ABSTRACT
The development of safe and effective vaccines against viruses is central to disease control. With advancements in DNA synthesis technology, the production of synthetic viral genomes has fueled many research efforts that aim to generate attenuated viruses by introducing synonymous mutations. Elucidation of the mechanisms underlying virus attenuation through synonymous mutagenesis is revealing interesting new biology that can be exploited for vaccine development. Here, we review recent advancements in this field of synthetic virology and focus on the molecular mechanisms of attenuation by genetic recoding of viruses. We highlight the action of the zinc finger antiviral protein (ZAP) and RNase L, two proteins involved in the inhibition of viruses enriched for CpG and UpA dinucleotides, that are often the products of virus recoding algorithms. Additionally, we discuss current challenges in the field as well as studies that may illuminate how other host functions, such as translation, are potentially involved in the attenuation of recoded viruses.
KEYWORDS: codon, RNA, virus, translation
INTRODUCTION
A core property of nucleic acids is the ability to store and communicate information encoded in the order of constituent nucleotides. Genes are composed of linear arrays of codons that are translated by ribosomes to produce polypeptides: a flow of information that is universal in both cells and viruses. Because a total of 61 codons can be translated into 20 amino acids (plus termination codons), redundancy in the genetic code allows organisms to develop and maintain coding biases. Coding biases occur when certain codons, codon pairs, or nucleotide combinations are enriched or depleted in protein-coding sequences. These biases can influence gene expression by altering either mRNA stability or translation efficiency. Consequently, viruses are subjected to host-imposed pressures on coding sequences, and some viruses appear to have nucleotide compositions that have been selected to optimize replication in particular hosts.
Recent advances in DNA synthesis have impacted research that aims to understand coding biases and how synonymous mutations might affect cellular and viral processes. Disrupting coding biases by large-scale genomic recoding of viruses can lead to substantial reductions in viral fitness. Thus, the recoding and synthesis of entire viral genomes have created a platform for the generation of live, attenuated vaccines, i.e., viruses that carry synonymous mutations and have suboptimal replication cycles both in cell culture and in vivo, as well as a platform to investigate the underlying molecular biology of viruses (1, 2).
The methods and approaches to virus recoding through codon or codon pair deoptimization approaches, manipulation of the dinucleotide content, and other genomic alterations of viral genomes have been extensively reviewed elsewhere (1, 2). Here, we review the biological mechanisms underlying the attenuation of recoded viruses. We highlight the activity of certain antiviral proteins and their links to the nucleotide composition of virus genomes. We also discuss how recoding approaches can impact translation efficiency and promote viral RNA degradation. Understanding the mechanisms underlying attenuation following large-scale genomic recoding of viruses should impact vaccine design and potentially other therapeutic approaches.
CODING BIASES IN CELLULAR AND VIRAL GENOMES
The nucleotide sequence does not solely specify the order of the amino acids to be translated but also can impact translation efficiency (3), mRNA stability (4), and the subcellular localization of an mRNA molecule (5). A prominent example of nucleotide compositional bias is the extreme suppression in human genomes—and, more generally, genomes of vertebrates—of CpG dinucleotides (6). The tendency to avoid CpG dinucleotides is not observed in the majority of invertebrates, some clades of plants, and bacteria, whose DNA genomes exhibit higher CpG dinucleotide frequencies (7). On the other hand, UpA dinucleotides (or TpA in DNA) are underrepresented in virtually all living forms (8). Variation in the degrees to which these dinucleotides are suppressed within genomes is observed: in humans, CpG dinucleotides are suppressed in all genomic DNA, while UpA dinucleotides are mostly suppressed in cytoplasmic RNA sequences (8, 9).
Two hypotheses might be advanced to explain compositional biases. The first posits that dinucleotide and codon biases arise from underlying mutagenic sources and that synonymous mutations arise without affecting the fitness of the gene or the organism (10). An example of such a mutational process is that which gives rise to the CpG-suppressed state of human genomes. In DNA, cytosines spontaneously undergo oxidative deamination at a low rate. Ordinarily, cytosine deamination generates uracil, which is recognized by DNA repair mechanisms and corrected to the original G:C base pair. However, cytosines in a 5′-CpG-3′ context are often substrates of DNA methyltransferases (11). The product of this reaction is a 5-methyl-cytosine whose deamination generates thymine, generating a G:T rather than a G:U mismatch, which is less frequently “correctly” repaired to the original G:C base pair. This process appears to be responsible for the naturally occurring depletion of CpG suppression (12). In fact, the absence of DNA methyltransferases in certain species of invertebrates correlates with the higher CpG frequencies observed in their genomes (13, 14). An alternative hypothesis that might drive compositional bias argues that synonymous mutations influence the fitness of an organism and are therefore selected. Evidence in support of this hypothesis includes the correlation between preferred codons in highly expressed genes and the abundance of the cognate tRNA (15, 16), suggesting that selection for translation efficiency might impact nucleotide composition. In reality, both hypotheses likely contribute to the generation and maintenance of codon or dinucleotide biases in host genes.
Viruses that infect eukaryotes mimic, to some extent, the compositional biases of their hosts (17, 18). For example, viruses that replicate in mammals have lower CpG dinucleotide contents than viruses that replicate in insects (9). Correspondingly, mammalian genomes have fewer CpG dinucleotides than expected, but insect genomes do not. Additionally, viruses that infect plants are profoundly suppressed in UpA dinucleotides, mimicking the low level of TpAs present in plant genomes (19). Indeed, using machine learning approaches and large viral ecological data sets, one study suggested that virus reservoirs can be predicted based on the coding biases of both viral and host RNA sequences (20). Key evidence in support of the notion that viral mimicry of host nucleotide composition is a selected property is provided by a longitudinal study of H1N1 influenza A viral genome sequences (21). H1N1 influenza A virus (IAV) is believed to have been transmitted from birds to humans, giving rise to the infamous 1918 pandemic. Initially, this H1N1 virus had a relatively large number of CpG dinucleotides, but viruses subsequently isolated from humans over the ensuing century have progressively fewer CpGs in their genomes. This observation suggests that host-specific selective pressures drive compositional biases in viral genomes.
APPROACHES TO VIRAL GENETIC RECODING
Generally, the main goal of genetic recoding of viruses is to identify intrinsic coding or compositional biases of viral genomes that facilitate replication and deliberately disrupt such biases. An effectively attenuated virus will ultimately have fitness that is reduced in vivo to the extent that it is apathogenic yet retain sufficient levels of replication to elicit a strong immune response. One of the advantages of using genomic recoding to generate attenuated viruses is the inherent stability of the introduced mutations, as the fitness deficits imposed by each nucleotide substitution are imperceptible, but their cumulative effect can be substantial. Conversely, live attenuated vaccines that carry a small number of amino acid substitutions might easily reacquire wild-type fitness following immunization. A well-known example is the live, attenuated poliovirus vaccine that encodes 3 amino acid substitutions that significantly reduce virus replication. However, reversion occurs in vivo, and neurovirulence is evident following transmission (22). Indeed, most current cases of poliovirus infection involve a reverted vaccine strain and not wild-type virus. Few studies examined the stability of recoded genomes. Several codon pair-deoptimized, temperature-sensitive respiratory syncytial virus (RSV) mutants were serially passaged under consecutively higher-temperature culture conditions; however, no virus adaptation or reversion was observed (23). Nonetheless, one mutant virus that contained a smaller number of mutations regained fitness, possibly by increasing the transcription of the recoded gene without losing any attenuating mutations. Another study showed that long-term passage of a codon-deoptimized poliovirus resulted in the progressive purging of introduced CpG dinucleotides from its genome (24). Thus, while a small number of introduced mutations may be insufficient to maintain virus attenuation, large-scale genetic recoding approaches of viruses are likely to generate stable attenuated viruses that may be used as vaccine candidates.
Modulation of codon and dinucleotide frequencies.
Most previous studies have employed “codon pair deoptimization” as an approach to attenuate viruses. Codon pair deoptimization is achieved by replacing frequently observed codon pairs with infrequently used codon pairs (Fig. 1). This approach has shown to be effective in a wide range of viruses, including picornaviruses (25–27), flaviviruses (28), orthomyxoviruses (29), pneumoviruses (30), herpesviruses (31), arenaviruses (32), and lentiviruses (33). Most of these viruses are human pathogens, and codon pair deoptimization gives various levels of attenuation, as assessed by replication in human cells. Attenuation has been observed both in vitro and in animal model systems (30, 34). In some studies, recoded viruses that are transmitted by mosquitos were shown to be significantly attenuated in human cells but not insect cells (35). This finding suggested that codon pair deoptimization has host-specific effects and is not a universal mechanism of attenuation. Although codon pair deoptimization was initially and reasonably thought to exert its effects through decreased translation efficiency, codon pair deoptimization also perturbs dinucleotide composition. Indeed, codon pair deoptimization results in viruses with elevated levels of CpG and UpA dinucleotides, primarily by enriching these dinucleotides at codon boundaries (Fig. 1). Increasing the frequency of these dinucleotides in viral genomes has been shown to attenuate viruses in hosts as divergent as humans and plants (19, 36–39). Recoding methodologies that disrupt codon usage, not codon pairs, have also been successful in generating attenuated viruses. Such studies aimed to mimic the codon usage of avian influenza viruses in human IAV (40) or to mimic the less frequent codons in humans (41). However, in these cases, the investigators inadvertently increased the frequency of CpG and UpA dinucleotides. Increased levels of CpG/UpA dinucleotides have been found to be deleterious for virus replication in both coding and noncoding regions of RNA molecules (36, 42, 43). Further investigation is required to tease apart the effects of codon and codon pair deoptimization and increases of CpG/UpA dinucleotides on viral replication. A systematic approach that uses identical recoded sequences in either translated or untranslated regions of viruses might be useful to elucidate the direct role of dinucleotides in RNA versus modified codons in a sequence.
FIG 1.
Codon substitution approaches used in genetic recoding of viruses. Several synonymous mutagenesis approaches may be applied to generate attenuated viruses. Codon deoptimization aims to replace highly frequent codons (such as GAG for glutamate and UGC for cysteine) with underrepresented codons (such as GAA and UGU, respectively). Codon substitution may also lead to increased CpG and/or UpA frequencies by introducing these dinucleotides either at the codon-codon boundary (e.g., replacing the GCA-GAG pair with GCG-GAG) or within a codon (e.g., AGA-to-CGU substitution). Replacing serine and leucine codons with “near-stop” codons (i.e., AGU and CUU to UCA and UUA) may also lead to viruses whose replication is aborted at unusually high frequencies through the frequent generation of mutants expressing truncated viral proteins.
Evolutionary space entrapment.
A consequence of genetically recoding viral genomes is the reframing of the “evolutionary space.” During the propagation of viral populations, nucleotide and protein sequences that contribute to high fitness are selected. An optimized nucleotide sequence results in a given set of protein variants that can be easily accessed by nucleotide substitutions (44). Recoding viral genomes may reframe the position of that viral sequence in evolutionary space, change the spectrum of protein variants that can be accessed by nucleotide substitutions, and sensitize a sequence to mutation. One approach to exploit this concept for the attenuation of viruses is the substitution of leucine and serine codons, the two most redundant codons in the genetic code, for “near-stop” codons, i.e., codons in which a single nucleotide substitution can generate a nonsense mutation (45) (Fig. 1). Indeed, coxsackie B3 viruses and IAVs that were recoded to contain numerous near-stop codons were shown to exhibit severe replication defects in the presence of mutagenic compounds. Analysis of infected cells revealed the presence of viral genomes with acquired stop codons. On the other hand, reframing of viral evolutionary space may also facilitate the acquisition of resistance to antiviral drugs. Indeed, a mutant of human immunodeficiency virus 1 (HIV-1) that carried several synonymous mutations in the HIV-1 protease-coding sequence (46) was found to more easily adapt to the presence of protease inhibitors than the parental, wild-type virus.
CpG DINUCLEOTIDE-INDUCED ATTENUATION
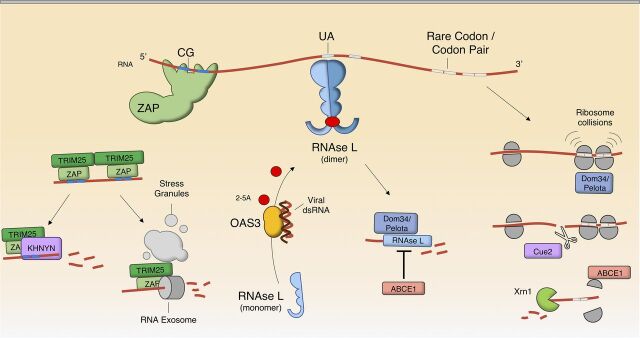
The introduction of CpG dinucleotides in the genomes of viruses, either deliberately or as a consequence of codon/codon pair deoptimization, frequently results in reduced viral fitness (37, 38, 43). The major mechanism by which CpG-enriched viruses are attenuated is through recognition by the zinc finger antiviral protein (ZAP) (Fig. 2). ZAP was initially identified as an inhibitor of Moloney murine leukemia virus (MLV) (47), and it has since been implicated in the inhibition of a broad range of viruses, including alphaviruses (48), filoviruses (49), flaviviruses (50), and hepadnaviruses (51). However, other viruses, such as yellow fever virus and vesicular stomatitis virus (VSV), were reported to be insensitive to ZAP (48). ZAP is composed of an N-terminal RNA-binding domain (comprised of four CCCH-type zinc fingers), two WWE domains, and a catalytically inactive poly(ADP-ribose) polymerase (PARP)-like domain. Early studies showed that the antiviral activity of ZAP was strictly dependent on its RNA-binding domain and suggested that ZAP recognized and bound to viral RNA (52, 53). Indeed, in some cases, the expression of the RNA-binding domain of ZAP alone was sufficient to inhibit virus replication (47). Early attempts to determine what RNA features or sequences were recognized by ZAP failed to identify any consensus sequence or structure (49, 53). Later, it became clear that CpG dinucleotide enrichment rendered HIV-1 sensitive to the expression of ZAP (43). Indeed, CpG-enriched HIV-1 mutants were found to be substantially attenuated in ZAP-expressing cells but replicated at near-wild-type levels when ZAP was absent. Similarly, CpG-enriched echovirus 7 mutants (42) were attenuated in a ZAP-dependent manner. Reanalysis of previously identified ZAP-responsive elements revealed that they, as well as viruses that were previously shown to be sensitive to ZAP, contained high frequencies of CpG dinucleotides (43). Using crosslinking immunoprecipitation coupled with RNA sequencing (CLIP-Seq) approaches, it was found that ZAP indeed specifically bound to CpG-rich RNA elements (Fig. 2). Moreover, structural analyses have recently demonstrated that ZAP selectively binds to CpG dinucleotides through residues located primarily in the second zinc finger: a hydrophobic pocket therein can accommodate only CpG dinucleotides (54, 55). Mutagenesis in and around the CpG-binding site in ZAP caused a loss of specific antiviral activity against CpG-enriched HIV-1 (54). A recent study reported that certain areas of HIV-1 genomes are more sensitive to the introduction of CpG dinucleotides, and, hence, to the activity of ZAP, than other regions (56). It is possible that sequence context and CpG distribution might affect sensitivity to ZAP, and further investigation is required to clarify this topic. Collectively, these data indicate that ZAP is responsible for the attenuation of CpG-enriched viruses, in at least some instances, and that ZAP recognition of CpG dinucleotides in viral genomes is essential for its antiviral activity.
FIG 2.
Molecular mechanisms that limit the replication of recoded viruses. Several elements present in viral RNA may be recognized and eliminated by various mechanisms. CpG-rich RNA is detected by ZAP, whose interaction with TRIM25 may facilitate the coalescence of other cellular proteins that determine its fate. Viral RNA that is recognized by ZAP can be degraded by the endonuclease KHNYN or relocalized to stress granules, where it may become a substrate of the RNA exosome. The presence of dsRNA, a frequent product of virus replication, is detected by OAS3, leading to the production of 2′-5′-oligoadenylate (2-5A) from ATP. The ankyrin repeats of RNase L interact with 2-5A, promoting the formation of a dimeric, active state of this protein. RNase L interacts with Dom34/Pelota and cleaves mRNA 3′ to UpA dinucleotides. This reaction is inhibited by ABCE1. The presence of rare codons or codon pairs may lead to slow ribosome translocation, causing ribosome collisions. Stalled ribosomes are sensed by Dom34/Pelota, recruiting Cue2/N4BP2 that cleaves the translating mRNA. Ribosome dissociation is promoted by ABCE1, while Xrn1 and other exonucleases degrade mRNA containing inhibitory codon pairs.
ZAP is ubiquitous in human tissues, but its levels can also be increased upon viral infection or stimulation with type I interferons (IFNs) (57). IFN-mediated ZAP induction is dependent on IFN regulatory factor 3 (IRF3) and IRF3-binding sites in the ZAP/ZC3HAV1 promoter region (58). The ZC3HAV1 gene encodes multiple ZAP isoforms, most prominently the so-called long (ZAP-L) and short (ZAP-S) isoforms, which differ from each other in the presence or absence of a PARP-like domain. Two additional isoforms, the medium (ZAP-M) and extralong (ZAP-XL) isoforms, resulting from the extended readthrough of exon 4 are also expressed in human cells (59). All ZAP isoforms have antiviral activity, but their potency varies somewhat (59). While little is known about the mechanism underlying the differential potencies of the isoforms, differences in cellular localizations may contribute (59, 60). How the expression of each isoform is regulated is not completely understood, but two cellular mRNA antiterminator proteins (SCAF4 and SCAF8) are reported to modulate the expression of ZAP-L and ZAP-S (61). These proteins bind a polyadenylation signal located 5′ to the terminal exon of ZC3HAV1, preventing transcription termination by masking this site, thus favoring the expression of ZAP-L. When SCAF4 and SCAF8 are absent, the otherwise masked polyadenylation site is used, favoring ZAP-S expression. Plausibly, the expression of different ZAP isoforms in different tissues may affect virus replication and the in vivo biological impact of recoded viruses.
Early studies demonstrated that viral RNA levels were reduced in the presence of ZAP (47, 53, 62). This apparent destabilization of viral RNA was shown to be limited to the cytoplasm, where ZAP is present, and nuclear viral RNA was not affected (43, 47). Some studies have suggested that ZAP can also destabilize certain retrotransposons and CpG-rich cellular mRNAs (63, 64). The process by which ZAP induces cytoplasmic RNA depletion is thought to involve the recruitment of nucleases (Fig. 2). Some studies have suggested that the RNA-binding domain of ZAP binds components of the RNA exosome such as EXOSC4 and EXOSC5 (65) and that such proteins are important for anti-MLV and anti-Japanese encephalitis virus activity as well as ZAP-dependent depletion of TRAILR4 mRNA (50, 63). ZAP has been found to colocalize with EXOSC5, recruiting both target RNA and exosome components to stress granules (64, 66, 67). However, other studies have reported that exosome component depletion did not appreciably rescue CpG-enriched HIV-1 replication (43). Moreover, pharmacological inhibition of the exosome only marginally increased hepatitis B virus RNA levels (51). Recently, a different putative endonuclease, KHNYN, was shown to bind ZAP and to be essential for ZAP-dependent inhibition of CpG-enriched HIV-1 replication (68). However, KHNYN knockout did not rescue Sindbis virus or MLV replication (68). One explanation for these discrepancies may be that ZAP recruits different RNases in different viral contexts (Fig. 2). Consistent with this idea, MLV and Sindbis virus infections lead to the formation of ZAP-positive stress granules (66, 69), while HIV-1 infection does not (70). Possibly, subcellular localization determines which RNases ZAP interacts with; perhaps, ZAP recruits exosome components to stress granules but binds KHNYN in the cytosol.
Another cofactor, TRIM25, an E3 ubiquitin and IFN-stimulated gene 15 (ISG15) ligase, was reported to be required for ZAP antiviral activity (71, 72) (Fig. 2). TRIM25 contains a RING domain, which mediates the transfer of ubiquitin to the target protein, and is known to ubiquitinate several substrates, most notably RIG-I and MDA5. The TRIM25 SPRY domain is known to interact with and provide specificity for E3 ubiquitin ligase substrates, such as the CARD domain of RIG-I (73). The SPRY domain was also shown to be important for ZAP binding and cofactor activity (71). While a few studies have demonstrated that certain lysine residues in ZAP are ubiquitinated by TRIM25 (71, 72, 74), ZAP ubiquitination does not seem to impact its antiviral activity (71, 72). Nevertheless, a truncated form of TRIM25 that lacks its RING domain or a catalytically inactive mutant (C50S,C53S) does not support ZAP antiviral activity. While these findings are consistent with the idea that the ubiquitin ligase activity of TRIM25 is important for antiviral effects, perturbing the RING domain, either by introducing mutations or by removing it entirely, also disrupts TRIM25 oligomerization. Homotypic interactions between the coiled-coil domains drive the formation of antiparallel TRIM25 dimers (75), while the interaction of two RING domains drives higher-order multimerization and is necessary for its ubiquitin ligase activity (76, 77). Additionally, two recent papers have reported that TRIM25 was also capable of binding RNA, despite disagreement over which TRIM25 domains mediated this interaction (74, 78). One report indicated that a patch of hydrophobic amino acids within the SPRY domain binds RNA and that RNA binding is essential for the in vitro ubiquitination of ZAP (74). In contrast, another report indicated that the SPRY domain alone cannot bind RNA in vitro and that seven lysine residues located between the coiled-coil domain and the SPRY domain drive RNA binding (78). Clearly, the ability of TRIM25 to bind RNA is likely to be important for its mechanistic role in the antiviral activity of ZAP. Given its propensity to form higher-order multimers, TRIM25 may function as a nucleation factor where several proteins, including ZAP and KHNYN, coalesce to degrade viral RNA. Evidently, further work is required to elucidate the role of TRIM25 in ZAP activity.
While the above-mentioned studies suggest a direct role for ZAP and associated proteins in the depletion of viral RNA from infected cells, an additional mode of action of ZAP might be through sensing and signaling. One study showed that ZAP overexpression in the presence of RIG-I agonists, or during IAV and Newcastle disease virus infections, caused increased production of IFN-α and IFN-β (79). ZAP-S, in particular, activated both the NF-κB and IRF3 transcriptional pathways in a manner that was dependent on RIG-I and MAVS (79). In contrast, ZAP-S was also reported to facilitate the resolution of innate immune responses (80). Specifically, ZAP-S was reported to limit the expression of certain IFN genes. This finding is concordant with previous results that indicated that Sindbis virus infection of ZAP-deficient mice led to high immune responses and survival rates compared to wild-type mice (81). Another study reported that CpG-enriched IAV had replication defects in mice, while host immune responses were substantially exacerbated compared to wild-type virus infection (37). Others have shown that treatment of primary human plasmacytoid dendritic cells with short CpG-rich RNAs increased the production of IFN-α (82). It is paradoxical that ZAP might both enhance an innate immune response and limit the duration of the expression of IFN genes, and in animal model systems, the level of viral replication is an obvious important confounding variable. Careful delineation of the spatial and temporal parameters with which these events take place may be key to understanding these putative roles of ZAP in the establishment and modulation of innate immune responses.
Together, these findings suggest that while ZAP has broad antiviral activity that is mobilized by CpG dinucleotides present in viral genomes, its mechanism of action may be complex. Fundamental to the understanding of ZAP activity is the elucidation of subcellular localization and its interaction partners. ZAP may restrict CpG-enriched virus replication by a variety of mechanisms, including RNA degradation, translation inhibition, and the induction of an innate immune response.
UpA DINUCLEOTIDE-INDUCED ATTENUATION
The dinucleotide UpA is almost universally suppressed in all life forms and viruses. The addition of UpA dinucleotides to viral genomes, which are frequently present only at low levels, can lead to a loss of viral fitness for picornaviruses, flaviviruses, and potyviruses (35, 36, 38, 39). Curiously, UpA-enriched echovirus 7 mutants were found to be selectively attenuated in unmanipulated cells but replicated with near-wild-type kinetics in ZAP knockout cells (42). This finding is unexpected because CLIP-Seq (43) and crystallographic studies (54, 55) have indicated that ZAP binds selectively to CpG dinucleotides in viral RNA. Even though UpA dinucleotides are unusually abundant in HIV-1, no specific interaction was observed between ZAP and these dinucleotides (43). One possible resolution of this apparent contradiction may be that U- and A-rich RNA molecules are less likely to form secondary structures. Thus, increasing the UpA content of viruses may increase the exposure of CpG dinucleotides, facilitating recognition by ZAP. In all published structures of the N-terminal domain of ZAP, several hydrophobic pockets were predicted, but only one of them interacted with CpG dinucleotides (55, 83). Perhaps, in some circumstances, those regions might interact with UpA dinucleotides; however, no evidence of that possibility was found in infected cells (43). Further investigation is required to assess the precise role of ZAP in the inhibition of UpA-enriched viruses.
Knockout of RNase L or 2′-5′-oligoadenylate synthase 3 (OAS3) has been reported to alleviate, to some extent, the replication defect in certain compositionally altered viruses (42). The OAS3/RNase L system that responds directly to the presence of double-strand RNA (dsRNA) has been reported to inhibit the replication of several viruses, including encephalomyocarditis virus (EMCV) (84), coxsackievirus B4 (85), and Theiler’s virus (86). Viral dsRNA formed through replication complexes of RNA molecules with opposite polarity or through stem-containing secondary structures can be recognized by different OAS paralogs (OAS1, OAS2, and OAS3). OAS proteins catalyze the production of 2′-5′-oligoadenylate from ATP (87–89), which is recognized via nine N-terminal ankyrin repeats, present in RNase L. The 2′-5′-oligoadenylate–ankyrin repeat interaction leads to the formation of an active RNase L dimer (90–92). Notably, OAS3 seems to be a more potent activator of RNase L than OAS1 and OAS2 during virus infection (93–95).
The RNase activity of RNase L targets RNA immediately 3′ to UpA and UpU dinucleotides (96, 97) (Fig. 2). Indeed, hepatitis C virus (HCV) genomes with higher frequencies of UpA and/or UpU dinucleotides were found to be more sensitive to RNase L (98), and circulating HCV genotypes that are more resistant to the effects of IFN contain lower levels of these dinucleotides (99). RNase L was also reported to preferentially cleave at UpA or UpU dinucleotides in poliovirus and IAV RNAs (100, 101). Thus, viruses with fewer UpA or UpU dinucleotides appear more resistant to RNase L. However, UpU dinucleotides are not underrepresented in human genomes or viruses that infect humans. Possibly, the UpU motif targeted by RNase L may be part of a more complex sequence that determines substrate specificity. Indeed, while increasing the frequency of UpA dinucleotides in viral RNA is likely to lead to virus attenuation via the OAS3/RNase L pathway, further investigation is required to address whether this effect is recapitulated by the elevation of the UpU dinucleotide frequency.
A cofactor for RNase L, termed Pelota/Dom34, has recently been identified (102) via its apparent ability to target EMCV RNA for degradation and thereby inhibit replication. Pelota directly binds RNase L, and it is a homolog of the gene Dom34 in Saccharomyces cerevisiae that is a component of the No-Go RNA decay pathway (103) (Fig. 2). In yeast and drosophila model systems, the no-go decay pathway is responsible for the degradation of mRNA with stalled or collided ribosomes that arise if translating mRNA contains rare codons, rare codon pairs, or stable secondary RNA structures (104–106). Additionally, an ATP-binding protein termed ABCE1 was shown to regulate the activity of RNase L. ABCE1 inhibits the antiviral activity of RNase L against EMCV (107) and was reported to be essential for infection by viruses that are naturally UpA rich (108, 109). Overall, the OAS3/RNase L/Pelota axis appears important for the attenuation of UpA-enriched viruses. Notably, however, viruses have evolved a plethora of mechanisms to counteract the activity of the OAS3/RNase L pathway (86), and this may explain why CpG suppression is more striking than UpA suppression in mammalian viruses (9).
Induction of the innate immune response upon the sensing of UpA-rich RNA may represent a further mechanism of UpA-induced viral attenuation. While OAS3 and RNase L are induced by IFN (110), they are constitutively present in most human tissues. Indeed, in experiments where fibroblasts were treated with 2′-5′-oligoadenylate or infected with Sendai virus, IFN-β production was dependent on RNase L expression (111). Moreover, short, UpA/UpU-containing RNAs, which are the products of the RNase L action on HCV genomes or host RNAs, activated RIG-I and MDA5-dependent signaling pathways (111, 112). Interestingly, while RNase L activation can cause translation arrest and global mRNA turnover, mRNAs encoding certain antiviral proteins, in particular IFNB and IFNL mRNAs, are resistant to this degradation (113, 114). Additionally, during IAV and VSV infections, the RNA products of RNase L were reported to activate the NLRP3 inflammasome in a MAVS-dependent but RIG-I/MDA5-independent manner, leading to the production of several cytokines, including interleukin-1β (IL-1β) (115). Together, these findings suggest that RNase L exerts antiviral activity both directly, by degrading UpA-rich RNA, and indirectly, by facilitating the establishment of an IFN-induced antiviral state.
CODON- AND CODON PAIR-DERIVED ATTENUATION
Approaches that aim to disrupt inherent codon or codon pair biases frequently result in virus attenuation. However, because of the confounding effects of enriching CpG and UpA dinucleotides that accompany these manipulations, it is difficult to disambiguate the underlying mechanisms (38). Nevertheless, it is plausible that additional mechanisms that are not directly related to CpG or UpA content contribute to virus attenuation following genetic recoding. Notably, not only codon pair deoptimization but also codon pair optimization was reported to diminish RSV replication (116), and both types of perturbation resulted in a temperature-sensitive phenotype, with reduced replication at 37°C but not at 32°C (30). Codon optimization of the adenovirus fiber protein can also lead to virus attenuation (117), even though codon optimization resulted in increased protein expression.
The study of codon usage has greatly improved our understanding of relationships between mRNA stability and translation (118). Genes that contain frequently used codons were thought to be more highly expressed due to a higher abundance of cognate tRNAs, as predicted by the number of tRNA genes present in a genome (119). However, tRNA gene number does not accurately predict tRNA abundance (120). In a study that assessed the ability of all possible codon pairs to promote or repress the expression of a fluorescent reporter gene in yeast (121), 17 codon pairs were found to be associated with reduced reporter expression. Of these, 16 contained at least one CpG dinucleotide. In this study, translation was sensitive to the order of codons; i.e., reversal of the codon order improved translation. Interestingly, the majority of codon pairs contained a codon that relied on wobble decoding, and mRNAs containing inhibitory codon pairs had elevated ribosome occupancy. Crucially, the inhibitory effects of certain codon pairs could be alleviated by the overexpression of a nonnative tRNA that precisely matched the target codon, while the overexpression of a native, wobble-decoding tRNA did not. Another study observed that certain codon pairs reduced translation elongation rates (122), an effect that could also be mitigated by overexpressing artificial, wobble-independent tRNAs. Thus, inhibitory codon pairs may reduce the rate at which the ribosome decodes wobble-containing codons, leading to ribosome stalling and collisions, relocation to stress granules, ribosome disassembly, and mRNA degradation via an RNase termed Cue2 (123) (termed N4BP2 in humans). While no link between codon- or codon pair-deoptimized viruses and ribosome stalling has yet been established, the disruption of coding biases may lead to virus attenuation via such a mechanism (Fig. 2).
Codon usage can also vary within individual genes and may be the result of selection against strong secondary structures near the translation initiation codon of mRNA molecules (124, 125). Some organisms appear to have evolved to repress codons that promote the formation of stable mRNA secondary structures since such structures can impact start codon recognition and translation initiation (126–128). Similarly, some viral genomes also exhibit apparent selection against strong secondary structures near initiation codons (129, 130). Another feature of 5′ regions of cellular genes is a “ramp” of poorly adapted codons whereby infrequently used codons that are putatively translated with low efficiency are enriched in the 5′ portion of open reading frames (ORFs), in contrast to 3′ regions (3). Indeed, ribosome density was found to be elevated in 5′ regions, implying slower transit. These biases may be a mechanism by which translation initiation is slowed and ribosomal “traffic jams” are avoided (131). The genomes of some viruses also show a preference for certain codons at the 5′ ends of genes (129, 132), but it is unclear whether selection pressures that impose these choices arise from the optimization of translation efficiency, GC content, or secondary structure. It is likely that current codon/codon pair deoptimization approaches could differentially impact translation depending on where in viral genomes or open reading frames they are applied.
Finally, another intragenic codon bias has been observed in eukaryotic ORFs, specifically the reuse of codons across an mRNA sequence, perhaps to increase the use of recycled tRNAs and facilitate translation (133). This pattern involves not only frequently used codons but also rare codons, and it is particularly apparent in rapidly induced genes, such as those involved in stress responses. It is possible that viruses might have evolved similar patterns to optimize the translation of their genes. Codon or codon pair deoptimization or optimization will likely disrupt these biases.
While mounting evidence in the field of RNA biology has described several protein complexes that detect and eliminate mRNA containing rare codons or codon pairs, it is unknown if the same pathways are involved in the recognition and degradation of recoded viruses. Approaches that combine specific genetic recoding of viruses and targeted gene perturbations, such as CRISPR-Cas9, will further our understanding of the role of these pathways in virus recoding-induced attenuation.
CONCLUDING REMARKS
The increasing use of bioinformatic algorithms and access to large-scale DNA synthesis technology have powered the study of genetic coding biases in viruses. Modifications involving CpG and UpA dinucleotide enrichment led to the discovery of important antiviral mechanisms, such as ZAP-mediated and RNase L-mediated viral RNA degradation, which appear to be central mechanisms for the attenuation of recoded viruses. Codon and codon pair deoptimizations were among the first successful attempts to reduce virus replication by the introduction of synonymous mutations. Nevertheless, how much viral attenuation is due to codon pair-specific effects and how much is related to dinucleotide frequency modifications are still not completely understood. One way to address this question is to systematically genetically modify viral genomes by codon pair deoptimization and assess virus replication in cells where ZAP and RNase L are absent. Studying the replication and codon biases of organisms that lack orthologs of ZAP and RNase L may also illuminate attenuation mechanisms. Another important question that remains unaddressed is why, in some cases, codon optimization approaches can also lead to decreased replication of viruses. It is possible that codon optimization disrupts intragenic biases, as discussed above, but further investigation is required. Assessing whether manipulations impact not only translation but also mRNA stability will further our understanding of how genetic recoding leads to viral attenuation.
This relatively new field of synthetic virology has already yielded important discoveries. Further development of this area will expand the range of viral genome manipulations that lead to attenuation via novel mechanisms. Ultimately, this knowledge will impact the development of prophylactic interventions and improve the control of infectious diseases.
Footnotes
Citation Gonçalves-Carneiro D, Bieniasz PD. 2021. Mechanisms of attenuation by genetic recoding of viruses. mBio 12:e02238-20. https://doi.org/10.1128/mBio.02238-20.
Contributor Information
Ben Berkhout, Academic Medical Center of the University of Amsterdam.
Vinayaka R. Prasad, Albert Einstein College of Medicine.
REFERENCES
- 1.Martínez MA, Jordan-Paiz A, Franco S, Nevot M. 2016. Synonymous virus genome recoding as a tool to impact viral fitness. Trends Microbiol 24:134–147. doi: 10.1016/j.tim.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 2.Martínez MA, Jordan-Paiz A, Franco S, Nevot M. 2019. Synonymous genome recoding: a tool to explore microbial biology and new therapeutic strategies. Nucleic Acids Res 47:10506–10519. doi: 10.1093/nar/gkz831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tuller T, Carmi A, Vestsigian K, Navon S, Dorfan Y, Zaborske J, Pan T, Dahan O, Furman I, Pilpel Y. 2010. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell 141:344–354. doi: 10.1016/j.cell.2010.03.031. [DOI] [PubMed] [Google Scholar]
- 4.Hia F, Yang SF, Shichino Y, Yoshinaga M, Murakawa Y, Vandenbon A, Fukao A, Fujiwara T, Landthaler M, Natsume T, Adachi S, Iwasaki S, Takeuchi O. 2019. Codon bias confers stability to human mRNAs. EMBO Rep 20:e48220. doi: 10.15252/embr.201948220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mordstein C, Savisaar R, Young RS, Bazile J, Talmane L, Luft J, Liss M, Taylor MS, Hurst LD, Kudla G. 2020. Codon usage and splicing jointly influence mRNA localization. Cell Syst 10:351–362.e8. doi: 10.1016/j.cels.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper DN, Gerber-Huber S. 1985. DNA methylation and CpG suppression. Cell Differ 17:199–205. doi: 10.1016/0045-6039(85)90488-9. [DOI] [PubMed] [Google Scholar]
- 7.Bird AP, Taggart MH. 1980. Variable patterns of total DNA and rDNA methylation in animals. Nucleic Acids Res 8:1485–1497. doi: 10.1093/nar/8.7.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beutler E, Gelbart T, Han JH, Koziol JA, Beutler B. 1989. Evolution of the genome and the genetic code: selection at the dinucleotide level by methylation and polyribonucleotide cleavage. Proc Natl Acad Sci U S A 86:192–196. doi: 10.1073/pnas.86.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simmonds P, Xia W, Baillie JK, McKinnon K. 2013. Modelling mutational and selection pressures on dinucleotides in eukaryotic phyla—selection against CpG and UpA in cytoplasmically expressed RNA and in RNA viruses. BMC Genomics 14:610. doi: 10.1186/1471-2164-14-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marais G, Mouchiroud D, Duret L. 2001. Does recombination improve selection on codon usage? Lessons from nematode and fly complete genomes. Proc Natl Acad Sci U S A 98:5688–5692. doi: 10.1073/pnas.091427698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jurkowska RZ, Jurkowski TP, Jeltsch A. 2011. Structure and function of mammalian DNA methyltransferases. Chembiochem 12:206–222. doi: 10.1002/cbic.201000195. [DOI] [PubMed] [Google Scholar]
- 12.Cooper DN, Krawczak M. 1989. Cytosine methylation and the fate of CpG dinucleotides in vertebrate genomes. Hum Genet 83:181–188. doi: 10.1007/BF00286715. [DOI] [PubMed] [Google Scholar]
- 13.Bewick AJ, Vogel KJ, Moore AJ, Schmitz RJ. 2017. Evolution of DNA methylation across insects. Mol Biol Evol 34:654–665. doi: 10.1093/molbev/msw264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falckenhayn C, Carneiro VC, de Mendonça Amarante A, Schmid K, Hanna K, Kang S, Helm M, Dimopoulos G, Fantappié MR, Lyko F. 2016. Comprehensive DNA methylation analysis of the Aedes aegypti genome. Sci Rep 6:36444. doi: 10.1038/srep36444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanaya S, Yamada Y, Kinouchi M, Kudo Y, Ikemura T. 2001. Codon usage and tRNA genes in eukaryotes: correlation of codon usage diversity with translation efficiency and with CG-dinucleotide usage as assessed by multivariate analysis. J Mol Evol 53:290–298. doi: 10.1007/s002390010219. [DOI] [PubMed] [Google Scholar]
- 16.Moriyama EN, Powell JR. 1997. Codon usage bias and tRNA abundance in Drosophila. J Mol Evol 45:514–523. doi: 10.1007/pl00006256. [DOI] [PubMed] [Google Scholar]
- 17.Kumar N, Kulkarni DD, Lee B, Kaushik R, Bhatia S, Sood R, Pateriya AK, Bhat S, Singh VP. 2018. Evolution of codon usage bias in henipaviruses is governed by natural selection and is host-specific. Viruses 10:604. doi: 10.3390/v10110604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang H, Liu S, Zhang B, Wei W. 2016. Analysis of synonymous codon usage bias of Zika virus and its adaption to the hosts. PLoS One 11:e0166260. doi: 10.1371/journal.pone.0166260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.González de Prádena A, Sánchez Jimenez A, San León D, Simmonds P, García JA, Valli AA. 2020. Plant virus genome is shaped by specific dinucleotide restrictions that influence viral infection. mBio 11:e02818-19. doi: 10.1128/mBio.02818-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Babayan SA, Orton RJ, Streicker DG. 2018. Predicting reservoir hosts and arthropod vectors from evolutionary signatures in RNA virus genomes. Science 362:577–580. doi: 10.1126/science.aap9072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greenbaum BD, Levine AJ, Bhanot G, Rabadan R. 2008. Patterns of evolution and host gene mimicry in influenza and other RNA viruses. PLoS Pathog 4:e1000079. doi: 10.1371/journal.ppat.1000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Famulare M, Chang S, Iber J, Zhao K, Adeniji JA, Bukbuk D, Baba M, Behrend M, Burns CC, Oberste MS. 2016. Sabin vaccine reversion in the field: a comprehensive analysis of Sabin-like poliovirus isolates in Nigeria. J Virol 90:317–331. doi: 10.1128/JVI.01532-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Nouën C, McCarty T, Brown M, Smith ML, Lleras R, Dolan MA, Mehedi M, Yang L, Luongo C, Liang B, Munir S, DiNapoli JM, Mueller S, Wimmer E, Collins PL, Buchholz UJ. 2017. Genetic stability of genome-scale deoptimized RNA virus vaccine candidates under selective pressure. Proc Natl Acad Sci U S A 114:E386–E395. doi: 10.1073/pnas.1619242114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burns CC, Shaw J, Campagnoli R, Jorba J, Vincent A, Quay J, Kew O. 2006. Modulation of poliovirus replicative fitness in HeLa cells by deoptimization of synonymous codon usage in the capsid region. J Virol 80:3259–3272. doi: 10.1128/JVI.80.7.3259-3272.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coleman JR, Papamichail D, Skiena S, Futcher B, Wimmer E, Mueller S. 2008. Virus attenuation by genome-scale changes in codon pair bias. Science 320:1784–1787. doi: 10.1126/science.1155761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song Y, Gorbatsevych O, Liu Y, Mugavero J, Shen SH, Ward CB, Asare E, Jiang P, Paul AV, Mueller S, Wimmer E. 2017. Limits of variation, specific infectivity, and genome packaging of massively recoded poliovirus genomes. Proc Natl Acad Sci U S A 114:E8731–E8740. doi: 10.1073/pnas.1714385114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsai Y-H, Huang S-W, Hsieh W-S, Cheng C-K, Chang C-F, Wang Y-F, Wang J-R. 2019. Enterovirus A71 containing codon-deoptimized VP1 and high-fidelity polymerase as next-generation vaccine candidate. J Virol 93:e02308-18. doi: 10.1128/JVI.02308-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li P, Ke X, Wang T, Tan Z, Luo D, Miao Y, Sun J, Zhang Y, Liu Y, Hu Q, Xu F, Wang H, Zheng Z. 2018. Zika virus attenuation by codon pair deoptimization induces sterilizing immunity in mouse models. J Virol 92:e00701-18. doi: 10.1128/JVI.00701-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mueller S, Coleman JR, Papamichail D, Ward CB, Nimnual A, Futcher B, Skiena S, Wimmer E. 2010. Live attenuated influenza virus vaccines by computer-aided rational design. Nat Biotechnol 28:723–726. doi: 10.1038/nbt.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le Nouën C, Brock LG, Luongo C, McCarty T, Yang L, Mehedi M, Wimmer E, Mueller S, Collins PL, Buchholz UJ, DiNapoli JM. 2014. Attenuation of human respiratory syncytial virus by genome-scale codon-pair deoptimization. Proc Natl Acad Sci U S A 111:13169–13174. doi: 10.1073/pnas.1411290111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eschke K, Trimpert J, Osterrieder N, Kunec D. 2018. Attenuation of a very virulent Marek’s disease herpesvirus (MDV) by codon pair bias deoptimization. PLoS Pathog 14:e1006857. doi: 10.1371/journal.ppat.1006857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng BYH, Nogales A, de la Torre JC, Martínez-Sobrido L. 2017. Development of live-attenuated arenavirus vaccines based on codon deoptimization of the viral glycoprotein. Virology 501:35–46. doi: 10.1016/j.virol.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martrus G, Nevot M, Andres C, Clotet B, Martinez MA. 2013. Changes in codon-pair bias of human immunodeficiency virus type 1 have profound effects on virus replication in cell culture. Retrovirology 10:78. doi: 10.1186/1742-4690-10-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mueller S, Stauft CB, Kalkeri R, Koidei F, Kushnir A, Tasker S, Coleman JR. 2020. A codon-pair deoptimized live-attenuated vaccine against respiratory syncytial virus is immunogenic and efficacious in non-human primates. Vaccine 38:2943–2948. doi: 10.1016/j.vaccine.2020.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen SH, Stauft CB, Gorbatsevych O, Song Y, Ward CB, Yurovsky A, Mueller S, Futcher B, Wimmer E. 2015. Large-scale recoding of an arbovirus genome to rebalance its insect versus mammalian preference. Proc Natl Acad Sci U S A 112:4749–4754. doi: 10.1073/pnas.1502864112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fros JJ, Dietrich I, Alshaikhahmed K, Passchier TC, Evans DJ, Simmonds P. 2017. CpG and UpA dinucleotides in both coding and non-coding regions of echovirus 7 inhibit replication initiation post-entry. Elife 6:e29112. doi: 10.7554/eLife.29112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gaunt E, Wise HM, Zhang H, Lee LN, Atkinson NJ, Nicol MQ, Highton AJ, Klenerman P, Beard PM, Dutia BM, Digard P, Simmonds P. 2016. Elevation of CpG frequencies in influenza A genome attenuates pathogenicity but enhances host response to infection. Elife 5:e12735. doi: 10.7554/eLife.12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tulloch F, Atkinson NJ, Evans DJ, Ryan MD, Simmonds P. 2014. RNA virus attenuation by codon pair deoptimisation is an artefact of increases in CpG/UpA dinucleotide frequencies. Elife 3:e04531. doi: 10.7554/eLife.04531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ibrahim A, Fros J, Bertran A, Sechan F, Odon V, Torrance L, Kormelink R, Simmonds P. 2019. A functional investigation of the suppression of CpG and UpA dinucleotide frequencies in plant RNA virus genomes. Sci Rep 9:18359. doi: 10.1038/s41598-019-54853-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fan RLY, Valkenburg SA, Wong CKS, Li OTW, Nicholls JM, Rabadan R, Peiris JSM, Poon LLM. 2015. Generation of live attenuated influenza virus by using codon usage bias. J Virol 89:10762–10773. doi: 10.1128/JVI.01443-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nogales A, Baker SF, Ortiz-Riaño E, Dewhurst S, Topham DJ, Martínez-Sobrido L. 2014. Influenza A virus attenuation by codon deoptimization of the NS gene for vaccine development. J Virol 88:10525–10540. doi: 10.1128/JVI.01565-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Odon V, Fros JJ, Goonawardane N, Dietrich I, Ibrahim A, Alshaikhahmed K, Nguyen D, Simmonds P. 2019. The role of ZAP and OAS3/RNAseL pathways in the attenuation of an RNA virus with elevated frequencies of CpG and UpA dinucleotides. Nucleic Acids Res 47:8061–8083. doi: 10.1093/nar/gkz581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takata MA, Gonçalves-Carneiro D, Zang TM, Soll SJ, York A, Blanco-Melo D, Bieniasz PD. 2017. CG dinucleotide suppression enables antiviral defence targeting non-self RNA. Nature 550:124–127. doi: 10.1038/nature24039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lauring AS, Acevedo A, Cooper SB, Andino R. 2012. Codon usage determines the mutational robustness, evolutionary capacity, and virulence of an RNA virus. Cell Host Microbe 12:623–632. doi: 10.1016/j.chom.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moratorio G, Henningsson R, Barbezange C, Carrau L, Bordería AV, Blanc H, Beaucourt S, Poirier EZ, Vallet T, Boussier J, Mounce BC, Fontes M, Vignuzzi M. 2017. Attenuation of RNA viruses by redirecting their evolution in sequence space. Nat Microbiol 2:17088. doi: 10.1038/nmicrobiol.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nevot M, Jordan-Paiz A, Martrus G, Andrés C, García-Cehic D, Gregori J, Franco S, Quer J, Martinez MA. 2018. HIV-1 protease evolvability is affected by synonymous nucleotide recoding. J Virol 92:e00777-18. doi: 10.1128/JVI.00777-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao G, Guo X, Goff SP. 2002. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science 297:1703–1706. doi: 10.1126/science.1074276. [DOI] [PubMed] [Google Scholar]
- 48.Bick MJ, Carroll J-WN, Gao G, Goff SP, Rice CM, MacDonald MR. 2003. Expression of the zinc-finger antiviral protein inhibits alphavirus replication. J Virol 77:11555–11562. doi: 10.1128/jvi.77.21.11555-11562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Müller S, Möller P, Bick MJ, Wurr S, Becker S, Günther S, Kümmerer BM. 2007. Inhibition of filovirus replication by the zinc finger antiviral protein. J Virol 81:2391–2400. doi: 10.1128/JVI.01601-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chiu H-P, Chiu H, Yang C-F, Lee Y-L, Chiu F-L, Kuo H-C, Lin R-J, Lin Y-L. 2018. Inhibition of Japanese encephalitis virus infection by the host zinc-finger antiviral protein. PLoS Pathog 14:e1007166. doi: 10.1371/journal.ppat.1007166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mao R, Nie H, Cai D, Zhang J, Liu H, Yan R, Cuconati A, Block TM, Guo J-T, Guo H. 2013. Inhibition of hepatitis B virus replication by the host zinc finger antiviral protein. PLoS Pathog 9:e1003494. doi: 10.1371/journal.ppat.1003494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang X, Lv F, Gao G. 2010. Mutagenesis analysis of the zinc-finger antiviral protein. Retrovirology 7:19. doi: 10.1186/1742-4690-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guo X, Carroll J-WN, MacDonald MR, Goff SP, Gao G. 2004. The zinc finger antiviral protein directly binds to specific viral mRNAs through the CCCH zinc finger motifs. J Virol 78:12781–12787. doi: 10.1128/JVI.78.23.12781-12787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meagher JL, Takata M, Gonçalves-Carneiro D, Keane SC, Rebendenne A, Ong H, Orr VK, MacDonald MR, Stuckey JA, Bieniasz PD, Smith JL. 2019. Structure of the zinc-finger antiviral protein in complex with RNA reveals a mechanism for selective targeting of CG-rich viral sequences. Proc Natl Acad Sci U S A 116:24303–24309. doi: 10.1073/pnas.1913232116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luo X, Wang X, Gao Y, Zhu J, Liu S, Gao G, Gao P. 2020. Molecular mechanism of RNA recognition by zinc-finger antiviral protein. Cell Rep 30:46–52.e4. doi: 10.1016/j.celrep.2019.11.116. [DOI] [PubMed] [Google Scholar]
- 56.Kmiec D, Nchioua R, Sherrill-Mix S, Stürzel CM, Heusinger E, Braun E, Gondim MVP, Hotter D, Sparrer KMJ, Hahn BH, Sauter D, Kirchhoff F. 2020. CpG frequency in the 5′ third of the env gene determines sensitivity of primary HIV-1 strains to the zinc-finger antiviral protein. mBio 11:e02903-19. doi: 10.1128/mBio.02903-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shaw AE, Hughes J, Gu Q, Behdenna A, Singer JB, Dennis T, Orton RJ, Varela M, Gifford RJ, Wilson SJ, Palmarini M. 2017. Fundamental properties of the mammalian innate immune system revealed by multispecies comparison of type I interferon responses. PLoS Biol 15:e2004086. doi: 10.1371/journal.pbio.2004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang N, Dong Q, Li J, Jangra RK, Fan M, Brasier AR, Lemon SM, Pfeffer LM, Li K. 2010. Viral induction of the zinc-finger antiviral protein is IRF3-dependent but NF-κB-independent. J Biol Chem 285:6080–6090. doi: 10.1074/jbc.M109.054486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li MMH, Aguilar EG, Michailidis E, Pabon J, Park P, Wu X, de Jong YP, Schneider WM, Molina H, Rice CM, MacDonald MR. 2019. Characterization of novel splice variants of zinc finger antiviral protein (ZAP). J Virol 93:e00715-19. doi: 10.1128/JVI.00715-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Charron G, Li MMH, MacDonald MR, Hang HC. 2013. Prenylome profiling reveals S-farnesylation is crucial for membrane targeting and antiviral activity of ZAP long-isoform. Proc Natl Acad Sci U S A 110:11085–11090. doi: 10.1073/pnas.1302564110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gregersen LH, Mitter R, Ugalde AP, Nojima T, Proudfoot NJ, Agami R, Stewart A, Svejstrup JQ. 2019. SCAF4 and SCAF8, mRNA anti-terminator proteins. Cell 177:1797–1813.e18. doi: 10.1016/j.cell.2019.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.MacDonald MR, Machlin ES, Albin OR, Levy DE. 2007. The zinc finger antiviral protein acts synergistically with an interferon-induced factor for maximal activity against alphaviruses. J Virol 81:13509–13518. doi: 10.1128/JVI.00402-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Todorova T, Bock FJ, Chang P. 2014. PARP13 regulates cellular mRNA post-transcriptionally and functions as a pro-apoptotic factor by destabilizing TRAILR4 transcript. Nat Commun 5:5362. doi: 10.1038/ncomms6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moldovan JB, Moran JV. 2015. The zinc-finger antiviral protein ZAP inhibits LINE and Alu retrotransposition. PLoS Genet 11:e1005121. doi: 10.1371/journal.pgen.1005121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guo X, Ma J, Sun J, Gao G. 2007. The zinc-finger antiviral protein recruits the RNA processing exosome to degrade the target mRNA. Proc Natl Acad Sci U S A 104:151–156. doi: 10.1073/pnas.0607063104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee H, Komano J, Saitoh Y, Yamaoka S, Kozaki T, Misawa T, Takahama M, Satoh T, Takeuchi O, Yamamoto N, Matsuura Y, Saitoh T, Akira S. 2013. Zinc-finger antiviral protein mediates retinoic acid inducible gene I-like receptor-independent antiviral response to murine leukemia virus. Proc Natl Acad Sci U S A 110:12379–12384. doi: 10.1073/pnas.1310604110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goodier JL, Pereira GC, Cheung LE, Rose RJ, Kazazian HH, Jr. 2015. The broad-spectrum antiviral protein ZAP restricts human retrotransposition. PLoS Genet 11:e1005252. doi: 10.1371/journal.pgen.1005252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ficarelli M, Wilson H, Pedro Galão R, Mazzon M, Antzin-Anduetza I, Marsh M, Neil SJD, Swanson CM. 2019. KHNYN is essential for the zinc finger antiviral protein (ZAP) to restrict HIV-1 containing clustered CpG dinucleotides. Elife 8:e46767. doi: 10.7554/eLife.46767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Law LMJ, Razooky BS, Li MMH, You S, Jurado A, Rice CM, MacDonald MR. 2019. ZAP’s stress granule localization is correlated with its antiviral activity and induced by virus replication. PLoS Pathog 15:e1007798. doi: 10.1371/journal.ppat.1007798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cinti A, Le Sage V, Ghanem M, Mouland AJ. 2016. HIV-1 Gag blocks selenite-induced stress granule assembly by altering the mRNA cap-binding complex. mBio 7:e00329-16. doi: 10.1128/mBio.00329-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li MMH, Lau Z, Cheung P, Aguilar EG, Schneider WM, Bozzacco L, Molina H, Buehler E, Takaoka A, Rice CM, Felsenfeld DP, MacDonald MR. 2017. TRIM25 enhances the antiviral action of zinc-finger antiviral protein (ZAP). PLoS Pathog 13:e1006145. doi: 10.1371/journal.ppat.1006145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zheng X, Wang X, Tu F, Wang Q, Fan Z, Gao G. 2017. TRIM25 is required for the antiviral activity of zinc finger antiviral protein. J Virol 91:e00088-17. doi: 10.1128/JVI.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gack MU, Shin YC, Joo C-H, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. 2007. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 74.Choudhury NR, Heikel G, Trubitsyna M, Kubik P, Nowak JS, Webb S, Granneman S, Spanos C, Rappsilber J, Castello A, Michlewski G. 2017. RNA-binding activity of TRIM25 is mediated by its PRY/SPRY domain and is required for ubiquitination. BMC Biol 15:105. doi: 10.1186/s12915-017-0444-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sanchez JG, Okreglicka K, Chandrasekaran V, Welker JM, Sundquist WI, Pornillos O. 2014. The tripartite motif coiled-coil is an elongated antiparallel hairpin dimer. Proc Natl Acad Sci U S A 111:2494–2499. doi: 10.1073/pnas.1318962111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Koliopoulos MG, Esposito D, Christodoulou E, Taylor IA, Rittinger K. 2016. Functional role of TRIM E3 ligase oligomerization and regulation of catalytic activity. EMBO J 35:1204–1218. doi: 10.15252/embj.201593741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sanchez JG, Chiang JJ, Sparrer KMJ, Alam SL, Chi M, Roganowicz MD, Sankaran B, Gack MU, Pornillos O. 2016. Mechanism of TRIM25 catalytic activation in the antiviral RIG-I pathway. Cell Rep 16:1315–1325. doi: 10.1016/j.celrep.2016.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sanchez JG, Sparrer KMJ, Chiang C, Reis RA, Chiang JJ, Zurenski MA, Wan Y, Gack MU, Pornillos O. 2018. TRIM25 binds RNA to modulate cellular anti-viral defense. J Mol Biol 430:5280–5293. doi: 10.1016/j.jmb.2018.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hayakawa S, Shiratori S, Yamato H, Kameyama T, Kitatsuji C, Kashigi F, Goto S, Kameoka S, Fujikura D, Yamada T, Mizutani T, Kazumata M, Sato M, Tanaka J, Asaka M, Ohba Y, Miyazaki T, Imamura M, Takaoka A. 2011. ZAPS is a potent stimulator of signaling mediated by the RNA helicase RIG-I during antiviral responses. Nat Immunol 12:37–44. doi: 10.1038/ni.1963. [DOI] [PubMed] [Google Scholar]
- 80.Schwerk J, Soveg FW, Ryan AP, Thomas KR, Hatfield LD, Ozarkar S, Forero A, Kell AM, Roby JA, So L, Hyde JL, Gale M, Daugherty MD, Savan R. 2019. RNA-binding protein isoforms ZAP-S and ZAP-L have distinct antiviral and immune resolution functions. Nat Immunol 20:1610–1620. doi: 10.1038/s41590-019-0527-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang X, Li MMH, Zhao J, Li S, MacDonald MR, Rice CM, Gao X, Gao G. 2016. Sindbis virus can exploit a host antiviral protein to evade immune surveillance. J Virol 90:10247–10258. doi: 10.1128/JVI.01487-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jimenez-Baranda S, Greenbaum B, Manches O, Handler J, Rabadán R, Levine A, Bhardwaj N. 2011. Oligonucleotide motifs that disappear during the evolution of influenza virus in humans increase alpha interferon secretion by plasmacytoid dendritic cells. J Virol 85:3893–3904. doi: 10.1128/JVI.01908-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen S, Xu Y, Zhang K, Wang X, Sun J, Gao G, Liu Y. 2012. Structure of N-terminal domain of ZAP indicates how a zinc-finger protein recognizes complex RNA. Nat Struct Mol Biol 19:430–435. doi: 10.1038/nsmb.2243. [DOI] [PubMed] [Google Scholar]
- 84.Li XL, Blackford JA, Hassel BA. 1998. RNase L mediates the antiviral effect of interferon through a selective reduction in viral RNA during encephalomyocarditis virus infection. J Virol 72:2752–2759. doi: 10.1128/JVI.72.4.2752-2759.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Flodström-Tullberg M, Hultcrantz M, Stotland A, Maday A, Tsai D, Fine C, Williams B, Silverman R, Sarvetnick N. 2005. RNase L and double-stranded RNA-dependent protein kinase exert complementary roles in islet cell defense during coxsackievirus infection. J Immunol 174:1171–1177. doi: 10.4049/jimmunol.174.3.1171. [DOI] [PubMed] [Google Scholar]
- 86.Drappier M, Jha BK, Stone S, Elliott R, Zhang R, Vertommen D, Weiss SR, Silverman RH, Michiels T. 2018. A novel mechanism of RNase L inhibition: Theiler’s virus L* protein prevents 2-5A from binding to RNase L. PLoS Pathog 14:e1006989. doi: 10.1371/journal.ppat.1006989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Donovan J, Whitney G, Rath S, Korennykh A. 2015. Structural mechanism of sensing long dsRNA via a noncatalytic domain in human oligoadenylate synthetase 3. Proc Natl Acad Sci U S A 112:3949–3954. doi: 10.1073/pnas.1419409112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lohöfener J, Steinke N, Kay-Fedorov P, Baruch P, Nikulin A, Tishchenko S, Manstein DJ, Fedorov R. 2015. The activation mechanism of 2′-5′-oligoadenylate synthetase gives new insights into OAS/cGAS triggers of innate immunity. Structure 23:851–862. doi: 10.1016/j.str.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 89.Donovan J, Dufner M, Korennykh A. 2013. Structural basis for cytosolic double-stranded RNA surveillance by human oligoadenylate synthetase 1. Proc Natl Acad Sci U S A 110:1652–1657. doi: 10.1073/pnas.1218528110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tanaka N, Nakanishi M, Kusakabe Y, Goto Y, Kitade Y, Nakamura KT. 2004. Structural basis for recognition of 2′,5′-linked oligoadenylates by human ribonuclease L. EMBO J 23:3929–3938. doi: 10.1038/sj.emboj.7600420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dong B, Silverman RH. 1995. 2-5A-dependent RNase molecules dimerize during activation by 2-5A. J Biol Chem 270:4133–4137. doi: 10.1074/jbc.270.8.4133. [DOI] [PubMed] [Google Scholar]
- 92.Castelli JC, Hassel BA, Maran A, Paranjape J, Hewitt JA, Li X, Hsu Y-T, Silverman RH, Youle RJ. 1998. The role of 2′-5′ oligoadenylate-activated ribonuclease L in apoptosis. Cell Death Differ 5:313–320. doi: 10.1038/sj.cdd.4400352. [DOI] [PubMed] [Google Scholar]
- 93.Li Y, Banerjee S, Wang Y, Goldstein SA, Dong B, Gaughan C, Silverman RH, Weiss SR. 2016. Activation of RNase L is dependent on OAS3 expression during infection with diverse human viruses. Proc Natl Acad Sci U S A 113:2241–2246. doi: 10.1073/pnas.1519657113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li Y, Dong B, Wei Z, Silverman RH, Weiss SR. 2019. Activation of RNase L in Egyptian Rousette bat-derived RoNi/7 cells is dependent primarily on OAS3 and independent of MAVS signaling. mBio 10:e02414-19. doi: 10.1128/mBio.02414-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lin R-J, Yu H-P, Chang B-L, Tang W-C, Liao C-L, Lin Y-L. 2009. Distinct antiviral roles for human 2′,5′-oligoadenylate synthetase family members against dengue virus infection. J Immunol 183:8035–8043. doi: 10.4049/jimmunol.0902728. [DOI] [PubMed] [Google Scholar]
- 96.Wreschner DH, McCauley JW, Skehel JJ, Kerr IM. 1981. Interferon action—sequence specificity of the ppp(A2′p)nA-dependent ribonuclease. Nature 289:414–417. doi: 10.1038/289414a0. [DOI] [PubMed] [Google Scholar]
- 97.Floyd-Smith G, Slattery E, Lengyel P. 1981. Interferon action: RNA cleavage pattern of a (2′-5′)oligoadenylate-dependent endonuclease. Science 212:1030–1032. doi: 10.1126/science.6165080. [DOI] [PubMed] [Google Scholar]
- 98.Han J-Q, Wroblewski G, Xu Z, Silverman RH, Barton DJ. 2004. Sensitivity of hepatitis C virus RNA to the antiviral enzyme ribonuclease L is determined by a subset of efficient cleavage sites. J Interferon Cytokine Res 24:664–676. doi: 10.1089/jir.2004.24.664. [DOI] [PubMed] [Google Scholar]
- 99.Washenberger CL, Han J-Q, Kechris KJ, Jha BK, Silverman RH, Barton DJ. 2007. Hepatitis C virus RNA: dinucleotide frequencies and cleavage by RNase L. Virus Res 130:85–95. doi: 10.1016/j.virusres.2007.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cooper DA, Jha BK, Silverman RH, Hesselberth JR, Barton DJ. 2014. Ribonuclease L and metal-ion-independent endoribonuclease cleavage sites in host and viral RNAs. Nucleic Acids Res 42:5202–5216. doi: 10.1093/nar/gku118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cooper DA, Banerjee S, Chakrabarti A, García-Sastre A, Hesselberth JR, Silverman RH, Barton DJ. 2015. RNase L targets distinct sites in influenza A virus RNAs. J Virol 89:2764–2776. doi: 10.1128/JVI.02953-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nogimori T, Nishiura K, Kawashima S, Nagai T, Oishi Y, Hosoda N, Imataka H, Kitamura Y, Kitade Y, Hoshino S. 2019. Dom34 mediates targeting of exogenous RNA in the antiviral OAS/RNase L pathway. Nucleic Acids Res 47:432–449. doi: 10.1093/nar/gky1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Becker T, Armache J-P, Jarasch A, Anger AM, Villa E, Sieber H, Motaal BA, Mielke T, Berninghausen O, Beckmann R. 2011. Structure of the no-go mRNA decay complex Dom34-Hbs1 bound to a stalled 80S ribosome. Nat Struct Mol Biol 18:715–720. doi: 10.1038/nsmb.2057. [DOI] [PubMed] [Google Scholar]
- 104.Simms CL, Yan LL, Zaher HS. 2017. Ribosome collision is critical for quality control during no-go decay. Mol Cell 68:361–373.e5. doi: 10.1016/j.molcel.2017.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Harigaya Y, Parker R. 2017. The link between adjacent codon pairs and mRNA stability. BMC Genomics 18:364. doi: 10.1186/s12864-017-3749-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ghoneim DH, Zhang X, Brule CE, Mathews DH, Grayhack EJ. 2019. Conservation of location of several specific inhibitory codon pairs in the Saccharomyces sensu stricto yeasts reveals translational selection. Nucleic Acids Res 47:1164–1177. doi: 10.1093/nar/gky1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Martinand C, Salehzada T, Silhol M, Lebleu B, Bisbal C. 1998. RNase L inhibitor (RLI) antisense constructions block partially the down regulation of the 2-5A/RNase L pathway in encephalomyocarditis-virus-(EMCV)-infected cells. Eur J Biochem 254:248–255. doi: 10.1046/j.1432-1327.1998.2540248.x. [DOI] [PubMed] [Google Scholar]
- 108.Martinand C, Montavon C, Salehzada T, Silhol M, Lebleu B, Bisbal C. 1999. RNase L inhibitor is induced during human immunodeficiency virus type 1 infection and down regulates the 2-5A/RNase L pathway in human T cells. J Virol 73:290–296. doi: 10.1128/JVI.73.1.290-296.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Anderson DE, Pfeffermann K, Kim SY, Sawatsky B, Pearson J, Kovtun M, Corcoran DL, Krebs Y, Sigmundsson K, Jamison SF, Yeo ZZJ, Rennick LJ, Wang L-F, Talbot PJ, Duprex WP, Garcia-Blanco MA, von Messling V. 2019. Comparative loss-of-function screens reveal ABCE1 as an essential cellular host factor for efficient translation of Paramyxoviridae and Pneumoviridae. mBio 10:e00826-19. doi: 10.1128/mBio.00826-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sadler AJ, Williams BRG. 2008. Interferon-inducible antiviral effectors. Nat Rev Immunol 8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Malathi K, Dong B, Gale M, Jr, Silverman RH. 2007. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature 448:816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Malathi K, Saito T, Crochet N, Barton DJ, Gale M, Jr, Silverman RH. 2010. RNase L releases a small RNA from HCV RNA that refolds into a potent PAMP. RNA 16:2108–2119. doi: 10.1261/rna.2244210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Burke JM, Moon SL, Matheny T, Parker R. 2019. RNase L reprograms translation by widespread mRNA turnover escaped by antiviral mRNAs. Mol Cell 75:1203–1217.e5. doi: 10.1016/j.molcel.2019.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chitrakar A, Rath S, Donovan J, Demarest K, Li Y, Sridhar RR, Weiss SR, Kotenko SV, Wingreen NS, Korennykh A. 2019. Real-time 2-5A kinetics suggest that interferons β and λ evade global arrest of translation by RNase L. Proc Natl Acad Sci U S A 116:2103–2111. doi: 10.1073/pnas.1818363116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chakrabarti A, Banerjee S, Franchi L, Loo Y-M, Gale M, Jr, Núñez G, Silverman RH. 2015. RNase L activates the NLRP3 inflammasome during viral infections. Cell Host Microbe 17:466–477. doi: 10.1016/j.chom.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Le Nouën C, Luongo CL, Yang L, Mueller S, Wimmer E, DiNapoli JM, Collins PL, Buchholz UJ. 2020. Optimization of the codon pair usage of human respiratory syncytial virus paradoxically resulted in reduced viral replication in vivo and reduced immunogenicity. J Virol 94:e01296-19. doi: 10.1128/JVI.01296-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Villanueva E, Martí-Solano M, Fillat C. 2016. Codon optimization of the adenoviral fiber negatively impacts structural protein expression and viral fitness. Sci Rep 6:27546. doi: 10.1038/srep27546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Plotkin JB, Kudla G. 2011. Synonymous but not the same: the causes and consequences of codon bias. Nat Rev Genet 12:32–42. doi: 10.1038/nrg2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Iben JR, Maraia RJ. 2014. tRNA gene copy number variation in humans. Gene 536:376–384. doi: 10.1016/j.gene.2013.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Torres AG, Reina O, Stephan-Otto Attolini C, Ribas de Pouplana L. 2019. Differential expression of human tRNA genes drives the abundance of tRNA-derived fragments. Proc Natl Acad Sci U S A 116:8451–8456. doi: 10.1073/pnas.1821120116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gamble CE, Brule CE, Dean KM, Fields S, Grayhack EJ. 2016. Adjacent codons act in concert to modulate translation efficiency in yeast. Cell 166:679–690. doi: 10.1016/j.cell.2016.05.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tesina P, Lessen LN, Buschauer R, Cheng J, Wu CC-C, Berninghausen O, Buskirk AR, Becker T, Beckmann R, Green R. 2020. Molecular mechanism of translational stalling by inhibitory codon combinations and poly(A) tracts. EMBO J 39:e103365. doi: 10.15252/embj.2019103365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.D’Orazio KN, Wu CC-C, Sinha N, Loll-Krippleber R, Brown GW, Green R. 2019. The endonuclease Cue2 cleaves mRNAs at stalled ribosomes during no go decay. Elife 8:e49117. doi: 10.7554/eLife.49117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gu W, Zhou T, Wilke CO. 2010. A universal trend of reduced mRNA stability near the translation-initiation site in prokaryotes and eukaryotes. PLoS Comput Biol 6:e1000664. doi: 10.1371/journal.pcbi.1000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bulmer M. 1991. The selection-mutation-drift theory of synonymous codon usage. Genetics 129:897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Eyre-Walker A, Bulmer M. 1993. Reduced synonymous substitution rate at the start of enterobacterial genes. Nucleic Acids Res 21:4599–4603. doi: 10.1093/nar/21.19.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ding Y, Shah P, Plotkin JB. 2012. Weak 5′-mRNA secondary structures in short eukaryotic genes. Genome Biol Evol 4:1046–1053. doi: 10.1093/gbe/evs082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Niimura Y, Terabe M, Gojobori T, Miura K. 2003. Comparative analysis of the base biases at the gene terminal portions in seven eukaryote genomes. Nucleic Acids Res 31:5195–5201. doi: 10.1093/nar/gkg701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhou T, Wilke CO. 2011. Reduced stability of mRNA secondary structure near the translation-initiation site in dsDNA viruses. BMC Evol Biol 11:59. doi: 10.1186/1471-2148-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Goz E, Tuller T. 2015. Widespread signatures of local mRNA folding structure selection in four dengue virus serotypes. BMC Genomics 16(Suppl 1):S4. doi: 10.1186/1471-2164-16-S10-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhang S, Goldman E, Zubay G. 1994. Clustering of low usage codons and ribosome movement. J Theor Biol 170:339–354. doi: 10.1006/jtbi.1994.1196. [DOI] [PubMed] [Google Scholar]
- 132.Clyde K, Harris E. 2006. RNA secondary structure in the coding region of dengue virus type 2 directs translation start codon selection and is required for viral replication. J Virol 80:2170–2182. doi: 10.1128/JVI.80.5.2170-2182.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Cannarozzi G, Schraudolph NN, Faty M, von Rohr P, Friberg MT, Roth AC, Gonnet P, Gonnet G, Barral Y. 2010. A role for codon order in translation dynamics. Cell 141:355–367. doi: 10.1016/j.cell.2010.02.036. [DOI] [PubMed] [Google Scholar]