Abstract
It is widely known that a quick disclosure of the COVID-19 can help to reduce its spread dramatically. Transcriptase polymerase chain reaction could be a more useful, rapid, and trustworthy technique for the evaluation and classification of the COVID-19 disease. Currently, a computerized method for classifying computed tomography (CT) images of chests can be crucial for speeding up the detection while the COVID-19 epidemic is rapidly spreading. In this article, the authors have proposed an optimized convolutional neural network model (ADECO-CNN) to divide infected and not infected patients. Furthermore, the ADECO-CNN approach is compared with pretrained convolutional neural network (CNN)-based VGG19, GoogleNet, and ResNet models. Extensive analysis proved that the ADECO-CNN-optimized CNN model can classify CT images with 99.99% accuracy, 99.96% sensitivity, 99.92% precision, and 99.97% specificity.
Keywords: Convolutional neural network (CNN), COVID-19, computed tomography (CT) images, deep learning, diagnostic imaging
I. Introduction
The novel Coronavirus is an infectious acute disease and can cause respiratory failure due to significant alveoli damage possibly leading to death. This disease was first recognized in Wuhan, China, as a pneumonia outbreak having no specific reasons in late December 2019. Later, it was confirmed as an infection as a result of the Coronavirus called 2019-nCoV. Specifically, February 11, 2020, the United Nations (by means of the World Health Organization) named it COVID-19 and later declared it a worldwide pandemic. Furthermore, the virus was classified as severe-acute-respiratory-syndrome Coronavirus 2 (in short called SARS-Cov-2) by an independent and supranational group of virologists called international committee on taxonomy of viruses (ICTV). This extremely contagious virus spreads from person to person and throughout the world [1]. According to WHO reports, this viral disease affected 192 countries/regions with 2.244.501 confirmed deaths and 103.597.957 confirmed positive cases on February 2, 2021 (source: COVID-19 Dashboard at the Johns Hopkins University).
Infection caused by COVID-19 can result in serious pneumonia and can become fatal like acute respiratory distress syndrome (ARDS) [2]. Clinical symptoms include fatigue, fever, and cough, while other patients can sometimes be asymptomatic. In severe cases, patients may develop ARDS and need ICU or oxygen assistance therapy. Laboratory diagnosis of cases having the SARS-Cov-2 includes elevation of the C-reaction protein level, lymphopenia, and erythrocyte sedimentation rate. The sequencing of the genome of the SARS-Cov-2 allowed to use the real-time tests called reverse transcriptase polymerase chain reaction (shortened as RT-PCR) as a diagnostic protocol to inhibit the spread of the COVID-19 disease. In an RT-PCR test, swabbed nucleic acid samples are taken from a patient's throat, nose, and lower respiratory tract [3]. The diagnosis of COVID-19 cases in patients is also derived from their anamnesis, etiological factors, their symptoms, as well as on medical images. Clinical sampling for nucleic acid testing is reliable criteria but has some limitations such as insufficient specimen, shortage of test kits, laboratory errors, uneven detection technology, erroneous sampling, and technical problems [4].
The cost of diagnostic tests is also causing financial problems for both patients and governments especially in countries with private health facilities. Health care professionals are considering radiology images as a diagnostic tool to identify COVID-19 patients due to challenges in pathological diagnosis on nucleic acid testing. Radiologists play important roles in the diagnosis and rapid identification of suspected cases; this can be beneficial not only for public but also health systems.
Early diagnosis of highly contagious COVID-19 patients can save many lives by isolating suspected patients to quarantine in separate spaces or in specialized hospitals for serious cases to control the outbreak. Chest radiographs (X-rays) are normally insensitive to make a preliminary detection of the infection [5]; however, as the disease progresses, radiographies are useful to spot pulmonary opacities, and in a severe case, may appear as whited out lungs. Pulmonary consolidation and opacities are observed over the time course of a disease outbreak on chest radiographs [6]. Chest computer tomography (CT) imaging is highly recommended in early detection of disease as it evaluates the extent and nature of the lesion. It also assesses changes that are not usually visible in chest X-rays. CT imaging focuses on lesion features such as shape, quantity, distribution, density, and concomitant symptoms [7].
Chest CT imaging provides a high effectiveness in early diagnosis of COVID-19 infections. Fang et al.
[8] carried out some experiments and comparisons between the RT-PCR and the information obtained by analyzing the chest CT images of 51 patients performed within three days. Fang et al.
[8] found that the effectiveness of a chest CT was higher when compared to the RT-PCR at an early stage, which is 98% versus 71% having  [8]. In patients with negative results of the RT-PCR but with characteristics features in chest CT images resulted positive after repeating the swab test. Chest CT images can be a vital warning signal for being a carrier and can be very helpful for patients with any suspicious symptoms of COVID-19 [9].
[8]. In patients with negative results of the RT-PCR but with characteristics features in chest CT images resulted positive after repeating the swab test. Chest CT images can be a vital warning signal for being a carrier and can be very helpful for patients with any suspicious symptoms of COVID-19 [9].
CT images can be utilized as one of the main diagnostic solution for the detection of the COVID-19 disease due to high rate of the false negative of the RT-PCR. In fact, it is well-known among researchers that the low sensitivity of RT-PCR diagnosis cannot be acceptable in extreme situations like the actual outbreak since an unidentified infected person can spread the virus to healthy persons. A team of radiologists is required to analyze the disease and the results can be influenced by individual bias and experience as it is a highly subjective task. Moreover, manual annotation of the infection is time consuming and tedious work as with a large number of patients increasing day by day. Hence, there is a need for some automated prediction techniques to analyze chest CT images with high sensitivity and speed. Since March 2020, publicly available medical images have been used for COVID-19 positive case detection. This enables researchers to study for the automatic identification of disease diagnosis by analyzing patterns in medical images.
In recent times, lot of deep learning schemes dealt with the COVID-19 infection detection via radiographic images [10]. The anomaly detection method was proposed for assisting the radiologist in analyzing a huge number of chest X-rays by Zhang et al. [11]. CT images have been used to separate COVID-19 from other lung diseases with the help of a deep learning model by Li et al. [12]. COVID-19 infection detection, using CT images with a high sensitivity and accuracy, is still a challenging job due to the variation in size, position, and texture of infections.
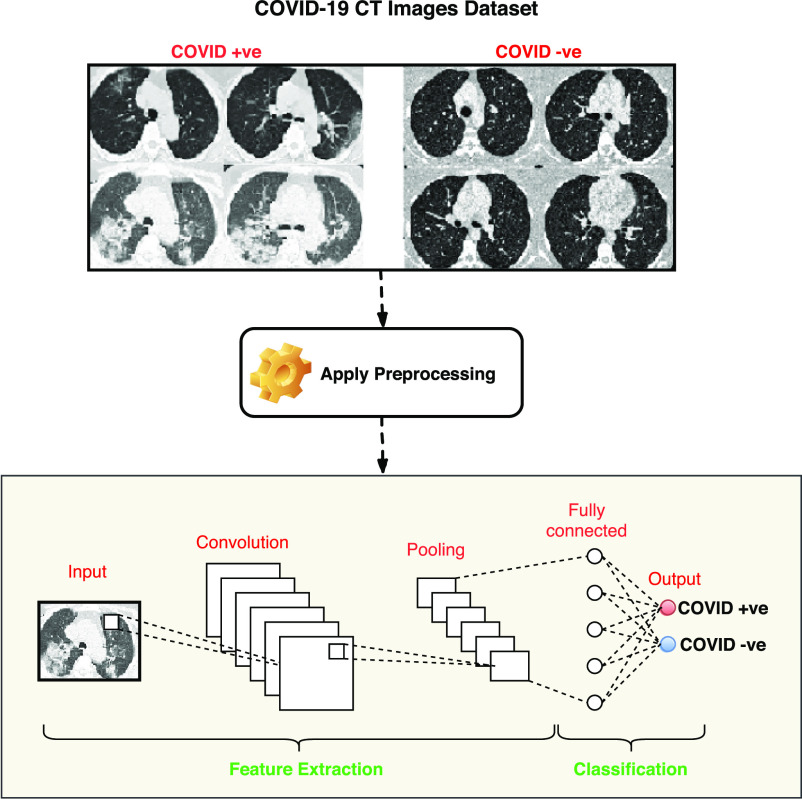
In order to address the aforementioned aspects, an optimized convolutional neural network (CNN) architecture has been designed to help (and automate) the detection of COVID-19 infections by means of pictures from thoracic CT. At first, image preprocessing steps are used for segmentation and accurate edge detection to contrast any normal tissues and infected tissues. For the performance evaluation of the proposed model (that we call ADECO-CNN within the whole manuscript, automatic detection of novel Coronavirus disease from CT images using optimized convolutional neural network) method, an open dataset containing CT images belonging to COVID-19 patients has been used. By considering specificity and sensitivity, the ADECO-CNN novel deep-learning-based model takes advantages of the CNN architecture to classify the preprocessed CT images. A sample sketch diagram of the ADECO-CNN methodology is shown in Fig. 1.
Fig. 1.
ADECO-CNN architecture to detect the Coronavirus disease.
A. Contribution and Organization of This Article
The major improvements introduced by this study are listed as follows.
-
1)
Techniques are introduced to make detection of the COVID-19 infection faster by analysis of CT images.
-
2)
Four steps image normalization approach is formulated to eliminate noise from lung images and to improve the image quality.
-
3)
ADECO-CNN-optimized CNN model is compared with the reference models (namely GoogleNet, VGG19, and ResNet). Such comparison has been performed with and without applying image normalization technique.
-
4)
ADECO-CNN in combination with image normalization technique classifies patients into COVID
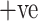 and COVID
and COVID 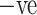 cases with 99.99% accuracy.
cases with 99.99% accuracy. -
5)
To ensure generalizability, the ADECO-CNN approach is validated on another CT images dataset and achieved robust results with 98.2% accuracy.
The rest of this article is organized as follows. Section II discussed the existing literature on COVID-19. Section III presented the adopted dataset. Section IV gives a short introduction of the deep learning models. Section V illustrates some discussions on experimental results. Finally, Section VI concludes this article.
II. Related Work
Recently, image patterns such as chest radiographs and chest CTs are being used for COVID-19 detection [12]. Cohen et al. [13] provided a dataset based on the COVID-19 public image collection of X-rays and CT scans. They extracted more than 125 images from online websites and publications. They further augmented images and used four pretrained CNN models (ResNet18, AlesNet, SqueezNet, and DenseNet201) for the categorization of COVID-19 cases using X-rays. SqueezNet achieved the highest result with 98% accuracy and 96% sensitivity. Bernheim et al. [14] explored the relationship between symptom onset and chest CT images. They found that imaging patterns not only help in understanding the pathophysiology of the infection but also in the prediction of complication development. Convolution-based deep learning schemes have been extensively utilized in studies involving medical images [15].
Internet-of-Things-based architecture for big data analysis was proposed in [16] and evaluation of green supply chain management (GSCM) in [17]. Huang et al. [18] adopted deep deterministic policy gradient to optimize resource allocation. They also proved the effectiveness of the performance aware resource allocation scheme. Chang et al. [19] simulated medical images by the fusion algorithm. They applied computational intelligence for an efficient and effective healthcare system. A deep recurrent neural network with power spectral density features applied on time-series data was proposed in [20]. An intrusion detection method using the functional-link neural network was proposed in [21]. A novel deep learning model CLSTM was proposed in [22] to predict the remaining useful life.
Segmentation facilitates the evaluation of radiological images and used as a preprocessing step to get the region of interest that is a lesion or infected region. Shan et al. [23] performed an automatic segmentation of complete lung and infectious sites using CT images by their proposed model, that is, a three-dimensional convolutional neural model called VB-Net. The dataset used in their experiment is not publically available. They used 249 infected patients for training and 300 new patients for testing. Their proposed model autocontour the infectious part and also estimates the volume, shape, and percentage of infection.
Many researchers have applied different modified transfer learning approaches to facilitate COVID-19 detection purposes. Wang et al. [24] used a modified transfer learning method that relies on a CNN-based model (M-inception) that was previously trained and studied the changes that can be seen within the CT images of patients that are infected by COVID-19. The accuracy achieved using a deep-learning-based prediction method was 89%. They used 453 CT images belonging to patients that were reported to be COVID-19 positive along with viral aggressive pneumonia cases. They achieved 83% classification accuracy by combining the AdaBoost and Decision tree.
Narin et al. [25] adopted a transfer learning scheme for processing X-ray chest images when dealing with the diagnosis of COVID-19 infected patients. The authors adopted three different models: the Inception ResNetV2, ResNet50, and InceptionV3. The last one outperformed with 98% accuracy in their experiment. After an extensive literature review, it is not difficult to realize that, recently, CT-images are more and more being adopted by researchers to get improved results in the classification of COVID-19. Therefore, this study also used CT images in the deep-learning-based computational models.
III. Material
A. COVID-19 Chest CT Images Dataset
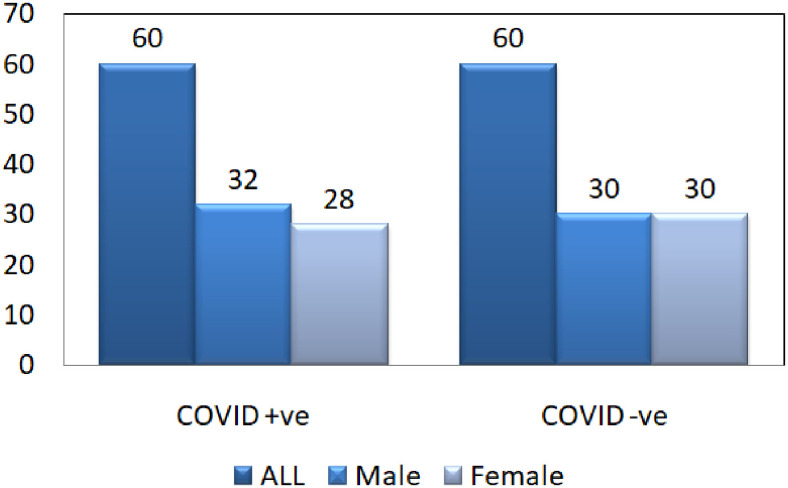
The data source [26] used in the experiment is available at.1 SARS-COV-2 CT-Scan dataset comprises of CT images belonging to several patients. It has 1252 CT images belonging to patients that were positive to SARS-CoV-2 (referred as COVID 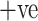 ) and 1230 CT images belonging to patients that were not infected (referred as COVID
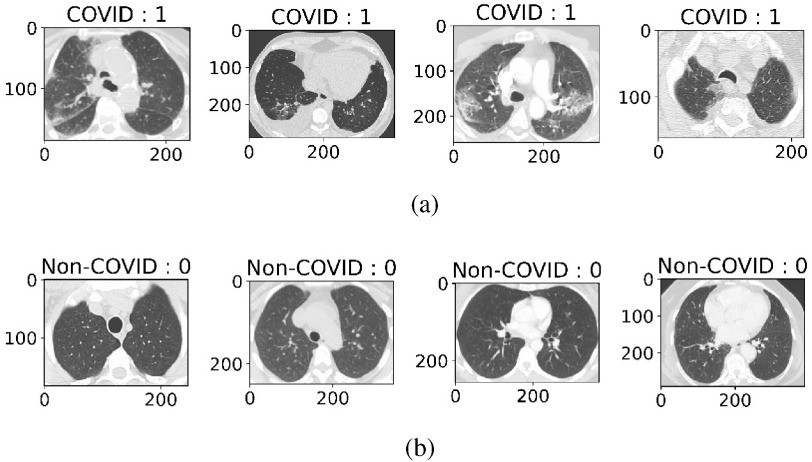
) and 1230 CT images belonging to patients that were not infected (referred as COVID 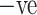 ). The entire dataset contains 2482 CT scans in total. A visual representation of the numbers is presented in Fig. 2, where it can be seen how many men and women are present in the dataset. COVID
). The entire dataset contains 2482 CT scans in total. A visual representation of the numbers is presented in Fig. 2, where it can be seen how many men and women are present in the dataset. COVID 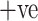 cases having infectious patches in the lungs are shown in Fig. 3(a) and COVID
cases having infectious patches in the lungs are shown in Fig. 3(a) and COVID 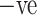 cases having no infection are shown in Fig. 3(b). The aforementioned dataset was composed of images taken from patients in hospitals in the Brazilian metropolitan area of Sao Paulo. The main idea behind this dataset was to foster researchers in setting up and tuning appropriate artificial intelligence algorithms aimed at discerning whether an individual has been infected by COVID-19 or not just by looking at the CT images coming from them.
cases having no infection are shown in Fig. 3(b). The aforementioned dataset was composed of images taken from patients in hospitals in the Brazilian metropolitan area of Sao Paulo. The main idea behind this dataset was to foster researchers in setting up and tuning appropriate artificial intelligence algorithms aimed at discerning whether an individual has been infected by COVID-19 or not just by looking at the CT images coming from them.
Fig. 2.
Number of patients in dataset. Among 60 COVID 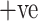 patients, 32 are male and 28 are female patients, and in COVID
patients, 32 are male and 28 are female patients, and in COVID 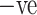 patients, 30 are male and 30 are female patients.
patients, 30 are male and 30 are female patients.
Fig. 3.
CT images of COVID 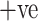 and COVID
and COVID 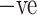 cases. (a) COVID +ve Images. (b) COVID
cases. (a) COVID +ve Images. (b) COVID 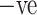 Images.
Images.
IV. Methods
This section discusses the preprocessing steps used in the experiments and proposes an optimized CNN model to classify CT chest images with the aim of identifying infected patients. The introduced model shows robust results when classifying COVID-19 infected patients. Then, the ADECO-CNN values are compared with transfer learning models namely VGG19, GoogleNet, and ResNet.
A. CT Images Preprocessing
Since the features directly derived from CT images showed different intensities and gray scales, it is necessary to apply a data preprocessing prior that such images were given as input to the classifier. Data preprocessing strategies involve standardization and normalization. Features in normalization are calculated as
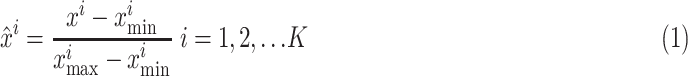 |
where 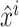 are normalized features extracted from feature
are normalized features extracted from feature 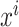 ,
, 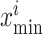 are minimum feature values, and
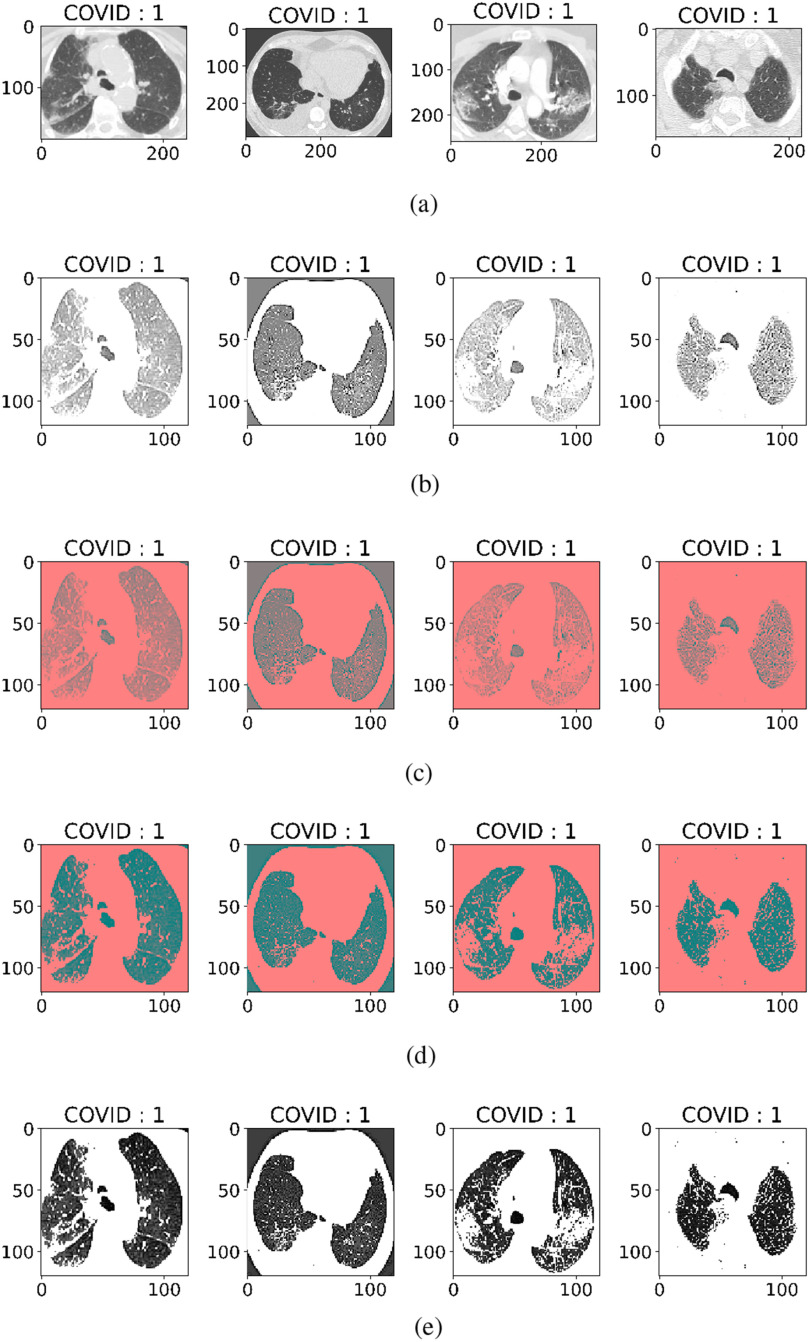
are minimum feature values, and 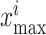 are maximum feature values. In this article, all images are preprocessed to extract lesion regions properly by removing noise in the CT images. Preprocessing involves the following four steps.
are maximum feature values. In this article, all images are preprocessed to extract lesion regions properly by removing noise in the CT images. Preprocessing involves the following four steps.
-
1)
All the CT images were of different intensity and sizes as shown in Fig. 4(a). In the first step, all images have been converted into a fixed size by reducing the dimensions to
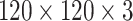 before training as shown in Fig. 4(b).
before training as shown in Fig. 4(b). -
2)
Second, a filter with values ([0,
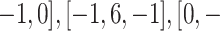 1,0]) is applied for edge detection. Images with edges can be seen in Fig. 4(c).
1,0]) is applied for edge detection. Images with edges can be seen in Fig. 4(c). -
3)
Then, at the third step, values of the luma component is extracted by converting the RGB image to YUV. Luminance is more important than color, so the resolution of U (blue projection) and V (red projection) is reduced but Y is kept at full resolution as shown in Fig. 4(d).
-
4)
Then, at the last step, the intensity values are equalized by converting the YUV image back to RGB by smoothing edges and histogram equalization as shown in Fig. 4(e).
Fig. 4.
Four steps of preprocessing. (a) CT scan images without preprocessing. (b) Detection of edges. (c) BGR-image to YUV-image conversion. (d) Equalization of image intensity. (e) YUV-image to BGR-image conversion.
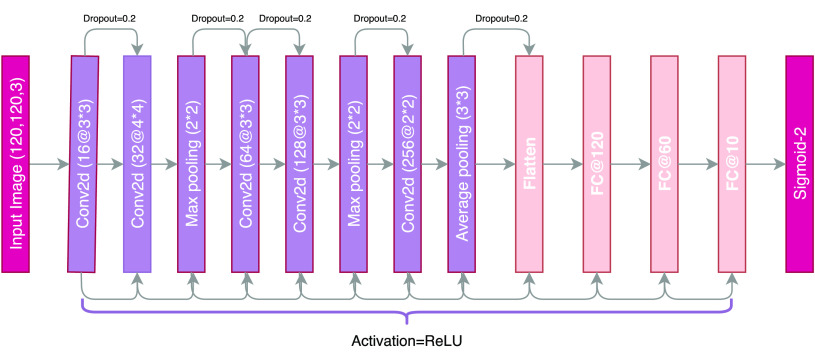
B. ADECO-CNN-Optimized CNN Model
The CNN has been widely extensively adopted in several tasks of image classification and is considered a very powerful tool. It has attracted considerable attention in a variety of domains such as image recognition, image analysis, object detection, and computer vision tasks. The CNN extracts features from image efficiently and its hierarchical structure makes it dynamic in dealing with images. Such layers are logically arranged in three dimensions: depth, width, and height. In such a representation, neurons of a layer are attached to the neurons of the next layer in a limited way. Finally, in the output layer, this number diminishes to score with a single vector probability. Extracted features from preprocessed CT scan images are used to correct the classification of COVID  and COVID
and COVID  cases. The architectural design of the ADECO-CNN is illustrated in Fig. 5. Predominantly this model involves the following steps.
cases. The architectural design of the ADECO-CNN is illustrated in Fig. 5. Predominantly this model involves the following steps.
Fig. 5.
Architectural diagram of the ADECO-CNN model.
1). Feature Extraction
In the first step, the CNN applies many convolutions and pooling layers to extract potential features. The spatial size of the extracted features is minimized by applying the max pooling layer. The pooling layer also helps in reducing the overfitting issue. Max pooling layer considers maximum values, while average pooling considers average values from the feature map obtained after the convolutional process. Stride refers to spacing in image pixels used with pooling. After each convolution step, a new “convolved” image is produced containing features extracted from the previous step. Let an image be represented as 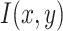 , and
, and 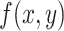 represents the filter that is used, then the resulting transformation is
represents the filter that is used, then the resulting transformation is
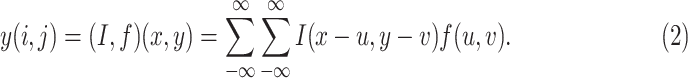 |
Here, the rectified linear unit (ReLu) is adopted as an activation function that is adopted as a complex feature mapping connecting output and input of each layer. The ReLu is a linear function providing positive input values directly or returning a zero value.
2). Classification
The flatten layer converts the data into one-dimensional array by creating a single long feature vector and feeding it to the dense fully connected layer. Dense layers carry out a classification by utilizing extracted features of an image obtained from convolutional layers. Usually, the dropout layer with the help of the activation function reduces the feature map and minimizes overfitting. Different weights are associated with fully connected layers in every row and need computational capabilities accordingly. Fig. 5 represents the complete example of our ADECO-CNN model. In the last layer of dense layers, sigmoid is used to predict the final output. The standard sigmoid function is
 |
C. Reference CNN-Models for Transfer Learning
In this section, CNN models for transfer learning used in experiments are illustrated, i.e., VGG19 [27], GoogleNET [28], and ResNet [29]. The pretrained CNN has the capability for transfer learning and uses training and testing dataset as input. Table I presents the details of the tuned parameters for transfer learning. The parameters are tuned after several experiments even though possible choices or alternatives have no limit and could be further explored to improve performances. The parameter referring to the number of untrained layers is called Layer Cutoff. Layers closer to the output are made more trainable for extracting the features coming from the convolutional layers. It is important to note that the initial layers focus on not specific features, while later ones capture specific ones. Some common hyperparameters are shared by all the CNN models such as ReLU for activation, Dropout Layer to prevent overfitting, and Adam optimizer for optimization. The general architecture of the networks is different among transfer learning models.
TABLE I. CNN Models Used in Experiment and Hyperparameter Values for Transfer Learning.
| Network | Parameter | Description |
|---|---|---|
| VGG19 | Layer Cutoff | 18 |
| Neural Network | 1024 nodes | |
| GoogleNet | Layer Cutoff | 249 |
| Neural Network | 1000 nodes | |
| ResNet v2 | Layer Cutoff | 730 |
| Neural Network | No |
1). VGG19
The VGG network architecture with 19 layers (VGG19) was the base of ImageNet challenge submission in 2014. VGG19 consists of five blocks of convolutional layers and three fully connected layers. Filter size 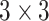 with stride 1 and padding 1 is used in convolution. The ReLU activation function is used after each convolution and spatial dimensions are reduced by max-pooling with
with stride 1 and padding 1 is used in convolution. The ReLU activation function is used after each convolution and spatial dimensions are reduced by max-pooling with 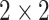 filter and stride 2 and no padding.
filter and stride 2 and no padding.
2). Googlenet
GoogleNet performs convolutions of different filter sizes by inception modules [30]. GoogleNet outperformed and won the ImageNet Large Scale Visual Recognition Competition (ILSVRC)2 in 2014. It has 22 layers and uses four million parameters. Layers go deeper with parallel different field sizes and achieved a 6.67% error rate. All the convolutions even inside of the inception module use the ReLu activation function and a 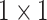 filter is used before
filter is used before 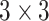 and
and 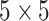 convolutions.
convolutions.
3). Resnet
ResNet, a winner of the ILSVRC in 2015, is a short form of residual network. Very deep models are being used for visual recognition tasks to improve the classification accuracy of complex tasks. As the network gets deeper, the training process becomes difficult and accuracy also starts to degrade [31]. Residual learning was introduced to solve this problem by using the concept of skip connections. Generally, in a convolutional-based deep neural network, layers are arranged for training and network learn features at the end part of these layers. The residual-based network contains a residual connection that jumps over two or more layers of a network. ResNet uses shortcut connections for this purpose by connecting the 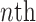 layer to the
layer to the 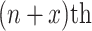 layer [32]. The problem of the degradation of accuracy in a deeper convolutional network is resolved by 34 layered ResNet and it is easy for training.
layer [32]. The problem of the degradation of accuracy in a deeper convolutional network is resolved by 34 layered ResNet and it is easy for training.
V. Experimental Results
Extensive experiments have been performed to evaluate the ADECO-CNN model on the CT images dataset. The dataset is divided into a 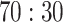 ratio randomly for the training set and test set, respectively. The hyperparameter summary of the ADECO-CNN model is presented in Table II. Furthermore, fivefold cross validation has been conducted to avoid overfitting. Experiments have been conducted on the ADECO-CNN model and other transfer learning models through original images and normalized images. The effect of preprocessing steps after normalizing images in the diagnosis of Coronavirus disease detection is presented in Table III. The performance of each model is very low using original images of a dataset, the main reason may be the large scale and intensity level difference between the images. Fortunately, all the models showed significant improvements to what concerns the specificity, sensitivity, and accuracy, after following the preprocessing image normalization steps.
ratio randomly for the training set and test set, respectively. The hyperparameter summary of the ADECO-CNN model is presented in Table II. Furthermore, fivefold cross validation has been conducted to avoid overfitting. Experiments have been conducted on the ADECO-CNN model and other transfer learning models through original images and normalized images. The effect of preprocessing steps after normalizing images in the diagnosis of Coronavirus disease detection is presented in Table III. The performance of each model is very low using original images of a dataset, the main reason may be the large scale and intensity level difference between the images. Fortunately, all the models showed significant improvements to what concerns the specificity, sensitivity, and accuracy, after following the preprocessing image normalization steps.
TABLE II. Summary of the Hyperparameter Values of the ADECO-CNN Model.
| Hyperparameter | Value |
|---|---|
| Input dimension | (120, 120, 3) |
| Epochs | 13 |
| Batch size | 32 |
| Pooling | 2*2 |
| Function | Binary cross entropy |
| Optimizer | Adam |
TABLE III. Evaluation of Preprocessing on ADECO-CNN and CNN-Based Transfer Learning Models.
| Method | Measure | Original | Normalized |
|---|---|---|---|
| VGG19 | ACC | 73.14  0.88 0.88 |
81.07 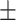 0.21 0.21 |
| SEN | 66.13 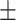 0.98 0.98 |
82.15 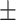 0.04 0.04 |
|
| PRE | 86.07 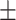 0.98 0.98 |
94.85 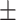 0.04 0.04 |
|
| SPE | 69.35 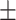 0.05 0.05 |
84.29 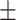 0.38 0.38 |
|
| GoogleNet | ACC | 79.24 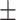 0.73 0.73 |
84.24 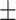 0.21 0.21 |
| SEN | 66.15 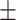 0.31 0.31 |
87.40 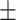 0.75 0.75 |
|
| PRE | 67.13 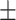 0.31 0.31 |
79.10 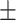 0.75 0.75 |
|
| SPE | 71.24 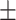 0.98 0.98 |
89.11 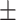 0.14 0.14 |
|
| ResNet | ACC | 81.88 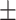 0.24 0.24 |
91.02 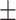 0.03 0.03 |
| SEN | 82.22 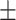 0.05 0.05 |
89.10 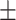 0.15 0.15 |
|
| PRE | 88.44 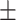 0.11 0.11 |
95.90 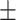 0.01 0.01 |
|
| SPE | 86.77 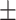 0.92 0.92 |
96.50 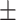 0.12 0.12 |
|
| ADECO-CNN | ACC | 67.17 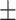 0.87 0.87 |
99.99 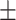 0.01 0.01 |
| SEN | 81.13 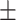 0.52 0.52 |
99.96 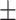 0.04 0.04 |
|
| PRE | 82.13 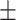 0.52 0.52 |
99.92 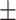 0.08 0.08 |
|
| SPE | 79.24 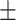 0.11 0.11 |
99.97 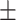 0.03 0.03 |
ACC = Accuracy, SEN = Sensitivity, PRE = Precision, and SPE = Specificity.
A. Performance Evaluation
The ADECO-CNN scheme is trained and run with 2482 CT images in which 1252 images of COVID-19 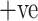 and 1230 images of COVID-19
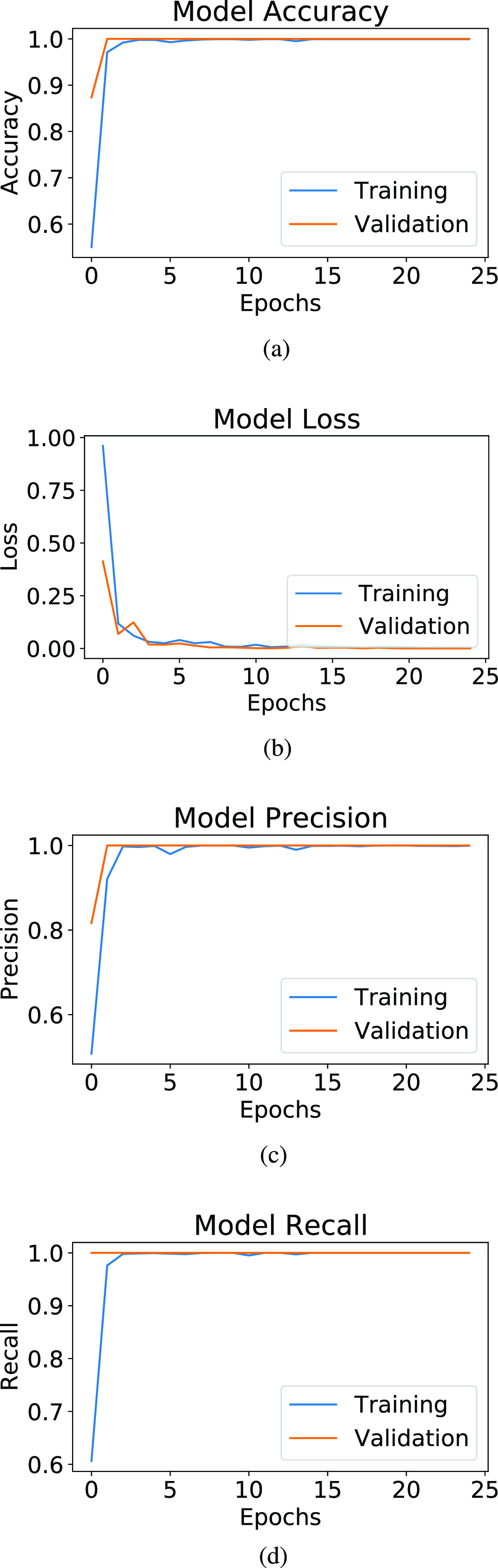
and 1230 images of COVID-19 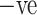 cases. It takes 1.5 h to execute 12 epochs over the CT images to be used in the binary classification. Fig. 6(a) represents the training and validation accuracy curve, while Fig. 6(b) represents the training and validation loss curve, and Fig. 6(c) represents the training and validation precision curve, while Fig. 6(d) represents the training and validation recall curve.
cases. It takes 1.5 h to execute 12 epochs over the CT images to be used in the binary classification. Fig. 6(a) represents the training and validation accuracy curve, while Fig. 6(b) represents the training and validation loss curve, and Fig. 6(c) represents the training and validation precision curve, while Fig. 6(d) represents the training and validation recall curve.
Fig. 6.
ADECO-CNN model experimental evaluation. (a) Accuracy. (b) Loss. (c) Precision. (d) Recall (sensitivity).
Table IV presents the results of the ADECO-CNN model and it proved that the optimized CNN outperformed with binary classification tasks. The result of the ADECO-CNN model indicate that it can discriminate COVID-19 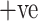 cases from COVID-19
cases from COVID-19 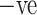 cases with 99.99% accuracy, which is the indispensable requirement of current pandemic disease. The adopted image normalization technique at the preprocessing step helps the ADECO-CNN model to achieve such good results. Edge detection and high contrast images make them simple for the training process of the model. The results of the ADECO-CNN model are equally good for other metrics.
cases with 99.99% accuracy, which is the indispensable requirement of current pandemic disease. The adopted image normalization technique at the preprocessing step helps the ADECO-CNN model to achieve such good results. Edge detection and high contrast images make them simple for the training process of the model. The results of the ADECO-CNN model are equally good for other metrics.
TABLE IV. Performance Comparison Between CNN-Based Transfer Learning Models.
| Network | Accuracy | Sensitivity | Precision | Specificity |
|---|---|---|---|---|
| VGG19 | 81.07 | 82.15 | 94.85 | 84.29 |
| GoogleNet | 84.24 | 87.40 | 79.10 | 89.11 |
| ResNET | 91.02 | 89.10 | 95.90 | 96.50 |
| ADECO-CNN | 99.99 | 99.96 | 99.92 | 99.97 |
B. Comparison With Different Models
The deep-learning-based transfer learning frameworks used during the comparison are ResNet, GoogleMet, and VGG19. All the state-of-the-art models performed well in the image recognition and classification tasks. Table IV shows that the ADECO-CNN model is more efficient for accuracy, sensitivity, precision, and specificity compared to the other models. Another very important aspect is that the ADECO-CNN model does not have false positive/false negative outcomes, so it can efficiently classify COVID-19 patients with 99.99% accuracy.
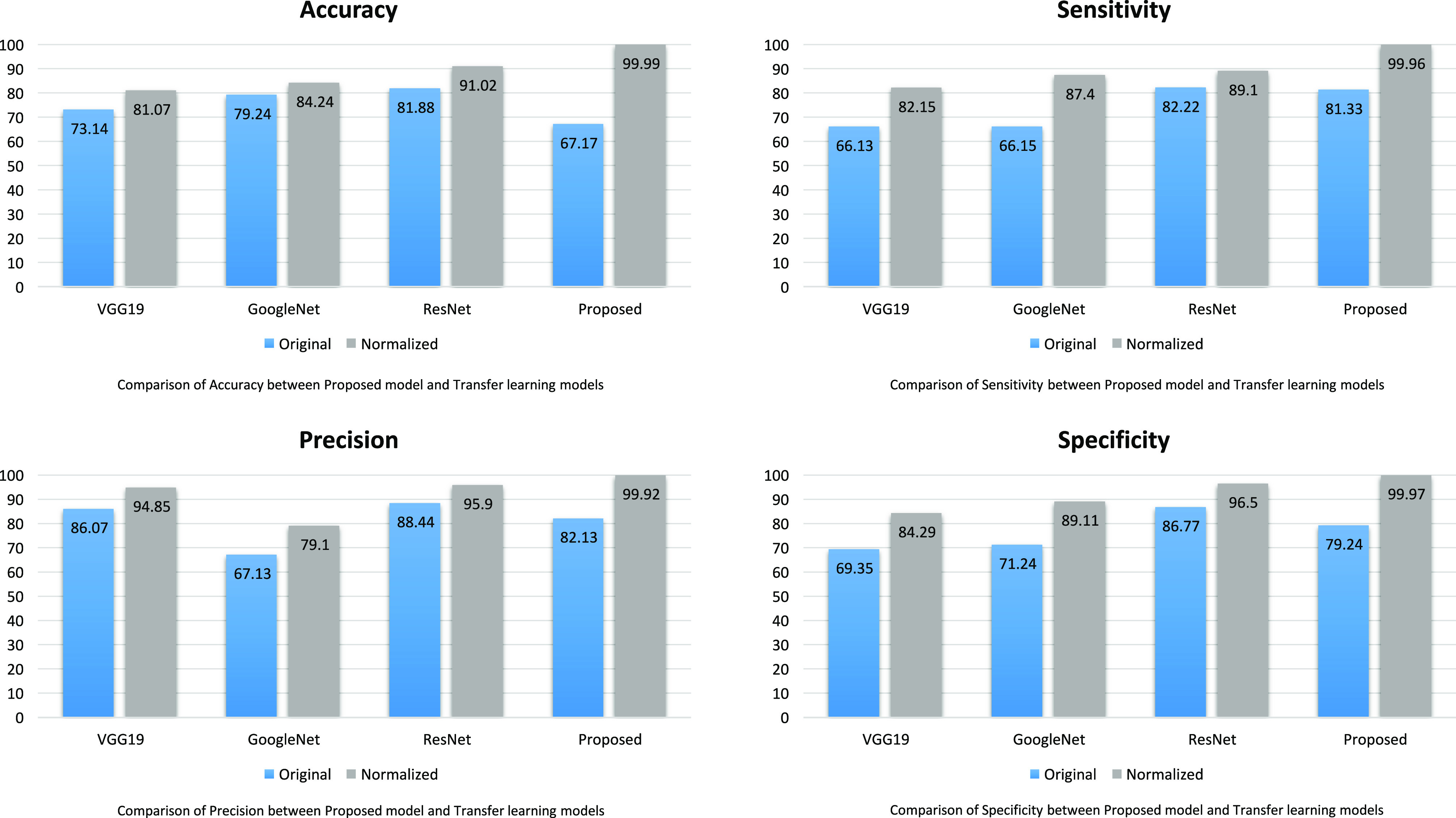
Accuracy is the primary measure to evaluate the performance of the classification model and is calculated by accurately classified classes by total classes. Fig. 7(a) shows the accuracy comparison between the model introduced in this article and other competitive transfer learning ones. It is not difficult to note that our proposed model substantially achieves a higher accuracy as compared to transfer learning models. The Accuracy of VGG19, GoogleNet, ResNet, and ADECO-CNN is 81.07%, 84.24%, 91.02%, and 99.99%, respectively.
Fig. 7.
Result comparison between ADECO-CNN and transfer learning models.
Sensitivity deals with the performance of discovering COVID-19 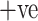 cases only. It is a very significant measure due to the pandemic nature of the disease and the main objective is to control its spread. This metric identifies every patient who is suffering from COVID-19. Fig. 7(b) illustrates the sensitivity analysis of all the models and it shows clearly that the ADECO-CNN model achieved the best sensitivity for COVID-19
cases only. It is a very significant measure due to the pandemic nature of the disease and the main objective is to control its spread. This metric identifies every patient who is suffering from COVID-19. Fig. 7(b) illustrates the sensitivity analysis of all the models and it shows clearly that the ADECO-CNN model achieved the best sensitivity for COVID-19 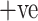 cases than any other transfer learning model. The sensitivity value of the ADECO-CNN approach is 99.96%. However, the sensitivity value of the existing approaches namely VGG19 is 82.15%, GoogleNet is 87.4%, and ResNet is 89.1%. Hence, the ADECO-CNN outperforms the existing works.
cases than any other transfer learning model. The sensitivity value of the ADECO-CNN approach is 99.96%. However, the sensitivity value of the existing approaches namely VGG19 is 82.15%, GoogleNet is 87.4%, and ResNet is 89.1%. Hence, the ADECO-CNN outperforms the existing works.
Precision evaluates the performance of COVID 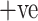 cases overall predicted positive cases. Fig. 7(c) analyzes the precision comparison of the ADECO-CNN model with the other competitive models and reveals that the ADECO-CNN achieved a significantly higher precision value than all the other models. The precision value of the ADECO-CNN model is 99.92%. However, the precision value of other transfer learning models namely VGG19 is 94.85%, GoogleNet is 79.1%, and ResNet is 95.9%. The ADECO-CNN approach outperforms the other transfer learning models in terms of precision.
cases overall predicted positive cases. Fig. 7(c) analyzes the precision comparison of the ADECO-CNN model with the other competitive models and reveals that the ADECO-CNN achieved a significantly higher precision value than all the other models. The precision value of the ADECO-CNN model is 99.92%. However, the precision value of other transfer learning models namely VGG19 is 94.85%, GoogleNet is 79.1%, and ResNet is 95.9%. The ADECO-CNN approach outperforms the other transfer learning models in terms of precision.
Specificity refers to COVID-19 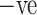 cases only. It identifies a normal and healthy patient who is not infected with COVID-19. Fig. 7(d) analyzes the specificity comparison of the ADECO-CNN model with the other competitive models and reveals that the ADECO-CNN achieved a significantly higher specificity value than all the other models. The specificity value of the ADECO-CNN model is 99.97%. The specificity value of transfer learning models, namely, VGG19 is 84.29%, GoogleNet is 89.11%, and ResNet is 96.5%.
cases only. It identifies a normal and healthy patient who is not infected with COVID-19. Fig. 7(d) analyzes the specificity comparison of the ADECO-CNN model with the other competitive models and reveals that the ADECO-CNN achieved a significantly higher specificity value than all the other models. The specificity value of the ADECO-CNN model is 99.97%. The specificity value of transfer learning models, namely, VGG19 is 84.29%, GoogleNet is 89.11%, and ResNet is 96.5%.
C. Importance of Image Normalization
Image normalization at the preprocessing step aims to reduce noise in CT images to improve the training of the ADECO-CNN model. It involves five extensive steps to normalize images. After fixing the image size to 120 × 120 × 3, edges are detected by filters ([0,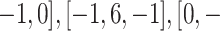 1,0]). Then, the RGB pictures are transformed into their corresponding version having their luma component, red projection, and blue projection (YUV) to keep luminance Y at full resolution and the resolution of channels U and V is reduced. At the last step, histogram normalization is performed to smooth the edges. The performance of the ADECO-CNN model has also been evaluated without applying any image normalization. It is evident from Table III that image normalization remarkably improved the performance of the ADECO-CNN approach and transfer learning models. Therefore, it is a very important step in the training process of the model.
1,0]). Then, the RGB pictures are transformed into their corresponding version having their luma component, red projection, and blue projection (YUV) to keep luminance Y at full resolution and the resolution of channels U and V is reduced. At the last step, histogram normalization is performed to smooth the edges. The performance of the ADECO-CNN model has also been evaluated without applying any image normalization. It is evident from Table III that image normalization remarkably improved the performance of the ADECO-CNN approach and transfer learning models. Therefore, it is a very important step in the training process of the model.
D. Validation of ADECO-CNN Model
To ensure the robustness and generalizability of the ADECO-CNN approach (optimized CNN with normalized images), it is validated using the dataset that can be found here [33], containing 348 COVID-19 and 396 non COVID-19 images. Table V presents the outcome of the ADECO-CNN on COVID19 CT dataset. It is proved that the ADECO-CNN shows robust results and can classify COVID-19 cases with 98.2% accuracy, 95.7% sensitivity, 97.9% precision, and 96.8% specificity.
TABLE V. Validation of the ADECO-CNN Approach on COVID19-CT Dataset.
| Dataset | Accuracy | Sensitivity | Precision | Specificity |
|---|---|---|---|---|
| COVID19-CT | 98.2 | 95.7 | 97.9 | 96.8 |
The accuracy of the ADECO-CNN on dataset 1 is also compared with other state-of-the-art models in literature. The results are presented in Table VI and proved that the ADECO-CNN has a robust result with 99.99% accuracy.
TABLE VI. Performance Evaluation With State-of-the-Art Models Using CT Images for COVID-19 Detection in Literature.
E. Stability of the ADECO-CNN Model
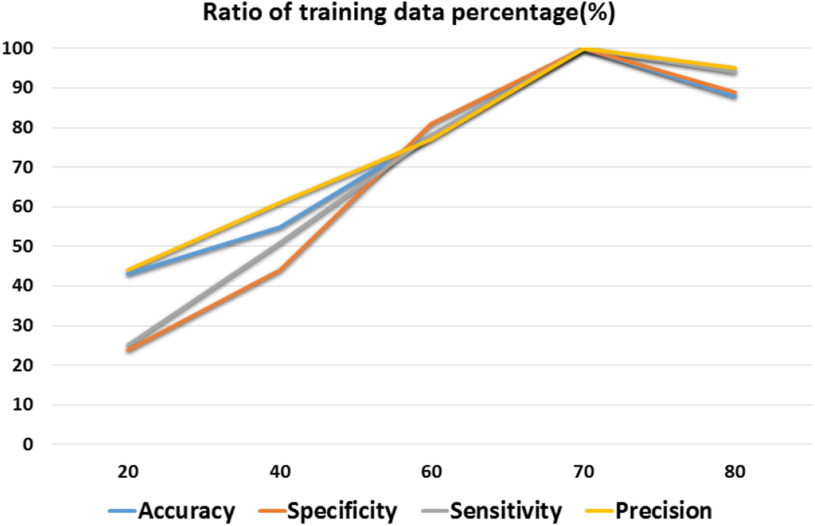
To verify the stability of the ADECO-CNN approach, it is tested under different data ratios of the training set. Testing size is fixed in each experiment. Fluctuation in the results with a change of the training data ratio from 20% to 80% is presented in Fig. 8. It can be observed from Fig. 8 that performance improves with the increase in the size of the training data. Afterwards, 40% stability can be observed. The model achieves the best result when 70% of the data are used in the training. Promising results and stable performance proved how the proposed solution is able to identify COVID-19  cases using a high stability and accuracy.
cases using a high stability and accuracy.
Fig. 8.
Stability of the ADECO-CNN approach with different training data ratio.
VI. Conclusion
This article proposed a novel classification model that can be very useful in predicting the COVID-19 disease by just analyzing chest CT images. Initially chest CT images from the COVID-19 dataset were normalized upon having applied four preprocessing phases. After that, the dataset of CT-images was split into two different parts: a test set and a training one. The training set was then utilized to classify COVID-19 infected patients and to build a model. Furthermore, fivefold cross validation was conducted to avoid overfitting and ensure generalizability.
To further validate the authors’ proposal, the ADECO-CNN approach was related to the existing transfer learning models namely VGG19, GoogleNet, and ResNet. Those models were considered the state of the art by the scientific community. Extensive experiments proved that the ADECO-CNN optimized CNN outperforms the other models in accuracy, sensitivity, precision, and specificity. Therefore, the ADECO-CNN approach is the best among any other COVID-19 disease prediction method and can be implemented in real-time disease classification from chest CT images at any place to control the early outbreak of the disease.
Moreover, there is the need to develop models capable of differentiating COVID-19 cases by additional similar diseases like pneumonia. Besides this medical imaging, other risk factors for the onset of the disease should be considered for a more holistic approach. Nonetheless, the ADECO-CNN work contributes to a low cost, rapid automatic early diagnosis of positive cases even before the appearance of any clear symptoms. Even though the deadly virus has no treatment yet, initial screening will not only help in the treatment, but also in taking timely action for quarantine of COVID-19  cases. This method of diagnosis of COVID
cases. This method of diagnosis of COVID  cases from medical images will also save the risk of exposure of medical staff and nurses to the outbreak.
cases from medical images will also save the risk of exposure of medical staff and nurses to the outbreak.
Acknowledgment
The authors declare no conflict of interest. The funding agency had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Footnotes
[Online]. Available: https://www.kaggle.com/plameneduardo/sarscov2-ctscan-dataset.
[Online]. Available: http://www.image-net.org/challenges/LSVRC/.
Contributor Information
Aniello Castiglione, Email: castiglione@ieee.org.
Pandi Vijayakumar, Email: vijibond2000@gmail.com.
Michele Nappi, Email: mnappi@unisa.it.
Saima Sadiq, Email: s.kamrran@gmail.com.
Muhammad Umer, Email: umersabir1996@gmail.com.
References
- [1].Song F. et al. , “Emerging 2019 novel coronavirus (2019-nCoV) pneumonia,” Radiology, vol. 295, no. 1, pp. 210–217, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Huang C. et al. , “Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China,” Lancet, vol. 395, no. 10223, pp. 497–506, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Yu F., Du L., Ojcius D. M., Pan C., and Jiang S., “Measures for diagnosing and treating infections by a novel coronavirus responsible for a pneumonia outbreak originating in Wuhan, China,” Microbes Infection, vol. 22, no. 2, pp. 74–79, Mar. 2020, doi: 10.1016/j.micinf.2020.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Qian M., Yi Q., Qihua F., and Ming G., “Understanding the influencing factors of nucleic acid detection of 2019 novel coronavirus,” Chin J. Lab Med., vol. 10, pp. 1–7, 2020. [Google Scholar]
- [5].Abdel-Basset M., Chang V., and Mohamed R., “HSMA_WOA: A hybrid novel slime mould algorithm with whale optimization algorithm for tackling the image segmentation problem of chest x-ray images,” Appl. Soft Comput., vol. 95, 2020, Art. no. 106642. [Online]. Available: http://www.sciencedirect.com/science/article/pii/S1568494620305809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Phan L. T. et al. , “Importation and human-to-human transmission of a novel coronavirus in vietnam,” New England J. Med., vol. 382, no. 9, pp. 872–874, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Jin Y.-H. et al. , “A rapid advice guideline for the diagnosis and treatment of 2019 novel coronavirus (2019-nCoV) infected pneumonia,” Mil. Med. Res., vol. 7, no. 1, pp. 1–23, 2020, doi: 10.1186/s40779-020-0233-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Fang Y. et al. , “Sensitivity of chest CT for COVID-19: Comparison to RT-PCR,” Radiology, vol. 296, no. 2, pp. E115–E117, 2020, Art. no. 200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Xie X., Zhong Z., Zhao W., Zheng C., Wang F., and Liu J., “Chest CT for typical 2019-nCoV pneumonia: Relationship to negative RT-PCR testing,” Radiology, vol. 296, no. 2, pp. E41–E45, 2020, Art. no. 200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Abdel-Basset M., Chang V., Hawash H., Chakrabortty R. K., and Ryan M., “FSS-2019-nCov: A deep learning architecture for semi-supervised few-shot segmentation of COVID-19 infection,” Knowl.-Based Syst., vol. 212, 2021, Art. no. 106647. [Online]. Available: http://www.sciencedirect.com/science/article/pii/S0950705120307760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zhang J., Xie Y., Li Y., Shen C., and Xia Y., “COVID-19 screening on chest x-ray images using deep learning based anomaly detection,” IEEE Trans. Med. Imag., vol. 40, no. 3, pp. 879–890, Mar. 2021, doi: 10.1109/TMI.2020.3040950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Li L. et al. , “Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT,” Radiology, vol. 296, no. 2, pp. E65–E71, 2020, Art. no. 200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Cohen J. P., Morrison P., and Dao L., “COVID-19 image data collection,” 2020, arXiv:2003.11597.
- [14].Bernheim A. et al. , “Chest CT findings in coronavirus disease-19 (COVID-19): Relationship to duration of infection,” Radiology, vol. 295, no. 3, pp. 685–691, 2020, Art. no. 200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Umer M., Sadiq S., Ahmad M., Ullah S., Choi G. S., and Mehmood A., “A novel stacked CNN for malarial parasite detection in thin blood smear images,” IEEE Access, vol. 8, pp. 93 782–93 792, 2020. [Google Scholar]
- [16].Moreno M. V. et al. , “Applicability of big data techniques to smart cities deployments,” IEEE Trans. Ind. Informat., vol. 13, no. 2, pp. 800–809, Apr. 2017. [Google Scholar]
- [17].Abdel-Baset M., Chang V., and Gamal A., “Evaluation of the green supply chain management practices: A novel neutrosophic approach,” Comput. Ind., vol. 108, pp. 210–220, 2019. [Google Scholar]
- [18].Huang B. et al. , “Deep reinforcement learning for performance-aware adaptive resource allocation in mobile edge computing,” Wireless Commun. Mobile Comput., vol. 2020, pp. 1–17, 2020, Art. no. 2765491. [Google Scholar]
- [19].Chang V., “Computational intelligence for medical imaging simulations,” J. Med. Syst., vol. 42, no. 1, pp. 1–12, 2018, doi: 10.1007/s10916-017-0861-x. [DOI] [PubMed] [Google Scholar]
- [20].Li X., Wang Y., Zhang B., and Ma J., “PSDRNN: An efficient and effective HAR scheme based on feature extraction and deep learning,” IEEE Trans. Ind. Informat., vol. 16, no. 10, pp. 6703–6713, Oct. 2020. [Google Scholar]
- [21].Naik B., Obaidat M. S., Nayak J., Pelusi D., Vijayakumar P., and Islam S. H., “Intelligent secure ecosystem based on metaheuristic and functional link neural network for edge of things,” IEEE Trans. Ind. Informat., vol. 16, no. 3, pp. 1947–1956, Mar. 2020. [Google Scholar]
- [22].Ma M. and Mao Z., “Deep convolution-based LSTM network for remaining useful life prediction,” IEEE Trans. Ind. Informat., vol. 17, no. 3, pp. 1658–1667, Mar. 2021. [Google Scholar]
- [23].Shan F. et al. , “Abnormal lung quantification in chest CT images of COVID-19 patients with deep learning and its application to severity prediction,” Med. Phys., 2020, doi: 10.1002/mp.14609. [DOI] [PMC free article] [PubMed]
- [24].Wang S. et al. , “A deep learning algorithm using CT images to screen for corona virus disease (COVID-19),” MedRxiv, pp. 1–17, 2020, doi: 10.1007/s00330-021-07715-1. [DOI] [PMC free article] [PubMed]
- [25].Narin A., Kaya C., and Pamuk Z., “Automatic detection of coronavirus disease (COVID-19) using x-ray images and deep convolutional neural networks,” 2020, arXiv:2003.10849. [DOI] [PMC free article] [PubMed]
- [26].Soares E., Angelov P., Biaso S., Froes M. H., and Abe D. K., “SARS-CoV-2 CT-Scan dataset: A large dataset of real patients CT scans for SARS-CoV-2 identification,” Medrxiv, pp. 1–8, 2020, doi: 10.1101/2020.04.24.20078584. [DOI] [Google Scholar]
- [27].Simonyan K. and Zisserman A., “Very deep convolutional networks for large-scale image recognition,” in Proc. 3rd Int. Conf. Learn. Representations, 2015. [Online]. Available: https://iclr.cc/archive/www/doku.php%3Fid=iclr2015:main.html [Google Scholar]
- [28].Szegedy C. et al. , “Going deeper with convolutions,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit., 2015, pp. 1–9. [Google Scholar]
- [29].He K., Zhang X., Ren S., and Sun J., “Deep residual learning for image recognition,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit., 2016, pp. 770–778. [Google Scholar]
- [30].Ballester P. and Araujo R. M., “On the performance of GoogLeNet and AlexNet applied to sketches,” in Proc. 30th AAAI Conf. Artif. Intell., 2016, pp. 1124–1128, doi: 10.5555/3015812.3015979. [DOI] [Google Scholar]
- [31].Li L.-J., Su H., Lim Y., and Fei-Fei L., “Objects as attributes for scene classification,” in Proc. Eur. Conf. Comput. Vis., 2010, pp. 57–69. [Google Scholar]
- [32].Krizhevsky A. et al. , “Learning multiple layers of features from tiny images,” Univ. Toronto, Toronto, ON, Canada, Tech. Rep. TR-2009, 2009. [Google Scholar]
- [33].Zhao J., Zhang Y., He X., and Xie P., “COVID-CT-Dataset: A CT scan dataset about COVID-19,” 2020, arXiv:2003.13865.
- [34].Song Y. et al. , “Deep learning enables accurate diagnosis of novel Coronavirus (COVID-19) with CT images,” IEEE/ACM Trans. Comput. Biol. Bioinf., to be published, doi: 10.1109/TCBB.2021.3065361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Barstugan M., Ozkaya U., and Ozturk S., “Coronavirus (COVID-19) classification using CT images by machine learning methods,” 2020, arXiv:2003.09424.