Abstract
Polycomb repression complex 1 (PRC1) is a multiprotein assembly that regulates transcription. The Polycomb group ring finger 1 protein (PCGF1) is central in the assembly of the noncanonical PRC1 variant called PRC1.1 through its direct interaction with BCOR (BCL-6-interacting corepressor) or its paralog, BCOR-like 1 (BCORL1). Previous structural studies revealed that the C-terminal PUFD domain of BCORL1 is necessary and sufficient to heterodimerize with the RAWUL domain of PCGF1 and, together, form a new protein–protein binding interface that associates with the histone demethylase KDM2B. Here, we show that the PUFD of BCOR and BCORL1 differ in their abilities to assemble with KDM2B. Unlike BCORL1, the PUFD of BCOR alone does not stably assemble with KDM2B. Rather, additional residues N-terminal to the BCOR PUFD are necessary for stable association. Nuclear magnetic resonance (NMR) structure determination and 15N T2 relaxation time measurements of the BCOR PUFD alone indicate that the termini of the BCOR PUFD, which are critical for binding PCGF1 and KDM2B, are disordered. This suggests a hierarchical mode of assembly whereby BCOR PUFD termini become structurally ordered upon binding PCGF1, which then allows stable association with KDM2B. Notably, BCOR internal tandem duplications (ITDs) leading to pediatric kidney and brain tumors map to the PUFD termini. Binding studies with the BCOR ITD indicate the ITD would disrupt PRC1.1 assembly, suggesting loss of the ability to assemble PRC1.1 is a critical molecular event driving tumorigenesis.
Graphical Abstract
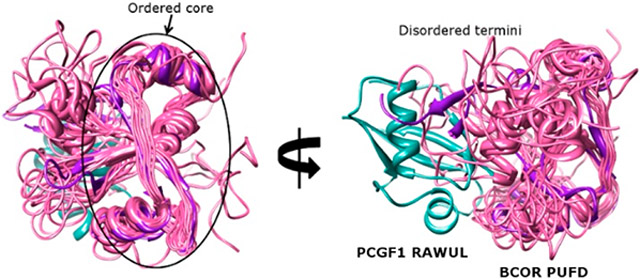
The Polycomb group (PcG) proteins make up a complex family of transcriptional regulators that play multiple roles in gene regulation, including the temporal and spatial control of gene expression.1,2 A multiprotein PcG assembly called Polycomb repression complex 1 (PRC1), first identified in Drosophila,3,4 has four core members with a limited number of homologues. In humans, the existence of multiple homologues for each of the core members has resulted in the existence of many PRC1 variants. Functional distinction of each of these variants is largely determined by the presence of one of six PCGF proteins.5 PRC1.2 and PRC1.4 (which contain PCGF2 and PCGF4, respectively) are most similar to Drosophila PRC1 and are considered the canonical forms of the complex. PRC1.1, which includes PCGF1, is a noncanonical form because PCGF1 directly interacts with non-PcG proteins BCOR or BCORL1 and KDM2B. KDM2B is a histone demethylase, one of two enzymatic properties of PRC1.1, the other being the histone H2A ubiquitination activity mediated by the heterodimer between the PcG proteins RING1 or RNF2 and PCGF1.
PRC1.1 complexes can be formed with either BCOR or the related BCORL1, suggesting that the two proteins might be redundant. While this may be true in some instances, the two proteins have regions of unique coding sequence and distinct expression profiles that suggest non-overlapping functions. For example, mutations in BCOR or BCORL1 in the germline cause distinct human syndromes6-8 while mutations in hematopoietic cells in either BCOR or BCORL1 have been shown to be associated with acute myelogenous leukemias.9,10 Of interest to this study is the identification of in-frame internal tandem duplications (ITDs) in BCOR observed in clear cell sarcoma of the kidney (CCSK),11,12 primitive neuroectodermal tumors of the central nervous system (CNS-PNET),13 and other tumors.14 Such alterations have not yet been reported for BCORL1.
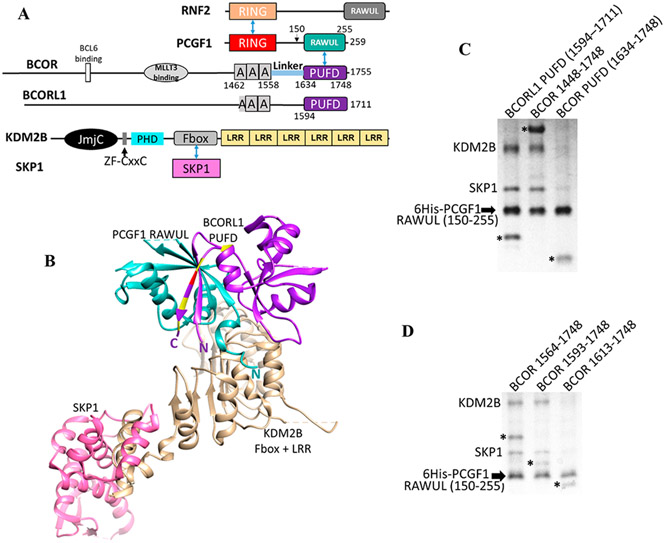
All of the tumor-associated ITDs result in the insertion of 20–40 residues within the PUFD domain of BCOR11-13 (Figure 1A). More specifically, the ITDs all occur within the N- or C-terminal β-strands of the PUFD15 (Figure 1B) that together form an intermolecular antiparallel β-sheet with the PCGF1 RAWUL.16 We have previously shown that hetero-dimerization between PCGF1 and the PUFD of BCORL1 is critical in PRC1.1 assembly because PCGF1 and BCORL1 together form the protein interaction surface that binds the leucine rich repeats (LRRs) of KDM2B17 (Figure 1B,C). Thus, assembly of PRC1.1 appears to proceed in an ordered manner whereby PCGF1 and BCORL1 interact and then, together, bind KDM2B.
Figure 1.
PRC1.1, BCOR, and BCORL1. (A) Domain structure of PRC1.1 components. Blue arrows indicate direct protein–protein interactions between domains. The “A” within BCOR and BCORL1 is an ankyrin domain. (B) Interaction of the PCGF1 RAWUL/BCORL1 PUFD with KDM2B17 (PDB entry 5JH5) showing the importance of the N-terminus of PCGF1 RAWUL and both termini of the BCORL1 PUFD (termini labeled N and C). Residue positions where ITDs occur within BCOR are colored yellow and red at the equivalent positions on the BCORL1 structure. The red residue is the site of the ITD used in this study. (C and D). PCGF1 RAWUL [residues 150–255 with an N-terminal hexahistidine (6His) tag] was co-expressed in bacteria with KDM2B/SKP1 and different BCOR proteins. Components were assembled on the Ni2+ resin via the 6His tag on PCGF1 RAWUL. The KDM2B used in all experiments in this study contains the Fbox and the LRR domains (residues 1059–1336). Full-length SKP1 binds the Fbox domain of KDM2B and was required for KDM2B stabilization. BCORL1 and BCOR proteins in the gels are denoted with an asterisk. (C) Comparison between the PUFDs of BCORL1 (lane 1, positive control) and BCOR (lane 3). The presence of the BCOR band in lane 3 indicates the ability of the BCOR PUFD to dimerize with PCGF1. The absence of KDM2B/SKP indicates the inability to assemble the tetramer. Though the BCORL1 and BCOR PUFDs have similar numbers of residues, the difference in migration for the BCOR and BCORL1 PUFDs can be ascribed to the BCOR PUFD being slightly smaller in mass (13.6 kDa) than BCORL1 PUFD (14.0 kDa) and having a lower pI (4.59 for the BCOR PUFD vs 5.26 for the BCORL1 PUFD). (D) Residues N-terminal to the BCOR PUFD are required for assembly with KDM2B. When BCOR 1593–1748 is co-expressed (lane 2), a positive assembly signal is indicated by the presence of KDM2B/SKP1, which is absent when BCOR 1613–1748 (lane 3) or BCOR 1634–1748 (C, lane 3) is expressed. Thus, BCOR 1593–1748 is necessary and sufficient for KDM2B assembly. All expression constructs were identical except for BCOR. Equal amounts of the lysate of bacteria expressing all components were introduced onto Ni2+ resin. The resin was washed; equal amounts of the eluted contents were subjected to SDS–PAGE.
Here, we sought to dissect the differences, if any, between the PUFDs of BCOR and BCORL1 and their ability to assemble with KDM2B. Our studies reveal the importance of a specific residue present in BCORL1, but not BCOR, and the PUFD in general in PRC1.1 assembly. In addition, we show that the BCOR PUFD termini are disordered and critical for PRC1.1 assembly whereas a BCOR PUFD domain with an ITD failed to assemble. These results further emphasize the important role of the RAWUL domain of PCGF1 in stabilizing the PUFD and thereby allowing the key contacts to be made with KDM2B.
EXPERIMENTAL METHODS
Protein Preparation.
All genes for proteins used in this study were cloned into and expressed using T7 promoter vectors (e.g., pET) and expressed in BL21-Gold(DE3) cells pretransformed with pRARE to provide additional tRNAs that are less available in bacteria. Cells were grown in LB, and harvested cells were typically resuspended in 50 mM Tris (pH 8.0), 150 mM NaCl, 10 mM imidazole (pH 7.5), 5 mM βME, and 1 mM PMSF and lysed by sonication. The proteins were purified from the soluble lysate using Ni2+ affinity chromatography. All PCGF1/BCOR and PCGF1/BCORL1 dimers were further purified using gel filtration chromatography. For all other proteins, a leader sequence housing the hexahistidine tag was proteolyzed by tobacco etch virus protease (TEV) followed by a second Ni2+ affinity chromatography step in which the nonbinding portion was collected. The proteins were further purified using ion exchange and gel filtration chromatography.
Ni2+ Pull-Down Assay.
For panels C and D of Figure 1 and Figure 2C, all components were co-expressed in BL21-Gold(DE3) cells pretransformed with pRARE. Inductions were performed at 15 °C. Harvested cells from 1.5 mL cultures were resuspended with 150 μL of lysis buffer [50 mM Tris (pH 8.0), 200 mM NaCl, 20 mM imidazole (pH 7.5), 1 mM CaCl2, 1 mM MgCl2, 1 mM PMSF, and 1.25 mg/mL hen egg white lysozyme], and lysed via several freeze/thaw cycles. DNaseI was used to shear the DNA. Soluble lysates were incubated with 15 μL of Ni2+-sepharose beads, washed with 50 mM Tris (pH 8.0), 200 mM NaCl, and 20 mM imidazole (pH 7.5), and then eluted with a 500 mM imidazole (pH 7.5), 200 mM NaCl buffer. Additional details of the experiments can be found in the text and figure legends.
Figure 2.
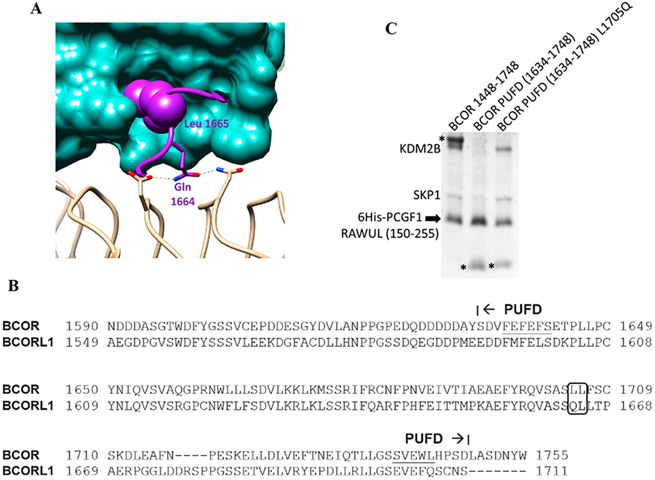
BCOR PUFD L1705Q rescues KDM2B binding. (A) Close-up view showing the important role of BCORL1 Gln 1664. PCGF1 is colored sea green, BCORL1 purple, and KDM2B beige. BCORL1 Leu 1665 inserts into the Leu cage within the PCGF1 RAWUL, thereby stabilizing and allowing Gln 1664 to contact KDM2B. Both Gln 1664 and Leu 1665 are necessary for stable interaction with KDM2B.17 (B) Sequence comparison of the BCOR and BCORL1 PUFD. The BCOR residue at the position equivalent to BCORL1 Gln 1664 is leucine (L1705). (C) BCOR PUFD with the Leu1705Gln mutation rescues its ability to assemble with KDM2B. Pull down performed as in panels C and D of Figure 1. Lane 1 serves as the positive control; a single-amino acid change within the BCOR PUFD (L1705Q) rescues PRC1.1 assembly as indicated by the presence of KDM2B/SKP1 in lane 3 and its absence from lane 2.
Biolayer Interferometry.
The Octet RED96e (Pall ForteBio) biolayer interferometry instrument was used to measure the affinities. The anti-penta-His1K biosensor was used to immobilize the KDM2B/SKP1 dimer via interaction with the hexahistidine tag on KDM2B. Titrations were performed using varying concentrations of PCGF1 RAWUL (150–259) bound to various BCOR/BCORL1 proteins. The running buffer consisted of 20 mM Tris (pH 8.0) and 150 mM NaCl. As a control to determine the effect, if any, of the orientation of the immobilized KDM2B/SKP1 dimer, the complementary experiment was performed by immobilizing a biotin-labeled PCGF1 150–259/BCOR 1593–1748 onto a streptavidin biosensor (SAX) and introducing KDM2B/SKP1 in the mobile phase. The two KD values were comparable (106 nM vs 84 nM). The BLItz biolayer interferometry instrument (Pall ForteBio) was used to measure the affinities between KDM2B/SKP1 and BCOR proteins (BCOR linker 1559–1634 and BCOR 1448–1748). The amino groups of the KDM2B/SKP1 dimer were labeled with biotin using EZ-Link NHs-PEG4-Biotin (Thermofisher), and the dimer was immobilized on the High Precision Streptavidin (SAX) biosensors.
BCOR PUFD Nuclear Magnetic Resonance (NMR).
The BCOR PUFD used for the NMR studies included BCOR residues 1634–1748 with Cys 1649 and 1682 mutated to Ser. Expression and purification were as described above. All NMR experiments were performed at 300 K using either a Bruker 700 MHz spectrometer fitted with a conventional 1H/13C/15N probe or a Bruker 600 MHz spectrometer fitted with a cryogenically cooled 1H/13C/15N probe. For heteronuclear NMR studies, the proteins were expressed in minimal medium containing M9 salts, MgSO4, CaCl2, biotin, thiamine hydrochloride, FeCl2, ZnCl2, yeast extract, 15NH4Cl, and either unlabeled glucose or [13C]glucose. Signal assignments were performed on 13C- and 15N-labeled BCOR PUFD in 10 mM Tris (pH 8.0), 50 mM NaCl, 1 mM TCEP, and a 5% D2O solution and prepared to a concentration of 3 mM. Resonance assignments for backbone nuclei (1HN, 15N, 13C′, 13Cα, and 13Cβ) were made using HNCACB and CBCA(CO)NH experiments. Backbone assignments were confirmed using HNCO and HN(CA)CO experiments. Side chain carbons were assigned using CCONH experiments, while side chain protons were assigned using HCCH TOCSY. Two-dimensional 1H–13C HSQC spectra of biosynthetic fractionally 13C-labeled protein were used for the stereospecific assignment of the side chain methyl groups (valine and leucine) as previously described.18 15N NOESY-HSQC and 13C NOESY-HSQC data were collected to provide structural restraints. All NMR data were processed using NMRPipe19 and analyzed using NMRView.20 Structure calculations were performed using CNS version 1.221 and Aria2.3.22,23 Structural calculations incorporated nuclear Overhauser effects (NOEs), dihedral angle restraints calculated by TALOS,24 and hydrogen bonds. For dynamics studies, backbone amide HSQC-based 15N transverse (T2) relaxation times were measured using standard CPMG-based pulse programs from Bruker with 8–10 15N T2 delay times. Relative peak intensities over the relaxation series were derived using the nonLinLS module of NMRPipe, and the resulting decaying exponentials were fit to derive the T2 values. Error estimates were derived by Monte Carlo analysis.
RESULTS
The PCGF1 RAWUL/BCOR PUFD Dimer Does Not Stably Assemble with KDM2B.
The PUFDs of BCOR and BCORL1 are each capable of forming a heterodimeric complex with the RAWUL of PCGF1.16 To test whether the BCOR PUFD, like the BCORL1 PUFD, can form a four-component BCOR/PCGF1/KDM2B/SKP1 complex, we performed in vitro pull-down experiments with bacterially produced proteins. We co-expressed a hexahistidine-tagged PCGF1 RAWUL domain, KDM2B 1059–1336, which includes the Fbox domain and six tandem LRRs (hereafter KDM2B), the Fbox binding protein SKP1 that directly binds and stabilizes KDM2B, and a variant of either BCOR or BCORL1 PUFD domains. Assembly of all four components into a single complex is indicated by the presence of the proteins in the SDS–PAGE gel of the Ni2+ affinity purification of hexahistidine-tagged PCGF1. The BCORL1 PUFD is capable of assembly with the three other proteins as previously shown (Figure 1C, lane 1),17 which is indicated by the appearance of all four proteins. However, when the BCOR PUFD (1634–1748) was co-expressed with the other components, dimerization with PCGF1 RAWUL was indicated by presence of the BCOR PUFD in the gel but the level of binding of KDM2B/SKP1 was significantly reduced (lane 3). When BCOR was extended to residues 1448–1748, which encompasses the ankyrin (ANK) repeats, the “linker”, and the PUFD, it is capable of assembly in the four-component complex (Figure 1C, lane 2). Given that the PCGF1 RAWUL/BCOR 1448–1748 heterodimer is capable of mediating assembly with KDM2B and thus SKP1, but not when only the BCOR PUFD is used, we sought to determine the minimal region in the C-terminus of BCOR that is required for KDM2B assembly. We tested BCOR deletion variants in the pull-down assay. BCOR 1564–1748 and 1593–1748 both were capable of assembly with KDM2B and SKP1, while neither was visible when using BCOR 1613–1748 (Figure 1D). This result indicates that unlike BCORL1, BCOR requires up to 50 additional residues N-terminal to the PUFD for stable association with KDM2B/SKP1.
BCOR PUFD Leu1705Gln Rescues the Ability to Assemble with KDM2B.
Structure analysis of the BCORL1 PUFD in complex with PCGF1 RAWUL and KDM2B revealed a potential basis for the reduced affinity of the BCOR PUFD for KDM2B.17 In the PUFD of BCORL1, the side chain of Leu 1665 is encased within a hydrophobic pocket of PCGF1 RAWUL, which helps to stabilize the adjacent Gln 1664 allowing its side chain to make hydrogen bond contacts with KDM2B (Figure 2A). Mutation of either BCORL1 Gln 1664 or Leu 1665 disrupts the binding of the BCORL1 PUFD to KDM2B.17 While the residue equivalent to BCORL1 Leu 1665 is conserved in BCOR (Leu 1706), the residue equivalent to BCORL1 Gln 1664 in BCOR is not (Leu 1705) (Figure 2B). We wondered whether the BCOR PUFD would assemble with KDM2B/SKP1 if Leu 1705 were changed to Gln, allowing the key contacts with KDM2B. BCOR PUFD Leu1705Gln was indeed able to rescue the assembly with KDM2B/SKP1 (Figure 2C). This is consistent with a model in which BCOR Leu 1705 reduces the binding affinity for KDM2B relative to that of BCORL1 but the general architecture of the binding surfaces is preserved. Combined with the results depicted in Figure 1D, the reduced affinity of the BCOR PUFD suggests that the contacts made by the residues N-terminal to the BCOR PUFD help compensate for this reduced affinity of the BCOR PUFD and still allow for assembly with KDM2B/SKP1.
BCOR ANK, a Linker, and the PUFD Contribute to the Binding of KDM2B.
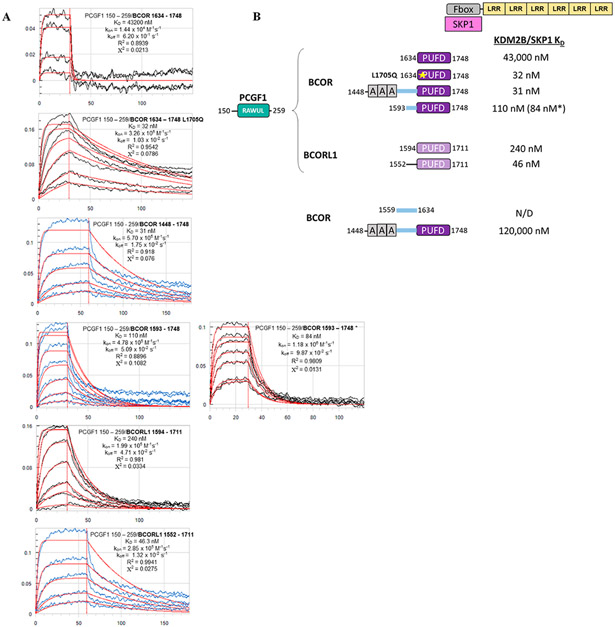
We wished to measure the contributions made by the different regions of BCOR to the assembly with KDM2B/SKP1. To determine the regions outside of the BCOR PUFD that provide KDM2B binding energy, we used biolayer interferometry (BLI) to measure the affinities of the PCGF1 RAWUL dimer for various BCOR/BCORL1 proteins binding to the KDM2B/SKP1 heterodimer (Figure 3A,B). The results of the affinity measurements indicate that the ANK repeats, the linker connecting the ANK repeats and the PUFD, and the PUFD itself, all contribute to the binding of KDM2B. In agreement with the pull-down experiments presented above, PCGF1 RAWUL/BCOR PUFD only exhibited a weak affinity for KDM2B/SKP1 with an equilibrium dissociation constant (KD) of 43000 nM. Including part of the linker with the PUFD (1593–1748) greatly increased the affinity for KDM2B/SKP1 (KD ~ 100 nM). This result is consistent with published data,25 which revealed the importance of the linker for KDM2B binding. Including the ANK repeats along with the linker region (1448–1748) further increased the affinity (KD ~ 31 nM). Consistent with the pull-down experiments depicted in Figure 2C, the BCOR PUFD Leu1705Gln mutant exhibited affinity for KDM2B/SKP1 binding comparable to that of BCOR region 1448–1748. As expected for BCORL1, its PUFD alone is able to stably associate with KDM2B/SKP1 (KD ~ 240 nM). Like BCOR, additional regions outside of the BCORL1 PUFD also make contributions to KDM2B/SKP1 binding as indicated by the lower KD for BCORL1 1552–1711 (region equivalent to BCOR 1593–1748). Thus, the interaction between BCOR and KDM2B/SKP1 is critically dependent on the BCOR linker region, while the interaction between BCORL1 and KDM2B/SKP1 is not.
Figure 3.
Binding affinity measurements between the dimer of PCGF1 RAWUL/PUFD-containing protein combinations and the KDM2B/SKP1 dimer using biolayer interferometry (BLI). (A) Raw BLI titration data. The KDM2B/SKP1 dimer was immobilized. The dimer of PCGF1 RAWUL bound to the indicated BCOR and BCORL1 variants introduced in the mobile phase. (B) Summary of the KD values recorded in panel A. KD values are consistent with Ni2+ pull-down data. For example, consistent with Figure 2B, PCGF1 RAWUL bound to BCOR PUFD exhibits a very low affinity (43000 nM) for KDM2B/SKP1 but affinity is regained with the L1705Q mutant (32 nM). An asterisk indicates the experiment was conducted in the reverse orientation; i.e., the biotin-tagged PCGF1/BCOR dimer was immobilized, and KDM2B/SKP1 introduced in the mobile phase, which helps to ensure the reliability of the method.
Our results showing the added contribution of the BCOR linker region to KDM2B affinity are consistent with prior coimmunoprecipitation (co-IP) studies in human embryonic stem cells.25 This study showed that BCOR with the linker deleted does not assemble with KDM2B. In addition, BCOR with the PUFD deleted (Δ1634–1748) was also capable of KDM2B binding. These results suggest that the linker by itself may be solely responsible for binding KDM2B. We attempted to measure the affinity between the BCOR linker (1559–1634) and KDM2B/SKP1 in the absence of PCGF1 RAWUL. We were unable to detect even nominal binding between the BCOR linker and KDM2B/SKP1. BCOR 1448–1748 without PCGF1 RAWUL does exhibit a stronger affinity with a measured KD of ~120 μM (R2 = 0.9899; X2 = 1.565) for the interaction with KDM2B/SKP1. While considerably stronger than that of the linker alone, this value remains far weaker compared to when PCGF1 is present and bound to BCOR 1448–1748 (31 nM).
Together, the affinity measurements are consistent with the model in which the PUFD, from either BCOR or BCORL1, binds the PCGF1 RAWUL and places the linker residues and ANK domains in place to contact KDM2B. Additional pull-down data presented below for a BCOR ITD further support this model.
The BCOR PUFD β-Sheet That Binds PCGF1 Is Disordered in the Absence of PCGF1.
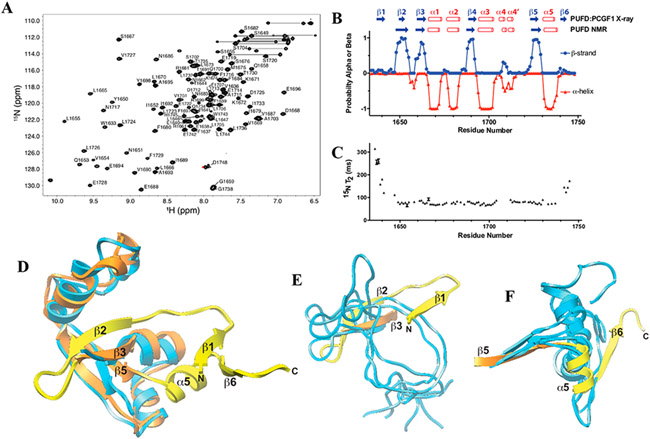
The inability of BCOR alone to bind KDM2B can in part be explained by the structure and dynamics studies of the BCOR PUFD using multidimensional NMR methods (Figure 4 and Table 1). The 1H–15N HSQC spectrum of the 15N-labeled BCOR PUFD exhibited widely dispersed backbone amide signals, which were indicative of a structured protein and were easily assigned (Figure 4A). The secondary structure predicted using the protein energetic conformational analysis from NMR chemical shifts (PECAN) server using the sequence information and assigned chemical shifts (Figure 4B) mostly matched that of the BCOR PUFD crystal structure.16 The exceptions were the N- and C-terminal β-strands, β1 and β6, for which the PECAN probabilities were close to zero (Figure 4B, top). The PUFD termini are critical for association with PCGF1 RAWUL. The crystal structure of PCGF1 RAWUL in complex with the PUFD of either BCOR or BCORL1 shows the PUFD termini form the major interface with PCGF1 RAWUL, whereby the β-sheet region of the RAWUL is augmented by the β-strands formed by the PUFD termini.16 The RNF2 RAWUL binding partners that create similar augmented β-sheets are disordered when not bound to RNF2 RAWUL.27,28 We measured the backbone 15N T2 relaxation times from the relative peak intensities across a series of two-dimensional (2D) 1H–15N shift correlation spectra with an increase in the Carr–Purcell–Meiboom–Gill (CPMG) relaxation times (Figure 4C). The T2 relaxation times are increased for the N-terminus (residues 1634–1648) and for the C-terminus (residues 1740—1748), and these regions correspond closely with the unstructured terminal regions predicted by PECAN. Structure determination of the BCOR PUFD, based on analysis of nuclear Overhauser effects (NOEs), torsion angles, and hydrogen bonds, supports the disordered nature of the BCOR PUFD termini. While the core of the BCOR PUFD aligns well between the NMR and crystal structure (Figure 4D), the termini of the lowest-energy NMR structures show the lack of a uniform structure, and none of these overlay with the termini in the BCOR PUFD:PCGF1 RAWUL crystal structure (in panels E and F of Figure 4, compare blue NMR structures to the yellow crystal structure).
Figure 4.
NMR structure and dynamics of BCOR PUFD. (A) Heteronuclear 1H–15N shift correlation spectrum of the BCOR PUFD (S1634–D1748) in 10 mM Tris-HCl, 50 mM NaCl, 1 mM TCEP (pH 8.0), and 5% D2O with labeled backbone amide signals. (B) Secondary structure probabilities deduced from the assigned chemical shifts using the program PECAN.26 A comparison with the BCOR PUFD secondary structure observed in the BCOR PUFD:PCGF1 RAWUL complex crystal structure (PDB entry 4HPL) is shown at the top. (C) BCOR PUFD backbone 15N T2 relaxation times derived by measuring relative peak intensities in 2D 1H–15N shift correlation spectra at eight different T2 relaxation times and by fitting these to a single exponential. Errors shown were estimated on the basis of Monte Carlo analysis of the measured relative intensities for each residue. The shorter relaxation times measured for residues in the core serve as internal controls for the longer times measured for the terminal residues. (D) Overlay of one representative model of the BCOR PUFD NMR solution structure (blue) on the BCOR component of the PCGF1 RAWUL:BCOR PUFD crystal structure over the structurally ordered core. The ensemble of 10 lowest-energy BCOR PUFD NMR structures has a pairwise RMSD over this region of 2.54 Å (Table 1) and as shown is in good agreement with the PCGF1 RAWUL:BCOR PUFD crystal structure (RMSD of 2.68 Å for the comparison shown). Regions of the BCOR PUFD of the crystal structure that extend beyond the core are colored yellow. (E and F) Same overlay of the BCOR PUFD cores as in panel D, but showing just the (E) N-terminus and (F) C-terminus of five representative BCOR PUFD solution structure models.
Table 1.
NMR Structure Statistics for the BCOR PUFD
| no. of NMR distance constraints | |
| total NOE | 1212 |
| intraresidue | 588 |
| sequential (∣i – j∣ = 1) | 308 |
| medium-range (∣i – j∣ < 4) | 132 |
| long-range (∣i – j∣ > 5) | 184 |
| statistics for the ensemble of accepted structures | |
| violations (mean and standard deviation) | |
| distance constraints (0.5 Å) | 1 ± 1 |
| J coupling constraints (1 Hz) | 0 ± 0 |
| maximum distance constraint violation (Å) | 0.54 ± 0.03 |
| deviations from idealized geometry | |
| bond lengths (Å) | 0.040 ± 0.0002 |
| bond angles (deg) | 0.59 ± 0.03 |
| impropers (deg) | 1.56 ± 0.09 |
| average pairwise RMSD (Å) | |
| structurally ordereda | |
| backbone N, Cα, CO | 3.37 |
| heavy atoms | 4.39 |
| secondary structure | |
| backbone N, Cα, CO | 2.54 |
| heavy atoms | 3.52 |
Structurally ordered residues were defined as residues with T2 relaxation times of <100 ms (residues 1653–1737).
The disordered BCOR PUFD termini have important consequences for PRC1.1 assembly. Upon dimerization with PCGF1 RAWUL, the structural stabilization of the PUFD termini could also affect the conformation of the linker region so that it is positioned for a more favorable KDM2B interaction. It is worth noting that 17 residues beyond the N-terminus of the PCGF1 RAWUL were also necessary for formation of a stable complex with KDM2B.16 Thus, for both PCGF1 and BCOR/BCORL1, regions N-terminal to the highly conserved RAWUL and PUFD domains contribute to the affinity for KDM2B.
The BCOR ITD Hinders PRC1.1 Assembly.
The ITDs observed in pediatric kidney and brain tumors11-13 occur within the disordered termini of the BCOR PUFD (Figure 1A,B). Given the location of the ITD and the importance of the PUFD termini in binding PCGF1 RAWUL, we tested whether the PUFD ITD disrupts the binding to PCGF1 RAWUL and, consequently, PRC1.1 assembly. We used the Ni2+ affinity pull-down assay to test binding. We mixed an equal volume of the bacterial lysate expressing just the 6His-PCGF1 RAWUL with different purified BCOR proteins (Figure 5A, left gel). This mixture was incubated on a Ni2+ resin and washed, and then the bound assembly eluted from the resin and was viewed via SDS–PAGE (Figure 5A, right gel). When no PRC1.1 component is added to the PCGF1 RAWUL-expressing lysate, only a small fraction of 6His-PCGF1 RAWUL binds to and is eluted from the Ni2+ resin (Figure 5A, lane 1). This reflects the mostly insoluble state of PCGF1 RAWUL when expressed alone in bacteria. The addition of the purified wild-type BCOR PUFD to the lysate results in induced stabilization of the RAWUL as indicated by the presence of the bound wild-type PUFD and the more abundant PCGF1 RAWUL (in Figure 5A, compare PCGF1 bands in lanes 1 and 2). For the ITD experiments, we used the ITD in which 30 BCOR residues (1712–1741) are duplicated and placed in tandem. This ITD has been observed in both CCSK (noted as CCSK1912 and 382T11) and CNS-PNET (noted as MB_150047 and CNS-PNET_15–031913). The addition of BCOR CCSK19 showed significantly decreased levels of binding and stabilization of PCGF1 RAWUL (in Figure 5A, compare lanes 2 and 3). Next, we tested whether the BCOR ITD also disrupts PRC1.1 assembly. Introducing KDM2B/SKP1 along with wild-type BCOR to the PCGF1 RAWUL lysate resulted in stronger synergistic stabilization of all PRC1.1 components (Figure 5A, lane 4) where all components showed greater intensity. BCOR CCSK19 was far less capable of this stabilization (in Figure 5A, compare lanes 4 and 5). These results provide support for the essential role the PUFD plays in PRC1.1 assembly. Despite using BCOR 1448–1748, which exhibits the highest affinity (Figure 3), perturbing just the BCOR PUFD with the ITD is sufficient to hinder PCGF1 binding and PRC1.1 assembly.
Figure 5.
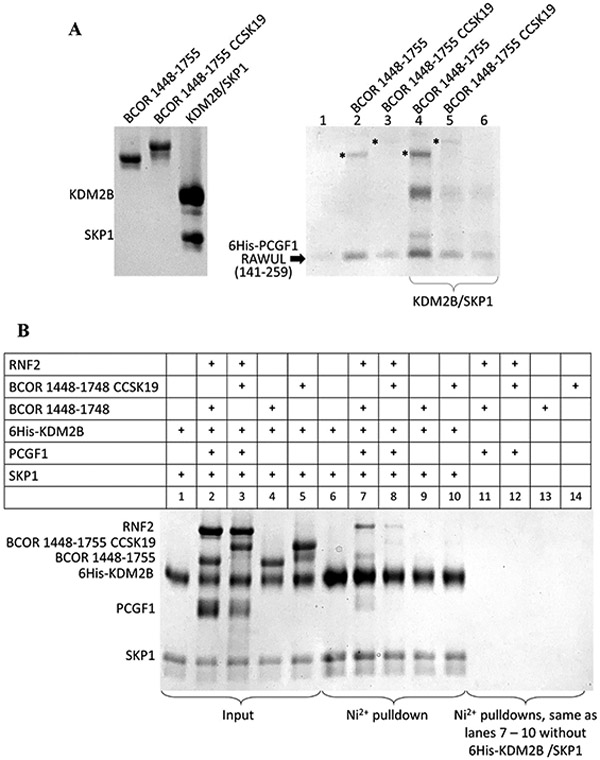
BCOR ITD disrupts PRC1.1 assembly. SDS–PAGE of Ni2+ affinity pull-down assays. (A) SDS–PAGE of the purified proteins (input) used for incubation with the 6His-PCGF1 RAWUL (140–255)-expressing bacterial lysate (left). SDS–PAGE of the Ni2+ affinity pull-down assay (right). Equal volumes of the bacterial lysate that expressed 6His-PCGF1 RAWUL were incubated with purified, non-6His-tagged proteins (labeled above the lanes and shown in the left gel) to a final concentration of 3 μM for 30 min prior to binding to the Ni2+-sepharose beads. Washing and elution were the same as described above. Because equal amounts of bacterial lysates were used for each of the lanes, any enrichment observed in the SDS–PAGE indicates a mutual stabilization of the entire assembly bound to the Ni2+-sepharose resin. For lanes 4–6, the purified KDM2B/SKP1 dimer (final concentration of 3 μM) was also added in addition to the proteins indicated above the gel. An asterisk indicates the BCOR proteins. (B) All proteins used were not from lysates but rather from purified protein solutions prepared at a final concentration of 5 μM in 120 μL of 50 mM Tris (pH 8.0), 200 mM NaCl, 50 mM imidazole (pH 7.5), and 10 mM βME and allowed to incubate at room temperature for 30 min. The protein solutions were then introduced onto 17 βL of Ni2+-sepharose beads, washed, and eluted as described above. The assembled proteins were immobilized on the Ni2+ column via the hexahistidine (6His)-tagged KDM2B/SKP1 dimer. Lanes 6–10 show elutions of the mixtures shown in lanes 1–5, respectively. The lower intensity of the BCOR band in lane 7 compared to the other pull-down figures presented in this study is likely the result of the alternative, non-optimal orientation of the Ni2+ immobilization utilized in this experiment where the hexahistine tag is at the N-terminus of KDM2B. The other pull-down experiments utilized the hexahistine tag on PCGF1 RAWUL.
To determine whether regions beyond the PCGF1 RAWUL affect PRC1.1 assembly, we next tested the assembly of the five-component complex using full-length PCGF1 and RNF2 proteins. We separately isolated the trimer of 6His-PCGF1/RNF2 along with BCOR 1448–1748 (RPB) and tested its binding to the KDM2B/SKP1 dimer. The wild-type RPB trimer was easily isolated from heterologous bacterial co-expression of all three components. We attempted similar expression and purification of RPB with BCOR 1448–1748 CCSK19. As with wild-type RPB, an initial Ni2+ affinity purification and cleavage of the hexahistidine-containing leader sequence on PCGF1 was performed. Unlike with wild-type RPB, however, the subsequent Ni2+ affinity purification to separate the cleaved and uncleaved fractions resulted in extensive separation of BCOR CCSK19 from the other components. Despite this, an elution peak isolated from the second Ni2+ affinity chromatography purification that contained all three components was isolated. We did not perform any further purification of this sample, i.e., size exclusion chromatography, fearing further disassembly of the trimer. Thus, while this isolation contained RNF2, PCGF1, and BCOR CCSK19, we could not confirm that the components were stably assembled in a single complex and may have resulted in smaller amounts of PCGF1 (Figure 5B, lane 3). Despite this, we proceeded to mix both the BCOR WT and the CCSK19 RPBs with the KDM2B/SKP1 dimer, which were then immobilized on a Ni2+ column via the N-terminal hexahistidine-tagged (6His) species on KDM2B. The resin was washed, and the assembled contents were eluted and then viewed via SDS–PAGE (Figure 5B). The more intense bands for RNF2, BCOR, and PCGF1 proteins in the pull-down study indicate far better assembly with KDM2B/SKP1 for wild-type BCOR compared to when using BCOR CCSK19 (in Figure 5B, compare lanes 7 and 8), although BCOR CCSK19 was still capable of some assembly. Though we cannot be certain that the less intense bands observed for BCOR CCSK19 are the result of smaller amounts of PCGF1 in the mixture, the inability of BCOR CCSK19 to stably form the trimer and its hindered ability to assemble on the Ni2+ column support the model in which BCOR CCSK19 hinders dimerization with the PCGF1 RAWUL, which, consequently, disrupts PRC1.1 assembly.
DISCUSSION
The key findings of this study are as follows. First, the PUFD of both BCOR and BCORL1 is critical for PRC1.1 assembly, though there are key structural differences in their interaction with PCGF1. Second, the PUFD termini of BCOR, which are the key structural components for binding PCGF1, are disordered in the absence of PCGF1. Third, a BCOR ITD occurring within the PUFD termini hinders binding to PCGF1 RAWUL and, consequently, PRC1.1 assembly.
Our data suggest that the PUFD of both BCOR and BCORL1 is necessary for the interaction with PCGF1 and KDM2B. While the PUFD alone of BCORL1 is necessary and sufficient to bind PCGF1 RAWUL and together they bind KDM2B,17 the BCOR PUFD lacks a key Gln side chain at residue 1705 that precludes BCOR PUFD alone from similarly assembling with KDM2B. The PUFD linker residues compensate for the lack of sufficient binding energy for interaction of KDM2B with the BCOR PUFD.
The importance of the C-terminal regions of both BCOR (ANK, linker, and PUFD) and KDM2B (Fbox and LRR) in PRC1.1 assembly has previously been demonstrated. In studies conducted in HEK293 cells, BCOR ANK, the linker, and the PUFD were necessary to immunoprecipitate both full-length KDM2B and a truncated KDM2B missing its N-terminal JmjC domain.29 In hESC cells, a BCOR construct housing just the ANK, the linker, and the PUFD could assemble with KDM2B while a full-length BCOR that lacked the same C-terminal region could not.25 Interestingly, BCOR with the PUFD deleted was still able to co-immunoprecipitate with KDM2B but did so in the absence of PCGF1 and RNF2. This result suggests the possibility that alternative binding modes may exist for the interaction between BCOR and KDM2B that does not involve the BCOR PUFD. To this point, when full-length PCGF1/RNF2 is expressed in insect cells, this dimer can bind to full-length KDM2B in the absence of BCOR.30 These discrepancies in the relative importance of the linker and PUFD of BCOR in PRC1.1 assembly could stem from additional cellular factors that affect PRC1.1 assembly that are absent in the in vitro studies.
For PRC1.1 assembly that does occur via the dimerization between the PUFD and PCGF1 RAWUL, the data presented here further indicate that PRC1 assembly occurs in an ordered manner. The lack of a structure to the PUFD termini would preclude the PUFD alone from even making nominal contacts with KDM2B. Thus, to bind KDM2B, the PUFD must first bind PCGF1 RAWUL, allowing the residues of the PUFD termini to be placed in conformations that are necessary to interact with the LRR of KDM2B. The interaction with PCGF1 RAWUL not only stabilizes the BCOR PUFD termini but also would better position the linker residues for KDM2B interactions. The importance of sequences beyond the strict boundaries associated with the structured PUFD also extends to the PCGF1 RAWUL as residues N-terminal to the RAWUL make direct contacts with KDM2B.17
The importance of the BCOR PUFD in PRC1.1 assembly has important clinical implications. The BCOR ITDs observed in CCSK and CNS-PNET all occur within the BCOR PUFD termini. The BCOR CCSK19 ITD studied here has 30 duplicated residues placed within the BCOR PUFD C-terminal strand required to bind PCGF1 (Figure 1B). We find that this BCOR ITD severely restricts PRC1.1 assembly by hindering binding to PCGF1. This in turn hinders KDM2B association. To the best of our knowledge, this is the first evidence of the molecular consequence of the BCOR ITDs that lead to pediatric tumors. Given the location of the other ITDs all occurring within or very near the BCOR PUFD termini, the inability to assemble PRC1.1 is likely to be the common molecular consequence for all other ITDs.
The disruption of PRC1.1 is also clinically relevant to lymphoma therapy. Malignant transformation of B cells leading to lymphoma is promoted by BCOR-associated repression of differentiation and cell cycle checkpoint genes. A recent study suggests that inhibiting BCL6 recruitment of PRC1.1 is a potential therapeutic strategy for the treatment of lymphoma.31 KDM2B, however, is also involved in recruiting PRC1.1. Thus, blocking assembly of PRC1.1 with a small molecule inhibitor may serve as an additional therapeutic strategy for lymphoma. Our biochemical and structural studies highlight two interaction interfaces that could be targeted for disruption: (i) the interface between the BCOR PUFD and PCGF1, which is required for KDM2B binding, and (ii) the interface between the BCOR linker region and KDM2B that provides additional affinity.
ACKNOWLEDGMENTS
The authors thank Richard Yip for assistance with the biolayer interferometry experiments.
Funding
S.J.W. was supported by the National Institutes of Health (NIH) (F31GM099418). C.A.K. was supported by the Robert A. Welch Foundation (AQ-1813) and the National Institute of General Medical Sciences of the NIH under Grant R01GM114338. V.J.B. was supported by the NIH (R01CA071540, R01HD084459) and funds from the Minnesota Masonic Charities, and the University of Minnesota Medical School. A.P.H. was supported by the NIH (GM58670) and the Robert A. Welch Foundation (AQ-1842). This study utilized the Biomolecular NMR Core Laboratory at the University of Texas Health Science Center at San Antonio, which is supported by the Office of the Vice President for Research and the Mays Cancer Center through National Cancer Institute Cancer Center Support Grant P30 CA054174.
ABBREVIATIONS
- BCOR
BCL6 corepressor
- BCORL1
BCL6 corepressor-like 1
- CBX
chromobox
- CCSK
clear cell sarcoma of the kidney
- H2Aub
ubiquitinated histone H2A
- KDM2B
lysine demethylase 2B
- ITD
internal tandem duplication
- PcG
Polycomb group
- PRC1
Polycomb repression complex 1
- PCGF
Polycomb group ring finger
- PDB
Protein Data Bank
- PUFD
PCGF1 ubiquitin fold discriminator
- RAWUL
RING finger- and WD40-associated ubiquitin-like
- RING
really interesting new gene
- RMSD
root-mean-square deviation
- SDS–PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- SKP1
S-phase kinase-associated protein 1
Footnotes
The authors declare no competing financial interest.
Accession Codes
The BCOR PUFD structure was deposited as entry 2N1L. Chemical shift assignments have been deposited in the Biological Magnetic Resonance Data Bank as entry 25565. BCOR, Q6W2J9; BCORL1, Q5H9F3; KDM2B, Q8NHM5; PCGF1, Q9BSM1; SKP1, P63208; RNF2, Q99496.
Contributor Information
Sarah J. Wong, Department of Biochemistry, University of Texas Health Science Center at San Antonio, San Antonio, Texas 78229-3990, United States
Olga Senkovich, Department of Biochemistry and Molecular Genetics, Midwestern University, Glendale, Arizona 85308, United States.
Jason A. Artigas, Department of Biochemistry and Molecular Genetics, Midwestern University, Glendale, Arizona 85308, United States
Micah D. Gearhart, Department of Genetics, Cell Biology and Development, Masonic Cancer Center and Developmental Biology Center, University of Minnesota, Minneapolis, Minnesota 55455, United States
Udayar Ilangovan, Department of Biochemistry, University of Texas Health Science Center at San Antonio, San Antonio, Texas 78229-3990, United States.
David W. Graham, Department of Biochemistry and Molecular Genetics, Midwestern University, Glendale, Arizona 85308, United States
Kelsey N. Abel, Department of Biochemistry and Molecular Genetics, Midwestern University, Glendale, Arizona 85308, United States
Tianrong Yu, Department of Biochemistry and Molecular Genetics, Midwestern University, Glendale, Arizona 85308, United States.
Andrew P. Hinck, Department of Structural Biology, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania 15260, United States
Vivian J. Bardwell, Department of Genetics, Cell Biology and Development, Masonic Cancer Center and Developmental Biology Center, University of Minnesota, Minneapolis, Minnesota 55455, United States
Chongwoo A. Kim, Department of Biochemistry and Molecular Genetics, Midwestern University, Glendale, Arizona 85308, United States.
REFERENCES
- (1).Schuettengruber B, Bourbon HM, Di Croce L, and Cavalli G (2017) Genome regulation by polycomb and trithorax: 70 years and counting. Cell 171, 34–57. [DOI] [PubMed] [Google Scholar]
- (2).Simon JA, and Kingston RE (2013) Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell 49, 808–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Shao Z, Raible F, Mollaaghababa R, Guyon JR, Wu CT, Bender W, and Kingston RE (1999) Stabilization of chromatin structure by PRC1, a polycomb complex. Cell 98, 37–46. [DOI] [PubMed] [Google Scholar]
- (4).Francis NJ, Saurin AJ, Shao Z, and Kingston RE (2001) Reconstitution of a functional core polycomb repressive complex. Mol. Cell 8, 545–556. [DOI] [PubMed] [Google Scholar]
- (5).Gao Z, Zhang J, Bonasio R, Strino F, Sawai A, Parisi F, Kluger Y, and Reinberg D (2012) PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol. Cell 45, 344–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Ng D, Thakker N, Corcoran CM, Donnai D, Perveen R, Schneider A, Hadley DW, Tifft C, Zhang L, Wilkie AO, van der Smagt JJ, Gorlin RJ, Burgess SM, Bardwell VJ, Black GC, and Biesecker LG (2004) Oculofaciocardiodental and lenz microphthalmia syndromes result from distinct classes of mutations in BCOR. Nat. Genet. 36, 411–416. [DOI] [PubMed] [Google Scholar]
- (7).Shukla A, Girisha KM, Somashekar PH, Nampoothiri S, McClellan R, and Vernon HJ (2019) Variants in the transcriptional corepressor BCORL1 are associated with an X-linked disorder of intellectual disability, dysmorphic features, and behavioral abnormalities. Am. J. Med. Genet., Part A 179, 870–874. [DOI] [PubMed] [Google Scholar]
- (8).Ragge N, Isidor B, Bitoun P, Odent S, Giurgea I, Cogne B, Deb W, Vincent M, Le Gall J, Morton J, Lim D, Study DDD, Le Meur G, Zazo Seco C, Zafeiropoulou D, Bax D, Zwijnenburg P, Arteche A, Swafiri ST, Cleaver R, McEntagart M, Kini U, Newman W, Ayuso C, Corton M, Herenger Y, Jeanne M, Calvas P, and Chassaing N (2019) Expanding the phenotype of the X-linked BCOR microphthalmia syndromes. Hum. Genet. 138, 1051–1069. [DOI] [PubMed] [Google Scholar]
- (9).Grossmann V, Tiacci E, Holmes AB, Kohlmann A, Martelli MP, Kern W, Spanhol-Rosseto A, Klein HU, Dugas M, Schindela S, Trifonov V, Schnittger S, Haferlach C, Bassan R, Wells VA, Spinelli O, Chan J, Rossi R, Baldoni S, De Carolis L, Goetze K, Serve H, Peceny R, Kreuzer KA, Oruzio D, Specchia G, Di Raimondo F, Fabbiano F, Sborgia M, Liso A, Farinelli L, Rambaldi A, Pasqualucci L, Rabadan R, Haferlach T, and Falini B (2011) Whole-exome sequencing identifies somatic mutations of BCOR in acute myeloid leukemia with normal karyotype. Blood 118, 6153–6163. [DOI] [PubMed] [Google Scholar]
- (10).Li M, Collins R, Jiao Y, Ouillette P, Bixby D, Erba H, Vogelstein B, Kinzler KW, Papadopoulos N, and Malek SN (2011) Somatic mutations in the transcriptional corepressor gene BCORL1 in adult acute myelogenous leukemia. Blood 118, 5914–5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Roy A, Kumar V, Zorman B, Fang E, Haines KM, Doddapaneni H, Hampton OA, White S, Bavle AA, Patel NR, Eldin KW, John Hicks M, Rakheja D, Leavey PJ, Skapek SX, Amatruda JF, Nuchtern JG, Chintagumpala MM, Wheeler DA, Plon SE, Sumazin P, and Parsons DW (2015) Recurrent internal tandem duplications of BCOR in clear cell sarcoma of the kidney. Nat. Commun. 6, 8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Ueno-Yokohata H, Okita H, Nakasato K, Akimoto S, Hata J, Koshinaga T, Fukuzawa M, and Kiyokawa N (2015) Consistent in-frame internal tandem duplications of BCOR characterize clear cell sarcoma of the kidney. Nat. Genet. 47, 861–863. [DOI] [PubMed] [Google Scholar]
- (13).Sturm D, Orr BA, Toprak UH, Hovestadt V, Jones DT, Capper D, Sill M, Buchhalter I, Northcott PA, Leis I, Ryzhova M, Koelsche C, Pfaff E, Allen SJ, Balasubramanian G, Worst BC, Pajtler KW, Brabetz S, Johann PD, Sahm F, Reimand J, Mackay A, Carvalho DM, Remke M, Phillips JJ, Perry A, Cowdrey C, Drissi R, Fouladi M, Giangaspero F, Lastowska M, Grajkowska W, Scheurlen W, Pietsch T, Hagel C, Gojo J, Lotsch D, Berger W, Slavc I, Haberler C, Jouvet A, Holm S, Hofer S, Prinz M, Keohane C, Fried I, Mawrin C, Scheie D, Mobley BC, Schniederjan MJ, Santi M, Buccoliero AM, Dahiya S, Kramm CM, von Bueren AO, von Hoff K, Rutkowski S, Herold-Mende C, Fruhwald MC, Milde T, Hasselblatt M, Wesseling P, Rossler J, Schuller U, Ebinger M, Schittenhelm J, Frank S, Grobholz R, Vajtai I, Hans V, Schneppenheim R, Zitterbart K, Collins VP, Aronica E, Varlet P, Puget S, Dufour C, Grill J, Figarella-Branger D, Wolter M, Schuhmann MU, Shalaby T, Grotzer M, Van Meter T, Monoranu CM, Felsberg J, Reifenberger G, Snuderl M, Forrester LA, Koster J, Versteeg R, Volckmann R, Van Sluis P, Wolf S, Mikkelsen T, Gajjar A, Aldape K, Moore AS, Taylor MD, Jones C, Jabado N, Karajannis MA, Eils R, Schlesner M, Lichter P, Von Deimling A, Pfister SM, Ellison DW, Korshunov A, and Kool M (2016) New brain tumor entities emerge from molecular classification of CNS-PNETs. Cell 164, 1060–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Astolfi A, Fiore M, Melchionda F, Indio V, Bertuccio SN, and Pession A (2019) BCOR involvement in cancer. Epigenomics 11, 835–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Kim CA (2017) The Role of RAWUL and SAM in Polycomb Repression Complex 1 Assembly and Function. In Polycomb Group Proteins (Pirrotta V, Ed.) pp 5, Elsevier, Boston. [Google Scholar]
- (16).Junco SE, Wang R, Gaipa JC, Taylor AB, Schirf V, Gearhart MD, Bardwell VJ, Demeler B, Hart PJ, and Kim CA (2013) Structure of the polycomb group protein PCGF1 in complex with BCOR reveals basis for binding selectivity of PCGF homologs. Structure 21, 665–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Wong SJ, Gearhart MD, Taylor AB, Nanyes DR, Ha DJ, Robinson AK, Artigas JA, Lee OJ, Demeler B, Hart PJ, Bardwell VJ, and Kim CA (2016) KDM2B recruitment of the polycomb group complex, PRC1.1, requires cooperation between PCGF1 and BCORL1. Structure 24, 1795–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Neri D, Szyperski T, Otting G, Senn H, and Wuthrich K (1989) Stereospecific nuclear magnetic resonance assignments of the methyl groups of valine and leucine in the DNA-binding domain of the 434 repressor by biosynthetically directed fractional 13C labeling. Biochemistry 28, 7510–7516. [DOI] [PubMed] [Google Scholar]
- (19).Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, and Bax A (1995) NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293. [DOI] [PubMed] [Google Scholar]
- (20).Johnson BA, and Blevins RA (1994) NMR view: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4, 603–614. [DOI] [PubMed] [Google Scholar]
- (21).Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, and Warren GL (1998) Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr., Sect. D: Biol. Crystallogr. 54 (Part 5), 905–921. [DOI] [PubMed] [Google Scholar]
- (22).Linge JP, O’Donoghue SI, and Nilges M (2001) Automated assignment of ambiguous nuclear overhauser effects with ARIA. Methods Enzymol 339, 71–90. [DOI] [PubMed] [Google Scholar]
- (23).Bardiaux B, Malliavin T, and Nilges M (2012) ARIA for solution and solid-state NMR. Methods Mol. Biol. 831, 453–483. [DOI] [PubMed] [Google Scholar]
- (24).Cornilescu G, Delaglio F, and Bax A (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol NMR 13, 289–302. [DOI] [PubMed] [Google Scholar]
- (25).Wang Z, Gearhart MD, Lee YW, Kumar I, Ramazanov B, Zhang Y, Hernandez C, Lu AY, Neuenkirchen N, Deng J, Jin J, Kluger Y, Neubert TA, Bardwell VJ, and Ivanova NB (2018) A non-canonical BCOR-PRC1.1 complex represses differentiation programs in human ESCs. Cell Stem Cell 22, 235–251.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Eghbalnia HR, Wang L, Bahrami A, Assadi A, and Markley JL (2005) Protein energetic conformational analysis from NMR chemical shifts (PECAN) and its use in determining secondary structural elements. J. Biomol. NMR 32, 71–81. [DOI] [PubMed] [Google Scholar]
- (27).Wang R, Ilangovan U, Robinson AK, Schirf V, Schwarz PM, Lafer EM, Demeler B, Hinck AP, and Kim CA (2008) Structural transitions of the RING1B C-terminal region upon binding the polycomb cbox domain. Biochemistry 47, 8007–8015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Wang R, Taylor AB, Leal BZ, Chadwell LV, Ilangovan U, Robinson AK, Schirf V, Hart PJ, Lafer EM, Demeler B, Hinck AP, McEwen DG, and Kim CA (2010) Polycomb group targeting through different binding partners of RING1B C-terminal domain. Structure 18, 966–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Gearhart MD, Corcoran CM, Wamstad JA, and Bardwell VJ (2006) Polycomb group and SCF ubiquitin ligases are found in a novel BCOR complex that is recruited to BCL6 targets. Mol. Cell. Biol 26, 6880–6889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Rose NR, King HW, Blackledge NP, Fursova NA, Ember KJ, Fischer R, Kessler BM, and Klose RJ (2016) RYBP stimulates PRC1 to shape chromatin-based communication between polycomb repressive complexes. eLife 5, e18591 DOI: 10.7554/eLife.18591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Beguelin W, Teater M, Gearhart MD, Calvo Fernandez MT, Goldstein RL, Cardenas MG, Hatzi K, Rosen M, Shen H, Corcoran CM, Hamline MY, Gascoyne RD, Levine RL, Abdel-Wahab O, Licht JD, Shaknovich R, Elemento O, Bardwell VJ, and Melnick AM (2016) EZH2 and BCL6 cooperate to assemble CBX8-BCOR complex to repress bivalent promoters, mediate germinal center formation and lymphomagenesis. Cancer Cell 30, 197–213. [DOI] [PMC free article] [PubMed] [Google Scholar]