Abstract
Chest computed tomography (CT) imaging has become indispensable for staging and managing coronavirus disease 2019 (COVID-19), and current evaluation of anomalies/abnormalities associated with COVID-19 has been performed majorly by the visual score. The development of automated methods for quantifying COVID-19 abnormalities in these CT images is invaluable to clinicians. The hallmark of COVID-19 in chest CT images is the presence of ground-glass opacities in the lung region, which are tedious to segment manually. We propose anamorphic depth embedding-based lightweight CNN, called Anam-Net, to segment anomalies in COVID-19 chest CT images. The proposed Anam-Net has 7.8 times fewer parameters compared to the state-of-the-art UNet (or its variants), making it lightweight capable of providing inferences in mobile or resource constraint (point-of-care) platforms. The results from chest CT images (test cases) across different experiments showed that the proposed method could provide good Dice similarity scores for abnormal and normal regions in the lung. We have benchmarked Anam-Net with other state-of-the-art architectures, such as ENet, LEDNet, UNet++, SegNet, Attention UNet, and DeepLabV3+. The proposed Anam-Net was also deployed on embedded systems, such as Raspberry Pi 4, NVIDIA Jetson Xavier, and mobile-based Android application (CovSeg) embedded with Anam-Net to demonstrate its suitability for point-of-care platforms. The generated codes, models, and the mobile application are available for enthusiastic users at https://github.com/NaveenPaluru/Segmentation-COVID-19.
Keywords: Abnormalities, coronavirus, coronavirus disease 2019 (COVID-19), deep learning, segmentation
I. Introduction
Coronavirus disease 2019 (COVID-19) is an illness caused by a novel coronavirus, formerly known as 2019-nCoV. It is a constituent of a spectrum of viruses that cause respiratory diseases, such as severe acute respiratory syndrome (SARS) and middle east respiratory syndrome (MERS) [1]. COVID-19 virus was first identified in Wuhan, Hubei, China. The virus causes respiratory disorders with common symptoms being fever, dry cough, and shortness of breath [2]. Currently, reverse transcription-polymerase chain reaction (RT-PCR) is the standard test for diagnosing COVID-19 [3]. However, imaging-based diagnosis [ultrasound, chest X-ray, and chest computed tomography (CT)] is also playing a crucial role in the identification and management of COVID-19 infection. Notably, a shortage of RT-PCR tests for the diagnosis of COVID-19 has led to considering chest CT as a screening and diagnostic tool [4]. The RT-PCR tests have high false negatives, which requires serial sampling, and in such cases, findings in CT were beneficial [5]. The CT-based conclusions on 76 asymptomatic individuals with confirmed COVID-19 from the “Diamond Princess” cruise ship were able to identify pneumonia in 54% of these cases [6]. Also, CT is more sensitive to parenchymal lung disease, disease progression, and alternative diagnoses, including acute heart failure from COVID-19 myocardial injury [7]. Even a multinational consensus statement that was recently released confirmed that chest CT would provide clinically actionable items in diagnosis, management, triage, and therapy of COVID-19 [3].
The common imaging features observed from a chest CT of COVID-19 patient include anomalies/abnormalities, such as ground-glass opacity (GGO), consolidation, and rare characteristics, such as pericardial effusion and pleural effusion [8]–[10], with GGO being a common feature among all chest CT images. In a retrospective study, Chung et al. [9] emphasized the identification of common features as mentioned above for better management of COVID-19. There are ongoing attempts to utilize learning-based methods to triage the patients based on chest CT. Li et al. [11] have proposed a 3-D deep learning model for using 2-D local and 3-D global features to identify COVID-19 disease. Hierarchical attention and spatial pyramid networks were introduced to capture the abnormal features in the Lungs of COVID-19 patients [12]. Pan et al. [13] and Wang et al. [14] conducted a systematic study on chest CT images for understanding the changes in the lung during recovery from COVID-19. Caruso et al. [10] investigated the chest CT features of patients with COVID-19 and compared the diagnostic performance of chest CT with the gold-standard RT-PCR. They reported a sensitivity of 97% while using chest CT. The work of Pan et al. [13] defined four stages of disease progression of COVID-19 and its recovery. The remission and recovery from COVID-19 had a strong correlation with reducing the size of abnormalities in chest CT [13]. Even attempts to use lung opacity (abnormality) as a deep learning feature [15] to divide COVID-19 patients into categories of mild, moderate, severe, and critical have significantly been successful. It has also been suggested to use lung opacification as an image feature to monitor COVID-19 progression to provide better clinical management [15].
Given that chest CT imaging has an essential role in diagnosing, monitoring, and managing COVID-19, quantifying anomalies in chest CT in an automated way is the need of the hour. Current practice is semiquantitative and performed by visual score. Manual segmentation of these abnormalities in chest CT images is a tedious job. Automated methods, such as the one proposed here, can run on embedded as well as mobile (point-of-care) platforms with minimal hardware, such as Raspberry Pi, NVIDIA Jetson Xavier, or an Android device, and will immensely help the clinicians to manage COVID-19 better, specifically when the patient load is very high. Needless to mention that performing annotations slice-by-slice is tedious and expensive. Furthermore, chest CT imaging is not a standard clinical protocol for COVID-19 patients; these data sets’ availability in sufficiently large numbers, including annotations to develop a deep learning-based methodology, will be a challenge. Given the variability in the chest CT protocols [16] and diversity in the patient population, a deep learning model that is easy to train and deployable for quantifying the abnormalities in chest CT images is of utmost importance.
Automatic segmentation of ground-glass opacities (GGOs) in lung CT images was investigated for diffuse parenchymal lung diseases (DPLDs). Zhu et al. [17] have proposed an algorithm based on a Markov random field for segmenting GGO. Zhou et al. [18] have shown that the K-NN classifier, when boosted with nonparametric density estimates, can segment the GGO nodules more accurately. Jung et al. [19] proposed an intensity-based segmentation followed by an asymmetric multiphase deforming model for the segmentation of GGOs. All these methods were based on statistical models to segment the GGOs; however, lung infection in COVID-19 is a manifestation of GGOs and consolidation. The abnormalities present in the COVID-19 other than GGOs, such as consolidation, can be as high as 45% of the total abnormality [14]. These statistical methods may not be applicable for segmenting the COVID-19 chest CT images as the imaging features differ significantly. Recently, Fan et al. [20] proposed a pseudolabel generating strategy within a deep semisupervised network known as Inf-Net for segmenting abnormalities in COVID-19. This work only focuses on lung infection segmentation of COVID-19 patients, leading to a drop in accuracy when considering noninfected slices. Even Ouyang et al. [21] proposed a novel dual sampling attention strategy for effective mitigation of the imbalanced learning in chest CT images with 3-D convolutional neural networks for automatic diagnosis of COVID-19 from community-acquired pneumonia.
This work proposes the utility of supervised deep learning-based fast and fully automated way of segmenting anomalies (primarily, GGO and consolidations) and also normal lung tissue in chest CT images of patients having COVID-19. The emphasis is also on this model being lightweight such that it can be deployed in point-of-care platforms to have better clinical utility. We call our approach anamorphic depth embedding-based lightweight convolutional neural network, shortened as Anam-Net. The key steps involved in segmenting COVID-19 anomalies have been outlined in Fig. 1. The anamorphic depth of feature embeddings was obtained by AD-block, as shown in Fig. 2. We also bring in a label-based weighting strategy for the network’s cost function for effective learning. In supervised learning, cost-sensitive networks penalize the classifier’s loss by a weighting factor found from the prior information of annotated data. Several works adapt cost-sensitive training for robust segmentation [22]–[24]. In short, the main contributions of this work are as follows.
-
1)
Development of novel lightweight deep learning-based robust feature learning algorithm designed for the COVID-19 anomaly segmentation in chest CT images, with the fully convolutional AD-block built within symmetric encoder–decoder architecture. This AD-block enabled efficient gradient flow in the network.
-
2)
The adapted label weighting scheme during training makes the model highly robust during the testing phase.
-
3)
The computational time required for training and testing the proposed Anam-Net to segment abnormalities is low, making it highly attractive in the clinic.
-
4)
The proposed network has very few parameters, thereby reducing the need for extensive annotated data for training the network, and making it easy to train using site-specific data or for a specific chest CT protocol. We deployed the proposed Anam-Net on Raspberry pi 4 and NVIDIA Jetson Xavier Agx modules enabling deep learning-based embedded systems that can provide a quick initial assessment of COVID-19 lung infection. We have also developed an Android application (CovSeg) that can run on mobile devices embedded with Anam-Net for segmenting COVID-19 Anomalies. The hardware setup has been shown in Fig. 3.
-
5)
Finally, Anam-Net was evaluated on three data sets under different experimental conditions and benchmarked against state-of-the-art heavy and lightweight deep learning models, including UNet [24], ENet [23], UNet++ [25], SegNet [26], Attention UNet [27], LEDNet [28], and DeepLabV3+ [29]. Also, to the best of our knowledge, the proposed approach Anam-Net, which involves an anamorphic transformation in the depth of embeddings, has been introduced for image segmentation for the first time, resulting in a highly versatile lightweight network to perform the segmentation.
Fig. 1.

Key steps of the proposed approach for automated segmentation of abnormalities in chest CT images. The details of the Anam-Net architecture are given in Fig. 2, and layerwise details are shown in Table I.
Fig. 2.

Anam-Net: network architecture utilized for segmenting abnormalities in COVID-19 chest CT images. Fully convolutional anamorphic depth blocks (AD-blocks) with depthwise squeezing and stretching have been incorporated after downsampling operation in the encoder and also after upsampling operation in the decoder. Each AD-block has 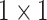 convolution for depthwise squeezing followed by
convolution for depthwise squeezing followed by 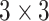 convolution and finally
convolution and finally 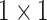 convolution for depthwise stretching. The layerwise details of encoder and decoder are shown in Table I and that of AD-block in Table II. Note that these AD-blocks are independent of each other and do not share parameters.
convolution for depthwise stretching. The layerwise details of encoder and decoder are shown in Table I and that of AD-block in Table II. Note that these AD-blocks are independent of each other and do not share parameters.
Fig. 3.

Hardware devices used for deploying the proposed Anam-Net to segment the COVID-19 anomalies in point-of-care platforms. The abnormalities in the lung region are indicated in red, and the normal lung region is in green. The Raspberry Pi 4 and the sample segmented slice #676 from the test cases (Experiment 1) are shown in (a). NVIDIA Jetson Xavier along with the sample segmented slice #676 from the test cases (Experiment 1) is shown in (b). One Euro coin was placed in these photographs to provide a form factor comparison. The snapshot of the developed Android application (CovSeg) showing the sample segmented slice #676 from test cases (Experiment 1) was provided in (c). The inference analysis was given in Table V.
The imaging data, along with ground truth annotations, were made available as an open-source [30]. The source code, trained models, and the mobile application for the proposed segmentation scheme along with consolidated results are available as open-source in [31]. The rest of this article is arranged as follows. Section II provides the details of the proposed method. This section is followed by Section III describing the data set utilized along with implementation details and figures of merit used for quantitatively assessing the performance of the proposed approach. The hardware deployment details are given in Section IV, and we provide the results in Section V. We present a detailed discussion of the results and the limitations of this study in Section VI. Finally, the conclusions were provided in Section VII.
II. Methods
The key steps involved in the proposed approach for segmenting the abnormalities in COVID-19 chest CT images along with the proposed Anam-Net architecture are given in Figs. 1 and 2, respectively. We discuss each of these steps in detail in the following.
A. Lung Extraction
The first step for segmenting abnormalities in chest CT images is to extract the lung region, the lung masks given in the data sets I and II (refer to Table III) were extracted using the method described in [32], and these masks were posted for easy usage at link [30]. These lung masks were obtained using a pretrained U-Net architecture [24] trained with batch normalization [32] consisting of 231 training samples obtained from a database of 5300 samples. Furthermore, the U-Net (R231) method was found to be more accurate in terms of lung segmentation compared to other trained models like chest imaging platform (CIP) and progressive holistically nested networks (P-HNNs) [32]. These 231 samples were obtained using random sampling, sampling from image phenotype, and manual selection of cases with different pathologies, such as fibrosis, trauma, and other pathologies [32]. Since the training samples used in U-Net (R231) were having a wide variety of lung pathology and organization, the U-Net (R231) method enabled accurate lung extraction on chest CT scans obtained from COVID-19 patients. Note that any process (including simple windowing) to extract the lung region is sufficient for the proposed work.
TABLE III. Details of the Data Sets Utilized in This Work.
B. Deep Learning for Medical Image Segmentation
Recently, techniques, such as neural architectural search (NAS), knowledge distillation, and cross-modality adaptation, were utilized for robust segmentation. Yu et al. [34] proposed a coarse to fine NAS strategy for 3-D biomedical segmentation by performing an architectural search at the macrolevel and systematic microlevel operations at each macrolevel topology. In a similar study, Guo et al. [35] implemented the crucial step of segmenting the organs at risk (head and neck) during radiotherapy treatment planning by adapting the NAS strategy. For the problem at hand related to COVID-19, the NAS approach may not be suited due to high computational complexity resulting in large training times. Li et al. [36] proposed a semisupervised system to address the lack of labeled data. This approach formulates the loss function as a weighted combination of supervised component for the labeled data and a regularized detail for the unlabelled data. In contrast to this, Clough et al. [37] proposed an unsupervised strategy based on topological loss derived from prior information of the object to be segmented. The knowledge transfer from heavy models for developing the lightweight models through model pruning was explored by Zhou et al. [38]. However, this kind of knowledge transfer requires a pretrained model. Wang et al. [39] proposed a user in the loop strategy for 2-D segmentation of the placenta from fetal MRI and 3-D segmentation of brain tumors from FLAIR images. In this work, the authors deployed user interactions as a hard constraint into a back propagatable conditional random field for end-to-end training. Gu et al. [40] proposed context encoding networks consisting of atrous convolution operations for 2-D medical image segmentation. Chen et al. [41] proposed a cross-modality adaption strategy between MR and CT images through a deep synergistic feature alignment module for robust medical image segmentation.
UNet architecture [24] has a deep symmetric encoder–decoder network with skip connections. Similarly, SegNet [26] also has an encoder–decoder structure embedded with a nonlinear upsampling mechanism for performing semantic segmentation. Zhou et al. [25] proposed an advanced version of UNet known as UNet++, which deploys nested dense connections [42] within the symmetric encoder–decoder framework. For enhancing the salience feature extraction, Oktay et al. [27] incorporated an attention module within the UNet framework. Chen et al. [29] introduced DeepLabV3+ formed with atrous separable convolutions for refining the segmentation results across the object boundaries. Jha et al. [43] proposed a deep residual UNet known as ResUNet++ for colonoscopic image segmentation. This architecture adds on a residual connection and attention module to the existing bottleneck of UNet. A cascade of UNet known as DoubleU-Net [44] was proposed, where the second UNet refines the segmentation results from the first UNet. However, these architectures, i.e., UNet, SegNet, UNet++, Attention UNet, DeepLabV3+, ResUNet++, and DoubleU-Net, involve many parameters resulting in heavy models (memory intensive) and have specialized compute requirements. On the other hand, an efficient neural network (ENet [23]) was developed to work with less number of parameters and was designed as a robust asymmetric encoder–decoder model for semantic segmentation in computer vision. The engineered design of ENet includes bottleneck layers similar to deep residual learning [45]. Furthermore, it has dilated convolutions [46] for avoiding higher downsampling rates and asymmetric convolutions [47] for achieving large speedups. Note that ENet architecture consists of a large encoder and a small decoder [23]. A deep encoder was deployed to operate on images with smaller resolution and enable filtering operations, while a shallow decoder was utilized to upsample the output [23]. In similar lines, another lightweight architecture LEDNet [28] also brings in an asymmetric encoder–decoder module having ResNet [45] as backbone along with a channelwise split and shuffle operation for fast and accurate segmentation.
C. Proposed Approach for Segmenting COVID-19 Anomalies
The proposed Anam-Net brings the best of UNet (symmetric encoder–decoder architecture) and ENet (fewer number of parameters), with a shallow symmetric network along with bottleneck layers. Primarily, the presented work proposed an AD-block (motivated from [45]) within a minisymmetric encoder–decoder segmentation module, as shown in Fig. 2. The AD-block consists of 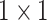 convolution for depthwise squeezing followed by
convolution for depthwise squeezing followed by 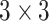 convolution and finally
convolution and finally 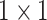 convolution for depthwise stretching (refer to Table II). The AD-block’s key idea is to squeeze (equivalent to the project) the feature space dimension (depthwise) before performing expensive
convolution for depthwise stretching (refer to Table II). The AD-block’s key idea is to squeeze (equivalent to the project) the feature space dimension (depthwise) before performing expensive 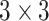 convolutions. Such a
convolutions. Such a 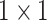 projection-based low-dimensional embeddings possess information about a relatively large input patch [48]. Following this low-dimensional projection, local feature extraction by
projection-based low-dimensional embeddings possess information about a relatively large input patch [48]. Following this low-dimensional projection, local feature extraction by 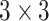 convolutions was performed without reducing the depthwise feature space dimension. Finally, the depthwise feature space dimension was stretched to the initial stage by another
convolutions was performed without reducing the depthwise feature space dimension. Finally, the depthwise feature space dimension was stretched to the initial stage by another 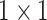 projection. The final output of AD-block denoted by
projection. The final output of AD-block denoted by  is obtained by adding the feature maps
is obtained by adding the feature maps  at the input of AD-block to the output of sequence of convolution operations
at the input of AD-block to the output of sequence of convolution operations  parameterized by
parameterized by 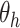 performing depth squeezing, feature extraction, and depth stretching. In short
performing depth squeezing, feature extraction, and depth stretching. In short
 |
and it is easier to optimize 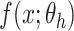 than to learn the underlying mapping
than to learn the underlying mapping 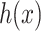 directly from
directly from 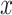 [45]. To summarize, for a given spatial resolution, the encoding operation in the proposed Anam-Net is
[45]. To summarize, for a given spatial resolution, the encoding operation in the proposed Anam-Net is
 |
where 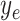 is the output of encoding operation,
is the output of encoding operation, 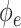 is the convolution operation parameterized by
is the convolution operation parameterized by 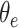 ,
,  is the input to be encoded, and
is the input to be encoded, and 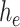 is the output of the AD-block in the encoder. Furthermore,
is the output of the AD-block in the encoder. Furthermore, 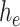 can be written as
can be written as
 |
where 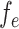 is the output of sequence of convolution operations parameterized by
is the output of sequence of convolution operations parameterized by 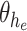 performing depth squeezing, feature extraction, and depth stretching. In similar lines, the decoding operation at same resolution, as mentioned earlier, in the encoding operation of the proposed Anam-Net is
performing depth squeezing, feature extraction, and depth stretching. In similar lines, the decoding operation at same resolution, as mentioned earlier, in the encoding operation of the proposed Anam-Net is
 |
where 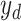 is the output of decoding operation,
is the output of decoding operation, 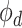 is the convolution operation parameterized by
is the convolution operation parameterized by 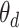 ,
, 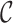 is the feature concatenation operation,
is the feature concatenation operation, 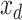 is the input to be decoded, and
is the input to be decoded, and 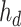 is the output of the AD-block in the decoder. As mentioned earlier,
is the output of the AD-block in the decoder. As mentioned earlier, 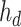 can be expressed as
can be expressed as
 |
where 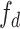 is the output of sequence of convolution operations parameterized by
is the output of sequence of convolution operations parameterized by 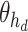 performing depth squeezing, feature extraction, and depth stretching. Note that the UNet embeddings can be seen as a special case of our proposed Anam-Net embeddings when
performing depth squeezing, feature extraction, and depth stretching. Note that the UNet embeddings can be seen as a special case of our proposed Anam-Net embeddings when 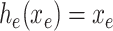 and
and 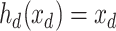 .
.
TABLE II. Architecture Details of the AD-Block With  Being the Minibatch Size,
Being the Minibatch Size,  the Depth of Embeddings (Feature Maps), and
the Depth of Embeddings (Feature Maps), and 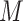 the Spatial Extent of Embeddings.
the Spatial Extent of Embeddings.
| Block | Input |
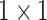 Convolution Convolution |
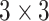 Convolution Convolution |
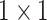 Convolution Convolution |
Output |
|---|---|---|---|---|---|
| Size | (N, Z, M, M) | (N, Z/4, M, M) | (N, Z/4, M, M) | (N, Z, M, M) | (N, Z, M, M) |
Overall, the proposed Anam-Net architecture consists of six such AD-blocks (three in the encoder and three in the decoder) to provide salience and robust feature learning. Each convolution layer in the proposed Anam-Net consists of convolution operation followed by batch normalization [49] and ReLU [50]. We provide the layerwise details of the encoder–decoder module in Table I and that of the AD-block in Table II. Given a minibatch of size 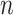 , we compute the loss as
, we compute the loss as
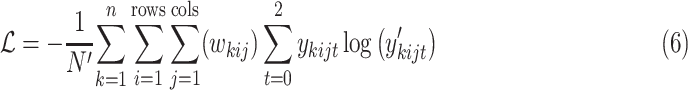 |
where 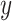 is the one hot encoded label (
is the one hot encoded label (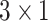 ),
), 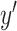 is the predicted softmax probabilities (
is the predicted softmax probabilities (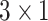 ),
), 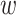 is the weight associated with label
is the weight associated with label 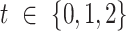 , and
, and 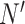 is
is 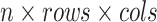 . The weight associated with each label in
. The weight associated with each label in  was given as
was given as
 |
where 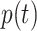 is the fraction of samples with label
is the fraction of samples with label  in the training set. After performing end to end training, the architecture was subjected to segment each pixel of the unseen test sample into three categories: background, abnormal-lung region, and normal-lung region. The model’s output is a probabilistic map (three maps, one each for background, abnormal, and normal) having the same spatial dimension as input. Depending on the maximal probabilistic score across these three categories, each pixel was assigned a label. It is important to note that the background (nonlung region) was identified, as outlined in Section II-A.
in the training set. After performing end to end training, the architecture was subjected to segment each pixel of the unseen test sample into three categories: background, abnormal-lung region, and normal-lung region. The model’s output is a probabilistic map (three maps, one each for background, abnormal, and normal) having the same spatial dimension as input. Depending on the maximal probabilistic score across these three categories, each pixel was assigned a label. It is important to note that the background (nonlung region) was identified, as outlined in Section II-A.
TABLE I. Architecture Details of Proposed Anam-Net, Where 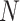 Is the Minibatch Size and the Flow of the Architecture Is From Left to Right in Each Row (Top to Bottom for Successive step). The Schematic Was Provided in Fig. 2. The Architecture Details of Anamorphic Depth (AD) Block Are Given in Table II.
Is the Minibatch Size and the Flow of the Architecture Is From Left to Right in Each Row (Top to Bottom for Successive step). The Schematic Was Provided in Fig. 2. The Architecture Details of Anamorphic Depth (AD) Block Are Given in Table II.
| Block | Input | Convolution | MaxPool | AD Block | Convolution | MaxPool |
| Size | (N, 1, 512, 512) | (N, 64, 512, 512) | (N, 64, 256, 256) | (N, 64, 256, 256) | (N, 128, 256, 256) | (N, 128, 128, 128) |
| Block | AD Block | Convolution | MaxPool | AD Block | Convolution | MaxPool |
| Size | (N, 128, 128, 128) | (N, 256, 128, 128) | (N, 256, 64, 64) | (N, 256, 64, 64) | (N, 256, 64, 64) | (N, 256, 32, 32) |
| Block | Trans Conv | AD Block | Concat | Convolution | Trans Conv | AD Block |
| Size | (N, 256, 64, 64) | (N, 256, 64, 64) | (N, 512, 64, 64) | (N, 256, 64, 64) | (N, 256, 128, 128) | (N, 256, 128, 128) |
| Block | Concat | Convolution | Trans Conv | AD Block | Concat | Convolution |
| size | (N, 512, 128, 128) | (N, 256, 128, 128) | (N, 128, 256, 256) | (N, 128, 256, 256) | (N, 256, 256, 256) | (N, 128, 256, 256) |
| Block | Trans Conv | Concat | Convolution | Output | ||
| Size | (N, 64, 512, 512) | (N, 128, 512, 512) | (N, 64, 512, 512) | (N, 3, 512, 512) | ||
III. Experiments and Implementation
The summary of the data sets utilized in this study is shown in Table III. We have conducted three experiments for evaluating the performance of all discussed models. The details of the experiments performed on these data sets are as follows.
-
Experiment 1:
The chest CT images that were utilized in this experiment had 929 axial chest CT images from approximately 49 patients with COVID-19 that were converted from openly accessible images provided by the Italian Society of Medical and Interventional Radiology [51] and radiopedia [52]. These images were made available in two sets. The data set I consist of 100 slices from >40 patients. From this set, we have selected 90 slices for training. To enhance robust learning, we performed data augmentation, such as horizontal flip and vertical flip on each training sample that resulted in a training set consisting of 270 samples. The Data set II consists of 829 slices from nine patient CT volumes. Out of these, we sampled 704 slices for testing. The remaining 125 slices were blank or had very minimal lung information. Note that Data set II was exclusively utilized for testing (none of the data was utilized in training). The testing performed on all models was with test data that the network has never seen at the patient level. All these images were annotated for abnormalities and have masks for GGO, consolidations, and pleural effusion. The detailed procedure for the annotations and data preparation can be found at this link [53]. These annotations masks were combined together to form a single abnormal mask. The annotations along with original chest CT slices and lung masks as NIFTI files can be found here [30]. These annotations (original input slice and ground truth), for sample test cases, have been presented in Fig. 4, the inference analysis of a sample test case on resource constraint platforms has been provided in Table V, and dice similarity scores for the abnormal region of sample test cases (Fig. 4) have been shown in Table VI. Finally, the averaged figures of merit over all 704 chest CT images (test cases) considered in this experiment were provided in Table VII.
-
Experiment 2:
The chest CT images that were utilized in this experiment consists of 3410 axial chest CT images from 20 patients with COVID-19 (refer to Data set III in Table III). As mentioned earlier, we excluded the chest CT slices that did not have any visible lung region (either completely collapsed or near the end slices) from testing and training. We split this data set at the patient level into four equal folds F1, F2, F3, and F4. The fold F4 with 545 CT images was explicitly used for testing, and we trained the deep models using threefold cross-validation on the folds F1, F2, and F3. The averaged figures of merit across three cross folds over all the 545 chest CT images (test cases) considered in this experiment were shown in Table VIII.
-
Experiment 3:
In this experiment, we performed cross data set examination, wherein the models trained in Experiment 2 were tested on the test cases in Experiment 1. This kind of cross-examination enabled us to study the generalizability of the deep models for practical application scenarios. The averaged figures of merit over all the 704 chest CT images (test cases) considered in this experiment were given in Table IX.
Fig. 4.
Representative segmentation results of Experiment 1 (key steps were given in Fig. 1). Randomly selected input slices (test cases) were shown in the top row. The annotations (ground truth) are shown in the second row. The abnormalities in the lung region are indicated in red, and the normal lung region is in green. Dice similarity scores of the abnormal lung regions for these test cases have been shown in Table VI.
TABLE V. Comparison Between the Hardware Devices Used for Deploying the Proposed Anam-Net in Terms of Cost, Available Memory, and Inference Time (in Seconds).
| Device | Cost | Available Memory | Inference |
|---|---|---|---|
| Raspberry Pi 4 | $50 | 4 GB RAM | Anam-Net: 23.3 s UNet: 43.3 s |
| NVIDIA Jetson | $700 | 512-core Volta GPU 32 GB RAM |
Anam-Net: 2.9 s UNet: 5.2 s |
| Nokia 5.1 Plus | $95 | 3 GB RAM | Anam-Net: 6.5 s UNet: 11.3 s |
TABLE VI. Dice Similarity Scores for the Abnormal Lung Region for Test Cases Shown in Fig. 4. The Last Row Represents the Average of These Results. The Best Results Are Shown in Bold.
| Model | UNet | ENet | UNet++ | SegNet | Attention UNet | LEDNet | DeepLabV3+ | Anam-Net (Proposed) |
|---|---|---|---|---|---|---|---|---|
| Slice No. | ||||||||
| 50 | 0.36 | 0.30 | 0.44 | 0.35 | 0.33 | 0.37 | 0.26 | 0.38 |
| 122 | 0.16 | 0.28 | 0.19 | 0.12 | 0.16 | 0.14 | 0.13 | 0.80 |
| 292 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.00 | 0.00 | 1.00 |
| 676 | 0.90 | 0.81 | 0.90 | 0.86 | 0.89 | 0.84 | 0.86 | 0.89 |
| Average | 0.36 | 0.35 | 0.39 | 0.33 | 0.60 | 0.34 | 0.31 | 0.77 |
TABLE VII. Averaged Figures of Merit Over All 704 Chest CT Images (Test Cases) Considered in the Experiment 1. The Best Results Are Shown in Bold.
| Model | Class | Sensitivity | Specificity | Accuracy | Dice Score |
|---|---|---|---|---|---|
| UNet | Abnormal | 0.932 | 0.983 | 0.982 | 0.608 |
| Normal | 0.902 | 0.997 | 0.981 | 0.943 | |
| ENet | Abnormal | 0.857 | 0.990 | 0.988 | 0.694 |
| Normal | 0.954 | 0.991 | 0.985 | 0.956 | |
| UNet++ | Abnormal | 0.954 | 0.986 | 0.986 | 0.674 |
| Normal | 0.924 | 0.997 | 0.985 | 0.955 | |
| SegNet | Abnormal | 0.936 | 0.981 | 0.980 | 0.587 |
| Normal | 0.897 | 0.995 | 0.978 | 0.935 | |
| Attention UNet | Abnormal | 0.951 | 0.988 | 0.987 | 0.695 |
| Normal | 0.932 | 0.997 | 0.986 | 0.960 | |
| LEDNet | Abnormal | 0.907 | 0.983 | 0.981 | 0.597 |
| Normal | 0.901 | 0.991 | 0.976 | 0.929 | |
| DeepLabV3+ | Abnormal | 0.850 | 0.958 | 0.956 | 0.366 |
| Normal | 0.762 | 0.993 | 0.954 | 0.850 | |
| Anam-Net (Proposed) | Abnormal | 0.900 | 0.993 | 0.991 | 0.755 |
| Normal | 0.959 | 0.997 | 0.990 | 0.972 |
TABLE VIII. Averaged Figures of Merit Across Three Cross Folds Over All 545 Chest CT Images (Test Cases) Considered in Experiment 2. The Best Results Are Shown in Bold.
| Model | Class | Sensitivity | Specificity | Accuracy | Dice Score |
|---|---|---|---|---|---|
| UNet | Abnormal | 0.907 | 0.990 | 0.988 | 0.791 |
| Normal | 0.913 | 0.995 | 0.987 | 0.938 | |
| ENet | Abnormal | 0.736 | 0.989 | 0.984 | 0.686 |
| Normal | 0.914 | 0.989 | 0.912 | 0.911 | |
| UNet++ | Abnormal | 0.951 | 0.990 | 0.988 | 0.805 |
| Normal | 0.906 | 0.995 | 0.987 | 0.937 | |
| SegNet | Abnormal | 0.657 | 0.988 | 0.980 | 0.612 |
| Normal | 0.888 | 0.987 | 0.977 | 0.885 | |
| Attention UNet | Abnormal | 0.966 | 0.987 | 0.987 | 0.783 |
| Normal | 0.884 | 0.997 | 0.986 | 0.930 | |
| LEDNet | Abnormal | 0.782 | 0.989 | 0.984 | 0.707 |
| Normal | 0.912 | 0.987 | 0.979 | 0.899 | |
| DeepLabV3+ | Abnormal | 0.521 | 0.992 | 0.981 | 0.563 |
| Normal | 0.930 | 0.983 | 0.978 | 0.895 | |
| Anam-Net (Proposed) | Abnormal | 0.918 | 0.990 | 0.988 | 0.798 |
| Normal | 0.911 | 0.997 | 0.988 | 0.941 |
TABLE IX. Averaged Figures of Merit Over All 704 Chest CT Images (Test Cases) Considered in Experiment 3. The Best Results Are Shown in Bold.
| Model | Class | Sensitivity | Specificity | Accuracy | Dice Score |
|---|---|---|---|---|---|
| UNet | Abnormal | 0.960 | 0.985 | 0.985 | 0.657 |
| Normal | 0.916 | 0.998 | 0.984 | 0.951 | |
| ENet | Abnormal | 0.864 | 0.972 | 0.970 | 0.464 |
| Normal | 0.845 | 0.994 | 0.968 | 0.902 | |
| UNet++ | Abnormal | 0.951 | 0.983 | 0.983 | 0.628 |
| Normal | 0.913 | 0.997 | 0.982 | 0.947 | |
| SegNet | Abnormal | 0.884 | 0.946 | 0.945 | 0.325 |
| Normal | 0.699 | 0.995 | 0.945 | 0.813 | |
| Attention UNet | Abnormal | 0.931 | 0.989 | 0.988 | 0.704 |
| Normal | 0.941 | 0.998 | 0.988 | 0.964 | |
| LEDNet | Abnormal | 0.872 | 0.981 | 0.979 | 0.559 |
| Normal | 0.906 | 0.983 | 0.970 | 0.912 | |
| DeepLabV3+ | Abnormal | 0.845 | 0.906 | 0.905 | 0.209 |
| Normal | 0.489 | 0.990 | 0.904 | 0.637 | |
| Anam-Net (Proposed) | Abnormal | 0.937 | 0.986 | 0.986 | 0.664 |
| Normal | 0.927 | 0.998 | 0.985 | 0.956 |
Implementation: The proposed Anam-Net was trained using PyTorch [54] with a minibatch of size 5. The cost function was optimized using Adam optimizer [55] with an initial learning rate of 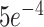 and gradually decayed by a factor of 0.1 once after every 33rd epoch. All computations performed in this work, including training of CNN, utilized a Linux workstation having i9 9900X (CPU) with 128-GB RAM and NVIDIA Quadro RTX 8000 GPU card. For fair comparison, the state-of-the-art methods, such as UNet [24], ENet [23], UNet++ [25], SegNet [26], Attention UNet [27], LEDNet [28], and DeepLabV3+ [29], were also trained with the same training data and tested on the same test data in all experiments. For testing, the Anam-Net in the Fig. 1 was replaced by these trained models. The number of parameters, model size, and typical training time along with the inference time were presented in Table IV for quick comparison. To quantitatively evaluate the performance of obtained segmentation results from all the models, we calculated the figures of merit, specificity, sensitivity, accuracy, and the Dice similarity score for both abnormal and normal classes. The computed value of figures of merit will be between 0 to 1, and in all cases, the higher value (close to 1) indicates better performance of a model.
and gradually decayed by a factor of 0.1 once after every 33rd epoch. All computations performed in this work, including training of CNN, utilized a Linux workstation having i9 9900X (CPU) with 128-GB RAM and NVIDIA Quadro RTX 8000 GPU card. For fair comparison, the state-of-the-art methods, such as UNet [24], ENet [23], UNet++ [25], SegNet [26], Attention UNet [27], LEDNet [28], and DeepLabV3+ [29], were also trained with the same training data and tested on the same test data in all experiments. For testing, the Anam-Net in the Fig. 1 was replaced by these trained models. The number of parameters, model size, and typical training time along with the inference time were presented in Table IV for quick comparison. To quantitatively evaluate the performance of obtained segmentation results from all the models, we calculated the figures of merit, specificity, sensitivity, accuracy, and the Dice similarity score for both abnormal and normal classes. The computed value of figures of merit will be between 0 to 1, and in all cases, the higher value (close to 1) indicates better performance of a model.
TABLE IV. Comparison of Deep Learning Models Utilized in This Work in Terms of Number of Training Parameters, Model Size, Training Time (for 100 Epochs in Experiment 1), and Inference Time.
| Model | Parameters | Model Size | Training time | Inference time |
|---|---|---|---|---|
| UNet | 31.07 M | 118.24 MB | 51 min | 531 ms |
| ENet | 343.7 K | 1.33MB | 15 min | 248 ms |
| UNet++ | 9.16 M | 34.95 MB | 58 min | 551 ms |
| SegNet | 29.44 M | 112.31 MB | 37 min | 528 ms |
| Attention UNet | 34.87 M | 133.05 MB | 63 min | 569 ms |
| LEDNet | 0.91 M | 3.8 MB | 16 min | 298 ms |
| DeepLabV3+ | 54.70 M | 208.66 MB | 42 min | 895 ms |
| Anam-Net (Proposed) | 4.47 M | 17.21 MB | 27 min | 362 ms |
IV. Hardware Deployment
Raspberry Pi 4 Model B is the latest version among the various raspberry pi tiny dual-display computers released to date. It is a low-cost embedded system with increased connectivity, memory capacity, and processor speed compared to its predecessor Raspberry Pi 3 Model B+. The total cost of the Raspberry Pi 4 Model B embedded platform is $50. To embed the Anam-net on Raspberry Pi 4, we converted the trained Anam-Net model from PyTorch into the Tensorflow Lite version. Tensorflow Lite is a variant of TensorFlow, which helps to run a deep learning model on the mobile, the Internet of Things (IoT), and embedded devices. It acts as an accelerator to reduce the inference time of models deployed on the embedded systems. The inference time of the TensorFlow Lite version of the Anam-net model on Raspberry Pi was 23.3 s, whereas the inference time of the TensorFlow Lite version of the UNet model on Raspberry Pi was 43.3 s. We currently have the PyTorch Lite versions for Android, and PyTorch does not have any official support for Raspberry Pi. We see that this high inference time in tens of seconds is due to the model conversion from PyTorch to Tensorflow using third-party tools. The hardware setup and the inference analysis are given Fig. 3 and Table V, respectively.
The Anam-Net model was also deployed on the NVIDIA Jetson AGX Xavier developer kit. NVIDIA Jetson AGX Xavier is the latest version among all the Jetson platforms released by NVIDIA. Jetson AGX Xavier is a deep learning model accelerator with 20 times more performance and ten times more energy efficiency than its predecessor Jetson TX2. Jetson AGX Xavier consists of an eight-core ARM processor CPU and 512-core Volta GPU with Tensor cores. The Anam-Net model deployed on Jetson AGX Xavier was able to perform inference within 2.9 s. In contrast, the UNet model, when deployed on the same, gave an inference time of 5.2 s. The hardware setup and the inference analysis are given Fig. 3 and Table V, respectively. The Anam-Net model and the UNet model were also trained on the Jetson AGX Xavier platform. The training time for an epoch with a batch size of 5 for the Anam-Net model was 1.49 min, whereas, for UNet, it was 3.19 min.
We have also developed an Android Application for the mobile platforms, called CovSeg, for segmenting the COVID-19 Anomalies. The PyTorch trained model was converted to its lite version as given here [56]. We developed the front end and the back end of the CovSeg application in Android Studio [57]. The Android application snapshot on Nokia 5.1 Plus mobile phone and the inference analysis are given Fig. 3 and Table V, respectively.
V. Results
As mentioned earlier, the segmentation results from all test cases of the three experiments were obtained and compared with the ground truth labels. The proposed method was effective in segmenting the abnormalities across all COVID-19 chest CT images. It is even useful when there is no abnormality; the proposed method provided a null result as expected. The example results, including the ground truth labels, were presented in Fig. 4, and the corresponding Dice score for the abnormal class is presented in Table VI. The proposed Anam-Net’s performance in terms of sensitivity, specificity, accuracy, and Dice score was superior. However, the performance of UNet++ was also on par with the proposed approach because of extensive dense connections in its design that result in hierarchical encoder–decoder modules enabling efficient feature propagation for accurate segmentation. The averaged results from Experiment 1 were presented in Table VII, and that of Experiments 2 and 3 are in Tables VIII and IX, respectively. Furthermore, higher specificity and higher accuracy for both the classes (abnormal and normal) are desirable in disease monitoring, especially in the remission of COVID-19. As it can be seen from Tables VII and VIII, the proposed Anam-Net with fewer parameters was able to provide accurate segmentation results compared to already existing models. As shown in Table IX, even in the cross data set examination (Experiment 3), the performance of Anam-Net was reasonably good (second-best) and was comparable with the best performing method Attention UNet.
As it can be seen from results in Fig. 4 and Tables VI and VII, the proposed Anam-Net provides superior performance with the utilization of anamorphic depth embeddings, which enabled the network to be lightweight. The patient-level (a sample test case from Experiment 2) segmentation results are shown in Fig. 5, wherein the average Dice score of the normal lung region was 0.95, and that of the abnormal region was 0.68. In most cases, while processing the initial slices of the lung, we observed that the Dice score for the abnormal region was minimal (< 0.5). However, on average, the accuracy of segmenting the anomalies was as high as 0.98 in all the experiments (refer to Tables VII–IX). Overall, the next best performing network was Attention UNet, which has at least 7.8 times more parameters (refer to Table IV) and required 2.3 times more training time compared to Anam-Net. Also, these heavy models (sizes being in hundreds of MB) may not be well suited for the point-of-care platforms for providing quick inference. Despite the proposed model having only 4.47 million parameters (third lightest), it was able to outperform the rest of the networks in all figures of merit (see Table VII). The lightweight networks performance was much inferior compared to heavy networks and among all networks DeepLabV3+ providing the lowest Dice score (see Tables VII–IX).
Fig. 5.
Representative segmentation results on a patient volume (test cases) from Experiment 2 (key steps were given in Fig. 1). The input slices (test cases) were shown in the first and fourth columns. The respective annotations (ground truth) are shown in the second and fifth columns. The predictions of the Anam-Net are in given the third and sixth columns. The abnormalities in the lung region are indicated in red, and the normal lung region is in green. Dice similarity scores of the abnormal (in red) and normal (in green) lung regions are given below the corresponding slices.
VI. Discussion
As can be seen from Table VII, the Dice similarity score of the proposed method (Anam-Net) was the highest compared to existing state-of-the-art architectures. Specifically, we observed that except for Attention UNet and the proposed Anam-Net, rest networks failed in identifying healthy individuals (slice 292 shown in Fig. 4). The Anam-Net successfully generated the null result; the same is evident when comparing the sensitivity values of the normal class indicated in Table VII. In essence, the training times required for Anam-Net is lesser (roughly about 43%) compared to Attention UNet (since the number of parameters being fewer; see Table IV), and Anam-Net was able to generate more accurate results compared to others. The improved accuracy of Anam-Net compared to the UNet model can be attributed to the AD-block, which was confirmed by the ablation study (refer to Table X). Without the AD-block, the performance in terms of the Dice score was comparable with UNet results (compare the second row in Table X with UNet results in Table VII).
TABLE X. Ablation Study for All AD-Blocks and the Label Weighting Strategy During the Training of Anam-Net. The Best Results Are Shown in Bold. Note That These Results Are Averaged Over 704 Chest CT Images (All Test Cases) From Experiment 1.
| Figure of Merit | AD Blocks | Label Weighting | Abnormal | Normal |
|---|---|---|---|---|
| Dice Similarity | ✓ | ✓ | 0.755 | 0.972 |
| Dice Similarity | 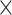 |
✓ | 0.666 | 0.955 |
| Dice Similarity | ✓ | 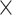 |
0.558 | 0.934 |
| Dice Similarity | 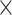 |
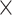 |
0.295 | 0.778 |
Obtaining accurate labels is important while developing deep learning models for the automatic segmentation of abnormalities. The labels can be classified into two major categories, namely strong annotations [wherein the radiologist has performed a proper segmentation of the region of interest (ROI)] and weak annotations (which can simply be scribbles, sparse dots, or noisy annotated labels) for pictorial representation (please check [58, Fig. 5]). Zheng et al.
[59] have used weak annotations in the form of patient-level labels, i.e., whether the patient is COVID-positive or COVID-negative to train the network for automatically detecting COVID-19 cases. Zheng et al.
[59] have used weakly supervised learning for COVID-19 detection, wherein a spatial global pooling layer and a temporal global pooling layer were introduced into the DeCovNet. Xu et al.
[60] have developed a model that can handle multiple classes with patient-level labels. Xu et al.
[60] included patient-level labels belonging to different classes, such as Influenza, COVID, and pneumonia, while training the deep learning model; however, these studies did not consider annotations (i.e., segmenting the abnormality in the CT image) for training the model. Obtaining these strong annotations to train a traditional CNN is time-consuming and expensive. The proposed Anam-Net can be seen as Mini-CNN with the number of trainable parameters being seven times less compared to UNet (or its variants) architecture. Anam-Net will also have a universal appeal to deploy for a site/protocol specific accurate segmentation network within adequate training time (see Table IV). We have also compared the proposed Anam-Net and UNet in terms of parameter sensitivity. The parameters (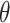 ) of the trained models (from Experiment 1) were perturbed by
) of the trained models (from Experiment 1) were perturbed by 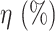 , and the perturbed models were tested on 704 chest CT images (all the test cases) from Experiment 1. The averaged results of the parameter sensitivity analysis are shown in Table XI. These results indicate that the proposed Anam-Net has the same stability (if not better) as UNet and is suitable for a critical application, such as the prognosis of COVID-19.
, and the perturbed models were tested on 704 chest CT images (all the test cases) from Experiment 1. The averaged results of the parameter sensitivity analysis are shown in Table XI. These results indicate that the proposed Anam-Net has the same stability (if not better) as UNet and is suitable for a critical application, such as the prognosis of COVID-19.
TABLE XI. Comparison Between the Proposed Anam-Net and UNet in Terms of Parameter Sensitivity. The Parameters (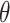 ) of the Trained Models (From Experiment 1) Were Perturbed by
) of the Trained Models (From Experiment 1) Were Perturbed by 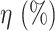 . Note That These Results Are Averaged Over 704 Chest CT Images (All Test Cases) From Experiment 1.
. Note That These Results Are Averaged Over 704 Chest CT Images (All Test Cases) From Experiment 1.
Perturbation 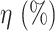
|
Anam-Net (Average Dice Score) | UNet (Average Dice Score) |
|---|---|---|
| −0.1 | 0.193 | 0.136 |
| −0.01 | 0.414 | 0.387 |
| −0.001 | 0.859 | 0.776 |
| 0.0 | 0.863 | 0.775 |
| 0.001 | 0.846 | 0.769 |
| 0.01 | 0.324 | 0.445 |
| 0.1 | 0.172 | 0.095 |
Shan et al. [61] have proposed a human-in-the-loop (HITL) strategy to improve the annotations (i.e., using image segmentation) required for training the network to quantify COVID-19 infection; this work initially used a small batch of segmentations (obtained from a radiologist) to train the VB-network. This approach utilized the trained network to generate a rough segmentation, which was then corrected by the radiologists, and the fixed segmentation improved the network performance in an iterative fashion [61]. The HITL strategy requires intervention from the radiologists for accurately training the deep learning model. Furthermore, this strategy is computationally expensive as opposed to the proposed Anam-Net, which takes about 27 min for an end to end training. Note that the number of parameters involved in the UNet model is 31.07 million as opposed to 4.47 million in the case of Anam-Net (see Table IV), making the proposed approach computationally efficient with an added advantage of being trainable with a smaller data set without compromising performance (see Table VII). The performance of other state-of-the-art lightweight or heavy models is subpar compared to the proposed Anam-Net, and the observed improvement was at least 1.24 times among lightweight networks and twice compared to other massive networks.
Even radiologist’s performance for differentiating COVID-19 pneumonia from non-COVID-19 pneumonia could be as low as 60% [62], and stand-alone chest CT images without any feature engineering might not reveal distinct patterns of COVID-19. The proposed method can quickly provide the most affected region in chest CT for a radiologist to enable faster diagnosis. Current evaluation of the COVID-19 or otherwise other pneumonia severity by the radiologist at best is semiquantitative and typically performed by visual scoring [15]. Any automated methods will always provide unbiased estimates, which is desirable in clinical practice. Techniques such as the proposed one will pave the way for effective and better management of COVID-19 and associated morbidity. As there is a push for low-dose chest CT [16], the variation in protocols demands to the retraining of CNNs; thus, the proposed method will be able to meet this need and provide the versatility without compromising the accuracy of the outcome.
The number of CT scans utilized for training a deep learning model to quantify the lung infection, as provided in [61], was 249, while Zheng et al. [59] and Xu et al. [60] utilized 499 CT scans and 618 CT slices, respectively. The proposed Anam-Net utilized only 90 chest CT images in Experiment 1 and resulted in an average Dice score of 0.87 averaged over 704 test images (which is almost eight times larger than the training set). Note that, in this experiment, we intentionally had a smaller training set and a larger test set, as, in the current pandemic situation, it is challenging to obtain accurate annotations from radiologists, which have increased clinical load. This work aimed to propose a novel network specifically designed for the task at hand and benchmark it among already existing state-of-the-art networks on large test data with a constrain of available training data being limited. In Experiment 3, we conducted a cross data set examination to further analyze the generalizability of Anam-Net, and even in this case, Anam-Net gave good Dice scores for abnormal and normal lung regions (refer to Table IX). The lightweight CNNs based on attentive hierarchical spatial pyramid modules were recently proposed to segment the abnormal regions in COVID-19 chest CT images [12]; the network had about 472.44k parameters. Even with ImageNet pretraining, this model gave only 0.84 sensitivity. Its performance was inferior to the standard UNet approach when test data were limited to only 40 chest CT images [12]. Note that Qiu et al. [12] utilize the same training data as that of Experiment 1 in this work. The proposed method in this work, Anam-Net, was stand-alone and did not require any pretraining; furthermore, Anam-Net was validated with larger test data (i.e., with 704 chest CT images in Experiment 1, 545 chest CT images in Experiment 2, and 704 chest CT images in Experiment 3). The Anam-Net showed improved specificity, accuracy, and Dice similarity score compared to standard UNet (see Tables VII–IX and other state-of-the-art deep learning models.
The study presented here has few limitations, the first one being the dimensionality of chest CT images being restricted to 2-D. The operations performed here can be applied in three-dimensions, and a detailed study in this respect will be taken up as future work. The analysis of the results on the chest CT images (test cases) indicate that the proposed Anam-Net is inherently biased to the peripheral part of the lung (can be observed in Fig. 4), and most COVID-19 chest CT images have the manifestation of peripheral abnormalities [8]–[10], but these peripheral abnormalities might be absent in few cases especially in asymptomatic and pediatric patients, which brings down the Dice similarity score. Low Dice scores can be further improved by incorporating spatial-semantic context into the Anam-Net; this will also be explored in the future. As there is an increased variation in chest CT protocols to reduce the effective dose to the patients without compromising the diagnostic accuracy [16], the amount of fully annotated chest CT data acquired under the same protocol is still a challenge. The work presented here provides a solution to this challenge explicitly by requiring significantly less training data without compromising the accuracy of the segmentation, making them attractive and easy to deploy in the clinic. These novel methods are critical for making deep learning methods more appealing for real-time COVID-19 imaging studies.
VII. Conclusion
This work presented an anamorphic depth embedding-based lightweight CNN, called Anam-Net, to segment anomalies in COVID-19 chest CT images. As chest CT imaging is becoming the main workhorse for staging and managing COVID-19, the methodology proposed here is the need of the hour. The results from the chest CT images (test cases) across the three experiments showed that the proposed method could provide good Dice similarity scores for abnormal and normal regions. Furthermore, the Anam-Net was benchmarked against other state-of-the-art lightweight and heavy networks, such as ENet, UNet++, SegNet, Attention UNet, LEDNet, and DeepLabV3+, and found to provide higher specificity, accuracy, and Dice score averaged over all the chest CT test images across three different experiments. The advantage of Anam-Net compared to other models is low computational complexity (requiring 50% of the training time compared to the next best performing network and the number of parameters being seven times fewer), making it attractive to be deployed in a clinical setting. Anam-Net’s model size is in the order of tens of megabytes (to be specific, 17.2 MB) and makes it easily deployable in mobile platforms to provide a quick assessment of the abnormalities in COVID-19 chest CT images. The deployment in mobile and embedded hardware platforms confirmed that the proposed Anam-Net is well suited for the point of care settings.
Acknowledgment
The authors are thankful to Dr. Johannes Hofmanninger for providing the lung masks as open-source, which were utilized in this work. Naveen Paluru acknowledges the Prime Ministers Research Fellowship (PMRF).
Biographies

Naveen Paluru received the Master’s by Research degree from the Department of Electrical Engineering, Indian Institute of Technology (IIT), Tirupati, India, in 2019. He is currently pursuing the Ph.D. degree with the Department of Computational and Data Sciences, Indian Institute of Science (IISc), Bengaluru, India.
His main research interests are machine learning and deep learning for automated medical image analysis.
Mr. Paluru has been awarded the Prime Ministers Research Fellowship (PMRF) in 2020, for the doctoral fellowship.

Aveen Dayal received the bachelor’s degree in computer science and engineering from BML Munjal University, Gurgaon, India, in 2020.
He is currently a Visiting Research Student with the Department of Information and Communication Technology, University of Agder, Grimstad, Norway. His main research interests are in machine learning and deep learning for autonomous cyber–physical systems.

Håvard Bjørke Jenssen received the M.B., B.Ch., and B.A.O. degrees from the Royal College of Surgeons in Ireland, Dublin, Ireland, in 2013.
He is currently a Radiologist with Oslo University Hospital, Oslo, Norway. His main research interests are in the segmentation of radiological imagery and the application of artificial intelligence in radiological practice.

Tomas Sakinis is currently pursuing the Ph.D. degree in a project involving automated segmentation within neuroradiology from the University of Oslo.
He is currently a Radiologist with Oslo University Hospital—Rikshospitalet, Oslo, Norway, where he completed his radiological residency. His main research interest is in the use of iterative and interactive AI-aided segmentation and the application of these methods in clinical practice. He is also leading the development of MedSeg—a free browser-based segmentation tool with AI capabilities.

Linga Reddy Cenkeramaddi (Senior Member, IEEE) received the master’s degree in electrical engineering from IIT Delhi, New Delhi, India, in 2004, and the Ph.D. degree in electrical engineering from the Norwegian University of Science and Technology (NTNU), Trondheim, Norway, in 2011.
He worked for Texas Instruments, Bengaluru, India, in mixed-signal circuit design before joining the Ph.D. program at NTNU. After finishing his Ph.D. degree, he worked in radiation imaging for an atmosphere space interaction monitor (ASIM mission to International Space Station) at the University of Bergen, Bergen, Norway, from 2010 to 2012. He is currently an Associate Professor with the University of Agder, Grimstad, Norway. His main scientific interests are in cyber–physical systems, autonomous systems, and wireless embedded systems.

Jaya Prakash received the B.Tech. degree in information technology from the Amrita School of Engineering, Bengaluru, India, in 2010, and the M.Sc. degree in engineering and the Ph.D. degree in medical imaging from the Indian Institute of Science, Bengaluru, in 2012 and 2014, respectively.
Prior to his current position as an Assistant Professor with the Department of Instrumentation and Applied Physics, Indian Institute of Science, he was the Group Leader of the Computational Data Analytics Group, Institute for Biological and Medical Imaging in Helmholtz Zentrum Munich, Oberschleißheim, Germany. His research interests are image reconstruction, inverse problems, optoacoustic imaging, biomedical instrumentation, and biomedical optics.

Phaneendra K. Yalavarthy (Senior Member, IEEE) received the M.Sc. degree in engineering from the Indian Institute of Science, Bengaluru, India, in 2004, and the Ph.D. degree in biomedical computation from the Dartmouth College, Hanover, NH, USA, in 2007.
He is currently an Associate Professor with the Department of Computational and Data Sciences, Indian Institute of Science. His research interests include medical image computing, medical image analysis, and biomedical optics.
Dr. Yalavarthy is a Senior Member of the Society of Photo-optical Instrumentation Engineers (SPIE) and Optical Society of America (OSA). He also serves as an Associate Editor of the IEEE Transactions on Medical Imaging.
Funding Statement
This work was supported in part by the WIPRO GECDS Collaborative Laboratory of Artificial Intelligence in Healthcare and Medical Imaging and the Indo-Norwegian Collaboration in Autonomous Cyber-Physical Systems (INCAPS) project, 287918 of the INTPART program supported by the Norwegian Research Council.
Contributor Information
Naveen Paluru, Email: naveenp@iisc.ac.in.
Håvard Bjørke Jenssen, Email: hbjenssen@gmail.com.
Tomas Sakinis, Email: sakinis.tomas@gmail.com.
Linga Reddy Cenkeramaddi, Email: linga.cenkeramaddi@uia.no.
Jaya Prakash, Email: jayap@iisc.ac.in.
Phaneendra K. Yalavarthy, Email: yalavarthy@iisc.ac.in.
References
- [1].Memish Z. A., Perlman S., Van Kerkhove M. D., and Zumla A., “Middle East respiratory syndrome,” The Lancet, vol. 395, no. 10229, pp. 1063–1077, 2020. [Online]. Available: https://www.thelancet.com/journals/lancet/article/PIIS0140-6736(19)33221-0/fulltext [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Chen N.et al. , “Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in wuhan, China: A descriptive study,” Lancet, vol. 395, no. 10223, pp. 507–513, Feb. 2020. [Online]. Available: http://www.sciencedirect.com/science/article/pii/S0140673620302117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Rubin G. D.et al. , “The role of chest imaging in patient management during the COVID-19 pandemic: A multinational consensus statement from the Fleischner society,” Radiology, vol. 296, no. 1, pp. 172–180, Jul. 2020, doi: 10.1148/radiol.2020201365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hope M. D., Raptis C. A., and Henry T. S., “Chest computed tomography for detection of coronavirus disease 2019 (COVID-19): Don’t rush the science,” Ann. Internal Med., vol. 173, no. 2, pp. 147–148, Jul. 2020. [Online]. Available: https://doi.org/10.7326/M20-1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Fang Y.et al. , “Sensitivity of chest CT for COVID-19: Comparison to RT-PCR,” Radiology, vol. 296, no. 2, pp. E115–E117, 2020, doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Inui S.et al. , “Erratum: Chest CT findings in cases from the cruise ship ‘diamond princess’ with coronavirus disease 2019 (COVID-19),” Radiol.: Cardiothoracic Imag., vol. 2, no. 2, 2020, Art. no. e200110, doi: 10.1148/ryct.2020200110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Driggin E.et al. , “Cardiovascular considerations for patients, health care workers, and health systems during the COVID-19 pandemic,” J. Amer. College Cardiol., vol. 75, no. 18, pp. 2352–2371, 2020. [Online]. Available: http://www.sciencedirect.Chest computed tomography for detection of coronavirus disease 2019 (COVID-19): Don’t rush the http://sciencecom/science/article/pii/S0735109720346374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kanne J. P., “Chest CT findings in 2019 novel coronavirus (2019-nCoV) infections from wuhan, China: Key points for the radiologist,” Radiology, vol. 295, no. 1, pp. 16–17, Apr. 2020, doi: 10.1148/radiol.2020200241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chung M.et al. , “CT imaging features of 2019 novel coronavirus (2019-nCoV),” Radiology, vol. 295, no. 1, pp. 202–207, 2020. [Online]. Available: https://pubs.rsna.org/doi/10.1148/radiol.2020200230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Caruso D.et al. , “Chest CT features of COVID-19 in Rome, Italy,” Radiology, vol. 296, no. 2, pp. 201–237, 2020, doi: 10.1148/radiol.2020201237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Li L.et al. , “Using artificial intelligence to detect COVID-19 and community-acquired pneumonia based on pulmonary CT: Evaluation of the diagnostic accuracy,” Radiology, vol. 296, no. 2, pp. E65–E71, Aug. 2020, doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Qiu Y., Liu Y., Li S., and Xu J., “MiniSeg: An extremely minimum network for efficient COVID-19 segmentation,” 2020, arXiv:2004.09750. [Online]. Available: http://arxiv.org/abs/2004.09750 [DOI] [PubMed]
- [13].Pan F.et al. , “Time course of lung changes at chest CT during recovery from coronavirus disease 2019 (COVID-19),” Radiology, vol. 295, no. 3, pp. 715–721, Jun. 2020, doi: 10.1148/radiol.2020200370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Wang Y.et al. , “Temporal changes of CT findings in 90 patients with COVID-19 pneumonia: A longitudinal study,” Radiology, vol. 296, no. 2, 2020, Art. no.200843, doi: 10.1148/radiol.2020200843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Huang L.et al. , “Serial quantitative chest CT assessment of COVID-19: A deep learning approach,” Radiol.: Cardiothoracic Imag., vol. 2, no. 2, 2020, Art. no. e200075, doi: 10.1148/ryct.2020200075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Iezzi R.et al. , “Tailoring protocols for chest CT applications: When and how?” Diagnostic Interventional Radiol., vol. 23, no. 6, pp. 420–427, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zhu Y., Tan Y., Hua Y., Zhang G., and Zhang J., “Automatic segmentation of ground-glass opacities in lung CT images by using Markov random field-based algorithms,” J. Digit. Imag., vol. 25, no. 3, pp. 409–422, 2012, doi: 10.1007/s10278-011-9435-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhou J., Chang S., Metaxas D. N., Zhao B., Schwartz L. H., and Ginsberg M. S., “Automatic detection and segmentation of ground glass opacity nodules,” in Proc. Int. Conf. Med. Image Comput. Comput.-Assist. Intervent. Berlin, Germany: Springer, 2006, pp. 784–791, doi: 10.1007/11866565_96. [DOI] [PubMed] [Google Scholar]
- [19].Jung J., Hong H., and Goo J. M., “Ground-glass nodule segmentation in chest CT images using asymmetric multi-phase deformable model and pulmonary vessel removal,” Comput. Biol. Med., vol. 92, pp. 128–138, Jan. 2018, doi: 10.1016/j.compbiomed.2017.11.013. [DOI] [PubMed] [Google Scholar]
- [20].Fan D.-P.et al. , “Inf-net: Automatic COVID-19 lung infection segmentation from CT images,” IEEE Trans. Med. Imag., vol. 39, no. 8, pp. 2626–2637, Aug. 2020. [DOI] [PubMed] [Google Scholar]
- [21].Ouyang X.et al. , “Dual-sampling attention network for diagnosis of COVID-19 from community acquired pneumonia,” IEEE Trans. Med. Imag., vol. 39, no. 8, pp. 2595–2605, Aug. 2020. [DOI] [PubMed] [Google Scholar]
- [22].Akkus Z., Kostandy P., Philbrick K. A., and Erickson B. J., “Robust brain extraction tool for CT head images,” Neurocomputing, vol. 392, pp. 189–195, Jun. 2020. [Google Scholar]
- [23].Paszke A., Chaurasia A., Kim S., and Culurciello E., “ENet: A deep neural network architecture for real-time semantic segmentation,” 2016, arXiv:1606.02147. [Online]. Available: http://arxiv.org/abs/1606.02147
- [24].Ronneberger O., Fischer P., and Brox T., “U-Net: Convolutional networks for biomedical image segmentation,” in Medical Image Computing and Computer-Assisted Intervention (MICCAI) (Lecture Notes in Computer Science), vol. 9351. Cham, Switzerland: Springer, 2015, pp. 234–241. [Online]. Available: https://arxiv.org/abs/1505.04597 and [Online]. Available: http://lmb.informatik.uni-freiburg.de/Publications/2015/RFB15a [Google Scholar]
- [25].Zhou Z., Siddiquee M. M. R., Tajbakhsh N., and Liang J., “UNet++: Redesigning skip connections to exploit multiscale features in image segmentation,” IEEE Trans. Med. Imag., vol. 39, no. 6, pp. 1856–1867, Jun. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Badrinarayanan V., Kendall A., and Cipolla R., “SegNet: A deep convolutional encoder-decoder architecture for image segmentation,” IEEE Trans. Pattern Anal. Mach. Intell., vol. 39, no. 12, pp. 2481–2495, Dec. 2017. [DOI] [PubMed] [Google Scholar]
- [27].Oktay O.et al. , “Attention U-net: Learning where to look for the pancreas,” 2018, arXiv:1804.03999. [Online]. Available: http://arxiv.org/abs/1804.03999
- [28].Wang Y.et al. , “Lednet: A lightweight encoder-decoder network for real-time semantic segmentation,” in Proc. IEEE Int. Conf. Image Process. (ICIP), Sep. 2019, pp. 1860–1864. [Google Scholar]
- [29].Chen L.-C., Zhu Y., Papandreou G., Schroff F., and Adam H., “Encoder-decoder with atrous separable convolution for semantic image segmentation,” in Proc. Eur. Conf. Comput. Vis. (ECCV), 2018, pp. 801–818. [Google Scholar]
- [30].COVID-19 CT Segmentation Dataset. Accessed: May 29, 2020. [Online]. Available: http://medicalsegmentation.com/covid19/
- [31].Github Page. Accessed: Jun. 7, 2020. [Online]. Available: https://github.com/NaveenPaluru/Segmentation-COVID-19
- [32].Hofmanninger J., Prayer F., Pan J., Rohrich S., Prosch H., and Langs G., “Automatic lung segmentation in routine imaging is primarily a data diversity problem, not a methodology problem,” 2020, arXiv:2001.11767. [Online]. Available: http://arxiv.org/abs/2001.11767 [DOI] [PMC free article] [PubMed]
- [33].Jun M.et al. , “COVID-19 CT lung and infection segmentation dataset (version 1.0) [Data set],” Zenodo, Tech. Rep., 2020, doi: 10.5281/zenodo.3757476. [DOI]
- [34].Yu Q.et al. , “C2FNAS: Coarse-to-fine neural architecture search for 3D medical image segmentation,” in Proc. IEEE/CVF Conf. Comput. Vis. Pattern Recognit. (CVPR), Jun. 2020, pp. 4126–4135. [Google Scholar]
- [35].Guo D.et al. , “Organ at risk segmentation for head and neck cancer using stratified learning and neural architecture search,” in Proc. IEEE/CVF Conf. Comput. Vis. Pattern Recognit. (CVPR), Jun. 2020, pp. 4223–4232. [Google Scholar]
- [36].Li X., Yu L., Chen H., Fu C.-W., Xing L., and Heng P.-A., “Transformation-consistent self-ensembling model for semisupervised medical image segmentation,” IEEE Trans. Neural Netw. Learn. Syst., early access, Jun. 1, 2020, doi: 10.1109/TNNLS.2020.2995319. [DOI] [PubMed]
- [37].Clough J., Byrne N., Oksuz I., Zimmer V. A., Schnabel J. A., and King A., “A topological loss function for deep-learning based image segmentation using persistent homology,” IEEE Trans. Pattern Anal. Mach. Intell., early access, Sep. 4, 2020, doi: 10.1109/TPAMI.2020.3013679. [DOI] [PMC free article] [PubMed]
- [38].Zhou Y., Yen G. G., and Yi Z., “Evolutionary compression of deep neural networks for biomedical image segmentation,” IEEE Trans. Neural Netw. Learn. Syst., vol. 31, no. 8, pp. 2916–2929, Aug. 2020. [DOI] [PubMed] [Google Scholar]
- [39].Wang G.et al. , “DeepIGeoS: A deep interactive geodesic framework for medical image segmentation,” IEEE Trans. Pattern Anal. Mach. Intell., vol. 41, no. 7, pp. 1559–1572, Jul. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Gu Z.et al. , “CE-net: Context encoder network for 2D medical image segmentation,” IEEE Trans. Med. Imag., vol. 38, no. 10, pp. 2281–2292, Oct. 2019. [DOI] [PubMed] [Google Scholar]
- [41].Chen C., Dou Q., Chen H., Qin J., and Heng P. A., “Unsupervised bidirectional cross-modality adaptation via deeply synergistic image and feature alignment for medical image segmentation,” IEEE Trans. Med. Imag., vol. 39, no. 7, pp. 2494–2505, Jul. 2020. [DOI] [PubMed] [Google Scholar]
- [42].Huang G., Liu Z., Van Der Maaten L., and Weinberger K. Q., “Densely connected convolutional networks,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit., Jul. 2017, pp. 4700–4708. [Google Scholar]
- [43].Jha D.et al. , “ResUNet++: An advanced architecture for medical image segmentation,” in Proc. IEEE Int. Symp. Multimedia (ISM), Dec. 2019, pp. 225–2255. [Google Scholar]
- [44].Jha D., Riegler M. A., Johansen D., Halvorsen P., and Johansen H. D., “DoubleU-net: A deep convolutional neural network for medical image segmentation,” 2020, arXiv:2006.04868. [Online]. Available: http://arxiv.org/abs/2006.04868
- [45].He K., Zhang X., Ren S., and Sun J., “Deep residual learning for image recognition,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit. (CVPR), Jun. 2016, pp. 770–778. [Google Scholar]
- [46].Yu F. and Koltun V., “Multi-scale context aggregation by dilated convolutions,” 2015, arXiv:1511.07122. [Online]. Available: http://arxiv.org/abs/1511.07122
- [47].Jin J., Dundar A., and Culurciello E., “Flattened convolutional neural networks for feedforward acceleration,” 2014, arXiv:1412.5474. [Online]. Available: http://arxiv.org/abs/1412.5474
- [48].Szegedy C.et al. , “Going deeper with convolutions,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit. (CVPR), Jun. 2015, pp. 1–9. [Google Scholar]
- [49].Ioffe S. and Szegedy C., “Batch normalization: Accelerating deep network training by reducing internal covariate shift,” 2015, arXiv:1502.03167. [Online]. Available: http://arxiv.org/abs/1502.03167
- [50].Krizhevsky A., Sutskever I., and Hinton G. E., “Imagenet classification with deep convolutional neural networks,” in Proc. Adv. Neural Inf. Process. Syst., 2012, pp. 1097–1105. [Google Scholar]
- [51].COVID-19 Database. Accessed: May 29, 2020. [Online]. Available: https://www.sirm.org/en/category/articles/covid-19-database/
- [52].COVID-19. Accessed: May 29, 2020. [Online]. Available: https://radiopaedia.org/articles/covid-19-3
- [53].COVID-19 Radiology Data Collection and Preparation for Artificial Intelligence. Accessed: May 29, 2020. [Online]. Available: https://bit.ly/34t8Ih4
- [54].Paszke A.et al. , “Pytorch: An imperative style, high-performance deep learning library,” in Proc. Adv. Neural Inf. Process. Syst., 2019, pp. 8026–8037. [Google Scholar]
- [55].Kingma D. P. and Ba J., “Adam: A method for stochastic optimization,” 2014, arXiv:1412.6980. [Online]. Available: http://arxiv.org/abs/1412.6980
- [56].Pytorch Mobile. Accessed: Sep. 15, 2020. [Online]. Available: https://pytorch.org/mobile/android/
- [57].Android Studio. Accessed: Sep. 15, 2020. [Online]. Available: https://developer.android.com/studio
- [58].Tajbakhsh N., Jeyaseelan L., Li Q., Chiang J. N., Wu Z., and Ding X., “Embracing imperfect datasets: A review of deep learning solutions for medical image segmentation,” Med. Image Anal., vol. 63, Jul. 2020, Art. no. 101693. [Online]. Available: http://www.sciencedirect.com/science/article/pii/S136184152030058X [DOI] [PubMed] [Google Scholar]
- [59].Wang X.et al. , “A weakly-supervised framework for COVID-19 classification and lesion localization from chest CT,” IEEE Trans. Med. Imag., vol. 39, no. 8, pp. 2615–2625, 2020, doi: 10.1109/TMI.2020.2995965. [DOI] [PubMed] [Google Scholar]
- [60].Xu X.et al. , “Deep learning system to screen coronavirus disease 2019 pneumonia,” 2020, arXiv:2002.09334. [Online]. Available: http://arxiv.org/abs/2002.09334
- [61].Shan F.et al. , “Lung infection quantification of COVID-19 in CT images with deep learning,” 2020, arXiv:2003.04655. [Online]. Available: http://arxiv.org/abs/2003.04655
- [62].Bai H. X.et al. , “Performance of radiologists in differentiating COVID-19 from non-COVID-19 viral pneumonia at chest CT,” Radiology, vol. 296, no. 2, 2020, Art. no.200823, doi: 10.1148/radiol.2020200823. [DOI] [PMC free article] [PubMed] [Google Scholar]




