ABSTRACT
Pseudomonas aeruginosa strains PA14 and PAO1 are among the two best-characterized model organisms used to study the mechanisms of biofilm formation while also representing two distinct lineages of P. aeruginosa. Previous work has shown that PA14 and PAO1 use different strategies for surface colonization; they also have different extracellular matrix composition and different propensities to disperse from biofilms back into the planktonic phase surrounding them. We expand on this work here by exploring the consequences of these different biofilm production strategies during direct competition. Using differentially labeled strains and microfluidic culture methods, we show that PAO1 can outcompete PA14 in direct competition during early colonization and subsequent biofilm growth, that they can do so in constant and perturbed environments, and that this advantage is specific to biofilm growth and requires production of the Psl polysaccharide. In contrast, P. aeruginosa PA14 is better able to invade preformed biofilms and is more inclined to remain surface-associated under starvation conditions. These data together suggest that while P. aeruginosa PAO1 and PA14 are both able to effectively colonize surfaces, they do so in different ways that are advantageous under different environmental settings.
IMPORTANCE Recent studies indicate that P. aeruginosa PAO1 and PA14 use distinct strategies to initiate biofilm formation. We investigated whether their respective colonization and matrix secretion strategies impact their ability to compete under different biofilm-forming regimes. Our work shows that these different strategies do indeed impact how these strains fair in direct competition: PAO1 dominates during colonization of a naive surface, while PA14 is more effective in colonizing a preformed biofilm. These data suggest that even for very similar microbes there can be distinct strategies to successfully colonize and persist on surfaces during the biofilm life cycle.
KEYWORDS: Pseudomonas aeruginosa, biofilm, exopolysaccharide, colonization, competition, dispersal, starvation, invasion
INTRODUCTION
Biofilms are surface-attached microbial communities that mediate long-term growth and persistence on substrates as varied as vegetable produce and indwelling medical devices (1, 2). Biofilms are typically polymicrobial in nature and exhibit an increased tolerance to antimicrobial agents, predation, and reactive oxygen species (3–7). The transition from planktonic to biofilm modes of growth is complex and uses signaling systems initiated by surface engagement that have downstream consequences for biofilm formation (8–10). To form a biofilm, motile bacteria such as Pseudomonas aeruginosa use appendages including flagella and pili to mediate early surface engagement (11, 12). Surface colonization and growth during biofilm formation are also linked to extracellular polysaccharide (EPS) production, which provides cell-to-surface and cell-cell adhesion (13). For some species, EPS matrix is crucial for early surface attachment (13); matrix material also strongly influences biofilm architecture and structural integrity (14, 15), collective biophysical properties (16–18), and the population dynamics of different strains and species competing for space and resources (19).
Our understanding of biofilm formation comes in part from the study of the PA14 and PAO1 strains of P. aeruginosa, a Gram-negative bacterium that forms prolific biofilms in aquatic environments, soils, and clinical settings (20–23). This species uses two distinct signaling systems to initiate biofilm formation (24, 25): the second messenger c-di-GMP, which is regulated through the Wsp system (26–28), and the second messenger cyclic AMP (cAMP), which is regulated through the Pil-Chp/Vfr system (29, 30). Prior work documenting variation in propensity of PAO1 and PA14 to commit to surface attachment have intimated that different strains of P. aeruginosa more heavily rely on different components of the signaling pathways controlling biofilm initiation. PAO1, for example, commits to surface attachment and extracellular matrix secretion relatively quickly via the Wsp system-mediated increases in c-di-GMP production (28, 31, 32). PA14 surface commitment control may instead be dominated by signaling via the Pil-Chp system initiated through type IV pili (TFP) engaging a surface, which in turn transmits this surface signal to the adenylate cyclase, CyaB. The activation of CyaB increases concentrations of cAMP, resulting in production and secretion of cell surface-localized PilY1. The PilY1 protein participates in the activation of diguanylate cyclase SadC by an unknown mechanism to enhance c-di-GMP production (30, 33–36).
P. aeruginosa PAO1 and PA14, both isolated from infected wounds (37, 38), represent two distinct lineages based on whole-genome phylogenetic analysis (39). As noted above, they differ in their pattern for committing to surface attachment; their matrix compositions differ as well: both produce Pel, but PAO1 produces a major matrix polysaccharide, Psl, that PA14 lacks (40–42). The overall result is that these two strains exhibit distinct surface association strategies: PAO1 commits relatively quickly to attachment and matrix production after surface contact, while PA14 shows a nonprocessive mode of surface commitment in which cells frequently disengage from surfaces after initial contact while their progeny are primed for subsequent reattachment through a cAMP-dependent signaling pathway (25, 30, 43, 44).
Although PAO1 and PA14 are both heavily studied as models for biofilm formation, there is relatively little work exploring the adaptive rationale for their distinct surface association and biofilm commitment strategies. Biofilm production is very expensive both in terms of metabolic cost and opportunity cost if poorly timed relative to environmental suitability, and the differences in biofilm production strategy between strains and species may often reflect adaptations to different ecological circumstances (45–47). These niche-specific conditions may include, for example, variation in the typical lifetime of a given location patch for growth, the longevity of important nutrient supplies, and whether encountered surfaces are usually bare or already occupied by other bacteria. On the basis of the mechanistic differences in how PAO1 and PA14 control surface attachment and commitment to matrix production, we explore how these two strains compete against each other in model biofilms, studying in particular their population dynamics and spatial structure with respect to each other in coculture versus monoculture. We find that their distinct biofilm production strategies do indeed have significant consequences for the outcome of competition and that PAO1 and PA14 are best suited for surface occupation under different environmental circumstances.
RESULTS
PAO1 outcompetes PA14 in dual-strain biofilms.
To test if the colonization strategies used by P. aeruginosa PA14 or PAO1 impart an advantage during the initial colonization of a clean surface, we used microfluidic devices in which bacteria can be monitored as they attach and grow on glass substrata. To allow for visualization of each strain by fluorescence microscopy, constitutive fluorescent protein expression constructs were introduced, with GFP and mKO-κ inserted at the att site of PAO1 and PA14, respectively (48).
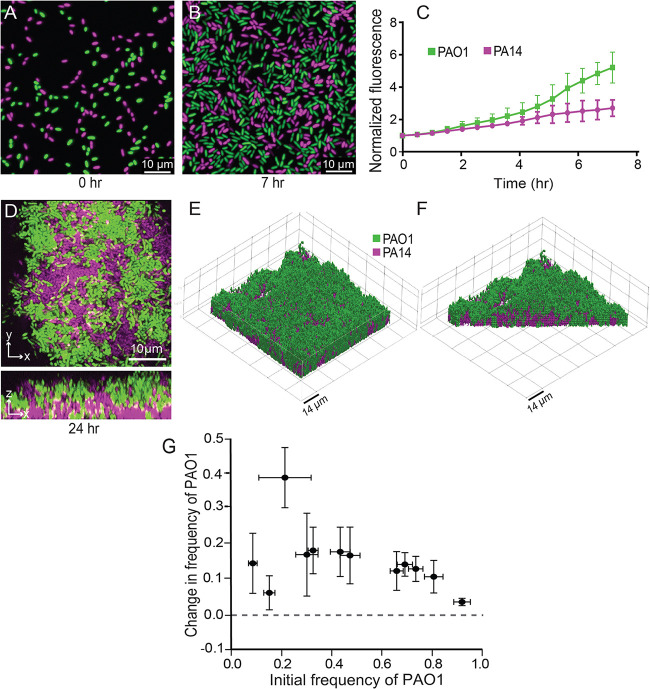
To assess competition during early biofilm formation, PAO1 and PA14 were inoculated into microfluidic chambers in a 1:1 ratio and imaged over 7 h of flow in a buffered minimal medium containing 1.0 mM K2HPO4, 0.6 mM MgSO4, and 0.4% arginine. We measured the fold change in fluorescence for each strain over time normalized to 0 h; PAO1 showed a significant advantage in biomass accumulation over PA14 as early as 4 h after surface inoculation. After 7 h of incubation, the number of PAO1 cells was ∼2-fold greater than that of PA14 (Fig. 1A to C).
FIG 1.
PA14-PAO1 dual strain biofilm population dynamics. (A, B, and D) Representative widefield fluorescence (A and B) and confocal (D) images of PA14-PAO1 coculture biofilms at the time points indicated under each image. Biofilms were inoculated with a 1:1 mixture of each strain at 0 h. The same color scheme for PAO1 (green) and PA14 (magenta) is used in all panels here and in all figures. (C) Quantification of early surface coverage by each strain (0 to 7 h). n = 3, P < 0.05 after 5.5 h by 2-way ANOVA with Sidak posttest. (E) 3D rendering of a PA14-PAO1 dual-strain biofilm at 24 h. (F) Cutaway 3D rendering of the same biofilm in panel E, showing its internal structure. (G) Change in frequency of strain WT PAO1 as a function of its initial frequency in dual-strain biofilms with WT PA14. n = 9 nonoverlapping image stacks from 4 separate microfluidic devices. Error bars denote the standard deviations.
If the observed advantage of PAO1 during biofilm competition was due to enhanced growth rate or antagonistic interactions with PA14 via diffusible secreted factors, one would expect a similar competitive outcome in mixed liquid culture conditions. We tested this possibility by growing planktonic PAO1 and PA14 in glass tubes containing sterile biofilm medium either in monoculture or 1:1 coculture. PAO1 and PA14 grew equally well when cultured separately (see Fig. S1A in the supplemental material), and neither strain outgrew the other in mixed planktonic cultures (Fig. S1B). Notably, the CFU counts of the strains in a coculture were lower by similar measures than their respective CFU counts when grown alone; this indicates neutral competition in which the two strains compete for limited resources in mixed liquid coculture, but neither has an advantage over the other. These results suggest that the advantage in biofilm competition for PAO1 is not due to higher basal growth capacity but rather to other root causes.
Biofilms often exhibit complex architectures that can vary significantly in mixed-strain or mixed-species contexts (49–53); in light of these observations, we next assessed the population dynamics and spatial structure of PAO1 and PA14 allowed to grow for longer periods in biofilm coculture. The two strains were grown together in microfluidic chambers for 24 h and then imaged by confocal microscopy. Consistent with the pattern observed in early-stage biofilms inoculated with a 1:1 ratio of PAO1 and PA14 (Fig. 1A to C), after 24 h PAO1 appeared to overgrow PA14 (Fig. 1D). Z-projections of confocal image stacks revealed that PAO1 grows across the top of the PA14 cell clusters (Fig. 1D to F), consistent with a previous study competing different strains of PAO1 that vary in their matrix production (54).
The above-described data provide evidence that when grown in the biofilm context, PAO1 has an advantage against PA14 during early colonization and 24 h of biofilm growth. Thus far, a 1:1 inoculation ratio of PAO1 to PA14 was used to assess dual-strain population dynamics. This condition does not account for the possibility of frequency-dependent selection, in which the outcome of competition may depend on the initial ratio of the two strains. To test if the competitive advantage of PAO1 was frequency dependent, we varied the starting PAO1 relative abundance and measured the change in frequency of PAO1 versus PA14 after 24 h. Regardless of the starting conditions, PAO1 consistently increased in frequency over PA14 (Fig. 1G), and PA14 biovolume accumulation was suppressed in coculture with PAO1 relative to when PA14 was cultivated on its own (Fig. S2A and B). The pattern shown in Fig. 1G is characteristic of uniform positive selection that would ultimately lead to complete displacement of PA14 by PAO1 during successive rounds of biofilm competition. We also tested whether changes in initial surface colonization density might affect these population dynamics and found that they did not (Fig. S2C). We conclude from these experiments that under the flow conditions used here, PAO1 outcompetes PA14 in a frequency- and density-independent fashion.
Psl is required for PAO1 to outcompete PA14 in biofilm competition.
We have provided evidence that PAO1 robustly outcompetes PA14 when both are grown together in biofilms under flow. To clarify the mechanism of these dynamics, we attempted to alter the competition outcome by manipulating the biofilm formation capacity of PAO1 or PA14. P. aeruginosa produces three extracellular polysaccharides (EPS) known to facilitate biofilm formation: Pel, Psl, and alginate (55). Alginate is broadly conserved in pseudomonads but only conditionally expressed in PAO1 and PA14 during periods of stress (41, 56, 57) and was shown previously to not contribute to in vitro biofilm formation by these strains (41). PAO1 and PA14 both produce Pel, while Psl is unique to PAO1 (58, 59). Previous work has also shown that Psl is a cooperative resource among secreting cells but that cells that do not produce it are excluded and outcompeted in a PAO1 background (54). We hypothesized that Psl provides an advantage for PAO1 not afforded to PA14 and tested a PAOl mutant with a clean deletion of the psl promoter (referred to as the PAO1 Δpsl mutant here) against PA14 during biofilm growth. Loss of Psl production eliminated the competitive advantage of PAO1 in biofilm coculture with PA14, and indeed the PAO1 Δpsl mutant decreased in relative abundance by over 40% in chambers inoculated with a 1:1 mixed culture (Fig. 2; see Fig. S3A for absolute initial and final frequency data). This result was entirely explained by the dynamics of competition during biofilm growth between PAO1 and the PAO1 Δpsl mutant rather than differences in colonization of the glass at the start of the experiment (Fig. S3A). As a baseline control, we also competed WT PAO1 against the PAO1 Δpsl strain, which recapitulated earlier work showing that WT PAO1 overgrows and outcompetes its isogenic Δpsl deletion mutant (Fig. S3B and C) (54).
FIG 2.
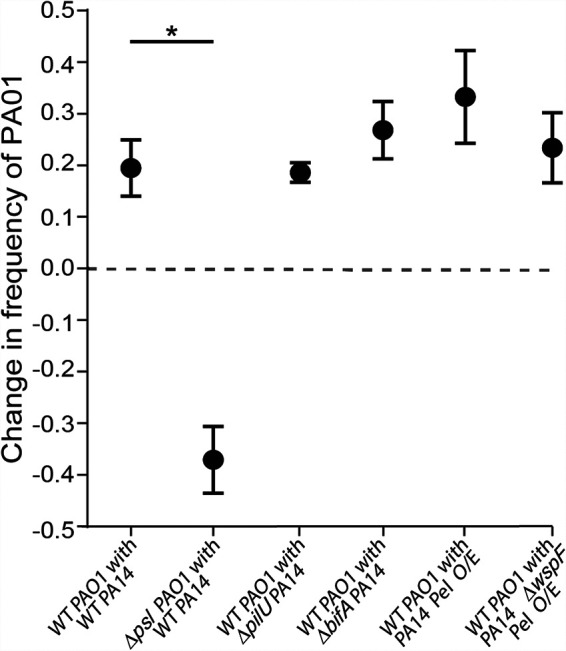
Competitive dynamics of PAO1 and PA14 with altered piliation or matrix production capacity. This figure reports the change in frequency of WT PAO1 and the PAO1 Δpsl mutant in coculture biofilms with WT PA14 as well as the change in frequency of WT PAO1 in a dual-strain biofilms with PA14ΔpilU, PA14 ΔbifA, and Pel-overproducing strains (Pel O/E) supplemented with arabinose. Biofilms were inoculated with a 1:1 mixture of each strain pair (n = 9 to 18 nonoverlapping image stacks from 4 to 6 separate microfluidic chambers). Error bars denote the standard deviations; Wilcoxon signed-rank tests with Bonferroni correction were used for pairwise comparisons; *, P < 0.05; all other comparisons were not significant.
We next explored whether the inverse manipulation of increasing PA14 biofilm production capacity could allow it to better compete against WT PAO1. First, we took advantage of the observation that type 4 pili (T4P) have been shown to mediate initial attachment by P. aeruginosa (12), and recent work showed that manipulation of T4P functions may enhance surface commitment by PA14 (44). Specifically, we tested if the PA14 ΔpilU mutant, which is hyperpiliated, shows high constitutive cAMP signaling, and rapidly colonizes surfaces, would gain a competitive advantage when grown in a biofilm with PAO1. This was not the case; the PA14 ΔpilU mutant was outcompeted by PAO1 by a margin comparable to that of wild-type (WT) PA14 (Fig. 2). Taking note of the absolute initial and final frequency data (Fig. S3A), whereas all other strain pairs initially colonized the chamber glass surfaces at ∼1:1, reflecting the 1:1 mixed liquid cultures introduced to the microfluidic chambers to initiate these experiments, the ΔpilU PA14 strain was outcompeted by PAO1 even at this initial colonization stage. Against the ΔpilU PA14 mutant, PAO1 was overrepresented on the surface after the inoculation period from a 1:1 mixed liquid culture and subsequently increased in relative abundance even further (compare red and blue data points in Fig. S3A).
Next, having shown that Psl plays an important role in the PAO1 strain’s ability to compete against PA14, we reasoned that PA14 might gain an advantage, or at least mitigate its disadvantage, through hyperinduction of its primary matrix polysaccharide, Pel. To test this hypothesis, we first used a ΔbifA mutant, which we showed previously displays increased c-di-GMP signaling and enhanced Pel polysaccharide production (60), but the PA14 ΔbifA mutant was not any more successful against PAO1 than WT PA14. As an alternative approach, we next used a strain that expresses the pel operon under the control of an arabinose-inducible promoter (designated Pel O/E). Interestingly, while induction of pel expression with 0.2% arabinose increased biofilm accumulation of this strain in monoculture (Fig. S3D), its competitive ability under either arabinose condition was not enhanced relative to that of WT PA14 when grown with PAO1 (Fig. 2). Finally, we used a variant of the Pel O/E strain that also overproduced c-di-GMP due to a mutation in the wspF gene (designated Pel O/E ΔwspF); again, this strain, when induced with 0.2% arabinose, did not show enhanced fitness against PAO1 compared to the PA14 parent strain (Fig. 2) despite enhanced biomass in monoculture (Fig. S3D). Taken together, these data show that Psl production is a key factor allowing PAO1 to outcompete PA14 and that enhancing PA14 biofilm production via several different strategies does not mitigate PA14’s disadvantage against Psl-producing PAO1 during biofilm formation.
PAO1 outcompetes PA14 during dispersal from one patch to another.
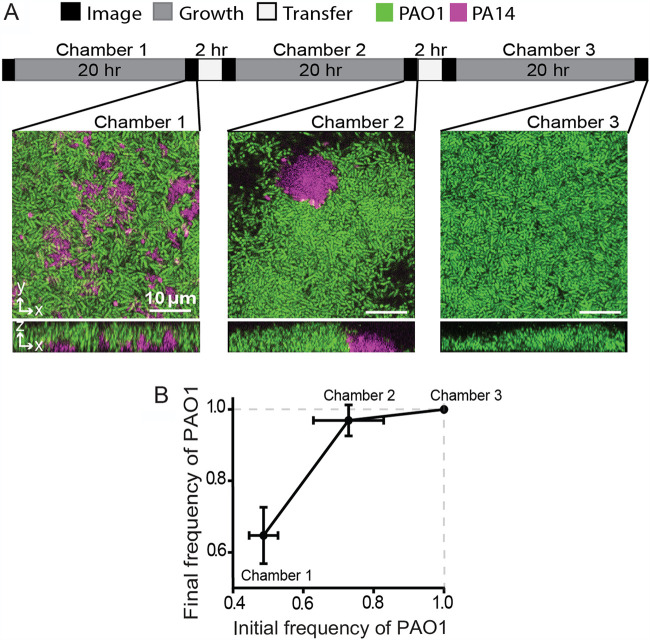
Competition for access to space and nutrients in one location is important for evolutionary fitness, as is the ability, when required, to disperse to new locations for future growth (61). Bacteria that commit strongly to biofilm production in a given location may experience a trade-off in their ability to disperse to new locations (45, 62), so we explored whether introduction of a simulated dispersal regime influences the outcome of competition between PA14 and PAO1. We grew a 1:1 mixture of PA14 and PAO1 in microfluidic chambers under flow for 20 h, after which we introduced a dispersal stage. For each such event, the outflow tube from the first microfluidic chamber was attached to a second, clean chamber, and the biofilm effluent was used to seed the downstream chamber for 2 h. The goal was to imitate the natural transition of P. aeruginosa from an existing biofilm to a new environment with an intervening planktonic phase (Fig. 3A, top).
FIG 3.
PAO1 outcompetes PA14 in a dispersal regime. (A) Graphical summary of dispersal experiment regime (top) and representative images of biofilms in serially inoculated chambers (bottom). (B) The frequency of PAO1 in dispersal experiments through serially inoculated chambers with intervening 20-h incubations. All error bars denote standard deviations (n = 9 nonoverlapping image stacks from 4 separate microfluidic chambers).
In biofilms coinoculated 1:1 with PAO1 and PA14, PAO1 accounted for ∼70% of the population at the end of the 20-h incubation (Fig. 3A and B), consistent with experiments in the sections described above. Following the first transfer to a new chamber and 20 h of incubation, PAO1 rose in relative abundance to an average of 97% of the total population. After the second transfer and 20 h of incubation, PAO1 had reached fixation; that is, it comprised 100% of the population in all sample image stacks (Fig. 3A and B). These data indicate that PAO1 suffers no disadvantage during dispersal steps and, as predicted by the population dynamics data in Fig. 1H, drives the PA14 strain entirely out of coinoculated biofilms after successive rounds of biofilm competition punctuated by dispersal events. The results reflect the net outcome of competition during surface colonization, subsequent biofilm growth, dispersal into the effluent liquid phase, recolonization of downstream sites, and new rounds of biofilm growth.
It is possible that in the experiments described above, PA14 dispersal out of biofilms incubated for 20 h was blocked due to PAO1 overgrowing and locking PA14 into place in the lower layers of the cocultured biofilms. We assessed this possibility by repeating the serial chamber inoculation experiment described above, but we allowed for only 3 h of incubation before each dispersal event. We chose 3-h incubations because PAO1 could not overgrow PA14 in this short of an interval. As there were relatively few cells departing the chambers after 3 h of coculture incubation, too few cells were found on the surface of downstream chambers to reliably quantify, so we instead calculated the strain frequencies observed in chambers after 3 h of incubation and then inoculated new chambers with the same strain frequencies but at ∼500-fold higher density. With this experimental regimen, PAO1 was again found to increase in frequency from one chamber to the next but had not yet reached 100% of the population by the end of the experiment (Fig. S4). Therefore, shortening the biofilm incubation periods between chamber transfers did not alter the qualitative result that PAO1 consistently displaces PA14, but it did slow the speed at which PAO1 does so.
Differential response of PAO1 and PA14 in biofilms subject to starvation.
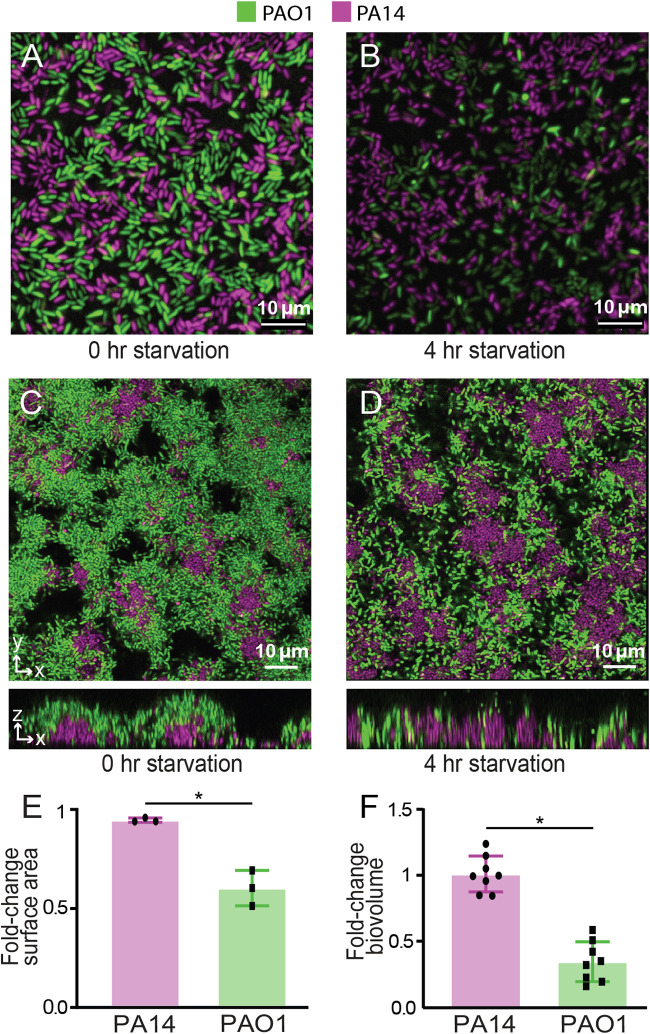
The experiments described in the previous section suggest that PAO1 exceeds PA14 in its dispersal during simulated disturbance events in which the cells leaving an existing biofilm under normal flow conditions must colonize a new location. Disturbance events can be more concrete, for example, when nutrient supplies are suddenly cut off at a given location due to depletion or change in flow conditions. Indeed, nutrient supply in many natural environments may more often occur in transient bursts rather than continuously. Considering these factors, we explored how PAO1 and PA14 in coculture biofilms would react to carbon starvation. We grew dual-strain biofilms for 12 h and then changed the influent flow to a carbon-free (but otherwise equivalent) biofilm medium for 4 h. Following the switch to nutrient-depleted medium flow, PA14’s surface coverage reduced by 5%, while that of PAO1 decreased by 40% (Fig. 4A, B, and E). To see if this result would hold for larger biofilms incubated for longer periods prior to starvation, we repeated this experiment after allowing the coinoculated biofilms to grow for 24 h. Once again the total amount of PA14 remained nearly unchanged following starvation, while that of PAO1 decreased in this case by nearly 60% (Fig. 4C, D, and F).
FIG 4.
Responses of biofilm-dwelling PAO1 and PA14 to nutrient depletion. Coculture biofilms of PAO1 and PA14 were grown for 12 h or 24 h and deprived of their carbon source (arginine) for 4 h. The surface area occupied by PAO1 and PA14 in 12-h-old biofilms (coinoculated at 1:1) was measured before (A) and after (B) carbon deprivation. (E) Fold changes in surface area occupied by each strain are shown. The total biovolumes of PAO1 and PA14 in 24-h-old biofilms (coinoculated at 1:1) were assessed before (C) and after (D) carbon deprivation. (F) Fold changes in the biovolume of each strain are shown. Error bars denote standard deviations (n = 3 for panel E; n = 8 nonoverlapping image stacks from 4 separate microfluidic chambers for panel F). Error bars denote the standard deviations. Wilcoxon signed-rank tests with Bonferroni correction were used for pairwise comparisons; *, P < 0.05.
To clarify whether cell death was occurring in biofilms following carbon depletion, we repeated these experiments once again, growing 1:1 coculture biofilms of PA14 and PAO1 for 24 h prior to starvation; in this case, the starvation media lacking carbon also contained propidium iodide (PI), whose diffusion into cells is an indicator of compromised membrane integrity and cell death. We found no PI staining until 3 h after the switch to carbon-free media, with somewhat more cell death in PA14 than PAO1 (Fig. S5A). A similar result was reflected in the cells collected from the liquid effluent of the chambers for the full duration of the starvation treatment (Fig. S5B).
These data and those of the previous section suggest that PAO1 is more inclined to disperse from existing cell monolayers and from biofilms several cell layers in depth, during both continual influx of carbon-replete media and following a sudden loss of a carbon source. PA14 is slower to commit to biofilm production but also more inclined to stay in place if nutrient supplies are cut off for short periods.
PA14 is more proficient than PAO1 at invading resident biofilms.
The data described above suggest that PAO1 is proficient at outcompeting PA14 during early surface colonization and subsequent biofilm growth as well as during successive rounds of serially connected new locations to inhabit. Following nutrient deprivation, PA14 mostly retains its occupied surface, while PAO1 is more inclined to disperse. So far, all experimental regimes in the preceding sections have challenged the two strains to colonize and compete in previously unoccupied locations, but in more natural contexts one might imagine that colonizing bacteria instead encounter surfaces that are already occupied by other strains and species. Our next experiments were designed to assess the relative abilities of PA14 and PAO1 to invade preexisting biofilms of the other strain.
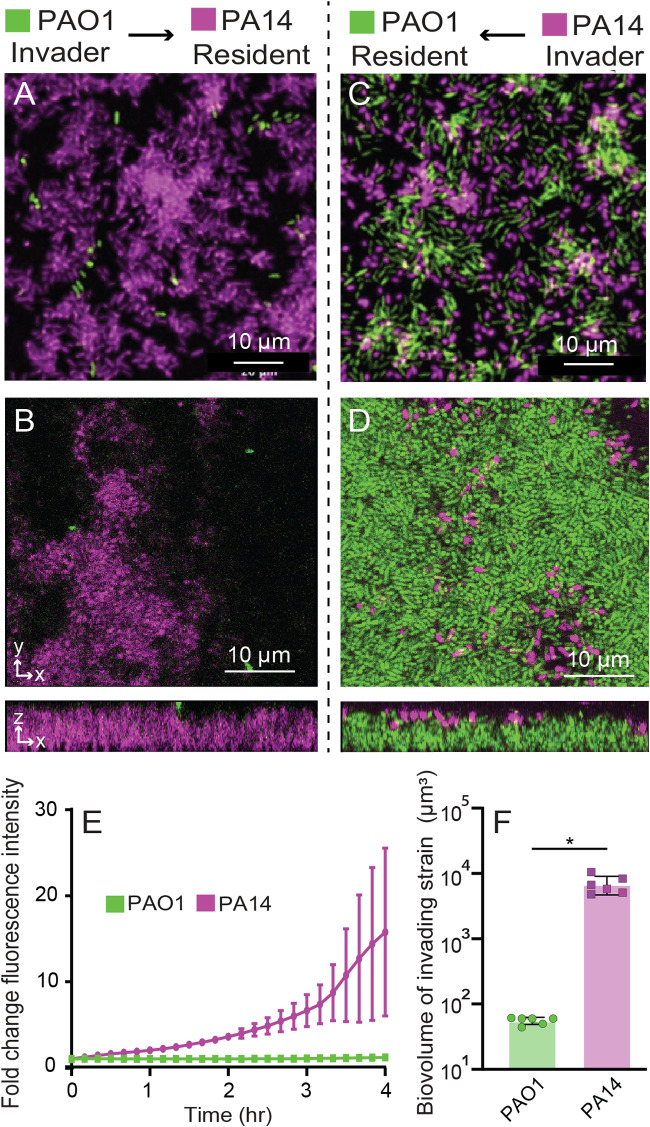
Here, we grew a biofilm of one of the strains in monoculture, which we refer to as the resident strain. After 12 h of growth of the resident strain, we introduced the second strain (the invader) for 4 h to assess its ability to colonize and integrate into the resident biofilm. By visual inspection alone it was evident that PAO1 showed minimal invasion into resident PA14 biofilms (Fig. 5A and B), while PA14 was considerably better able to invade preformed biofilms of PAO1 (Fig. 5C and D). To quantify invasion efficiency by PAO1 and PA14, we measured the fold change in total fluorescence of the invading strain over the 4-h invasion assay. PA14 invades resident PAO1 biofilms rapidly, whereas PAO1 hardly invades resident PA14 biofilms at all (Fig. 5E). In separate experiments that repeated this invasion protocol using resident biofilms that had been incubated for 24 h, we also measured the total biovolume of the invading strain after 4 h. By the end of the assay, PA14 invading biovolume was ∼100-fold higher than that of PAO1 invading a resident PA14 biofilm, as measured by confocal microscopy (Fig. 5F).
FIG 5.
Reciprocal invasion dynamics of resident biofilms. Resident biofilms of PA14 were grown for 12 h and invaded by PAO1 for 4 h (A, widefield fluorescence image; B, confocal optical section and z-projection). Resident biofilms of PAO1 were grown for 12 h (A, C, and E) or 24 h (B, D, and F) and invaded by PA14 for 4 h (C, widefield fluorescence image; D, confocal optical section and z-projection). (E) The invasion efficiency of PA14 (purple) and PAO1 (green) was measured over time by normalizing the change in fluorescence intensity from the start of the assay through 4 h of invasion. n = 3, P < 0.05 after 3 h by 2-way ANOVA with Sidak posttest. (F) The total invading strain biovolume of PAO1 and PA14 at the end of the invasion assay (n = 6 nonoverlapping image stacks from 4 separate microfluidic chambers). Error bars denote the standard deviations. P < 0.05 by Wilcoxon signed-rank test.
DISCUSSION
Biofilms are spatially and physiologically heterogenous environments in which bacteria both cooperate and compete intensely with each other for access to space and resources. Biofilm formation itself is increasingly conceptualized as a core response to ecological competition (19, 63, 64), and here we investigated reference strains P. aeruginosa PAO1 and PA14 for their ability to compete with each other in the context of biofilm growth. Long studied as models of biofilm formation, these two strains of P. aeruginosa vary substantially in their regulatory mechanisms of surface attachment and compositions of secreted extracellular matrix (16, 25, 42, 44, 65). We show that under microfluidic biofilm culture conditions, PAO1 quickly outcompetes and overgrows PA14 in a density- and frequency-independent manner. PAO1 and PA14 were also found to spatially segregate over 24 h, with PA14 mostly limited to the substratum and PAO1 overgrowing PA14.
That PAO1 begins to outnumber PA14 in the early (2 to 7 h) stages of surface occupation is most likely attributable to the difference in their patterns of surface attachment. Prior work has intimated that PAO1 commits quickly to surface adhesion and extracellular matrix secretion relative to PA14 (25, 44). Following surface colonization, subsequent differences between PAO1 and PA14 in their extracellular matrix begin to contribute to their population dynamics. This interpretation is reinforced by the fact that the competitive advantage of PAO1 is specific to the biofilm mode of growth, during which PAO1 produces matrix containing Pel and Psl, while PA14 only produces Pel. Psl production by PAO1 was essential for its advantage against PA14 under flow and was the dominant controlling factor (of those we examined here) for the outcome of competition; no other physiological manipulation that we tested could alter the outcome of competition between the two.
Others have highlighted Psl production as fundamental to competition in biofilm environments for P. aeruginosa, including when isogenic strains are competed against each other (54) and when PAO1 cohabits biofilms with other species, such as P. protegens and Klebsiella pneumoniae (65). Extensive work has explored the relative roles of the Pel and Psl polysaccharides of the P. aeruginosa extracellular matrix (42, 66), indicating that they offer some structural redundancy to one another in strains that can produce both, such as PAO1. There is also evidence that Pel and Psl make different contributions to the viscoelastic properties and spatial structure of Pseudomonas biofilms. Of particular note, Chew et al. (16) found that Psl is important for extracellular matrix cross-linking and overall matrix elasticity, strengthening biofilms and promoting microcolony formation extending from basal substrata. Pel, on the other hand, tends to increase biofilm viscosity, encourage streamer formation (67, 68), and facilitate lateral spreading (16). Paralleling the observations we made here, Chew et al. also found that Psl-producing PAO1 formed mixed-species biofilms in which it grew over the top of Staphylococcus aureus, while Psl-deficient PAO1 was less able to overgrow S. aureus in coculture (16).
An additional important element of fitness for any biofilm-producing microbe is balancing investment into local competition versus dispersal (69). Previous work has linked relative investment in matrix production to trade-offs in this balance (62). Here, we explored how PAO1 and PA14 disperse in two different ways, measuring the propensity of each to leave biofilms passively during normal flow of nutrient-replete medium and the propensity of each strain to disperse during a starvation event. We found that PAO1 was more prone to dispersal under both conditions. In serially colonized chambers during flow of nutrient-replete medium, PAO1 ultimately displaced the PA14 strain. Under starvation conditions in which we only studied single chambers subjected to nutrient deprivation, we saw that PA14 was far more strongly inclined to stay in place. Thus, PA14 may be more suited to retaining a grip on space it has occupied when nutrients run low, even as some of its population dies under starvation, while PAO1 is more inclined overall to disperse. The relative advantage of staying in place under starvation conditions depends on the prevailing environmental conditions: if local nutrient supply fluctuates, then remaining in place may be the better strategy, but if nutrient supply does not return once depleted, then dispersal will be optimal.
Our final series of experiments showed that PA14 is markedly better able to colonize a previously established biofilm of PAO1 than vice versa, by 2 orders of magnitude, over a relatively short colonization and growth time scale of 4 h. This suggests that PA14 is better suited to exploiting previously colonized environments, while PAO1 is superior in dispersing to and then competing for space and resources on unoccupied surfaces. Recent genomic analysis has found that strains similar to PA14 predominate among CF-derived isolates, while the PAO1-like strains are more likely to be encountered among environmental isolates (70) (noting, though, that the original PA14 and PAO1 isolates both came from wounds). We speculate on the basis of our results that PA14 has evolved a surface occupation strategy best suited to taking advantage of previously colonized surfaces and commitment to staying in place under fluctuating nutrient conditions, while PAO1 is better suited to rapid exploitation of unoccupied surfaces followed by rapid dispersal under nutrient limitation to find new locations for future growth. Due to the enormous diversity of environments in which bacterial biofilms grow, we imagine these are just two among many possible strategies that evolved to optimize surface occupation among Pseudomonas and other bacterial species.
MATERIALS AND METHODS
Strains and media.
Strains of P. aeruginosa are all derivatives of PAO1 or PA14 and were constructed by standard allelic exchange. The biofilm growth substrate was buffered minimal medium containing 1.0 mM K2HPO4, 0.6 mM MgSO4, and 0.4% arginine. Starvation medium omitted arginine and, for the cell death assays, contained propidium iodide at 2 μg/ml.
Microfluidic device assembly.
The microfluidic devices were made by bonding polydimethylsiloxane (PDMS) chamber molds to size number 1.5 cover glass slips (60 mm by 36 mm [length by width]; Thermo-Fisher, Waltham, MA) using standard soft lithography techniques (71). Each PDMS mold contained 4 chambers, each of which measured 3,000 μm by 500 μm by 75 μm (length by width by depth). To establish flow in these chambers, medium was loaded into 1-ml BD plastic syringes with 25-gauge needles. These syringes were joined to number 30 Cole-Parmer PTFE tubing (inner diameter, 0.3 mm), which was connected to prebored holes in the microfluidic device. Tubing was also placed on the opposite end of the chamber to direct the effluent to a waste container. Syringes were mounted to syringe pumps (Pico plus elite; Harvard Apparatus), and flow was maintained at 0.5 μl/min for all experiments. Note that while this flow rate is 100-fold lower than that used for some other biofilm growth protocols with P. aeruginosa (25), it results in similar average flow velocities through our PDMS microfluidic chambers, which are likewise ∼100-fold smaller in cross-sectional area than commercial flow devices like those used in Lee et al. (44).
Biofilm growth.
Overnight cultures of P. aeruginosa PAO1 and PA14 were grown at 37°C with shaking in lysogeny broth (LB) prior to the start of biofilm experiments. Cultures were normalized to an optical density at 600 nm (OD600) of 1.0 in KA biofilm medium containing 50 mM Tris-HCl (pH 7.4), 0.6 mM MgSO4, 1.0 K2HPO4, and 0.4% arginine (72). If coculture biofilms were being grown, equal volumes of cultures adjusted to an OD600 of 1.0 were mixed and used as the inoculum for the microfluidic chamber (completely filling its inner volume), and then the bacteria were allowed to rest for 1 h at room temperature to permit cells to attach to the glass surface. For the experiments with varied initial frequencies, after cultures of each strain were adjusted to an OD600 of 1.0, different ratios of the two cultures were added to obtain the desired frequency prior to inoculation. After resting for 1 h to allow surface attachment, the devices were run at 0.5 μl/min at 37°C and imaged by widefield or confocal microscopy (see below) at time intervals that varied per experiment, as noted in Results. At every sampling time point, images were acquired from 3 or more nonoverlapping locations within each biofilm chamber. All experiments were repeated with at least 3 biological replicates with 3 or more technical replicates on different days. Total replicates for each experiment are noted in the figure legends for each data set in the text and supplemental material.
For the serial chamber inoculation experiments, dual-strain biofilms (1:1 initial frequency of PA14 to PAO1) were incubated for 24 h or 3 h (Fig. 3; see also Fig. S4 in the supplemental material), and a 0.5-cm length of tubing was connected to the outlet channel. At every sampling time point, images were acquired from nonoverlapping locations within each biofilm chamber. The outlet chamber was then allowed to seed a new chamber for 2 h, initiating chamber 2. Chamber 2 was then incubated for 20 h, imaged, and used to seed chamber 3 in a similar manner. The same procedure was performed for the version of this experiment in which biofilms grew for only 3 h prior to being used to seed the next downstream chamber, with one exception. Because very few cells exited chambers incubated for only 3 h, there were too few of them found in the connected downstream chambers to quantify. To compensate for this issue, we quantified the relative abundance of PAO1 and PA14 at the end of chamber 1 after 3 h of incubation and then inoculated a new chamber with the same strain relative abundances, except with about a 500-fold increased density, to allow sufficient biomass to accumulate to determine the relative frequency of each strain at the beginning of biofilm growth in the downstream chamber.
For invasion experiments, we grew the resident biofilm for 12 h at a medium flow rate of 0.5 μl per/min at 37°C, after which we introduced the invading strain (adjusted to an OD600 of 1.0) at the same flow rate for 4 h by switching the chamber inlets to new tubing connected to new syringes containing the invading strain. At every sampling time point, at least 3 images were acquired from nonoverlapping locations within each biofilm chamber. For the version of this experiment for which we used confocal microscopy, we grew resident biofilms for 24 h prior to invasion to ensure that the resident biofilms completely covered the glass substratum before the invading strain was introduced to the chambers.
Microscopy and image analysis.
Biofilms in the microfluidic chambers were imaged using a Zeiss LSM 880 microscope with a 40×/1.2-numeric-aperture (NA) or 10×/0.4 NA water objective (confocal imaging) or a Nikon Eclipse Ti inverted microscope with a Plan Apochromat 100× DM oil objective (widefield imaging). For confocal imaging, a 543-nm laser line was used to excite mKO-κ, and a 488-nm laser line was used to excite GFP; for widefield imaging, mKO-κ and GFP were imaged using standard mCherry and fluorescein isothiocyanate filter sets. All quantitative analysis of confocal microscopy data was performed using BiofilmQ (73). Confocal two-dimensional (2D) sections and Z-projections were generated using Zeiss Zen software, and 3D renderings of biofilms in Fig. 1 were generated using Paraview. Widefield images were generated using the native Nikon Elements software and analyzed using ImageJ.
Starvation assay.
For the biofilm starvation studies, dual-strain biofilms (1:1 initial frequency of PA14 to PAO1) were incubated for 12 h or 24 h, after which inflow ports were reconnected to new tubing connected to new syringes containing medium identical to the original incubation medium but without the carbon source (arginine). For the version of this experiment with 12-h biofilms, images were taken by widefield microscopy every 10 min for 4 h. For the version of this experiment with 24-h biofilms, images were taken immediately after the start of starvation (0 h) and then once more at the end of the starvation period (4 h). In another version of this experiment for Fig. S5 with 24-h biofilms inoculated with 1:1 PAO1-PA14, propidium iodide was added to the starvation medium (2 μg/ml) that lacked arginine, and images were taken every 1 h for 4 h to monitor propidium iodide uptake by PAO1 and PA14 as a proxy for cell death. Effluent during the course of the 4-h starvation was collected into a 1.5-ml Eppendorf tube on ice (to stop any further cell division). Cells in the effluent were then collected and placed under agar pads for imaging to determine the fraction of dead cells that had dispersed from the biofilm during the starvation treatment.
Statistics.
All statistical analyses were performed in GraphPad Prism. Pairwise comparisons were performed using Wilcoxon signed-rank tests with Bonferroni correction. The comparisons in Fig. 1C and 5E were performed by 2-way analysis of variance (ANOVA) with a Sidak posttest. All error bars indicate standard deviations unless otherwise noted.
ACKNOWLEDGMENTS
We note the collaborative effort of both first authors for the collection of data for this project. We are grateful to members of the Nadell and O’Toole labs for comments on the project as well as three anonymous referees for comments on earlier drafts of the manuscript.
This work was supported by funding from the NIH (R37-AI83256-06) to G.A.O. C.D.N. was supported by the Cystic Fibrosis Foundation (STANTO19RO), NSF (MCB-1817342), and NIH (P30-DK117469).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Jahid IK, Ha SD. 2012. A review of microbial biofilms of produce: future challenge to food safety. Food Sci Biotechnol 21:299–316. 10.1007/s10068-012-0041-1. [DOI] [Google Scholar]
- 2.Donlan RM. 2001. Biofilms and device-associated infections. Emerg Infect Dis 7:277–281. 10.3201/eid0702.010226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Acker H, Van Dijck P, Coenye T. 2014. Molecular mechanisms of antimicrobial tolerance and resistance in bacterial and fungal biofilms. Trends Microbiol 22:326–333. 10.1016/j.tim.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Matz C, McDougald D, Moreno AM, Yung PY, Yildiz FH, Kjelleberg S. 2005. Biofilm formation and phenotypic variation enhance predation-driven persistence of Vibrio cholerae. Proc Natl Acad Sci USA 102:16819–16824. 10.1073/pnas.0505350102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gambino M, Cappitelli F. 2016. Mini-review: biofilm responses to oxidative stress. Biofouling 32:167–178. 10.1080/08927014.2015.1134515. [DOI] [PubMed] [Google Scholar]
- 6.Vidakovic L, Singh PK, Hartmann R, Nadell CD, Drescher K. 2018. Dynamic biofilm architecture confers individual and collective mechanisms of viral protection. Nat Microbiol 3:26–31. 10.1038/s41564-017-0050-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wucher BR, Elsayed M, Adelman JS, Kadouri DE, Nadell CD. 2021. Bacterial predation transforms the landscape and community assembly of biofilms. Curr Biol 31:2643–2651. 10.1016/j.cub.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valentini M, Filloux A. 2016. Biofilms and Cyclic di-GMP (c-di-GMP) signaling: lessons from Pseudomonas aeruginosa and other bacteria. J Biol Chem 291:12547–12555. 10.1074/jbc.R115.711507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fong JCN, Yildiz FH. 2008. Interplay between cyclic AMP-cyclic AMP receptor protein and cyclic di-GMP signaling in Vibrio cholerae biofilm formation. J Bacteriol 190:6646–6659. 10.1128/JB.00466-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Passos da Silva D, Schofield MC, Parsek MR, Tseng BS. 2017. An update on the sociomicrobiology of quorum sensing in Gram-negative biofilm development. Pathogens 6:1–9. 10.3390/pathogens6040051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toutain CM, Caizza NC, Zegans ME, O'Toole GA. 2007. Roles for flagellar stators in biofilm formation by Pseudomonas aeruginosa. Res Microbiol 158:471–477. 10.1016/j.resmic.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 12.O'Toole GA, Kolter R. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30:295–304. 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 13.Zhao K, Tseng BS, Beckerman B, Jin F, Gibiansky ML, Harrison JJ, Luijten E, Parsek MR, Wong GCL. 2013. Psl trails guide exploration and microcolony formation in Pseudomonas aeruginosa biofilms. Nature 497:388–391. 10.1038/nature12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colvin KM, Gordon VD, Murakami K, Borlee BR, Wozniak DJ, Wong GCL, Parsek MR. 2011. The pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog 7:e1001264. 10.1371/journal.ppat.1001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghafoor A, Hay ID, Rehm BHA. 2011. Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture. Appl Environ Microbiol 77:5238–5246. 10.1128/AEM.00637-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chew SC, Kundukad B, Seviour T, Van der Maarel JRC, Yang L, Rice SA, Doyle P, Kjelleberg S. 2014. Dynamic remodeling of microbial biofilms by functionally distinct exopolysaccharides. mBio 5:e01536-11. 10.1128/mBio.01536-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartmann R, Singh PK, Pearce P, Mok R, Song B, Díaz-Pascual F, Dunkel J, Drescher K. 2019. Emergence of three-dimensional order and structure in growing biofilms. Nat Phys 15:251–256. 10.1038/s41567-018-0356-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cont A, Rossy T, Al-Mayyah Z, Persat A. 2020. Biofilms deform soft surfaces and disrupt epithelia. Elife 9:1–22. 10.7554/eLife.56533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nadell CD, Drescher K, Foster KR. 2016. Spatial structure, cooperation and competition in biofilms. Nat Rev Microbiol 14:589–600. 10.1038/nrmicro.2016.84. [DOI] [PubMed] [Google Scholar]
- 20.Pellett S, Bigley DV, Grimes DJ. 1983. Distribution of Pseudomonas aeruginosa in a riverine ecosystem. Appl Environ Microbiol 45:328–332. 10.1128/aem.45.1.328-332.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Green SK, Schroth MN, Cho JJ, Kominos SD, Vitanza-Jack VB. 1974. Agricultural Plants and Soil as a Reservoir for Pseudomonas aeruginosa. Appl Microbiol 28:987–991. 10.1128/am.28.6.987-991.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Romling U, Wingender J, Muller H, Tummler B. 1994. A major Pseudomonas aeruginosa clone common to patients and aquatic habitats. Appl Environ Microbiol 60:1734–1738. 10.1128/aem.60.6.1734-1738.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moreau-Marquis S, Stanton BA, O'Toole GA. 2008. Pseudomonas aeruginosa biofilm formation in the cystic fibrosis airway. Pulm Pharmacol Ther 21:595–599. 10.1016/j.pupt.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Armbruster CR, Parsek MR. 2018. New insight into the early stages of biofilm formation. Proc Natl Acad Sci USA 115:4317–4319. 10.1073/pnas.1804084115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pa P, Parsek MR, Toole GAO, Golestanian R, Wong CL. 2020. Social cooperativity of bacteria during reversible surface attachment in young biofilms: a quantitative comparison of Pseudomonas aeruginosa PA14 and PAO1. mBio 11:e02644-19. 10.1128/mBio.02644-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7:263–273. 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 27.Jones CJ, Utada A, Davis KR, Thongsomboon W, Zamorano Sanchez D, Banakar V, Cegelski L, Wong GCL, Yildiz FH. 2015. C-di-GMP regulates motile to sessile transition by modulating MshA pili biogenesis and near-surface motility behavior in Vibrio cholerae. PLoS Pathog 11:e1005068. 10.1371/journal.ppat.1005068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Güvener ZT, Harwood CS. 2007. Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol Microbiol 66:1459–1473. 10.1111/j.1365-2958.2007.06008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDonough KA, Rodriguez A. 2011. The myriad roles of cyclic AMP in microbial pathogens: from signal to sword. Nat Rev Microbiol 10:27–38. 10.1038/nrmicro2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo Y, Zhao K, Baker AE, Kuchma SL, Coggan KA, Wolfgang MC, Wong GCL, O’Toole GA. 2015. A hierarchical cascade of second messengers regulates Pseudomonas aeruginosa Surface Behaviors. mBio 6:e02456-14. 10.1128/mBio.02456-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Connor JR, Kuwada NJ, Huangyutitham V, Wiggins PA, Harwood CS. 2012. Surface sensing and lateral subcellular localization of WspA, the receptor in a chemosensory-like system leading to c-di-GMP production. Mol Microbiol 86:720–729. 10.1111/mmi.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hickman JW, Tifrea DF, Harwood CS. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci USA 102:14422–14427. 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Treuner-Lange A, Chang YW, Glatter T, Herfurth M, Lindow S, Chreifi G, Jensen GJ, Søgaard-Andersen L. 2020. PilY1 and minor pilins form a complex priming the type IVa pilus in Myxococcus xanthus. Nat Commun 11:5054. 10.1038/s41467-020-18803-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuchma SL, Ballok AE, Merritt JH, Hammond JH, Lu W, Rabinowitz JD, O'Toole GA. 2010. Cyclic-di-GMP-mediated repression of swarming motility by Pseudomonas aeruginosa: the pilY1 gene and its impact on surface-associated behaviors. J Bacteriol 192:2950–2964. 10.1128/JB.01642-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Webster SS, Lee CK, Schmidt WC, Wong GCL. 2020. Fine tuning cyclic-di-GMP signaling in Pseudomonas aeruginosa using the type 4 pili alignment complex. bioRxiv 10.1101/2020.10.17.343988. [DOI]
- 36.Persat A, Inclan YF, Engel JN, Stone HA, Gitai Z. 2015. Type IV pili mechanochemically regulate virulence factors in Pseudomonas aeruginosa. Proc Natl Acad Sci USA 112:7563–7568. 10.1073/pnas.1502025112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holloway BW. 1955. Genetic recombination in Pseudomonas aeruginosa. J Gen Microbiol 13:572–581. 10.1099/00221287-13-3-572. [DOI] [PubMed] [Google Scholar]
- 38.Holloway BW, Morgan AF. 1986. Genome organization in Pseudomonas. Annu Rev Microbiol 40:79–105. 10.1146/annurev.mi.40.100186.000455. [DOI] [PubMed] [Google Scholar]
- 39.Freschi L, Vincent AT, Jeukens J, Emond-Rheault JG, Kukavica-Ibrulj I, Dupont MJ, Charette SJ, Boyle B, Levesque RC. 2019. The Pseudomonas aeruginosa pan-genome provides new insights on its population structure, horizontal gene transfer, and pathogenicity. Genome Biol Evol 11:109–120. 10.1093/gbe/evy259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryder C, Byrd M, Wozniak DJ. 2007. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr Opin Microbiol 10:644–648. 10.1016/j.mib.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wozniak DJ, Wyckoff TJO, Starkey M, Keyser R, Azadi P, O'Toole GA, Parsek MR. 2003. Alginate is not a significant component of the extracellular polysaccharide matrix of PA14 and PAO1 Pseudomonas aeruginosa biofilms. Proc Natl Acad Sci USA 100:7907–7912. 10.1073/pnas.1231792100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Colvin KM, Irie Y, Tart CS, Urbano R, Whitney JC, Ryder C, Howell PL, Wozniak DJ, Parsek MR. 2012. The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ Microbiol 14:1913–1928. 10.1111/j.1462-2920.2011.02657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Armbruster CR, Lee CK, Parker-Gilham J, de Anda J, Xia A, Tseng BS, Hoffman LR, Jin F, Harwood CS, Wong GCL, Parsek MR. 2019. Heterogeneity in surface sensing produces a division of labor in Pseudomonas aeruginosa populations. Elife 8:1–29. 10.7554/eLife.45084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee CK, De Anda J, Baker AE, Bennett RR, Luo Y, Lee EY, Keefe JA, Helali JS, Ma J, Zhao K, Golestanian R, O’Toole GA, Wong GCL. 2018. Multigenerational memory and adaptive adhesion in early bacterial biofilm communities. Proc Natl Acad Sci USA 115:4471–4476. 10.1073/pnas.1720071115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yan J, Nadell CD, Bassler BL. 2017. Environmental fluctuation governs selection for plasticity in biofilm production. ISME J 11:1569–1577. 10.1038/ismej.2017.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yawata Y, Nguyen J, Stocker R, Rusconi R. 2016. Microfluidic studies of biofilm formation in dynamic environments. J Bacteriol 198:2589–2595. 10.1128/JB.00118-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grinberg M, Orevi T, Kashtan N. 2019. Bacterial surface colonization, preferential attachment and fitness under periodic stress. PLoS Comput Biol 15:e1006815. 10.1371/journal.pcbi.1006815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Choi KH, Schweizer HP. 2006. mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat Protoc 1:153–161. 10.1038/nprot.2006.24. [DOI] [PubMed] [Google Scholar]
- 49.Parsek MR, Tolker-Nielsen T. 2008. Pattern formation in Pseudomonas aeruginosa biofilms. Curr Opin Microbiol 11:560–566. 10.1016/j.mib.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 50.Teal TK, Lies DP, Wold BJ, Newman DK. 2006. Spatiometabolic stratification of Shewanella oneidensis biofilms. Appl Environ Microbiol 72:7324–7330. 10.1128/AEM.01163-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burmølle M, Ren D, Bjarnsholt T, Sørensen SJ. 2014. Interactions in multispecies biofilms: do they actually matter? Trends Microbiol 22:84–91. 10.1016/j.tim.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 52.Liu W, Jacquiod S, Brejnrod A, Russel J, Burmølle M, Sørensen SJ. 2019. Deciphering links between bacterial interactions and spatial organization in multispecies biofilms. ISME J 13:3054–3066. 10.1038/s41396-019-0494-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mitri S, Xavier JB, Foster KR. 2011. Social evolution in multispecies biofilms. Proc Natl Acad Sci USA 108(Suppl 2):10839–10846. 10.1073/pnas.1100292108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Irie Y, Roberts AEL, Kragh KN, Gordon VD, Hutchison J, Allen RJ, Melaugh G, Bjarnsholt T, West SA, Diggle SP. 2017. The Pseudomonas aeruginosa PSL polysaccharide is a social but noncheatable trait in biofilms. mBio 8:e00374-17. 10.1128/mBio.00374-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Franklin MJ, Nivens DE, Weadge JT, Lynne Howell P. 2011. Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, alginate, Pel, and Psl. Front Microbiol 2:167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goldberg JB, Gorman WL, Flynn JL, Ohman DE. 1993. A mutation in algN permits trans activation of alginate production by algT in Pseudomonas species. J Bacteriol 175:1303–1308. 10.1128/jb.175.5.1303-1308.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sabra W, Kim EJ, Zeng AP. 2002. Physiological responses of Pseudomonas aeruginosa PAO1 to oxidative stress in controlled microaerobic and aerobic cultures. Microbiology (Reading) 148:3195–3202. 10.1099/00221287-148-10-3195. [DOI] [PubMed] [Google Scholar]
- 58.Friedman L, Kolter R. 2004. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J Bacteriol 186:4457–4465. 10.1128/JB.186.14.4457-4465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Friedman L, Kolter R. 2004. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol Microbiol 51:675–690. 10.1046/j.1365-2958.2003.03877.x. [DOI] [PubMed] [Google Scholar]
- 60.Kuchma SL, Brothers KM, Merritt JH, Liberati NT, Ausubel FM, O'Toole GA. 2007. BifA, a cyclic-di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol 189:8165–8178. 10.1128/JB.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McDougald D, Rice SA, Barraud N, Steinberg PD, Kjelleberg S. 2011. Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat Rev Microbiol 10:39–50. 10.1038/nrmicro2695. [DOI] [PubMed] [Google Scholar]
- 62.Nadell CD, Bassler BL. 2011. A fitness trade-off between local competition and dispersal in Vibrio cholerae biofilms. Proc Natl Acad Sci USA 108:14181–14185. 10.1073/pnas.1111147108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oliveira NM, Oliveria NM, Martinez-Garcia E, Xavier J, Durham WM, Kolter R, Kim W, Foster KR. 2015. Biofilm formation as a response to ecological competition. PLoS Biol 13:e1002191. 10.1371/journal.pbio.1002191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. 2010. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol 8:15–25. 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Periasamy S, Nair HAS, Lee KWK, Ong J, Goh JQJ, Kjelleberg S, Rice SA. 2015. Pseudomonas aeruginosa PAO1 exopolysaccharides are important for mixed species biofilm community development and stress tolerance. Front Microbiol 6:851. 10.3389/fmicb.2015.00851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mann EE, Wozniak DJ. 2012. Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol Rev 36:893–916. 10.1111/j.1574-6976.2011.00322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rusconi R, Lecuyer S, Guglielmini L, Stone HA. 2010. Laminar flow around corners triggers the formation of biofilm streamers. J R Soc Interface 7:1293–1299. 10.1098/rsif.2010.0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Drescher K, Shen Y, Bassler BL, Stone HA. 2013. Biofilm streamers cause catastrophic disruption of flow with consequences for environmental and medical systems. Proc Natl Acad Sci USA 110:4345–4350. 10.1073/pnas.1300321110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yawata Y, Cordero OX, Menolascina F, Hehemann J-H, Polz MF, Stocker R. 2014. Competition–dispersal tradeoff ecologically differentiates recently speciated marine bacterioplankton populations. Proc Natl Acad Sci USA 111:5622–5627. 10.1073/pnas.1318943111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ozer EA, Nnah E, DIdelot X, Whitaker RJ, Hauser AR, Ochman H. 2019. The population structure of Pseudomonas aeruginosa Is characterized by genetic isolation of exoU+ and exoS+ lineages. Genome Biol Evol 11:1780–1796. 10.1093/gbe/evz119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ng JMK, Gitlin I, Stroock AD, Whitesides GM. 2002. Review components for integrated poly (dimethylsiloxane) microfluidic systems. Electrophoresis 23:3461–3473. . [DOI] [PubMed] [Google Scholar]
- 72.Collins AJ, Pastora AB, Jarrod Smith T, Dahlstrom K, O’Toole GA. 2020. MapA, a second large RTX adhesin, contributes to biofilm formation by Pseudomonas fluorescens. J Bacteriol 202:e00277-20. 10.1128/JB.00277-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hartmann R, Jeckel H, Jelli E, Singh PK, Vaidya S, Bayer M, Rode DKH, Vidakovic L, Díaz-Pascual F, Fong JCN, Dragoš A, Lamprecht O, Thöming JG, Netter N, Häussler S, Nadell CD, Sourjik V, Kovács ÁT, Yildiz FH, Drescher K. 2021. Quantitative image analysis of microbial communities with BiofilmQ. Nat Microbiol 6:151–156. 10.1038/s41564-020-00817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S5. Download JB.00265-21-s0001.pdf, PDF file, 1.5 MB (1.5MB, pdf)