Abstract
A recent and extensive research activity highlighted the process behind the attack and spread of COVID-19 in the human body. What emerged is that the SARS-CoV-2 virus makes use of both the ACE2 receptor, expressed by pneumocytes in the ephitelial alveolar lining, and by the endothelium to spread the disease and to replicate itself. Since the endothelium is an extended tissue lying in the circulatory system, this may lead to a large state of diffuse endothelial inflammation with serious clinical consequences. This situation may be further compromised by the immune system, that may generate pro-inflammatory cytokines (IL-6) as a consequence of the infection. In this paper we propose and analyze a molecular communication system, designed for the detection of excessive IL-6 level, that allows monitoring its evolution in the blood vessels. The proposed analysis was performed by using the BiNS2 simulator, which is suitable for the numerical analysis of flow-based molecular communications in blood vessels, as well as Markov models of the endothelium.
Keywords: SARS-CoV-2, COVID-19, cytokine storm, flow-based molecular communications, blood vessel simulations, monitoring system
I. Introduction
An extensive research activity recently carried out highlighted the process behind the attack and spread of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in the human body [1], [2], [3], leading to the so-called COVID-19 disease. The SARS-CoV-2 virus makes use of the ACE2 receptor expressed by pneumocytes in the epithelial alveolar lining to penetrate the host and replicate itself. This is the process behind significant lung lesions observed. In addition, the ACE2 receptor is also present on endothelial cells, present in other organs such as kidneys and heart, thus making the situation even more complex. The endothelium is essential for the regulation of vascular tone and the maintenance of vascular homoeostasis. Therefore, the process could end with a large state of diffuse endothelial inflammation, which includes impaired microcirculatory function in different vascular beds, with serious clinical consequences. The observed situation is further compromised by the immune response and the related generation of pro-inflammatory cytokines. Observations in cases of critical infections led to the identification of the so-called “cytokine storm,” that is, a violent immune response due to the massive generation of Interleukin-6 (IL-6) [2]. Precisely, high levels of IL-6 have been linked to the emergence of the worst consequences of the infection.
Starting from these experimental observations, the purpose of the research that was conducted is twofold. On the one hand, it prefigures a system for monitoring the presence and evolution of the IL-6 level in blood vessels, which makes use of the vascular molecular communication mechanisms, recently studied in different case studies [4], [5], [6]. The second objective is to evaluate the expected effect of clinical attempts to contain the state of inflammation by making use of antibodies which are intended to occupy IL-6 receptors (IL-6R) and therefore contain the violence of inflammation.
The research approach followed in this paper leverages flow-based molecular communications in blood vessels. This communication environment was recently considered for some purposes having common features with the present research. In particular, communications based in the vascular environment were proposed for monitoring biological parameters and controlling specific processes, such as hyperviscosity [6]. In this paper we propose and analyze a molecular communication system designed for the detection of excessive IL-6 levels. This approach is expected to reduce the time needed to detect the dangerous evolution of the infection and allows a timely treatment of it. After having defined the entities involved in the communication chain, we analyzed the resulting system through numerical simulations and theoretical models. For this purpose, we used BiNS2 [4], [7], which is a well assessed simulator for molecular communications, as well as queuing theory, which is a theoretical tool frequently used in molecular communications.
The emulated communication environment analyzes a portion of a blood vessel. The presence of IL-6 particles is emulated though an emission of a bursts of particles, modeling the intrinsic discrete emission of cytokines, which combines at reception site as a continuous flow of particles. This emulates the cytokine storm mentioned above, having the potential of hitting the endothelium at the reception section. Hence, the emulated receiving process consists of particle absorption by the vessel wall. The simulation of this phenomenon, together with a theoretical analysis of absorption process based on Markov models, allows dimensioning the detection process through receptors having the properties to bind with IL-6 cytokines. Examples of synthetic receptors able to absorb cytokines and further transmit biological signals are shown in [8] and [9]. Furthermore, having the possibility to numerically evaluate the reception process at the receiver site, it is also possible to evaluate the concentration of antibodies necessary to effectively counteract the inflammatory process by occupying IL-6R receptors exposed by the endothelium. This approach is the basis for designing external devices used for monitoring purposes. We emphasize that, due to the increasing diffusion of personalized medicine, patients should be constantly monitored, especially during prophylaxis administration as well as during randomized, controlled trials to assess efficacy and safety [10].
To sum up, the main contribution of this paper consists of the definition of a monitoring system for cytokines storm detection in patients affected by the COVID-19 disease. Its pillars are the analysis of the mechanisms underlying the absorption of cytokines at molecular level, and the identifications of key enabling technologies that could be used in the future to implement such devices.
The paper is organized as follows. Section II includes some background on molecular communications in the cardiovascular system and some details about the COVID-19 disease. In Section III, we present a monitoring system leveraging on mathematical models as well as some enabling technologies in the medical field. Numerical results are presented in Section IV, using both simulations and analytical models for estimating of the assimilation levels. Conclusions and future outlooks are presented in Section V.
II. Biological Background
The SARS-CoV-2 outbreak highlighted the need for fast methods to detect infections and track their evolution continuously. To this aim, it is urgent to provide any clinical support to professionals working to slow down the rapid spread of this infectious disease. The most accurate known methods for executing diagnostic tests for SARS-CoV-2 are based on direct analysis of diagnostic specimens. They can be taken from the upper or lower respiratory tract. However, some issues exist, such as the latency for obtaining accurate results, also due to the need of repeating tests several times when the reliability of results is not satisfactory.
Furthermore, some experimental results show that the accuracy of Reverse transcriptase-polymerase chain reaction (RT-PCR) may vary by specimen type [11] revealing that some test results were discordant for some specimen pairs taken from the same patient [12], [13], thus the infectiousness cannot be clearly detected.
From the current studies on SARS-CoV-2 emerged that this infection is a systemic disease. It affects some important organs, such as heart, lung, liver and kidney, due to the ACE2 receptor which is commonly exposed by organs and tissues. This is an entry receptor for the virus and makes it possible the spreading of the infection. Indeed, the infection drives and develops the inflammation, triggering the cytokine storm, which causes both significant consequences in disease progression [14] and damage to organs and tissues [15], thus it is a major cause of deaths in COVID-19 [16].
The mechanism underneath the internalization of the SARS-CoV-2 inside the host cells exploits the high affinity between the ACE2 receptor and the virus spike glycoprotein (S). The glycoprotein (S) consists of two subunits: S1 attaches on ACE2, and S2 leads to the virus internalization via their membranes fusion [17]. The virus internalization results in the release of its RNA, which is translated and processed by the infected cell to assemble virus progeny promoting viral replication [18].
The second relevant consequence concerns the binding to ACE2 receptor. Its internalization triggers a cascade of effects, from the down-regulation of both ACE2 receptors and angiotensin-(1–7) levels, up to the increase of angiotensin-II (AngII), causing free AngII accumulation in plasma. AngII interacts on both immune cells and tissue-resident cells. The activated synthesis of AngII from tissue-resident cells promotes the productions of proinflammatory factors. In more detail, the combination of the above effects activates NF-
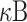 pathway, triggering the transcription of interleukin-6 (IL-6) cytokine on both infected and immune cells [19]. As a consequence, the IL-6 concentration is upregulated towards abnormal levels by the virus itself [20] while the anti-inflammatory cytokines, such as IL-10, are downregulated, thus enforcing the cytokine storm [21].
pathway, triggering the transcription of interleukin-6 (IL-6) cytokine on both infected and immune cells [19]. As a consequence, the IL-6 concentration is upregulated towards abnormal levels by the virus itself [20] while the anti-inflammatory cytokines, such as IL-10, are downregulated, thus enforcing the cytokine storm [21].
In the early stage of infections (not just the viral ones), different types of cytokines are synthesized in order to activate a rapid immune response. The IL-6 is one of the principal pro-inflammatory cytokines, which plays an important role in cytokine storm [10]. Indeed, high levels of IL-6 both activate the coagulation system and increase vascular permeability, promoting the rapid spread of inflammation [22], [23].
Moreover, in normal inflammatory responses IL-6 stimulates the immune response towards the classic-signaling pathway, exploiting the IL-6 membrane receptor (IL-6R) exposed by immune cells only, whereas the majority of cells do not expose it. The membrane-bound IL-6R does not possess intracellular signaling capabilities, but signals instead through interaction with membrane-bound glycoprotein 130 (gp130). The complex IL-6-IL-6R binds to gp130, which then initiates signaling through its intracellular domain. However, IL-6R can be found also in its soluble form (sIL-6R). Its concentration during infections and inflammations increases considerably due to AngII accumulation in plasma, caused by ACE2 down-regulation due to SARS-CoV-2 binding, thus facilitating the formation of IL-6/sIL-6R compound [1]. This enables the activation of the pro-inflammatory trans-signaling pathway that allows the binding of IL-6/sIL-6R compound with gp130, which acts as another type of IL-6R and it is exposed by a wide range of cells, thus expanding its range towards cells that normally are not its target (including the endothelial ones) [19], [24], [25]. When the IL-6/IL-6R/gp130 complex is formed, the IL-6 signal is transmitted via multiple intracellular signaling pathways. This results in systemic hyperinflammation involving the production of additional cytokines, including IL-6, as well as increased vascular endothelial growth factor, which contribute to vascular hyperpermeability, leakiness, hypotension, and pulmonary dysfunction, as reported in [10]. In more detail, lung tissue damages may be worsen by high levels of IL-6, which are induced by the SARS-CoV-2 infection [26], [27]. Severe viral damages were observed also in blood vessels, where the inflammatory progression hurts the endothelial cells, increasing the cardiac load and promoting the coagulation activation, which may also lead to thrombosis on thin vessels [28].
Although an inflammatory response is usually a useful effect to counteract harmful stimuli (i.e., pathogens, cell injuries, etc.) [29], an overproduction of a wide range of inflammatory cytokines by the infected cells, due to the cytokine storm, causes the continuous recruitment of immune cells on the sites of inflammation, thus amplifying the inflammatory response. This exponential growth is due to the loss of the negative feedback on the immune system, leading to repeated attacks of the autoimmune system which could cause multiple organs failure [21].
What emerges from the current biomedical research approaches and results is that the clinical activities can highly benefit from the ICT research results. Such contribution is not aimed at replacing the usage of diagnostic specimens, but rather to contribute in order to obtain more accurate diagnoses.
The proposal shown in this paper supports the approaches presented above with the aim of contributing to the detection of COVID-19 infection. The approach consists of detecting the biomarkers in blood vessels associated with COVID-19. Thus, the proposal leverages the research activities aiming to identify such biomarkers, even those able to correctly identify both convalescent COVID-19 patients and those who could not survive. For example, [30] shows the identification of a number of biomarkers including the immunological interleukin (IL-6). The identification of effective laboratory biomarkers capable of classifying patients according to their risk is essential to ensure timely treatment. Our proposal contributes to pursue this objective through an approach that can be included in the family of detection techniques based on molecular communication systems [5], [6], along with all others defined for monitoring body parameters (i.e., blood pressure, blood viscosity, glucose level, inflammation level and so on) [31], [32], [33].
III. System Model
The analyzed scenario focuses on the signal detection in a short section of a blood vessel, that we call monitored section, which is covered by a number of receptors. Essentially, receptors are distributed over the endothelial cells, which cover the inner surface of blood vessels. Those receptors are compliant with IL-6 molecules and, upon the absorption of such molecules, may trigger a cascade of secondary events, such as the increase in STAT3 phosphorylation and the upregulation of intercellular adhesion molecule-1 (ICAM-1) [34], [35], [36], [37], which are involved in monocyte recruitment to the endothelium [38]. Basically, the idea is to exploit the IL-6-induced ICAM-1 expression mediated via the JAK/Stat3 signaling pathway, monitoring the exposition of such adhesion molecules in order to estimate the level of absorbed IL-6 molecules in the monitored section.
A. Enabling Technologies for the Monitoring System
The complete design of the monitoring system is beyond the scope of this paper. However, we refer to some recent papers that illustrate the enabling technologies that can be used for injecting and detecting specific molecules across the endothelium, making it possible to monitor some sections.
For example, a graphene patch for diabetics is proposed in [39]. It can be used to both analyze the body sweat (for glucose and pH values monitoring) and to trigger the release of drugs through an array of microneedles. In [40], [41] a lot of flexible and noninvasive chemical sensors for health monitoring are presented. Those sensors use some electrodes for driving the propagation of ions and glucose by means of iontophoresis. The epidermal device presented in [42] allows performing a continuous mapping of the blood flow by means of thermal sensors. In [43], [44], [45], the microneedles array for both sensing chemicals and releasing drugs and vaccines through the skin are presented.
The available technologies summarized so far allow implementing of a monitoring system capable of injecting molecules and monitoring their binding along a short section of a blood vessel.
In fact, the injection of molecules can be realized by the well known transdermal drug delivery processes, based on microneedles or iontophoresis [46]. The molecules released in this way activate the portion of the blood vessel that should be monitored.
The candidate molecules to be used for this purpose are aptamers [47]. Due to their similarity with antibodies, they expose an innate targeted recognition capability, and can easily bind to specific molecules [48], [49], [50]. Moreover, aptamers can be engineered in labs and quickly produced by chemical synthesis [49], [50]. Their binding capability makes the dynamic tracking of cellular molecules possible, where one arm binds to a fluorescent protein and the others to the desired targets [48]. After the injection/release, the aptamer-fluorescent protein compound perfuses towards the blood vessel in order to bind to the target molecules (i.e., the ICAM-1 as introduced above).
Finally, fluorophores and green fluorescent proteins (GFP) can emit light signals of specific wavelengths that can be easily detected [51].
To sum up, the in-vivo fluorescent imaging techniques based on GFP allow detecting the level of fluorescence emitted by the overall aptamer-GFP compounds, effectively hooked to the exposed ICAM-1 in the monitored section. The larger the intensity of the fluorescence, the larger the amount of ICAM-1 expressed, which is in turn related to the amount of absorbed IL-6 cytokines. This fluorescence should be detected by an external optical sensor.
B. Endothelium Absorption Dynamics
The considered monitoring system includes fixed receivers, represented by the endothelial cells, as sketched at the beginning of Section III-A. In this scenario, we do not consider a canonical transmitter for a localized molecule release. We assume that the ligands (IL-6 cytokines) compliant with the surface receptors (IL-6R), exposed on specific cells or with its soluble version in the plasma (sIL-6R), are already circulating in the blood circulatory system. The secondary IL-6 receptors (i.e., gp130) are commonly exposed on endothelial cells. The goal of the monitoring system is to discriminate between normal and abnormal concentration values of these molecules, in order to trigger a timely reaction, thus limiting the spread of infection. This can be done by the systemic release of a number of antibodies compliant with secondary IL-6 receptors, which can limit the absorption of IL-6 cytokines and thus the overall progress of the inflammatory disease. Thus, in the considered environment, the following chemical reactions can take place:
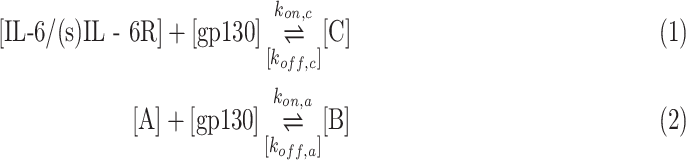 |
where
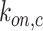 and
and
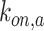 denote the on-rate constant of cytokines and antibodies, respectively, whereas the
denote the on-rate constant of cytokines and antibodies, respectively, whereas the
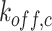 and
and
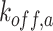 denote the relevant off-rates. C is the complex obtained by the reaction between cytokines and compliant receptor, whereas B is that obtained by the antibody reaction.
denote the relevant off-rates. C is the complex obtained by the reaction between cytokines and compliant receptor, whereas B is that obtained by the antibody reaction.
The on-rates depends on the frequency of collision between a ligand and a free compliant receptor. In more detail, in [52] the constant
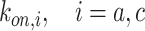 , is defined as the product of
, is defined as the product of
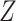 , which represents the hitting rate of a ligand on an unbound receptor, and
, which represents the hitting rate of a ligand on an unbound receptor, and
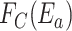 , which is the fraction of collisions having an energy large enough to form a bond, that is higher than the threshold
, which is the fraction of collisions having an energy large enough to form a bond, that is higher than the threshold
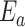 . Thus,
. Thus,
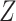 depends in some way on the number of receptors. In our model, we assume that each collision between a ligand and a receptor generates an assimilation, that is
depends in some way on the number of receptors. In our model, we assume that each collision between a ligand and a receptor generates an assimilation, that is
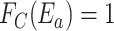 .
.
Instead, the off-rates depend on the average lifetime of obtained complex [53]
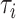 ,
,
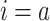 ,
,
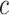 , that is
, that is
 |
The value of off-rates were estimated by means of lab experiments and can be found in literature. As for the IL-6 reaction, a typical value of
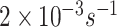 was found in [54], although the authors of [55] point out that this value can be either increased or decreased by about one order of magnitude (from
was found in [54], although the authors of [55] point out that this value can be either increased or decreased by about one order of magnitude (from
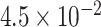 to
to
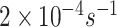 , depending on the presence of other substances.
, depending on the presence of other substances.
In order to describe our system, we consider the main drivers leading to molecules movement and the consequent absorption. In particular, being the bloodstream the considered environment, we have to consider three main forces moving molecules and contributing to hitting the vessel walls and potentially compliant receptors:
-
1)The Brownian motion of the particles [56], which is characterized by the diffusion coefficient, given by
where
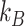 is the Boltzmann constant,
is the Boltzmann constant,
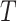 is the temperature expressed in kelvins,
is the temperature expressed in kelvins,
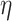 is the viscosity of the medium, and
is the viscosity of the medium, and
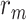 is the radius of the considered molecules;
is the radius of the considered molecules; -
2)
The drag force of the blood, which in real vessels is known to assume the so-called Casson profile [57], with a nearly constant velocity in the center of the vessel and velocity values vanishing when approaching to the vessel walls;
-
3)
Collisions with blood cells, mainly red blood cells (RBCs), which cause bounces and, as stated in [58], marginalization towards endothelium.
We follow an approach similar to that already adopted in [59], where the analysis is done by considering first a single receptor modeled as a pure loss system with a single server, and then it is extended to the whole receiver. In this case it is the set of endothelial cells in the monitored region. According to general model presented in [52], the status of the receiver at any time
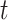 can be represented by the value of a random variable
can be represented by the value of a random variable
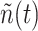 , which denotes the number of ligand-receptor bonds at time
, which denotes the number of ligand-receptor bonds at time
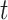 . Thus, the receiver behavior is modeled by the random process
. Thus, the receiver behavior is modeled by the random process
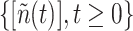 . In general,
. In general,
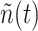 increases when new ligand-receptor bonds are created, and decreases due to the internalization of these bonds [60], or upon their rupture, which makes the receptors available again. Our model inherits some concepts from both [52] and [59], but extends the latter with a multi-service approach [61]. In fact, in the model considered in this paper, each receptor can create bonds with two types of ligands, that is the IL-6 and an agonist antibody, with the constraint that each receptor can create only a single bond at a time. Thus, our receptor can be modeled as a vector of random processes
increases when new ligand-receptor bonds are created, and decreases due to the internalization of these bonds [60], or upon their rupture, which makes the receptors available again. Our model inherits some concepts from both [52] and [59], but extends the latter with a multi-service approach [61]. In fact, in the model considered in this paper, each receptor can create bonds with two types of ligands, that is the IL-6 and an agonist antibody, with the constraint that each receptor can create only a single bond at a time. Thus, our receptor can be modeled as a vector of random processes
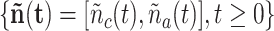 , that is a bi-dimensional, birth-death, discrete-valued, continuous-time Markov process, with
, that is a bi-dimensional, birth-death, discrete-valued, continuous-time Markov process, with
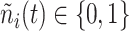 , for
, for
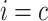 ,
,
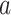 . At any time
. At any time
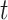 , a state
, a state
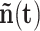 of this process is acceptable only if
of this process is acceptable only if
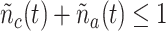 , since the receptor can form a bond with the IL-6 or its antagonist antibody. Hence, in this system the following states are the only acceptable at any time:
, since the receptor can form a bond with the IL-6 or its antagonist antibody. Hence, in this system the following states are the only acceptable at any time:
-
•
State (0,0): The receptor if free. The probability that the Markov chain is in this state at time
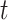 is denoted as
is denoted as
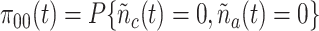 .
. -
•
State (1,0): The receptor formed a bond with the cytokine IL-6. The probability that the Markov chain is in this state at time
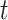 is denoted as
is denoted as
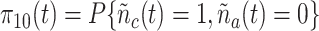 .
. -
•
State (0,1): The receptor formed a bond with the antagonist antibody of cytokine IL-6. The probability that the Markov chain is in this state at time
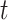 is denoted as
is denoted as
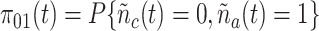 .
.
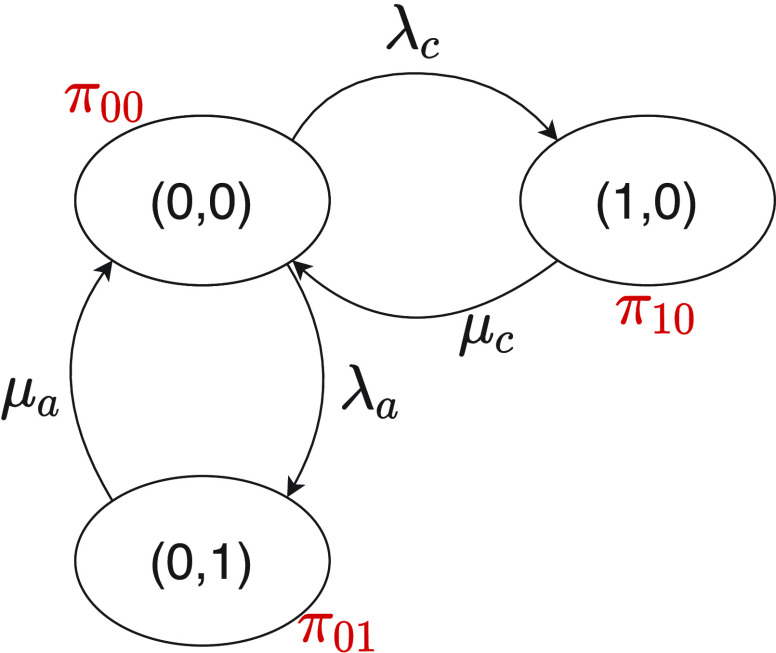
The relevant state transition diagram is depicted in Fig. 1, where
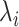 indicates the bond formation rate for each ligand
indicates the bond formation rate for each ligand
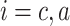 , whereas
, whereas
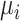 indicates the release rate for each bond, that is
indicates the release rate for each bond, that is
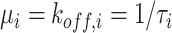 . Then the dynamical equations governing the state probabilities are the following:
. Then the dynamical equations governing the state probabilities are the following:
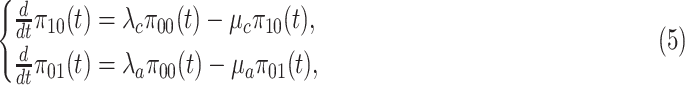 |
Fig. 1.
State transition diagram for the reception process at each receptor. State variables are indicated inside the oval, whereas state probability are indicated in red close to it.
In the steady state, considering also that
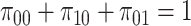 , the probability of having a bond with an IL-6 molecule results
, the probability of having a bond with an IL-6 molecule results
 |
A quantity of high interest to be estimated is the arrival rate, that is the rate at which a ligand can form a bond with a compliant receptor. In [52], the arrival rate
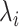 can be expressed as the product of
can be expressed as the product of
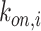 and the concentration of ligands of type
and the concentration of ligands of type
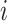 close to the considered receptor,
close to the considered receptor,
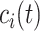 . We assume that this concentration is uniform in the region close to the vessel wall, thus it is not dependent on the specific endothelial cell or receptor. Taking into account what mentioned above about the definition of
. We assume that this concentration is uniform in the region close to the vessel wall, thus it is not dependent on the specific endothelial cell or receptor. Taking into account what mentioned above about the definition of
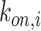 , then it follows that
, then it follows that
 |
where
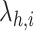 is the rate at which a molecule of type
is the rate at which a molecule of type
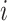 hits the endothelium in the monitored area, whereas
hits the endothelium in the monitored area, whereas
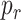 is the probability that a molecule will hit a receptor when a collision with the endhotelium happens. The value of
is the probability that a molecule will hit a receptor when a collision with the endhotelium happens. The value of
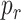 can be estimated as
can be estimated as
 |
where
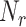 is the number of receptors per endothelial cell,
is the number of receptors per endothelial cell,
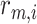 is the radius of the ligand
is the radius of the ligand
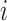 ,
,
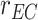 is the radius of the receptor, and
is the radius of the receptor, and
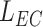 is the side of the endothelial cell, which we model as a square. This means that
is the side of the endothelial cell, which we model as a square. This means that
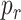 is the fraction of the surface of each endothelial cell covered by receptors. It also depends on the radius of the molecule (
is the fraction of the surface of each endothelial cell covered by receptors. It also depends on the radius of the molecule (
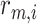 ), since we assume that the reception process occurs also upon a minimal contact between the receptor and the ligand.
), since we assume that the reception process occurs also upon a minimal contact between the receptor and the ligand.
At the cell level, the number of receptors
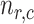 that established a bond with an IL-6 molecule can be modeled through a Binomial distribution
that established a bond with an IL-6 molecule can be modeled through a Binomial distribution
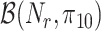 , since each cell has
, since each cell has
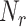 receptors and each of them is occupied by an IL-6 molecules with probability
receptors and each of them is occupied by an IL-6 molecules with probability
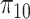 . Thus, the average number of receptors busy with a IL-6 bond (
. Thus, the average number of receptors busy with a IL-6 bond (
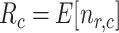 ) in each endothelial cell is simply equal to
) in each endothelial cell is simply equal to
 |
In order to keep the value of
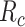 below a safety threshold value
below a safety threshold value
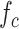 , so that the resulting inflammatory process due to cytokines’ storm may be controlled, it is necessary to vary the concentration of antagonist antibodies
, so that the resulting inflammatory process due to cytokines’ storm may be controlled, it is necessary to vary the concentration of antagonist antibodies
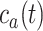 and to select antibodies with a suitable value of
and to select antibodies with a suitable value of
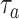 . We analyze the effect of concentration and average bond lifetime of antibodies in the performance evaluation section.
. We analyze the effect of concentration and average bond lifetime of antibodies in the performance evaluation section.
IV. Performance Evaluation
A. Simulation Setup
The analysis of the proposed system is based on an extensive simulation campaign performed by the experimentally assessed simulator BiNS2. It is a particle-based simulator developed in Java [4], [5], able to leverage NVIDIA CUDA libraries to significantly reduce the computational time by partitioning the collision detection workload over several GPU devices [7]. Its main features are:
-
•
Definition of the internal state of each simulated object in order to perform specific functions/behaviors (i.e., emission of molecules according to specific pattern/modulation schemes, reception schemes by exposing different type of receptors with different properties, computational/pre-processing features, etc.);
-
•
Tracking the position of the simulated objects (i.e., cells and molecules released in the simulated scenario) which are mapped by spherical shapes;
-
•
Fine grained algorithm for detecting and managing any interaction between cells/molecules by means of collisions or absorption with surface receptors;
-
•
Customized 3D environment composed of a set of simple containers (i.e., spheres, cylinders, cubes);
-
•
Customizable propagation rules for both the whole simulated space (i.e., free diffusion, flow-based diffusion) and for each moving object (i.e., an active or passive propulsion system).
The simulated blood vessel section is modelled as a cylindrical object filled by spherical elements of different sizes, which emulate the behavior of the blood cells (i.e., RBC, white blood cells, and platelets), as detailed in [4].
The simulated scenario is characterized by the following parameters: plasma viscosity, blood flow velocity, number of released molecules, release points, reception areas, number and size of receptors. For the purposes of this analysis we have set their values in the range of healthy conditions, as reported in Table I. The simulated blood vessel portion was filled by proper concentration of blood cells [4].
TABLE I. Simulation Parameters.
| Symbols | Value | Description |
|---|---|---|
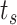 |
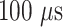 |
Simulation time step |
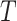 |
310°
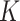
|
Plasma temperature |
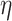 |
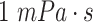 |
Plasma viscosity |
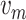 |
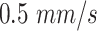 |
Blood flow velocity |
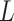 |
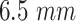 |
Blood vessel length |
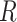 |
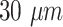 |
Blood vessel radius |
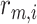 |
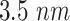 |
Molecule radius |
 |
[5, 10, 25, 50]
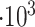
|
Molecules released |
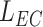 |
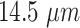 |
Endothelial cell (EC) size |
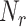 |
[2000, 1500, 1000, 500] | Number of receptors/EC |
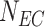 |
897 | Number of ECs |
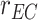 |
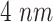 |
EC receptor radius |
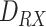 |
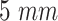 |
TX-Monitored section distance |
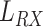 |
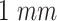 |
Monitored section length |
Since we consider small blood vessels, the corresponding velocity profile exhibits a laminar profile (see [4] and references therein). This condition can be modeled by the Poiseuille’s equation:
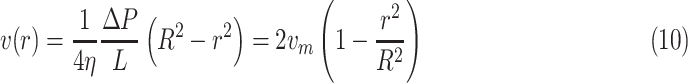 |
where
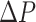 is the pressure drop along a vessel section of length
is the pressure drop along a vessel section of length
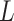 ,
,
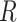 is the vessel radius,
is the vessel radius,
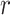 is the distance from the longitudinal axis of the vessel, and
is the distance from the longitudinal axis of the vessel, and
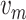 is average flow velocity. This equation shows that the flow velocity assumes a parabolic profile, with its maximum on the longitudinal axis, decreasing to zero close to the vessel walls. This profile is important in order to maintain blood physical integrity and carry out cardiovascular functions, such as margination. It consists of blood cells that, in small vessels, where the mean velocity is low, adhere to the endothelium and migrate outside the vessel passing between endothelial cells. In our simulator, we implemented a drag force enforcing the velocity profile in (10) for all moving objects. However, as it happens in real setting, also in the simulation RBCs tend to occupy the central portion of the vessel. Thus, they aggregate into the Rouleaux form [62], and form a plug flow region along the center of the vessel causing a flattened parabolic velocity profile rather than the expected parabolic velocity profile typical of a Newtonian fluid [57], [62]. This effect, known as Casson profile, is modeled also in our simulations. In addition to the advection mechanism described above, all moving objects in the simulator are subject to diffusion, with a diffusion coefficient
is average flow velocity. This equation shows that the flow velocity assumes a parabolic profile, with its maximum on the longitudinal axis, decreasing to zero close to the vessel walls. This profile is important in order to maintain blood physical integrity and carry out cardiovascular functions, such as margination. It consists of blood cells that, in small vessels, where the mean velocity is low, adhere to the endothelium and migrate outside the vessel passing between endothelial cells. In our simulator, we implemented a drag force enforcing the velocity profile in (10) for all moving objects. However, as it happens in real setting, also in the simulation RBCs tend to occupy the central portion of the vessel. Thus, they aggregate into the Rouleaux form [62], and form a plug flow region along the center of the vessel causing a flattened parabolic velocity profile rather than the expected parabolic velocity profile typical of a Newtonian fluid [57], [62]. This effect, known as Casson profile, is modeled also in our simulations. In addition to the advection mechanism described above, all moving objects in the simulator are subject to diffusion, with a diffusion coefficient
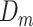 computed as in (4). The effect of diffusion is negligible for large cells (RBCs, white blood cells, and platelets), but it is important for molecules, on both radial and longitudinal directions. For each dimension in a 3D environment, it is implemented by a Gaussian displacement with zero mean and variance equal to
computed as in (4). The effect of diffusion is negligible for large cells (RBCs, white blood cells, and platelets), but it is important for molecules, on both radial and longitudinal directions. For each dimension in a 3D environment, it is implemented by a Gaussian displacement with zero mean and variance equal to
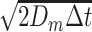 , modeling the Brownian motion. Finally, the simulator also models collisions between moving objects. This phenomenon is particularly important since it is responsible for pushing small objects (i.e., molecules) towards vessel walls, and it is correctly represented by our simulations.
, modeling the Brownian motion. Finally, the simulator also models collisions between moving objects. This phenomenon is particularly important since it is responsible for pushing small objects (i.e., molecules) towards vessel walls, and it is correctly represented by our simulations.
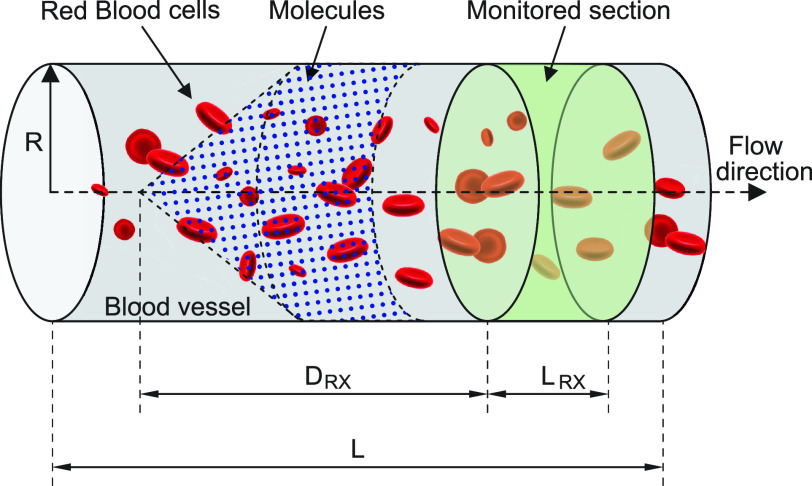
The molecules are emitted on a point position along the longitudinal axis of the vessel (as shown in Fig. 2), with different burst values
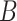 , as reported in Table I. Molecules propagate along the bloodstream without any absorption or reaction (except for collisions among themselves, with vessel walls, or with RBCs) until they reach the monitored section of the vessel, located at distance
, as reported in Table I. Molecules propagate along the bloodstream without any absorption or reaction (except for collisions among themselves, with vessel walls, or with RBCs) until they reach the monitored section of the vessel, located at distance
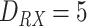 mm from the emission point with a length
mm from the emission point with a length
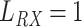 mm.
mm.
Fig. 2.
Simulated scenario in BiNS2 framework. The molecules are released on a pointwise location along the vessel axis. The monitored section is located to the right end of the blood vessel section, at a distance
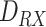 from the release point.
from the release point.
In general, the inner surface of blood vessels are covered by endothelial cells that expose a few thousand receptors on their surface compliant with a subset of cytokines produced by the body. From a simulation point of view, although the simulator can model all surface receptors, the large number of elements to be instantiated as software objects in the simulations (i.e., cells, molecules, receptors) for this analysis may generate a huge volume of interactions among them, which reduces the simulation speed. For this reason we have decided to model the surface receptors of the vessel walls through a probability
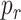 of absorbing the colliding molecules, evaluated according to (8), without any appreciable loss in the simulation accuracy, limiting at the same time both computational load and processing time.
of absorbing the colliding molecules, evaluated according to (8), without any appreciable loss in the simulation accuracy, limiting at the same time both computational load and processing time.
In this specific set of simulations, only the inner surface of the monitored section (see Fig. 2) is covered by endothelial cells, modeled as squares of side
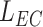 . This means that for each collision of the released molecules with these cells a probability of absorption
. This means that for each collision of the released molecules with these cells a probability of absorption
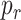 , given by (8), exists. In this analysis we have considered different values of
, given by (8), exists. In this analysis we have considered different values of
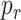 , related to the amount of available receptors
, related to the amount of available receptors
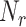 for each endothelial cell, ranging from 500 to 2000. This allows obtaining a preliminary insight about what happens when not all receptors are available to form an IL-6/(s)IL-6R/gp130 complex, because receptors have already formed a ligand-receptor binding with other compatible compounds. It means that the antibodies compliant with the secondary IL-6R can lead to a reduction of IL-6 transcription at cellular level which finally contributes to a reduction of the inflammation level due to the abnormal IL-6 upregulation induced by the SARS-CoV-2 [10], [14]. In fact, therapies with antibodies are still being tested for the treatment of patients [10].
for each endothelial cell, ranging from 500 to 2000. This allows obtaining a preliminary insight about what happens when not all receptors are available to form an IL-6/(s)IL-6R/gp130 complex, because receptors have already formed a ligand-receptor binding with other compatible compounds. It means that the antibodies compliant with the secondary IL-6R can lead to a reduction of IL-6 transcription at cellular level which finally contributes to a reduction of the inflammation level due to the abnormal IL-6 upregulation induced by the SARS-CoV-2 [10], [14]. In fact, therapies with antibodies are still being tested for the treatment of patients [10].
Our analysis assumes that the molecules displacement is already in a steady state when they cross the monitored region. This means that their radial positions does not depend on either the distance from the emission point or its position in the vessel section. To this aim, we resorted to literature results [63], [64]. It results that in microfluidic systems the movement of a burst of emitted particles can be governed by two regimes. The first one is the dispersion regime, where the movement is mainly due to diffusion and the particle distribution is independent of the point or uniform release. The second one is the flow-dominated regime, when advection dominates on diffusion. In our simulation setting, since it results that
 |
then we can assume to be in the dispersion regime, thus without any dependency on the emission point. In addition, the presence of RBCs helps the system to converge to the steady state condition and push molecules towards vessel walls, as discussed in [58].
B. Numerical Results
The simulation results allowed collecting the number of absorptions carried out by surface receptors on the endothelium for each discrete time step
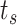 .
.
However, in order to estimate the arrival rate
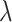 of molecules to any single receptor on the endothelium, we preliminary run a number of simulations while disabling the absorption from the surface receptors (
of molecules to any single receptor on the endothelium, we preliminary run a number of simulations while disabling the absorption from the surface receptors (
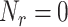 ). During these simulations, we recorded the number of collisions between molecules and endothelium
). During these simulations, we recorded the number of collisions between molecules and endothelium
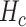 , and monitored the average time
, and monitored the average time
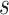 needed by molecules to cross the monitored region, whose length
needed by molecules to cross the monitored region, whose length
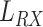 equals 1 mm. The estimated value
equals 1 mm. The estimated value
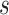 is slightly higher than the average time needed by the blood flow to cross the monitored section (
is slightly higher than the average time needed by the blood flow to cross the monitored section (
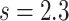 s), which is
s), which is
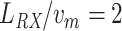 s. This can be explained by the fact that the RBCs tend to push the smaller particles towards the regions of the vessel close to the walls [58], the so-called cell-free layer (CFL) [65], where the presence of RBCs is lower. This behavior is particularly evident in small vessels, which are those of greatest interest for the phenomena we investigate. In this region the flow velocity is significantly reduced due to the parabolic/Casson profile of the flow velocity [57]. Nevertheless, the drag effect of the flow is still present, and it is exerted also by means of collisions between RBCs and molecules, which are both pushed forward and/or towards the vessels wall, thus causing collisions and, potentially, absorption. This is particularly significant in small vessels where the size of CFL tends to vanish. Consequently, it is expected that molecules, which are mainly concentrated in the CFL, have an average velocity slightly smaller than the flow velocity.
s. This can be explained by the fact that the RBCs tend to push the smaller particles towards the regions of the vessel close to the walls [58], the so-called cell-free layer (CFL) [65], where the presence of RBCs is lower. This behavior is particularly evident in small vessels, which are those of greatest interest for the phenomena we investigate. In this region the flow velocity is significantly reduced due to the parabolic/Casson profile of the flow velocity [57]. Nevertheless, the drag effect of the flow is still present, and it is exerted also by means of collisions between RBCs and molecules, which are both pushed forward and/or towards the vessels wall, thus causing collisions and, potentially, absorption. This is particularly significant in small vessels where the size of CFL tends to vanish. Consequently, it is expected that molecules, which are mainly concentrated in the CFL, have an average velocity slightly smaller than the flow velocity.
By using the
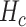 value it is possible to estimate the average number of collisions experienced by a single molecule, which is simply
value it is possible to estimate the average number of collisions experienced by a single molecule, which is simply
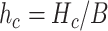 . Then, the collision rate in the monitored area for each molecule is obtained by dividing the average number of collisions per molecule by the average crossing time
. Then, the collision rate in the monitored area for each molecule is obtained by dividing the average number of collisions per molecule by the average crossing time
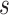 :
:
 |
Finally, the average rate of collisions close to the area occupied by each receptor
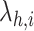 can be estimated by the sum of the collision rates
can be estimated by the sum of the collision rates
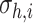 of the molecules simultaneously present in the monitored area, which is a number
of the molecules simultaneously present in the monitored area, which is a number
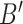 slightly lower than the burst size due to dispersion dynamics (
slightly lower than the burst size due to dispersion dynamics (
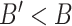 ), scaled by the total number of receptors, that is:
), scaled by the total number of receptors, that is:
 |
Thus, the average arrival rate
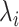 at each receptor on the endothelium of the monitored area can be obtained by using (7).
at each receptor on the endothelium of the monitored area can be obtained by using (7).
We assessed these assumptions by running a number of simulations with different values of
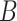 and
and
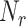 , in order to evaluate the behavior of the system for different concentration values. In these simulations, we used the following widely used model of absorbing receptors. Essentially, a molecule can occupy a receptor for a single discrete simulation time step
, in order to evaluate the behavior of the system for different concentration values. In these simulations, we used the following widely used model of absorbing receptors. Essentially, a molecule can occupy a receptor for a single discrete simulation time step
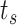 . After this, the molecule is removed from simulation and the receptor returns available. Although this model does not fit well the system described in Section III-B, it allowed us to validate some of our assumptions, such as the fact that molecules are close to the vessel walls, as well as the existence of a linear relationship between
. After this, the molecule is removed from simulation and the receptor returns available. Although this model does not fit well the system described in Section III-B, it allowed us to validate some of our assumptions, such as the fact that molecules are close to the vessel walls, as well as the existence of a linear relationship between
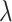 and molecule concentration. In fact, although it is possible to simulate the model described in Section III-B, the usage of realistic value for
and molecule concentration. In fact, although it is possible to simulate the model described in Section III-B, the usage of realistic value for
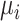 would make this type of simulations incredibly time consuming, since the simulation time step, evaluating collision dynamics, is equal to
would make this type of simulations incredibly time consuming, since the simulation time step, evaluating collision dynamics, is equal to
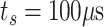 .
.
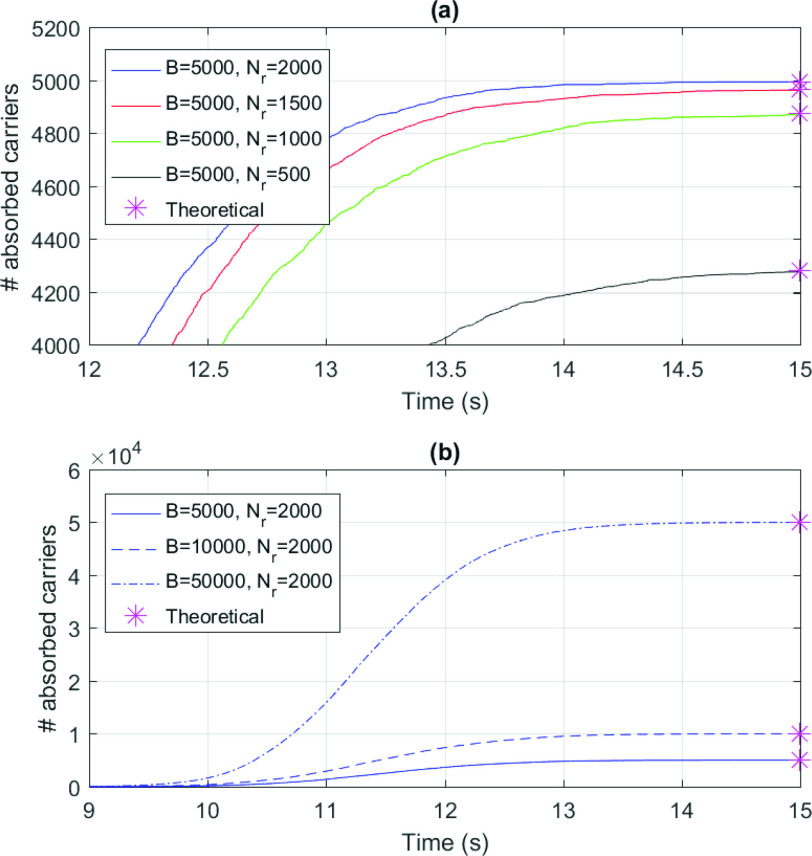
Fig. 3 shows a comparison of the results of our simulations with the theoretical predictions determined as follows. By recalling that
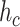 is the average number of collisions experienced by a molecule with vessels walls, it could be absorbed only in case of collision with a receptor, as stated in Section III-B. This event can happen after a maximum of
is the average number of collisions experienced by a molecule with vessels walls, it could be absorbed only in case of collision with a receptor, as stated in Section III-B. This event can happen after a maximum of
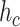 attempts, thus giving the following absorbing probability
attempts, thus giving the following absorbing probability
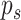 in the monitored region:
in the monitored region:
 |
Fig. 3.
Number of absorbed molecules in numerical simulations for different numbers or surface receptors
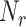 (a) and size of the emitted burst
(a) and size of the emitted burst
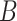 (b). Asterisks indicated theoretical predictions.
(b). Asterisks indicated theoretical predictions.
The number of absorbed molecules can be estimated as
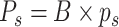 . An excellent match between theory and simulations can be observed. The lesson learned from these simulations is that results in Fig. 3 also confirms our previous assumptions done in the system model. In particular, we assumed that the massive presence of RBCs pushes much smaller molecules towards the vessels wall, thus favoring their absorption. This was confirmed for both small and large concentration of molecules, thus proving the correctness of our model. In fact, when the number of available receptors is large enough (
. An excellent match between theory and simulations can be observed. The lesson learned from these simulations is that results in Fig. 3 also confirms our previous assumptions done in the system model. In particular, we assumed that the massive presence of RBCs pushes much smaller molecules towards the vessels wall, thus favoring their absorption. This was confirmed for both small and large concentration of molecules, thus proving the correctness of our model. In fact, when the number of available receptors is large enough (
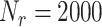 ), all molecules have a chance of being assimilated by receptors on the endothelium. This stimulates a further consideration, which is central for our approach: if there are enough free receptors, also large concentrations of molecules can be absorbed. When this number decreases, also modest concentrations of molecules could not be completely absorbed. Thus, the next step is to include in the model the presence of antagonist antibodies to better model this phenomenon. Clearly, in order to consider a realistic duration of bond lifetime
), all molecules have a chance of being assimilated by receptors on the endothelium. This stimulates a further consideration, which is central for our approach: if there are enough free receptors, also large concentrations of molecules can be absorbed. When this number decreases, also modest concentrations of molecules could not be completely absorbed. Thus, the next step is to include in the model the presence of antagonist antibodies to better model this phenomenon. Clearly, in order to consider a realistic duration of bond lifetime
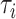 , simulations modeling particle collisions are not the most suitable approach. Thus, we resort to our theoretical model, that is based on the same assumptions of the simulations.
, simulations modeling particle collisions are not the most suitable approach. Thus, we resort to our theoretical model, that is based on the same assumptions of the simulations.
Let us consider a scenario in which the arrival rate of cytokines, that was obtained by simulations, is
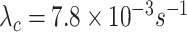 , whereas the average bond lifetime
, whereas the average bond lifetime
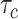 is
is
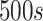 [54]. As for antibodies, we used the following values:
[54]. As for antibodies, we used the following values:
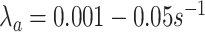 , and
, and
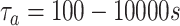 . The number of surface receptors is
. The number of surface receptors is
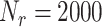 .
.
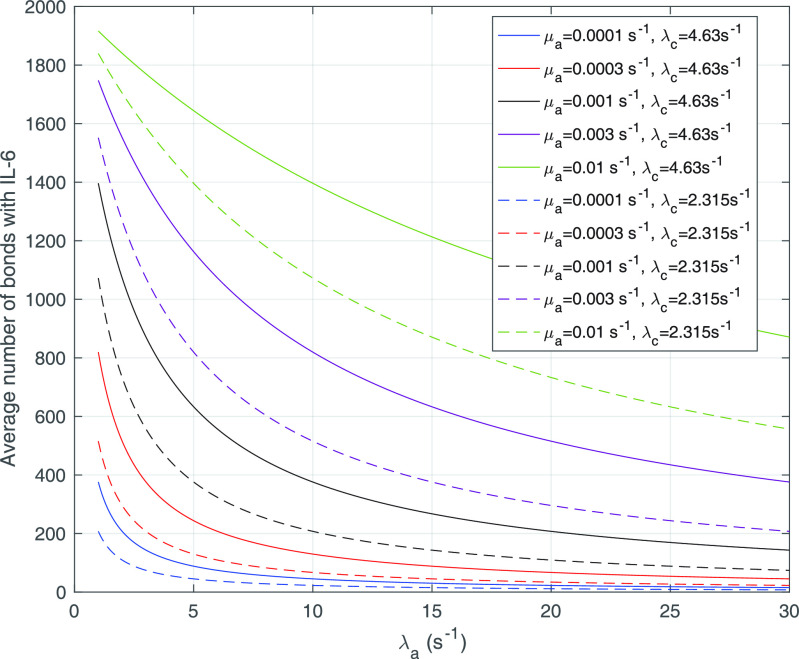
Fig. 4 shows the average number of bonds (
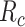 , see (9)) with IL-6 cytokines as a function of the arrival rate of antagonist antibodies, for different values of their bond lifetimes with IL-6R receptors. It emerges that the factor of major impact is the average lifetime of antibody bonds, whereas their concentration is important but less significant. In fact, large values of
, see (9)) with IL-6 cytokines as a function of the arrival rate of antagonist antibodies, for different values of their bond lifetimes with IL-6R receptors. It emerges that the factor of major impact is the average lifetime of antibody bonds, whereas their concentration is important but less significant. In fact, large values of
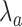 are necessary for enforcing the same response to cytokines for lower values of
are necessary for enforcing the same response to cytokines for lower values of
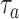 . In order deepen the analysis, we also give the
. In order deepen the analysis, we also give the
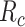 values for a halved value of
values for a halved value of
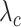 . From the analysis of the results we can infer that, for the same value of
. From the analysis of the results we can infer that, for the same value of
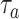 , approximately a half dose of antibodies should be administered to achieve the same effect, which confirm our expectations.
, approximately a half dose of antibodies should be administered to achieve the same effect, which confirm our expectations.
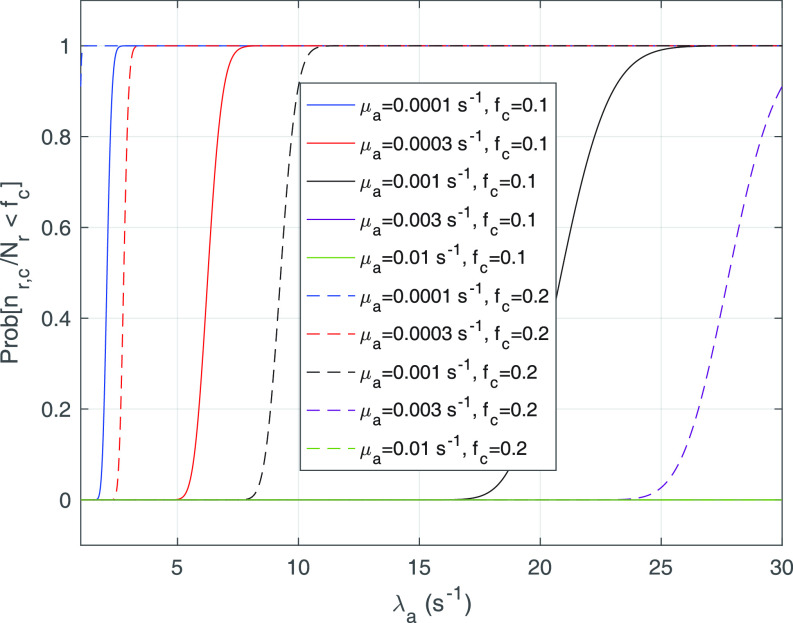
Fig. 5.
Probability of having a number of bonds between the IL-6/sIL-6R complex and the gp130 receptor at least equal to the activation threshold, that is
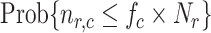 , for
, for
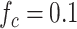 and 0.2, as a function of the arrival rate of antibody molecules
and 0.2, as a function of the arrival rate of antibody molecules
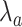 . Different values of average lifetime of antibody’s bonds are considered.
. Different values of average lifetime of antibody’s bonds are considered.
Fig. 4.
Average number of bonds with IL-6 (
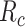 ) as a function of the arrival rate of antibody molecules, for different values of the average lifetime of antibodies bonds with IL-6R.
) as a function of the arrival rate of antibody molecules, for different values of the average lifetime of antibodies bonds with IL-6R.
Finally, we analyze the probability to keep the fraction of receptors engaged with IL-6 bonds (
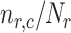 ) below the activation threshold
) below the activation threshold
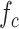 . We considered two threshold values, equal to 10% and 20%, respectively. In fact, larger numbers of IL-6-IL-6R bonds can trigger the immunity response we want to avoid. In the figure it emerges a sort of threshold behavior with respect to the concentration of antibodies, which depends on the lifetime of antibody bonds. This phenomenon is less evident for lower values of
. We considered two threshold values, equal to 10% and 20%, respectively. In fact, larger numbers of IL-6-IL-6R bonds can trigger the immunity response we want to avoid. In the figure it emerges a sort of threshold behavior with respect to the concentration of antibodies, which depends on the lifetime of antibody bonds. This phenomenon is less evident for lower values of
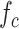 , when the problem is more challenging. In fact, the lower the value of
, when the problem is more challenging. In fact, the lower the value of
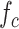 , the easier the system can enter an active state triggered by the cytokines storm. As discussed in [66], low values of the fraction of busy receptors are often enough to trigger a specific response. It is also interesting to see that, for small values of the bonds lifetime of antibodies, also large values of antibody concentration cannot counteract the cytokines storm, since for
, the easier the system can enter an active state triggered by the cytokines storm. As discussed in [66], low values of the fraction of busy receptors are often enough to trigger a specific response. It is also interesting to see that, for small values of the bonds lifetime of antibodies, also large values of antibody concentration cannot counteract the cytokines storm, since for
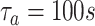 the probability of not overcoming the threshold is always 0.
the probability of not overcoming the threshold is always 0.
V. Conclusion
In this paper we show a proposal aiming to support the clinical activities in the detection of SARS-Cov-2 infection and the relevant monitoring of the disease evolution. Our solution is not an alternative to currently used detection approach based on diagnostic specimens, but it is rather to be considered a contribution aiming to obtain more accurate diagnoses. The proposal is based on a molecular communication system in blood vessels, designed for the detection of excessive concentration of IL-6 levels through an absorption process by receptors located on the inner surface of vessels. An extensive numerical analysis based on simulation allowed dimensioning the detection process through receptors having the properties to bind with IL-6 cytokines and to determine the possibility to counteract the resulting inflammatory process through antagonist antibodies. For this purpose we modeled each receptor with twofold bond capabilities, with two types of ligands, that is the IL-6 and an agonist antibody and analyzed the effect of concentration and average bond lifetime of antibodies by simulation. The bonding process of receptors with an IL-6 molecules was modeled through a Binomial distribution at the endothelial cell level, used to find the conditions for controlling the resulting inflammatory process due to IL-6 cytokine storm by a suitable concentration of antagonist antibodies. One of the main lessons learned is that the average lifetime of their bonds with IL-6R receptors has a very significant impact on the action of antibodies. Only for suitable values of this parameter the cytokines storm can be counteracted with a reasonable concentration (dose) of antibodies.
The future research activities on this subject consist of the usage of machine learning algorithm to combine the outcome of different detection systems in order to improve the degree of confidence of the combined detection process.
Biographies

Luca Felicetti received the master’s degree in information and communication engineering from the University of Perugia in 2011, and the Ph.D. degree in information engineering in 2015. He is currently a Postdoctoral Researcher with the Department of Engineering, University of Perugia. His current research interests focus on nanoscale networking and communications, and network and service management architectures for 5G networks.

Mauro Femminella (Member, IEEE) received the master’s and Ph.D. degrees in electronic engineering from the University of Perugia in 1999 and 2003, respectively. Since 2006, he has been an Assistant Professor with the Department of Engineering, University of Perugia. He has coauthored about 100 papers in international journals and refereed international conferences. His current research interests focus on nanoscale networking and communications, big data systems, and network and service management architectures and protocols for 5G networks.

Gianluca Reali (Member, IEEE) received the Ph.D. degree in telecommunications from the University of Perugia, Italy, in 1997, where he was a Researcher with the Department of Electronic and Information Engineering from 1997 to 2004. In 1999, he visited the Computer Science Department, UCLA. He has been an Associate Professor with the Department of Engineering, University of Perugia since 2005. His research activities include resource allocation over packet networks, wireless networking, network management, multimedia services, big data management, and nanoscale communications.
References
- [1].Magro G., “SARS-CoV-2 and COVID-19: Is interleukin-6 (IL-6) the ‘culprit lesion’ of ARDS onset? What is there besides Tocilizumab? SGP130Fc,” Cytokine X, vol. 2, no. 2, 2020, Art. no. 100029. [Online]. Available: http://www.sciencedirect.com/science/article/pii/S2590153220300094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Coomes E. A. and Haghbayan H.. (2020). Interleukin-6 in COVID-19: A Systematic Review and Meta-Analysis. [Online]. Available: https://www.medrxiv.org/content/early/2020/04/03/2020.03.30.20048058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Varga Z.et al. , “Endothelial cell infection and endotheliitis in COVID-19,” Lancet, vol. 395, no. 10234, pp. 1417–1418, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Felicetti L., Femminella M., and Reali G., “Simulation of molecular signaling in blood vessels: Software design and application to atherogenesis,” Nano Commun. Netw., vol. 4, no. 3, pp. 98–119, 2013. [Google Scholar]
- [5].Felicetti L., Femminella M., Reali G., and Liò P., “A molecular communication system in blood vessels for tumor detection,” in Proc. ACM 1st Annu. Int. Conf. Nanoscale Comput. Commun., 2014, pp. 1–9. [Google Scholar]
- [6].Felicetti L., Femminella M., and Reali G., “A molecular communications system for live detection of hyperviscosity syndrome,” IEEE Trans Nanobiosci., vol. 19, no. 3, pp. 410–421, Jul. 2020. [DOI] [PubMed] [Google Scholar]
- [7].Stroobant P.et al. , “Parallel algorithms for simulating interacting carriers in nanocommunication,” Nano Commun. Netw., vol. 20, pp. 20–30, Jun. 2019. [Online]. Available: http://www.sciencedirect.com/science/article/pii/S1878778918300991 [Google Scholar]
- [8].Engelowski E.et al. , “Synthetic cytokine receptors transmit biological signals using artificial ligands,” Nat. Commun., vol. 9, no. 1, p. 2034, May 2018. [Online]. Available: https://doi.org/10.1038/s41467-018-04454-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Floss D. M. and Scheller J., “Naturally occurring and synthetic constitutive-active cytokine receptors in disease and therapy,” Cytok. Growth Factor Rev., vol. 47, pp. 1–20, Jun. 2019. [Online]. Available: http://www.sciencedirect.com/science/article/pii/S135961011930053X [DOI] [PubMed] [Google Scholar]
- [10].Fajgenbaum D. C. and June C. H., “Cytokine storm,” New England J. Med., vol. 383, no. 23, pp. 2255–2273, 2020. [Online]. Available: https://doi.org/10.1056/NEJMra2026131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wang W.et al. , “Detection of SARS-CoV-2 in different types of clinical specimens,” JAMA, vol. 323, no. 18, pp. 1843–1844, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kujawski S. A.et al. , “Clinical and virologic characteristics of the first 12 patients with coronavirus disease 2019 (COVID-19) in the United States,” Nat. Med., vol. 26, no. 6, pp. 861–868, 2020. [DOI] [PubMed] [Google Scholar]
- [13].Wolfel R.et al. , “Virological assessment of hospitalized patients with COVID-2019,” Nature, vol. 581, no. 7809, pp. 465–469, 2020. [DOI] [PubMed] [Google Scholar]
- [14].Song P., Li W., Xie J., Hou Y., and You C., “Cytokine storm induced by SARS-CoV-2,” Clinica Chimica Acta, vol. 509, pp. 280–287, Oct. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wang D.et al. , “Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China,” JAMA, vol. 323, no. 11, pp. 1061–1069, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Tang L., Yin Z., Hu Y., and Mei H., “Controlling cytokine storm is vital in COVID-19,” Front. Immunol., vol. 11, Nov. 2020, Art. no. 570993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Saxena S. K., Kumar S., Baxi P., Srivastava N., Puri B., and Ratho R. K., “Chasing COVID-19 through SARS-CoV-2 spike glycoprotein,” Virusdisease, vol. 31, no. 4, pp. 399–407, Dec. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kumar S., Nyodu R., Maurya V. K., and Saxena S. K., “Morphology, genome organization, replication, and pathogenesis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2),” in Medical Virology: From Pathogenesis to Disease Control. Singapore: Springer, 2020, pp. 23–31. [Online]. Available: https://doi.org/10.1007/978-981-15-4814-7_3 [Google Scholar]
- [19].Iwasaki M., Saito J., Zhao H., Sakamoto A., Hirota K., and Ma D., “Inflammation triggered by SARS-CoV-2 and ACE2 augment drives multiple organ failure of severe COVID-19: Molecular mechanisms and implications,” Inflammation, vol. 44, no. 1, pp. 13–34, Oct. 2020. [Online]. Available: https://doi.org/10.1007/s10753-020-01337-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wang W.et al. , “Up-regulation of IL-6 and TNF-a induced by SARS-coronavirus spike protein in murine macrophages via NF-kappaB pathway,” Virus Res., vol. 128, nos. 1–2, pp. 1–8, Sep. 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ding Y.et al. , “The clinical pathology of severe acute respiratory syndrome (SARS): A report from China,” J. Pathol., vol. 200, no. 3, pp. 282–289, Jul. 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Tisoncik J. R., Korth M. J., Simmons C. P., Farrar J., Martin T. R., and Katze M. G., “Into the eye of the cytokine storm,” Microbiol. Mol. Biol. Rev., vol. 76, no. 1, pp. 16–32, Mar. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Tanaka T., Narazaki M., and Kishimoto T., “Immunotherapeutic implications of IL-6 blockade for cytokine storm,” Immunotherapy, vol. 8, no. 8, pp. 959–970, 2016. [DOI] [PubMed] [Google Scholar]
- [24].Kang S.et al. , “Il-6 trans-signaling induces plasminogen activator inhibitor-1 from vascular endothelial cells in cytokine release syndrome,” Proc. Nat. Acad. Sci., vol. 117, no. 36, pp. 22351–22356, 2020. [Online]. Available: https://www.pnas.org/content/117/36/22351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Tanaka T., Narazaki M., and Kishimoto T., “IL-6 in inflammation, immunity, and disease,” Cold Spring Harbor Perspect. Biol., vol. 6, no. 10, Sep. 2014, Art. no. a016295. [Online]. Available: https://pubmed.ncbi.nlm.nih.gov/25190079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Huang Y.et al. , “Clinical characteristics of laboratory confirmed positive cases of SARS-CoV-2 infection in Wuhan, China: A retrospective single center analysis,” Travel Med. Infect. Dis., vol. 36, Jul./Aug. 2020, Art. no. 101606. [Online]. Available: https://doi.org/10.1016/j.tmaid.2020.101606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Huang C.et al. , “Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China,” Lancet, vol. 395, no. 10223, pp. 497–506, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Geng Y. J., Wei Z. Y., Qian H. Y., Huang J., Lodato R., and Castriotta R. J., “Pathophysiological characteristics and therapeutic approaches for pulmonary injury and cardiovascular complications of coronavirus disease 2019,” Cardiovasc. Pathol., vol. 47, Jul./Aug. 2020, Art. no. 107228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Medzhitov R., “Recognition of microorganisms and activation of the immune response,” Nature, vol. 449, no. 7164, pp. 819–826, 2007. [DOI] [PubMed] [Google Scholar]
- [30].Ponti G., Maccaferri M., Ruini C., Tomasi A., and Ozben T., “Biomarkers associated with COVID-19 disease progression,” Crit. Rev. Clin. Lab. Sci., vol. 57, no. 6, pp. 389–399, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Dubach J. M., Harjes D. I., and Clark H. A., “Fluorescent ion-selective nanosensors for intracellular analysis with improved lifetime and size,” Nano Lett., vol. 7, no. 6, pp. 1827–1831, Jun. 2007. [Online]. Available: https://doi.org/10.1021/nl0707860 [DOI] [PubMed] [Google Scholar]
- [32].Li J., Peng T., and Peng Y., “A cholesterol biosensor based on entrapment of cholesterol oxidase in a silicic sol-gel matrix at a prussian blue modified electrode,” Electroanalysis, vol. 15, no. 12, pp. 1031–1037, 2003. [Online]. Available: https://onlinelibrary.wiley.com/doi/abs/10.1002/elan.200390124 [Google Scholar]
- [33].Tallury P., Malhotra A., Byrne L. M., and Santra S., “Nanobioimaging and sensing of infectious diseases,” Adv. Drug Deliv. Rev., vol. 62, nos. 4–5, pp. 424–437, Mar. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hou T.et al. , “Roles of IL-6-gp130 signaling in vascular inflammation,” Current Cardiol. Rev., vol. 4, no. 3, pp. 179–192, Aug. 2008, [Online]. Available: https://pubmed.ncbi.nlm.nih.gov/19936194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Bagca B. G. and Avci C. B., “The potential of JAK/ STAT pathway inhibition by ruxolitinib in the treatment of COVID-19,” Cytok. Growth Factor Rev., vol. 54, pp. 51–61, Aug. 2020, [Online]. Available: https://pubmed.ncbi.nlm.nih.gov/32636055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wung B. S., Ni C. W., and Wang D. L., “ICAM-1 induction by TNFα and IL-6 is mediated by distinct pathways via Rac in endothelial cells,” J. Biomed. Sci., vol. 12, no. 1, pp. 91–101, Jan. 2005. [Online]. Available: https://doi.org/10.1007/s11373-004-8170-z [DOI] [PubMed] [Google Scholar]
- [37].Chen S.-C., Chang Y.-L., Wang D. L., and Cheng J.-J., “Herbal remedy magnolol suppresses IL-6-induced STAT3 activation and gene expression in endothelial cells,” Brit. J. Pharmacol., vol. 148, no. 2, pp. 226–232, May 2006. [Online]. Available: https://pubmed.ncbi.nlm.nih.gov/16520748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Moshapa F. T., Riches-Suman K., and Palmer T. M., “Therapeutic targeting of the proinflammatory IL-6-JAK/ STAT signalling pathways responsible for vascular restenosis in type 2 diabetes mellitus,” Cardiol. Res. Pract., vol. 2019, Jan. 2019, Art. no. 9846312. [Online]. Available: https://pubmed.ncbi.nlm.nih.gov/30719343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lee H.et al. , “A graphene-based electrochemical device with thermoresponsive microneedles for diabetes monitoring and therapy,” Nat. Nanotechnol., vol. 11, no. 6, pp. 566–572, 2016. [DOI] [PubMed] [Google Scholar]
- [40].Liu Y., Pharr M., and Salvatore G. A., “Lab-on-Skin: A review of flexible and stretchable electronics for wearable health monitoring,” ACS Nano, vol. 11, no. 10, pp. 9614–9635, 2017. [DOI] [PubMed] [Google Scholar]
- [41].Chen Y.et al. , “Skin-like biosensor system via electrochemical channels for noninvasive blood glucose monitoring,” Sci. Adv., vol. 3, no. 12, Dec. 2017, Art. no. e1701629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Webb R. C.et al. , “Epidermal devices for noninvasive, precise, and continuous mapping of macrovascular and microvascular blood flow,” Sci. Adv., vol. 1, no. 9, Oct. 2015, Art. no. e1500701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Donnelly R. and Douroumis D., “Microneedles for drug and vaccine delivery and patient monitoring,” Drug Deliv. Transl. Res., vol. 5, no. 4, pp. 311–312, Aug. 2015. [DOI] [PubMed] [Google Scholar]
- [44].Kim Y. C., Park J. H., and Prausnitz M. R., “Microneedles for drug and vaccine delivery,” Adv. Drug Deliv. Rev., vol. 64, no. 14, pp. 1547–1568, Nov. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Cahill E. M. and O’Cearbhaill E. D., “Toward biofunctional microneedles for stimulus responsive drug delivery,” Bioconjugate Chem., vol. 26, no. 7, pp. 1289–1296, Jul. 2015. [DOI] [PubMed] [Google Scholar]
- [46].Waghule T.et al. , “Microneedles: A smart approach and increasing potential for transdermal drug delivery system,” Biomed. Pharmacotherapy, vol. 109, pp. 1249–1258, Jan. 2019. [Online]. Available: http://www.sciencedirect.com/science/article/pii/S0753332218348091 [DOI] [PubMed] [Google Scholar]
- [47].De Franciscis V. L.et al. , “ICAM-1 APTAMERS, diagnostic and therapeutic uses thereof,” U.S. Patent WO2 020 020 947 A1, Jan. 2020.
- [48].Shui B.et al. , “RNA aptamers that functionally interact with green fluorescent protein and its derivatives,” Nucleic Acids Res., vol. 40, no. 5, p. e39, Mar. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Zhou J. and Rossi J., “Aptamers as targeted therapeutics: Current potential and challenges,” Nat. Rev. Drug Disc., vol. 16, no. 3, pp. 181–202, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Nozari A. and Berezovski M. V., “Aptamers for CD Antigens: From cell profiling to activity modulation,” Mol. Therapy Nucleic Acids, vol. 6, pp. 29–44, Mar. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Amoh Y., Katsuoka K., and Hoffman R. M., “Color-coded fluorescent protein imaging of angiogenesis: The AngioMouse models,” Curr. Pharm. Des., vol. 14, no. 36, pp. 3810–3819, 2008. [DOI] [PubMed] [Google Scholar]
- [52].Pierobon M. and Akyildiz I., “Noise analysis in ligand-binding reception for molecular communication in nanonetworks,” IEEE Trans. Signal Process., vol. 59, no. 9, pp. 4168–4182, Sep. 2011. [Google Scholar]
- [53].Corzo J., “Time, the forgotten dimension of ligand binding teaching,” Biochem. Mol. Biol. Educ., vol. 34, no. 6, pp. 413–416, Nov. 2006. [DOI] [PubMed] [Google Scholar]
- [54].Hammacher A., Simpson R. J., and Nice E. C., “The interleukin-6 (IL-6) partial antagonist (Q159E,T162P)IL-6 interacts with the IL-6 receptor and gp130 but fails to induce a stable hexameric receptor complex,” J. Biol. Chem., vol. 271, no. 10, pp. 5464–5473, Mar. 1996. [DOI] [PubMed] [Google Scholar]
- [55].Adams R.et al. , “Discovery of a junctional epitope antibody that stabilizes IL-6 and gp80 protein: Protein interaction and modulates its downstream signaling,” Sci. Rep., vol. 7, Jan. 2017, Art. no. 37716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Philibert J., “One and a half century of diffusion: Fick, Einstein, before and beyond,” Diff. Fundam., vol. 4, no. 11, pp. 1–19, 2006. [Google Scholar]
- [57].Das B.et al. , “Red blood cell velocity profiles in skeletal muscle venules at low flow rates are described by the Casson model,” Clin. Hemorheol. Microcirc., vol. 36, no. 3, pp. 217–233, 2007. [PubMed] [Google Scholar]
- [58].Tan J., Thomas A., and Liu Y., “Influence of red blood cells on nanoparticle targeted delivery in microcirculation,” Soft Matter, vol. 8, pp. 1934–1946, Dec. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Femminella M., Reali G., and Vasilakos A. V., “A molecular communications model for drug delivery,” IEEE Trans. Nanobiosci., vol. 14, no. 8, pp. 935–945, Dec. 2015. [DOI] [PubMed] [Google Scholar]
- [60].Lauffenburger D. and Linderman J., Receptors: Models for Binding, Trafficking, and Signalling. Oxford, U.K.: Oxford Univ. Press, 1996. [Google Scholar]
- [61].Collings T. W. R., “A queueing problem in which customers have different service distributions,” J. Royal Stat. Soc. C., vol. 23, no. 1, pp. 75–82, Mar. 1974. [Online]. Available: https://ideas.repec.org/a/bla/jorssc/v23y1974i1p75-82.html [Google Scholar]
- [62].Caro C. G., Pedley T. J., Schroter R. C., Seed W. A., and Parker K. H., The Mechanics of the Circulation, 2nd ed. Cambridge, U.K.: Cambridge Univ. Press, 2011. [Google Scholar]
- [63].Wicke W., Schwering T., Ahmadzadeh A., Jamali V., Noel A., and Schober R., “Modeling duct flow for molecular communication,” in Proc. IEEE Global Commun. Conf. (GLOBECOM), 2018, pp. 206–212. [Google Scholar]
- [64].Dinc F., Akdeniz B. C., Pusane A. E., and Tugcu T., “A general analytical approximation to impulse response of 3-D microfluidic channels in molecular communication,” IEEE Trans. Nanobiosci., vol. 18, no. 3, pp. 396–403, Jul. 2019. [DOI] [PubMed] [Google Scholar]
- [65].Bagchi P., “Mesoscale simulation of blood flow in small vessels,” Biophys. J., vol. 92, no. 6, pp. 1858–1877, Mar. 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Lambert D., “Drugs and receptors,” Contin. Educ. Anaesthesia Crit. Care Pain, vol. 4, no. 6, pp. 181–184, 2004. [Google Scholar]