Abstract
Named for its assembly near exon-exon junctions during pre-mRNA splicing, the exon junction complex (EJC) regulates multiple aspects of RNA biochemistry, including export of spliced mRNAs from the nucleus and translation. Transcriptome analyses have revealed broad EJC occupancy of spliced metazoan transcripts, yet inhibition of core subunits has been linked to surprisingly specific phenotypes and a growing number of studies support gene-specific regulatory roles. Here we report results from a classroom-based RNAi screen revealing the EJC is necessary for regeneration in the planarian flatworm Schmidtea mediterranea. RNAi animals rapidly lost the stem and progenitor cells that drive formation of new tissue during both regeneration and cell turnover, but exhibited normal amputation-induced changes in gene expression in differentiated tissues. Together with previous reports that partial loss of EJC function causes stem cell defects in Drosophila and mice, our observations implicate the EJC as a conserved, posttranscriptional regulator of gene expression in stem cell lineages. This work also highlights the combined educational and scientific impacts of discovery-based research in the undergraduate biology curriculum.
Keywords: EJC, RNAi, Posttranscriptional regulation, Regeneration
1. Introduction
RNA-binding proteins (RBPs) play key roles in controlling the splicing, stability, subcellular localization, and translation of their target transcripts (Mitchell and Parker, 2014). One important regulator of all these aspects of RNA biochemistry is the exon junction complex (EJC). The core of this ribonucleoprotein structure is made up of four conserved subunits – Magoh (Mago nashi), Y14 (RBM8/Tsunagi), the DEAD-box RNA helicase eIF4A3, and MLN51 (CASC3/Barentsz) (Andersen et al., 2006; Bono et al., 2006). Magoh and Y14 form a heterodimer that is brought together with eIF4A3 by the spliceosome. The resulting ‘pre-EJC’ is deposited on nascent transcripts in a sequence-independent manner, typically 20–24 nucleotides upstream of exon-exon junctions. It remains associated with mature mRNAs during nuclear export, and is joined by the predominantly cytoplasmic protein MLN51 to complete the tetrameric core (Bono and Gehring, 2011). EJCs are eventually displaced by the ribosome during the pioneering round of translation, but not before heavily influencing the fate of bound transcripts. This entails transient interactions between the EJC core and ‘peripheral’ subunits that carry out specific biochemical functions, including splicing, nuclear export, translation, and nonsense-mediated mRNA decay (Le Hir et al., 2016).
Intriguingly, while transcriptome-wide analyses have shown EJCs associate with the large majority of exon-exon junctions (Hauer et al., 2016; Saulière et al., 2012; Singh et al., 2012), multiple lines of evidence indicate they function in more than just ‘housekeeping’ capacities. Not only is there substantial heterogeneity in the extent and distribution of EJC binding, but numerous cases of gene-specific regulation have been documented. For example, the EJC controls splicing of genes such as MAPK, BCLX, DLG1, and piwi (Ashton-Beaucage et al., 2010; Hayashi et al., 2014; Liu et al., 2016; Malone et al., 2014; Michelle et al., 2012; Roignant and Treisman, 2010). EJC subunits also regulate localization of oskar mRNA to the posterior of the Drosophila oocyte (Ghosh et al., 2012; Hachet and Ephrussi, 2001; Mohr et al., 2001; Newmark and Boswell, 1994; Palacios et al., 2004; van Eeden et al., 2001), and expression levels of the germline protein PIE-1 in the C. elegans embryo, likely via translational control of maternally deposited transcripts (Gauvin et al., 2018). Non-housekeeping roles for the EJC are further supported by the specificity of magoh, Y14, and eIF4A3 loss-of-function phenotypes. These include defects in axis formation in the Drosophila oocyte (Micklem et al., 1997; Mohr et al., 2001; Newmark et al., 1997), masculinization of the germline in C. elegans (Li et al., 2000), and heart looping abnormalities in Xenopus (Haremaki et al., 2010). In summary, it appears the EJC may function as much as a transcript-specific regulator of genes with specialized developmental functions as a global effector of processes such as maturation and export of mRNAs from the nucleus.
One emerging role for the EJC in development is the regulation of stem and progenitor cell populations. In Drosophila, mago nashi and Y14 are required to restrict oocyte fate to a single descendent of the germline stem cells (GSCs), and mago nashi functions independently of Y14 to drive GSC differentiation (Parma et al., 2007). In mice, haploinsufficiency of magoh, Y14, or eIF4A3 results in neurogenesis defects and microcephaly (reduced brain size), due to depletion of neural stem cells and transit-amplifying neural progenitors (Mao et al., 2016, 2015; Silver et al., 2010). Inhibition of EJC subunits also causes hypopigmentation in both frogs and mice, resulting from decreased numbers of melanocyte/melanophore progenitors derived from the neural crest (Haremaki et al., 2010; Silver et al., 2013). The direct RNA targets of the EJC in these undifferentiated cell types are currently unknown.
We now extend these observations by reporting a functional analysis of the EJC in the planarian flatworm Schmidtea mediterranea. Planarians are a classic model for the study of regeneration (Reddien, 2018), and recent single-cell sequencing efforts have dramatically enhanced our understanding of the somatic stem cell lineage that drives formation of new tissue at sites of amputation (Fincher et al., 2018; Molinaro and Pearson, 2016; Plass et al., 2018; van Wolfswinkel et al., 2014; Wurtzel et al., 2015; Zeng et al., 2018). At the top of this hierarchy are pluripotent stem cells, also known as ‘clonogenic neoblasts,’ that are capable of rescuing viability and regenerative potential when injected into lethally irradiated animals (Wagner et al., 2011; Zeng et al., 2018). These cells give rise to fate-determined division progeny, or progenitors, during physiological cell turnover as well as regeneration. Progenitor cells retain an undifferentiated morphology, but adopt lineage-specific gene expression profiles as they acquire their specialized fates (Fincher et al., 2018; Plass et al., 2018; Scimone et al., 2014; Zeng et al., 2018).
Posttranscriptional control mechanisms have emerged as key regulators of the planarian stem cell lineage (Krishna et al., 2019). Many RBPs show enriched expression in stem and/or progenitor cells, including homologs of the piRNA-binding protein PIWI (Palakodeti et al., 2008; Reddien et al., 2005; Rouhana et al., 2014), the PUF family protein Pumilio (Salvetti et al., 2005), the DEAD-box helicases Vasa and RCK/p54/Me31B (Rouhana et al., 2010; Shibata et al., 1999; Wagner et al., 2012; Yoshida-Kashikawa et al., 2007), the translational regulators Bruno and MEX-3 (Guo et al., 2006; Solana et al., 2016; Zhu et al., 2015), the dimethylarginine-associated factors Tudor and PRMT5 (Rouhana et al., 2012; Solana et al., 2009), components of the snRNA-processing Integrator complex (Schmidt et al., 2018), LSm splicing factors (Fernandez-Taboada et al., 2010), and the deadenylating complex subunit CCR4-NOT (Solana et al., 2013). Together with RBPs exhibiting constitutive expression, these factors help to orchestrate gene expression, cell fate specification, and differentiation (Bansal et al., 2017; Krishna et al., 2019; Lakshmanan et al., 2016; Palakodeti et al., 2008; Reddien et al., 2005; Rouhana et al., 2010; Shibata et al., 2016; Solana et al., 2016, 2013; Zhu et al., 2015).
Here, we show RNAi knockdown of the broadly expressed EJC core subunits leads to rapid loss of stem and multiple progenitor cell sub-populations in S. mediterranea. By extension, this blocks regeneration, but not amputation-induced changes in gene expression or re-patterning of the body axes in differentiated tissues. Our results further support the importance of posttranscriptional control mechanisms in planarian neoblasts and establish the EJC as an evolutionarily conserved regulator of metazoan stem cell lineages.
2. Materials and methods
2.1. Planarian maintenance
A clonal, asexual population of S. mediterranea (strain CIW4) was maintained under standard laboratory conditions on a diet of homogenized calf liver, as previously described (Oviedo et al., 2008). All animals were fed one to three times per week and then starved between five days and two weeks prior to initiation of RNAi feedings.
2.2. Cloning and RNA interference
cDNAs were amplified by RT-PCR (see Table S1 for primer sequences) and cloned into the dsRNA expression vector pT4P as previously described (Accorsi et al., 2017). The C. elegans gene unc-22 was used as a negative control. RNAi clones were transformed into E. coli strain HT115, and transformants were used to grow dsRNA-expressing cultures. These were pelleted, mixed with homogenized calf liver, and fed to animals according to a standard protocol (Newmark et al., 2003; Accorsi et al., 2017). For the RNAi screen, animals were fed six to eight times, except in cases where tissue homeostasis phenotypes prevented completion of feedings (see Supplementary Materials and Methods for further screen details). For remaining experiments, animals were administered four RNAi feedings on days 0, 2, 4, and 6 (with the exception of CAM-1 knockdown, which entailed a single feeding). Any animals that failed to eat were removed. RNAi animals were then amputated and/or fixed as indicated in figure legends.
RNAi knockdown of magoh, Y14, eIF4A3, and MLN51 was confirmed by qRT-PCR and in situ hybridization (Fig. S1). Extended knockdown (20 feedings interspersed with multiple rounds of amputation and regeneration, over a total period of nearly three months) did not result in a visible phenotype for MLN51. Several magoh(RNAi), Y14(RNAi), and eIF4A3(RNAi) animals were amputated to confirm the presence of a regeneration phenotype in experiments where RNAi knockdown of these genes had no apparent effects on differentiated tissues.
2.3. WISH, TUNEL, and H3P immunostaining
Whole-mount in situ hybridization (WISH) and whole-mount TUNEL were performed as previously described (Pearson et al., 2009; Pellettieri et al., 2010). For H3P immunostaining, animals were killed in 5% n-acetyl cysteine in PBS, fixed for 20 min in 4% formaldehyde in PBS with 0.3% Triton X-100 (PBST), and bleached overnight in 6% H2O2 in PBST. Fixed animals were incubated in 1:300 Anti-Phospho-Histone H3 (Ser 10) primary antibody (EMD Millipore, Cat. No. 05–817R–I) and 1:300 Goat Anti-Rabbit HRP secondary antibody (ThermoFisher Scientific, Cat. No. 65–6120); both antibody incubations were overnight at room temperature and were followed by eight 15-min washes in PBST. Labeling was visualized using tyramide signal amplification (TSA) with 1:3000 FITC-tyramide, as previously described (Pearson et al., 2009). Following TSA development, animals were again washed in PBST and mounted in Vectashield (Vector Laboratories). All animals labeled by WISH, TUNEL, and H3P were imaged and analyzed as described below.
2.4. Microscopy, image acquisition, and image analysis
All RNAi phenotypes (or lack thereof) were documented with an Olympus SZX16 microscope equipped with a DP72 digital camera. Live animals were photographed to show gross phenotypes. Labeled specimens were mounted on glass slides under coverslips. For each experiment, all images were acquired using identical camera settings. Images were processed by aligning cropped photographs of representative animals in panels with uniform black or white backgrounds, and then applying the same brightness and contrast adjustments evenly across all RNAi conditions. TUNEL and H3P results were quantified using ImageJ (http://rsb.info.nih.gov/ij), with the number of TUNEL-positive or H3P-positive nuclei normalized for animal area.
2.5. qRT-PCR
qRT-PCR was performed in technical and biological triplicate. Following RNAi feedings, total RNA was isolated from 10 pooled RNAi animals per condition by homogenization in TRIzol Reagent (ThermoFisher Scientific). Samples were DNase treated with the TURBO DNA-free Kit (ThermoFisher Scientific) and quantified using a NanoDrop 2000 spectrophotometer. 1 μg purified RNA per sample was then reverse transcribed using an oligo (dT)20 primer and the SuperScript III First-Strand Synthesis System (ThermoFisher Scientific). cDNA products were diluted 1:10 in molecular biology-grade water and used as templates in triplicate, 20 μL qPCRs, with 5 μL 1:10 cDNA, 10 μL iTaq Universal SYBR Green Supermix (BIO-RAD), and 1.2 μL forward and reverse primers added to each reaction (final primer concentrations of 0.6 μM; see Table S1 for sequences and amplicon sizes). Reactions were cycled in an Applied Biosystems StepOnePlus Real-Time PCR System (95 °C for 15 s; 60 °C for 60 s; 40 cycles), with specificity verified by gel electrophoresis, melt curve analysis, and inclusion of no-template controls. Results were normalized by the delta-delta CT method, using the ubiquitously expressed GAPDH gene as a reference (Eisenhoffer et al., 2008).
2.6. RT-PCR splicing analysis
To analyze splicing, total RNA and cDNA were prepared from neg. con.(RNAi) and magoh(RNAi) animals using Trizol Reagent and the SuperScript III First-Strand Synthesis System, respectively (ThermoFisher Scientific). Reverse transcriptase was omitted from negative control (-RT) reactions. cDNA and -RT control samples were used as templates in PCRs (see Table S1 for primer sequences). Reaction products were analyzed on 1% agarose gels.
3. Results and discussion
3.1. A classroom-based RNAi screen for stem cell and regeneration genes
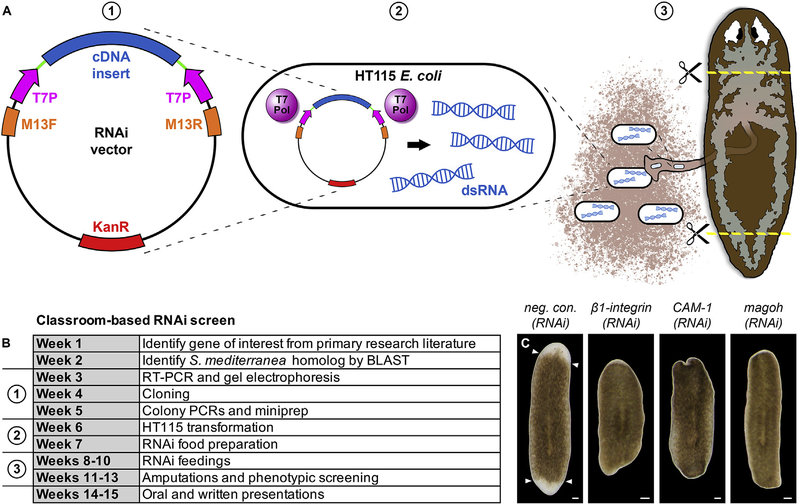
This line of research originated with a discovery in a semester-long lab conducted as part of an undergraduate developmental biology course. Students in this class clone and characterize novel S. mediterranea genes using RNA interference, simultaneously advancing their personal knowledge of key concepts in the field and our collective understanding of stem cell biology and regeneration (Fig. 1A and B; Supplementary Materials and Methods). To date, this screen has identified three genes required for formation of the blastema, the stem cell-derived mass of new tissue that forms at sites of amputation (Fig. 1C; Table S2). One of these, Smed-β1-integrin (Smed prefixes are hereafter omitted for brevity), was independently observed to be necessary for blastema outgrowth (Bonar and Petersen, 2017; Seebeck et al., 2017). A second, magoh (mago nashi homolog), was known to be expressed in the ovaries of sexually reproducing S. mediterranea hermaphrodites (Zayas et al., 2005), but had not previously been analyzed in the asexual strain we used here.
Fig. 1.
Identification of genes required for regeneration through an in-class RNAi screen. (A,B) RNAi screen approach and schedule. Students in a 15-week, undergraduate developmental biology course chose genes of interest from the primary research literature and identified their planarian homologs by conducting BLAST searches against S. mediterranea EST databases (SmedGD and PlanMine) (Brandl et al., 2016; Robb et al., 2015). Partial cDNAs were amplified by RT-PCR, cloned into an RNAi vector, and transformed into the HT115 E. coli strain with inducible T7 RNA polymerase to generate double-stranded RNA (dsRNA). dsRNA-expressing bacteria were then mixed with homogenized calf liver and fed to planarians, which were amputated and screened for regeneration phenotypes. See Supplementary Materials and Methods for details. Schematic diagram adapted from (Newmark et al., 2003). (C) We identified 3 genes (out of 29 screened) required for formation of the regeneration blastema: β1-integrin (Bonar and Petersen, 2017; Seebeck et al., 2017), a putative cell adhesion molecule (CAM-1), and a homolog of Drosophila mago nashi (magoh). Images show representative animals amputated the day after the final RNAi feeding and photographed 5 days post-amputation. Arrowheads denote approximate positions of original amputation planes. Scale bars: 100 μm.
3.2. The EJC is required for regeneration and tissue homeostasis
Given that magoh encodes a core subunit of the EJC (Fig. 2A), we also identified, cloned, and characterized S. mediterranea homologs of Y14, eIF4A3, and MLN51 (Fig. S2). Like magoh, Y14 and eIF4A3 were required for blastema formation (Fig. 2B). RNAi knockdown of MLN51 did not generate a visible phenotype, despite the fact that its expression was reduced to approximately the same extent as the other EJC subunits in our RNAi experiments (Fig. S1A). Our negative results for this gene are consistent with reports that it is dispensable for EJC function in some other systems, including the developing mouse brain (Mao et al., 2017). However, we cannot exclude the possibility that stronger knockdown would have elicited a phenotype. In uncut animals, RNAi knockdown of magoh, Y14, or eIF4A3, but not MLN51, led to head regression, ventral curling, and eventual lysis (Fig. 2C and E). Variability in the level of gene knockdown is likely to account for at least part of the variable penetrance of these phenotypes; it is also possible that magoh, Y14, and/or eIF4A3 have at least some EJC-independent functions. A small number of magoh(RNAi), Y14(RNAi), and eIF4A3(RNAi) animals exhibited one or more dorsal lesions (Fig. 2D and E). We conclude that the EJC is necessary for both regeneration and adult tissue homeostasis in planarians.
Fig. 2.
The EJC is required for regeneration and tissue homeostasis. (A) Schematic model of EJC assembly. Magoh and Y14 form a heterodimer that is brought together with eIF4A3 by the spliceosome in the nucleus and deposited just upstream of exon-exon junctions. MLN51 is the last subunit to join the complex, possibly after nuclear export of spliced mRNAs, and is dispensable for many EJC functions. (B–D) RNAi knockdown of magoh, Y14, or eIF4A3, but not MLN51, resulted in failure to form a blastema (B), head regression and ventral curling (C), and occasional dorsal lesions (D). For analysis of regeneration phenotypes, animals were amputated the day after the final RNAi feeding and trunk fragments were then photographed 5 days post-amputation. Similar results were observed for head and tail fragments. Representative tissue homeostasis phenotypes are shown in uncut animals photographed between 4 and 7 days after the final RNAi feeding [4 days: Y14(RNAi) dorsal lesion; 5 days: eIF4A3(RNAi) head regression and all ventral curling; 7 days: magoh(RNAi) and Y14(RNAi) head regression, neg. con.(RNAi), and MLN51(RNAi)]. (E) Percentage of animals exhibiting tissue homeostasis phenotypes 5 and 7 days after the final RNAi feeding. H.R. = head regression; V.C. = ventral curling; Les. = one or more dorsal lesions. Multiple phenotypes were sometimes present simultaneously (e.g., head regression and ventral curling). 100% of neg. con.(RNAi) and MLN51(RNAi) animals exhibited no discernible phenotypes. Results were compiled from 3 independent experiments with a sum total of over 50 animals per condition (neg. con.: n = 56; magoh: n = 59; Y14: n = 63; eIF4A3: n = 61; MLN51: n = 53). Scale bars: 200 μm (B,C); 100 μm (D).
3.3. The EJC is required for stem and progenitor cell maintenance
Failure to form a blastema, head regression, and ventral curling are all hallmarks of stem cell loss or dysfunction in S. mediterranea (Reddien et al., 2005). We therefore directly assessed the effects of EJC inhibition on stem cell number using an immunostaining approach with an antibody to a phosphorylated form of histone H3 (H3P) (Newmark and Sánchez Alvarado, 2000). H3P+ cells rapidly decreased following magoh knockdown, reaching near zero by five days following the final RNAi feeding, with a similar decline observed in Y14(RNAi) and eIF4A3(RNAi) animals (Fig. 3A and B; Fig. S3A). The decline in H3P+ cells was associated with a significant increase in whole-mount TUNEL (Fig. 3C and D), which may indicate stem cell depletion occurs through cell death. This approach does not distinguish between undifferentiated and differentiated cell types, however, so we cannot presently exclude alternative possibilities, such as a defect in self-renewal.
Fig. 3.
EJC inhibition results in loss of cycling stem cells and increased cell death. (A) H3P labeling of cycling stem cells. Images show representative animals labeled 5 days after the final RNAi feeding (see Fig. S3A for timecourse). (B) Quantitative analysis of H3P labeling. Boxes denote interquartile range with median. Whiskers denote minimum and maximum values. Asterisks indicate p-values for two-tailed T-tests in comparison with negative controls < 1 × 10−30; N.S. = not significant. Results were compiled from 3 independent experiments with a sum total of over 25 animals per condition (neg. con.: n = 58; magoh: n = 29; Y14: n = 37; eIF4A3: n = 54; MLN51: n = 55). (C,D) Stem cell loss was associated with increased cell death, as visualized by whole-mount TUNEL. Images (C) show representative animals labeled 3 days after the final RNAi feeding. TUNEL levels (D) were determined from 3 independent experiments with a sum total of over 15 animals per condition (neg. con.: n = 23; magoh: n = 27; Y14: n = 25; eIF4A3: n = 17; MLN51: n = 21). Boxes denote interquartile range with median. Whiskers denote minimum and maximum values. Asterisks indicate p-values for two-tailed T-tests in comparison with negative controls <0.005; N.S. = not significant. Scale bars: 100 μm (A,C).
Planarian stem cells and their differentiating division progeny have traditionally been defined by morphology, but can now be categorized according to expression of specific lineage markers (Baguñà, 2012; Reddien, 2018). The piwi homolog smedwi-1 is highly expressed in pluripotent neoblasts (Reddien et al., 2005; Wagner et al., 2011; Zeng et al., 2018). We conducted whole-mount in situ hybridization (WISH) with a probe for smedwi-1, and this confirmed stem cell loss following knockdown of EJC subunits (Fig. 4A). smedwi-1 qRT-PCR yielded similar results (Fig. S3B). Fate-determined stem cell division progeny adopt lineage-specific gene expression profiles as they become incorporated into differentiated tissues (Fincher et al., 2018; Plass et al., 2018; Scimone et al., 2014; Zeng et al., 2018). To assess the effects of EJC inhibition on these progenitor populations, we conducted WISH with probes for prog-1 and dd_554, markers for epidermis and pharynx precursors, respectively (Eisenhoffer et al., 2008; Zhu et al., 2015). Both were strongly reduced following magoh, Y14, or eIF4A3 knockdown, indicating these cell types are also eliminated by inhibition of EJC function (Fig. 4A).
Fig. 4.
The EJC is required for stem and progenitor cell maintenance. (A) WISH for stem and progenitor cell markers. Images show representative animals labeled 5 days after the final RNAi feeding. (B) Schematic model of epidermal lineage, adapted from (Tu et al., 2015). Note that marker co-expression can occur during transition states and that epidermal cells at the dorsal-ventral boundary arise from a divergent lineage (Plass et al., 2018). (C) WISH timecourse for stem and progenitor cell markers from 2 to 4 days after the final RNAi feeding. Expression patterns for neg. con.(RNAi) animals did not vary over this timeframe. Scale bars: 200 μm (A,C).
We completed a more detailed, temporal analysis of stem and progenitor cell loss in magoh(RNAi) animals using probes for smedwi-1 and prog-1, as well as agat-1 and zpuf-6, which are expressed during later stages of epidermal differentiation (Fig. 4B) (Eisenhoffer et al., 2008; Tu et al., 2015). smedwi-1+, prog-1+, and agat-1+ cells all decreased rapidly, with agat-1 expression showing an especially pronounced decline (Fig. 4C). This represents a marked contrast from the sequential loss of these markers in lethally irradiated animals (Eisenhoffer et al., 2008), where progenitor depletion is thought to be a secondary consequence of stem cell ablation. Our results therefore suggest the EJC is directly required for maintenance of both undifferentiated stem cells and differentiating progenitors. zpuf-6+ cells were more resistant to EJC inhibition, displaying only a modest reduction by the time cells expressing the other markers were almost completely eliminated (Fig. 4C). prog-1 and agat-1 label mesenchymal progenitors, whereas zpuf-6 marks cells beginning to intercalate into the mature epidermis (Tu et al., 2015). Taken together, then, our results are consistent with a model in which stem cells and their early division progeny are particularly reliant on EJC-regulated gene expression, with cells becoming more resistant to the effects of EJC inhibition as they approach a terminally differentiated state.
3.4. The EJC is not required for differentiated cells to respond to amputation
magoh, Y14, eIF4A3, and MLN51 all exhibited broad expression (Fig. 5), raising the possibility the EJC plays a predominantly house-keeping role, with stem and progenitor cell loss in RNAi animals merely reflecting greater susceptibility of these cell types to global perturbation of gene expression. To begin addressing this possibility, we first analyzed the expression of markers for fully differentiated epidermal and pharynx cells. In contrast to the progenitor markers in these lineages, neither the mature epidermal markers PRSS12 and ifb (Molina et al., 2011; Wurtzel et al., 2017) nor the pharynx marker VIT (Fincher et al., 2018) exhibited altered expression in magoh(RNAi) animals (Fig. 6A). ifb expression was also indistinguishable in Y14(RNAi), eIF4A3(RNAi), and negative control(RNAi) animals (Fig. S4A).
Fig. 5.
EJC subunits are broadly expressed. (A) t-SNE plots illustrating relative gene expression levels determined by single-cell sequencing (Fincher et al., 2018). Unlike smedwi-1, EJC subunits do not exhibit enriched expression in any particular cluster/cell type. (B,C) Representative WISH patterns in control (B) and irradiated (C) animals (the latter were fixed 24 h after exposure to a 10, 000 rad dose). Irradiation ablated smedwi-1+ cells, as expected, but did not change the expression patterns of EJC subunits. Scale bars: 100 μm (B,C).
Fig. 6.
The EJC is not required for differentiated cells to respond to amputation. (A) WISH for differentiated epidermal and pharynx markers in uncut animals labeled 5 days after the final RNAi feeding. PRSS12 is expressed throughout the dorsal and ventral epidermis, whereas ifb expression is restricted to the dorsal-ventral boundary. (B) Wound-induced gene expression analyzed by WISH in trunk fragments fixed immediately after amputation (0 h) and 6 h later. (C) WISH for the respective anterior and posterior positional control genes sFRP-1 and wntP-2 (wnt 11–5) in uncut animals (top), and head, trunk, and tail fragments fixed 48 h post-amputation (bottom). Arrowheads denote sFRP-1 induction at the new anterior pole in trunk and tail fragments (left), wntP-2 induction at the new posterior pole in head and trunk fragments (right), and retraction of the wntP-2 expression domain toward the posterior pole in tail fragments (lower right). (D) Localized (arrowheads) and systemic cell death responses in head fragments labeled 4 and 72 h post-amputation, respectively. Amputations were conducted 5 days (B,C) or 3 days (D) after the final RNAi feeding. Scale bars: 200 μm (A–C); 100 μm (D).
We next analyzed the effects of EJC inhibition on a series of regenerative responses known to occur in fully differentiated tissues (Pellettieri, 2019; Reddien, 2018). Amputation, like other types of injury, leads to rapid induction of gene expression and elevated cell death near the wound site (Pellettieri et al., 2010; Scimone et al., 2017; Wenemoser et al., 2012; Wurtzel et al., 2015). A subsequent ‘missing-tissue’ response, specific to regeneration, encompasses remodeling of uninjured tissues to restore anatomical scale and proportion. This process reestablishes normal patterning information, including regionalized expression of ‘positional-control genes’ (PCGs) in the body-wall muscle, and is associated with systemic induction of cell death (Gurley et al., 2010; Pellettieri et al., 2010; Petersen and Reddien, 2009; Scimone et al., 2017; Witchley et al., 2014). magoh(RNAi) animals exhibited no apparent defects in these events, displaying normal transcriptional activation of the wound-induced genes egr-2, fos-1, nlg1, and wntless (Fig. 6B), restoration of graded PCG expression along the anterior-posterior axis (Fig. 6C), and both localized and systemic cell death responses (Fig. 6D). Similar results were observed in Y14(RNAi) and eIF4A3(RNAi) animals (Fig. S4B–D). Importantly, these analyses were conducted in animals unable to form a blastema, confirming efficacy of gene knockdown (Materials and Methods).
3.5. magoh knockdown fails to prevent splicing of piwi family transcripts
Disruption of EJC subunits in Drosophila causes intron retention in piwi transcripts (Hayashi et al., 2014; Malone et al., 2014). In S. mediterranea, the piwi homologs smedwi-1–3 are key stem cell regulators, and RNAi knockdown of smedwi-2 results in a stem cell loss phenotype similar to the one reported here (Palakodeti et al., 2008; Reddien et al., 2005). These observations prompted us to test whether magoh knockdown impacts smedwi-1–3 splicing using an RT-PCR assay (Materials and Methods). We failed to observe any evidence of intron retention (Fig. S5). While we cannot exclude the possibility that our assay missed some aberrant splicing events, these results suggest unbiased approaches may be more effective in identifying EJC target transcripts in planarians (Conclusions).
3.6. Conclusions
Our results indicate the EJC is not required for gene expression per se in planarians, or for all aspects of the regenerative response, but rather for maintaining the population of undifferentiated cells that give rise to new tissue. It remains a formal possibility that RNAi knockdown of core subunits broadly alters gene expression, without detectably influencing the ability of differentiated tissues to respond to amputation, but we favor the alternative that the EJC has a specialized role in regulating stem and progenitor cell types. Indeed, previous observations that germline and somatic stem cells are highly sensitive to inhibition of magoh, Y14, and eIF4A3 in other organisms (Introduction) suggest this role may be evolutionarily conserved.
A key challenge for the future will be identifying direct RNA targets of the EJC in stem cell lineages, and determining whether those transcripts are regulated at the level of splicing, stability, subcellular localization, or translation. Transcriptomic approaches like CLIP-seq have proven effective in other organisms and represent one promising strategy (Hauer et al., 2016; Saulière et al., 2012). It is also noteworthy that a subset of the RBPs enriched in planarian stem cells localize to chromatoid bodies, perinuclear structures thought to be analogous to germ granules (Krishna et al., 2019). As these have been linked to the regulation of mRNA localization, stability, and translational control in the germline (Strome and Updike, 2015), it will be interesting to examine whether EJC subunits and their target RNAs might associate with chromatoid bodies or other subcellular structures, and how that localization is affected by differentiation. Research on these and related questions in the experimentally tractable stem cell system in planarians promises to further expand our growing knowledge of developmental roles of the EJC.
Supplementary Material
Acknowledgements
We thank Stuart Nelson, Amber Poirier, Aimée Joyce, Sarah Anderson, Derek Starkey, Alexandra Abbate, Leanna Landfair, Ryan Woodcock, Spencer Lynch, Brian Stevens, Allie Tolles, Callum Yule, and Adam Hayward for assistance with experiments and data analysis. We also recognize the collective efforts of students in Keene State College’s Developmental Biology course in the RNAi screen.
Funding
This work was supported by grants from the National Science Foundation (1656793), and New Hampshire-INBRE through an Institutional Development Award (P20GM103506) from the National Institute of General Medical Sciences of the National Institutes of Health.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ydbio.2019.09.010.
References
- Accorsi A, Williams MM, Ross EJ, Robb SMC, Elliott SA, Tu KC, Sánchez Alvarado A, 2017. Hands-on classroom activities for exploring regeneration and stem cell biology with planarians. Am. Biol. Teach. 79, 208–223. [Google Scholar]
- Andersen CBF, Ballut L, Johansen JS, Chamieh H, Nielsen KH, Oliveira CLP, Pedersen JS, Séraphin B, Le Hir H, Andersen GR, 2006. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science 313, 1968–1972. [DOI] [PubMed] [Google Scholar]
- Ashton-Beaucage D, Udell CM, Lavoie H, Baril C, Lefrançois M, Chagnon P, Gendron P, Caron-Lizotte O, Bonneil E, Thibault P, Therrien M, 2010. The exon junction complex controls the splicing of MAPK and other long intron-containing transcripts in Drosophila. Cell 143, 251–262. [DOI] [PubMed] [Google Scholar]
- Baguñà J, 2012. The planarian neoblast: the rambling history of its origin and some current black boxes. Int. J. Dev. Biol. 56, 19–37. [DOI] [PubMed] [Google Scholar]
- Bansal D, Kulkarni J, Nadahalli K, Lakshmanan V, Krishna S, Sasidharan V, Geo J, Dilipkumar S, Pasricha R, Gulyani A, Raghavan S, Palakodeti D, 2017. Cytoplasmic poly (A)-binding protein critically regulates epidermal maintenance and turnover in the planarian Schmidtea mediterranea. Development 144, 3066–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonar NA, Petersen CP, 2017. Integrin suppresses neurogenesis and regulates brain tissue assembly in planarian regeneration. Development 144, 784–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono F, Ebert J, Lorentzen E, Conti E, 2006. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell 126, 713–725. [DOI] [PubMed] [Google Scholar]
- Bono F, Gehring NH, 2011. Assembly, disassembly and recycling: the dynamics of exon junction complexes. RNA Biol. 8, 24–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandl H, Moon H, Vila-Farré M, Liu S-Y, Henry I, Rink JC, 2016. PlanMine–a mineable resource of planarian biology and biodiversity. Nucleic Acids Res. 44, D764–D773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhoffer GT, Kang H, Sánchez Alvarado A, 2008. Molecular analysis of stem cells and their descendants during cell turnover and regeneration in the planarian Schmidtea mediterranea. Cell Stem Cell 3, 327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Taboada E, Moritz S, Zeuschner D, Stehling M, Scholer HR, Salo E, Gentile L, 2010. Smed-SmB, a member of the LSm protein superfamily, is essential for chromatoid body organization and planarian stem cell proliferation. Development 137, 1055–1065. [DOI] [PubMed] [Google Scholar]
- Fincher CT, Wurtzel O, de Hoog T, Kravarik KM, Reddien PW, 2018. Cell type transcriptome atlas for the planarian Schmidtea mediterranea. Science 360, eaaq1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauvin TJ, Han B, Sun MJ, Griffin EE, 2018. PIE-1 translation in the germline lineage contributes to PIE-1 asymmetry in the early Caenorhabditis elegans embryo. G3 8, 3791–3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Marchand V, Gáspár I, Ephrussi A, 2012. Control of RNP motility and localization by a splicing-dependent structure in oskar mRNA. Nat. Struct. Mol. Biol. 19, 441–449. [DOI] [PubMed] [Google Scholar]
- Guo T, Peters AH, Newmark PA, 2006. A Bruno-like gene is required for stem cell maintenance in planarians. Dev. Cell 11, 159–169. [DOI] [PubMed] [Google Scholar]
- Gurley KA, Elliott SA, Simakov O, Schmidt HA, Holstein TW, Sánchez Alvarado A, 2010. Expression of secreted Wnt pathway components reveals unexpected complexity of the planarian amputation response. Dev. Biol. 347, 24–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachet O, Ephrussi A, 2001. Drosophila Y14 shuttles to the posterior of the oocyte and is required for oskar mRNA transport. Curr. Biol. 11, 1666–1674. [DOI] [PubMed] [Google Scholar]
- Haremaki T, Sridharan J, Dvora S, Weinstein DC, 2010. Regulation of vertebrate embryogenesis by the exon junction complex core component Eif4a3. Dev. Dynam. 239, 1977–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauer C, Sieber J, Schwarzl T, Hollerer I, Curk T, Alleaume A-M, Hentze MW, Kulozik AE, 2016. Exon junction complexes show a distributional bias toward alternatively spliced mRNAs and against mRNAs coding for ribosomal proteins. Cell Rep. 16, 1588–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi R, Handler D, Ish-Horowicz D, Brennecke J, 2014. The exon junction complex is required for definition and excision of neighboring introns in Drosophila. Genes Dev. 28, 1772–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna S, Palakodeti D, Solana J, 2019. Post-transcriptional regulation in planarian stem cells. Semin. Cell Dev. Biol. 87, 69–78. [DOI] [PubMed] [Google Scholar]
- Lakshmanan V, Bansal D, Kulkarni J, Poduval D, Krishna S, Sasidharan V, Anand P, Seshasayee A, Palakodeti D, 2016. Genome-wide analysis of polyadenylation events in Schmidtea mediterranea. G3 6, 3035–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Saulière J, Wang Z, 2016. The exon junction complex as a node of post-transcriptional networks. Nat. Rev. Mol. Cell Biol. 17, 41–54. [DOI] [PubMed] [Google Scholar]
- Li W, Boswell R, Wood WB, 2000. mag-1, a homolog of Drosophila mago nashi, regulates hermaphrodite germ-line sex determination in Caenorhabditis elegans. Dev. Biol. 218, 172–182. [DOI] [PubMed] [Google Scholar]
- Liu M, Li Y, Liu A, Li R, Su Y, Du J, Li C, Zhu AJ, 2016. The exon junction complex regulates the splicing of cell polarity gene dlg1 to control Wingless signaling in development. Elife 5, e17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Mestdagh C, Akhtar J, Kreim N, Deinhard P, Sachidanandam R, Treisman J, Roignant J-Y, 2014. The exon junction complex controls transposable element activity by ensuring faithful splicing of the piwi transcript. Genes Dev. 28, 1786–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao H, Brown HE, Silver DL, 2017. Mouse models of Casc3 reveal developmental functions distinct from other components of the exon junction complex. RNA 23, 23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao H, McMahon JJ, Tsai Y-H, Wang Z, Silver DL, 2016. Haploinsufficiency for core exon junction complex components disrupts embryonic neurogenesis and causes p53-mediated microcephaly. PLoS Genet. 12, e1006282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao H, Pilaz L-J, McMahon JJ, Golzio C, Wu D, Shi L, Katsanis N, Silver DL, 2015. Rbm8a haploinsufficiency disrupts embryonic cortical development resulting in microcephaly. J. Neurosci. 35, 7003–7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelle L, Cloutier A, Toutant J, Shkreta L, Thibault P, Durand M, Garneau D, Gendron D, Lapointe E, Couture S, Le Hir H, Klinck R, Elela SA, Prinos P, Chabot B, 2012. Proteins associated with the exon junction complex also control the alternative splicing of apoptotic regulators. Mol. Cell. Biol. 32, 954–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micklem DR, Dasgupta R, Elliott H, Gergely F, Davidson C, Brand A, González-Reyes A, St Johnston D, 1997. The mago nashi gene is required for the polarisation of the oocyte and the formation of perpendicular axes in Drosophila. Curr. Biol. 7, 468–478. [DOI] [PubMed] [Google Scholar]
- Mitchell SF, Parker R, 2014. Principles and properties of eukaryotic mRNPs. Mol. Cell 54, 547–558. [DOI] [PubMed] [Google Scholar]
- Mohr SE, Dillon ST, Boswell RE, 2001. The RNA-binding protein Tsunagi interacts with Mago Nashi to establish polarity and localize oskar mRNA during Drosophila oogenesis. Genes Dev. 15, 2886–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina MD, Neto A, Maeso I, Gómez-Skarmeta JL, Saló E, Cebrià F, 2011. Noggin and noggin-like genes control dorsoventral axis regeneration in planarians. Curr. Biol. 21, 300–305. [DOI] [PubMed] [Google Scholar]
- Molinaro AM, Pearson BJ, 2016. In silico lineage tracing through single cell transcriptomics identifies a neural stem cell population in planarians. Genome Biol. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newmark PA, Boswell RE, 1994. The mago nashi locus encodes an essential product required for germ plasm assembly in Drosophila. Development 120, 1303–1313. [DOI] [PubMed] [Google Scholar]
- Newmark PA, Mohr SE, Gong L, Boswell RE, 1997. Mago nashi mediates the posterior follicle cell-to-oocyte signal to organize axis formation in Drosophila. Development 124, 3197–3207. [DOI] [PubMed] [Google Scholar]
- Newmark PA, Reddien PW, Cebria F, Sánchez Alvarado A, 2003. Ingestion of bacterially expressed double-stranded RNA inhibits gene expression in planarians. Proc. Natl. Acad. Sci. U.S.A. 100, 11861–11865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newmark PA, Sánchez Alvarado A, 2000. Bromodeoxyuridine specifically labels the regenerative stem cells of planarians. Dev. Biol. 220, 142–153. [DOI] [PubMed] [Google Scholar]
- Oviedo NJ, Nicolas CL, Adams DS, Levin M, 2008. Establishing and maintaining a colony of planarians. Cold Spring Harb. Protoc. 3, 896–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios IM, Gatfield D, St Johnston D, Izaurralde E, 2004. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature 427, 753–757. [DOI] [PubMed] [Google Scholar]
- Palakodeti D, Smielewska M, Lu Y-C, Yeo GW, Graveley BR, 2008. The PIWI proteins SMEDWI-2 and SMEDWI-3 are required for stem cell function and piRNA expression in planarians. RNA 14, 1174–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parma DH, Bennett PE, Boswell RE, 2007. Mago Nashi and Tsunagi/Y14, respectively, regulate Drosophila germline stem cell differentiation and oocyte specification. Dev. Biol. 308, 507–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson BJ, Eisenhoffer GT, Gurley KA, Rink JC, Miller DE, Sánchez Alvarado A, 2009. A formaldehyde-based whole mount in situ hybridization method for planarians. Dev. Dynam. 238, 443–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellettieri J, 2019. Regenerative tissue remodeling in planarians – The mysteries of morphallaxis. Semin. Cell Dev. Biol. 87, 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellettieri J, Fitzgerald P, Watanabe S, Mancuso J, Green DR, Sánchez Alvarado A, 2010. Cell death and tissue remodeling in planarian regeneration. Dev. Biol. 338, 76–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen CP, Reddien PW, 2009. A wound-induced Wnt expression program controls planarian regeneration polarity. Proc. Natl. Acad. Sci. 106, 17061–17066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plass M, Solana J, Wolf FA, Ayoub S, Misios A, Glažar P, Obermayer B, Theis FJ, Kocks C, Rajewsky N, 2018. Cell type atlas and lineage tree of a whole complex animal by single-cell transcriptomics. Science 360, eaaq1723. [DOI] [PubMed] [Google Scholar]
- Reddien PW, 2018. The cellular and molecular basis for planarian regeneration. Cell 175, 327–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Oviedo NJ, Jennings JR, Jenkin JC, Sánchez Alvarado A, 2005. SMEDWI-2 Is a PIWI-like protein that regulates planarian stem cells. Science 310, 1327–1330. [DOI] [PubMed] [Google Scholar]
- Robb SMC, Gotting K, Ross E, Sánchez Alvarado A, 2015. SmedGD 2.0: the Schmidtea mediterranea genome database. Genesis 53, 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roignant JY, Treisman JE, 2010. Exon junction complex subunits are required to splice Drosophila MAP Kinase, a large heterochromatic gene. Cell 143, 238–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhana L, Shibata N, Nishimura O, Agata K, 2010. Different requirements for conserved post-transcriptional regulators in planarian regeneration and stem cell maintenance. Dev. Biol. 341, 429–443. [DOI] [PubMed] [Google Scholar]
- Rouhana L, Vieira AP, Roberts-Galbraith RH, Newmark PA, 2012. PRMT5 and the role of symmetrical dimethylarginine in chromatoid bodies of planarian stem cells. Development 139, 1083–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhana L, Weiss JA, King RS, Newmark PA, 2014. PIWI homologs mediate Histone H4 mRNA localization to planarian chromatoid bodies. Development 141, 2592–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvetti A, Rossi L, Lena A, Batistoni R, Deri P, Rainaldi G, Locci MT, Evangelista M, Gremigni V, 2005. DjPum, a homologue of Drosophila Pumilio, is essential to planarian stem cell maintenance. Development 132, 1863–1874. [DOI] [PubMed] [Google Scholar]
- Saulière J, Murigneux V, Wang Z, Marquenet E, Barbosa I, Le Tonquèze O, Audic Y, Paillard L, Roest Crollius H, Le Hir H, 2012. CLIP-seq of eIF4AIII reveals transcriptome-wide mapping of the human exon junction complex. Nat. Struct. Mol. Biol. 19, 1124–1131. [DOI] [PubMed] [Google Scholar]
- Schmidt D, Reuter H, Hüttner K, Ruhe L, Rabert F, Seebeck F, Irimia M, Solana J, Bartscherer K, 2018. The Integrator complex regulates differential snRNA processing and fate of adult stem cells in the highly regenerative planarian Schmidtea mediterranea. PLoS Genet. 14, e1007828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scimone ML, Cote LE, Reddien PW, 2017. Orthogonal muscle fibres have different instructive roles in planarian regeneration. Nature 551, 623–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scimone ML, Kravarik KM, Lapan SW, Reddien PW, 2014. Neoblast specialization in regeneration of the planarian Schmidtea mediterranea. Stem Cell Reports 3, 339–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seebeck F, März M, Meyer A-W, Reuter H, Vogg MC, Stehling M, Mildner K, Zeuschner D, Rabert F, Bartscherer K, 2017. Integrins are required for tissue organization and restriction of neurogenesis in regenerating planarians. Development 144, 795–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata N, Kashima M, Ishiko T, Nishimura O, Rouhana L, Misaki K, Yonemura S, Saito K, Siomi H, Siomi MC, Agata K, 2016. Inheritance of a nuclear PIWI from pluripotent stem cells by somatic descendants ensures differentiation by silencing transposons in planarian. Dev. Cell 37, 226–237. [DOI] [PubMed] [Google Scholar]
- Shibata N, Umesono Y, Orii H, Sakurai T, Watanabe K, Agata K, 1999. Expression of vasa (vas)-related genes in germline cells and totipotent somatic stem cells of planarians. Dev. Biol. 206, 73–87. [DOI] [PubMed] [Google Scholar]
- Silver DL, Leeds KE, Hwang H-W, Miller EE, Pavan WJ, 2013. The EJC component Magoh regulates proliferation and expansion of neural crest-derived melanocytes. Dev. Biol. 375, 172–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver DL, Watkins-Chow DE, Schreck KC, Pierfelice TJ, Larson DM, Burnetti AJ, Liaw H-J, Myung K, Walsh CA, Gaiano N, Pavan WJ, 2010. The exon junction complex component Magoh controls brain size by regulating neural stem cell division. Nat. Neurosci. 13, 551–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Kucukural A, Cenik C, Leszyk JD, Shaffer SA, Weng Z, Moore MJ, 2012. The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell 151, 750–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solana J, Gamberi C, Mihaylova Y, Grosswendt S, Chen C, Lasko P, Rajewsky N, Aboobaker AA, 2013. The CCR4-NOT complex mediates deadenylation and degradation of stem cell mRNAs and promotes planarian stem cell differentiation. PLoS Genet. 9, e1004003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solana J, Irimia M, Ayoub S, Orejuela MR, Zywitza V, Jens M, Tapial J, Ray D, Morris Q, Hughes TR, Blencowe BJ, Rajewsky N, 2016. Conserved functional antagonism of CELF and MBNL proteins controls stem cell-specific alternative splicing in planarians. Elife 5, e16797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solana J, Lasko P, Romero R, 2009. Spoltud-1 is a chromatoid body component required for planarian long-term stem cell self-renewal. Dev. Biol. 328, 410–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S, Updike D, 2015. Specifying and protecting germ cell fate. Nat. Rev. Mol. Cell Biol. 16, 406–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu KC, Cheng L-C, T K Vu H, Lange JJ, McKinney SA, Seidel CW, Sánchez Alvarado A, 2015. Egr-5 is a post-mitotic regulator of planarian epidermal differentiation. Elife 4, e10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eeden FJ, Palacios IM, Petronczki M, Weston MJ, St Johnston D, 2001. Barentsz is essential for the posterior localization of oskar mRNA and colocalizes with it to the posterior pole. J. Cell Biol. 154, 511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wolfswinkel JC, Wagner DE, Reddien PW, 2014. Single-cell analysis reveals functionally distinct classes within the planarian stem cell compartment. Cell Stem Cell 15, 326–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner DE, Ho JJ, Reddien PW, 2012. Genetic regulators of a pluripotent adult stem cell system in planarians identified by RNAi and clonal analysis. Cell Stem Cell 10, 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner DE, Wang IE, Reddien PW, 2011. Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science 332, 811–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenemoser D, Lapan SW, Wilkinson AW, Bell GW, Reddien PW, 2012. A molecular wound response program associated with regeneration initiation in planarians. Genes Dev. 26, 988–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witchley JN, Mayer M, Wagner DE, Owen JH, Reddien PW, 2014. Muscle cells provide instructions for planarian regeneration. Cell Rep. 4, 633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzel O, Cote LE, Poirier A, Satija R, Regev A, Reddien PW, 2015. A generic and cell-type-specific wound response precedes regeneration in planarians. Dev. Cell 35, 632–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzel O, Oderberg IM, Reddien PW, 2017. Planarian epidermal stem cells respond to positional cues to promote cell-type diversity. Dev. Cell 40, 491–504 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida-Kashikawa M, Shibata N, Takechi K, Agata K, 2007. DjCBC-1, a conserved DEAD box RNA helicase of the RCK/p54/Me31B family, is a component of RNA-protein complexes in planarian stem cells and neurons. Dev. Dynam. 236, 3436–3450. [DOI] [PubMed] [Google Scholar]
- Zayas RM, Hernandez A, Habermann B, Wang Y, Stary JM, Newmark PA, 2005. The planarian Schmidtea mediterranea as a model for epigenetic germ cell specification: analysis of ESTs from the hermaphroditic strain. Proc. Natl. Acad. Sci. U.S.A. 102, 18491–18496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng A, Li H, Guo L, Gao X, McKinney S, Wang Y, Yu Z, Park J, Semerad C, Ross E, Cheng L-C, Davies E, Lei K, Wang W, Perera A, Hall K, Peak A, Box A, Sánchez Alvarado A, 2018. Prospectively isolated Tetraspanin+ neoblasts are adult pluripotent stem cells underlying planaria regeneration. Cell 173, 1593–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu SJ, Hallows SE, Currie KW, Xu C, Pearson BJ, 2015. A mex 3 homolog is required for differentiation during planarian stem cell lineage development. Elife 4, e07025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Further reading
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC, 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, Lopez R, 2019. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan S, Goldman N, 2001. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 18, 691–699. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.