Abstract
Borrelia burgdorferi sensu lato A14S was cultured from a skin biopsy specimen of a patient with erythema migrans in The Netherlands. This isolate had a unique DNA fingerprint pattern compared to 135 other B. burgdorferi sensu lato isolates. In this study, the isolate A14S was further characterized by protein analysis with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and reactivity with various monoclonal antibodies. In addition, the 16S rRNA, ospA, and ospC genes, as well as the 5S-23S rRNA intergenic spacer DNA, were amplified by PCR, cloned, and sequenced. SDS-PAGE protein profiles and phylogenetic analysis based on all of the analyzed genes confirmed that B. burgdorferi sensu lato A14S was phenotypically and genetically different from the three human pathogenic species B. burgdorferi sensu stricto, Borrelia garinii, and Borrelia afzelii, as well as from other B. burgdorferi sensu lato species. Our findings indicate that Borrelia genomic groups or isolates other than the three well-known human pathogenic species may also cause human Lyme borreliosis.
Lyme borreliosis (LB) is a worldwide multisystemic infection transmitted to humans by the bite of an infected tick of the genus Ixodes. Erythema migrans, an early characteristic skin lesion, is relatively consistent and is found in about 50% of LB cases (17). Borrelia burgdorferi sensu lato, the causative bacterium of LB, is genetically divergent and has been divided into several species or genomic groups. These include B. burgdorferi sensu stricto, Borrelia garinii, Borrelia afzelii (1, 2), Borrelia japonica (6), Borrelia valaisiana (21), Borrelia lusitaniae (8), Borrelia andersonii (10), Borrelia tanukii, Borrelia turdi (5), and Borrelia bissettii (formerly genomic group DN127) (14). B. burgdorferi sensu stricto, B. garinii, and B. afzelii have been cultured from patients and are thought to be responsible for human LB. Recently, Picken et al. (12, 18) reported that nine B. burgdorferi isolates from patients in Slovenia were closely related to the North American isolate 25015, a member of B. bissettii. Thus, strains belonging to B. bissettii may also cause human disease.
In 1992, we cultured a B. burgdorferi sensu lato isolate, A14S, from the skin biopsy specimen of a Dutch patient with erythema migrans who had contracted the disease in The Netherlands. This isolate could not be typed based on its reactivity with various monoclonal antibodies (MAbs) against B. burgdorferi sensu lato (20). Also, it showed a unique pattern in ribotyping, differing from B. burgdorferi sensu stricto, B. garinii, and B. afzelii (20). In addition, a striking genetic difference between this isolate and 135 other LB-related Borrelia isolates was noted in the randomly amplified polymorphic DNA (RAPD) fingerprinting analysis (22). Therefore, Borrelia isolate A14S could well represent a novel B. burgdorferi sensu lato strain causing human LB.
In the present study, isolate A14S at low passage (passages 5 to 10) was further analyzed by protein profiling with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and reactivity with MAbs against different outer surface proteins of B. burgdorferi sensu lato. The 16S rRNA, ospA, and ospC genes, as well as the 5S-23S rRNA intergenic spacer from this isolate, were amplified by PCR, sequenced, and compared to those from B. burgdorferi sensu stricto, B. garinii, and B. afzelii, the three pathogenic Borrelia species for humans, and other species in the B. burgdorferi sensu lato complex.
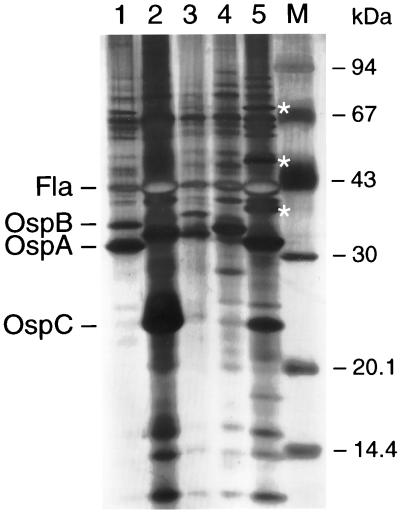
SDS-PAGE was performed as described previously (20). The protein profile of isolate A14S was clearly different from those of the representative isolates of B. burgdorferi sensu stricto, B. garinii, B. afzelii, and B. valaisiana (Fig. 1). For example, isolate A14S contained at least three unique protein bands with molecular masses of 37, 49, and 72 kDa (Fig. 1, asterisks). Apart from its reactivity with B. burgdorferi sensu stricto-specific MAb H3TS (20), this isolate reacted with the OspA-specific MAb LA26, which recognizes both B. burgdorferi sensu stricto and B. afzelii. No reactivity was observed to the OspB-specific MAb 84C and the OspC-specific MAb L22 C11, two MAbs reactive with OspB and OspC proteins, respectively, from B. burgdorferi sensu stricto, B. garinii, and B. afzelii (data not shown).
FIG. 1.
Silver-stained SDS-PAGE gel of whole-cell lysates of B. burgdorferi sensu lato strains. Lanes 1 to 5, B. burgdorferi sensu stricto B31, B. garinii 20047, B. afzelii VS461, B. valaisiana AR-2, and Borrelia isolate A14S, respectively. The positions of flagellin (Fla), OspA, OspB, and OspC are indicated by arrows. The unique protein bands of isolate A14S are marked with asterisks. Molecular mass standards (Pharmacia) are shown on the right.
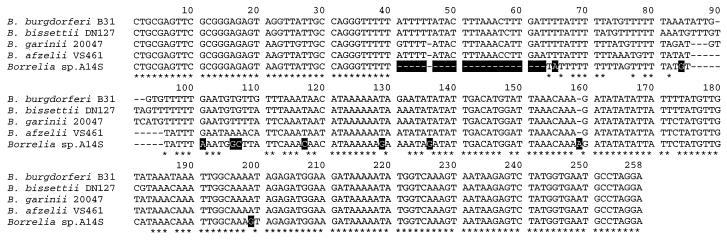
The 5S-23S spacer PCR-restriction fragment length polymorphism (RFLP) analysis has been widely applied to assess the genetic diversity of B. burgdorferi sensu lato strains and to identify Borrelia genomic groups (13). In our study, a 225-bp 5S-23S intergenic spacer from Borrelia isolate A14S was amplified by PCR. The size of this spacer was notably different in comparison to those of other European isolates from five different Borrelia species, which ranged from 246 to 257 bp (13). The PCR amplicon of isolate A14S was sequenced directly as described previously (21). Digestion of this amplicon with MseI resulted in three fragments with sizes of 106, 68, and 51 bp, respectively (data not shown). This MseI restriction pattern was different from previously reported patterns A to Q from other LB-related Borrelia species (11, 13) and was designated pattern R. Sequence analysis of the 5S-23S intergenic spacer DNA of isolate A14S showed only 91.6, 92.9, 92.4, and 93.3% identity to the type strains of B. burgdorferi sensu stricto B31, B. garinii 20047, B. afzelii VS461, and B. bissettii DN127, respectively. At 10 positions, nucleotides differing between all type strains of these B. burgdorferi sensu lato species and isolate A14S could be identified (Fig. 2).
FIG. 2.
Sequences of the 5S-23S rRNA intergenic spacers of Borrelia sp. strain A14S and the type strains of B. burgdorferi species known to be pathogenic to humans. Gaps were introduced to obtain maximum levels of homology. The nucleotides identical among all isolates are indicated with asterisks under the sequences, and the unique nucleotide positions of isolate A14S are shaded in black. The accession numbers of the 5S-23S rRNA intergenic spacer sequences of Borrelia strains used for comparison are L30127 (B. burgdorferi sensu stricto B31), L30119 (B. garinii 20047), L30135 (B. afzelii VS461), and L30126 (B. bissettii DN127).
For further evaluation of the phylogeny of Borrelia isolate A14S, the highly conserved 16S rRNA gene from this isolate was amplified by PCR with primer BRNA8 (5′ ACGCTGGCAGTCGTCTTA 3′, positions 33 to 55, B. burgdorferi B31 numbering [3]) (22) and primer UniB (5′ T[A/C]AAGGAGGTGATCCAGC 3′, positions 1540 to 1522) as described previously (22). The PCR amplicon was cloned directly into the PCR2.1 vector and transformed into Escherichia coli INV"F′ cells by following the manufacturer’s instructions (Invitrogen BV, Leek, The Netherlands). Subsequently, plasmid DNA from three recombinant colonies containing the amplified fragment was prepared by using a plasmid minipreparation kit (Qiagen GmbH, Hilden, Germany). DNA sequence was determined by the dideoxy chain-termination technique in an ABI 373A sequencer by using either the Dye terminator or the Dye primer cycle sequencing kit (Applied Biosystems, Inc., Foster City, Calif.) with additional custom-synthesized primers. A phylogenetic tree was constructed by using a neighbor-joining method with Kimura’s two-parameter distance algorithm in the MEGA program (7).
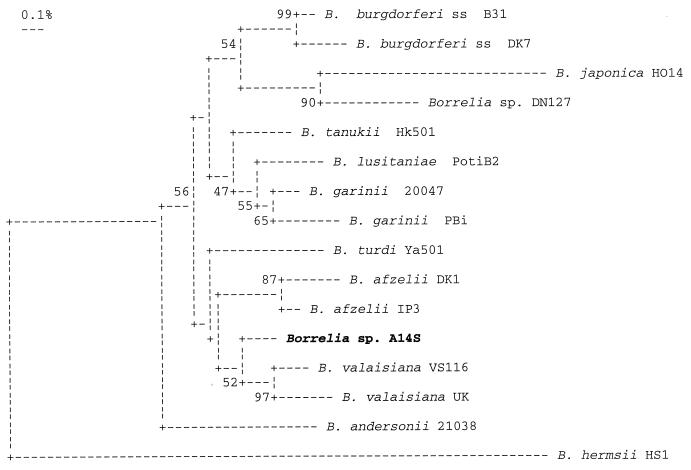
A nearly complete (1,503-bp) 16S cDNA was amplified from isolate A14S by PCR. Sequence analysis showed that it possessed more than 98.5% similarity with the five LB Borrelia species occurring in Europe, B. burgdorferi sensu stricto (B31, 98.9%), B. garinii (20047, 99.2%). B. afzelii (DK1, 99.0%), B. valaisiana (VS116, 98.7%), and B. lusitaniae (99.1%), but less than 97% similarity with B. japonica (HO14, 95.5%), B. andersonii (19857, 96.6%), and B. bissettii (DN127, 96.3%), which have been isolated mainly in Japan (B. japonica) and North America (B. andersonii and B. bissettii). This finding suggested that Borrelia isolate A14S was closely related to the European Borrelia species. Only 13, 11, and 17 nucleotide substitutions in Borrelia isolate A14S were identified when its 16S rRNA gene was compared to the corresponding genes from B. burgdorferi sensu stricto (B31), B. garinii (20047), and B. afzelii (DK1), respectively. It is noteworthy that some of these nucleotide substitutions in the 16S rRNA gene of Borrelia isolate A14S occurred in the BfaI restriction site, which would allow easy distinction of A14S from the three Borrelia species causing human LB (8). Like other newly described Borrelia species such as B. lusitaniae (8), B. tanukii, and B. turdi (5), isolate A14S constituted an independent branch in the phylogenetic tree based on its 16S rRNA gene sequence (Fig. 3), indicating that isolate A14S represents a new Borrelia genomic group in the B. burgdorferi sensu lato complex.
FIG. 3.
Phylogenetic tree of B. burgdorferi sensu lato strains based on 1,330 bp of 16S rRNA gene sequences (positions 34 to 1364). B. hermsii HS1 was used as an out-of-group control. Numbers at each of the branch nodes indicate results from bootstrap analysis. The accession numbers (in parentheses) of the 16S rRNA gene sequences of Borrelia strains used for comparison are as follows: B31 (U03396), DK7 (X85195), 20047 (D67018), PBi (X85199), DK1 (X85190), IP3 (M84815), HO14 (L40597), VS116 (X98232), UK (X98233), PotiB2 (X98228), 21038 (L46701), DN127 (L40596), Hk501 (D67023), Ya501 (D67022), and HS1(U42292).
The ospA gene of Borrelia isolate A14S was amplified from genomic DNA by PCR with primer OspA1 (5′ GGAGAATATATTATGAAA 3′, positions −12 to 6) and OspA2 (5′ CTCCTTATTTTAAAGCG 3′, positions 826 to 809) as described by Dykhuizen et al. (3). Five recombinant colonies were selected and sequenced. An open reading frame of 822 bp was identified from each of the recombinants. No sequence discrepancy was found among these colonies. The nucleotide sequence of the ospA gene of isolate A14S had 87.6, 88.0, 88.6, and 87.3% identity to the ospA sequences of B. burgdorferi sensu stricto B31, B. garinii PBi, B. afzelii VS461, and B. bissettii 25015, respectively. The protein in isolate A14S encoded by this gene contained 273 amino acids and had a deduced molecular mass of 29.5 kDa. Phylogenetic analysis based on the deduced amino acid sequences of the OspA proteins of isolate A14S and isolates from other B. burgdorferi sensu lato species was well in accordance with results from 16S rRNA gene sequence analysis (data not shown). Borrelia isolate A14S constituted a separate branch in both phylogenetic trees. Each of the other clusters in these phylogenetic trees corresponded to one of the earlier-defined Borrelia species.
Previous studies showed that among Borrelia strains ospC is more polymorphic than ospA and could be divided into at least 35 different ospC RFLP patterns (9). Despite the high genetic heterogeneity of ospC, however, a species-specific motif has been identified in the deduced amino acid sequences of OspC from a number of B. burgdorferi sensu lato strains (4, 9). In this study, the ospC gene from Borrelia isolate A14S was amplified by PCR with primer OspC-N (5′ CACAAATTAATGAAAAAGAATACA 3′) and primer OspC-C (5′ CCAGTTACTTTTTTAAAACAAATTA 3′), which were predicted to yield an approximately 650-bp amplicon (9). Unexpectedly, a predominant large fragment with a size between 1.0 and 1.1 kb was obtained in our PCR, while a very weak band at 0.65 kb was also observed. Cloning and sequencing of the larger fragment confirmed that it consisted of a 633-bp complete ospC gene as well as a 403-bp flanking sequence at the 3′ end. An 8-bp deletion occurring at the original primer binding site in the flanking sequence of ospC caused a mismatch of primer OspC-C with the sequence of the ospC gene of A14S. The nucleotide sequence of the ospC gene of isolate A14S showed 83.1, 83.3, 82.9, and 78.6% identity to the ospC sequences of B. burgdorferi sensu stricto B31, B. garinii 20047, B. afzelii VS461, and B. bissettii 25015, respectively. Digestion of the ospC gene from Borrelia isolate A14S with DraI yielded two fragments with sizes of 154 and 479 bp. This RFLP pattern, as well as the RFLP pattern after DpnII digestion, differed from all of the ospC RFLP patterns from B. burgdorferi sensu stricto, B. garinii, and B. afzelii reported by Livey et al. (9).
The deduced translation product (OspC) from the 633-bp ospC gene of Borrelia isolate A14S contained 210 amino acids and had a molecular mass of 22.2 kDa. Its amino acid sequence from positions 23 to 34 of Borrelia isolate A14S differed from that of the species-specific motifs described for other Borrelia species or genomic groups. A nucleotide T-to-G conversion at position 90 of the ospC gene of isolate A14S would result in an amino acid change from an Asn to a Lys residue. Such a conversion has been reported previously for only one B. garinii isolate from Denmark (strain DK35) (19), but the difference in amino acids at positions 29 and 35 distinguishes the two strains.
It seems unlikely that the Borrelia isolate A14S is a mixture of multiple Borrelia species, since this isolate possessed some unique nucleotides in its 16S rRNA and ospC genes which were definitely different from those of Borrelia species in the B. burgdorferi sensu lato complex. It is also highly unlikely that the differences between isolate A14S and other Borrelia species are a result of in vitro passage. In vitro passage of B. burgdorferi can lead to plasmid loss, as well as changes in antigenic expression, and subsequently to differences in SDS-PAGE patterns and reactivities with MAbs (15, 16). However, the 16S rRNA, ospA, and ospC genes, as well as the 5S-23S intergenic spacer sequences, are not subject to major alteration by in vitro passage. On the basis of our present study, as well as of findings obtained by ribotyping (20) and RAPD fingerprinting analysis (22), we conclude that Borrelia isolate A14S does not belong to B. burgdorferi sensu stricto, B. garinii, and B. afzelii, the three Borrelia species pathogenic to humans. Based on the ospA, ospC, and 16S rRNA gene sequence analysis, 5S-23S rRNA intergenic spacer RFLP pattern (this study), and RAPD fingerprinting (22), isolate A14S also clearly differs from B. bissettii, the fourth genomic group recently shown to be able to cause human LB, as well as from other LB-associated Borrelia species. Therefore, this isolate most likely represents a new Borrelia genomic group, which is the fifth Borrelia genomic group with culture-confirmed pathogenic potential for causing human LB, besides B. burgdorferi sensu stricto, B. garinii, B. afzelii, and B. bissettii.
Nucleotide sequence accession numbers.
The 16S rRNA, ospA, ospC, and 5S-23S intergenic spacer sequences of isolate A14S that we determined in this study have been assigned GenBank accession no. AF102056, AF102057, AF102058, and U76616, respectively.
Acknowledgments
We thank Erol Fikrig for critical reading of the manuscript and Wim van Est for photography.
REFERENCES
- 1.Baranton G, Postic D, Saint Girons I, Boerlin P, Piffaretti J C, Assous M, Grimont P A. Delineation of Borrelia burgdorferi sensu stricto, Borrelia garinii sp. nov., and group VS461 associated with Lyme borreliosis. Int J Syst Bacteriol. 1992;42:378–383. doi: 10.1099/00207713-42-3-378. [DOI] [PubMed] [Google Scholar]
- 2.Canica M M, Nato F, du Merle L, Mazie J C, Baranton G, Postic D. Monoclonal antibodies for identification of Borrelia afzelii sp. nov. associated with late cutaneous manifestations of Lyme borreliosis. Scand J Infect Dis. 1993;25:441–448. doi: 10.3109/00365549309008525. [DOI] [PubMed] [Google Scholar]
- 3.Dykhuizen D E, Polin D S, Dunn J J, Wilske B, Preac-Mursic V, Dattwyler R J, Luft B J. Borrelia burgdorferi is clonal: implications for taxonomy and vaccine development. Proc Natl Acad Sci USA. 1993;90:10163–10167. doi: 10.1073/pnas.90.21.10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fukunaga M, Hamase A, Okada K, Inoue H, Tsuruta Y, Miyamoto K, Nakao M. Characterization of spirochetes isolated from ticks (Ixodes tanuki, Ixodes turdus, and Ixodes columnae) and comparison of the sequences with those of Borrelia burgdorferi sensu lato strains. Appl Environ Microbiol. 1996;62:2338–2344. doi: 10.1128/aem.62.7.2338-2344.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fukunaga M, Hamase A, Okada K, Nakao M. Borrelia tanukii sp. nov. and Borrelia turdae sp. nov. found from ixodid ticks in Japan: rapid species identification by 16S rRNA gene-targeted PCR analysis. Microbiol Immunol. 1996;40:877–881. doi: 10.1111/j.1348-0421.1996.tb01154.x. [DOI] [PubMed] [Google Scholar]
- 6.Kawabata H, Masuzawa T, Yanagihara Y. Genomic analysis of Borrelia japonica sp. nov. isolated from Ixodes ovatus in Japan. Microbiol Immunol. 1993;37:843–848. doi: 10.1111/j.1348-0421.1993.tb01714.x. [DOI] [PubMed] [Google Scholar]
- 7.Kumar S, Tamura K, Masatoshi N. MEGA: molecular evolutionary genetics analysis, version 1.01. University Park, Pa: Pennsylvania State University; 1993. [Google Scholar]
- 8.Le Fleche A, Postic D, Girardet K, Peter O, Baranton G. Characterization of Borrelia lusitaniae sp. nov. by 16S ribosomal DNA sequence analysis. Int J Syst Bacteriol. 1997;47:921–925. doi: 10.1099/00207713-47-4-921. [DOI] [PubMed] [Google Scholar]
- 9.Livey I, Gibbs C P, Schuster R, Dorner F. Evidence for lateral transfer and recombination in OspC variation in Lyme disease Borrelia. Mol Microbiol. 1995;18:257–269. doi: 10.1111/j.1365-2958.1995.mmi_18020257.x. [DOI] [PubMed] [Google Scholar]
- 10.Marconi R T, Liveris D, Schwartz I. Identification of novel insertion elements, restriction fragment length polymorphism patterns, and discontinuous 23S rRNA in Lyme disease spirochetes: phylogenetic analyses of rRNA genes and their intergenic spacers in Borrelia japonica sp. nov. and genomic group 21038 (Borrelia andersonii sp. nov.) isolates. J Clin Microbiol. 1995;33:2427–2434. doi: 10.1128/jcm.33.9.2427-2434.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Masuzawa T, Komikado T, Iwaki A, Suzuki H, Kaneda K, Yanagihara Y. Characterization of Borrelia sp. isolated from Ixodes tanuki, I. turdus, and I. columnae in Japan by restriction fragment length polymorphism of rrf (5S)-rrl (23S) intergenic spacer amplicons. FEMS Microbiol Lett. 1996;142:77–83. doi: 10.1111/j.1574-6968.1996.tb08411.x. [DOI] [PubMed] [Google Scholar]
- 12.Picken R N, Cheng Y, Strle F, Picken M M. Patient isolates of Borrelia burgdorferi sensu lato with genotypic and phenotypic similarities of strain 25015. J Infect Dis. 1996;174:1112–1115. doi: 10.1093/infdis/174.5.1112. [DOI] [PubMed] [Google Scholar]
- 13.Postic D, Assous M V, Grimont P A D, Baranton G. Diversity of Borrelia burgdorferi sensu lato evidenced by restriction fragment length polymorphism of rrf (5S)- rrl (23S) intergenic spacer amplicons. Int J Syst Bacteriol. 1994;44:743–752. doi: 10.1099/00207713-44-4-743. [DOI] [PubMed] [Google Scholar]
- 14.Postic D, Ras N M, Lane R S, Hendson M, Baranton G. Expanded diversity among Californian Borrelia isolates and description of Borrelia bissettii sp. nov. (formerly Borrelia group DN127) J Clin Microbiol. 1998;36:3497–3504. doi: 10.1128/jcm.36.12.3497-3504.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwan T G, Burgdorfer W. Antigenic changes of Borrelia burgdorferi as a result of in vitro cultivation. J Infect Dis. 1987;156:852–853. doi: 10.1093/infdis/156.5.852-a. [DOI] [PubMed] [Google Scholar]
- 16.Schwan T G, Burgdorfer W, Garon C F. Changes in infectivity and plasmid profile of the Lyme disease spirochete, Borrelia burgdorferi, as a result of in vitro cultivation. Infect Immun. 1988;56:1831–1836. doi: 10.1128/iai.56.8.1831-1836.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steere A C. Lyme disease. N Engl J Med. 1989;263:201–205. doi: 10.1056/NEJM198908313210906. [DOI] [PubMed] [Google Scholar]
- 18.Strle F, Picken R N, Cheng Y, Cimperman J, Maraspin V, Lotric-Furlan S, Ruzic-Sabljic E, Picken M M. Clinical findings for patients with Lyme borreliosis caused by Borrelia burgdorferi sensu lato with genotypic and phenotypic similarities to strain 25015. Clin Infect Dis. 1997;25:273–280. doi: 10.1086/514551. [DOI] [PubMed] [Google Scholar]
- 19.Theisen M, Borre M, Mathiesen M J, Mikkelsen B, Lebech A M, Hansen K. Evolution of the Borrelia burgdorferi outer surface protein OspC. J Bacteriol. 1995;177:3036–3044. doi: 10.1128/jb.177.11.3036-3044.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Dam A P, Kuiper H, Vos K, Widjojokusumo A, de Jongh B M, Spanjaard L, Ramselaar A C, Kramer M D, Dankert J. Different genospecies of Borrelia burgdorferi are associated with distinct clinical manifestations of Lyme borreliosis. Clin Infect Dis. 1993;17:708–717. doi: 10.1093/clinids/17.4.708. [DOI] [PubMed] [Google Scholar]
- 21.Wang G, van Dam A P, Le Fleche A, Postic D, Peter O, Baranton G, de Boer R, Spanjaard L, Dankert J. Genetic and phenotypic analysis of Borrelia valaisiana sp. nov. (Borrelia genomic groups VS116 and M19) Int J Syst Bacteriol. 1997;47:926–932. doi: 10.1099/00207713-47-4-926. [DOI] [PubMed] [Google Scholar]
- 22.Wang G, van Dam A P, Spanjaard L, Dankert J. Molecular typing of Borrelia burgdorferi sensu lato by randomly amplified polymorphic DNA fingerprinting analysis. J Clin Microbiol. 1998;36:768–776. doi: 10.1128/jcm.36.3.768-776.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]