Abstract
Given the outbreak of COVID-19 pandemic and the shortage of medical resource, extensive deep learning models have been proposed for automatic COVID-19 diagnosis, based on 3D computed tomography (CT) scans. However, the existing models independently process the 3D lesion segmentation and disease classification, ignoring the inherent correlation between these two tasks. In this paper, we propose a joint deep learning model of 3D lesion segmentation and classification for diagnosing COVID-19, called DeepSC-COVID, as the first attempt in this direction. Specifically, we establish a large-scale CT database containing 1,805 3D CT scans with fine-grained lesion annotations, and reveal 4 findings about lesion difference between COVID-19 and community acquired pneumonia (CAP). Inspired by our findings, DeepSC-COVID is designed with 3 subnets: a cross-task feature subnet for feature extraction, a 3D lesion subnet for lesion segmentation, and a classification subnet for disease diagnosis. Besides, the task-aware loss is proposed for learning the task interaction across the 3D lesion and classification subnets. Different from all existing models for COVID-19 diagnosis, our model is interpretable with fine-grained 3D lesion distribution. Finally, extensive experimental results show that the joint learning framework in our model significantly improves the performance of 3D lesion segmentation and disease classification in both efficiency and efficacy.
Keywords: Multi-task learning, CT scans, COVID-19, deep neural networks
I. Introduction
After being first identified in December 2019, COVID-19 has emerged as a pandemic of global health concern, causing unprecedented social and economic disruption [42], [44]. According to the WHO report [54], as of March 29, 2021, there were a total of 126,890,643 infected patients, 2,778,619 of whom died. The worldwide outbreak of COVID-19 has placed enormous pressure on healthcare systems and led to an extreme shortage of medical resources [41]. A feasible way to control the COVID-19 pandemic is to identify and isolate the infected cases [19], which requires an effective screening method with high sensitivity to detect infected people and their close contacts. Real-time reverse transcription polymerase chain reaction (RT-PCR) [43] is a common method for COVID-19 detection; however, it suffers from a high false negative rate [1], [14] in the early stages of the disease. Recently, the antigen test has been developed for the rapid diagnosis of COVID-19; however, it still suffers from relatively low specificity and sensitivity. As reported in [46], the sensitivity of the antigen test is only 30.2%. Chest computed tomography (CT) has been demonstrated to have better sensitivity for detecting COVID-19, especially in regions with severe epidemic situations [56], [58]. Unfortunately, it is a time-consuming process for doctors to interpret and make a diagnosis during COVID-19 outbreak from each CT scan with hundreds of slices. Even an experienced radiologist can only interpret 4–10 chest CT scans per hour [3], [10]. Therefore, an automatic CT interpretation model is highly desired for accurate, efficient and trustworthy COVID-19 diagnosis.
There are three great challenges in developing an automatic CT interpretation model for COVID-19 diagnosis. (1) Although the tasks of 3D lesion segmentation and disease classification are highly correlated with each other for COVID-19 diagnosis, they cannot be simultaneously learned in the existing deep learning (DL) models [26], [34], [48], [51], [53], [61]. Hence, it is challenging to develop a joint deep learning model of 3D lesion segmentation and disease classification. (2) Despite being a new disease, COVID-19 has similar imaging manifestations as other types of pneumonia, e.g., community acquired pneumonia (CAP) [5]. Thus, it is a challenging task for the model to produce a differential diagnosis between COVID-19 and other similar types of pneumonia. (3) Most automatic diagnosis models [27], [30], [31], [40] are based on “black box” deep neural networks (DNNs) [12], [20], [25], [52], which lack sufficient explainability to assist radiologists in making credible diagnoses. The explainability of DNNs is another challenge in the design of automatic CT interpretation models for COVID-19 diagnosis.
To tackle the above challenges, we establish a large-scale CT database, called 3DLSC-COVID, which is the first CT database with fine-grained 3D lesion segmentation and classification labels of COVID-19, CAP and non-pneumonia. Based on the lesion characteristics found from this database, we propose a joint DL model, namely DeepSC-COVID, for accurate 3D lesion segmentation and the diagnosis of COVID-19. Specifically, the DeepSC-COVID model consists of three subnets, i.e., cross-task feature, 3D lesion segmentation and disease classification subnets, and is able to simultaneously generate the 3D segmented lesion and the classification results of COVID-19, CAP or non-pneumonia. In the classification subnet, a new multi-layer visualization mechanism is developed to generate the evidence masks that contain small and indistinct lesions for disease diagnosis. In this way, the process of COVID-19 diagnosis in our model is explainable. Besides, in the training phase, a novel task-aware loss is proposed on the basis of our visualization mechanism for efficient interaction between the tasks of segmentation and classification. With the guidance of the segmented lesions, the classification subnet is able to focus on the lesions, such that the diagnosis of COVID-19 can be significantly accelerated with higher classification accuracy. Note that, different from the single-scale attention constrained mechanism [37], our task-aware loss has multi-scale attention constraint to generate more fine-grained visualization maps. Additionally, the task-aware loss is used in our method to optimize both tasks of segmentation and classification, thus being able to interact the information between these two tasks and to boost the performance of both tasks. In conclusion, the developed DeepSC-COVID model can provide the rapid, accurate and explainable diagnosis of COVID-19, meanwhile visualizing the fine-grained lesions for doctors.
To the best of our knowledge, our method is one of the pioneering works in joint learning of 3D lesion segmentation and disease classification based on 3D CT scans, especially for the disease of COVID-19. The main contributions of this paper are as follows. (1) We establish a large-scale database of CT scans, with fine-grained lesion annotations, for the diagnosis of COVID-19 and CAP. (2) We thoroughly analyze the new database, and yield 4 important findings about the lesion differences between the diseases. (3) We propose an explainable deep multi-task learning model for both tasks of 3D lesion segmentation and disease classification of COVID-19.
II. Related Work
A. Imaging-Based COVID-19 Databases
Although many people infected by COVID-19, it is still not easy to build a large-scale imaging-based COVID-19 databases, due to the privacy of the patients and hospitals. Table I summarizes the representative CT/X-rays based COVID-19 databases that are public online. As can be seen from this table, most existing public databases lack fine-grained lesion annotation, and only a few of them have small scale of lesion segmentation labels. This is probably because of the lack of the experts with rich experience in diagnosing COVID-19. In contrast, this paper establishes a large-scale database of 1,805 CT scans with 458,730 slices, in which 157,696 slices are annotated with lesions. It is worth mentioning that the lesion-annotated slices of our database are around 17 times more than those of the largest 3D lesion segmentation database [48].
TABLE I. Summary of the Existing COVID-19 Databases.
| Database | Type | # Slices | #Cases | Lesion Annotation* |
|---|---|---|---|---|
| [9] | X-rays | 761 | 412 | – |
| [8] | X-rays | 2,905 | – | – |
| [49] | X-rays | 16,756 | 13,645 | – |
| [13] | X-rays+CT | 5,381 | 1,311 | – |
| [59] | CT | 349 | 216 | – |
| [7] | CT | 1,103 | 64(videos) | – |
| [36] | CT | 1,110 | – | – |
| [4] | CT | 2,482 | 120 | – |
| [50] | CT | 453 | 99 | – |
| [61] | CT | 361,221 | 2,246 | 2D (4,695 slices) |
| [2] | CT | 144,167 | 750 | 2D (3,855 slices) |
| [32] | CT | 3,520 | 20 | 3D (1,844 slices) |
| [48] | CT | 76,250 | 558 | 3D (9,015 slices) |
| Ours | CT | 458,730 | 1,805 | 3D (157,696 slices) |
“2D” means that only part of the slices of one CT scan are annotated, while “3D” denotes that all the slices with lesions of a CT scan are annotated.
B. Automatic COVID-19 Diagnosis on CT Scans
In the past few months, many DL-based methods were developed for COVID-19 diagnosis on CT scans [17], [26], [37], [48], [61], [63]. They mainly focus on two tasks: disease classification and lesion segmentation. In order to automatically diagnose COVID-19, Li et al. [26] proposed a COVID-19 detection neural network (COVNet) using ResNet-50 [18] as the backbone. With a series of CT slices as inputs, COVNet generates a classification result for each CT scan. Similarly, Ouyang et al. [37] designed a dual-sampling attention network for classifying COVID-19 and CAP. Specifically, they proposed an online attention module with a 3D convolutional network to focus on the infection regions in lungs for the diagnosis. Different from the disease classification methods, other works [48], [63] focused on COVID-19 lesion segmentation. Specifically, Zhou et al. [63] proposed a fully automatic machine-agnostic method that can segment and quantify the infection regions on CT scans from different sources. Wang et al. [48] designed a noise-robust framework for automatic segmentation of COVID-19 pneumonia lesions from CT scans. Unfortunately, all above methods neglect the correlation between disease classification and lesion segmentation. In fact, the lesion segmentation results act as explainable diagnostic evidence for disease classification; meanwhile, the classification results are able to further improve the accuracy of lesion segmentation.
Only a few DL-based methods [23], [33], [61] have been developed to perform both tasks of lesion segmentation and disease classification for COVID-19. Specifically, Mahmud et al. [33] proposed a hybrid attention based network for lesion segmentation, diagnosis, and severity prediction of COVID-19. In their training stage, the lesion segmentation network is optimized firstly and is then integrated into the training of diagnosis and severity prediction. Similarly, Jin et al. [23] proposed a sequential optimization pipeline, in which they first train the lesion segmentation network alone, and then use the segmentation results to train the classification network. However, all these methods cannot be seen as multi-task learning according to the definition of [45], since they separately learn the two tasks, ignoring the information sharing between two tasks. In contrast, our DeepSC-COVID method is a multi-task deep learning work, as it can jointly learn the two tasks of 3D lesion segmentation and classification for COVID-19, achieving task-aware information sharing through the proposed cross-task feature subnet and the novel task-aware loss. This way, the tasks of lesion segmentation and disease classification can boost each other to achieve better performance.
III. Database and Analysis
A. Database Establishment
This retrospective study was performed in accordance with the Declaration of Helsinki of the World Medical Association and was approved by the medical ethics committee of Liyuan Hospital, Tongji Medical College, Huazhong University of Science and Technology. Besides, all data were anonymized.
For establishing our 3DLSC-COVID database,1. a total of 1,805 3D chest CT scans with more than 570,000 CT slices were collected from 2 standard CT scanners of Liyuan Hospital, i.e., UIH uCT 510 and GE Optima CT600. Among all CT scans, there were 794 positive cases of COVID-19, which were further confirmed by clinical symptoms and RT-PCR from January 16 to April 16, 2020. Additionally, 540 positive cases of CAP and 471 non-pneumonia cases were randomly selected from the same hospital between November 5, 2016 and April 28, 2020. In contrast to existing CT-based COVID-19 databases [57], [59], [63], our 3DLSC-COVID database is the first CT database with both fine-grained 3D lesion segmentation and disease classification labels for the COVID-19 and CAP diagnosis. More details about patients and CT scans of the 3DLSC-COVID database are summarized in the supplementary material.
For lesion segmentation, we recruited 2 resident radiologists with over 2 years of experience to annotate the areas and boundaries of the lesions in each 2D CT slice. Then, for each CT scan, the 2D annotated lesions of all CT slices were merged to obtain the 3D lesions. Subsequently, the 2 resident radiologists were asked to further refine the segmented lesions in 3D viewing mode. At last, both 2D and 3D lesions were reviewed and corrected by a senior radiologist with over 10 years of experience in thoracic radiology. Some examples of the annotated CT scans for COVID-19, CAP and non-pneumonia individuals are shown in Fig. 1.
Fig. 1.
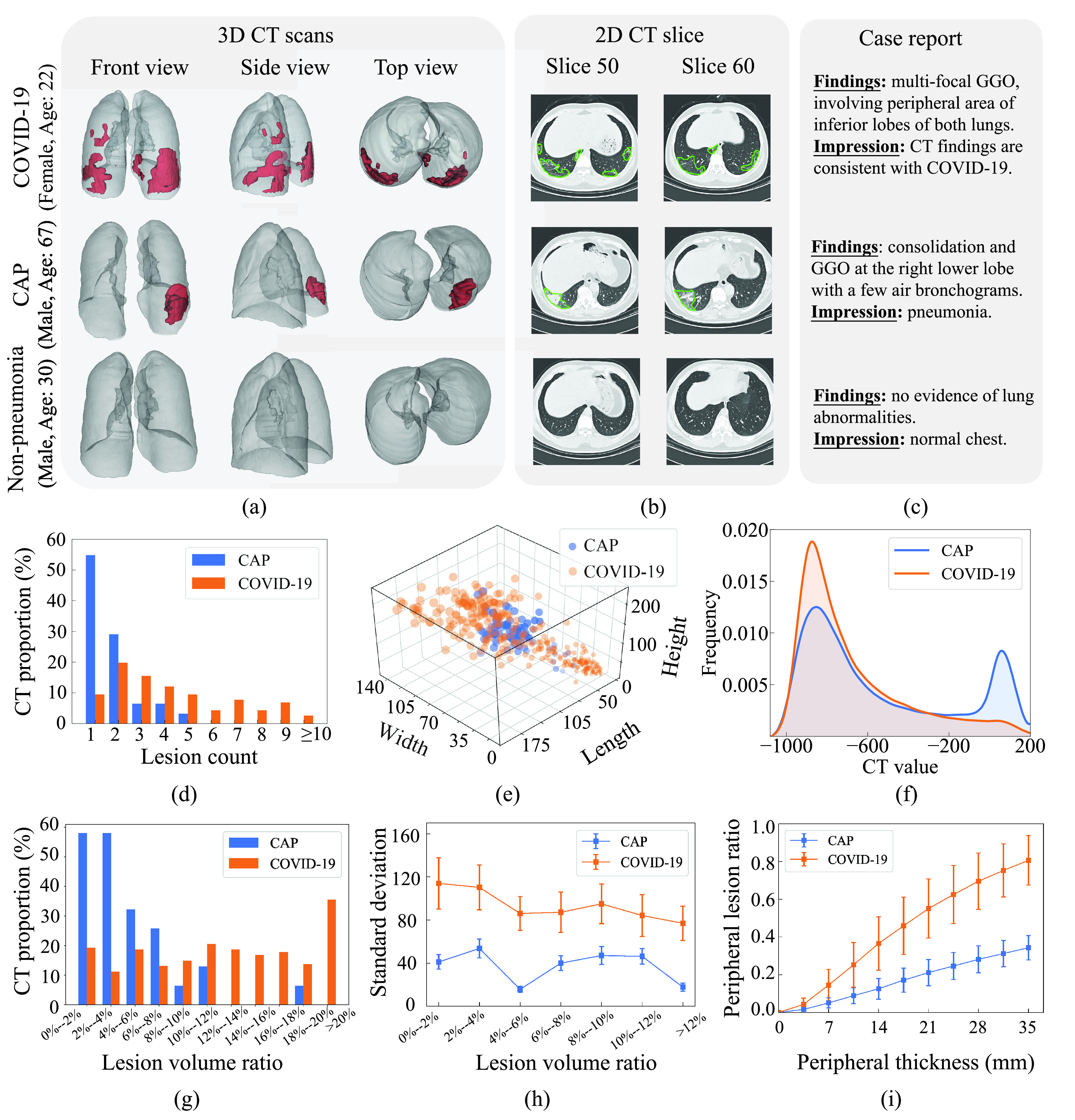
Illustration and statistical analysis of our database. (a) Front, side and top views of 3D chest CT scans with 3D lesion segmentation for one COVID-19 (upper row), one CAP (middle row) and one non-pneumonia (lower row) individuals. Note that the lesions in lungs are marked in red. (b), Two selected 2D CT slices, corresponding to 3D CT scan in the same row. The lesions in each slice are encircled by green lines. (c) Case reports of the three individuals. (d) Histogram of lesion counts in the CT scans for all COVID-19 and CAP individuals in our 3DLSC-COVID database. (e) Width, length and height of each lesion in 3D CT scans for COVID-19 and CAP, respectively. For better visualization, only 372 lesions are randomly selected from our database. (f) Distribution curves of CT values in the lesions of CAP and COVID-19 CT scans, respectively. (g) Histograms of lesion count in the CT scans for CAP and COVID-19, respectively. (h) Standard deviations of lesion distribution for CAP and COVID-19 with varied lesion volume ratios. (i) Changes of peripheral lesion ratio with peripheral thickness varying from 0 to 35 mm for CAP and COVID-19, respectively. Note that the results of these charts (d,f-i) are obtained upon all CAP and COVID-19 CT scans in our 3DLSC-COVID database.
B. Analysis of Characteristics of 3D Lesion
We characterize the 3D lesions of COVID-19 and CAP via thoroughly analyzing the lesion annotations in our 3DLSC-COVID database. Four important findings are obtained in terms of the count, size, CT value and spatial distribution of 3D lesions, which are briefly introduced as follows.
1). Finding 1: The Number of Lesions for COVID-19 is Considerably More Than That for CAP:
Analysis: As shown in Fig. 1 (d), the number of lesions for COVID-19 is around 2.6 times than that for CAP, i.e., averagely 4.4 lesions per CT scan for COVID-19 versus 1.7 for CAP. To be more specific, 54.8% CT scans of CAP in our database contain only one lesion, while around 55.2% CT scans for COVID-19 have 4 or more lesions. Fig. 1 (a) visualizes the segmented lesions of COVID-19 and CAP, which also indicate the obvious difference of lesion counts between COVID-19 and CAP.
2). Finding 2: The Overall Lesion Volume in COVID-19 CT Scans is Significantly Larger Than That for CAP. Additionally, Compared With CAP, the Lesions of COVID-19 Vary Significantly in Size:
Analysis: Fig. 1 (g) shows the histogram of the lesion volume ratio (LVR) for all COVID-19 and CAP individuals in the 3DLSC-COVID database. Here, LVR indicates the proportion of lesions to the whole lung. As shown in this figure, the average LVR for COVID-19 is around 3.3 times higher than that for CAP per CT scan, i.e., the average LVR is 14.3% for COVID-19 versus 4.4% for CAP. The CAP cases with LVR larger than 12% accounts for only 5%, while the COVID-19 cases with LVR larger than 12% accounts for above 50%.
In addition to the lesion volume, we compare the 3D size of lesions for COVID-19 and CAP. The 3D size is measured by drawing a bounding box for each lesion, which is defined as the minimum cuboid to wrap the lesion. Fig. 1 (e) shows the 3-D scatter diagram with axes of width, length and height, drawn on 372 randomly selected lesions from our database. As can be seen in this figure, the 3D size of lesions for CAP is concentrated. Specifically, the width, length and height of over 90% CAP lesions are densely distributed in the range of [35 mm, 105 mm] (span = 70 mm), [42 mm, 105 mm] (span = 63 mm) and [85 mm, 160 mm] (span = 75 mm), respectively. In contrast, the 3D lesion size of COVID-19 is with a larger span, i.e., [15 mm, 140 mm] (span = 125 mm) in width, [15 mm, 170 mm] (span = 155 mm) in length and [30 mm, 240 mm] (span = 210 mm) in height, respectively. This verifies that the lesions of COVID-19 vary significantly in size compared to those of CAP.
3). Finding 3: Compared to the CAP Lesions Which can Either Display Low or High Density in CT Images, the COVID-19 Lesions Tend to Mainly Display Low Density (Darker):
Analysis: The densities of CAP and COVID-19 lesions are investigated in terms of CT values. Fig. 1 (f) shows the distribution curves of CT values in the lesions of CAP and COVID-19, respectively. Note that smaller CT values indicate lower density. As can be seen, for COVID-19 lesions, the distribution curve only has one peak, with more than 70% of the CT values concentrated between −960 Hounsfield unit (HU) and −600 HU. In contrast, for CAP lesions, the CT value distribution has two primary peaks, i.e., over 75% of the CT values are distributed in [−970 HU, −580 HU] and [−70 HU, 140 HU]. As such, the COVID-19 lesions tend to mainly display low density, while the CAP lesions can either display low or high density. A possible medical explanation for this finding lies in the lesion types. Specifically, the COVID-19 lesions are mainly ground-glass opacity (GGO) [6], [22], which is a pattern of hazy increased lung opacity that shows low contrast with surrounding regions. In addition to GGO, CAP has another type of lesion called consolidation. The consolidation is a typical pneumonia lesion that has the homogeneous increase in lung parenchymal attenuation of CT scans, which is in highly contrast with surrounding regions. Some examples of the segmented lesions in 2D CT slices for COVID-19 and CAP can be seen in Fig. 1 (b).
4). Finding 4: The COVID-19 Lesions are Mostly Scattered in the Peripheral Area of Lungs. In Contrast, the CAP Lesions are More Concentrated, Which are Mainly Distributed in the Central Area of Lungs:
Analysis: The spatial distribution of the lesions is evaluated for CAP and COVID-19, by measuring the standard deviation of the lesion centers in all CT scans. Here, the lesion center is the central point of the lesion bounding box, and a larger standard deviation indicates more scattered lesion distribution. For specific analysis, we divide all CT scans into different groups upon their lesion volume ratios. Fig. 1 (h) shows the standard deviations of lesion distribution in different groups of CAP and COVID-19. As can be seen in this figure, the standard deviation of lesions in COVID-19 is significantly larger than that in CAP for all CT groups. In particular, for the CT group with lesion volume ratio from 4% to 6%, the standard deviation is 86.0 on average for COVID-19, compared with only 15.6 for CAP. This demonstrates that the spatial distribution of COVID-19 lesions is more scattered than that of CAP.
Next, the lesion distribution areas in CT scans are analyzed for CAP and COVID-19, by calculating the proportion of lesions within the peripheral lung areas to the overall lesions, denoted as peripheral lesion ratio (PLR). To calculate PLR, given a CT scan, we first generate the 3D binary masks of the lung areas by a state-of-the-art lung segmentation algorithm [21]. For the CT scan with slice
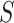 , width
, width
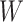 and height
and height
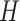 , the lung and lesion masks are denoted as
, the lung and lesion masks are denoted as
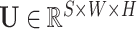 and
and
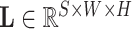 , respectively. Then, PLR is defined as follows:
, respectively. Then, PLR is defined as follows:
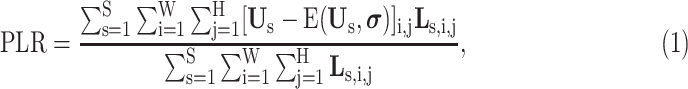 |
where
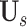 is the
is the
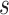 -th slice of the lung mask
-th slice of the lung mask
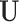 , and
, and
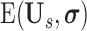 is the erosion operation with the erosion kernel of
is the erosion operation with the erosion kernel of
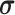 in radius. Note that the difference between the lung mask and its erosion result
in radius. Note that the difference between the lung mask and its erosion result
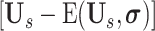 can be regard as the peripheral lung areas, which is controlled by the hyper-parameter of
can be regard as the peripheral lung areas, which is controlled by the hyper-parameter of
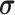 denoted as peripheral thickness in the following. Fig. 1 (i) shows the PLR with different peripheral thickness for the CT scans of COVID-19 and CAP in the 3DLSC-COVID database. As shown, the COVID-19 lesions are more possibly distributed in the peripheral area of the lung, e.g., PLR = 62.4% for COVID-19 lesions versus 24.5% for CAP lesions. This indicates the significant difference of lesion distribution between CAP and COVID-19 in CT scans.
denoted as peripheral thickness in the following. Fig. 1 (i) shows the PLR with different peripheral thickness for the CT scans of COVID-19 and CAP in the 3DLSC-COVID database. As shown, the COVID-19 lesions are more possibly distributed in the peripheral area of the lung, e.g., PLR = 62.4% for COVID-19 lesions versus 24.5% for CAP lesions. This indicates the significant difference of lesion distribution between CAP and COVID-19 in CT scans.
The above findings reveal the typical characteristics of lesions for COVID-19, and are used as guidance to design our DeepSC-COVID model for automatic CT interpretation in COVID-19 diagnosis.
IV. Methodology
A. Framework of DeepSC-COVID
As illustrated in Fig. 2, the proposed DeepSC-COVID model2 consists of 3 subnets: cross-task feature, 3D lesion and classification subnets. For 3D lesion segmentation, due to the limited GPU memory, it is difficult to input the full-sized CT scans. As such, the original CT scan is cropped into smaller non-overlapping 3D patches. For classification, the 3D CT scan is preprocessed by slice sampling at an average interval to remove the redundancy between adjacent slices, in order to improve the classification efficiency.
Fig. 2.
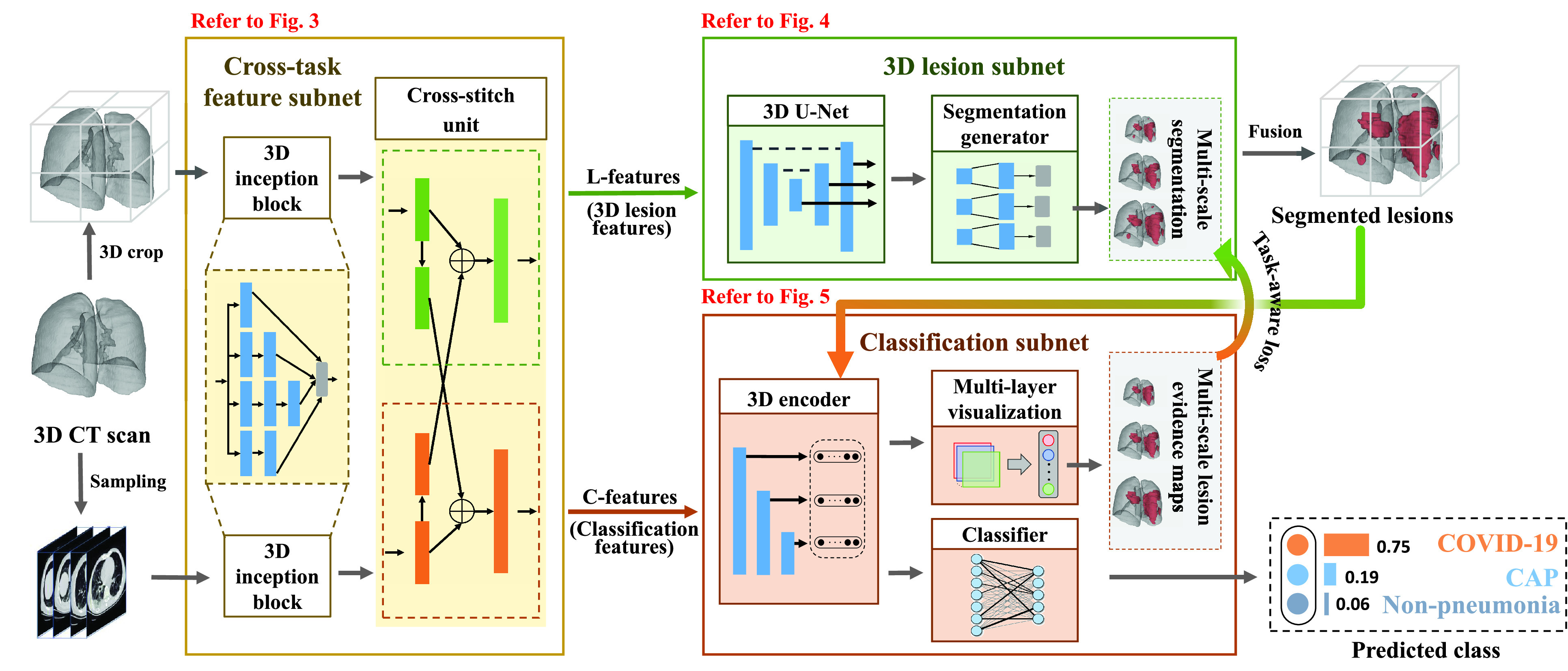
Framework of the proposed DeepSC-COVID model.
After preprocessing, both the cropped 3D CT patch and sampled 2D CT slices are fed into the cross-task feature subnet with 3D inception blocks and cross-stitch unit. Specifically, based on the classic 2D inception block [47], the 3D inception block is designed to extract the multi-scale 3D features from the cropped 3D CT patch and sampled 2D CT slices, respectively. Then, the cross-stitch unit is developed to mix the features to generate 3D lesion features (L-features) and classification features (C-features). These two features are fed into the 3D lesion and classification subnets, respectively. In the 3D lesion subnet, a 3D U-Net and a segmentation generator are designed to segment the multi-scale 3D lesions of COVID-19 or CAP. In the classification subnet, a 3D encoder and a classifier are developed to predict the probability scores for COVID-19, CAP and non-pneumonia. Besides, the task-aware loss is proposed for learning the task interaction across the 3D lesion and classification subnets. To obtain the evidence masks of the classification subnet, we propose a multi-layer visualization method for extracting the pathological regions for disease diagnosis. Finally, according to the predicted probabilities, the input 3D CT scan can be classified as COVID-19, CAP or non-pneumonia.
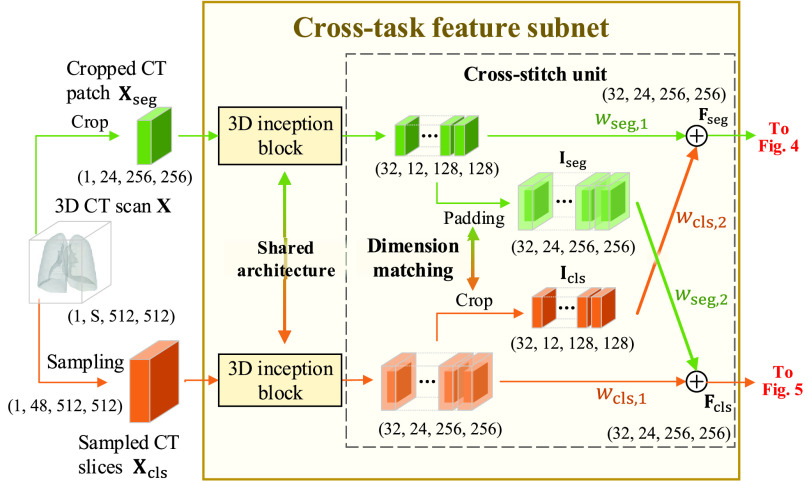
B. Cross-Task Feature Subnet
Let
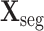 and
and
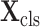 denote the cropped 3D patch and the sampled CT slices after preprocessing, the details of which is introduced in Section V-A. Given
denote the cropped 3D patch and the sampled CT slices after preprocessing, the details of which is introduced in Section V-A. Given
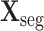 and
and
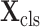 , the cross-task feature subnet is designed to jointly extract the 3D features for the subsequent 3D lesion segmentation and classification subnets. The structure of the cross-task feature subnet is shown in Fig. 3, which consists of 2 cascaded components, i.e., the 3D inception block and the cross-stitch unit. The structure details about these 2 components are described in the following paragraphs.
, the cross-task feature subnet is designed to jointly extract the 3D features for the subsequent 3D lesion segmentation and classification subnets. The structure of the cross-task feature subnet is shown in Fig. 3, which consists of 2 cascaded components, i.e., the 3D inception block and the cross-stitch unit. The structure details about these 2 components are described in the following paragraphs.
Fig. 3.
Structure of the cross-task feature subnet in the proposed DeepSC-COVID model.
1). 3D Inception Block:
Based on the classic 2D inception block [47], the 3D inception block is developed to extract the multi-scale 3D features. The 3D inception block has 4 branches with cascaded 3D convolutional layers. Benefiting from the multiple receptive fields of different branches, the multi-scale 3D features are extracted, followed by the group normalization [55] and rectified linear unit (ReLU) activation. The specific kernel size, stride and output channel for each 3D convolutional layer are shown in supplementary material. The
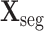 and
and
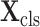 are input to two different 3D inception blocks, to extract 3D features for the segmentation and classification tasks, respectively. These two 3D inception blocks do not share parameters, allowing for inception blocks to extract more efficient features for each single task.
are input to two different 3D inception blocks, to extract 3D features for the segmentation and classification tasks, respectively. These two 3D inception blocks do not share parameters, allowing for inception blocks to extract more efficient features for each single task.
2). Cross-Stitch Unit:
Next, the cross-stitch unit is proposed to enhance the information interaction between the segmentation and classification tasks. Specifically, given the extracted 3D features from the 3D inception blocks, dimension matching is first conducted to unify the receptive field of the extracted features via zero-padding and 3D cropping. Let
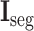 and
and
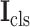 denote the dimension-matched features for segmentation and classification, respectively. Then, for enhancing the information interaction,
denote the dimension-matched features for segmentation and classification, respectively. Then, for enhancing the information interaction,
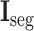 and
and
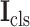 are linearly combined to generate the final cross-task 3D lesion features (L-features)
are linearly combined to generate the final cross-task 3D lesion features (L-features)
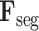 and classification features (C-features)
and classification features (C-features)
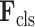 via the following formulation:
via the following formulation:
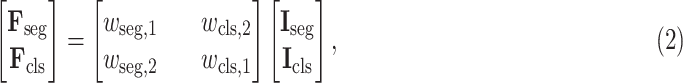 |
where
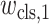 ,
,
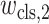 ,
,
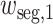 and
and
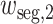 are learnable weights. Then,
are learnable weights. Then,
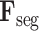 and
and
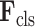 are fed into the subsequent 3D lesion and classification subnets for further processing.
are fed into the subsequent 3D lesion and classification subnets for further processing.
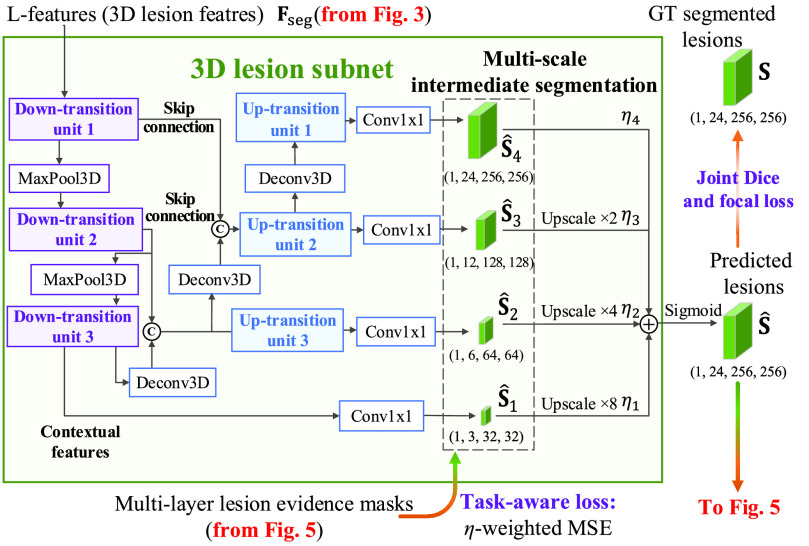
C. 3D Lesion Subnet
Given L-features
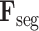 extracted from the cross-task feature subnet, the 3D lesion subnet is developed to segment the 3D lesions of CT scans. The structure of the 3D lesion subnet is shown in Fig. 4. In the 3D lesion subnet, a U-shaped 3D structure, which is composed of three down-transition units and three up-transition units, is designed to extract the features for precisely localizing 3D lesions. Specifically, the input L-features
extracted from the cross-task feature subnet, the 3D lesion subnet is developed to segment the 3D lesions of CT scans. The structure of the 3D lesion subnet is shown in Fig. 4. In the 3D lesion subnet, a U-shaped 3D structure, which is composed of three down-transition units and three up-transition units, is designed to extract the features for precisely localizing 3D lesions. Specifically, the input L-features
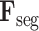 are progressively contracted and down-sampled through three down-transition units followed by 3D max pooling layers with stride of 2. In this way, the contextual information of 3D CT scans can be captured in the outputs of the last down-transition units, namely contextual features. Subsequently, the contextual features are progressively expanded and up-sampled through three up-transition units followed by deconvolutional layers [60] with stride of 2. Note that the skip connections are adopted between the up-transition unit and its corresponding down-transition unit, in order to provide boundary information during the up-sampling process. Detailed structures of down-transition and up-transition units can be found in supplementary material.
are progressively contracted and down-sampled through three down-transition units followed by 3D max pooling layers with stride of 2. In this way, the contextual information of 3D CT scans can be captured in the outputs of the last down-transition units, namely contextual features. Subsequently, the contextual features are progressively expanded and up-sampled through three up-transition units followed by deconvolutional layers [60] with stride of 2. Note that the skip connections are adopted between the up-transition unit and its corresponding down-transition unit, in order to provide boundary information during the up-sampling process. Detailed structures of down-transition and up-transition units can be found in supplementary material.
Fig. 4.
Structure of the 3D lesion subnet in the proposed DeepSC-COVID model.
Then, the outputs of the last down-transition unit and each up-transition unit are further processed by the 3D convolution layers to generate the multi-scale intermediate segmentation. Assuming that
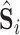 is the segmentation result at the
is the segmentation result at the
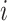 -th scale, the final segmentation lesion
-th scale, the final segmentation lesion
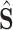 is calculated as follows:
is calculated as follows:
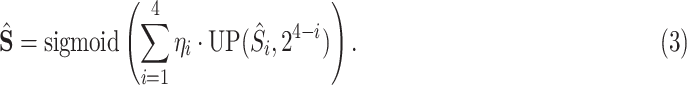 |
In the above equation, {
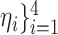 are the hyper-parameters to balance the intermediate segmentation at different scales, and
are the hyper-parameters to balance the intermediate segmentation at different scales, and
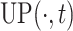 is the
is the
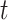 -time upscale operation. During the training stage, segmented lesion
-time upscale operation. During the training stage, segmented lesion
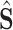 is supervised by its corresponding ground-truth lesion. Furthermore, the intermediate segmentation result is also supervised by the multi-scale lesion evidence masks from the classification subnet, with minimization on the task-aware loss. The details of the evidence masks and the task-aware loss are introduced in the following sections.
is supervised by its corresponding ground-truth lesion. Furthermore, the intermediate segmentation result is also supervised by the multi-scale lesion evidence masks from the classification subnet, with minimization on the task-aware loss. The details of the evidence masks and the task-aware loss are introduced in the following sections.
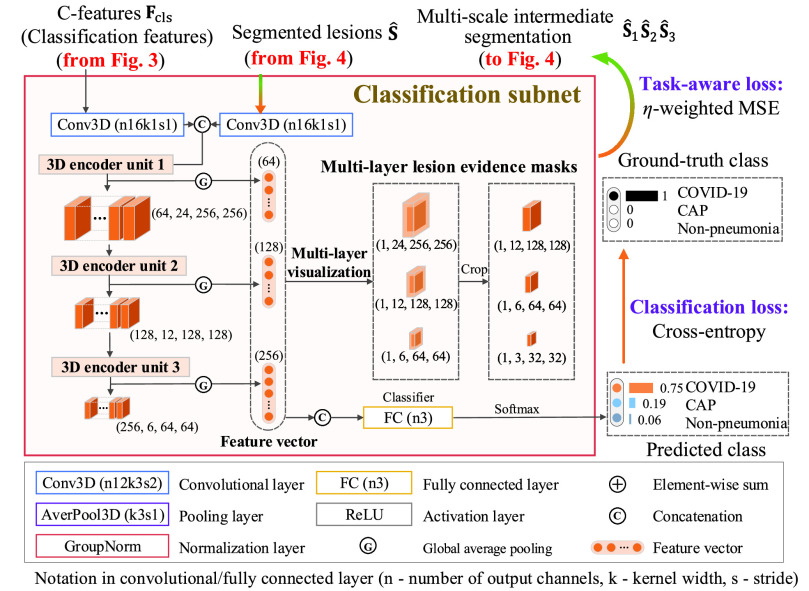
D. Classification Subnet
The classification subnet is developed to classify the input CT scan into 3 classes: COVID-19, CAP and non-pneumonia. The structure of the classification subnet is illustrated in Fig. 5. For focusing on lesions during classification, segmented lesions
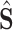 , together with C-features
, together with C-features
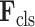 , are input to the classification subnet. Specifically, the segmented lesions and C-features are concatenated after convolutional layers. Then, the concatenated features are hierarchically encoded into small scales by the 3D encoder units, which are designed in a residual mechanism (see supplementary material for more details).
, are input to the classification subnet. Specifically, the segmented lesions and C-features are concatenated after convolutional layers. Then, the concatenated features are hierarchically encoded into small scales by the 3D encoder units, which are designed in a residual mechanism (see supplementary material for more details).
Fig. 5.
Structure of the classification subnet in the proposed DeepSC-COVID model.
Let
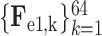 3 denote the 64-channel feature maps generated from the first 3D encoder unit. Subsequently, the channel-wise global average pooling is conducted on
3 denote the 64-channel feature maps generated from the first 3D encoder unit. Subsequently, the channel-wise global average pooling is conducted on
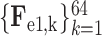 , outputting the spatial average of each feature map as a 64-element feature vector
, outputting the spatial average of each feature map as a 64-element feature vector
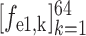 . Similarly, the encoded feature maps and their corresponding feature vectors are denoted as (
. Similarly, the encoded feature maps and their corresponding feature vectors are denoted as (
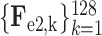 ,
,
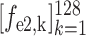 ) and (
) and (
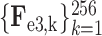 ,
,
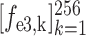 ) for the second and third encoder units, respectively. Finally, feature vectors
) for the second and third encoder units, respectively. Finally, feature vectors
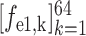 ,
,
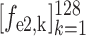 and
and
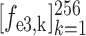 are concatenated for predicting the probability scores of COVID-19, CAP and non-pneumonia, through a fully connected layer. This classification process can be formulated as
are concatenated for predicting the probability scores of COVID-19, CAP and non-pneumonia, through a fully connected layer. This classification process can be formulated as
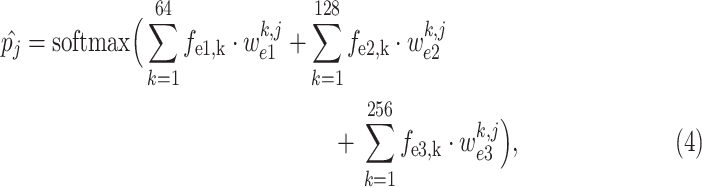 |
where
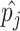 is the predicted probability of the
is the predicted probability of the
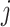 -th class, corresponding to COVID-19 (
-th class, corresponding to COVID-19 (
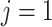 ), CAP (
), CAP (
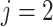 ) or non-pneumonia (
) or non-pneumonia (
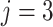 ). Additionally,
). Additionally,
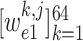 ,
,
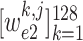 and
and
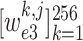 are the learnable weights in the fully connected layer, corresponding to the
are the learnable weights in the fully connected layer, corresponding to the
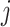 -th class. Consequently, the class with maximal probability score is regarded as the final classification result.
-th class. Consequently, the class with maximal probability score is regarded as the final classification result.
Next, inspired by the visualization algorithm [62], we mainly focus on multi-layer network visualization in the classification subnet for generating lesion evidence masks, which are utilized to supervise the intermediate segmentation results in the 3D lesion subnet, through the task-aware loss. Similar to equation (4), the lesion evidence masks of each class can be calculated as the weighted sum of the encoded feature maps, i.e.,
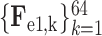 ,
,
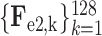 or
or
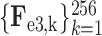 . Assume that
. Assume that
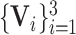 are the multi-layer lesion evidence masks of a predicted class (taking the
are the multi-layer lesion evidence masks of a predicted class (taking the
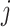 -th class as an example). Then, mathematically, they can be formulated as
-th class as an example). Then, mathematically, they can be formulated as
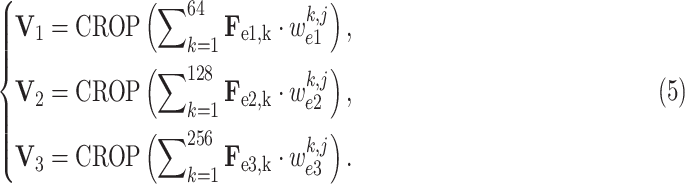 |
Recall that
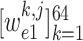 ,
,
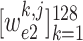 and
and
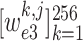 are learned weights in equation (4), and
are learned weights in equation (4), and
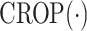 denotes the crop function. It is worth mentioning that the network visualization is only conducted in the training stage when calculating the task-aware loss, which is defined in the next section.
denotes the crop function. It is worth mentioning that the network visualization is only conducted in the training stage when calculating the task-aware loss, which is defined in the next section.
E. Loss Functions
The loss functions are introduced for training the DeepSC-COVID model, including segmentation, classification and task-aware loss. Specifically, segmentation and classification loss are developed for separately training 3D lesion and classification subnets. The task-aware loss is proposed to guide the 3D lesion subnet for precise segmentation upon the visualization masks learned from the classification subnet. The details about the proposed loss functions are introduced as follows.
1). Segmentation Loss:
For the segmentation task, the Dice loss [15] is adopted to measure the overlapping area between the predicted segmentation map
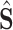 and its ground-truth lesion mask
and its ground-truth lesion mask
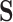 as follows:
as follows:
 |
where
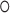 denotes the Hadamard product. In addition to the Dice loss, we utilize the focal loss [29] to reduce the effect of the class imbalance between the lesion and background regions:
denotes the Hadamard product. In addition to the Dice loss, we utilize the focal loss [29] to reduce the effect of the class imbalance between the lesion and background regions:
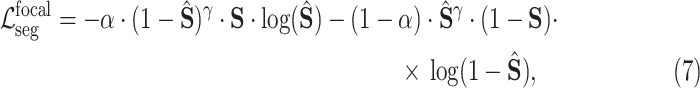 |
where
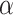 is a hyper-parameter to balance the training samples of lesions and background, and
is a hyper-parameter to balance the training samples of lesions and background, and
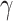 is a hyper-parameter controlling the degree of loss focus on hard samples. Consequently, the segmentation loss
is a hyper-parameter controlling the degree of loss focus on hard samples. Consequently, the segmentation loss
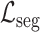 in our DeepSC-COVID can be calculated as
in our DeepSC-COVID can be calculated as
 |
where
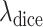 and
and
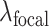 are the hyper-parameters to balance the Dice and focal loss.
are the hyper-parameters to balance the Dice and focal loss.
2). Classification Loss:
For the disease classification task, we develop the weighted cross-entropy loss to measure the distance between the predicted class and the ground-truth label. Mathematically, the classification loss
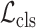 for training the classification subnet can be formulated as
for training the classification subnet can be formulated as
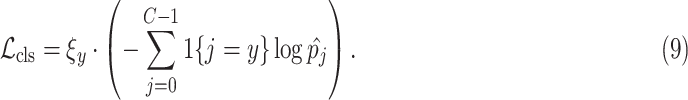 |
In the above equation,
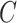 is the number of the classes (3 in this paper),
is the number of the classes (3 in this paper),
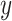 is the ground-truth label,
is the ground-truth label,
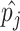 is the predicted probability of the
is the predicted probability of the
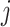 -th class, and
-th class, and
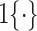 denotes the indicator function. It is worth noting that
denotes the indicator function. It is worth noting that
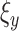 in equation (9) is the inverse frequency [29] of class
in equation (9) is the inverse frequency [29] of class
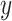 , which is counted over all training samples. This way, the class imbalance of the training samples can be relieved.
, which is counted over all training samples. This way, the class imbalance of the training samples can be relieved.
3). Task-Aware Loss:
In addition to the segmentation and classification loss, we further propose the task-aware loss, which guides the classification and segmentation task to focus on the task relevant regions of each other. Specifically, the multi-scale intermediate segmentation results
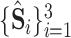 in the 3D lesion subnet are constrained to be similar with the multi-layer lesion evidence masks
in the 3D lesion subnet are constrained to be similar with the multi-layer lesion evidence masks
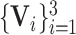 in the classification subnet through the task-aware loss
in the classification subnet through the task-aware loss
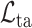 defined as follows:
defined as follows:
 |
Recall that
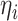 is defined in equation (3) for the 3D lesion subnet. Benefiting from the proposed task-aware loss, the 3D lesion subnet offers potential in segmenting some tiny lesions of the CT scans, which may be neglected by radiologists. For more details, see the ablation study of the task-aware loss in Section V-E.
is defined in equation (3) for the 3D lesion subnet. Benefiting from the proposed task-aware loss, the 3D lesion subnet offers potential in segmenting some tiny lesions of the CT scans, which may be neglected by radiologists. For more details, see the ablation study of the task-aware loss in Section V-E.
4). Total Loss:
By combining the segmentation, classification and task-aware loss, the total loss function for our DeepSC-COVID model can be formulated as follows:
 |
where
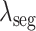 ,
,
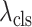 and
and
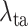 are the hyper-parameters to balance the corresponding loss.
are the hyper-parameters to balance the corresponding loss.
V. Experiments
A. Implementation Details
1). Database Split:
All experiments are conducted with data from the proposed 3DLSC-COVID database. Specifically, the CT scans in the 3DLSC-COVID database are divided into training and test sets. The training set contains 1,353 CT scans (595 for COVID-19, 405 for CAP and 353 for non-pneumonia) and the test set has 452 CT scans (199 for COVID-19, 135 for CAP and 118 for non-pneumonia). We employ 10-fold cross-validation strategy on the training data for adjusting hyper-parameters. The comparisons among our model, other models and human expertise with respect to lesion segmentation and disease classification are all conducted on the test set.
2). Preprocessing:
For efficient training, the 3D CT scans are preprocessed before inputting to the DeepSC-COVID model. The preprocessing phase includes two steps. First, for highlighting the anatomical structures, the original CT values of the CT scans are truncated into [−1,400 HU, 200 HU] [11]. Then, the CT scans are further normalized to [0, 1]. To focus on the lung areas, the CT scans are masked with the lung binary masks generated by a state-of-the-art lung segmentation algorithm [21].
Second, for conservation of the limited computational resources, the size of 3D CT scans is reduced to generate two inputs to the model, corresponding to the respective tasks of segmentation and classification. Let
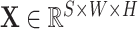 denote the 3D CT scan with slice number
denote the 3D CT scan with slice number
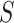 , width
, width
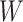 and height
and height
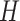 . In the 3DLSC-COVID database, the slice number
. In the 3DLSC-COVID database, the slice number
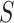 is within the range of
is within the range of
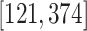 , and the width
, and the width
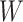 and the height
and the height
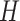 are both 512. For the segmentation task, the CT scan
are both 512. For the segmentation task, the CT scan
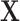 is cropped into smaller non-overlapping 3D patches
is cropped into smaller non-overlapping 3D patches
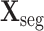 with size
with size
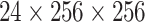 as the input. The 3D segmented patches are aligned as the final segmentation result. For the classification task,
as the input. The 3D segmented patches are aligned as the final segmentation result. For the classification task,
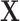 is processed by slice sampling to select
is processed by slice sampling to select
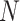 (
(
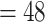 in this paper) slices spaced equidistantly, which can reduce the redundancy between consecutive slices for further accelerating the inference process. Mathematically, the sampled CT scan
in this paper) slices spaced equidistantly, which can reduce the redundancy between consecutive slices for further accelerating the inference process. Mathematically, the sampled CT scan
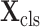 can be formulated as
can be formulated as
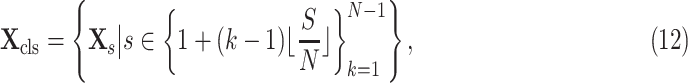 |
where
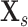 is the
is the
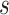 -th slice of the CT scan
-th slice of the CT scan
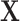 , and
, and
 is a floor function. Consequently, the size of
is a floor function. Consequently, the size of
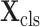 is
is
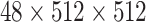 for the classification task.
for the classification task.
3). Model Training:
We follow the two-stage training scheme [28] to train our joint learning model. In the first stage, we separately pre-train the corresponding subnets for segmentation and classification tasks. For segmentation, we pre-train part of the cross-task feature and the 3D lesion segmentation subnets, and for classification, we pre-train the other part of the cross-task feature and the disease classification subnets. In the second stage, all the 3 subnets are simultaneously fine-tuned based on the pre-trained models, over both tasks of segmentation and classification. At both stages, the parameters are updated using the Adam optimizer [24], with a first-order momentum of 0.9 and a second-order momentum of 0.999. The initial learning rates are set to 0.001 for the pre-training stage and 0.0001 for the fine-tuning stage, which are adjusted by linear decay for stable training. The values of the key hyper-parameters for training can be found in the supplementary material. Our DeepSC-COVID model is implemented on PyTorch [38] with the Python environment. All experiments are conducted on a computer with an Intel(R) Core(TM) i7-6900 CPU@3.20 GHz, 128 GB RAM and 4 Nvidia GeForce GTX 1080 TI GPUs. For fair comparison, all compared methods are reimplemented and timed using the same computer as ours.
B. 3D Lesion Segmentation Results
We qualitatively and quantitatively evaluate the lesion segmentation performance of our DeepSC-COVID model. Table II reports the 3D lesion segmentation results of our DeepSC-COVID and other state-of-the-art segmentation models. As shown in this table, our model achieves high accuracy in 3D lesion segmentation, i.e., 73.3%, 80.2%, 95.6%, 71.8%, and 2.8 mm in terms of Dice similarity coefficient (DSC), sensitivity, specificity, normalized surface Dice (NSD) and root mean square symmetric surface distance (RMSD), respectively. In contrast to our model, the accuracy of other segmentation models is relatively low, e.g., the DSC scores are only 61.2%, 65.3%, 63.7% and 67.2% for UNet
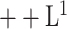 [64], UNet
[64], UNet
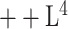 [64], DenseVNet [16] and COPLE-Net [48], respectively. Note that UNet
[64], DenseVNet [16] and COPLE-Net [48], respectively. Note that UNet
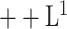 and UNet
and UNet
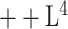 are the lightest and heaviest versions in [64]. Similar results can be found for other metrics, including sensitivity, specificity, RMSD and NSD. Additionally, Fig. 6 visualizes the segmentation results of our and the comparison models. As shown, our DeepSC-COVID model can locate both the COVID-19 and CAP lesions with higher accuracy than other models. In addition to the segmentation accuracy, we outperform most compared models in terms of segmentation efficiency, i.e., it takes 2.2 seconds for our model to segment a 3D CT scan, while other models require 1.0 to 10.8 seconds to process one 3D CT scan.
are the lightest and heaviest versions in [64]. Similar results can be found for other metrics, including sensitivity, specificity, RMSD and NSD. Additionally, Fig. 6 visualizes the segmentation results of our and the comparison models. As shown, our DeepSC-COVID model can locate both the COVID-19 and CAP lesions with higher accuracy than other models. In addition to the segmentation accuracy, we outperform most compared models in terms of segmentation efficiency, i.e., it takes 2.2 seconds for our model to segment a 3D CT scan, while other models require 1.0 to 10.8 seconds to process one 3D CT scan.
TABLE II. The Segmentation and Classification Performance of Our Model, Human Expert and Other Models. The Metrics are Presented in the Format of Mean (Standard Deviation). Note That the 3D Lesion Segmentation and Classification Subnets in Our Model Are Denoted as Seg. and Cls., Respectively.
| Modality | Time (s) | DSC (%) | Sensitivity (%) | Specificity (%) | NSD (%) | RMSD (mm) | ||
|---|---|---|---|---|---|---|---|---|
| Segmentation | Human expert | 3D | 869.2 | 60.3 (10.8) | 61.2 (7.2) | 88.7 (1.2) | 60.2 (3.6) | 12.8 (4.1) |
| U-Net++L1 | 2D | 1.0 | 61.2 (5.7) | 66.7 (6.4) | 90.9 (0.8) | 62.4 (3.6) | 13.6 (2.3) | |
| U-Net++L4 | 2D | 8.2 | 65.3 (6.2) | 71.3 (5.8) | 92.8 (0.6) | 66.8 (3.1) | 9.6 (2.6) | |
| DenseVNet | 3D | 2.8 | 63.7 (7.4) | 69.2 (5.9) | 92.1 (0.7) | 63.6 (2.7) | 7.8 (3.1) | |
| COPLE-Net | 2D | 10.8 | 67.2 (6.8) | 73.4 (7.2) | 93.2 (0.6) | 68.3 (1.9) | 5.2 (2.2) | |
| FSS-2019-nCov | 2D | 8.7 | 67.9(7.6) | 74.1 (5.2) | 93.8 (0.7) | 68.8 (2.1) | 5.4 (3.0) | |
| DeepSC-COVID w/o Cls. | 3D | 1.1 | 66.2 (7.7) | 72.7 (4.9) | 92.7 (0.8) | 64.3 (2.8) | 6.2 (2.5) | |
| DeepSC-COVID | 3D | 2.2 | 73.3 (8.5) | 80.2 (6.8) | 95.6 (0.7) | 71.8 (2.6) | 2.8 (1.6) | |
| Modality | Time (s) | Accuracy (%) | Sensitivity (%) | Specificity (%) | F1-score (%) | AUC (%) | ||
| Classification | Human expert | 3D | 378.7 | 95.8 (0.4) | 96.0 (0.4) | 97.9 (0.7) | 95.7 (0.3) | − |
| ResNet-50 | 2D | 9.2 | 77.7 (0.7) | 79.2 (0.2) | 87.7 (0.6) | 77.9 (0.8) | 92.0 (0.1) | |
| 3D ResNet-50 | 3D | 0.6 | 82.5 (0.2) | 82.5 (0.7) | 90.6 (0.2) | 82.0 (0.5) | 94.5 (0.2) | |
| COVNet | 2D | 5.3 | 87.2 (0.8) | 87.6 (0.3) | 93.7 (0.4) | 87.5 (0.3) | 97.4 (0.1) | |
| DeCoVNet | 3D | 2.3 | 89.2 (0.5) | 89.0 (0.4) | 94.3 (0.2) | 88.9 (0.8) | 98.3 (0.1) | |
| DeepSC-COVID w/o Seg. | 3D | 0.7 | 85.2(0.4) | 85.3 (0.2) | 91.9 (0.3) | 84.9 (0.1) | 94.4 (0.2) | |
| DeepSC-COVID | 3D | 2.2 | 94.5 (0.3) | 94.7 (0.2) | 97.3 (0.5) | 94.2 (0.6) | 98.8 (0.1) | |
Fig. 6.
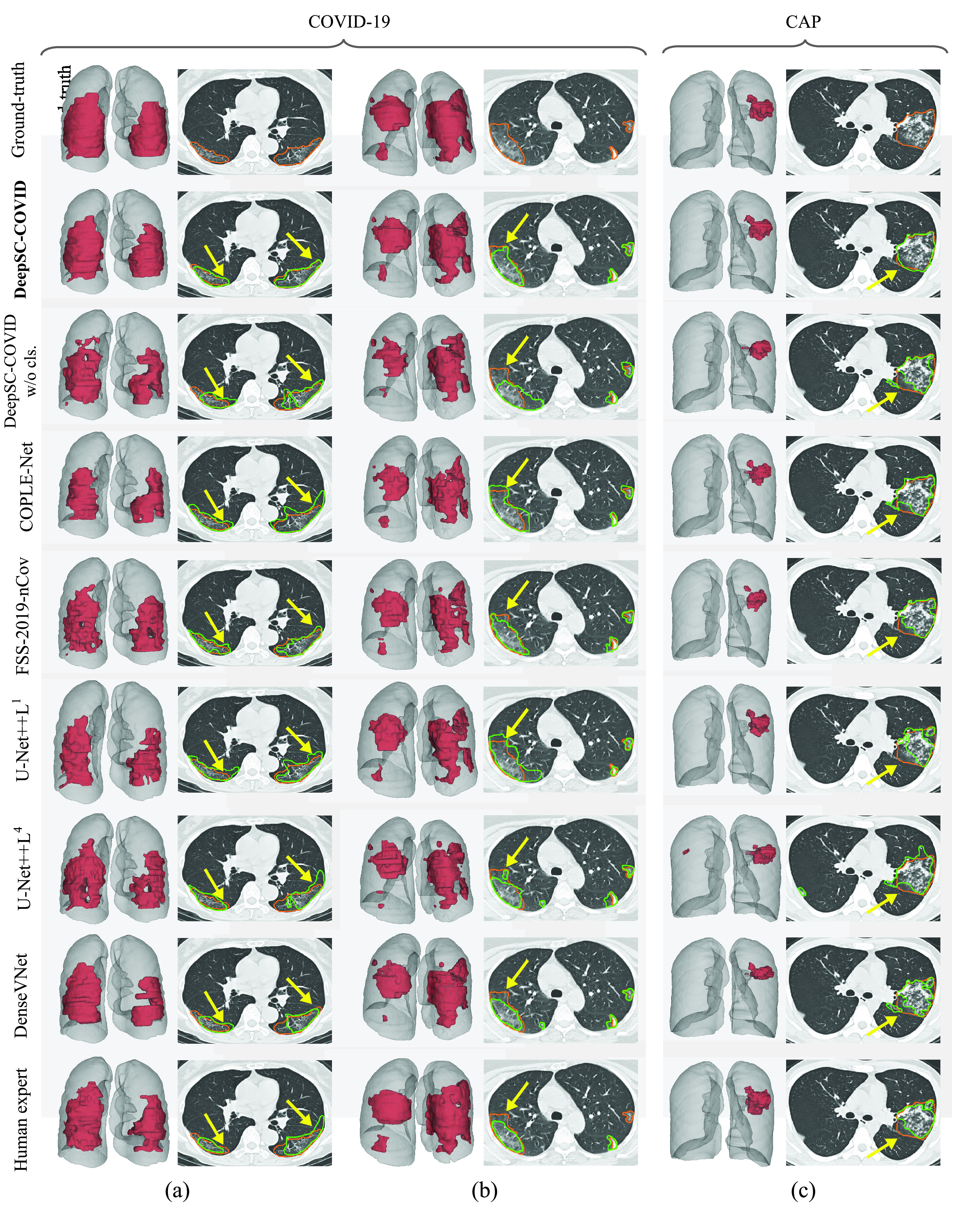
Visual comparison of 3D and 2D lesion segmentation results. (a-b) Segmentation results of two COVID-19 samples. (c) Segmentation results of CAP sample. For 3D visualizations, the lesions and lungs are shown in red and grey for better view. For 2D visualizations, orange and green curves indicate the ground-truth segmentation results and the results generated by different methods.
To further show the superiority of our DeepSC-COVID model, we compare the segmentation performance between our model and a human expert. Here, the human expert is a radiologist with 5 years of working experience. As shown in Table II, our model significantly outperforms the human expert in 3D lesion segmentation, with an improvement of 14%, 19%, 6.9%, 11.6%, and 10 mm in terms of DSC, sensitivity, specificity, NSD and RMSD, respectively. It is not surprising to see the low dice score of the human expert, since lesion segmentation in medical images is known to suffer from high inter-reader variability [35]. To conclude, our DeepSC-COVID model performs considerably better than the other segmentation models and the human expert for 3D lesion segmentation.
C. Disease Classification Results
Table II shows the classification results of our DeepSC-COVID model and 4 other state-of-the-art models for classifying COVID-19, CAP and non-pneumonia individuals. As can be seen, the classification accuracy (94.5%) of our model is considerably higher than those of the alternative methods, i.e., ResNet-50 [48] (77.7%), 3D ResNet-50 [39] (82.5%), COVNet [26] (87.2%) and DeCoVNet [51] (89.2%). Moreover, the sensitivity, specificity and area under the receiver operating characteristic (ROC) curve (AUC) of our model are the highest among all models. Table II also compares F1-score between our and other models. Our model has an F1-score of 94.2%, while ResNet-50, 3D ResNet-50, COVNet and DeCoVNet only yield values of 77.9%, 82.0%, 87.5% and 88.9%, respectively. In addition, Fig. 7 shows the ROC curves for each category, which visualize the tradeoff between sensitivity and specificity. Compared with other four models [26], [39], [48], [51], our ROC curve is closer to the upper-left corner, indicating that our model achieves better classification results than do the 4 other models. To summarize, our DeepSC-COVID model is considerably better than 4 other models with respect to classifying COVID-19, CAP and non-pneumonia.
Fig. 7.
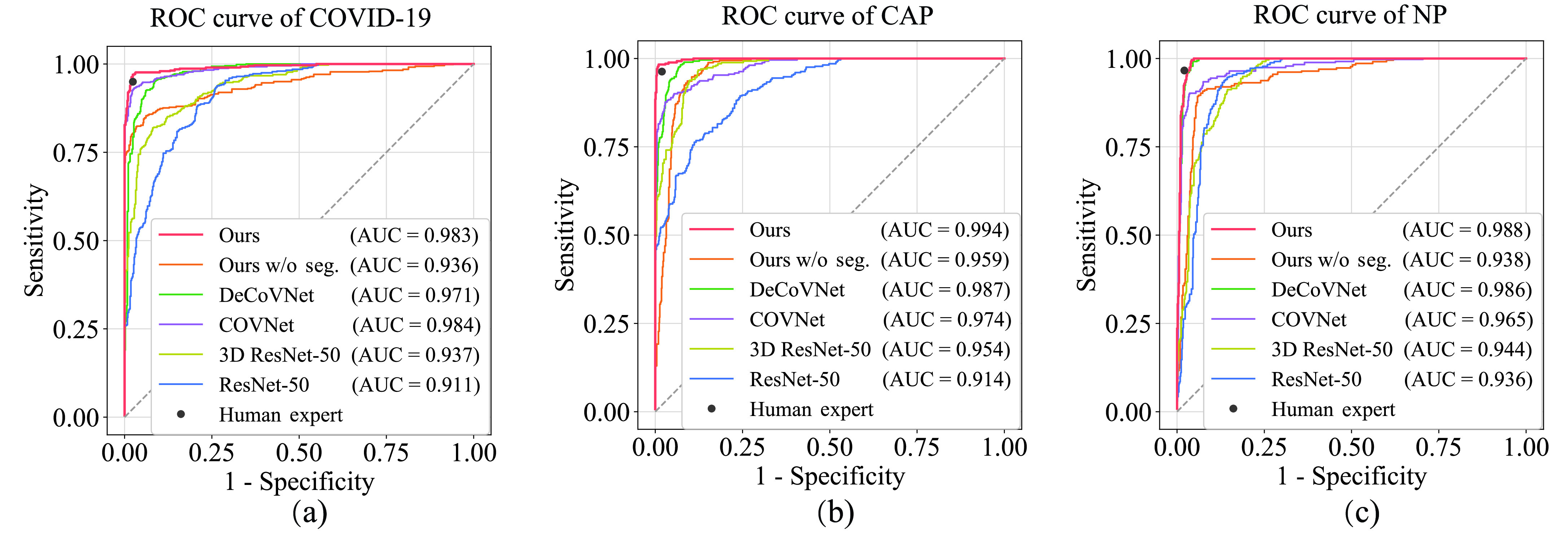
The ROC curves of our DeepSC-COVID model, other models and human expert in the identification of COVID-19 (a), CAP (b) and NP (c).
Compared to the human expert, the proposed DeepSC-COVID model offers a great advantage in diagnosis speed, i.e., the classification speed of our model is 2.2 seconds, which is significantly faster than the human expert (378.7 seconds). In addition, our model is comparable to the human expert in diagnosis accuracy, i.e., the average sensitivity, specificity and F1-score of our model are only around 1.0% lower than those of the human expert. Fig. 7 plots the classification performance of the human expert on sensitivity-specificity plane. As can be seen, for both COVID-19 and CAP classification, the point of the human expert is located in the lower-right areas of our ROC curves, which indicates that given the same specificity of the human expert, our model can achieve higher sensitivity by adjusting the classification threshold. All of these results indicate that the DeepSC-COVID model offers high classification accuracy and speed, which offers capability for auxiliary medical diagnosis and large-scale COVID-19 screening.
D. Multi-Task Gain
To evaluate the gain of multi-task learning, additional experiments are conducted with single tasks of segmentation and classification. Specifically, we first remove the classification subnet from our model for the single segmentation task, and then remove the segmentation subnet for single classification task. Table II reports the results of single-task learning. As reported, the accuracy of single-task learning is lower than that of multi-task learning for both tasks. Specifically, for segmentation, the multi-task gain values are 7.1%, 7.5%, 2.9%, 7.5% and 3.4 mm in terms of DSC, sensitivity, specificity, NSD and RMSD, respectively. For classification, the multi-task gain achieves values of 9.3%, 9.4%, 5.4%, 9.3% and 4.4% in terms of accuracy, sensitivity, specificity, F1-score and AUC, respectively. Additionally, the results of the single segmentation task are visualized in Fig. 6. The ROC curve of single classification task are shown in Fig. 7.
E. Ablation Study
Here, we analyze the effectiveness of different components in the proposed DeepSC-COVID model on the tasks of 3D lesion segmentation and disease classification through ablation study.
1). Effectiveness of 3D Inception Block:
We first analyze the impact of 3D inception block on 3D lesion segmentation and disease classification. Specifically, we replace the 3D inception block by conventional 3D convolutional layer, in which the kernel size, stride and output channel are the same as the 3D inception block. Fig. 8 shows the segmentation and classification results with and without the 3D inception block. As shown, the performance of both segmentation and classification tasks significantly degrades after replacing the 3D inception block. This indicates the effectiveness of our 3D inception block in extracting effective multi-scale 3D features for both tasks.
Fig. 8.
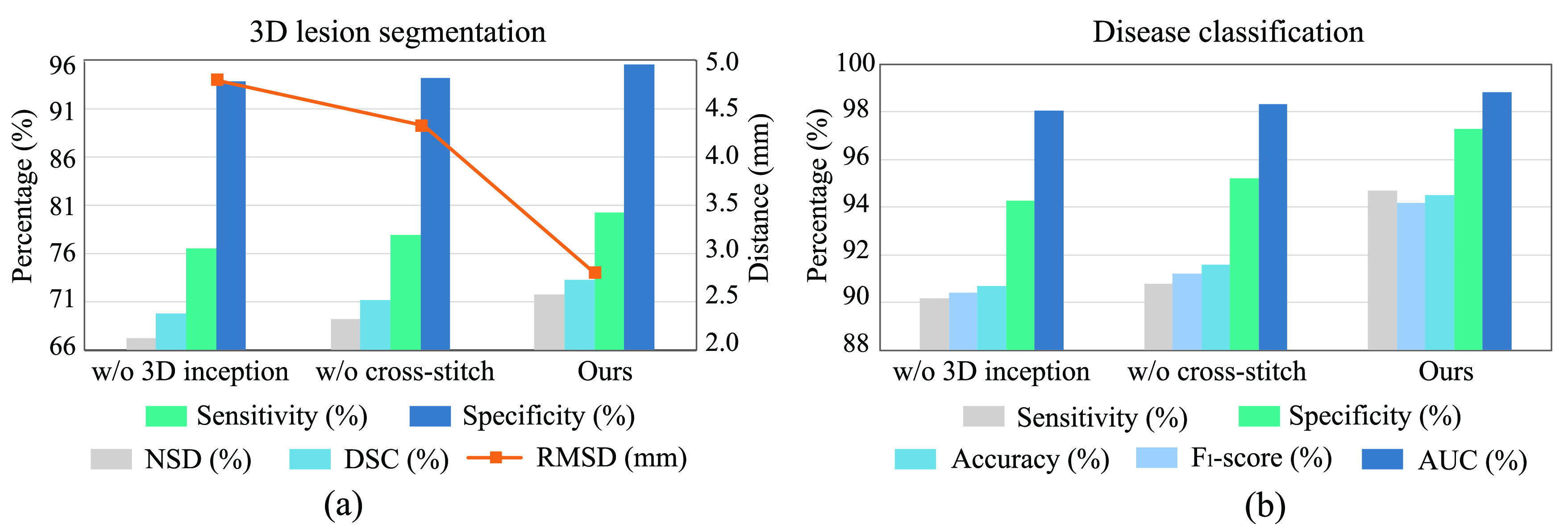
The impacts of 3D inception block and cross-stitch unit on the performance of segmentation and classification. (a) Results of 3D lesion segmentation, in terms of sensitivity, specificity, NSD, DSC and RMSD. (b) Results of disease classification, in terms of sensitivity, specificity, accuracy, F1-score and AUC.
2). Effectiveness of Cross-Stitch Unit:
We further conduct the ablation experiment to evaluate the impact of the cross-stitch unit on segmentation and classification performance, by removing it from the cross-task feature subnet in the proposed DeepSC-COVID model. Fig. 8 shows the segmentation and classification results with and without the cross-stitch unit. We can see from this figure that the performance of both the segmentation and classification degrades, when the cross-stitch unit is removed. This validates the positive contribution of cross-stitch unit to our model.
3). Effectiveness of Task-Aware Loss:
Finally, we evaluate the impact of the proposed task-aware loss. To be specific, we train the DeepSC-COVID model with different weights
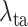 on the task-aware loss, i.e.,
on the task-aware loss, i.e.,
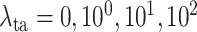 in equation (11) of the main text. Note that
in equation (11) of the main text. Note that
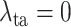 indicates that the task-aware loss is fully removed. Fig. 9 shows the segmentation and classification results with different
indicates that the task-aware loss is fully removed. Fig. 9 shows the segmentation and classification results with different
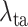 . As shown, the DeepSC-COVID model performs the worst, when the task-aware loss is fully removed (i.e.,
. As shown, the DeepSC-COVID model performs the worst, when the task-aware loss is fully removed (i.e.,
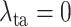 ). Besides, the performance of both segmentation and classification reduces, when
). Besides, the performance of both segmentation and classification reduces, when
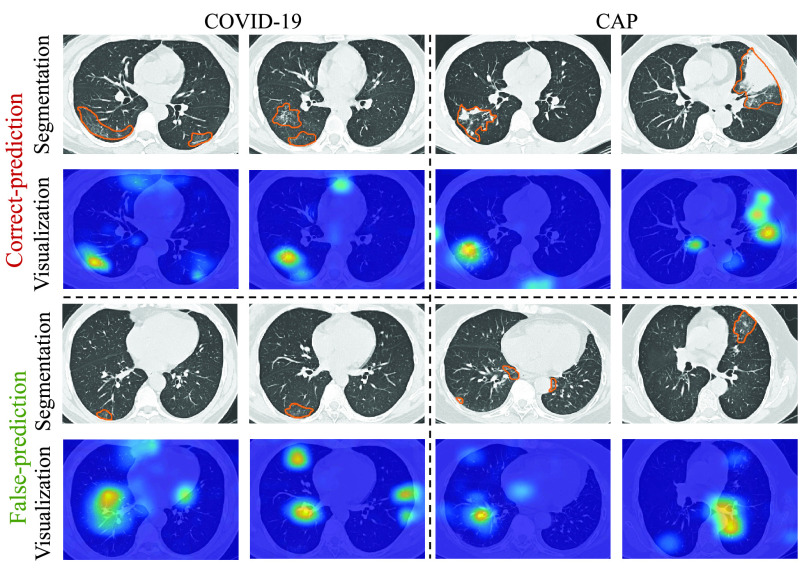
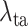 is either smaller or larger than 101. This implies that the under- or over-weighted task-aware loss degrades the performance of the DeepDC-COVID model. In summary, the task-aware loss has a positive impact on the proposed DeepDC-COVID model for both segmentation and classification tasks. In addition, Fig. 10 shows some examples of visualization maps and their corresponding segmentation results for both correct-prediction and false-prediction cases. As can be seen, in the correct-prediction case, the visualization map is consistent with the lesion segmentation result. By contrast, in the false-prediction case, there exists an obvious difference between the visualization map and segmented lesions. This verifies the effectiveness of the proposed task-aware loss. Moreover, these visualization results are consistent with the clinical experience, which further demonstrates the explainability of our method.
is either smaller or larger than 101. This implies that the under- or over-weighted task-aware loss degrades the performance of the DeepDC-COVID model. In summary, the task-aware loss has a positive impact on the proposed DeepDC-COVID model for both segmentation and classification tasks. In addition, Fig. 10 shows some examples of visualization maps and their corresponding segmentation results for both correct-prediction and false-prediction cases. As can be seen, in the correct-prediction case, the visualization map is consistent with the lesion segmentation result. By contrast, in the false-prediction case, there exists an obvious difference between the visualization map and segmented lesions. This verifies the effectiveness of the proposed task-aware loss. Moreover, these visualization results are consistent with the clinical experience, which further demonstrates the explainability of our method.
Fig. 9.
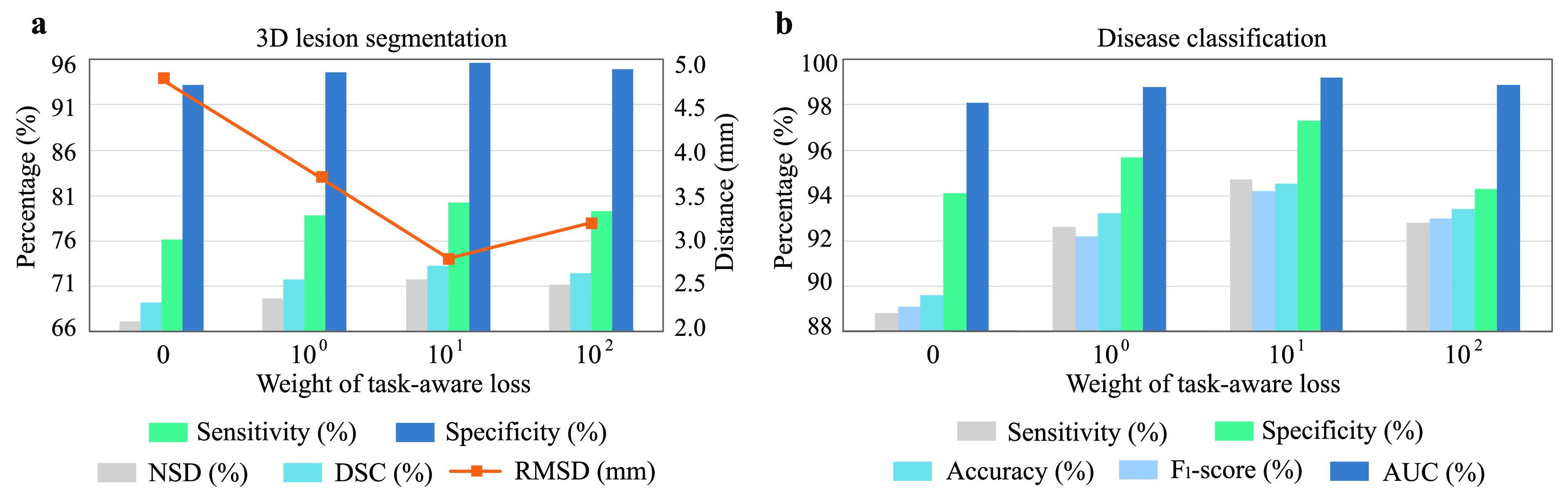
Segmentation and classification results with different weights on the task-aware loss. (a) Results of 3D lesion segmentation, in terms of sensitivity, specificity, NSD, DSC and RMSD. (b) Results of disease classification, in terms of sensitivity, specificity, accuracy, F1-score and AUC.
Fig. 10.
Visualization maps and segmentation results of both correct-prediction and false-prediction cases.
To evaluate the effectiveness of the multi-scale setting in the task-aware loss, we conduct an ablation study to replace the multi-scale setting with single-scale setting. As shown in Table III, compared with the multi-scale setting, the performance of the single-scale setting degrades by 3.6% in accuracy for classification and 3.0% in DSC for segmentation. This verifies the effectiveness of the multi-scale setting of our task-aware loss.
TABLE III. Mean Values in Terms of Percentage for Lesion Segmentation and Disease Classification Metrics on Multi-Scale and Single-Scale Settings of Our Task-Aware Loss.
| Task | Classification | Segmentation | ||||
|---|---|---|---|---|---|---|
| Acc. | Sen. | AUC | DSC | Sen. | NSD | |
| Single-scale | 90.9 | 90.3 | 98.6 | 70.3 | 77.9 | 69.3 |
| Multi-scale (ours) | 94.5 | 94.7 | 99.2 | 73.3 | 80.2 | 71.8 |
VI. Conclusions
In this study, we have proposed a CT interpretation model, namely DeepSC-COVID, for rapid, accurate and explainable screening of COVID-19. First, we built and released an large-scale database, called 3DLSC-COVID, which is the first database containing both 3D lesion segmentation and disease labels for the diagnosis of COVID-19, CAP and non-pneumonia. Besides, we obtained four important findings through qualitative and quantitative analysis over our 3DLSC-COVID database. Second, a novel multi-task learning architecture is proposed in DeepSC-COVID, for simultaneous learning of 3D lesion segmentation and disease classification. Benefiting from the multi-task learning architecture, our DeepSC-COVID model can segment the lesions more accurately with the knowledge acquired from the classification task. Finally, extensive experiments verified that our method advanced the state-of-the-art in 3D lesion segmentation and disease classification.
There is still room for improvement in our model as the future work. First, since our database only contains Chinese patients, it may exhibit limited diagnostic performance for other races. Future study should involve enlarging the database with multi-ethnic cases to enable robust interpretation performance. Second, our model only uses chest CT scan as the basis of diagnosis for COVID-19. Although the CT scan is validated as effective diagnostic evidence, other clinical tests, e.g., symptom records and disease history, can also contribute to the diagnosis of COVID-19. Hence, another future research direction is to take advantage of multiple inputs for more comprehensive interpretation. Third, this study only focuses on the immediate screening of COVID-19, i.e., the diagnosis result is either positive or negative. The graded diagnosis of COVID-19 is desirable for the CT interpretation model, for example, grading the suspected patients into negative, mild, moderate, severe and critical cases. As a result, both clinical diagnosis and prognosis can be significantly improved.
Funding Statement
This work was supported in part by the Beijing Natural Science Foundation under Grant JQ20020; in part by the NSFC project under Grant 61922009, Grant 61876013, and Grant 62050175; and in part by the Fundamental Research Funds for the Central Universities under Grant 2020kfyXGYJ097.
Footnotes
The 3DLSC-COVID database is available at IEEE Dataport https://dx.doi.org/10.21227/mxb3-7j48
The source codes of our DeepSC-COVID model are available at Github (https://github.com/XiaofeiWang2018/DeepSC-COVID)
In this section, subscripts
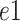 ,
,
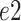 ,
,
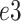 indicate the first, second and third 3D encoder unit. Subscript
indicate the first, second and third 3D encoder unit. Subscript
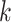 is the channel index of the corresponding feature.
is the channel index of the corresponding feature.
Contributor Information
Xiaofei Wang, Email: xfwang@buaa.edu.cn.
Lai Jiang, Email: jianglai.china@buaa.edu.cn.
Liu Li, Email: liu.li20@imperial.ac.uk.
Mai Xu, Email: maixu@buaa.edu.cn.
Xin Deng, Email: cindydeng@buaa.edu.cn.
Lisong Dai, Email: m201876035@hust.edu.cn.
Xiangyang Xu, Email: 1993ly0538@hust.edu.cn.
Tianyi Li, Email: tianyili@buaa.edu.cn.
Yichen Guo, Email: 16711024@buaa.edu.cn.
Zulin Wang, Email: wzulin@buaa.edu.cn.
Pier Luigi Dragotti, Email: p.dragotti@imperial.ac.uk.
References
- [1].Ai T.et al. , “Correlation of chest CT and RT-PCR testing for coronavirus disease 2019 (COVID-19) in China: A report of 1014 cases,” Radiology, vol. 262, Aug. 2020, Art. no. 200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Amyar A., Modzelewski R., Li H., and Ruan S., “Multi-task deep learning based CT imaging analysis for COVID-19 pneumonia: Classification and segmentation,” Comput. Biol. Med., vol. 126, Nov. 2020, Art. no. 104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].An P., Ye Y., Chen M., Chen Y., Fan W., and Wang Y., “Management strategy of novel coronavirus (COVID-19) pneumonia in the radiology department: A chinese experience,” Diagnostic Interventional Radiol., vol. 26, no. 3, p. 200, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Angelov P. and Almeida Soares E., “SARS-CoV-2 CT-scan dataset: A large dataset of real patients CT scans for SARS-CoV-2 identification,” Medrxiv, Apr. 2020.
- [5].Bartlett J. G. and Mundy L. M., “Community-acquired pneumonia,” New England J. Med., vol. 333, no. 24, pp. 1618–1624, 1995. [DOI] [PubMed] [Google Scholar]
- [6].Bernheim A.et al. , “Chest CT findings in coronavirus disease-19 (COVID-19): Relationship to duration of infection,” Radiology, vol. 295, no. 3, Jun. 2020, Art. no. 200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Born J.et al. , “POCOVID-net: Automatic detection of COVID-19 from a new lung ultrasound imaging dataset (POCUS),” 2020, arXiv:2004.12084. [Online]. Available: http://arxiv.org/abs/2004.12084
- [8].Chowdhury M. E.et al. , “Can ai help in screening viral and COVID-19 pneumonia?” IEEE Access, vol. 8, pp. 132665–132676, 2020. [Google Scholar]
- [9].Paul Cohen J., Morrison P., Dao L., Roth K., Duong T. Q, and Ghassemi M., “COVID-19 image data collection: Prospective predictions are the future,” 2020, arXiv:2006.11988. [Online]. Available: http://arxiv.org/abs/2006.11988
- [10].Cowan I. A., MacDonald S. L., and Floyd R. A., “Measuring and managing radiologist workload: Measuring radiologist reporting times using data from a radiology information system,” J. Med. Imag. Radiat. Oncol., vol. 57, no. 5, pp. 558–566, Oct. 2013. [DOI] [PubMed] [Google Scholar]
- [11].Dangis A.et al. , “Accuracy and reproducibility of low-dose submillisievert chest CT for the diagnosis of COVID-19,” Radiol., Cardiothoracic Imag., vol. 2, no. 2, Apr. 2020, Art. no. e200196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].De Fauw J.et al. , “Clinically applicable deep learning for diagnosis and referral in retinal disease,” Nature Med., vol. 24, no. 9, pp. 1342–1350, Sep. 2018. [DOI] [PubMed] [Google Scholar]
- [13].de la Iglesia Vayá M.et al. , “BIMCV COVID-19+: A large annotated dataset of RX and CT images from COVID-19 patients,” 2020, arXiv:2006.01174. [Online]. Available: http://arxiv.org/abs/2006.01174
- [14].Fang Y.et al. , “Sensitivity of chest ct for COVID-19: Comparison to RT-PCR,” Radiology, vol. 262, Aug. 2020, Art. no. 200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Fidon L.et al. , “Generalised wasserstein dice score for imbalanced multi-class segmentation using holistic convolutional networks,” in Proc. Int. MICCAI Brain Lesion Workshop. Cham, Switzerland: Springer, 2017, pp. 64–76. [Google Scholar]
- [16].Gibson E.et al. , “Automatic multi-organ segmentation on abdominal CT with dense V-Networks,” IEEE Trans. Med. Imag., vol. 37, no. 8, pp. 1822–1834, Aug. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Goel T., Murugan R., Mirjalili S., and Chakrabartty D. K., “Automatic screening of COVID-19 using an optimized generative adversarial network,” Cognit. Comput., vol. 4, pp. 1–16, Jan. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].He K., Zhang X., Ren S., and Sun J., “Deep residual learning for image recognition,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit., Mar. 2016, pp. 770–778. [Google Scholar]
- [19].Hellewell J.et al. , “Feasibility of controlling COVID-19 outbreaks by isolation of cases and contacts,” Lancet Global Health, vol. 8, no. 4, pp. e488–e496, Apr. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hinton G., “Deep learning-a technology with the potential to transform health care,” J. Amer. Med. Assoc., vol. 320, no. 11, pp. 1101–1102, 2018. [DOI] [PubMed] [Google Scholar]
- [21].Hofmanninger J., Prayer F., Pan J., Rohrich S., Prosch H., and Langs G., “Automatic lung segmentation in routine imaging is primarily a data diversity problem, not a methodology problem,” 2020, arXiv:2001.11767. [Online]. Available: http://arxiv.org/abs/2001.11767 [DOI] [PMC free article] [PubMed]
- [22].Michael D. H., Constantine A. R., Amar S., Mark M. H., and Travis S. H.. A Role for CT in COVID-19 What Data Really Tell Us so Far. Accessed: Apr. 2020. [Online]. Available: http://www.thelancet.com/article/S0140673620307285/pdf [Google Scholar]
- [23].Jin S.et al. , “Ai-assisted ct imaging analysis for COVID-19 screening: Building and deploying a medical ai system in four weeks,” MedRxiv, 2020. [DOI] [PMC free article] [PubMed]
- [24].Kingma D. P. and Ba J., “Adam: A method for stochastic optimization,” 2014, arXiv:1412.6980. [Online]. Available: http://arxiv.org/abs/1412.6980
- [25].LeCun Y., Bengio Y., and Hinton G., “Deep learning,” Nature, vol. 521, pp. 436–444, May 2015. [DOI] [PubMed] [Google Scholar]
- [26].Li L.et al. , “Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT,” Radiology, vol. 4, May 2020, Art. no. 200905. [Google Scholar]
- [27].Li L., Xu M., Wang X., Jiang L., and Liu H., “Attention based glaucoma detection: A large-scale database and CNN model,” in Proc. IEEE/CVF Conf. Comput. Vis. Pattern Recognit. (CVPR), Jun. 2019, pp. 10571–10580. [Google Scholar]
- [28].Li Y., Chen J., and Zheng Y., “A multi-task self-supervised learning framework for scopy images,” in Proc. Int. Symp. Biomed. Imag., 2020, pp. 2005–2009. [Google Scholar]
- [29].Lin T.-Y., Goyal P., Girshick R., He K., and Dollar P., “Focal loss for dense object detection,” in Proc. IEEE Int. Conf. Comput. Vis. (ICCV), Oct. 2017, pp. 2980–2988. [DOI] [PubMed] [Google Scholar]
- [30].Long E.et al. , “An artificial intelligence platform for the multihospital collaborative management of congenital cataracts,” Nature Biomed. Eng., vol. 1, no. 2, pp. 1–8, Feb. 2017. [Google Scholar]
- [31].Lu D., Disease Neuroimaging Initiative A., Popuri K., Ding G. W., Balachandar R., and Beg M. F., “Multimodal and multiscale deep neural networks for the early diagnosis of Alzheimer’s disease using structural MR and FDG-PET images,” Sci. Rep., vol. 8, no. 1, Dec. 2018, Art. no. 5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ma J.et al. , “Towards data-efficient learning: A benchmark for COVID-19 CT lung and infection segmentation,” 2020, arXiv:2004.12537. [Online]. Available: http://arxiv.org/abs/2004.12537 [DOI] [PubMed]
- [33].Mahmud T.et al. , “CovTANet: A hybrid tri-level attention based network for lesion segmentation, diagnosis, and severity prediction of COVID-19 chest CT scans,” 2021, arXiv:2101.00691. [Online]. Available: http://arxiv.org/abs/2101.00691 [DOI] [PMC free article] [PubMed]
- [34].Mei X.et al. , “Artificial intelligence–enabled rapid diagnosis of patients with COVID-19,” Nature Med., vol. 26, no. 8, pp. 1224–1228, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Menze B. H.et al. , “The multimodal brain tumor image segmentation benchmark (BRATS),” IEEE Trans. Med. Imag., vol. 34, no. 10, pp. 1993–2024, Oct. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Morozov S. P.et al. , “MosMedData: Chest CT scans with COVID-19 related findings dataset,” 2020, arXiv:2005.06465. [Online]. Available: http://arxiv.org/abs/2005.06465
- [37].Ouyang X.et al. , “Dual-sampling attention network for diagnosis of COVID-19 from community acquired pneumonia,” IEEE Trans. Med. Imag., vol. 39, no. 8, pp. 2595–2605, Aug. 2020. [DOI] [PubMed] [Google Scholar]
- [38].Paszke A.et al. , “Pytorch: An imperative style, high-performance deep learning library,” in Proc. Adv. Neural Inf. Process. Syst., 2019, pp. 8026–8037. [Google Scholar]
- [39].Qiu Z., Yao T., and Mei T., “Learning spatio-temporal representation with pseudo-3D residual networks,” in Proc. IEEE Int. Conf. Comput. Vis. (ICCV), Oct. 2017, pp. 5533–5541. [Google Scholar]
- [40].Rajalakshmi R., Subashini R., Anjana R. M., and Mohan V., “Automated diabetic retinopathy detection in smartphone-based fundus photography using artificial intelligence,” Eye, vol. 32, no. 6, pp. 1138–1144, Jun. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ranney M. L., Griffeth V., and Jha A. K., “Critical supply shortages — The need for ventilators and personal protective equipment during the COVID-19 pandemic,” New England J. Med., vol. 382, no. 18, p. e41, Apr. 2020. [DOI] [PubMed] [Google Scholar]
- [42].Remuzzi A. and Remuzzi G., COVID-19 and Italy: What Next. London, U.K.: The Lancet, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Rio D. C., “Reverse transcription–polymerase chain reaction,” Cold Spring Harbor Protocols, vol. 2014, no. 11, 2014, Art. no. prot080887. [DOI] [PubMed] [Google Scholar]
- [44].Rothan H. A. and Byrareddy S. N., “The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak,” J. Autoimmunity, vol. 109, May 2020, Art. no. 102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Ruder S., “An overview of multi-task learning in deep neural networks,” 2017, arXiv:1706.05098. [Online]. Available: http://arxiv.org/abs/1706.05098
- [46].Scohy A., Anantharajah A., Bodéus M., Kabamba-Mukadi B., Verroken A., and Rodriguez-Villalobos H., “Low performance of rapid antigen detection test as frontline testing for COVID-19 diagnosis,” J. Clin. Virol., vol. 129, Aug. 2020, Art. no. 104455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Szegedy C.et al. , “Going deeper with convolutions,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit. (CVPR), Jun. 2015, pp. 1–9. [Google Scholar]
- [48].Wang G.et al. , “A noise-robust framework for automatic segmentation of COVID-19 pneumonia lesions from CT images,” IEEE Trans. Med. Imag., vol. 39, no. 8, pp. 2653–2663, Aug. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Wang L., Lin Z. Q., and Wong A., “COVID-net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images,” Sci. Rep., vol. 10, no. 1, Dec. 2020, Art. no. 19549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Wang S.et al. , “A deep learning algorithm using CT images to screen for corona virus disease (COVID-19),” Eur. Radiol., pp. 1–9, 2021. [DOI] [PMC free article] [PubMed]
- [51].Wang X.et al. , “A weakly-supervised framework for COVID-19 classification and lesion localization from chest CT,” IEEE Trans. Med. Imag., vol. 39, no. 8, pp. 2615–2625, Aug. 2020. [DOI] [PubMed] [Google Scholar]
- [52].Wang X., Xu M., Li L., Wang Z., and Guan Z., “Pathology-aware deep network visualization and its application in glaucoma image synthesis,” in Proc. Int. Conf. Med. Image Comput. Comput.-Assisted Intervent. Cham, Switzerland: Springer, 2019, pp. 423–431. [Google Scholar]
- [53].Wang X., Xu M., Zhang J., Jiang L., and Li L., “Deep multi-task learning for diabetic retinopathy grading in fundus images,” in Proc. Assoc. Adv. Artif. Intell., 2021, pp. 1–5. [Google Scholar]
- [54].Coronavirus disease 2019 (COVID-19), W. H. Organization, Geneva, Switzerland, 2020. [Google Scholar]
- [55].Wu Y. and He K., “Group normalization,” in Proc. Eur. Conf. Comput. Vis., 2018, pp. 3–19. [Google Scholar]
- [56].Xu B.et al. , “Chest CT for detecting COVID-19: A systematic review and meta-analysis of diagnostic accuracy,” Eur. Radiol., vol. 30, no. 10, pp. 5720–5727, Oct. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Yan L.et al. , “An interpretable mortality prediction model for COVID-19 patients,” Nature Machine Intelligence, vol. 2, no. 5, pp. 283–288, 2020. [Google Scholar]
- [58].Yang W.et al. , “The role of imaging in 2019 novel coronavirus pneumonia (COVID-19),” Eur. Radiol., vol. 4, pp. 1–9, Oct. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Yang X., He X., Zhao J., Zhang Y., Zhang S., and Xie P., “COVID-CT-dataset: A CT scan dataset about COVID-19,” 2020, arXiv:2003.13865. [Online]. Available: http://arxiv.org/abs/2003.13865
- [60].Zeiler M. D., Krishnan D., Taylor G. W., and Fergus R., “Deconvolutional networks,” in Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit., Jun. 2010, pp. 2528–2535. [Google Scholar]
- [61].Zhang K.et al. , “Clinically applicable ai system for accurate diagnosis, quantitative measurements, and prognosis of COVID-19 pneumonia using computed tomography,” Cell, vol. 181, no. 6, pp. 1423–1433, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Zhou B., Khosla A., Lapedriza A., Oliva A., and Torralba A., “Learning deep features for discriminative localization,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit. (CVPR), Jun. 2016, pp. 2921–2929. [Google Scholar]
- [63].Zhou L.et al. , “A rapid, accurate and machine-agnostic segmentation and quantification method for CT-based COVID-19 diagnosis,” IEEE Trans. Med. Imag., vol. 39, no. 8, pp. 2638–2652, Aug. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Zhou Z., Siddiquee M. M. R., Tajbakhsh N., and Liang J., “UNet++: Redesigning skip connections to exploit multiscale features in image segmentation,” IEEE Trans. Med. Imag., vol. 39, no. 6, pp. 1856–1867, Jun. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]