Abstract
Background
Severe trauma and burns accompanied by sepsis are associated with high morbidity and mortality. Little is known about the transcriptional similarity between trauma, burns, sepsis, and systemic inflammatory response syndrome (SIRS). Uncovering key genes and molecular networks is critical to understanding the biology of disease. Conventional analysis of gene changes (fold change) analysis is difficult for time-serial data such as post-injury blood transcriptome.
Methods
Weighted gene co-expression network analysis (WGCNA) was applied to the trauma dataset to identify modules and hub genes. Module stability was tested by half sampling. Module preservations of burns, sepsis, and SIRS were calculated using trauma as reference. Module functional enrichment was analyzed in gProfiler server. Candidate drugs were screened using Connectivity Map based on hub genes. The modules were visualized in Cytoscape.
Results
Seventeen modules were identified. The modules were robust to the exclusion of half the sample. They were involved in lymphocyte and platelet activation, erythrocyte differentiation, cell cycle, translation, and interferon signaling. In addition, highly connected hub genes were identified in each module, such as GUCY1B1, BCL11B, HMMR, and CEACAM6. High BCL11B (M13) or low CEACAM6 (M27) expression indicates better survival prognosis in sepsis patients regardless of endotype class and age. Network preservation in burns, sepsis, and SIRS showed a general similarity between trauma and burns. M4, M5, M13, M16, M20, and M27 were significantly associated with injury time in trauma and burns. High M13 (T cell activation), low M15 (cell cycle), and low M27 (neutrophil activation) indicate better survival of sepsis patients, regardless of endotype class and age. Moreover, the modules can efficiently separate patients with different diseases. Some modules and hub genes have good prognostic performance in sepsis. Based on the hub genes of each module, six candidate drugs were screened.
Conclusion
This study first compared the gene co-expression modules in trauma, burns, sepsis, and SIRS. The identified modules are useful for disease prognosis and drug discovery. BCL11B and CEACAM6 may be promising biomarkers for sepsis risk assessment.
Keywords: WGCNA, SIRS, Connectivity Map, hub gene
Introduction
Trauma accounts for approximately 10% of deaths and 16% of disabilities worldwide.1 Billions of dollars have been spent on research into new biological therapeutics for severe injuries, as well as post-traumatic sepsis and septic shock.2 Burn injuries cause unpredictable trauma and sepsis is a complication associated with high morbidity and mortality.3 The mechanisms of the complex host response to trauma remain unclear, which in turn is a barrier to the development of effective therapies.
With the development of high-throughput technologies, microarray has become a mature method that measures thousands of genes in one experiment. It has been used to study the temporal dynamics of gene expression in human and animal models.2,4 These studies focused on changes in gene expression and signaling pathways. A “genomic storm” was observed in the blood transcriptome following severe blunt trauma. The early genomic response of leukocytes is consistent with concomitant increased expression of genes involved in systemic inflammation, innate immune, and compensatory anti-inflammatory response, as well as suppression of genes involved in adaptive immunity.2 In a second study, the transcriptomes of human trauma, burns, and murine models were integrated, and the correlation of gene changes between human subjects and mouse models was analyzed.4 The study found that genomic responses in mouse models poorly mimicked human inflammatory diseases.4 However, the molecular differences between trauma, burns, sepsis, and systemic inflammatory response syndrome (SIRS) in humans have not been presented.
Here, we used systems biology methods to analyze these valuable datasets. The state-of-the-art method can help identify important gene clusters (modules) and hub genes in trauma. A module is a cluster of co-expressed genes, where genes in one module have a low correlation with genes in other modules. Co-expressed genes respond to trauma coordinately, so a module-level analysis is more robust than an analysis of individual genes. Hub genes are genes with a high correlation with other genes in a module. Key modules and hub genes were identified. We also first analyzed the similarities between trauma, burns, sepsis, and SIRS at the module level. Modules can effectively separate different disease samples. Modules and hub genes have good prognostic performance in a sepsis dataset containing survival data. Finally, based on these hub genes, we discovered six candidate drugs that merit further experimental validation.
Materials and Methods
Datasets Preparation
Three datasets were analyzed in the study. Trauma dataset GSE36809 is the blood transcriptome of 167 patients with severe blunt trauma.2 Burns dataset GSE37069 is the blood transcriptome of 244 patients with severe burns.4 Sepsis and SIRS blood dataset GSE74224 is the blood transcriptome of 74 sepsis and 31 SIRS patients.5 GSE65682 is the blood transcriptome of 802 critically ill patients and healthy controls.6 All these datasets are publicly and freely available in NCBI GEO database.7 The Institutional Ethics Committee of Cangzhou Medical College, Cangzhou, Hebei, has approved the current research project.
Weighted Gene Co-Expression Network Analysis (WGCNA)
The trauma dataset which contains 857 samples was used for WGCNA. Briefly, the raw data was analyzed in the vendor-provided software Expression Console v1.4.1.46. Probe set level expression values were filtered by coefficient of variation (CV). For genes with multiple probe sets, the single probe set with the highest CV was retained. Because WGCNA was a memory-intensive process, only genes with an average expression intensity greater than 100 were used for network construction. Module identification was performed according to the package manual.8 Parameters were set as follows: softPower = 16, networkType = “signed”, minModuleSize =30, deepSplit = 4, MEDissThres = 0.2. Module stability was tested by sampling half of the samples. Stability was represented by correlation of intra-module connectivity between the original one and the sampled one in terms of mean ± standard deviation.9 After identifying the reference modules, the burns, sepsis, and SIRS datasets were projected onto the reference to analyze module preservation. The prameters for module preservation were set as: networkType = “signed”, nPermutations = 100.
Functional Annotation of the Modules
Functional annotation of the modules was performed in gProfiler.10 For simplicity, only informative and the key terms were recorded. Hub genes in a module were screened by intra-connectivity value. The modules were visualized in Cytoscape.11
Connectivity Map Analysis
Drug candidates are associated with trauma, burns, and sepsis according to the hub genes of differential modules. The top 3 hub genes in each module were extracted and categorized into two groups of up-regulation or down-regulation. The two groups of genes were submitted to Connectivity Map v2. The significance threshold for candidate drugs was set at 0.01. Only the candidates with negative enrichment scores were recorded. The top 2 drugs for each disease were recorded.
Statistical Analysis
One-way ANOVA was used to compare differences between multiple groups. The Student’s t-test was used to compare the means of two groups. Statistical significance was set at 0.01 unless otherwise stated. When multiple comparisons were made, P values were adjusted using the Bonferroni method. The AUC of ROC was calculated using the ROCR package. Survival analysis was performed in the SurvivalAnalysis package. We set a strict statistical significance of 0.01 throughout the manuscript to obtain more reliable results.
Results
Seventeen Modules Were Identified in the Trauma Dataset
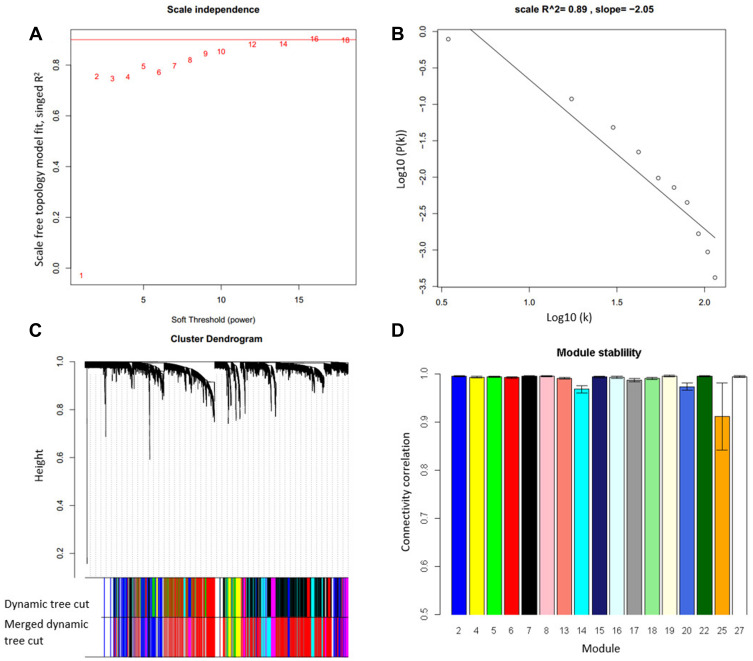
Gene modules in the trauma dataset were identified by WGCNA. The softPower was set to 16, resulting in a scale-free network, a common property for a biological network (Figure 1A and B). Based on the network, seventeen modules were identified by WGCNA (Figure 1C). To test the module stability, we used only half of the samples and analyzed the connectivity after sampling. The result showed that all modules were stable and had a mean connectivity correlation larger than 0.91 (Figure 1D). Thus, they are suitable for downstream analysis.
Figure 1.
Seventeen gene co-expression modules were identified in the trauma dataset. (A) The softPower was set at 14 to get a scale-free network. (B) Power law distribution test to check if the constructed network is scale-free network. (C) Seventeen modules were identified from the network and each module was assigned a color. (D) The modules were robust to 50% sample exclusion indicated by the correlation of original gene connectivity and the gene connectivity calculated from half samples.
Functional Enrichment Analysis Revealed Modules Function
To decipher the biological function of the identified modules, the genes were submitted to gProfiler for functional enrichment analysis. All modules were enriched with specific functions. For simplicity, the top term with the highest significance in each module was recorded (Table 1). Some small modules were also significant. For example, M15 contains 108 genes, and 75 of which are involved in the cell cycle. M27 contains 33 genes, and 24 of them are involved in neutrophil activation.
Table 1.
Functional Enrichment for the Seventeen Modules Identified in the Trauma Dataset
| Module | Size | Top Term (P ≤) | Hub Gene |
|---|---|---|---|
| 2 | 1235 | Factor: hdac2 (5E-48) vesicle-mediated transport (5E-28) | GNB2 |
| 4 | 625 | Factor: ETF (5E-13) Endomembrane system (3E-12) | SERPINB1 |
| 5 | 565 | Translation (1E-22) Mitochondrion (4E-18) | MRPL57 |
| 6 | 323 | Translation (4E-39) | RPL8 |
| 7 | 163 | Platelet activation (7E-15) | GUCY1B1 |
| 8 | 241 | Factor: GABP-alpha (1E-8) Response to stress (6E-6) | MCL1 |
| 13 | 121 | T cell activation (5E-16) | BCL11B |
| 14 | 215 | Factor: ETF (9E-10) Protein transport (1E-6) | MIER1 |
| 15 | 108 | Cell cycle (1E-49) | HMMR |
| 16 | 733 | Myeloid leukocyte activation (2E-29) | FLOT2 |
| 17 | 105 | Factor: BTEB3 (1E-4) Nuclear lumen (3E-10) | AKAP13 |
| 18 | 1774 | Factor: ZF5 (6E-41) Nucleus (1E-42) | SRSF11 |
| 19 | 85 | Erythrocyte differentiation (1E-7) | SELENBP1 |
| 20 | 85 | hsa-miR-576-3p (0.005) | ANAPC13 |
| 22 | 74 | Factor: IRF1 (6E-27) defense response to virus (7E-29) | IFI44L |
| 25 | 53 | Neutrophil degranulation (1E-4) | BASP1 |
| 27 | 33 | Neutrophil activation (3E-28) | CEACAM6 |
Notes: Hub gene is the top gene with high intra-module connectivity in each module. Size: the number of genes in a module. Module: the numerical label for each module. Top term: the informative and significant functional terms for each module.
Module Preservation Analysis for Trauma, Burns, Sepsis, and SIRS
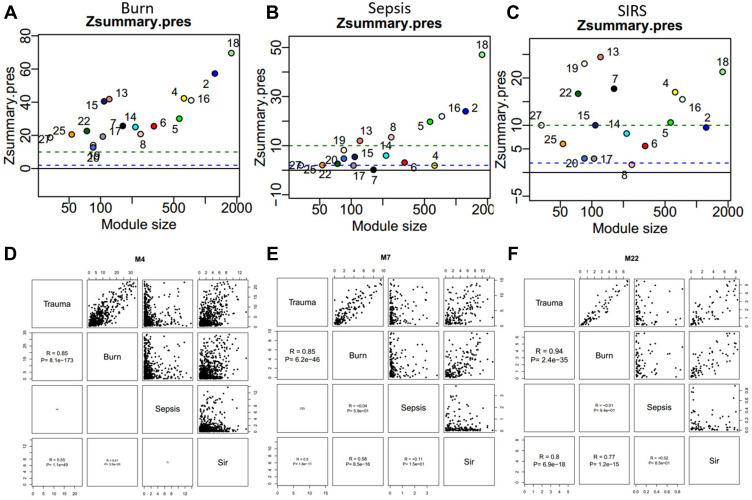
To compare the similarity at the module level between trauma, burns, sepsis and SIRS, we downloaded additional datasets from NCBI GEO and projected these datasets to trauma. The module preservation analysis revealed that the seventeen modules were all highly preserved in the burns dataset (Figure 2A). The preservation was low in the sepsis dataset. In sepsis, M4, M7, M17, M22, and M25 showed no preservation, while M2, M5, M8, M13, M16, and M18 showed strong preservation (Figure 2B). In SIRS, M8 showed no preservation, and M17 and M20 showed marginal preservation, and M4, M5, M7, M13, M16, M18, M19, and M22 and M25 showed strong preservation (Figure 2C). We also compared the four diseases by network connectivity. It showed that global connectivity is well correlated between trauma and burns but not trauma and other two diseases. To check if any intra-module connectivity is correlated, we performed a pair-wise comparison between two groups of diseases. We found that some of the modules are well correlated, such as M4, M7, and M22 (Figure 2D–F).
Figure 2.
Module preservation analysis and gene connectivity correlation analysis suggest the similarities and differences between trauma, burns, sepsis and SIRS. (A–C) The identified modules are preserved in the burns, sepsis, and SIRS, using trauma as reference. Zsummary.press statistic indicates -log10 statistical significance of the module. The green dashed line indicates strong preservation. The blue dashed line indicates moderate preservation. (D–F) Some modules have a high correlation of intra-module connectivity among trauma, sepsis, burns, and SIRS.
Abbreviations: R, Spearman correlation; SIRS, systemic inflammatory response syndrome.
Hub Genes are Important in Trauma, Burns, Sepsis and SIRS
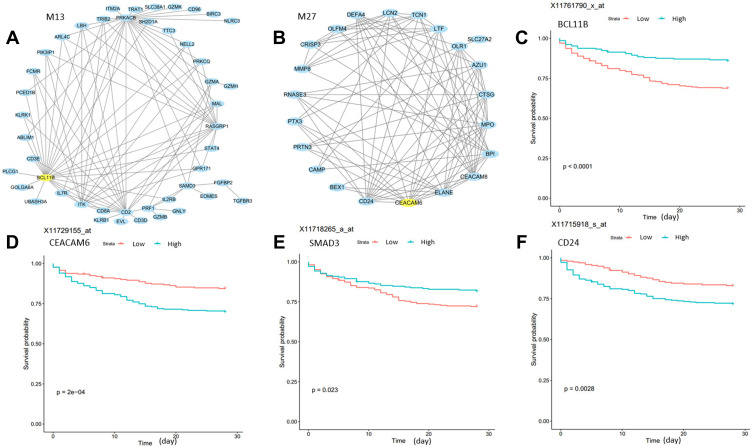
Connectivity, the sum of the correlation of a gene with all other genes in a module, is an important metric for network biology. We listed the top hub genes of the identified modules in Table 1. These genes were highly connected with other genes in the module it locates. To provide a systematic overview of the module, we selected the top 100 connections of each module and visualized two of the modules (Figure 3A and B). To demonstrate the utility of the hubs, we used the M13 and M27 hub genes BCL11B and CEACAM6 to stratify sepsis patients in GSE65682. Interestingly, all probe sets of the two genes in the microarray data can very significantly infer patient survival (Figure 3C and D). BCL11B is involved in the progression of neonatal sepsis.12 CEACAM6 gene expression in cultured epithelial cells was upregulated by wounding and neutrophil elastase.13 However, the role of CEACAM6 in trauma and sepsis has not yet been reported. In addition to these hub genes, other highly connected genes can also be found in these modules. For example, PRKACB in M13 was recently reported as a potential biomarker in sepsis.14 PRKACB, encoding the catalytic subunits of PKA, was down-regulated in chronic traumatic encephalopathy.15 SMAD3 in M13 is prognostic for survival (Figure 3E). Highly connected gene CD24 in M27 may protect the host against the exaggerated inflammatory response in sepsis.16 CD24 levels are elevated in patients and mice with traumatic brain injury.17 CD24 is also a prognostic factor for survival (Figure 3F).
Figure 3.
Hub genes in M13 and M27 are prognostic for sepsis patient survival. (A and B) Module visualization for the top 100 connections within M13 and M27 by using Cytoscape. Yellow nodes indicate the hub gene of modules. (C–F) Kaplan–Meier curves show that hub genes of M13 and M27, BCL11B and CEACAM6, and highly connected genes SMAD3 and CD24 can stratify sepsis patients. The P values are computed using Log rank test. Greenline: high expression; Red line: low expression.
Modules Were Associated with Injury-Time
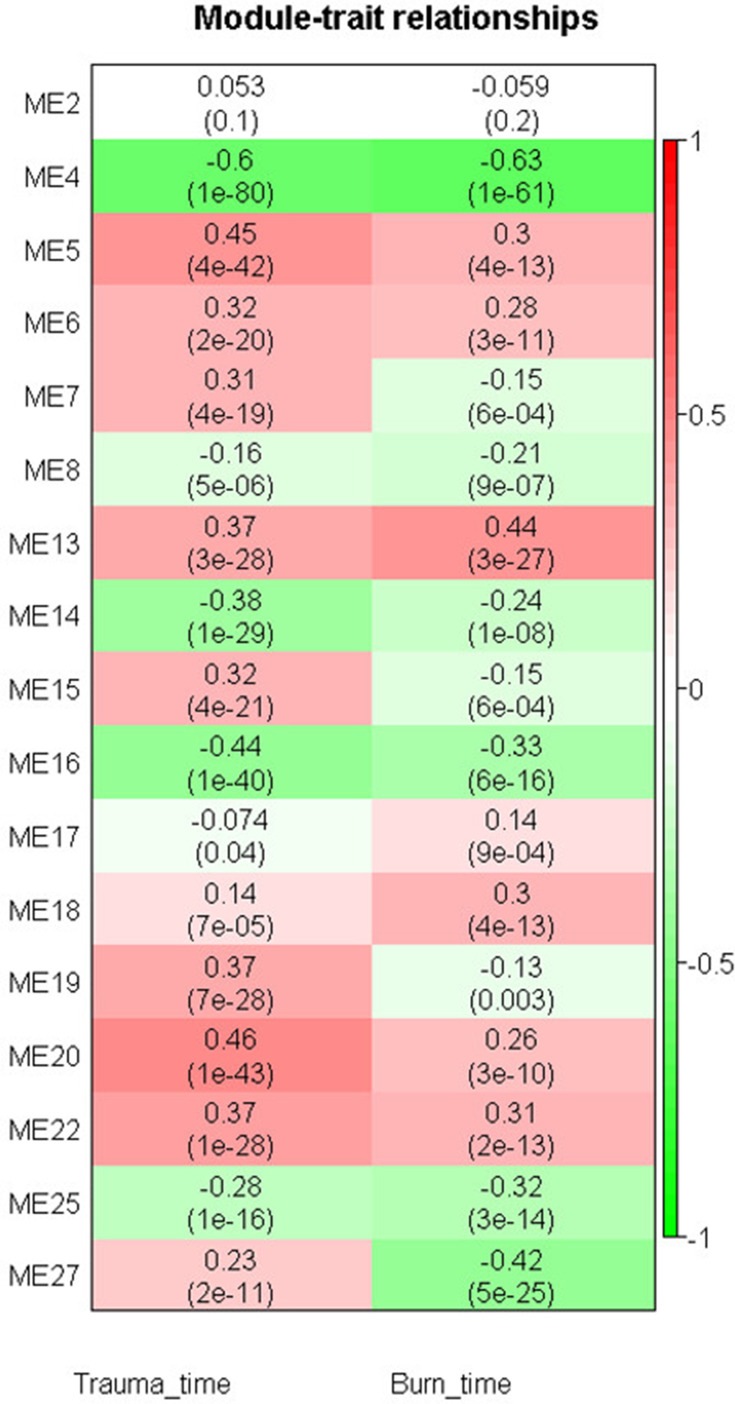
To determine whether module expression is associated with injury time, we correlated injury time with module eigengenes, which represents the gene expression profiles of each module. M4 was found to be most significantly and negatively correlated with injury time in both trauma and burns (r = −0.60, P ≤ 1E-80 and (r = 0.63, P ≤ 1E-61, respectively). M5 and M20 were positively correlated with injury-time (r = 0.45, P ≤ 4E-42 and r = 0.46, P ≤ 1E-43, respectively), whereas M16 was negatively correlated with injury-time in trauma (r = 0.44, P ≤ 1E-40). M13 was positively correlated with injury-time (r = 0.44, P ≤ 3E-27), whereas M27 was negatively correlated with injury-time (r = −0.42, P ≤ 5E-25) in burns (Figure 4).
Figure 4.
Module-trait relationship heat map for the trauma and burns datasets. Cells represent correlation between modules expression and trauma or burn time. Numbers in the bracket indicate the statistical significance.
Modules Can Efficiently Separate Patients with Different Diseases
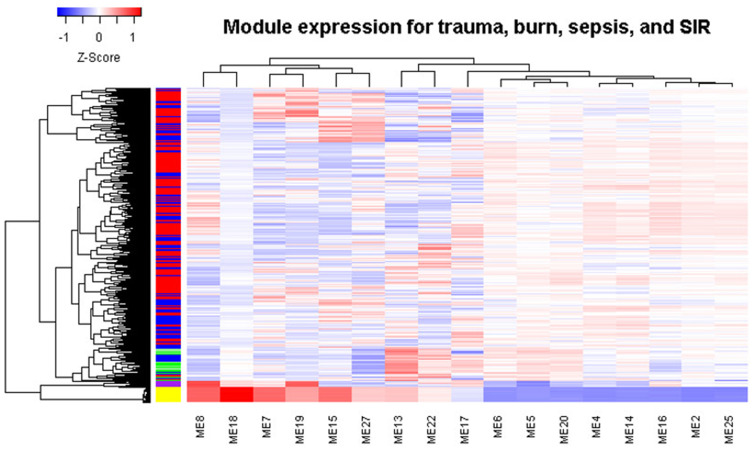
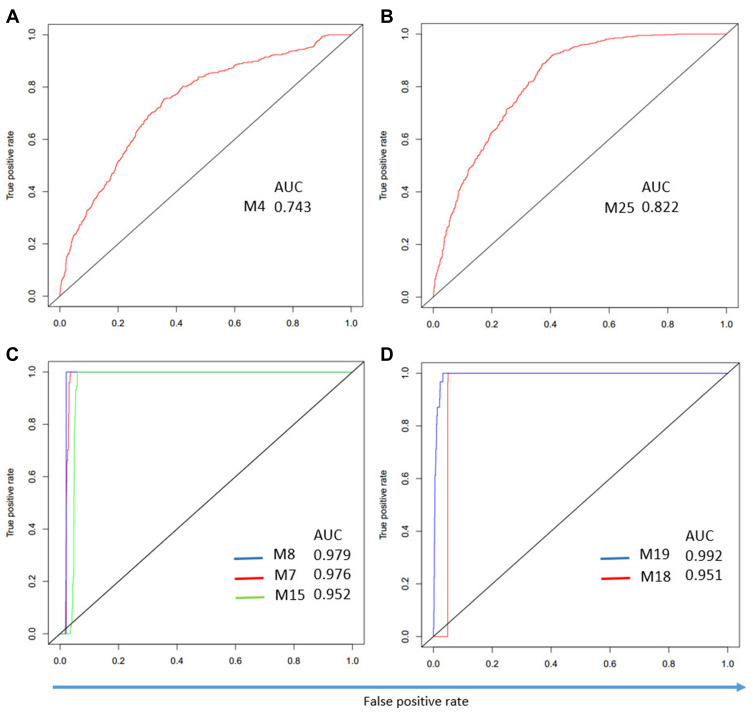
We found that the identified modules have different expression patterns in trauma, burns, sepsis, and SIRS (Figure 5). Sepsis forms a distinct cluster, while the control samples are clustered with SIRS. Therefore, we performed ROC analysis to test whether the modules can separate samples from different disease groups. For burns, M4 has the highest AUC of 0.74 (Figure 6A). This is consistent with our findings that trauma and burns are similar overall. For trauma, M25 has the highest AUC of 0.82 (Figure 6B). For sepsis, M7, M8, and M15 have an AUC greater than 0.91 (Figure 6C). For SIRS, both M18 and M19 have an AUC greater than 0.95 (Figure 6D). Thus, these modules can be used to discriminate different diseases.
Figure 5.
Module expression clustering heat map shows the different expression patterns across trauma, burns, sepsis and SIRS. In the left color bar, yellow indicates sepsis, green indicates normal samples, red indicates trauma, blue indicates burns, and purple indicates SIRS.
Abbreviation: SIRS, systemic inflammatory response syndrome.
Figure 6.
Modules can discriminate patients with different diseases. (A) ROC curve for M4 in the burns. (B) ROC curve for M25 in trauma. (C) ROC curve for M7, M8, and M15 in sepsis. (D) ROC curve for M18 and M19 in SIRS.
Abbreviations: ROC, receiver operating characteristic; AUC, area under the ROC curve.
Clinical Implication of the Identified Modules in Critically-Ill Patients
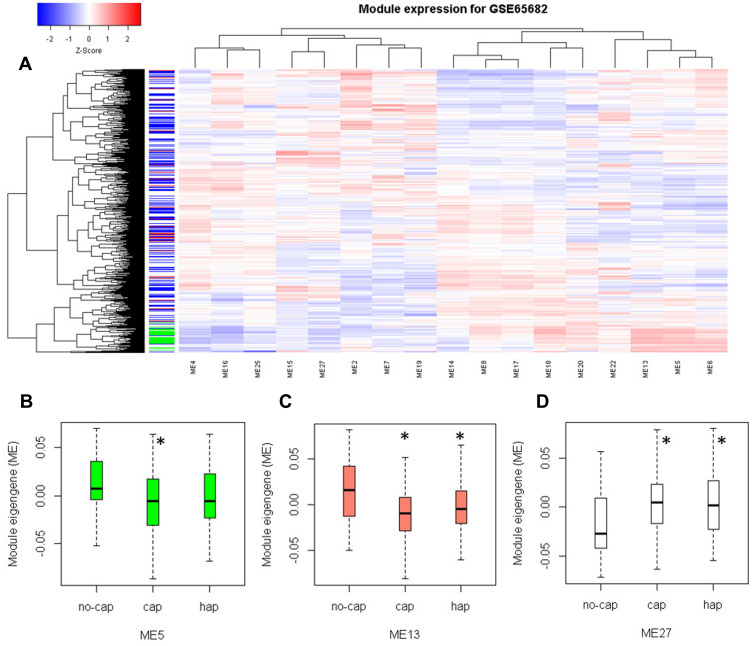
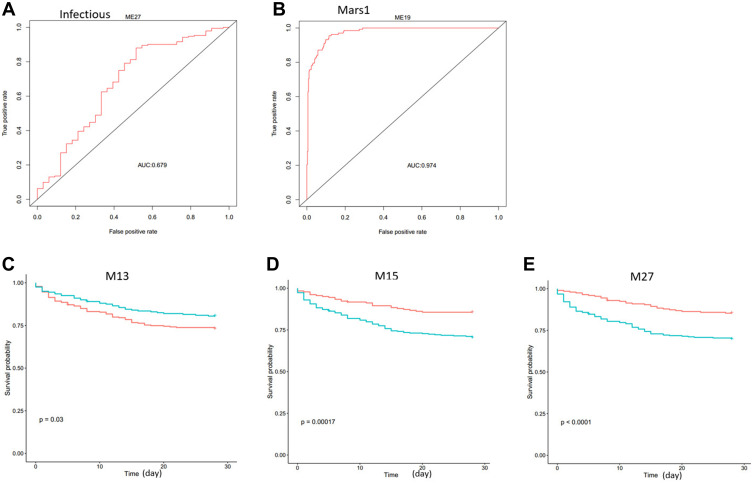
To examine if the modules work in the clinic, we downloaded an additional dataset GSE65682, which contains blood samples of 42 healthy and 760 critically ill patients. Module clustering analysis showed that healthy subjects had higher expression of M5, M6, and M13 (Figure 7A). Differential module expression analysis showed that M5 and M13 were down-regulated, and M27 was up-regulated in patients with community-acquired pneumonia (cap) compared to patients without cap (Figure 7B–D). We used module expression to distinguish patients from different groups. M27 has the highest ability to discriminate sepsis from those with non-infectious causes of disease (Figure 8A). As for the Molecular Diagnosis and Risk Stratification of Sepsis (MARS) classification, M19 can perfectly discriminate mars1 patients from other patients (Figure 8B). We also tested the prognostic ability of module gene expression for patient survival. In univariate analysis step, M13, M15, M19, M20, and M27 were identified to be associated with survival. In multivariate analysis, age and endotype were also included in the model. We found that M13, M15, and M27 could still stratify patients with different survival in the Cox proportional hazard model (Table 2, Figure 8C–E). Low M13, high M15, and high M27 indicate poor survival in sepsis patients. These results may indicate that M13, M15, and M27 are independent prognostic factors for survival in sepsis patients. This information suggests that cap patients may have a higher risk of death compared with no cap as cap patients have low M13 and high M27 (Figure 7C and D).
Figure 7.
Module expression is different in different groups of sepsis patients. (A) Module expression heat map for sepsis dataset GSE65682. (B–D) The box plot shows the module expression changed in M5, M13, and M27 in control, cap, and no-cap samples. In the left color bar, green indicates normal samples, white indicates no-cap, blue indicates hap, and red indicates cap. *Statistical significance < 0.01.
Abbreviations: Cap, community-acquired pneumonia; hap, hospital-acquired pneumonia.
Figure 8.
Modules are discriminative for infectious and mars1, and are prognostic for survival in sepsis dataset GSE65682. (A) M27 expression can separate samples with infectious sepsis and no infections. (B) M19 expression can distinguish samples with different MARS staging. (C–E) M13, M15 and M27 can significantly separate patients with different prognoses. The P values are computed using Log rank test.
Abbreviations: AUC, area under the ROC Curve; Mars, molecular diagnosis and risk stratification of sepsis.
Table 2.
Significant Modules in Univariate and Multivariate Analysis for Sepsis Dataset GSE65682
| Module | Univariate Analysis | Multivariate Analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |
| M13 | 2.6E-4 | 3.2E-7, 0.206 | 0.015 | 2.3E-4 | 1.3E-07, 0.411 | 0.028 |
| M15 | 4E+3 | 55, 3.0E+5 | 1.5E-4 | 6348 | 61, 6.6E+5 | 2.2E-4 |
| M19 | 1329 | 6.1, 2.9E+5 | 8.8E-3 | |||
| M20 | 4.3E-3 | 2.2E-6, 0.083 | 3.9E-3 | |||
| M27 | 4E+4 | 154, 1E+7 | 2.3E-4 | 1.1E+4 | 26, 4.6E+6 | 2.5E-3 |
Notes: In multivariate Cox regression analysis, endotype_class and age were also included in the survival model.
Abbreviations: HR, hazard ratios; CI, confidence intervals.
Connectivity Map Analysis Identified Candidate Drugs for Trauma
To identify drug candidates for trauma, we submitted hub genes from up-regulated modules in each disease to the Connectivity Map tool. Six drug candidates were identified and are listed in Table 3. Trichostatin A can reduce systemic inflammation and improves survival in mice with sepsis.18 Tanespimycin is a heat shock protein 90 inhibitor that can prolong survival, attenuate inflammation, and reduce lung injury in mice with sepsis.19 Other drugs need to be verified in future studies.
Table 3.
Candidate Drugs Identified by Connectivity Map
| Disease | Rank | Name | P |
|---|---|---|---|
| Sepsis | 1 | Trichostatin A | 5E-9 |
| 2 | Tanespimycin | 2E-8 | |
| Burn | 1 | Tanespimycin | 1E-5 |
| 2 | Monastrol | 3E-4 | |
| Trauma | 1 | LY-294002 | 7E-6 |
| 2 | Prochlorperazine | 4E-3 |
Notes: Top 3 hub genes in differential modules for each disease were submitted to the Connectivity Map tool. Only the top 2 candidate drugs were recorded.
Discussion
To our knowledge, this is the first report in which systems biology network analysis has been applied to the trauma dataset and compared with sepsis, burns, and SIRS.3,20 Traumatic injuries with their potential for infection were likely a common cause of death.2 Burn injuries are associated with the highest risk of sepsis.21 However, the relationship between these diseases is still unclear. Although high-throughput technologies, such as Next Generation Sequencing and microarray are widely used in disease research, trauma-related datasets are still precious. Traditional gene differential analysis or gene fold change methods have been applied to the transcriptome of trauma. Unfortunately, it is difficult to apply these methods because trauma, burns, or sepsis data are usually time-serial. A gene may have different directions of fluctuation at different time points. Fold change results will not be consistent if measured at different time points or different time points are chosen as controls. The module-based analysis will give more consistent results because the function of the module is always more stable than that of a single gene. Here, we first identified gene co-expression modules in trauma and compared them with those in burns, sepsis, and SIRS. Based on the modules, candidate drugs for trauma were implicated.
The identified modules and hub genes are important in trauma, burns, sepsis, and SIRS. We found that M13 is associated with lymphocyte activation, is up-regulated in trauma, and correlates positively with injury duration. In the original paper, the authors found that injury leads to early activation of genes involving innate and concomitant suppression of genes involving adaptive immunity.2 Therefore, lymphocyte activation is a sustained process. M25 was associated with neutrophil granules and was also up-regulated, but correlated negatively with injury time. This result may suggest that it is an acute response that is up-regulated early but decreases with duration of injury. Previous results also showed that granulocyte colony-stimulating factor (G-CSF) was elevated on day 1, but quickly decreased within 7 days.22 Recent studies have shown the adverse role of G-CSF after severe trauma.23 Therefore, it is difficult to perform fold-change analysis in time-series data because the direction of fold-change changes when different time points are chosen as controls. HMMR is the hub gene in M15 (cell cycle), which plays a role in wound repair in trauma.24 However, we identified HMMR as a risk gene in sepsis. Higher HMMR expression indicates poorer survival in sepsis (HR=1.38, P=0.0014). HMMR is up-regulated across the pediatric systemic inflammatory response syndrome, sepsis, and septic shock spectrum in children.25 Thus, the hub genes of these modules may be potential biomarkers or targets for therapy. There is also supportive evidence for other modules. M7 is associated with platelet activation. Severe injury usually results in increased platelet activation and function. However, the combination of increased platelet activation and decreased function was associated with increased mortality.26 Top M22 terms include interferon signaling (P =3E-26). The M22 hub gene interferon-induced protein 44-like (IFI44L) can distinguish viral from bacterial infections in febrile children.27 In traumatic brain injury, interferon is a risk factor contributing to the neuroinflammatory process.28
We also compared trauma with other inflammatory conditions, such as burns, sepsis, and SIRS. Module-level analysis confirmed the similarity between trauma and burns. In a previous work based on correlation analysis of gene alterations, human trauma had very a high similarity (r = 0.91) with burns.4 Some modules were not preserved in sepsis and SIRS. Currently, there are few reports analyzing the differences between these diseases. It has been shown that persistent coagulation disorders occurred in sepsis patients.29 Experimental human endotoxemia, a model of SIRS, showed a moderate similarity to trauma or burns (r = 0.47).4,30 In our result, the distribution of SIRS preservation is similar to that of burns, but SIRS preservation is marginal in 3 modules. These results may be indicative of the different mechanisms involved in trauma and sepsis/ SIRS.
Finally, the tool Connectivity Map was used to find candidate drugs. Of the six drugs with significant P values, some are already being used to treat disease. Others may be validated in future work.
This analysis is limited by the fact that WGCNA generates an undirected network. Therefore, current results provide only gene correlation information and lack direction information concerning gene or protein interactions. Further experiments will be needed to confirm the regulation relationship between genes.
Conclusion
A total of 17 gene co-expression modules were identified in the trauma dataset. These modules were stable and were involved in lymphocyte and platelet activation, erythrocyte differentiation, cell cycle, neutrophil degranulation, and interferon signaling. These modules were preserved in burns and SIRS, but less in sepsis. M4, M5, M13, M16, M20, and M27 were significantly associated with injury time in trauma and burns. Modules can efficiently separate patients with different diseases. Modules and hub genes have good prognosis performance in sepsis. The six candidate drugs for trauma treatment merit further investigation.
Funding Statement
This work was supported by the Science and Technology Research Development Guidance Plan of Cangzhou, China (Nos. 162302158 and 172301001) and the Key Research and Development Projects of Hebei Province, China (No.19272404D).
Abbreviations
SIRS, systemic inflammatory response syndrome; WGCNA, weighted gene co-expression network analysis; CV, coefficient of variation; ANOVA, analysis of variance; ROC, receiver operating characteristic; AUC, Area Under the ROC curve; NCBI, National Center for Biotechnology Information; GEO, Gene Expression Omnibus; cap, community-acquired pneumonia; MARS, Molecular Diagnosis and Risk Stratification of Sepsis.
Data Sharing Statement
The datasets are available at https://www.ncbi.nlm.nih.gov/geo/.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Lord JM, Midwinter MJ, Chen YF, et al. The systemic immune response to trauma: an overview of pathophysiology and treatment. Lancet. 2014;384(9952):1455–1465. doi: 10.1016/S0140-6736(14)60687-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xiao W, Mindrinos MN, Seok J, et al. A genomic storm in critically injured humans. J Exp Med. 2011;208(13):2581–2590. doi: 10.1084/jem.20111354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hazeldine J, Hampson P, Lord JM. The diagnostic and prognostic value of systems biology research in major traumatic and thermal injury: a review. Burns Trauma. 2016;4:33. doi: 10.1186/s41038-016-0059-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seok J, Warren HS, Cuenca AG, et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci USA. 2013;110(9):3507–3512. doi: 10.1073/pnas.1222878110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McHugh L, Seldon TA, Brandon RA, et al. A molecular host response assay to discriminate between sepsis and infection-negative systemic inflammation in critically ill patients: discovery and validation in independent cohorts. PLoS Med. 2015;12(12):e1001916. doi: 10.1371/journal.pmed.1001916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bos LDJ, Scicluna BP, Ong DSY, Cremer O, van der Poll T, Schultz MJ. Understanding heterogeneity in biologic phenotypes of acute respiratory distress syndrome by leukocyte expression profiles. Am J Respir Crit Care Med. 2019;200(1):42–50. doi: 10.1164/rccm.201809-1808OC [DOI] [PubMed] [Google Scholar]
- 7.Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41(Database issue):D991–995. doi: 10.1093/nar/gks1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 2008;9(1):559. doi: 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu W, Wang Y, He H. CoFly: a gene coexpression database for the fruit fly Drosophila melanogaster. Arch Insect Biochem Physiol. 2020;105(1):e21693. doi: 10.1002/arch.21693 [DOI] [PubMed] [Google Scholar]
- 10.Reimand J, Arak T, Adler P, et al. g:Profiler-a web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res. 2016;44(W1):W83–89. doi: 10.1093/nar/gkw199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang L, Qiao L, Zhu H, Jiang L, Yin L. Genomics of neonatal sepsis: has-miR-150 targeting BCL11B functions in disease progression. Ital J Pediatr. 2018;44(1):145. doi: 10.1186/s13052-018-0575-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shikotra A, Choy DF, Siddiqui S, Arthur G. A CEACAM6-high airway neutrophil phenotype and CEACAM6-high epithelial cells are features of severe asthma. J Immunol. 2017;198(8):3307–3317. [DOI] [PubMed] [Google Scholar]
- 14.Zeng X, Feng J, Yang Y, Zhao R, Yu Q. Screening of key genes of sepsis and septic shock using bioinformatics analysis. J Inflamm Res.. 2021;14:829–841. doi: 10.2147/JIR.S301663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho H, Hyeon SJ, Shin J-Y, et al. Alterations of transcriptome signatures in head trauma-related neurodegenerative disorders. Sci Rep. 2020;10(1):8811. doi: 10.1038/s41598-020-65916-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parlato M, Souza-Fonseca-Guimaraes F, Philippart F, Misset B, Adib-Conquy M, Cavaillon JM. CD24-triggered caspase-dependent apoptosis via mitochondrial membrane depolarization and reactive oxygen species production of human neutrophils is impaired in sepsis. J Immunol. 2014;192(5):2449–2459. doi: 10.4049/jimmunol.1301055 [DOI] [PubMed] [Google Scholar]
- 17.Li W, Ling H-P, You W-C, et al. Elevated cerebral cortical CD24 levels in patients and mice with traumatic brain injury: a potential negative role in nuclear factor κb/inflammatory factor pathway. Mol Neurobiol. 2014;49(1):187–198. doi: 10.1007/s12035-013-8509-4 [DOI] [PubMed] [Google Scholar]
- 18.Cui SN, Chen ZY, Yang XB, et al. Trichostatin A modulates the macrophage phenotype by enhancing autophagy to reduce inflammation during polymicrobial sepsis. Int Immunopharmacol. 2019;77:105973. doi: 10.1016/j.intimp.2019.105973 [DOI] [PubMed] [Google Scholar]
- 19.Chatterjee A, Dimitropoulou C, Drakopanayiotakis F, et al. Heat shock protein 90 inhibitors prolong survival, attenuate inflammation, and reduce lung injury in murine sepsis. Am J Respir Crit Care Med. 2007;176(7):667–675. doi: 10.1164/rccm.200702-291OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joachim RB, Altschuler GM, Hutchinson JN, Wong HR, Hide WA, Kobzik L. The relative resistance of children to sepsis mortality: from pathways to drug candidates. Mol Syst Biol. 2018;14(5):e7998. doi: 10.15252/msb.20177998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thornhill R, Strong D, Vasanth S, Mackenzie I. Trauma sepsis. Trauma. 2010;12(1):31–49. doi: 10.1177/1460408609356828 [DOI] [Google Scholar]
- 22.Tanaka H, Ishikawa K, Nishino M, Shimazu T, Yoshioka T. Changes in granulocyte colony-stimulating factor concentration in patients with trauma and sepsis. J Trauma Acute Care Surg. 1996;40(5):718–726. doi: 10.1097/00005373-199605000-00006 [DOI] [PubMed] [Google Scholar]
- 23.Cook KM, Sifri ZC, Baranski GM, Mohr AM, Livingston DH. The role of plasma granulocyte colony stimulating factor and bone marrow dysfunction after severe trauma. J Am Coll Surg. 2013;216(1):57–64. doi: 10.1016/j.jamcollsurg.2012.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tolg C, McCarthy JB, Yazdani A, Turley EA. Hyaluronan and RHAMM in wound repair and the “Cancerization” of stromal tissues. Biomed Res Int. 2014;2014:103923. doi: 10.1155/2014/103923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong HR, Cvijanovich N, Allen GL, et al. Genomic expression profiling across the pediatric systemic inflammatory response syndrome, sepsis, and septic shock spectrum. Crit Care Med. 2009;37(5):1558–1566. doi: 10.1097/CCM.0b013e31819fcc08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacoby RC, Owings JT, Holmes J, Battistella FD, Gosselin RC, Paglieroni TG. Platelet activation and function after trauma. J Trauma Acute Care Surg. 2001;51(4):639–647. doi: 10.1097/00005373-200110000-00003 [DOI] [PubMed] [Google Scholar]
- 27.Gómez-Carballa A, Cebey-López M, Pardo-Seco J, et al. A qPCR expression assay of IFI44L gene differentiates viral from bacterial infections in febrile children. Sci Rep. 2019;9(1):11780. doi: 10.1038/s41598-019-48162-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lau LT, Yu AC. Astrocytes produce and release interleukin-1, interleukin-6, tumor necrosis factor alpha and interferon-gamma following traumatic and metabolic injury. J Neurotrauma. 2001;18(3):351–359. doi: 10.1089/08977150151071035 [DOI] [PubMed] [Google Scholar]
- 29.Boldt J, Papsdorf M, Rothe A, Kumle B, Piper S. Changes of the hemostatic network in critically ill patients-Is there a difference between sepsis, trauma, and neurosurgery patients? Crit Care Med. 2000;28(2):445–450. doi: 10.1097/00003246-200002000-00026 [DOI] [PubMed] [Google Scholar]
- 30.Calvano SE, Coyle SM. Experimental human endotoxemia: a model of the systemic inflammatory response syndrome? Surg Infect. 2012;13(5):293–299. doi: 10.1089/sur.2012.155 [DOI] [PMC free article] [PubMed] [Google Scholar]