Abstract
Serotonin (5-HT) is a phylogenetically conserved monoamine neurotransmitter modulating important processes in the brain. To directly visualize the release of 5-HT, we developed a genetically encoded GPCR-Activation-Based 5-HT (GRAB5-HT) sensor with high sensitivity, selectivity, subsecond kinetics, and subcellular resolution. GRAB5-HT detects 5-HT release in multiple physiological and pathological conditions in both flies and mice, and provides new insights into the dynamics and mechanisms of 5-HT signaling.
Introduction
Serotonergic signaling in the brain plays a critical role in a wide range of physiological processes, including mood control, reward processing, and sleep-wake homeostatic regulation1–3. Indeed, drugs targeting central serotonergic activity have been used to treat virtually every psychiatric disorder, with the best example being the use of selective serotonin reuptake inhibitors (SSRIs) for depression4. Yet, despite the importance of 5-HT, our understanding of cell-specific 5-HT signaling during behaviors is very much lacking, in part due to our inability to measure 5-HT in vivo with high sensitivity and spatiotemporal resolution5,6,7. Here, using molecular engineering, we developed a genetically encoded fluorescent sensor for directly measuring extracellular 5-HT.
Results
Development and characterization of the GRAB5-HT1.0 in cultured cells
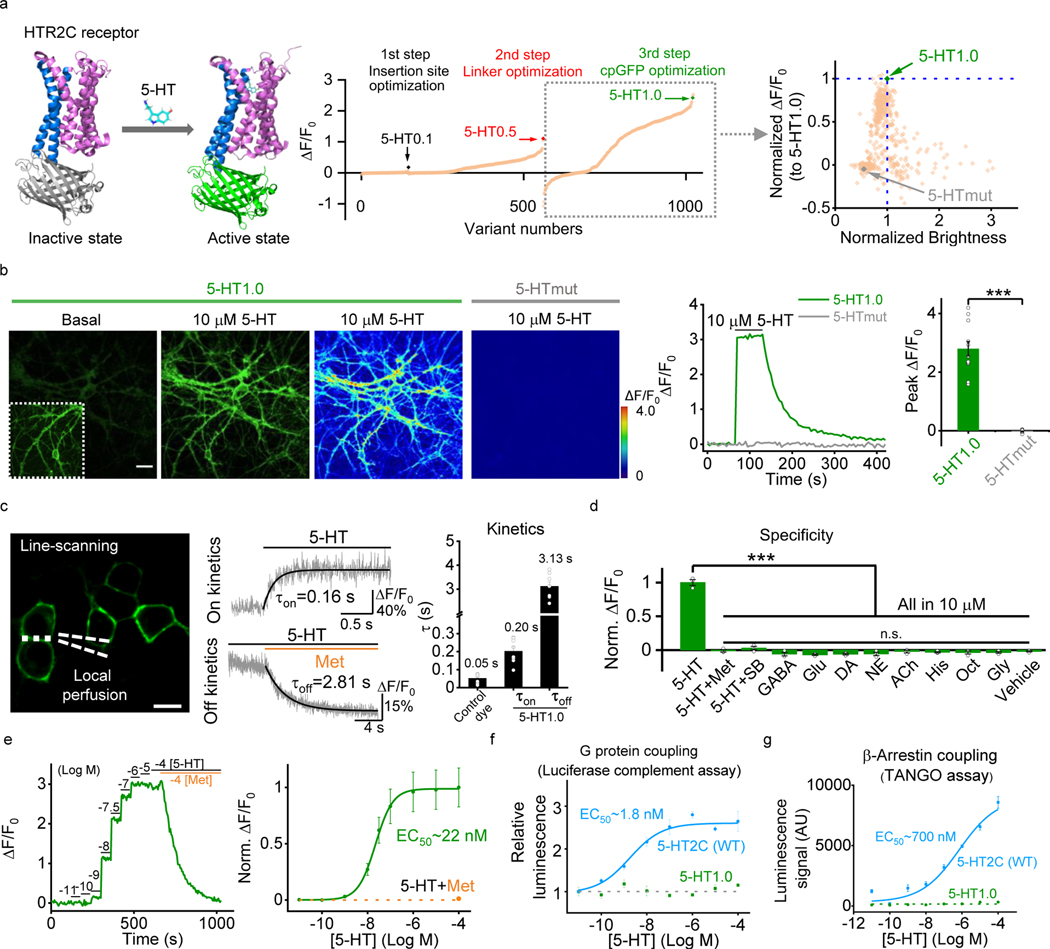
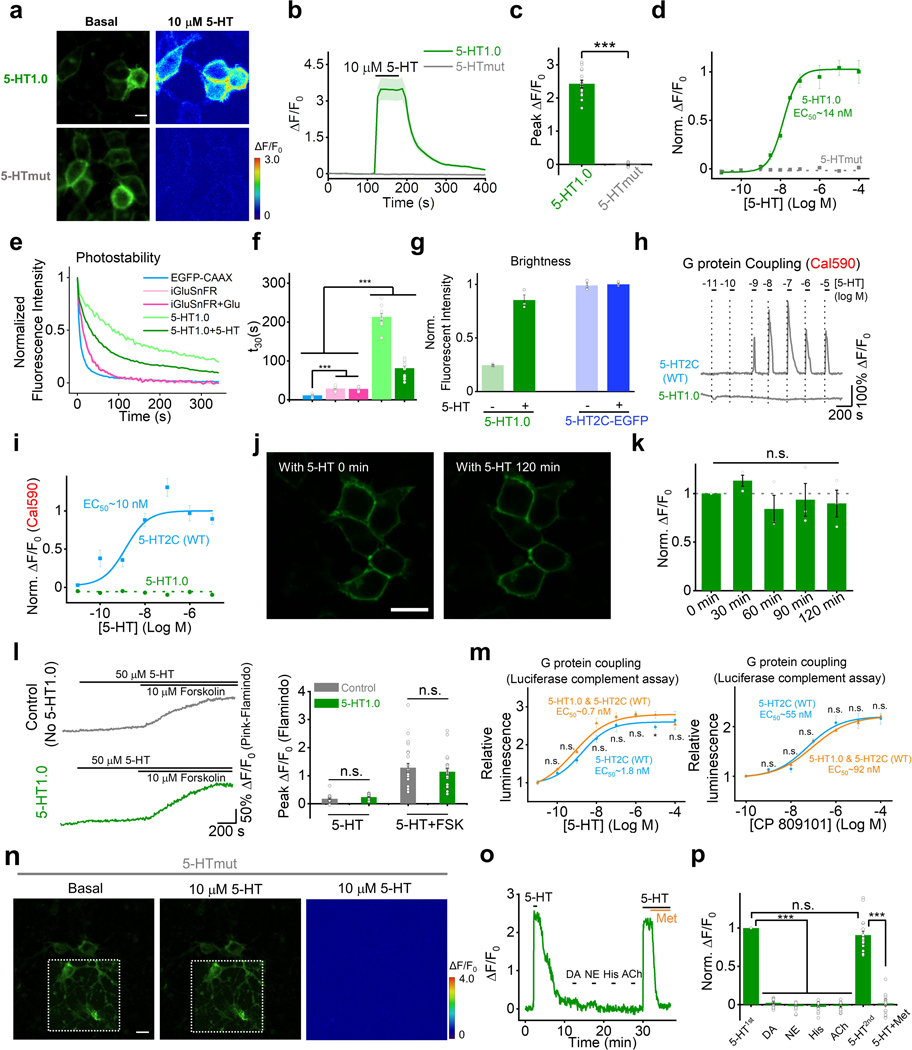
Previously, we and others have independently developed GPCR activation based (GRAB) sensors for detecting different neurotransmitters by converting the conformational change in the respective GPCR to a sensitive fluorescence change in the circular permutated GFP (cpGFP)8, 9,10,11. Using a similar strategy, we initiated the engineering of 5-HT-specific sensors by inserting a cpGFP into the third intracellular loop (ICL3) of various 5-HT receptors. Based on the performance of membrane trafficking and affinity of receptor-cpGFP chimeras, we selected the 5-HT2C receptor-based chimera for further optimization (Extended Data Fig. 1a,b). Our previous experience in optimizing GRAB sensors8, 10, 11 shows that the N- and C-terminus linkers between GPCR and cpGFP are critical for the sensor’s performance. Therefore, we randomly mutated 5-sites in both the N-term and C-term linkers between 5-HT2C receptor and cpGFP to improve the 5-HT sensor’s response. Moreover, we introduced mutations into the cpGFP, focusing on sites that are potentially critical for fast GFP folding and high brightness12, 13 (Extended Data Fig. 2). Mutagenesis and screening in the linker regions and cpGFP moiety resulted in a sensor with a 250% fluorescence change (ΔF/F0) in response to 5-HT in cultured HEK293T cells, which we named GRAB5-HT1.0 (referred to hereafter as simplified 5-HT1.0; Fig. 1a and Extended Data Fig. 3). In addition, we generated a 5-HT-insensitive version of this sensor by introducing D1343.32Q mutation into the receptor14, 15, GRAB5-HTmut (referred to hereafter as 5-HTmut; Fig. 1 and Extended Data Fig. 4). This mutant sensor showed similar membrane trafficking as 5-HT1.0 (Extended Data Fig. 4a and Extended Data Fig. 4n), but <2% ΔF/F0 even in the presence of 100 μM 5-HT (Fig. 1a and Extended Data Fig. 4a–d). In cultured rat cortical neurons, the 5-HT1.0 sensor produced a robust fluorescence increase (280% ΔF/F0) in both the soma and neurites in response to 5-HT bath application, whereas the 5-HTmut sensor had no measurable change in fluorescence (Fig. 1b).
Fig. 1: Design, optimization, and characterization of a novel genetically encoded 5-HT sensor.
a, Left, schematic representation illustrating the principle behind the GRAB5-HT sensor. The crystal structures are from the Protein Data Bank (PDB) archive (PDB ID: 6BQH and 6BQG for the inactive and active states of the 5-HT2C receptor, respectively14, and PDB ID: 3EVP for cpGFP32). Middle, the 5-HT sensor was optimized over three main steps, including the cpGFP insertion site, the linker between cpGFP and 5-HT2C receptor, and critical amino acids in cpGFP. Right, optimization of cpGFP and the engineering of 5-HTmut. The fluorescence change in each candidate sensor is plotted against the brightness, with both axes normalized to 5-HT1.0.
b, Representative images (left), fluorescence response traces (middle), and group data (right) of the fluorescence response in cultured rat cortical neurons expressing 5-HT1.0 (green) or 5-HTmut (gray); where indicated, 10 μM 5-HT was applied. The inset in the left fluorescence image shows the region with increased contrast; n = 12/3 (12 cells from 3 cultures) for each group. Two-tailed Student’s t-test was performed. P = 1.65 ×10−7 between 5-HT1.0 and 5-HTmut. Scale bar, 20 μm.
c, Kinetic analysis of the 5-HT1.0 sensor in cultured HEK293T cells. Left, a representative image showing the experiment protocol, in which the line-scanning mode was used to record the fluorescence change in HEK293T cells expressing 5-HT1.0 in response to local application of 5-HT, followed by metergoline (Met) in the continued presence of 5-HT. Middle, representative traces showing the rise and decay of 5-HT1.0 fluorescence in response to 5-HT (top) followed by Met (bottom). Right, a summary of on and off kinetics of 5-HT1.0; n = 10/3, 8/3,8/3 for control dye, τon and τoff group. Scale bar, 10 μm.
d, Summary of the change in fluorescence of 5-HT1.0 in response to 5-HT alone, 5-HT together with Met or SB 252084 (SB), and eight additional neurotransmitters and neuromodulators. GABA, gamma-aminobutyric acid; Glu, glutamate; DA, dopamine; NE, norepinephrine; ACh, acetylcholine; His, histamine; Oct, octopamine; and Gly, glycine; n = 3 wells per group with 300–500 cells per well. Two-tailed Student’s t-test was performed. P = 2.67 ×10−5, 4.44 ×10−5, 3.01 ×10−5, 1.73 ×10−5, 1.89 ×10−5, 2. 97 ×10−5, 2. 74 ×10−5, 2.00 ×10−5, 2.91 ×10−5, 2.16 ×10−5, and 2.09 ×10−5 between 5-HT and 5-HT + Met, 5-HT + SB, GABA, Glu, DA, NE, ACh, His, Oct, Gly and vehicle, respectively. F10,22 = 2.25, P = 0.0546 for 5-HT + Met, 5-HT + SB, GABA, Glu, DA, NE, ACh, His, Oct, Gly and vehicle by one-way ANOVA.
e, The dose-response curve was measured in neurons expressing 5-HT1.0 in response to increasing concentrations of 5-HT, followed by Met; n = 18/4 for each group.
f,g, G-protein coupling (f) and β-arrestin coupling (g) were measured for the 5-HT1.0 sensor and 5-HT2C receptor using a luciferase complementation assay and TANGO assay, respectively; n = 3 wells per group with 100–300 cells per well.
Data are shown as the mean ± s.e.m. in b-g, with the error bars or shaded regions indicating s.e.m., ***p<0.001, and n.s., not significant.
Next, we characterized the properties of the 5-HT1.0 sensor in detail. Specifically, we measured the brightness, photostability, the dose-response curve, response kinetics, signal specificity, and downstream coupling of this sensor. We found that the 5-HT1.0 had similar brightness and better photostability compared to EGFP in the presence of 5-HT (Extended Data Fig. 4e–g). Regarding the sensor’s kinetics, the τon and τoff values were around 0.2 s and 3.1 s, respectively, as measured by applying 5-HT (to measure the on-rate, τon) followed by the 5-HT receptor antagonist metergoline (Met, to measure the off-rate, τoff) in cultured HEK293T cells (Fig. 1c). In addition, the 5-HT1.0 was highly sensitive to 5-HT, with a half-maximal effective concentration (EC50) of 22 nM (Fig. 1e). None of the other neurotransmitters and neuromodulators tested elicited a detectable fluorescence change. Importantly, the 5-HT-induced signal was eliminated by the specific 5-HT2C receptor antagonist SB 242084 (SB) (Fig. 1d and Extended Data Fig. 4o,p), indicating a high specificity to 5-HT. Moreover, unlike the native 5-HT2C receptor, which couples to the intracellular G-protein and β-arrestin signaling pathways, the 5-HT1.0 sensor showed no detectable coupling to either of these two pathways, as measured by calcium imaging, cAMP imaging16, G-protein-dependent luciferase complementation assay17, TANGO assay18, and long-term measurements of membrane fluorescence in the presence of 5-HT (Fig. 1f,g and Extended Data Fig. 4h–l). Additionally, the expression of 5-HT1.0 did not alter the 5-HT2C receptor coupling with the G protein (Extended Data Fig. 4m), indicating the 5-HT1.0 has little effect on the activation of the wild-type 5-HT2C receptor.
Detecting 5-HT release in mouse brain slices and Drosophila with the GRAB5-HT1.0
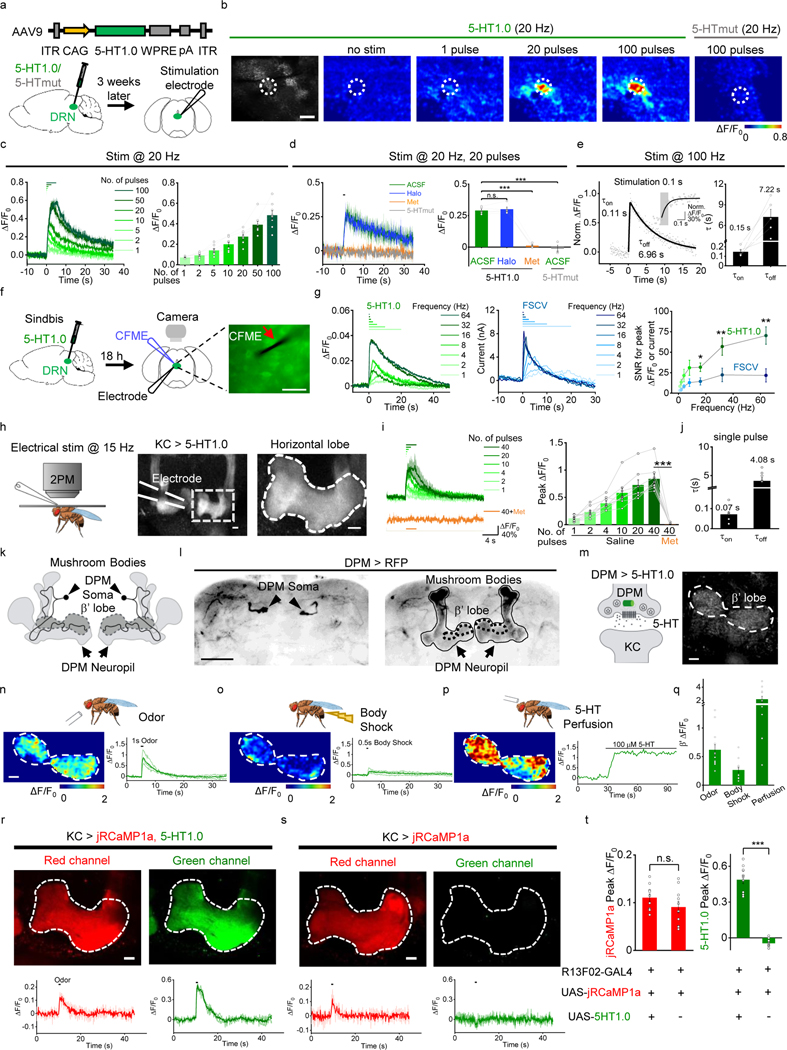
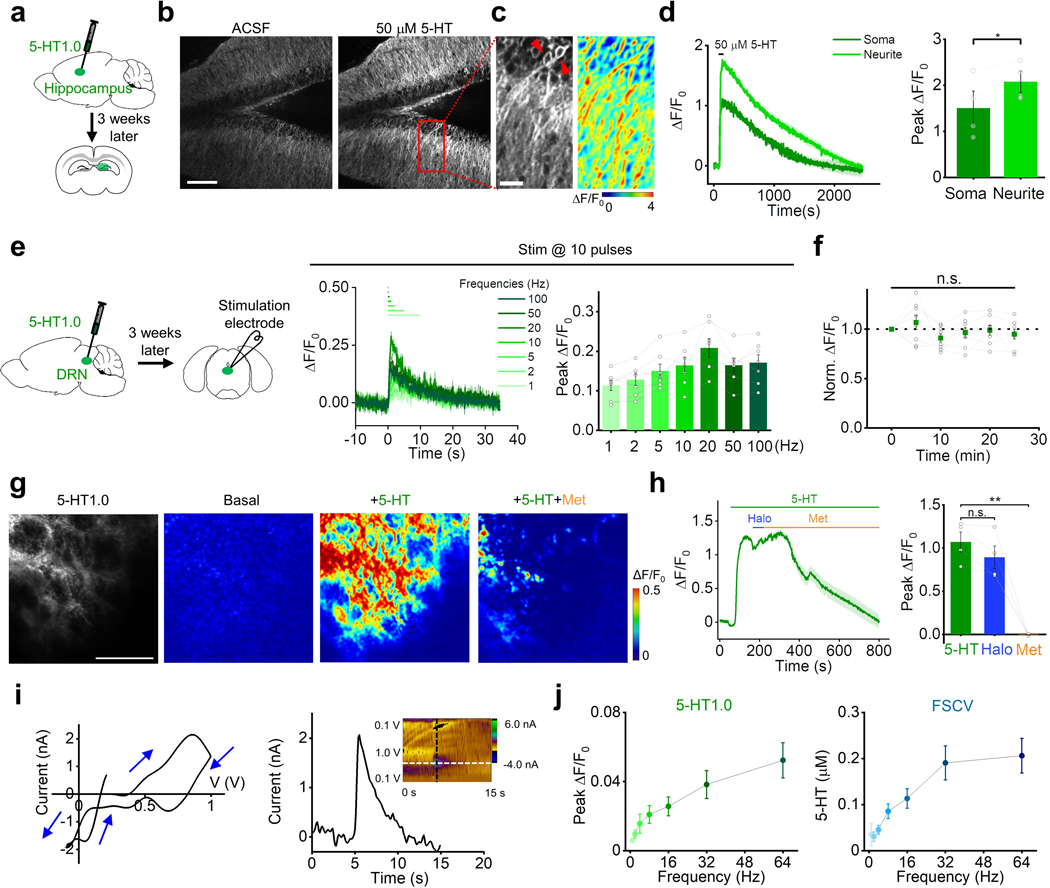
Having validated the sensor in cultured cells, to see whether 5-HT1.0 could function well in the mouse brain slices, we expressed 5-HT1.0 using adeno-associated virus (AAV) in the mouse hippocampus, which receives innervation from the serotonergic neurons. The 5-HT1.0 sensor showed more than 100% fluorescence increase to exogenously applied 5-HT in individual somata and neurites (Extended Data Fig. 5a–d). Besides, we also expressed either 5-HT1.0 or 5-HTmut in the mouse dorsal raphe nucleus (DRN) by AAVs (Fig. 2a). Dorsal raphe nucleus (DRN) is the largest serotonergic nucleus in the brain, and provides extensive projections to various brain regions19. In DRN slices expressing 5-HT1.0, a single electrical pulse evoked detectable fluorescence increases, and the response progressively enhanced with the increase in pulse number or frequency (Fig. 2b,c and Extended Data Fig. 5e). The stimulation evoked-response was repeatable for up to 25 min (Extended Data Fig. 5f) and blocked by the 5-HT receptor antagonist Met, but not the dopamine receptor antagonist haloperidol (Halo; Fig. 2d and Extended Data Fig. 5g,h). In contrast, the same electrical stimuli did not affect fluorescence in DRN expressing the 5-HTmut sensor (Fig. 2b,d). We also measured the kinetics of the fluorescence change in response to 100 ms electrical stimulation and found τon and τoff values of approximately 0.15 s and 7.22 s (Fig. 2e). We further compared the 5-HT1.0 sensor with existing fast-scan cyclic voltammetry (FSCV) in recording 5-HT by simultaneously conducting fluorescence imaging and electrochemical recording in DRN slices (Fig. 2f). Both methods could sensitively detect the single pulse-evoked 5-HT signal and the increase of response following incremental frequencies (Fig. 2g and Extended Data Fig. 5i,j). The 5-HT1.0 showed better signal-to-noise ratio (SNR) compared with FSCV (Fig. 2g). Taken together, these results showed that the 5-HT1.0 has the ability to measure the electrically induced endogenous 5-HT release ex vivo.
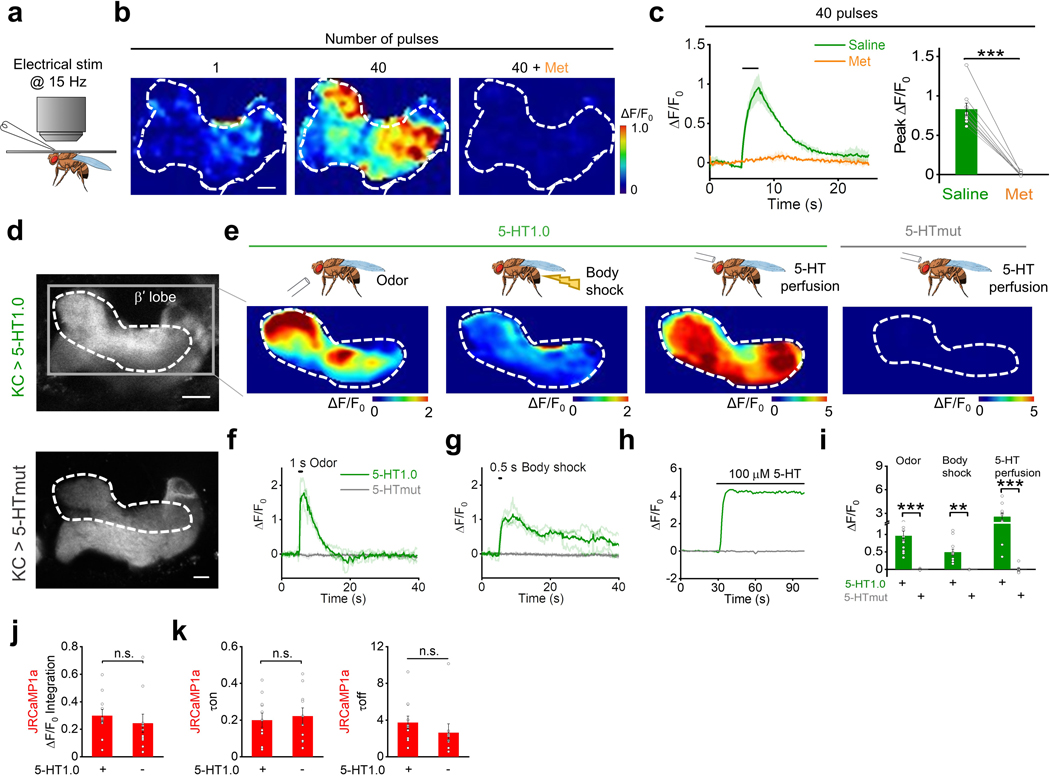
Fig. 2: GRAB5-HT can report the release of endogenous 5-HT in acute mouse brain slices and Drosophila.
a, Schematic illustration depicting the mouse brain slice experiments. Top, The AAV vector was used to express the 5-HT1.0 sensor. Bottom, AAV expressing either 5-HT1.0 or 5-HTmut was injected in the mouse DRN, after which acute brain slices were prepared and recorded.
b, Representative fluorescence image of 5-HT1.0, pseudocolor images of the fluorescence change in 5-HT1.0, and 5-HTmut in response to the indicated electrical stimuli delivered at 20 Hz. The duration of one pulse is 1 ms. The white dotted circle (50 μm diameter) indicates the ROI used for further analysis. Similar results were observed from 2–5 mice. Scale bar, 100 μm.
c, Individual traces (left) and quantification (right) of 5-HT1.0 fluorescence change in response to the indicated electrical stimuli delivered at 20 Hz; n = 8 slices from 5 mice.
d, Representative traces (left) and group data of 5-HT1.0 and 5-HTmut fluorescence change in response to electrical stimuli in slices treated with the dopamine receptor antagonist haloperidol (Halo) or the 5-HT receptor antagonist Met; n = 5 slices from 4 mice for 5-HT1.0 group, n = 3 slices from 1 mouse for 5-HTmut group. Two-tailed Student’s t-test was performed. P = 0.340 between 5-HT1.0 in ACSF and Halo; P = 1.47 ×10−5 between 5-HT1.0 in ACSF and Met; P = 2.18 ×10−5 between 5-HT1.0 in ACSF and 5-HTmut in ACSF.
e, Left, normalized fluorescence change of 5-HT1.0 in response to 10 electrical stimuli delivered at 100 Hz. The rise and decay phases are fitted with single-exponential functions (black traces). A magnified view of the on kinetics in the inset. Right, summary of τon and τoff; n = 5 slices from 4 mice.
f, Schematic drawing outlines the design of simultaneous imaging and fast-scan cyclic voltammetry (FSCV) experiments in mouse DRN slice preparation. The red arrow indicates that the carbon-fiber microelectrode (CFME) is placed near the neuron expressing 5-HT1.0. Scale bar, 20 μm.
g, Left, representative fluorescence traces of a DRN neuron expressing 5-HT1.0 to electrical stimuli consisting of a train of 20 pulses at varied frequency. Middle, current vs time traces of evoked 5-HT release at varied stimulating frequencies. Right, group summary of the signal to noise ratio (SNR) of 5-HT1.0 and FSCV; n = 11 neurons from 9 mice. Two-tailed Student’s t-test was performed. P = 0.0784, 0.0507, 0.112, 0.0936, 0.0252, 0.00647, and 0.00272 for 1 Hz, 2 Hz, 4 Hz, 8 Hz, 16 Hz, 32 Hz, and 64 Hz.
h, Left, a schematic drawing showing in vivo two-photon imaging of a Drosophila, with the stimulating electrode positioned near the mushroom body (MB). Middle and right, representative images of a fly expressing 5-HT1.0 in the Kenyon cells (KCs) of the MB; the image on the right is a magnified view of the dashed rectangle. The duration of one pulse is 1 ms. Similar results were observed from 8 flies. Scale bar, 10 μm.
i, Representative traces (left) and group analysis (right) of 5-HT1.0 fluorescence in response to the indicated electrical stimuli in either saline (control) or 10 μM Met; n = 8 flies for each group. Two-tailed Student’s t-test was performed. P = 2.36 ×10−5 between saline and Met.
j, Summary of 5-HT1.0 τon and τoff in response to a single electrical stimulation; n = 8 flies for each group.
k,l, Cartoon and Immunostaining images of DPMs. Cartoon of the DPMs in the MB (k), and immunostaining images of DPMs’ soma (l, left) and neuropil (l, right) in the MB region. Similar results were observed from 5 flies. Scale bar, 50 μm.
m, Representative image of a fly expressing 5-HT1.0 in the single serotonergic DPM neuron in the fly MB region of one hemisphere. Similar results were observed for more than 10 flies. Scale bar, 10 μm.
n-q, Representative pseudocolor images (left), traces (right) of 5-HT1.0 in the MB β’ lobe in response to a 1-s odor application (n), a 0.5-s body shock (o), and perfusion 100 μM of 5-HT (p). (q) Group analysis of 5-HT1.0 in response to different stimuli. n= 13 flies for odor and body shock group, n= 11 flies for perfusion group.
r-t, Representative fluorescence images (top), fluorescence response traces (bottom), and group summary (t) of jRCaMP1a in fly MB with co-expression of 5-HT1.0 (r) or expressing jRCaMP1a alone (s) in the KCs in response to 1-s odorant stimulation. Scale bar, 10 μm. n = 11 and 10 flies for jRCaMP1a response in 5-HT1.0+ and 5-HT1.0- group; n = 11 and 9 flies for 5-HT1.0 response in 5-HT1.0+ and 5-HT1.0- group. Two-tailed Student’s t-test was performed. P = 0.179 and P = 2.15 ×10−9 for jRCaMP1a and 5-HT1.0 peak response.
Data are shown as the mean ± s.e.m. in c-e, g, i, j, q-t, with the error bars or shaded regions indicating s.e.m., *p<0.05, ** p<0.01, ***p<0.001, and n.s., not significant.
We next tested whether the 5-HT1.0 sensor could be used to measure sensory-relevant changes in 5-HT signaling in vivo. To this end, we first used the Drosophila model, as 5-HT is known to be released from single serotoninergic dorsal paired medial (DPM) neurons that innervate Kenyon cells (KCs) in the mushroom body (MB) per hemisphere20, 21 and serotonergic signaling in the MB has been implicated in odor-related memory consolidation22. We expressed the 5-HT1.0 sensor in the KCs and found that the sensor reliably reported 5-HT release evoked by electrical stimulation of the horizontal lobe of MB with rapid on and off kinetics of ~0.07 s and ~4.08 s, respectively. Moreover, the signal was blocked by applying Met (Fig. 2h–j and Extended Data Fig. 6a–c). To see whether the sensor could report the physiological relevant 5-HT release from a single cell, we directly expressed the 5-HT1.0 sensor either in the single presynaptic serotonergic DPM neuron or the postsynaptic KCs. Consistent with the previous calcium imaging study in the DPM23, the 5-HT1.0 sensor in either DPM or KCs could report the 5-HT release in the MB β’ lobe in response to odor application or body shock (Fig. 2k–q, Extended Data Fig. 6d–i, and Supplementary Video 1). In contrast, no fluorescence change was detected in flies expressing the 5-HTmut sensor (Extended Data Fig. 6d–i, and Supplementary Video 1). Neither odor application nor body shock produced a saturated response of the 5-HT1.0 sensor, as the application of exogenous 100 μM 5-HT in the same flies elicited much larger responses (Extended Data Fig. 6d–i). When we co-expressed the 5-HT1.0 sensor together with a red fluorescent Calcium sensor jRCaMP1a in KCs, two-color imaging showed that the odorant application increased both the 5-HT1.0-mediated green 5-HT fluorescence and the jRCaMP1a-mediated red Calcium fluorescence responses (Fig. 2r–t). Moreover, jRCaMP1a-expressing flies with or without co-expression of 5-HT1.0 had similar odorant-evoked Calcium signals, suggesting little effect of the 5-HT1.0 sensor expression on neuronal Calcium activities (Fig. 2r–t and Extended Data Fig. 6j,k).
Monitoring endogenous 5-HT dynamics in mice in vivo
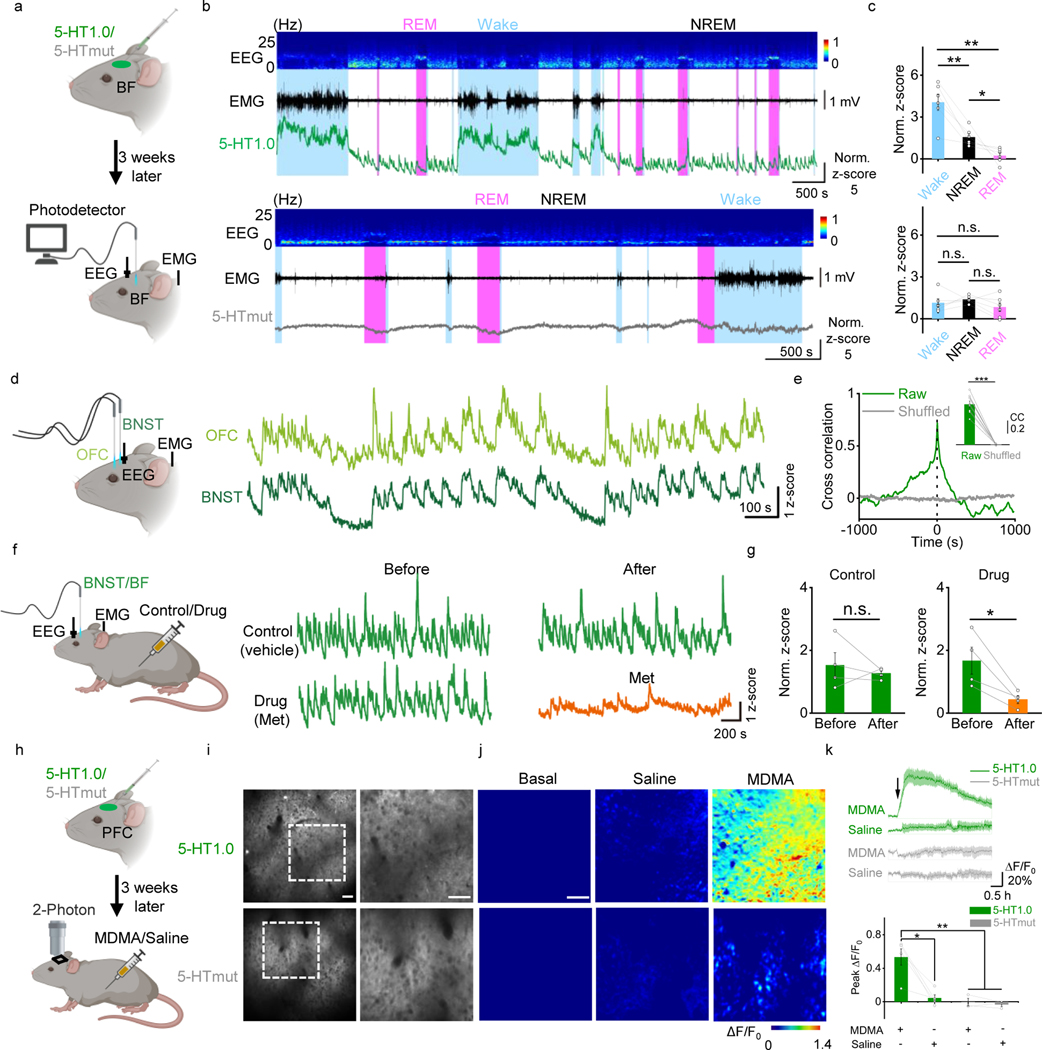
Following this, we examined whether the 5-HT1.0 sensor could measure the dynamics of serotonergic activity under physiological conditions in mice, such as during the sleep-wake cycle. The basal forebrain (BF), orbital frontal cortex (OFC), and the bed nucleus of the stria terminalis (BNST) not only participate in the regulation of sleep and wake cycles but also receive extensive DRN serotonergic projections24, 25. Hence, we expressed the 5-HT1.0 sensor in these several brain regions, and then performed simultaneous fiber-photometry and EEG/EMG recordings in freely behaving mice. The BF 5-HT1.0 sensor signal was generally higher during wake than that during sleep. Between rapid-eye-movement (REM) sleep and non-REM (NREM) sleep, the signal is lower during REM sleep, consistent with previous findings26 (Fig. 3a–c). As expected, we found no substantial change in fluorescence in mice expressing the 5-HTmut sensor during the sleep-wake cycle. Interestingly, simultaneous recordings of 5-HT1.0 in OFC and BNST revealed a tight correlation in fluorescence during NREM sleep (Fig. 3d,e), suggesting global synchrony of the 5-HT signaling despite the region-specific innervation by different subpopulations of the serotonergic neurons in DRN25, 27. Lastly, consistent with our previous findings, we found that treating mice with the 5-HT receptor antagonist Met largely blocked the fluorescence change of the 5-HT1.0 sensor (Fig. 3f, g), validating the specificity of measured signals in vivo.
Fig. 3: GRAB5-HT can report endogenous serotonergic activity in freely behaving mice.
a, Schematic diagram illustrating the use of fiber-photometry for measuring 5-HT1.0 and 5-HTmut fluorescence in the basal forebrain (BF) of freely behaving mice during the sleep-wake cycle. EEG and EMG were also measured.
b, Representative EEG, EMG, and 5-HT1.0 (top) and 5-HTmut (bottom) fluorescence measured during the sleep-wake cycle. The REM sleep state was shaded with pink color, and the waking state was shaded with a light blue color. Similar results were observed from 3 mice.
c, Summary of 5-HT1.0 (top) and 5-HTmut (bottom) fluorescence measured in awake mice and during waking, NREM, and REM sleep; n = 3 mice in two sessions for each group. Two-tailed Student’s t-test was performed. For 5-HT1.0, P = 0.00382 between wake and NREM; P = 0.00374 between wake and REM; P = 0.0334 between NREM and REM. For 5-HTmut, P = 0.474 between wake and NREM; P = 0.255 between wake and REM; P =0.107 between NREM and REM.
d, e, Same as in (a), except the 5-HT1.0 sensor was expressed in both the orbital frontal cortex (OFC; light green) and the bed nucleus of the stria terminalis (BNST; dark green), and the fluorescence response in each nucleus was recorded and analyzed. Similar results were observed from 4 mice. The cross-correlation between the signals in the OFC and BNST is shown in (e); n = 4 mice in two sessions for each group. Two-tailed Student’s t-test was performed. P = 8.72 ×10−6 between raw and shuffled group.
f, g, 5-HT1.0 fluorescence was measured in the BNST and BF as in (f), where indicated, the mice received an injection of saline (control) or Met. The normalized responses in the BNST (n = 3 mice) and BF (n = 1 mouse) were combined for the group summary. Two-tailed Student’s t-test was performed. P = 0.533 and P = 0.0361 for control and drug group.
h, Schematic diagram illustrating the use of two-photon imaging to measure 5-HT1.0 and 5-HTmut fluorescence in the prefrontal cortex (PFC) of head-fixed mice; MDMA or saline was injected intraperitoneally.
i, Representative fluorescence images of 5-HT1.0 (top, left) and 5-HTmut (bottom, left) measured in the mouse PFC. The images on the right are the corresponding magnified views of the dashed box on the left. Similar results were observed for more than 3 mice. Scale bar, 50 μm.
j, k, Representative pseudocolor images (j), averaged fluorescence traces (k, top), and group summary (k, bottom) showing 5-HT1.0 (top, green) and 5-HTmut (bottom, gray) fluorescence changes after an i.p. injection of saline (middle) or 10 mg/kg MDMA (right); n = 5 mice for 5-HT1.0 group, n = 3 mice for 5-HTmut group. Two-tailed Student’s t-test was performed. P = 0.0106 between 5-HT1.0 with MDMA and saline; P = 0.00669 between 5-HT1.0 with MDMA and 5-HTmut with MDMA; P = 0.00477 between 5-HT1.0 with MDMA and 5-HTmut with saline. Scale bar, 50 μm.
Data are shown as the mean ± s.e.m. in c, e, g, k, with the error bars or shaded regions indicating s.e.m., *p<0.05, ** p<0.01, ***p<0.001, and n.s., not significant.
Finally, to see whether the sensor can reveal the action of psychostimulant drugs, we used the Methylenedioxymethamphetamine (MDMA), which is a synthetically addictive compound that can alter mood and perception, and its effects have been partially attributed to an increase in extracellular 5-HT concentrations in the brain28. We examined MDMA’s effect in vivo by two-photon imaging in mice expressing the sensor in the prefrontal cortex (PFC), a higher cognitive region that receives abundant serotonergic innervation29. Intraperitoneal (i.p.) injection of MDMA caused a gradual increase in 5-HT1.0 fluorescence, which peaked after 1 hour and then gradually decayed over the following 3 hours (Fig. 3h–k). The time course is comparable with that of the MDMA’s psychostimulation effects in both human30 and mouse31. Meanwhile, MDMA had no effect on the fluorescence of the 5-HTmut sensor (Fig. 3h–k). These results together suggest that the 5-HT1.0 sensor is suitable for stable and long-term imaging in vivo.
Discussion
In summary, we report the development and application of a novel genetically encoded fluorescent GRAB sensor for measuring extracellular 5-HT dynamics. The newly developed GRAB5-HT1.0 sensor shows a high affinity to 5-HT (EC50 ~22 nM), relatively fast on kinetics (τon ~70 ms), high specificity and spatial resolution, and minimal impact on cellular physiology and thus is well suited for detecting physiologically relevant endogenous 5-HT release. Indeed, we demonstrated the utility of the GRAB5-HT1.0 sensor in detecting endogenous 5-HT release in response to a variety of stimuli and under various behavioral conditions in different animal models. Our finding that 5-HT changes dynamically throughout the sleep-wake cycle in mice provides new insights into the functional contribution of 5-HT in sleep regulation.
Regarding the potential buffering by the 5-HT1.0 sensor, we measured the G-protein coupling by exogenously applied 5-HT with a broad range of concentrations in the 5-HT2C and 5-HT2C+5-HT1.0 group in cultured cells and did not observe a significant difference of the 5-HT2C receptor mediated G-protein signaling between these two groups. In addition, the Drosophila in vivo imaging data also showed that the expression of 5-HT1.0 has little effect on the odor-evoked neuronal calcium signals. Nevertheless, one still need to be mindful about the buffering effect of the 5-HT1.0 sensor, especially under the conditions where the local 5-HT concentration is very low or the expression level of 5-HT1.0 is very high. One way to ameliorate the buffering effect is to balance the expression level and the photon (signal) generated from the 5-HT1.0 sensor. Future efforts could be applied to tune the sensor’s affinity while improving its brightness and ΔF/F0.
Of note, there is a tradeoff between a sensor’s affinity and off kinetics. Given the high affinity of 5-HT1.0 sensor, it inevitably shows a slightly slow off kinetics. These kinetic features could be beneficial as the rapid on kinetics reports the initial timing of the endogenous 5-HT release precisely (e.g., in local synaptic transmission) while the slow off kinetics is helpful to accumulate more photons and contribute to the high SNR. Nevertheless, to fully capture the dynamics of the fast endogenous serotonergic signaling awaits future improvement of better sensors with both fast on and off kinetics.
Methods
Primary cultures
Male and female postnatal day 0 (P0) Sprague-Dawley rat pups were obtained from (Beijing Vital River) and used to prepare cortical neurons. The cortex was dissected, and neurons were dissociated using 0.25% Trypsin-EDTA (GIBCO), plated on 12-mm glass coverslips coated with poly-D-lysine (Sigma-Aldrich), and cultured in neurobasal medium (GIBCO) containing 2% B-27 supplement (GIBCO), 1% GlutaMax (GIBCO), and 1% penicillin-streptomycin (GIBCO). The neurons were cultured at 37°C in a humidified atmosphere in air containing 5% CO2.
Cell lines
HEK293T cells were purchased from ATCC and verified based on their morphology under microscopy and an analysis of their growth curve. Stable cell lines expressing either 5-HT2C receptor or 5-HT1.0 were generated by co-transfecting cells with the PiggyBac plasmid carrying the target genes together with Tn5 transposase into a stable HEK293T cell line. Cells expressing the target genes were selected using 2 μg/ml Puromycin (Sigma). A HEK293 cell line stably expressing a tTA-dependent luciferase reporter and the β-arrestin2-TEV fusion gene used in the TANGO assay was a generous gift from Bryan L. Roth33. All cell lines were cultured at 37°C in 5% CO2 in DMEM (GIBCO) supplemented with 10% (v/v) fetal bovine serum (GIBCO) and 1% penicillin-streptomycin (GIBCO).
Drosophila
UAS-GRAB5-HT1.0 (attp40, UAS-GRAB5-HT1.0/CyO) and UAS-GRAB5-HTmut (attp40, UAS-GRAB5-HTmut/CyO) flies were generated in this study. The coding sequences of GRAB5-HT1.0 or GRAB5-HTmut were inserted into pJFRC2834 (Addgene plasmid #36431) using Gibson assembly. These vectors were injected into embryos and integrated into attp40 via PhiC31 by the Core Facility of Drosophila Resource and Technology, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences. The following fly stocks were used in this study: R13F02-Gal4 (BDSC:49032), VT64246-Gal4 (VDRC:204311) and UAS-jRCaMP1a (BDSC: 63792)35. Flies were raised on standard cornmeal-yeast medium at 25°C, with 70% relative humidity and a 12h/12h light/dark cycle. In Fig. 2h–j and Extended Data Fig. 6a–c, fly UAS-GRAB5-HT1.0/CyO; R13F02-Gal4/TM2 was used; in Fig. 2k–l UAS-CsChrimson-mCherry/CyO; VT64246-Gal4/TM2 was used; in Fig. 2m–q UAS-GRAB5-HT1.0/CyO; VT64246-Gal4/TM2 was used; in Fig. 2r–t and Extended Data Fig. 6j–k UAS-GRAB5-HT1.0/+; R13F02-Gal4/UAS-jRCaMP1a and fly R13F02-Gal4/UAS-jRCaMP1a were used; in Extended Data Fig. 6d–i UAS-GRAB5-HT1.0/CyO; R13F02-Gal4/TM2 and UAS-GRAB5-HTmut/+; R13F02-Gal4/+ were used.
Mouse
Wild-type male and female C57BL/6 (P25–60) mice were used to prepare the acute brain slices and for the in vivo mouse experiments. All mice were group-housed in a temperature-controlled room (21.5 degree centigrade) with a 12h/12h light/dark cycle, with a humidity controlled as 55%, provided with food and water ad libitum. All procedures for animal surgery and maintenance were performed using protocols that were approved by the Animal Care & Use Committees at Peking University, the Chinese Academy of Sciences, University of Virginia, and were performed following the guidelines established by the US National Institutes of Health.
Molecular biology
Plasmids were generated using the Gibson assembly method36, and DNA fragments were amplified by PCR using primers (Thermo Fisher Scientific) with 25–30 bp overlap. DNA fragments were assembled using T5-exonuclease (New England Biolabs), Phusion DNA polymerase (Thermo Fisher Scientific), and Taq ligase (iCloning). Sanger sequencing was performed at the Sequencing Platform in the School of Life Sciences of Peking University to verify plasmid sequences. cDNAs encoding various 5-HT receptors (HTR1E, HTR2C, HTR5A, and HTR6) were generated using PCR amplification of the full-length human GPCR cDNA library (hORFeome database 8.1). For optimizing the 5-HT sensor, cDNAs encoding the candidates in step 1 and step 2 were cloned into the pDisplay vector (Invitrogen) with an IgK leader sequence in the sensor upstream. In step 3, in addition to upstream IgK peptide, the IRES-mCherry-CAAX cascade was fused downstream of the sensor to calibrate the membrane signal. For optimizing the linker sequence and cpGFP, site-directed mutagenesis was performed using primers containing NNB codons (48 codons, encoding 20 possible amino acids). For characterization in cultured rat cortical neurons, GRAB5-HT1.0 and GRAB5-HTmut were cloned into the pAAV vector under the hSyn, TRE, or CAG promoter. In downstream coupling experiments, the GRAB5-HT sensor and the 5-HT2C receptor were cloned into the pTango and pPiggyBac vectors, respectively. Two mutations were introduced into pCS7-PiggyBAC to generate hyperactive piggyBac transposase (ViewSolid Biotech)37. The GRAB5-HT1.0-SmBit and 5-HT2C-SmBit constructs were derived from β2AR-SmBit17 using a BamHI site incorporated upstream of the GGSG linker. LgBit-mGq was a generous gift from Nevin A. Lambert.
Expression of GRAB5-HT in cultured cells and in vivo
HEK293T cells were plated on 12-mm glass coverslips in 24-well plates and grown to 70% confluence for transfection with PEI (1 mg DNA and 3 mg PEI per well); the medium was replaced after 4–6 hours, and cells were used for imaging 24 hours after transfection. Cultured rat cortical neurons were infected with AAVs expressing TRE-GRAB5-HT1.0 (titer: 3.8×10^13 particles/ml) and hSyn-tTA (titer: 1.3×10^14 particles/ml) or hSyn-GRAB5-HTmut (titer: 1×10^13 particles/ml) at 7–9 day of in vitro (DIV), and imaging was performed 7–14 days after infection.
For in vivo expression, mice were deeply anesthetized with an i.p. injection of 2,2,2-tribromoethanol (Avertin, 500 mg/kg, Sigma-Aldrich) or ketamine (10 mg/kg) and xylazine (2 mg/kg), placed in a stereotaxic frame, and AAVs were injected using a microsyringe pump (Nanoliter 2000 Injector, WPI). For the experiments shown in Extended Data Fig. 5a–d, AAV expressing hSyn-GRAB5-HT1.0 (titer: 4.6×10^13 particles/ml) was injected (volume: 400 nl) into the hippocampus at the following coordinates relative to Bregma: AP: −2.0 mm, ML: 1.5 mm (depth: 2.3 mm). For the experiments shown in Fig. 2a–e and Extended Data Fig. 5e–h, AAVs expressing CAG-GRAB5-HT1.0(titer: 1.3×10^13 particles/ml) or hSyn-GRAB5-HTmut (titer: 1×10^13 particles/ml) were injected (volume: 400 nl) into the DRN at the following coordinates relative to Bregma: AP: −4.3 mm, ML: 1.1 mm (depth: 2.85 mm, with a 20° ML angle). For the experiments shown in Fig. 2f,g, and Extended Data Fig. 5i,j, Sindbis virus expressing GRAB5-HT1.0 was injected (volume: 50 nl) into the DRN at the following coordinates relative to Bregma: AP: −4.3 mm, ML: 0.0 mm (depth: 3.00 mm). For the experiments shown in Fig. 3a–g, AAVs expressing CAG-GRAB5-HT1.0 or hSyn-GRAB5-HTmut were injected (volume: 400 nl) into the BF at the following coordinates relative to Bregma: AP: 0 mm, ML: 1.3 mm (depth: 5.0 mm), the OFC (AP: +2.6 mm, ML: 1.7 mm, depth: 1.7 mm), and the BNST (AP: +0.14 mm, ML: 0.8 mm, depth: 3.85 mm). For the experiments shown in Fig. 3h–k, AAVs expressing hSyn-GRAB5-HT1.0 (titer: 4.6×10^13 particles/ml) or hSyn-GRAB5-HTmut (titer: 1×10^13 particles/ml) were injected (volume: 400 nl) into the PFC at the following coordinates relative to Bregma: AP: +2.8 mm, ML: 0.5 mm (depth: 0.5 mm).
Fluorescence imaging of HEK293T cells and cultured rat cortical neurons
An inverted Ti-E A1 confocal microscope (Nikon) and an Opera Phenix high-content screening system (PerkinElmer) were used for imaging. The confocal microscope was equipped with a 40x/1.35 NA oil-immersion objective, a 488-nm laser, and a 561-nm laser. The GFP signal was collected using a 525/50-nm emission filter, and the RFP signal was collected using a 595/50-nm emission filter. Cultured cells expressing GRAB5-HT1.0 or GRAB5-HTmut were either bathed or perfused with Tyrode’s solution containing (in mM): 150 NaCl, 4 KCl, 2 MgCl2, 2 CaCl2, 10 HEPES, and 10 glucose (pH 7.4). Drugs were delivered via a custom-made perfusion system or bath application. The chamber was cleaned thoroughly between experiments using 75% ethanol. Photostability was measured using confocal microscopy (1-photon illumination) with the 488-nm laser at a laser power of 350 μW, and the Opera Phenix high-content screening system was equipped with a 40x/1.1 NA water-immersion objective, a 488-nm laser, and a 561-nm laser; the GFP signal was collected using a 525/50-nm emission filter, and the RFP signal was collected with a 600/30-nm emission filter. For imaging, the culture medium was replaced with 100 μl of Tyrode’s solution, and drugs (at various concentrations) were applied in Tyrode’s solution. The fluorescence signal of the GRAB5-HT sensors was calibrated using the GFP/RFP ratio. In order to measure the response kinetics of the 5-HT1.0 sensor, the line scanning mode of confocal was used to record the rapid fluorescence changes. A glass pipette filled with 10 μM 5-HT or Met placed near the 5-HT1.0 sensor expressing HEK293T cells. 5-HT or Met was puffed by the pipette to measure the on and off response kinetics, respectively. For calculating the on and off kinetics, different F0 was used. In detail, we used the basal fluorescent intensity before 5-HT applications as F0 when calculating the “on” response; while, when calculating the “off” response, we used the fluorescence intensity of 5-HT1.0 under saturating 5-HT (10 μM) as F0.
TANGO assay
5-HT at various concentrations was applied to 5-HT2C-expressing or 5-HT1.0-expressing HTLA cells33. The cells were then cultured for 12 h to allow expression of firefly luciferase (Fluc). Bright-Glo reagent (Fluc Luciferase Assay System, Promega) was then added to a final concentration of 5 μM, and luminescence was measured using a Victor X5 multilabel plate reader (PerkinElmer).
Luciferase complementation assay
The luciferase complementation assay was performed as previously described17. In brief, 48 h after transfection, cells were washed with PBS, harvested by trituration, and transferred to opaque 96-well plates containing 5-HT at various concentrations. Furimazine (NanoLuc Luciferase Assay, Promega) was then added quickly to each well, followed by measurement with Nluc.
Fluorescence imaging of GRAB5-HT in mouse brain slices
AAVs or Sindbis virus expressing GRAB5-HT1.0 or GRAB5-HTmut were injected into the mouse hippocampus or DRN as described above. Three weeks after AAV injection or 18 hours after Sindbis virus injection, mice were deeply anesthetized by an i.p. injection of Avertin or xylazine-ketamine and then transcardially perfused with 10 ml oxygenated slicing buffer consisting of (in mM): 110 choline-Cl, 2.5 KCl, 1 NaH2PO4, 25 NaHCO3, 7 MgCl2, 25 glucose, and 0.5 CaCl2. Mice were then decapitated, and brains were removed and placed in cold (0–4°C) oxygenated slicing buffer for an additional 1 min. Then, brains were rapidly mounted on the cutting stage of a VT1200 vibratome (Leica) for coronal sectioning at 300 μm thickness. Brain slices containing the hippocampus or DRN were initially allowed to recover for ≥40 min at 34°C in oxygen-saturated Ringer’s buffer consisting of (in mM): 125 NaCl, 2.5 KCl, 1 NaH2PO4, 25 NaHCO3, 1.3 MgCl2, 25 glucose, and 2 CaCl2. For two-photon imaging, slices were transferred to a recording chamber that was continuously perfused with 34°C oxygen-saturated Ringer’s buffer and placed in an FV1000MPE two-photon microscope (Olympus) equipped with a 25x/1.05 NA water-immersion objective. 5-HT1.0 or 5-HTmut fluorescence was excited using a mode-locked Mai Tai Ti: Sapphire laser (Spectra-Physics) at a wavelength of 920-nm and collected via a 495–540-nm filter. For electrical stimulation, a bipolar electrode (cat. WE30031.0A3, MicroProbes) was positioned near the DRN in the slice, and imaging and stimulation were synchronized using an Arduino board with a custom-written program. The parameters of the frame scan were set to a size of 128 × 96 pixels with a speed of 0.1482 s/frame for electrical stimulation and a size of 512 × 512 pixels with a speed of 1.109 s/frame for drug perfusion experiments. For the kinetics measurements, line scans were performed with a rate of 800–850 Hz. The stimulation voltage was set at 4–6 V, and the duration of each stimulation was set at 1 ms. Drugs were bath-applied by perfusion into the recording chamber in pre-mixed Ringer’s buffer. For the Sindbis virus-infected mouse brain slices, wide-field epifluorescence imaging was performed using Hamamatsu ORCA FLASH4.0 camera (Hamamatsu Photonics, Japan), and 5-HT1.0-expressing cells in acutely prepared brain slices are excited by a 460-nm ultrahigh-power low-noise LED (Prizmatix, Givat-Shmuel, Israel). The frame rate of the FLASH4.0 camera was set to 10 Hz. To synchronize image capture with electrical stimulation, and fast-scan cyclic voltammetry, the camera was set to external trigger mode and triggered by a custom-written IGOR Pro 6 program (WaveMetrics, Lake Oswego, OR). For electrical stimulation, a home-made bipolar electrode was positioned near the DRN in the slice, and the stimulation current was set at 350 μA, and the duration of each stimulation was set at 1 ms.
Fast-scan cyclic voltammetry (FSCV)
Carbon-fiber microelectrodes (CFME) were fabricated as described previously38. Briefly, cylindrical CFMEs (7 μm in radius) were constructed with T-650 carbon fiber (Cytec Engineering Materials), which was aspirated into a glass capillary (1.2 mm O.D and 0.68 mm I.D, A-M system) and the capillary pulled using the PE-22 puller (Narishige Int.). The carbon fiber was trimmed to 50 to 70 μm in length from the pulled glass tip and sealed with Epon epoxy which was cured at 100 °C for 2 hours followed by 150 °C overnight. CFMEs were cleaned in isopropyl alcohol for 30 min before the Nafion electrodeposition. Nafion was electrochemically deposited by submerging CFME tip in Nafion® solution (5 wt% 1100 EW Nafion® in methanol, Ion Power), and a constant potential of 1.0 V vs Ag/AgCl was applied to the electrode for 30 seconds. Then, Nafion coated electrodes were air-dried for 10 seconds, and then at 70 °C for 10 minutes. For electrochemical detection of 5-HT, a Jackson waveform was applied to the electrode by scanning the potential from 0.2 V to 1.0 V to – 0.1 V and back to 0.2 V at 1000 V/s using a ChemClamp potentiostat (Dagan). For data collection and analysis, TarHeel CV (provided by R.M. Wightman, University of North Carolina) was used. For the electrode calibrations, phosphate buffer (PBS) solution was used which consisting of (in mM): 131.25 NaCl, 3.0 KCl, 10.0 NaH2PO4, 1.2 MgCl2, 2.0 Na2SO4, and 1.2 CaCl2 (pH 7.4). A 5-HT stock solution was prepared in 0.1 M HClO4 and diluted to 500 nM with PBS for calibrations prior to the experiment.
Fluorescence imaging of transgenic flies
Fluorescence imaging in flies was performed using an Olympus two-photon microscope FV1000 equipped with a Spectra-Physics Mai Tai Ti: Sapphire laser. A 930-nm excitation laser was used for one-color imaging of 5-HT1.0 or 5-HTmut, and a 950-nm excitation laser was used for two-color imaging with 5-HT1.0 and jRCaMP1a. For detection, 495–540-nm filter for green channel and 575–630-nm filter for red channel. Adults male flies within 2 weeks post eclosion were used for imaging. To prepare the fly for imaging, adhesive tape was affixed to the head and wings. The tape above the head was excised, and the chitin head-shell, air sacs, and fat bodies were carefully removed to expose the central brain. The brain was bathed continuously in an adult hemolymph-like solution (AHLS) composed of (in mM): 108 NaCl, 5 KCl, 5 HEPES, 5 trehalose, 5 sucrose, 26 NaHCO3, 1 NaH2PO4, 2 CaCl2, and 1–2 MgCl2. For electrical stimulation, a glass electrode (resistance: 0.2 MΩ) was placed in proximity to the MB medial lobe, and voltage for stimulation was set at 10–30 V. For odorant stimulation, the odorant isoamyl acetate (cat. 306967, Sigma-Aldrich) was first diluted 200-fold in mineral oil, then diluted 5-fold with air, and delivered to the antenna at a rate of 1000 ml/min. For body shock, two copper wires were attached to the fly’s abdomen, and a 500-ms electrical stimulus was delivered at 50–80 V. For 5-HT application, the blood-brain barrier was carefully removed, and 5-HT was applied at a final concentration 100 μM. An Arduino board was used to synchronize the imaging and stimulation protocols. The sampling rate during electrical stimulation, odorant stimulation, body shock stimulation, and 5-HT perfusion was 12 Hz, 6.8 Hz, 6.8 Hz, and 1 Hz, respectively.
Immunostaining in transgenic flies
For immunofluorescence, adult flies within 5–12 days were dissected in PBS, then subject in 4% PFA and fixed on ice for 1–4 hours. The brains were washed with the Washing Buffer (3% NaCl, 1% Triton X-100 in PBS) for three times, with 10 min for each time. Samples were transferred to Penetration/Blocking Buffer (2% Triton-X100, 10% Normal Goat Serum in PBS) for 20 hours at 4 °C on the shaker. Samples were then transferred into solution with primary antibody diluted with Dilution Buffer (0.25% Triton-X100, 1% Normal Goat Serum in PBS) for 24 hours at 4 °C. After washing three times, samples were incubated with the secondary antibody for 14–20 hours at 4 °C on the shaker. At last, samples were washed three times and mounted with 50% glycerol. The primary and secondary antibodies were rabbit anti-mCherry (1:500; Abcam Cat#ab167453, RRID: AB_2571870) and AlexFlour647 goat anti-rabbit (1:500; AAT Bioquest, Cat#16710). An inverted Ti-E A1 confocal microscope (Nikon) was used for immunofluorescence imaging. The confocal microscope was equipped with a 20×/0.75 NA air objective. A 640nm laser and 663/738 nm emission filter were used for this experiment.
Two-photon imaging in mice
Fluorescence imaging in mice was performed using an Olympus two-photon microscope FV1000 equipped with a Spectra-Physics Mai Tai Ti: Sapphire laser. The excitation wavelength was 920-nm, and fluorescence was collected using a 495–540-nm filter. To perform the imaging in head-fixed mice, part of the mouse scalp was removed, and the underlying tissues and muscles were carefully removed to expose the skull. A metal recording chamber was affixed to the skull surface with glue followed by a thin layer of dental cement to strengthen the connection. One to two days later, the skull above the prefrontal cortex was carefully removed, taking care to avoid the major blood vessels. AAVs expressing GRAB5-HT1.0 or GRAB5-HTmut was injected as above described. A custom-made 4 mm x 4 mm square coverslip was placed over the exposed PFC and secured with glue. After surgery, mice were allowed to recover for at least three weeks. The mice were then fixed to the base and allowed to habituate for 2–3 days. During the experiment, drugs were administered by i.p. injection and the sampling rate was 0.1 Hz.
Fiber-photometry recording in mice
To monitor 5-HT release in various brain regions during the sleep-wake cycle, AAVs expressing GRAB5-HT1.0 or GRAB5-HTmut were injected via a glass pipette into the BF, OFC, and BNST using a Nanoject II (Drummond Scientific). An optical fiber (200 μm, 0.37 NA) with FC ferrule was carefully inserted at the same coordinates used for virus injection. The fiber was affixed to the skull surface using dental cement. After surgery, the mice were allowed to recover for at least one week. The photometry rig was constructed using parts obtained from Doric Lens, including a fluorescence optical mini cube (FMC4_AE(405)_E(460–490)_F(500–550)_S), a blue LED (CLED_465), an LED driver (LED_2), and a photoreceiver (NPM_2151_FOA_FC). To record GRAB5-HT1.0 and GRAB5-HTmut fluorescence signals, a beam of excitation light was emitted from an LED at 20 μW, and the optical signals from GRAB5-HT1.0 and GRAB5-HTmut were collected through optical fibers. For the fiber-photometry data, a software-controlled lock-in detection algorithm was implemented in the TDT RZ2 system using the fiber-photometry “Gizmo” in the Synapse software program (modulation frequency: 459 Hz; low-pass filter for demodulated signal: 20 Hz, 6th order). The photometry data were collected with a sampling frequency of 1017 Hz. The recording fiber was bleached before recording to eliminate autofluorescence from the fiber, and the background fluorescence intensity was recorded and subtracted from the recorded signal during data analysis.
EEG and EMG recordings
Mice were anesthetized with isoflurane (5% induction; 1.5–2% maintenance) and placed on a stereotaxic frame with a heating pad. For EEG, two stainless steel miniature screws were inserted in the skull above the visual cortex, and two additional steel screws were inserted in the skull above the frontal cortex. For EMG, two insulated EMG electrodes were inserted in the neck musculature, and a reference electrode was attached to a screw inserted in the skull above the cerebellum. The screws in the skull were affixed using thick dental cement. All experiments were performed at least one week after surgery. TDT system-3 amplifiers (RZ2 and PZ5) were used to record the EEG and EMG signals; the signal was passed through a 0.5-Hz high-pass filter at and digitized at 1526 Hz.
Quantification and statistical analysis
Animals or cells were randomly assigned into control or experimental groups. Data collection and analysis were not performed blind to the conditions of the experiments and no data were excluded for the analysis. No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those reported in previous publications (ref 8–11). Imaging data from cultured HEK293T cells, cultured rat cortical neurons, acute mouse brain slices, transgenic flies, and head-fixed mice were processed using ImageJ (1.52p) software (NIH) and analyzed using custom-written MATLAB (R2020a) programs. Traces were plotted using Origin 2020 (2020b). Exponential function fitting in Origin was used to correct for slight photobleaching of the traces in Fig. 2i,n,o,p,r,s and Extended Data Fig. 6c,f–h. In Fig. 2n–p and Extended Data Fig. 6b,e the background levels measured outside the ROI of the pseudocolor images were removed using ImageJ. Imaging data in head-fixed mice were corrected using motion correction algorism (EZcalcium) to correct the movement artifacts 39.
For the fiber-photometry data analysis, the raw data were binned into 1-Hz bins (i.e., down-sampled by 1000), and background autofluorescence was subtracted. For calculating ΔF/F0, a baseline value was obtained by fitting the autofluorescence-subtracted data with a 2nd order exponential function. Slow drift was removed from the z-score‒transformed ΔF/F0 using the MATLAB script “BEADS” with a cut-off frequency of 0.00035 cycles/sample (https://www.mathworks.com/matlabcentral/fileexchange/49974-beads-baseline-estimation-and-denoising-with-sparsity). To quantify the change in 5-HT fluorescence across multiple animals, the z-score‒transformed ΔF/F0 was further normalized using the standard deviation of the signal measured during REM sleep (when there was no apparent fluctuation in the signal), yielding a normalized z-score. This normalized z-score was used for the analysis in Fig. 3a–g.
For EEG and EMG data analysis, Fast Fourier transform (FFT) was used to perform spectral analysis with a frequency resolution of 0.18 Hz. Brain state was classified semi-automatically in 5-s epochs using a MATLAB GUI and then validated manually by trained experimenters. Wakefulness was defined as desynchronized EEG activity combined with high EMG activity, and NREM sleep was defined as synchronized EEG activity combined high-amplitude delta activity (0.5–4 Hz) combined with low EMG activity; and REM sleep was defined as high power at theta frequencies (6–9 Hz) combined with low EMG activity.
Except where indicated otherwise, all summary data are reported as the mean ± s.e.m. The signal to noise ratio (SNR) was calculated as the peak response divided by the standard deviation of the baseline fluorescence. Data distribution was assumed to be normal, and equal variances were formally tested. Two-tailed Student’s t-test, one-way ANOVA test were performed. *p<0.05, **p<0.01, ***p<0.001, n.s. p>0.05. The exact p-value is specified in the legends. Cartoons in Fig. 3a,d,f,h are created with BioRender.com.
Reporting Summary
Further information on research design is available in the Life Sciences Reporting Summary linked to this article.
Data Availability
Plasmids for expressing the sensors used in this study were deposited at Addgene (https://www.addgene.org/140552/). Human GPCR cDNA library (hORFeome database 8.1, http://horfdb.dfci.harvard.edu/index.php?page=home). Source data for Figures 1–3 and Extended Data Figures 1 and 4–6 are provided with this paper.
Code Availability
EZcalcium algorism and BEADS baseline estimation and denoising with sparsity algorism are available from https://github.com/porteralab/EZcalcium, https://www.mathworks.com/matlabcentral/fileexchange/49974-beads-baseline-estimation-and-denoising-with-sparsity.
Extended Data
Extended Data Fig. 1.
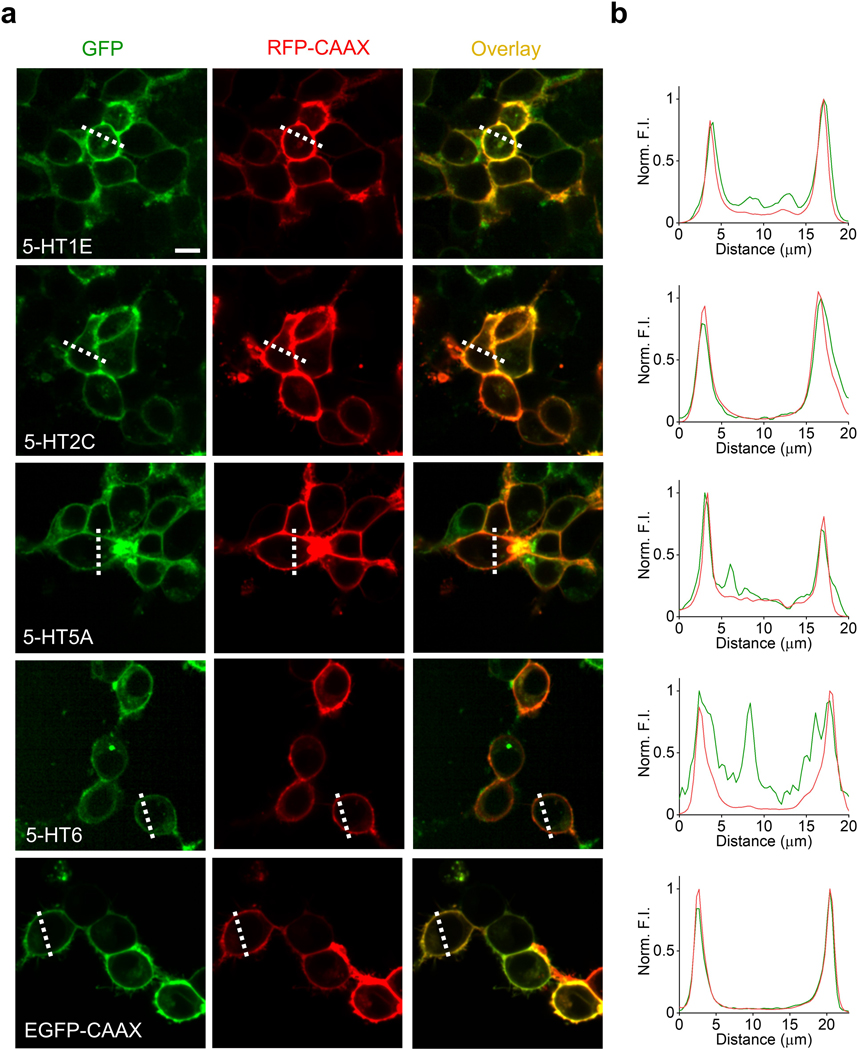
Characterization of the membrane trafficking for 5-HT receptor-based chimeras. a, Representative fluorescence images of HEK293T cells co-expressing the indicated 5-HT receptors fused with cpGFP (green) and RFP-CAAX (red); EGFP-CAAX was used as a positive control. Similar results were observed for more than 100 cells. Scale bar, 10 μm. b, Normalized fluorescence intensity measured at the white dashed lines shown in (a) for each candidate sensor.
Extended Data Fig. 2.
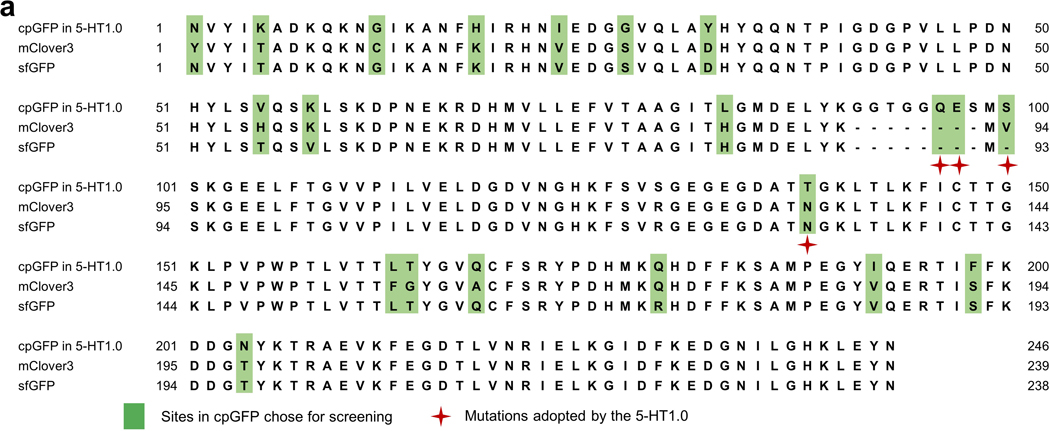
Sequence alignment of cpGFP from 5-HT1.0 sensor, sfGFP, and mClover3.a, The sequence of cpGFP from the 5-HT1.0 sensor, sfGFP, and mClover3 are aligned. Amino acids in the cpGFP chose for optimization are labeled with light green color, and the mutations adopted by the 5-HT1.0 sensor are indicated with red stars.
Extended Data Fig. 3.
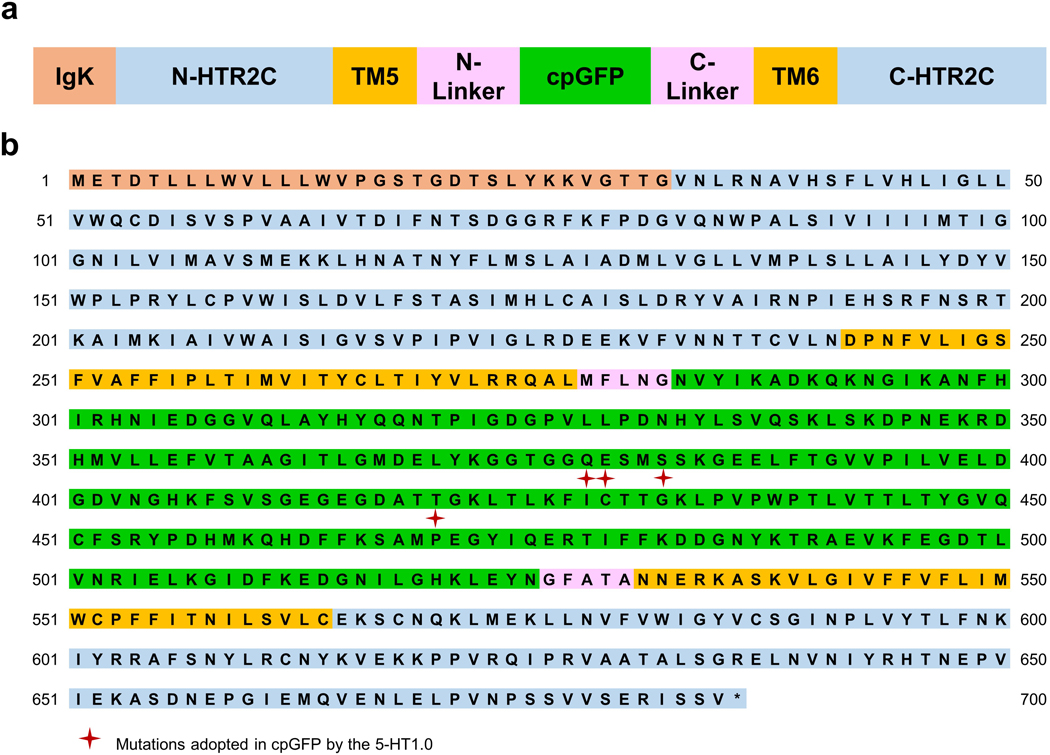
The amino acid sequence of 5-HT1.0 a, Schematic representation of the 5-HT1.0 structure. For simplicity, TM1–4, TM7, and H8 are not shown.
b, The amino acid sequence of the 5-HT1.0 sensor after three steps of evolution. The mutated amino acids in cpGFP (cpGFP from GCaMP6s, see Chen, T.W., et al. 2013.) are indicated with red stars.
Extended Data Fig. 4.
Further characterization of GRAB5-HT in cultured HEK293T cells and rat cortical neurons. a, Representative fluorescence and pseudocolor images of HEK293T cells expressing 5-HT1.0 or 5-HTmut before (left) and after (right) application of 10 μM 5-HT. Similar results were observed for more than 10 cells. Scale bar, 20 μm.
b,c, Representative fluorescence traces and group summary of the peak response in HEK293T cells expressing 5-HT1.0 or 5-HTmut; n = 14 and15 cells from 3 cultures for 5-HT1.0 and 5-HTmut group. Two-tailed Student’s t-test was performed. P = 8.18 ×10−12 between 5-HT1.0 and 5-HTmut group.
d, 5-HT dose-response curves measured in cells expressing 5-HT1.0 or 5-HTmut, the EC50 for 5-HT1.0 is shown. n = 3 wells per group with 300–500 cells per well.
e, Representative normalized fluorescence measured in HEK293T cells expressing 5-HT1.0, EGFP-CAAX, or iGluSnFR during continuous exposure to 488-nm laser (power: 350 μW).
f, Summary of the decay time constant calculated from the photobleaching curves shown in (e). n = 10/3, 14/3, and 12/3 for 5-HT1.0, EGFP-CAAX, and iGluSnFR, respectively. Two-tailed Student’s t-test was performed. P = 2.45 ×10−9, 1.90 ×10−9, 3.05 ×10−8, and 7.22 ×10−7 between EGFP-CAAX and iGluSnFR without or with Glu, and 5-HT1.0 without or with 5-HT. P = 4.43 ×10−8 and 7.78 ×10−6 between iGluSnFR without or with Glu and 5-HT1.0 without 5-HT. P = 4.62 ×10−8 and 7.05 ×10−6 between iGluSnFR without or with Glu and 5-HT1.0 with 5-HT.
g, Summary of the brightness measured in HEK293T cells expressing 5-HT1.0 or 5-HT2C -EGFP in the absence or presence of 10 μM 5-HT, normalized to the 5-HT2C-EGFP+5-HT group; n = 3 wells per group with 300–500 cells per well.
h,i, Intracellular calcium was measured in cells expressing 5-HT1.0 or the 5-HT2C receptor and loaded with the red fluorescent calcium dye Cal590. Representative traces are shown in (h), and the peak responses are plotted against 5-HT concentration in (i); n = 15/3 for each group.
j,k, Fluorescence response of 5-HT1.0 expressing cells to 5-HT perfusion for two hours. Representative fluorescence images (j) and the summary data (k) showing the response to 10 μM 5-HT applied at 30 min intervals to cells expressing 5-HT1.0; n = 3 wells per group with 100–300 cells per well. Scale bar, 20 μm. F4,10 = 0.888, P = 0.505 for 0 min, 30 min, 60 min, 90 min and 120 min by one-way ANOVA.
l, Left, the Gs-coupled cAMP level was detected by pink-Flamindo with or without 5-HT1.0 sensor expression. The exemplar fluorescence response traces of pink-Flamindo without (top) or with 5-HT1.0 sensor (bottom) expression, when treated with 50 μM 5-HT or 50 μM 5-HT+10 μM Forskolin. Right, quantification data for left. n=23/3, 23 cells from 3 cultures for each group. Two-tailed Student’s t-test was performed. P = 0.084 and P = 0.488 for 5-HT and 5-HT+FSK group.
m, Buffering effects of the 5-HT1.0 sensor by luciferase complementation assay. Luminescence signals were measured when treated with different concentrations of 5-HT (left) or 5-HT2C receptor specific agonist CP809101 (right) with or without co-expression of 5-HT1.0 sensor with 5-HT2C receptor. The luminescence signal of cells treated with the control buffer is normalized to 1. Data of 5-HT induced G-protein signaling in 5-HT2C receptor expression group were re-plotted from Fig. 1f. n = 3 wells per group with 100–300 cells per well. Two-tailed Student’s t-test was performed. P = 0.693, 0.0402, 0.993, 0.340, 0.0618, 0.0691 and 0.127 between 5-HT1.0 and 5-HT1.0 + 5-HT2C with 10−4, 10−5, 10−6, 10−7, 10−8, 10−9, and 10−10 M 5-HT. P = 0.733, 0.801, 0.346, 0.998, 0.304 and 0.380 between 5-HT1.0 and 5-HT1.0 + 5-HT2C with 10−4, 10−5, 10−6, 10−7, 10−8 and 10−9 M CP809101.
n, Cultured rat cortical neurons expressing the 5-HTmut sensor were imaged before (left) and after (middle) 5-HT application. These insets in the left and middle fluorescence images show the region with increased contrast. The pseudocolor image on the right shows the change in fluorescence of 5-HTmut in response to 10 μM 5-HT. Similar results were observed for more than 10 neurons. Scale bar, 20 μm.
o,p, Representative trace (o) and group summary (p) of cultured neurons expressing 5-HT1.0 in response to indicated compounds at 10 μM each; in (p), Met was applied where indicated; n = 9/3. Two-tailed Student’s t-test was performed. P = 6.74 ×10−22, 1.09 ×10−22, 1.27 ×10−21, 3.33 ×10−22, and 0.0939 between 5-HT1st and DA, NE, His, ACh and 5-HT2nd. P = 1.97 ×10−11 between 5-HT2nd and Met.
Data are shown as the mean ± s.e.m. in b-d, f, g, i, k-m, p, with the error bars or shaded regions indicating s.e.m., ∗p<0.05, ** p<0.01, ***p<0.001, and n.s., not significant.
Extended Data Fig. 5.
Probing endogenous 5-HT release in mouse brain slices. a, Schematic diagram depicting the acute mouse brain slice preparation, with AAV-mediated expression of 5-HT1.0 in the hippocampus.
b, Representative fluorescence images of the 5-HT1.0 sensor expressed in the mouse hippocampal neurons of brain slices in ACSF (left) and 50 μM 5-HT (right). Similar results were observed from 4 slices. Scale bar, 50 μm.
c, A magnified view of the rectangular region in (b) showing the 5-HT1.0 sensor response to exogenously applied 50 μM 5-HT; left, fluorescence image; right, corresponding pseudocolor image indicating ΔF/F0. The arrowheads indicate somata. Scale bar, 25 μm.
d, Representative traces (left) and quantification (right) of peak ΔF/F0 of the 5-HT1.0 sensor in response to 50 μM 5-HT from a single soma or neurite (n=4 slices from 1 mouse). Two-tailed Student’s t-test was performed. P = 0.0226 between soma and neurite.
e, Left, schematic diagram depicting the acute mouse brain slice preparation, with AAV-mediated expression of 5-HT1.0 in the DRN. Middle and right, fluorescence traces (middle) and group data (right) of the change in 5-HT1.0 fluorescence in response to 10 electrical stimuli applied at the indicated frequencies; n = 7 slices from 5 mice.
f, Summary of the change in 5-HT1.0 fluorescence in response to 6 trains of electrical stimuli (20 pulses at 20 Hz) delivered at 5-min intervals. The responses are normalized to the first train; n = 8 slices from 5 mice. F5,42 = 1.18, P = 0.335 for 0 min, 5 min, 10 min, 15 min, 20 min, and 25 min by one-way ANOVA.
g,h, Representative fluorescence image, pseudocolor images (g), fluorescence traces (h, left), and group data (h, right) of 5-HT1.0 fluorescence in response to perfusion of 5-HT, 5-HT+Halo, and 5-HT+Met; n = 4 slices from 3 mice for each group. Two-tailed Student’s t-test was performed. P = 0.0816 between 5-HT and Halo. P = 0.00297 between 5-HT and Met.
i, Left, representative FSCV data of 5-HT release in DRN. A specific 5-HT waveform (0.2 V to 1.0 V and ramped down to −0.1 V, and back to 0.2 V at a scan rate of 1000 V/s) was applied to the CFME at a frequency of 10 Hz. Right, current vs time traces are extracted at a horizontal white dashed line shows an immediate increase in 5-HT response after electrical stimulation (20 pulses, 2 ms pulse width, 64 Hz). A cyclic voltammogram (inset) is extracted at the vertical black dashed line shows oxidation and reduction peaks at 0.8 V and 0 V, respectively.
j, Left, group data of fluorescence response in 5-HT1.0-expressing DRN neurons to electrical stimuli with varied frequencies delivered at 20 pulses. Right, average data of peak 5-HT concentration measured by FSCV at varied stimulating frequencies delivered at 20 pulses; n = 11 neurons from 9 mice.
Data are shown as the mean ± s.e.m. in d-f, h, j, with the error bars or shaded regions indicating s.e.m., *p<0.05, ** p<0.01, ***p<0.001, and n.s., not significant.
Extended Data Fig. 6.
Probing endogenous 5-HT release in Drosophila in vivo a, Schematic drawing showing in vivo two-photon imaging of a Drosophila, with the stimulating electrode positioned near the mushroom body (MB).
b,c, Representative pseudocolor images (b), fluorescence traces, and group summary (c) of the change in 5-HT1.0 fluorescence in the MB horizontal lobe in response to 40 electrical stimuli at 15 Hz in control (saline) or 10 μM Met; n = 9 flies for each group. Two-tailed Student’s t-test was performed. P = 2.36 ×10–5 between saline and Met. Scale bar, 10 μm.
d, Fluorescence images measured in the MB of flies expressing 5-HT1.0 or 5-HTmut; the β’ lobe is indicated. Scale bar, 10 μm.
e-i, Representative pseudocolor images (e), fluorescence traces (f-h), and group summary (i) of 5-HT1.0 and 5-HTmut in the MB β’ lobe measured in response to a 1-s odor application, a 0.5-s body shock, and application of 100 μM 5-HT; n = 14, 12 and10 flies for 5-HT1.0 group under odor, body shock and perfusion conditions; n = 9, 5 and 9 flies for 5-HTmut group under odor, body shock and perfusion conditions. Two-tailed Student’s t-test was performed. P = 1.14 ×10−5, P = 0.00273, P = 8.93 ×10−5 between 5-HT1.0 and 5-HT mut under odor, body shock and perfusion conditions.
j,k, Quantification data of the τon, τoff (j), and area under the calcium transient curves (k) in the main Fig. 2r; n = 11 and 10 flies for 5-HT1.0+ and 5-HT1.0- group. Two-tailed Student’s t-test was performed. P = 0.497 for calcium signal between two groups. P = 0.710 for τon and P = 0.307 for τoff.
Data are shown as the mean ± s.e.m. in c, i-k, with the error bars or shaded regions indicating s.e.m., *p<0.05, ** p<0.01, ***p<0.001, and n.s., not significant.
Supplementary Material
Statistical Source Data
Statistical Source Data
Statistical Source Data
Statistical Source Data
Statistical Source Data
Statistical Source Data
Statistical Source Data
Acknowledgments
We thank Y. Rao for providing the two-photon microscope, X. Lei at PKU-CLS and the National Center for Protein Sciences at Peking University for support and assistance with the Opera Phenix high-content screening system. We thank D. Lin and X. Xu for the critical reading of the Manuscript. This work was supported by the Beijing Municipal Science & Technology Commission (Z181100001318002), the Beijing Brain Initiative of Beijing Municipal Science & Technology Commission (Z181100001518004), Guangdong Grant “Key Technologies for Treatment of Brain Disorders” (2018B030332001), the General Program of National Natural Science Foundation of China (projects 31671118, 31871087, and 31925017), Science Fund for Creative Research Groups of the National Natural Science Foundation of China (81821092), the NIH BRAIN Initiative (NS103558), grants from the Peking-Tsinghua Center for Life Sciences and the State Key Laboratory of Membrane Biology at Peking University School of Life Sciences (to Y.L.), the Shanghai Municipal Science and Technology Major Project (2018SHZDZX05 to M.X.), and the Shanghai Pujiang Program (18PJ1410800 to M.X.), Peking University Postdoctoral Fellowship (J.F.), Alzheimer’s Association Postdoctoral Research Fellowship (AARF 19 619387 to P.Z.), Peking-Tsinghua Center Excellence Postdoctoral Fellowship (Y.Z.), and Beijing Nova Program (Z201100006820100 to M.J.).
Footnotes
Competing Interests Statement
The authors declare competing financial interests. J.W., M.J., J.F, and Y. L have filed patent applications whose value might be affected by this publication.
References
- 1.Lesch KP, et al. Association of anxiety-related traits with a polymorphism in the serotonin transporter gene regulatory region. Science 274, 1527–1531 (1996). [DOI] [PubMed] [Google Scholar]
- 2.Li Y, et al. Serotonin neurons in the dorsal raphe nucleus encode reward signals. Nat Commun 7, 10503 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Portas CM, et al. On-line detection of extracellular levels of serotonin in dorsal raphe nucleus and frontal cortex over the sleep/wake cycle in the freely moving rat. Neuroscience 83, 807–814 (1998). [DOI] [PubMed] [Google Scholar]
- 4.Vaswani M, Linda FK & Ramesh S. Role of selective serotonin reuptake inhibitors in psychiatric disorders: a comprehensive review. Prog Neuropsychopharmacol Biol Psychiatry 27, 85–102 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Fuller RW Uptake inhibitors increase extracellular serotonin concentration measured by brain microdialysis. Life Sci 55, 163–167 (1994). [DOI] [PubMed] [Google Scholar]
- 6.Bunin MA, Prioleau C, Mailman RB & Wightman RM Release and uptake rates of 5-hydroxytryptamine in the dorsal raphe and substantia nigra reticulata of the rat brain. J Neurochem 70, 1077–1087 (1998). [DOI] [PubMed] [Google Scholar]
- 7.Candelario J & Chachisvilis M. Mechanical Stress Stimulates Conformational Changes in 5-Hydroxytryptamine Receptor 1B in Bone Cells. Cellular and Molecular Bioengineering 5, 277–286 (2012). [Google Scholar]
- 8.Jing M, et al. A genetically encoded fluorescent acetylcholine indicator for in vitro and in vivo studies. Nat Biotechnol 36, 726–737 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patriarchi T, et al. Ultrafast neuronal imaging of dopamine dynamics with designed genetically encoded sensors. Science 360 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun F, et al. A Genetically Encoded Fluorescent Sensor Enables Rapid and Specific Detection of Dopamine in Flies, Fish, and Mice. Cell 174, 481–496 e419 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng J, et al. A Genetically Encoded Fluorescent Sensor for Rapid and Specific In Vivo Detection of Norepinephrine. Neuron 102, 745–761 e748 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bajar BT, et al. Improving brightness and photostability of green and red fluorescent proteins for live cell imaging and FRET reporting. Sci Rep 6, 20889 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pedelacq JD, Cabantous S, Tran T, Terwilliger TC & Waldo GS Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol 24, 79–88 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Peng Y, et al. 5-HT2C Receptor Structures Reveal the Structural Basis of GPCR Polypharmacology. Cell 172, 719–730 e714 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ballesteros JA & Weinstein H.J.M.i.N. [19] Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. 25, 366–428 (1995). [Google Scholar]
- 16.Harada K, et al. Red fluorescent protein-based cAMP indicator applicable to optogenetics and in vivo imaging. Sci Rep 7, 7351 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wan Q, et al. Mini G protein probes for active G protein-coupled receptors (GPCRs) in live cells. J Biol Chem 293, 7466–7473 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barnea G, et al. The genetic design of signaling cascades to record receptor activation. Proc Natl Acad Sci U S A 105, 64–69 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishimura K, et al. Quantitative analysis of the distribution of serotonin-immunoreactive cell bodies in the mouse brain. Neurosci Lett 91, 265–270 (1988). [DOI] [PubMed] [Google Scholar]
- 20.Waddell S, Armstrong JD, Kitamoto T, Kaiser K & Quinn WG The amnesiac gene product is expressed in two neurons in the Drosophila brain that are critical for memory. Cell 103, 805–813 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Lee PT, et al. Serotonin-mushroom body circuit modulating the formation of anesthesia-resistant memory in Drosophila. Proc Natl Acad Sci U S A 108, 13794–13799 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keene AC, et al. Diverse odor-conditioned memories require uniquely timed dorsal paired medial neuron output. Neuron 44, 521–533 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Yu D, Keene AC, Srivatsan A, Waddell S & Davis RL Drosophila DPM neurons form a delayed and branch-specific memory trace after olfactory classical conditioning. Cell 123, 945–957 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Xu M, et al. Basal forebrain circuit for sleep-wake control. Nat Neurosci 18, 1641–1647 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ren J, et al. Anatomically Defined and Functionally Distinct Dorsal Raphe Serotonin Sub-systems. Cell 175, 472–487 e420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oikonomou G, et al. The Serotonergic Raphe Promote Sleep in Zebrafish and Mice. Neuron 103, 686–701 e688 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ren J, et al. Single-cell transcriptomes and whole-brain projections of serotonin neurons in the mouse dorsal and median raphe nuclei. Elife 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rudnick G & Wall SC The molecular mechanism of “ecstasy” [3,4-methylenedioxy-methamphetamine (MDMA)]: serotonin transporters are targets for MDMA-induced serotonin release. Proc Natl Acad Sci U S A 89, 1817–1821 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bicks LK, Koike H, Akbarian S & Morishita H. Prefrontal Cortex and Social Cognition in Mouse and Man. Front Psychol 6, 1805 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liechti ME, Saur MR, Gamma A, Hell D & Vollenweider FX Psychological and Physiological Effects of MDMA (“Ecstasy”) after Pretreatment with the 5-HT2 Antagonist Ketanserin in Healthy Humans. Neuropsychopharmacology 23, 396–404 (2000). [DOI] [PubMed] [Google Scholar]
- 31.Hagino Y, et al. Effects of MDMA on Extracellular Dopamine and Serotonin Levels in Mice Lacking Dopamine and/or Serotonin Transporters. Curr. Neuropharmacol. 9, 91–95 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Q, Shui B, Kotlikoff MI & Sondermann H. Structural Basis for Calcium Sensing by GCaMP2. Structure 16, 1817–1827 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kroeze WK, et al. PRESTO-Tango as an open-source resource for interrogation of the druggable human GPCRome. Nat Struct Mol Biol 22, 362–369 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pfeiffer BD, Truman JW & Rubin GM Using translational enhancers to increase transgene expression in Drosophila. Proc Natl Acad Sci U S A 109, 6626–6631 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dana H, et al. Sensitive red protein calcium indicators for imaging neural activity. Elife 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6, 343–345 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Yusa K, et al. Targeted gene correction of alpha1-antitrypsin deficiency in induced pluripotent stem cells. Nature 478, 391–394 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shin M & Venton BJ Electrochemical Measurements of Acetylcholine-Stimulated Dopamine Release in Adult Drosophila melanogaster Brains. Anal Chem 90, 10318–10325 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cantu DA, et al. EZcalcium: Open-Source Toolbox for Analysis of Calcium Imaging Data. Front Neural Circuits 14, 25 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Statistical Source Data
Statistical Source Data
Statistical Source Data
Statistical Source Data
Statistical Source Data
Statistical Source Data
Statistical Source Data
Data Availability Statement
Plasmids for expressing the sensors used in this study were deposited at Addgene (https://www.addgene.org/140552/). Human GPCR cDNA library (hORFeome database 8.1, http://horfdb.dfci.harvard.edu/index.php?page=home). Source data for Figures 1–3 and Extended Data Figures 1 and 4–6 are provided with this paper.