ABSTRACT
Mycobacteria spatially organize their plasma membrane, and many enzymes involved in envelope biosynthesis associate with a membrane compartment termed the intracellular membrane domain (IMD). The IMD is concentrated in the polar regions of growing cells and becomes less polarized under nongrowing conditions. Because mycobacteria elongate from the poles, the observed polar localization of the IMD during growth likely supports the localized biosynthesis of envelope components. While we have identified more than 300 IMD-associated proteins by proteomic analyses, only a few of these have been verified by independent experimental methods. Furthermore, some IMD-associated proteins may have escaped proteomic identification and remain to be identified. Here, we visually screened an arrayed library of 523 Mycobacterium smegmatis strains, each producing a Dendra2-FLAG-tagged recombinant protein. We identified 29 fusion proteins that showed polar fluorescence patterns characteristic of IMD proteins. Twenty of these had previously been suggested to localize to the IMD based on proteomic data. Of the nine remaining IMD candidate proteins, three were confirmed by biochemical methods to be associated with the IMD. Taken together, this new colocalization strategy is effective in verifying the IMD association of proteins found by proteomic analyses while facilitating the discovery of additional IMD-associated proteins.
IMPORTANCE The intracellular membrane domain (IMD) is a membrane subcompartment found in Mycobacterium smegmatis cells. Proteomic analysis of purified IMD identified more than 300 proteins, including enzymes involved in cell envelope biosynthesis. However, proteomics on its own is unlikely to detect every IMD-associated protein because of technical and biological limitations. Here, we describe fluorescent protein colocalization as an alternative, independent approach. Using a combination of fluorescence microscopy, proteomics, and subcellular fractionation, we identified three new proteins associated with the IMD. Such a robust method to rigorously define IMD proteins will benefit future investigations to decipher the synthesis, maintenance, and functions of this membrane domain and help delineate a more general mechanism of subcellular protein localization in mycobacteria.
KEYWORDS: fluorescence microscopy, Mycobacterial Systems Resource, mycobacterium, membrane domain, membrane proteins
INTRODUCTION
The cell envelope of mycobacteria comprises the plasma membrane, cell wall, and the mycomembrane, an outer membrane component that differs extensively from the classical outer membrane of Gram-negative bacteria. This diderm structure is unusually complex, with a peptidoglycan layer covalently linked to arabinogalactan, which, in turn, is covalently linked to mycolic acids. Mycolic acids are a component of the mycomembrane that endows mycobacteria with their characteristic waxy coat (1–5).
Mycobacteria must retain their complex cell envelope as they grow and elongate their rod-shaped cell. New components for cell envelope extension are added at the subpolar regions (6–12). The intracellular membrane domain (IMD) is a membrane structure that has been proposed to form laterally discrete domains within the conventional plasma membrane and is found enriched in the subpolar regions of actively growing cells (13, 14). The IMD is spatially positioned to support polar cell wall synthesis and growth, and its subpolar enrichment is reduced when cells are in a stressed environment (15, 16). The IMD can be biochemically isolated from the cell lysate of Mycobacterium smegmatis by sucrose density gradient sedimentation (14). During fractionation, the IMD sediments to a region of lighter density than the plasma membrane. The plasma membrane is purified in tight association with the cell wall components, and the complex is designated the plasma membrane-cell wall (PM-CW). Proteomic analysis of the IMD identified more than 300 candidate proteins associated with the IMD. Well-characterized examples of the IMD proteins include cell envelope biosynthetic enzymes such as PimB′, Ppm1, and GlfT2 (13). PimB′ is an essential mannosyltransferase that catalyzes the transfer of a second mannose to form phosphatidylinositol dimannosides, a component of the cell envelope (17, 18). Ppm1 is a subunit of the polyprenol phosphate mannnose (PPM) synthase, involved in synthesizing lipid-linked mannose donors, and is another essential enzyme for the synthesis of mannose-containing glycolipids (19–22). GlfT2 is a galactosyltransferase that extends the galactan chain of the arabinogalactan layer (23, 24). The identification of these essential biosynthetic enzymes, among others involved in cell envelope biosynthesis, highlights the critical roles of the IMD in cell envelope biosynthesis.
Although more than 300 putative IMD-associating proteins have been identified, we predicted that there would be additional IMD proteins that were not detected by proteomics due to its limitations. For example, proteins (small proteins in particular) that are not annotated in the protein database will not be identified by this method. Proteins that do not have trypsin cleavage sites will also be missed. Membrane proteins are difficult to detect by mass spectroscopy due to the hydrophobicity of the peptide fragments. Furthermore, sample preparation may also introduce a bias. For example, peripheral membrane proteins weakly associated with a membrane may dissociate from the membrane during cell lysate preparation. In addition to these false negatives, there may be false positives among those predicted IMD proteins due, for example, to nonspecific binding of non-IMD proteins after cell lysis. In this study, we took advantage of the recently established Mycobacterial Systems Resource (25), which is focused on 1,153 genes that are conserved across five different Mycobacterium species and are considered to be the core mycobacterial genome. This resource includes a library of Mycobacterium smegmatis strains, each producing one of 1,118 proteins tagged with C-terminal Dendra2-FLAG. We visually screened Dendra2 fluorescence patterns of 523 images for IMD-like fluorescence and identified 29 putative IMD proteins that exhibited colocalization patterns similar to known IMD proteins. Consistent with their colocalization, 20 of the identified proteins had been previously found in the IMD proteome. Here, we provide biochemical validation of five previous candidate IMD proteins as well as three new IMD proteins that were not identified previously by the proteomic approach.
RESULTS
Screening of fluorescence microscope images identifies candidate IMD-associated proteins.
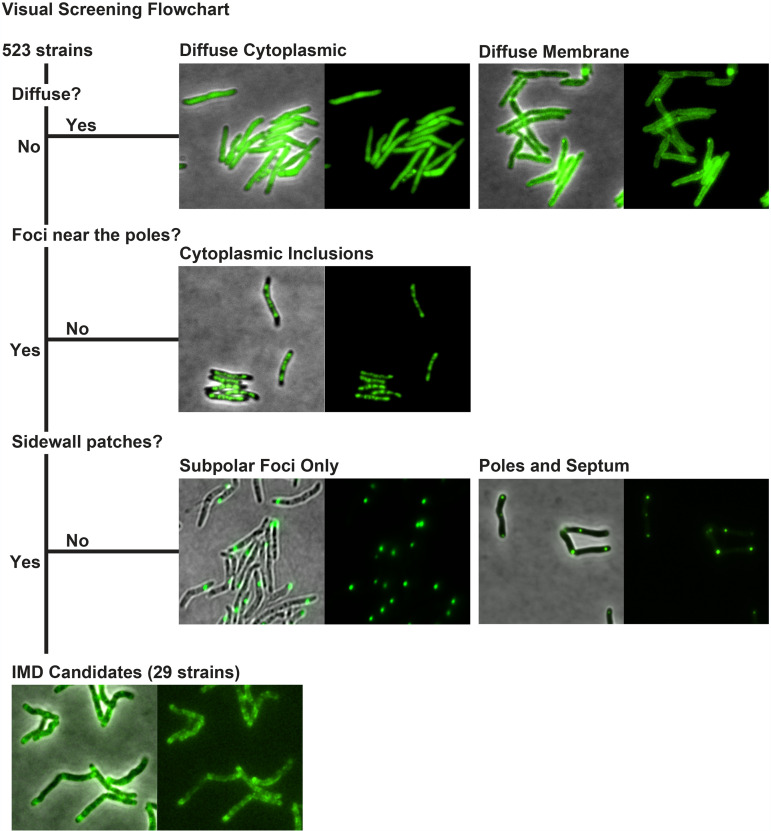
As an alternative approach to discover additional IMD-associated proteins, we screened fluorescence microscopy images of 523 M. smegmatis strains, each producing a protein tagged with Dendra2-FLAG at the C terminus (see Table S1 in the supplemental material and Materials and Methods). We had previously defined an IMD-like fluorescence pattern as fluorescence enrichment in the polar regions with weaker fluorescence patches along the sidewalls of the cell (13). Through visual screening, we identified 29 strains showing fluorescence patterns of Dendra2-FLAG-tagged proteins that were similar to the typical localization patterns of known IMD-associated proteins (Fig. 1). The identified proteins were then cross-referenced against the published comparative proteome of the IMD and the PM-CW (13). We found that 20 of the 29 proteins had been previously identified in the IMD proteome (Table 1). One of the remaining nine proteins, MenA (MSMEG_0988), was not listed in our proteome (13), but we have shown in a separate study that it is a PM-CW-associated protein (26). Therefore, we considered this a false positive and did not investigate this protein further. Among the eight remaining candidates, five proteins were previously associated with the PM-CW proteome by proteomic studies but had not been validated further (13), and three candidate proteins had not been detected in either of the published IMD or PM-CW proteomes (Table 1) (13).
FIG 1.
Visual screening of IMD protein candidates from 523 fluorescence images deposited in the Mycobacterial Systems Resource. Fluorescence images were visually screened for proteins that show fluorescence patterns that are similar to those of known IMD proteins. Excluding those fluorescence patterns that were diffuse, cytoplasmic inclusions, subpolar foci only, or polar foci and septum without sidewall patches, a total of 29 proteins were identified as the candidates of IMD proteins, showing subpolar foci and sidewall patches.
TABLE 1.
IMD protein candidates identified from visual screening of fluorescence microscope images
| MSMEG_a | Rv homolog | Gene | Annotation | Proteome | Biochemical (source or reference) |
|---|---|---|---|---|---|
| 0825 | Rv0422c | thiD | Phosphomethylpyrimidine kinase | PM-CW | IMD (this study) |
| 0861 | Rv0437c | psd | Phosphatidylserine decarboxylase | IMD | IMD (14) |
| 0863 | Rv0439c | Short-chain dehydrogenase | IMD | ||
| 0876 | Rv0068 | Short-chain dehydrogenase | IMD | IMD (this study) | |
| 0949 | Rv0505c | HAD-superfamily hydrolase | IMD | Cb (this study) | |
| 0972 | Rv0527 | ccdA | Cytochrome c biogenesis protein transmembrane region | NFc | IMD (this study) |
| 0988d | Rv0534c | menA | 1,4-Dihydroxy-2-naphthoate octaprenyltransferase | NF | PM-CW (26) |
| 1011 | NA | Short-chain dehydrogenase | IMD | ||
| 1055 | NA | Hexapeptide transferase | IMD | ||
| 1115 | Rv0558 | menG | Ubiquinone/menaquinone biosynthesis methyltransferase | IMD | IMD (26) |
| 1352 | Rv0646c | Alpha/beta hydrolase | IMD | ||
| 1954 | Rv3197 | ABC transporter | PM-CW | C (this study) | |
| 2080 | Rv3140 | fadE23 | Acyl coenzyme A dehydrogenase | IMD | |
| 2329 e | Rv3038c | UbiE/COQ5 family methyltransferase | IMD | C (this study) | |
| 2335 e | Rv3034c | Hexapeptide transferase | IMD | IMD (this study) | |
| 2698 | Rv2740 | Hypothetical protein MSMEG_2698 | IMD | ||
| 2934 | Rv2611c | patA | Lipid A biosynthesis lauroyl acyltransferase | IMD | IMD (this study) |
| 2975 | Rv2581c | Metallo-beta-lactamase | NF | ||
| 3863 | Rv2061c | Pyridoxamine 5'-phosphate oxidase | IMD | ||
| 4198 | Rv2139 | pyrD | Dihydroorotate dehydrogenase 2 | IMD | IMD (this study) (13) |
| 4227 | Rv2153c | murG | UDP diphospho-muramoyl pentapeptide beta-N-acetylglucosaminyltransferase | IMD | |
| 4254 | Rv2187 | fadD15 | AMP-binding protein | PM-CW | C (this study) |
| 4284 | Rv2216 | Hypothetical protein MSMEG_4284 | IMD | ||
| 4479 | Rv2342 | Hypothetical protein MSMEG_4479 | NF | IMD (this study) | |
| 4632 | Rv2449c | Saccharopine dehydrogenase | IMD | ||
| 4722 | Rv2509 | cmrA | Short-chain dehydrogenase | PM-CW | C (this study) |
| 6385 | Rv3791 | Short-chain dehydrogenase | IMD | ||
| 6403 | Rv3808c | glfT2 | Bifunctional UDP-galactofuranosyl transferase GlfT | IMD | IMD (this study) (13) |
| 6928 | Rv3909 | Hypothetical protein MSMEG_6928 | PM-CW | PM-CW (this study) |
Boldface type indicates the proteins that were previously found in the IMD proteome.
C, cytoplasm.
NF, not found.
This protein was previously characterized as a PM-CW protein (26) and was not investigated further in this study.
MSMEG_2329 and _2335 are identical to MSMEG_1049 and _1055, respectively, at the amino acid level and are found in the IMD proteome.
Confirmation of previously identified IMD proteins.
Among the 20 previously identified IMD proteins, GlfT2, PyrD, MenG, and MurG have already been examined by biochemical and fluorescence image analysis (8, 13, 14, 26), exhibiting the characteristic features of IMD-associated proteins. We chose two of these proteins, GlfT2 (MSMEG_6403) and PyrD (MSMEG_4198), to confirm the Dendra2-FLAG tag fusions are biochemically associated with the IMD fraction. GlfT2, a galactosyltransferase involved in galactan synthesis, was previously studied with four different fusion proteins: (i) GlfT2-HA, produced from a heterologous genomic locus under the control of a strong promoter; (ii) mTurquoise-GlfT2-FLAG, produced from a heterologous genomic locus under the control of a strong promoter; (iii) HA-mCherry-GlfT2, produced from its native locus (13); and (iv) GlfT2-GFP, produced from its native locus (8). All fluorescent protein fusions were shown previously to have subcellular localization characteristic of IMD-associated proteins. GlfT2-HA, mTurquoise-GlfT2-FLAG, and HA-mCherry-GlfT2 have also been shown to fractionate with the IMD biochemically.
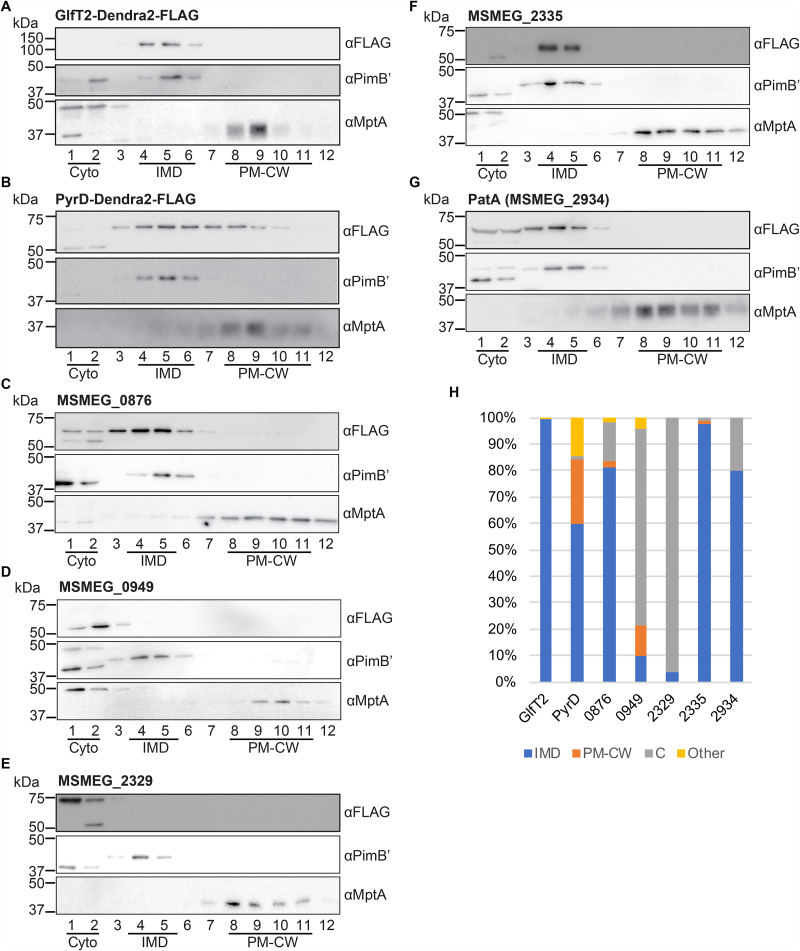
The GlfT2-Dendra2-FLAG fusion described in this report represents the fifth fusion construct tested for subcellular localization. To examine its localization, we grew the strain producing GlfT2-Dendra2-FLAG to log phase, lysed the cells by nitrogen cavitation, and subjected the lysate to sucrose density gradient fractionation. Equal volumes of each collected fraction were separated by SDS-PAGE, and the fusion protein was detected by immunoblotting. We confirmed that GlfT2-Dendra2-FLAG, like other previously tested fusion constructs, was at the predicted molecular weight of 100.2 kDa and specifically associated with the IMD (Fig. 2A). We defined the IMD enrichment as (i) at least 60% of the total band intensity across all gradient fractions is present in the IMD fractions (fractions [Fr.] 3 to 6) and (ii) the protein is enriched more than 2-fold in the IMD fractions (Fr. 3 to 6) relative to other subcellular compartments (i.e., the cytoplasmic fractions [Fr. 1 and 2] and the PM-CW fractions [Fr. 8 to 11]). Quantification of the immunoblot bands indicated that 99.6% of GlfT2-Dendra2-FLAG was associated with the IMD (Fig. 2C), confirming that this fusion protein behaves as an IMD-associated protein.
FIG 2.
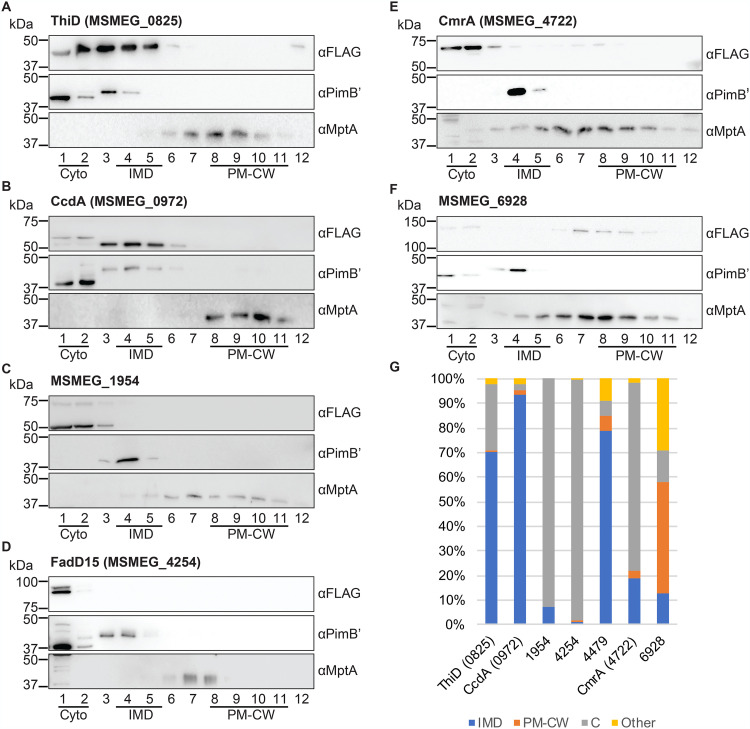
Validation of known IMD protein candidates by density gradient fractionation. (A to G) Among IMD protein candidates identified by the image screening, seven proteins, which were previously found in the IMD proteome, were examined by sucrose density gradient fractionation. Lysate was prepared from cells producing these proteins and was fractionated by sucrose density gradient sedimentation. An equal volume of each gradient fraction was separated by SDS-PAGE, and the FLAG epitope was detected by anti-FLAG immunoblotting. PimB′ (41.1 kDa), IMD marker; MptA (54.3 kDa), PM-CW marker. The apparent molecular weight of MptA on SDS-PAGE gel is about 40 kDa (32, 35). (H) Quantification of anti-FLAG immunoblot bands shown in panels A to G. C, cytoplasm. See Materials and Methods for details of the quantitative analysis.
The IMD association of PyrD, a dihydroorotate reductase involved in pyrimidine synthesis, has also been biochemically verified by the specific association of PyrD-HA with the IMD fractions of a sucrose gradient (13). Consistent with the previous finding, PyrD-Dendra2-FLAG was detected in the IMD fraction (Fig. 2B). Quantification of PyrD-Dendra2-FLAG bands revealed an IMD-specific enrichment pattern: 60.0% was found in the IMD, representing a more than 2-fold enrichment over the cytoplasm (1.75%) and the PM-CW (23.9%) fractions (Fig. 2C). This is consistent with our previous data in which a PyrD-HA was 78% associated with the IMD (13).
Validation of previously predicted IMD proteins.
In addition to these two previously characterized proteins, we examined the biochemical partitioning of five additional proteins, MSMEG_0876 (short-chain dehydrogenase), MSMEG_0949 (HAD-superfamily hydrolase), MSMEG_2329 (UbiE/COQ5 family methyltransferase), MSMEG_2335 (hexapeptide transferase), and MSMEG_2934 (PatA), that were previously predicted as IMD proteins based on mass spectroscopy of IMD fractions but were not validated by independent biochemical assays. The precise functions of these proteins are unknown, except for PatA, which is an acyltransferase involved in the synthesis of phosphatidylinositol mannosides (27). Our previous study suggested that PatA is an IMD-associated enzyme because a cell-free enzymatic assay showed the enriched production of AcPIM2 in the IMD fraction (14); however, there has been no biochemical confirmation of protein enrichment in the IMD. As shown in Fig. 2C, PatA and two other proteins, MSMEG_0876 and MSMEG_2335, were highly enriched in the IMD fractions of the density gradient. In contrast, MSMEG_0949 and MSMEG_2329 were found to be more enriched in the cytoplasmic fraction (Fr. 1 and 2) (Fig. 2C) despite their previous assignment as IMD proteins by proteomics and their Dendra2 fluorescence appearing as IMD associated. These analyses underscore the importance of using both microscopic and biochemical analyses of Dendra2-FLAG fusion proteins to independently validate previously predicted IMD proteins.
Verifying the production of newly identified IMD candidates.
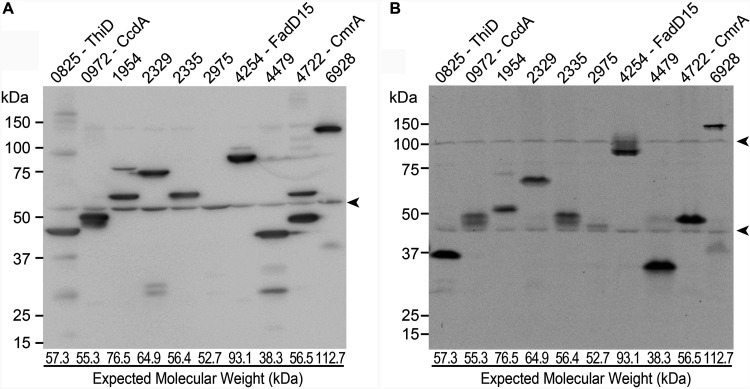
Next, we examined the eight proteins that had not been predicted to be IMD associated in our previous proteomic analysis. We grew each strain producing a Dendra2-FLAG fusion product to log phase, prepared a cell lysate, and analyzed the SDS-PAGE migration rate of the fusion protein by immunoblotting. The immunoblot image of the eight proteins demonstrates that all but ThiD and MSMEG_2975 migrated at the expected molecular weight. We noted, however, the relative protein levels differed, and in many cases there were smaller proteins that presumably correspond to stable proteolytic products or proteins translated from open reading frame-internal start codons (Fig. 3A). We could not detect MSMEG_2975 by immunoblotting. As an alternative approach, we detected Dendra2 protein by fluorescence imaging of nondenaturing seminative-PAGE. As shown in Fig. 3B, all of the proteins showed migration patterns similar to those observed by standard denaturing SDS-PAGE and immunoblot (compare with Fig. 3A), except that ThiD and MSMEG_4479 migrated slightly faster. A faint MSMEG_2975-Dendra2-FLAG band was visualized on the seminative gel, suggesting that the fluorescence imaging method was more sensitive for detecting this fusion protein relative to immunoblotting.
FIG 3.
Immunoblot analysis of newly discovered IMD protein candidates. (A) Crude cell lysates of the eight new IMD protein candidates (as well as MSMEG_2329 and MSMEG_2335, analyzed in Fig. 2) were analyzed by SDS-PAGE, and FLAG epitope was detected by immunoblotting. Expected molecular weight of each protein is indicated at the bottom of the lane. (B) Crude cell lysates were analyzed by PAGE under a nondenaturing condition, and Dendra2 fluorescence was detected in gel by a fluorescence imager. Arrowheads indicate nonspecific proteins.
MSMEG_4479 is an IMD-associated protein.
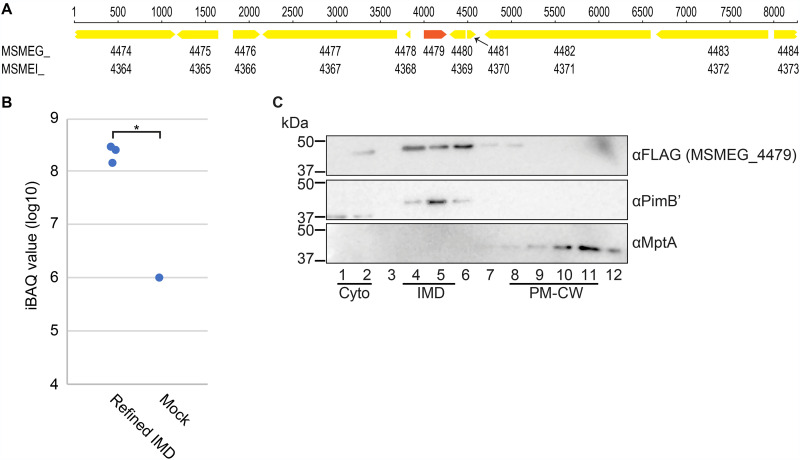
Among the eight candidates that were not found in the IMD proteome, we realized that MSMEG_4479 was missing from the list of annotated genes in the genome sequence (NC_018289, MSMEI_ locus tag prefix), which was used in our proteomic analysis (13). There are two frequently used complete genome sequences for M. smegmatis mc2155 strains in the NCBI Reference Sequence Database: NC_008596, submitted by TIGR on 20 November 2006, and NC_018289, submitted by the Institute of Genetics and Molecular and Cellular Biology (IGMCB) on 3 August 2012. There are 6,716 and 6,693 proteins annotated in the TIGR and IGMCB sequences, respectively. MSMEG_ is the locus tag prefix for the TIGR sequence, and when we cross-referenced MSMEG_4479 against the IGMCB sequence, we could not find the corresponding gene with the MSMEI_ locus tag prefix. We retrieved the genome region from these two complete sequences and searched for open reading frames. The DNA sequence was completely identical between the two sequences in the region 4,000 bp upstream and downstream of the MSMEG_4479 gene (Fig. 4A); however, the annotation of MSMEG_4479 was missing in the IGMCB sequence.
FIG 4.
MSMEG_4479 is identified in the IMD proteome. (A) The genome region surrounding MSMEG_4479. The nucleotide sequence in the region is identical between the two commonly used genome sequences (TIGR, NC_008596, MSMEG_ locus tag prefix; IGMCB, NC_018289, MSMEI_ locus tag prefix). Note that MSMEG_4479 does not have a corresponding MSMEI_ locus number while all other genes surrounding the region have a locus number with both locus tag prefixes. (B) Enrichment of MSMEG_4479 in the IMD, revealed by the reanalysis of previously published proteome data (refined IMD versus mock; see reference 13) using the TIGR genome sequence. iBAQ values were used to compare the protein abundance. Both IMD and mock preparations were analyzed in technical triplicate, but MSMEG_4479 was detected from the mock preparation in only one analysis. *, P < 0.05 by t test (two-tailed). (C) Subcellular localization of MSMEG_4479 was analyzed by density gradient fractionation. See the legend to Fig. 2 for details. Representative data of a biological duplicate are shown.
We reanalyzed our previous proteomic data using the TIGR sequence (NC_008596) as a reference genome. We found a normalized iBAQ value of 6.43 × 108 for MSMEG_4479 from the immunoprecipitated IMD vesicles, which was 700 times more enriched than that in the nonspecific binding fraction, which served as a negative control (Fig. 4B). The iBAQ value and the IMD enrichment were substantially above our previous threshold (13). Thus, our proteomic analysis agrees with the colocalization pattern, suggesting that MSMEG_4479 is an IMD protein, further underscoring the value of the microscopic colocalization study.
Next, we examined the subcellular localization of MSMEG_4479-Dendra2-FLAG by density gradient fractionation. The fusion protein was separated by SDS-PAGE and detected by immunoblotting (Fig. 4C). We measured the band intensity of the fusion protein from each fraction and determined the relative distribution across the density gradient (Fig. 5G). The enrichment of MSMEG_4479 in the IMD was 78.8%, and the protein enrichment in the cytoplasm and the PM-CW was less than 10%. Thus, we have biochemically confirmed MSMEG_4479 as an IMD protein.
FIG 5.
Subcellular fractionation of newly discovered IMD protein candidates. (A to F) Six proteins, which showed Dendra2 fluorescence patterns suggestive of IMD localization but were not previously identified in the IMD proteome, were analyzed by density gradient fractionation. See the legend to Fig. 2 for details. Representative data of two independent experiments are shown (see panel G for details). (G) Immunoblot bands of each protein, as shown in panels A to F and Fig. 4C, were quantified across density gradient fractions and calculated for the percent subcellular localization as described in Materials and Methods. For the new IMD proteins (ThiD, CcdA, and MSMEG_4479), cell culture, cell lysis, density gradient fractionation, and immunoblotting were repeated twice by independent researchers. Average percent localizations of the biological duplicates are shown. For other proteins, immunoblotting was repeated twice by independent researchers, and average percent localization of the technical duplicates is shown.
The IMD association of other candidate proteins determined by density gradient.
We examined the remaining six new candidates by sucrose density gradient (Fig. 5). MSMEG_2975 was not further analyzed due to its low steady-state levels in our extracts (Fig. 3). Based on the definition of IMD association described above, we classify ThiD and CcdA as two additional IMD-associated proteins.
ThiD (MSMEG_0825) is a phosphomethylpyrimidine kinase involved in thiamine biosynthesis (28). As mentioned above, the apparent molecular weight of ThiD was less than the expected molecular weight (Fig. 3). Cell lysates were prepared using a bead-beating cell disruptor for this initial experiment. In contrast, nitrogen cavitation was used to prepare cell lysates for sucrose gradient fractionation. Interestingly, when using a nitrogen cavitation lysate, ThiD migrated with an apparent molecular weight that is closer to its predicted size of 57.3 kDa (Fig. 5A), suggesting that ThiD is more sensitive/exposed to proteases particularly when the cell lysate is prepared by bead beating. The fractionation pattern of ThiD indicates that it is enriched in the IMD (70.3%) relative to the PM-CW (0.9%) and the cytoplasm (26.9%) (Fig. 5G). We conclude that ThiD is an IMD-associated protein, based on its colocalization and biochemical fractionation properties.
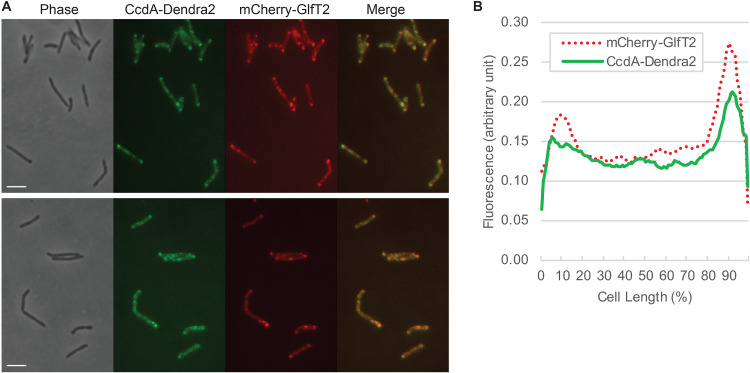
CcdA (MSMEG_0972) is a putative reductase that is predicted to regenerate the reduced form of the periplasmic thioredoxin CcsX, which is essential for the maturation of cytochrome c (29). The amino acid sequence of CcdA suggests that it is a polytopic transmembrane protein. When analyzed by density gradient sedimentation, CcdA-Dendra2-FLAG was highly enriched in the IMD fraction (93.3%) (Fig. 5B and G). To further test the IMD localization of CcdA, we expressed the gene encoding CcdA in a strain where the endogenous gene encoding GlfT2 is replaced with a fusion encoding HA-mCherry-GlfT2 (13). We found that CcdA colocalized with HA-mCherry-GlfT2, a well-established IMD marker (Fig. 6).
FIG 6.
Colocalization of CcdA-Dendra2-FLAG and HA-mCherry-GlfT2, visualized by fluorescence microscopy. (A) HA-mCherry-GlfT2 is produced from the endogenous locus and previously confirmed as an IMD marker (13). Two different fields are shown as representative images. Bar, 4 μm. (B) Fluorescence intensity profile along the long axis of the cell. Cell length is shown as percent, and cells were aligned so that a more intense pole is positioned to the right (100%). Average intensity profiles of 14 cells are shown.
The other four proteins, MSMEG_1954, FadD15, CmrA, and MSMEG_6928, were previously assigned to the PM-CW by proteomic analysis (13). However, our analysis shows that MSMEG_6928 is the only protein that is partially (45%) associated with the PM-CW in the density gradient fractionation (Fig. 5). The other three proteins were found in the cytoplasmic fraction. Similar to ThiD, full-length CmrA protein was predominant after lysis by nitrogen cavitation, in contrast to bead-beating lysis that accumulated a truncated protein (Fig. 5). However, CmrA did not associate with the IMD and was highly enriched in the cytoplasmic fraction (Fig. 5). We note that a small amount of the protein is detectable in the PM-CW fractions, suggesting that this protein partially associates with the PM-CW. In contrast to CmrA, both full-length (76.5 kDa) and truncated (∼50 kDa) versions of MSMEG_1954 protein remained detectable even after nitrogen cavitation cell lysis, and all forms of MSMEG_1954 were enriched in the cytoplasmic fraction (Fig. 5). Therefore, these biochemical characterizations were not consistent with the prediction that these proteins are associated with the IMD based on fluorescence imaging or associated with the PM-CW based on our proteomic analysis.
DISCUSSION
In our previous study, we purified the IMD by density gradient fractionation and immunoprecipitation and analyzed it by proteomics. We reported 309 proteins as IMD-associated proteins. In the current study, we used the Mycobacterial Systems Resource to visually screen 523 images of fluorescent protein localizations in M. smegmatis and identified 29 IMD candidates. Among them, 20 proteins were previously reported as a part of the IMD proteome, while three proteins were newly validated as IMD-associated proteins. Thus, fluorescence image screening is an independent method to identify additional IMD-associated proteins that escaped proteomic discovery.
We analyzed 7 out of 20 previously identified IMD proteins to validate the subcellular localization biochemically. Five of them were not characterized previously, and among them MSMEG_0876, MSMEG_2335, and PatA were enriched in the IMD fraction after density gradient fractionation. In contrast, two proteins, MSMEG_0949 and MSMEG_2329, appeared cytoplasmic and were not associated with the IMD. It remains possible that these proteins are associated with the IMD in vivo but were released from the membrane upon cell disruption. Alternatively, they may be cytoplasmic or lipid droplet associated (see below). Taken together, image screening is effective in identifying proteins that could be false positives of proteomic analysis, highlighting the importance of combining microscopic and biochemical validations for proteomics data.
Strikingly, we could identify only three additional IMD proteins (ThiD, CcdA, and MSMEG_4479) among nine new candidates. Identification of MSMEG_4479 highlights the importance of accurate gene annotation in the database we use for proteomic analysis. ThiD and CcdA are the only two truly new IMD proteins that were not identified by the proteomic analysis thus far. CcdA is an integral membrane protein with six predicted transmembrane segments. We suspect that the hydrophobic nature of the protein is one of the reasons why this protein evaded detection by proteomic studies.
We found five proteins to be cytoplasmic: FadD15, CmrA, MSMEG_0949, MSMEG_1954, and MSMEG_2329. Interestingly, all of these proteins were previously associated with a lipid droplet proteome (30). It remains to be determined why fluorescence imaging of these proteins revealed localization patterns that are similar to those of IMD-associated proteins. It should also be noted that the cytoplasmic localization of the fusion proteins by density gradient fractionation could be artifacts of fusion constructs rather than representing the true subcellular localization of these proteins.
How many IMD-associated proteins could there be in the M. smegmatis proteome? Considering that we found three new proteins from screening 523 proteins, we predict that an additional ∼30 proteins might be IMD associated (based on a genome encoding 6,716 proteins). However, we must also consider the possibility that the list of proteins in the IMD proteome contains additional false positives. Thus, the total number of IMD-associated proteins may not be much greater than 300 proteins. This represents up to 5% of the total known proteins produced by M. smegmatis. In a broader perspective, 2, 6, and 11% of proteins in human cells associate with the endoplasmic reticulum, Golgi apparatus, and plasma membrane, respectively (31). Therefore, the estimated percentage of IMD-associated proteins in M. smegmatis is reasonable for proteins associating with a subcellular membrane compartment.
In conclusion, the combination of biochemical and microscopy approaches described here has increased the rigor of assigning proteins to a subcellular factory, the mycobacterial IMD. The Mycobacterial Systems Resource offers a valuable platform to integrate knowledge on the atlas of proteins in mycobacterial cells.
MATERIALS AND METHODS
Construction of expression vectors and creation of fluorescence image library.
Expression vectors were constructed as a part of the Mycobacterial Systems Resource project (http://msrdb.org/) (25), creating plasmids to produce a Dendra2-FLAG fusion of 1,118 M. smegmatis proteins that represent the core genome conserved across five species of mycobacteria. Briefly, Mycobacterium smegmatis open reading frames were amplified from the start codon through the penultimate codon, excluding the respective stop codon, to allow C-terminal fusion to a glycine/alanine linker leading to the Dendra2-FLAG open reading frame. The plasmids insert into the L5 attB site in mycobacteria, and transformants were selected by resistance to apramycin. Cloned expression cassettes were constitutively expressed from the smyc promoter and translation initiation guided by a canonical Shine-Dalgarno sequence in the 5′ untranslated region. We analyzed fluorescence images of 523 strains that produced sufficient-quality images in the preliminary stage of image acquisition (see Table S1 in the supplemental material). Fluorescence localization was categorized as “IMD-like” by satisfying both of the following two criteria: (i) intense enrichment in the polar regions of the cell and (ii) less intense sidewall patches. Images of 523 strains were visually screened for IMD-like fluorescent localization (Fig. 1).
Preparation of crude cell lysates.
Cells were grown in Middlebrook 7H9 broth supplemented with 11 mM glucose, 14.5 mM NaCl, 0.05% Tween 80, and 12.5 μg/ml apramycin at 30°C with shaking until reaching a log phase (optical density at 600 nm [OD600] of 0.5 to 1.0). Cells were spun at 3,220 × g at 4°C, washed in 50 mM HEPES-NaOH (pH 7.4), and resuspended in a lysis buffer containing 25 mM HEPES-NaOH (pH 7.4), 15% glycerol, 2 mM EGTA, and a protease inhibitor cocktail (ThermoFisher). Cells were lysed by bead beating (acid washed, 106 μm; Millipore-Sigma), and lysate was stored frozen at −20°C.
Bicinchoninic acid assay.
Protein concentration was determined by following the manufacturer's instructions (ThermoFisher) using bovine serum albumin as a standard. Each sample was analyzed in duplicate, and the average was used to calculate the protein concentration.
Sucrose density gradient fractionation.
Sucrose density gradient fractionation was done as described before (13). Briefly, log-phase cells were washed in 50 mM HEPES-NaOH (pH 7.4) and resuspended in a lysis buffer containing 25 mM HEPES (pH 7.4), 20% sucrose, 2 mM EGTA, and a protease inhibitor mix (ThermoFisher). Nitrogen cavitation was repeated three times with 1,850 to 2,200 lb/in2 for 30 min each. Lysates were placed on top of a 20 to 50% sucrose gradient and sedimented at 35,000 rpm (218,000 × g) for 6 h at 4°C on an SW40Ti rotor (Beckman). Twelve 1-ml fractions were collected using a fraction collector (BioComp) and stored at −80°C.
SDS-PAGE and immunoblotting.
Cell lysates or sucrose gradient fractions were denatured in a reducing sample loading buffer containing 62 mM Tris-HCl (pH 6.8), 50 mM dithiothreitol, 2% (wt/vol) SDS, 12.5% (wt/vol) glycerol, and 100 μg/ml bromophenol blue at 95°C for 5 min and analyzed by SDS-PAGE (10% gel) using a running buffer containing 25 mM Tris-HCl (pH 8.3), 192 mM glycine, and 0.1% SDS (wt/vol). Precision plus protein Kaleidoscope (Bio-Rad) was used as a molecular weight standard. Proteins were transferred to an Immun-Blot polyvinylidene difluoride (PVDF) membrane (Bio-Rad), blocked in 5% skim milk in phosphate-buffered saline (PBS) supplemented with 0.05% Tween 20 (PBST20), and incubated with primary antibodies (mouse anti-FLAG [Millipore-Sigma]; rabbit anti-PimB′ [32]; or rabbit anti-MptA [32]). Membrane was washed with PBST20 and then incubated with secondary antibodies (horseradish peroxidase-conjugated anti-mouse IgG [GE Healthcare] or horseradish peroxidase-conjugated anti-rabbit IgG [GE Healthcare]). The membrane was washed with PBST20 and imaged by chemiluminescence using ImageQuant LAS-4000 mini (GE Healthcare) or Amersham ImageQuant 800 (Cytiva). For sucrose density gradient fractions, an equal volume of each fraction was loaded into each well. Immunoblot bands were quantified by ImageQuant TL (GE Healthcare) or Fiji (33). Subcellular localization was calculated as follows and expressed as percent localization of anti-FLAG immunoblot bands:
where xi denotes the anti-FLAG band intensity of fraction i.
Seminative PAGE and fluorescence.
Protein samples were incubated with nonreducing SDS-free sample loading buffer, containing 62 mM Tris-HCl (pH 6.8), 12.5% (wt/vol) glycerol, and 100 μg/ml bromophenol blue on ice for 30 min. The nondenatured samples were analyzed by standard SDS-PAGE (10% gel) in a running buffer containing 25 mM Tris-HCl (pH 8.3), 192 mM glycine, and 0.1% (wt/vol) SDS. Green fluorescence of Dendra2 was visualized using ImageQuant LAS-4000 mini (GE).
Electroporation of plasmid constructs.
An M. smegmatis strain producing HA-mCherry-GlfT2 from the endogenous locus (13) was electroporated with plasmid DNA at 2.8 kV, 99 μs, five times at 1-s intervals using a square wave electroporation system (BTX ECM830).
Fluorescence microscopy and image analysis.
M. smegmatis cells coproducing HA-mCherry-GlfT2 and CcdA-Dendra2-FLAG were grown in Middlebrook 7H9 broth to log phase (OD600 = 0.5 to 1.0), and 10 μl of cells was placed onto a 1% agar pad made of Middlebrook 7H9 broth. All images were taken using a fluorescence microscope (Nikon Eclipse E600) with a 100× Plan Fluor 1.30 (oil) lens objective, equipped with a cooled charge-coupled device Spot-RT digital camera (Diagnostic Instruments). Cell contours were outlined and fluorescence intensities along the long axis of the cell were measured using Oufti and custom-written MATLAB codes as described previously (12, 34). The cell length was normalized by taking the length of the long axis of the cell as 100%. Cells were oriented so that the brighter pole is on the right side of the graph.
ACKNOWLEDGMENTS
This work was supported by NIH R03 AI140259 to Y.S.M. and NIH R01 AI097191 to K.M.D., S.M.F., T.A.G., and J.T.W. C.A.Z.R. was a recipient of the American Society for Microbiology Undergraduate Research Fellowship. J.P. was a recipient of the Science Without Borders Fellowship from CAPES-Brazil (0328-13-8). M.P. was partially supported by a fellowship from the University of Massachusetts as part of the Chemistry-Biology Interface Training Program (National Research Service Award T32 GM139789).
Footnotes
Supplemental material is available online only.
Contributor Information
Yasu S. Morita, Email: ymorita@microbio.umass.edu.
Conrad W. Mullineaux, Queen Mary University of London
REFERENCES
- 1.Daffé M, Marrakchi H. 2019. Unraveling the structure of the mycobacterial envelope. Microbiol Spectr 10.1128/microbiolspec.GPP3-0027-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackson M. 2014. The mycobacterial cell envelope-lipids. Cold Spring Harb Perspect Med 4:a021105. 10.1101/cshperspect.a021105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaur D, Guerin ME, Skovierová H, Brennan PJ, Jackson M. 2009. Chapter 2: biogenesis of the cell wall and other glycoconjugates of Mycobacterium tuberculosis. Adv Appl Microbiol 69:23–78. 10.1016/S0065-2164(09)69002-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dulberger CL, Rubin EJ, Boutte CC. 2020. The mycobacterial cell envelope–a moving target. Nat Rev Microbiol 18:47–59. 10.1038/s41579-019-0273-7. [DOI] [PubMed] [Google Scholar]
- 5.Rahlwes KC, Sparks IL, Morita YS. 2019. Cell walls and membranes of Actinobacteria. Subcell Biochem 92:417–469. 10.1007/978-3-030-18768-2_13. [DOI] [PubMed] [Google Scholar]
- 6.Thanky NR, Young DB, Robertson BD. 2007. Unusual features of the cell cycle in mycobacteria: polar-restricted growth and the snapping-model of cell division. Tuberculosis (Edinb) 87:231–236. 10.1016/j.tube.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 7.Aldridge BB, Fernandez-Suarez M, Heller D, Ambravaneswaran V, Irimia D, Toner M, Fortune SM. 2012. Asymmetry and aging of mycobacterial cells lead to variable growth and antibiotic susceptibility. Science 335:100–104. 10.1126/science.1216166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meniche X, Otten R, Siegrist MS, Baer CE, Murphy KC, Bertozzi CR, Sassetti CM. 2014. Subpolar addition of new cell wall is directed by DivIVA in mycobacteria. Proc Natl Acad Sci USA 111:E3243–E3251. 10.1073/pnas.1402158111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santi I, Dhar N, Bousbaine D, Wakamoto Y, McKinney JD. 2013. Single-cell dynamics of the chromosome replication and cell division cycles in mycobacteria. Nat Commun 4:2470. 10.1038/ncomms3470. [DOI] [PubMed] [Google Scholar]
- 10.Singh B, Nitharwal RG, Ramesh M, Pettersson BMF, Kirsebom LA, Dasgupta S. 2013. Asymmetric growth and division in Mycobacterium spp.: compensatory mechanisms for non-medial septa. Mol Microbiol 88:64–76. 10.1111/mmi.12169. [DOI] [PubMed] [Google Scholar]
- 11.Joyce G, Williams KJ, Robb M, Noens E, Tizzano B, Shahrezaei V, Robertson BD. 2012. Cell division site placement and asymmetric growth in mycobacteria. PLoS One 7:e44582. 10.1371/journal.pone.0044582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.García-Heredia A, Pohane AA, Melzer ES, Carr CR, Fiolek TJ, Rundell SR, Chuin Lim H, Wagner JC, Morita YS, Swarts BM, Siegrist MS. 2018. Peptidoglycan precursor synthesis along the sidewall of pole-growing mycobacteria. Elife 7:e37243. 10.7554/eLife.37243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayashi JM, Luo C-Y, Mayfield JA, Hsu T, Fukuda T, Walfield AL, Giffen SR, Leszyk JD, Baer CE, Bennion OT, Madduri A, Shaffer SA, Aldridge BB, Sassetti CM, Sandler SJ, Kinoshita T, Moody DB, Morita YS. 2016. Spatially distinct and metabolically active membrane domain in mycobacteria. Proc Natl Acad Sci USA 113:5400–5405. 10.1073/pnas.1525165113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morita YS, Velasquez R, Taig E, Waller RF, Patterson JH, Tull D, Williams SJ, Billman-Jacobe H, McConville MJ. 2005. Compartmentalization of lipid biosynthesis in mycobacteria. J Biol Chem 280:21645–21652. 10.1074/jbc.M414181200. [DOI] [PubMed] [Google Scholar]
- 15.Hayashi JM, Richardson K, Melzer ES, Sandler SJ, Aldridge BB, Siegrist MS, Morita YS. 2018. Stress-induced reorganization of the mycobacterial membrane domain. mBio 9:e01823-17. 10.1128/mBio.01823-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.García-Heredia A, Kado T, Sein CE, Puffal J, Osman SH, Judd J, Gray TA, Morita YS, Siegrist MS. 2021. Membrane-partitioned cell wall synthesis in mycobacteria. Elife 10:e60263. 10.7554/eLife.60263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guerin ME, Kaur D, Somashekar BS, Gibbs S, Gest P, Chatterjee D, Brennan PJ, Jackson M. 2009. New insights into the early steps of phosphatidylinositol mannoside biosynthesis in mycobacteria: PimB’ is an essential enzyme of Mycobacterium smegmatis. J Biol Chem 284:25687–25696. 10.1074/jbc.M109.030593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lea-Smith DJ, Martin KL, Pyke JS, Tull D, McConville MJ, Coppel RL, Crellin PK. 2008. Analysis of a new mannosyltransferase required for the synthesis of phosphatidylinositol mannosides and lipoarbinomannan reveals two lipomannan pools in Corynebacterineae. J Biol Chem 283:6773–6782. 10.1074/jbc.M707139200. [DOI] [PubMed] [Google Scholar]
- 19.Rana AK, Singh A, Gurcha SS, Cox LR, Bhatt A, Besra GS. 2012. Ppm1-encoded polyprenyl monophosphomannose synthase activity is essential for lipoglycan synthesis and survival in mycobacteria. PLoS One 7:e48211. 10.1371/journal.pone.0048211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gurcha SS, Baulard AR, Kremer L, Locht C, Moody DB, Muhlecker W, Costello CE, Crick DC, Brennan PJ, Besra GS. 2002. Ppm1, a novel polyprenol monophosphomannose synthase from Mycobacterium tuberculosis. Biochem J 365:441–450. 10.1042/BJ20020107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baulard AR, Gurcha SS, Engohang-Ndong J, Gouffi K, Locht C, Besra GS. 2003. In vivo interaction between the polyprenol phosphate mannose synthase Ppm1 and the integral membrane protein Ppm2 from Mycobacterium smegmatis revealed by a bacterial two-hybrid system. J Biol Chem 278:2242–2248. 10.1074/jbc.M207922200. [DOI] [PubMed] [Google Scholar]
- 22.Gibson KJC, Eggeling L, Maughan WN, Krumbach K, Gurcha SS, Nigou J, Puzo G, Sahm H, Besra GS. 2003. Disruption of Cg-Ppm1, a polyprenyl monophosphomannose synthase, and the generation of lipoglycan-less mutants in Corynebacterium glutamicum. J Biol Chem 278:40842–40850. 10.1074/jbc.M307988200. [DOI] [PubMed] [Google Scholar]
- 23.Mikusová K, Yagi T, Stern R, McNeil MR, Besra GS, Crick DC, Brennan PJ. 2000. Biosynthesis of the galactan component of the mycobacterial cell wall. J Biol Chem 275:33890–33897. 10.1074/jbc.M006875200. [DOI] [PubMed] [Google Scholar]
- 24.Kremer L, Dover LG, Morehouse C, Hitchin P, Everett M, Morris HR, Dell A, Brennan PJ, McNeil MR, Flaherty C, Duncan K, Besra GS. 2001. Galactan biosynthesis in Mycobacterium tuberculosis. Identification of a bifunctional UDP-galactofuranosyltransferase. J Biol Chem 276:26430–26440. 10.1074/jbc.M102022200. [DOI] [PubMed] [Google Scholar]
- 25.Judd JA, Canestrari J, Clark R, Joseph A, Lapierre P, Lasek-Nesselquist E, Mir M, Palumbo M, Smith C, Stone M, Upadhyay A, Wirth SE, Dedrick RM, Meier CG, Russell DA, Dills A, Dove E, Kester J, Wolf ID, Zhu J, Rubin ER, Fortune S, Hatfull GF, Gray TA, Wade JT, Derbyshire KM. 2021. A Mycobacterial Systems Resource for the research community. mBio 12:e02401-20. 10.1128/mBio.02401-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Puffal J, Mayfield JA, Moody DB, Morita YS. 2018. Demethylmenaquinone methyl transferase is a membrane domain-associated protein essential for menaquinone homeostasis in Mycobacterium smegmatis. Front Microbiol 9:3145. 10.3389/fmicb.2018.03145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kordulakova J, Gilleron M, Puzo G, Brennan PJ, Gicquel B, Mikusova K, Jackson M. 2003. Identification of the required acyltransferase step in the biosynthesis of the phosphatidylinositol mannosides of Mycobacterium species. J Biol Chem 278:36285–36295. 10.1074/jbc.M303639200. [DOI] [PubMed] [Google Scholar]
- 28.Tyagi G, Singh P, Varma-Basil M, Bose M. 2017. Role of vitamins B, C, and D in the fight against tuberculosis. Int J Mycobacteriol 6:328–332. 10.4103/ijmy.ijmy_80_17. [DOI] [PubMed] [Google Scholar]
- 29.Small JL, Park SW, Kana BD, Ioerger TR, Sacchettini JC, Ehrt S. 2013. Perturbation of cytochrome c maturation reveals adaptability of the respiratory chain in Mycobacterium tuberculosis. mBio 4:e00475-13. 10.1128/mBio.00475-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Armstrong RM, Carter DC, Atkinson SN, Terhune SS, Zahrt TC. 2018. Association of Mycobacterium proteins with lipid droplets. J Bacteriol 200:e00240-18. 10.1128/JB.00240-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thul PJ, Åkesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, Alm T, Asplund A, Björk L, Breckels LM, Bäckström A, Danielsson F, Fagerberg L, Fall J, Gatto L, Gnann C, Hober S, Hjelmare M, Johansson F, Lee S, Lindskog C, Mulder J, Mulvey CM, Nilsson P, Oksvold P, Rockberg J, Schutten R, Schwenk JM, Sivertsson Å, Sjöstedt E, Skogs M, Stadler C, Sullivan DP, Tegel H, Winsnes C, Zhang C, Zwahlen M, Mardinoglu A, Pontén F, von Feilitzen K, Lilley KS, Uhlén M, Lundberg E. 2017. A subcellular map of the human proteome. Science 356:eaal3321. 10.1126/science.aal3321. [DOI] [PubMed] [Google Scholar]
- 32.Sena CBC, Fukuda T, Miyanagi K, Matsumoto S, Kobayashi K, Murakami Y, Maeda Y, Kinoshita T, Morita YS. 2010. Controlled expression of branch-forming mannosyltransferase is critical for mycobacterial lipoarabinomannan biosynthesis. J Biol Chem 285:13326–13336. 10.1074/jbc.M109.077297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paintdakhi A, Parry B, Campos M, Irnov I, Elf J, Surovtsev I, Jacobs-Wagner C. 2016. Oufti: an integrated software package for high-accuracy, high-throughput quantitative microscopy analysis. Mol Microbiol 99:767–777. 10.1111/mmi.13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fukuda T, Matsumura T, Ato M, Hamasaki M, Nishiuchi Y, Murakami Y, Maeda Y, Yoshimori T, Matsumoto S, Kobayashi K, Kinoshita T, Morita YS. 2013. Critical roles for lipomannan and lipoarabinomannan in cell wall integrity of mycobacteria and pathogenesis of tuberculosis. mBio 4:e00472-12. 10.1128/mBio.00472-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Download JB.00419-21-s0001.xlsx, XLSX file, 0.02 MB (17.5KB, xlsx)