marshall CJ, Springer CJ, Barford D, Marais R. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–867. doi: 10.1016/S0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- Wan et al. (2020).Wan ML, Wang Y, Zeng Z, Deng B, Zhu BS, Cao T, Li YK, Xiao J, Han Q, Wu Q. Colorectal cancer (CRC) as a multifactorial disease and its causal correlations with multiple signaling pathways. Bioscience Reports. 2020;40:20200265. doi: 10.1042/BSR20200265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2020b).Wang Q, Chen X, Jiang Y, Liu S, Liu H, Sun X, Zhang H, Liu Z, Tao Y, Li C, Hu Y, Liu D, Ye D, Liu Y, Wang M, Zhang X. Elevating H3K27me3 level sensitizes colorectal cancer to oxaliplatin. Journal of Molecular Cell Biology. 2020b;12:125–137. doi: 10.1093/jmcb/mjz032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2020a).Wang L, Hui H, Agrawal K, Kang Y, Li N, Tang R, Yuan J, Rana TM. m 6 A RNA methyltransferases METTL3/14 regulate immune responses to anti-PD-1 therapy. The EMBO Journal. 2020a;39 doi: 10.15252/embj.2020104514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2019).Wang X, Ghareeb WM, Zhang Y, Yu Q, Lu X, Huang Y, Huang S, Sun Y, Chi P. Hypermethylated and downregulated MEIS2 are involved in stemness properties and oxaliplatin-based chemotherapy resistance of colorectal cancer. Journal of Cellular Physiology. 2019;234:18180–18191. doi: 10.1002/jcp.28451. [DOI] [PubMed] [Google Scholar]
- Wang et al. (2018).Wang Q, Shi YL, Zhou K, Wang LL, Yan ZX, Liu YL, Xu LL, Zhao SW, Chu HL, Shi TT, Ma QH, Bi J. PIK3CA mutations confer resistance to first-line chemotherapy in colorectal cancer. Cell Death and Disease. 2018;9:739. doi: 10.1038/s41419-018-0776-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Zhang & Chen (2019).Wang X, Zhang H, Chen X. Drug resistance and combating drug resistance in cancer. Cancer Drug Resistance. 2019;2:141–160. doi: 10.20517/cdr.2019.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2020c).Wang X, Zhang H, Yang H, Bai M, Ning T, Deng T, Liu R, Fan Q, Zhu K, Li J, Zhan Y, Ying G, Ba Y. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Molecular Oncology. 2020c;14:539–555. doi: 10.1002/1878-0261.12629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber et al. (2019).Weber S, Koschade SE, Hoffmann CM, Dubash TD, Giessler KM, Dieter SM, Herbst F, Glimm H, Ball CR. The notch target gene HEYL modulates metastasis forming capacity of colorectal cancer patient-derived spheroid cells in vivo. BMC Cancer. 2019;19:1181. doi: 10.1186/s12885-019-6396-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks et al. (2012).Weeks JC, Catalano PJ, Cronin A, Finkelman MD, Mack JW, Keating NL, Schrag D. Patients’ expectations about effects of chemotherapy for advanced cancer. New England Journal of Medicine. 2012;367:1616–1625. doi: 10.1056/NEJMoa1204410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei & Liu (2002).Wei Z, Liu HT. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Research. 2002;12:9–18. doi: 10.1038/sj.cr.7290105. [DOI] [PubMed] [Google Scholar]
- Wilhelm et al. (2006).Wilhelm S, Carter C, Lynch M, Lowinger T, Dumas J, Smith RA, Schwartz B, Simantov R, Kelley S. Discovery and development of sorafenib: a multikinase inhibitor for treating cancer. Nature Reviews Drug Discovery. 2006;5:835–844. doi: 10.1038/nrd2130. [DOI] [PubMed] [Google Scholar]
- Wolpin & Mayer (2008).Wolpin BM, Mayer RJ. Systemic treatment of colorectal cancer. Gastroenterology. 2008;134:1296–1310. doi: 10.1053/j.gastro.2008.02.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolston et al. (2019).Woolston A, Khan K, Spain G, Barber LJ, Griffiths B, Gonzalez-Exposito R, Hornsteiner L, Punta M, Patil Y, Newey A, Mansukhani S, Davies MN, Furness A, Sclafani F, Peckitt C, Jiménez M, Kouvelakis K, Ranftl R, Begum R, Rana I, Thomas J, Bryant A, Quezada S, Wotherspoon A, Khan N, Fotiadis N, Marafioti T, Powles T, Lise S, Calvo F, Guettler S, Loga K von, Rao S, Watkins D, Starling N, Chau I, Sadanandam A, Cunningham D, Gerlinger M. Genomic and transcriptomic determinants of therapy resistance and immune landscape evolution during Anti-EGFR treatment in colorectal cancer. Cancer Cell. 2019;36:35–50. doi: 10.1016/j.ccell.2019.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu et al. (2009).Wu G, Huang H, Abreu JG, He X. Inhibition of GSK3 phosphorylation of β-catenin via phosphorylated PPPSPXS motifs of Wnt coreceptor LRP6. PLOS ONE. 2009;4 doi: 10.1371/journal.pone.0004926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Chen & Fang (2020a).Xie YH, Chen YX, Fang JY. Comprehensive review of targeted therapy for colorectal cancer. Signal Transduction and Targeted Therapy. 2020a;5:22. doi: 10.1038/s41392-020-0116-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie et al. (2013).Xie M, He CS, Wei SH, Zhang L. Notch-1 contributes to epidermal growth factor receptor tyrosine kinase inhibitor acquired resistance in non-small cell lung cancer in vitro and in vivo. European Journal of Cancer. 2013;49:3559–3572. doi: 10.1016/j.ejca.2013.07.007. [DOI] [PubMed] [Google Scholar]
- Xie et al. (2020b).Xie ZY, Wang FF, Xiao ZH, Liu SF, Tang SL, Lai YL. Overexpressing microRNA-34a overcomes ABCG2-mediated drug resistance to 5-FU in side population cells from colon cancer via suppressing DLL1. Journal of Biochemistry. 2020b;167:557–564. doi: 10.1093/jb/mvaa012. [DOI] [PubMed] [Google Scholar]
- Xu et al. (2017).Xu JM, Wang Y, Wang YL, Wang Y, Liu T, Ni M, Li MS, Lin L, Ge FJ, Gong C, Gu JY, Jia R, Wang HF, Chen YL, Liu RR, Zhao CH, Tan ZL, Jin Y, Zhu YP, Ogino S, Qian ZR. PIK3CA mutations contribute to acquired cetuximab resistance in patients with metastatic colorectal cancer. Clinical Cancer Research. 2017;23:4602–4616. doi: 10.1158/1078-0432.CCR-16-2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu et al. (2019).Xu Z, Zhang Y, Xu M, Zheng X, Lin M, Pan J, Ye C, Deng Y, Jiang C, Lin Y, Lu X, Chi P. Demethylation and overexpression of CSF2 are involved in immune response, chemotherapy resistance, and poor prognosis in colorectal cancer. OncoTargets and Therapy. 2019;12:11255–11269. doi: 10.2147/OTT.S216829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yachida et al. (2019).Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, Watanabe H, Masuda K, Nishimoto Y, Kubo M, Hosoda F, Rokutan H, Matsumoto M, Takamaru H, Yamada M, Matsuda T, Iwasaki M, Yamaji T, Yachida T, Soga T, Kurokawa K, Toyoda A, Ogura Y, Hayashi T, Hatakeyama M, Nakagama H, Saito Y, Fukuda S, Shibata T, Yamada T. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nature Medicine. 2019;25:968–976. doi: 10.1038/s41591-019-0458-7. [DOI] [PubMed] [Google Scholar]
- Yadav et al. (2020).Yadav VK, Huang YJ, George TA, Wei PL, Sumitra MR, Ho CL, Chang TH, Wu ATH, Huang HS. Preclinical evaluation of the novel small-molecule msi-n1014 for treating drug-resistant colon cancer via the lgr5/β-catenin/mir-142-3p network and reducing cancer-associated fibroblast transformation. Cancer. 2020;12:1–15. doi: 10.3390/cancers12061590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada et al. (2020).Yamada T, Matsuda A, Takahashi G, Iwai T, Takeda K, Ueda K, Kuriyama S, Koizumi M, Shinji S, Yokoyama Y, Ohta R, Yoshida H. Emerging RAS, BRAF, and EGFR mutations in cell-free DNA of metastatic colorectal patients are associated with both primary and secondary resistance to first-line anti-EGFR therapy. International Journal of Clinical Oncology. 2020;25:1523–1532. doi: 10.1007/s10147-020-01691-0. [DOI] [PubMed] [Google Scholar]
- Yarden & Pines (2012).Yarden Y, Pines G. The ERBB network: at last, cancer therapy meets systems biology. Nature Reviews Cancer. 2012;12:553–563. doi: 10.1038/nrc3309. [DOI] [PubMed] [Google Scholar]
- Yi et al. (2020).Yi H, Li G, Long Y, Liang W, Cui H, Zhang B, Tan Y, Li Y, Shen L, Deng D, Tang Y, Mao C, Tian S, Cai Y, Zhu Q, Hu Y, Chen W, Fang L. Integrative multi-omics analysis of a colon cancer cell line with heterogeneous Wnt activity revealed RUNX2 as an epigenetic regulator of EMT. Oncogene. 2020;39:5152–5164. doi: 10.1038/s41388-020-1351-z. [DOI] [PubMed] [Google Scholar]
- Yonesaka et al. (2011).Yonesaka K, Zejnullahu K, Okamoto I, Satoh T, Cappuzzo F, Souglakos J, Ercan D, Rogers A, Roncalli M, Takeda M, Fujisaka Y, Philips J, Shimizu T, Maenishi O, Cho Y, Sun J, Destro A, Taira K, Takeda K, Okabe T, Swanson J, Itoh H, Takada M, Lifshits E, Okuno K, Engelman JA, Shivdasani RA, Nishio K, Fukuoka M, Varella-Garcia M, Nakagawa K, Jänne PA. Activation of ERBB2 signaling causes resistance to the EGFR-directed therapeutic antibody cetuximab. Science Translational Medicine. 2011;3:99ra86. doi: 10.1126/scitranslmed.3002442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon et al. (2019).Yoon HH, Shi Q, Heying EN, Muranyi A, Bredno J, Ough F, Djalilvand A, Clements J, Bowermaster R, Liu WW, Barnes M, Alberts SR, Shanmugam K, Sinicrope FA. Intertumoral heterogeneity of CD3þand CD8þT-cell densities in the microenvironment of DNA mismatch-repair–deficient colon cancers: implications for prognosis. Clinical Cancer Research. 2019;25:125–133. doi: 10.1158/1078-0432.CCR-18-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu et al. (2017).Yu TC, Guo F, Yu Y, Sun T, Ma D, Han J, Qian Y, Kryczek I, Sun D, Nagarsheth N, Chen Y, Chen H, Hong J, Zou W, Fang JY. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell. 2017;170:548–563. doi: 10.1016/j.cell.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu et al. (2021).Yu J, Hu D, Cheng Y, Guo J, Wang Y, Tan Z, Peng J, Zhou H. Lipidomics and transcriptomics analyses of altered lipid species and pathways in oxaliplatin-treated colorectal cancer cells. Journal of Pharmaceutical and Biomedical Analysis. 2021;200:114077. doi: 10.1016/j.jpba.2021.114077. [DOI] [PubMed] [Google Scholar]
- Yuan et al. (2018).Yuan LL, Li L, Liu JN, Mei J, Lei CJ. Down-regulation of miR-29a facilitates apoptosis of colorectal carcinoma cell SW480 and suppresses its Paclitaxel resistance. European Review for Medical and Pharmacological Sciences. 2018;22:5499–5507. doi: 10.26355/eurrev_201809_15810. [DOI] [PubMed] [Google Scholar]
- Yue et al. (2016).Yue B, Cai D, Liu C, Fang C, Yan D. Linc00152 functions as a competing endogenous RNA to confer oxaliplatin resistance and holds prognostic values in colon cancer. Molecular Therapy. 2016;24:2064–2077. doi: 10.1038/mt.2016.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue et al. (2018).Yue B, Liu C, Sun H, Liu M, Song C, Cui R, Qiu S, Zhong M. A positive feed-forward loop between LncRNA-CYTOR and Wnt/β-Catenin signaling promotes metastasis of colon cancer. Molecular Therapy. 2018;26:1287–1298. doi: 10.1016/j.ymthe.2018.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurgelun et al. (2017).Yurgelun MB, Kulke MH, Fuchs CS, Allen BA, Uno H, Hornick JL, Ukaegbu CI, Brais LK, McNamara PG, Mayer RJ, Schrag D, Meyerhardt JA, Ng K, Kidd J, Singh N, Hartman AR, Wenstrup RJ, Syngal S. Cancer susceptibility gene mutations in individuals with colorectal cancer. Journal of Clinical Oncology. 2017;35:1086–1095. doi: 10.1200/JCO.2016.71.0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang et al. (2014a).Zhang L, Castanaro C, Luan B, Yang K, Fan L, Fairhurst JL, Rafique A, Potocky TB, Shan J, Delfino FJ, Shi E, Huang T, Martin JH, Chen G, MacDonald D, Rudge JS, Thurston G, Daly C. ERBB3/HER2 signaling promotes resistance to EGFR blockade in head and neck and colorectal cancer models. Molecular Cancer Therapeutics. 2014a;13:1345–1355. doi: 10.1158/1535-7163.MCT-13-1033. [DOI] [PubMed] [Google Scholar]
- Zhang et al. (2014b).Zhang L, Dong Y, Zhu N, Tsoi H, Zhao Z, Wu CW, Wang K, Zheng S, Ng SSM, Chan FKL, Sung JJY, Yu J. MicroRNA-139-5p exerts tumor suppressor function by targeting NOTCH1 in colorectal cancer. Molecular Cancer. 2014b;13:1–12. doi: 10.1186/1476-4598-13-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang et al. (2017a).Zhang C, Li C, Chen X, Zhou Y, Yin B, Ni R, Zhang Y, Liu J. Overexpression of dishevelled 2 is involved in tumor metastasis and is associated with poor prognosis in hepatocellular carcinoma. Clinical and Translational Oncology. 2017a;19:1507–1517. doi: 10.1007/s12094-017-1697-z. [DOI] [PubMed] [Google Scholar]
- Zhang et al. (2017b).Zhang K, Li M, Huang H, Li L, Yang J, Feng L, Gou J, Jiang M, Peng L, Chen L, Li T, Yang P, Yang Y, Wang Y, Peng Q, Dai X, Zhang T. Dishevelled1-3 contribute to multidrug resistance in colorectal cancer via activating Wnt/β-catenin signaling. Oncotarget. 2017b;8:115803–115816. doi: 10.18632/oncotarget.23253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou et al. (2017).Zou ZZ, Nie PP, Li YW, Hou BX, Rui-Li, Shi XP, Ma ZK, Han BW, Luo XY. Synergistic induction of apoptosis by salinomycin and gefitinib through lysosomal and mitochondrial dependent pathway overcomes gefitinib resistance in colorectal cancer. Oncotarget. 2017;8:22414–22432. doi: 10.18632/oncotarget.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao et al. (2020).Zhao Q, Zhuang K, Han K, Tang H, Wang Y, Si W, Yang Z. Silencing DVL3 defeats MTX resistance and attenuates stemness via Notch Signaling Pathway in colorectal cancer. Pathology Research and Practice. 2020 doi: 10.1016/j.prp.2020.152964. [DOI] [PubMed] [Google Scholar]
- Zhong et al. (2013).Zhong S, Li W, Chen Z, Xu J, Zhao J. MiR-222 and miR-29a contribute to the drug-resistance of breast cancer cells. Gene. 2013;531:8–14. doi: 10.1016/j.gene.2013.08.062. [DOI] [PubMed] [Google Scholar]
- Zhou et al. (2020).Zhou Y, Cheng X, Zhang F, Chen Q, Chen X, Shen Y, Lai C, Kota VG, Sun W, Huang Q, Yuan Y, Wang J, Lai M, Zhang D. Integrated multi-omics data analyses for exploring the co-occurring and mutually exclusive gene alteration events in colorectal cancer. Human Mutation. 2020;41:1588–1599. doi: 10.1002/humu.24059. [DOI] [PubMed] [Google Scholar]
- Zouggar, Haebe & Benoit (2020).Zouggar A, Haebe JR, Benoit YD. Intestinal microbiota influences dna methylome and susceptibility to colorectal cancer. Gene. 2020;11:1–6. doi: 10.3390/genes11070808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The following information was supplied regarding data availability:
There are no raw data