ABSTRACT
Multidrug-resistant (MDR) pathogens pose a significant public health threat. A major mechanism of resistance expressed by MDR pathogens is β-lactamase-mediated degradation of β-lactam antibiotics. The diazabicyclooctane (DBO) compounds zidebactam and WCK 5153, recognized as β-lactam “enhancers” due to inhibition of Pseudomonas aeruginosa penicillin-binding protein 2 (PBP2), are also class A and C β-lactamase inhibitors. To structurally probe their mode of PBP2 inhibition as well as investigate why P. aeruginosa PBP2 is less susceptible to inhibition by β-lactam antibiotics compared to the Escherichia coli PBP2, we determined the crystal structure of P. aeruginosa PBP2 in complex with WCK 5153. WCK 5153 forms an inhibitory covalent bond with the catalytic S327 of PBP2. The structure suggests a significant role for the diacylhydrazide moiety of WCK 5153 in interacting with the aspartate in the S-X-N/D PBP motif. Modeling of zidebactam in the active site of PBP2 reveals a similar binding mode. Both DBOs increase the melting temperature of PBP2, affirming their stabilizing interactions. To aid in the design of DBOs that can inhibit multiple PBPs, the ability of three DBOs to interact with P. aeruginosa PBP3 was explored crystallographically. Even though the DBOs show covalent binding to PBP3, they destabilized PBP3. Overall, the studies provide insights into zidebactam and WCK 5153 inhibition of PBP2 compared to their inhibition of PBP3 and the evolutionarily related KPC-2 β-lactamase. These molecular insights into the dual-target DBOs advance our knowledge regarding further DBO optimization efforts to develop novel potent β-lactamase-resistant, non-β-lactam PBP inhibitors.
KEYWORDS: Pseudomonas aeruginosa, antibiotic resistance, penicillin-binding proteins, structural biology
INTRODUCTION
Multidrug resistance (MDR) is a serious clinical threat demonstrated by Gram-negative bacterial pathogens such as Pseudomonas aeruginosa. The MDR phenotype is often multifactorial and can include production of β-lactamases, increased expression of efflux pumps, and mutations in porins and penicillin-binding proteins (PBPs). The World Health Organization (WHO) classifies carbapenem-resistant P. aeruginosa as a “top priority pathogen” (1), as there are limited treatment options. Recently, a novel combination developmental compound, WCK 5222 (cefepime and zidebactam [Fig. 1A]), showed promise against carbapenem-resistant P. aeruginosa (2–5), including against MDR and metallo-β-lactamase-expressing P. aeruginosa (6–8). Cefepime targets PBP3 whereas zidebactam is a dual-target inhibitor: it inhibits PBP2, thus being an “enhancer” for β-lactams, and also acts as a β-lactamase inhibitor (6). This “enhancer” activity is anticipated to be more prominent for β-lactams targeting PBPs other than PBP2, but β-lactams do often target more than one PBP. PBP2 and PBP3 are part of the elongase and divisome complex, respectively (9). Zidebactam and WCK 5153 are diazabicyclooctanes (DBOs) and are well differentiated from other DBO members in terms of their direct antibacterial activity against both Enterobacterales and P. aeruginosa (10) (Fig. 1A). The structural, kinetic, and microbiological determinants of zidebactam and analogs thereof were previously characterized as serine β-lactamase inhibitors (11). Here, we focus on the β-lactam “enhancer” activity of zidebactam and WCK 5153 that involves binding and inhibiting PBPs.
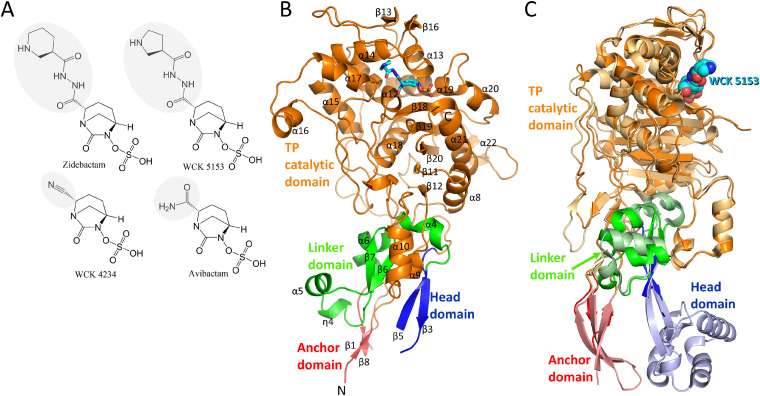
FIG 1.
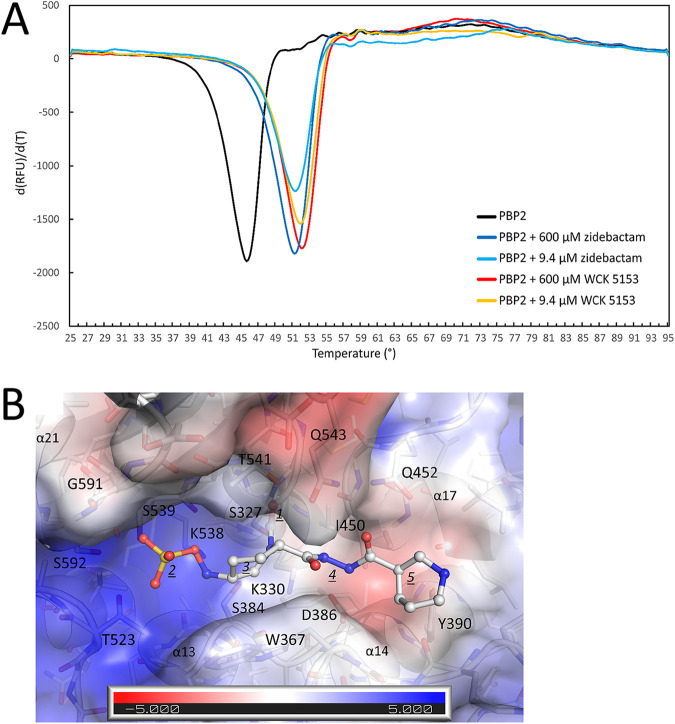
Structures of DBO inhibitors and P. aeruginosa PBP2. (A) Chemical structures of DBOs zidebactam, WCK 5153, WCK 4234, and avibactam. The R1-groups of the DBOs are shaded gray. (B) Co-crystal structure of P. aeruginosa PBP2 in complex with WCK 5153 (the latter is shown in stick representation with cyan-colored carbon atoms). The PBP2 anchor (red), head (blue), linker (green), and catalytic (orange) domains are labeled, as well as most of the secondary structure elements. (C) Superpositioning of the P. aeruginosa PBP2:WCK 5153 complex and E. coli PBP2 structure. The domain color coding for P. aeruginosa PBP2 is the same as in panel B with WCK 5153 depicted in spheres with cyan-colored carbon atoms. E. coli PBP2 is shown in similar but paler colors for its respective domains.
PBPs have an indispensable role in bacterial cell wall biosynthesis. These enzymes cross-link the peptidoglycan layer via its transpeptidation reaction to strengthen the cell wall. PBPs are targeted by β-lactam antibiotics. A limitation of β-lactams is that MDR pathogens can hydrolyze β-lactams by expressing β-lactamases. Therefore, developing non-β-lactam PBP-targeting antibiotics is highly desirable. The DBOs, as a novel class of β-lactamase and PBP inhibitors, were recently identified as part of β-lactam “enhancer” development efforts. The initial insights were obtained with avibactam, the first DBO β-lactamase inhibitor approved for clinical use (combined with ceftazidime), which binds P. aeruginosa PBPs (50% inhibitory concentration [IC50] in the range of 1.1 to 1.8 μg/ml) (12). Interestingly, zidebactam and WCK 5153 inhibit P. aeruginosa PBP2 with greater affinity (IC50 values of 0.26 and 0.14 μg/ml, respectively (13)) (Fig. 1A). These DBOs elicit antibacterial effects by themselves (i.e., in the absence of a β-lactam), yielding relatively low MICs against P. aeruginosa (11). This antibacterial effect is ascribed to their inhibition of P. aeruginosa PBP2.
The precise molecular details of zidebactam and WCK 5153 inhibition of P. aeruginosa PBP2 are not known. Here, we present the crystal structure of P. aeruginosa PBP2 in complex with WCK 5153; this is also the first crystal structure reported for P. aeruginosa PBP2. Additionally, we modeled zidebactam binding to PBP2. Next, PBP2 binding of WCK 5153 is compared with avibactam binding to Escherichia coli PBP2. Lastly, we describe crystal structures of zidebactam, WCK 4234, and avibactam bound to PBP3. These crystallographic studies are complemented with thermal shift binding assays. Our results detail the interactions of the DBOs in the acyl-enzyme complex state when bound to P. aeruginosa PBPs, and these insights could aid in further structure-based DBO optimization to develop novel antimicrobial therapeutics.
RESULTS AND DISCUSSION
Structural analysis of PBP2.
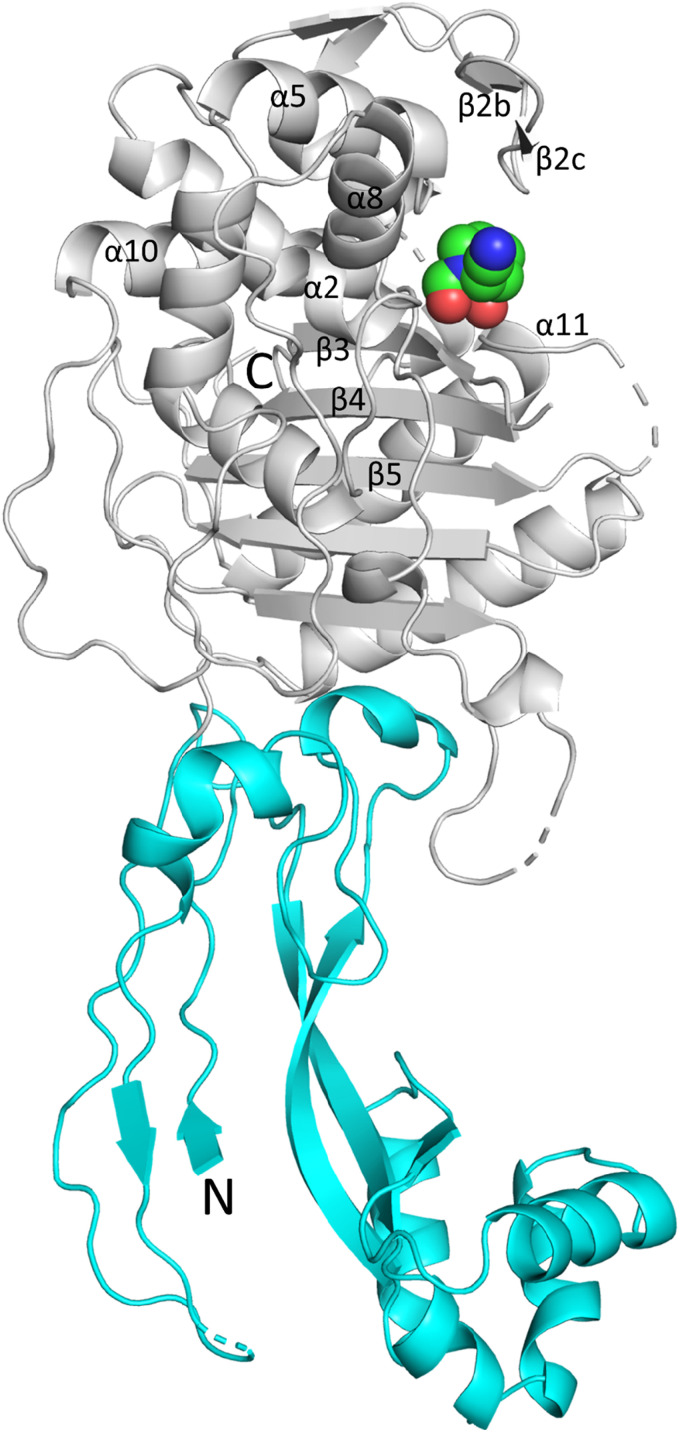
The P. aeruginosa PBP2 adopts the class B fold of a monofunctional PBP. This fold includes four distinct regions that protrude into the periplasm: (i) an anchor domain, (ii) a linker domain, (iii) a head domain, and (iv) the large C-terminal catalytic transpeptidase (TP) domain which cross-links the nascent peptidoglycan strand (this excludes the N-terminal transmembrane anchoring region which is not included in the construct). This PBP2 structure was resolved to 2.87-Å resolution and contains two non-crystallographically related molecules. These monomers are very similar, and our analysis will, therefore, mostly be limited to molecule A (Fig. 1B).
Notably, the PBP2 structure lacks the head domain, which could not be modeled in either of the two monomers (Fig. 1B; see also Fig. S1 in the supplemental material). Also, the anchor domain was modeled partially. Electron density for these unmodeled regions was poor, likely due to their inherent disorder or mobility. Structures of homologous PBP2s from Escherichia coli, Helicobacter pylori, and Neisseria gonorrhoeae show a similar fold (14–17); their sequence identities with P. aeruginosa PBP2 are 45%, 32%, and 27%, respectively (Fig. S1 shows their structure-based sequence alignment). Similarly, the head domain also could not be modeled in the N. gonorrhoeae PBP2 structure (PDB ID 5KSH) (17). Furthermore, the head domain in the E. coli and H. pylori structures also does not superimpose well, again indicating that its position can vary (14, 15). The sequence and structure of P. aeruginosa PBP2 are most like those of E. coli PBP2; their superpositioning yields a root mean square deviation (RMSD) of 1.04 Å for 426 Cα atoms, indicating their structural similarity. Figure 1C represents this superposition and also shows the portions of the head domain and the anchor domain that are better resolved in the E. coli PBP2 structure compared to the P. aeruginosa PBP2 structure. The catalytic domain (residues 242 to 646) contains a central β-sheet flanked by several α-helices and an additional small β-sheet. Within this catalytic domain, the highly flexible loop region comprising residues 545 to 560 and the C-terminal residues 623 to 646 are too disordered and could not be modeled (Fig. 1B and Fig. S1). The catalytic serine residue S327 possesses a covalently bound cocrystallized WCK 5153 molecule attached (Fig. 2A).
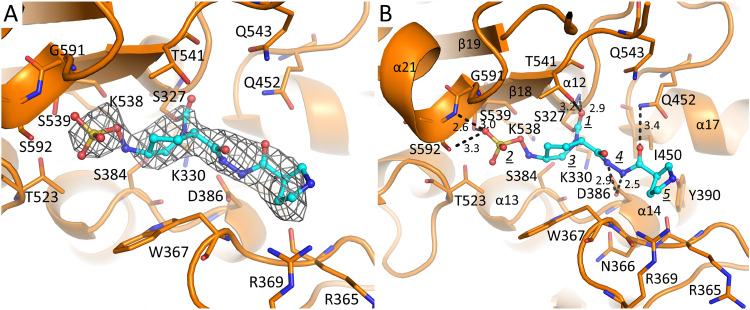
FIG 2.
WCK 5153 bound in the PBP2 active site. (A) |Fo|-|Fc| electron density difference map contoured at 2.5 σ level. WCK 5153 is depicted with cyan-colored carbon atoms. The unbiased density map was calculated after removing the ligand from the coordinates and performing 10 cycles of crystallographic refinement using Refmac. (B) Interactions of WCK 5153 in the active site of PBP2. Hydrogen bonds are depicted as dashed lines, and distances are shown (in Å). The following moieties of WCK 5153 are labeled: carbonyl oxygen (1), sulfate (2), piperidine ring of the DBO scaffold (3), diacylhydrazide (4), and the pyrrolidine ring (5). WCK 5153 is covalently attached to the catalytic S327. Key secondary structure elements are labeled.
Structure-based sequence alignment of PBP2 sequences from P. aeruginosa, E. coli, H. pylori, and N. gonorrhoeae. Assigned secondary elements in P. aeruginosa PBP2 are indicated above the sequences (α and β; 310 helices are labeled as “η”). Residues not included in the crystallized construct or too disordered to be modeled are indicated by a gray interrupted line above the sequence. Residues either conserved or similar to the respective residue in P. aeruginosa are shown as red and orange, respectively. Fully conserved residues in all four sequences are indicated by an asterisk below the sequence. Active site residues are indicated by green boxes. Download FIG S1, TIF file, 2.6 MB (2.6MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
WCK 5153 occupies the active site of PBP2.
The active sites of PBPs possess several conserved motifs, and some of those are shared with homologous serine β-lactamases. The nucleophilic serine (S327) is located in the conserved motif S-X-X-K (S327-T328-V329-K330) situated at the beginning of a short 310 helix at the start of the α12 helix (Fig. 2B and Fig. S1). Here, S327 forms a covalent bond with WCK 5153, and the backbone nitrogen interacts with the carbonyl oxygen. In molecule B, the carbonyl oxygen of WCK 5153 is shifted 0.5 Å, such that the backbone nitrogen of T541 now also forms a hydrogen bond interaction (Fig. S2). The covalent bond between S327 and WCK 5153, with the carbonyl oxygen positioned in the oxyanion hole, is a common acyl-enzyme inhibited state for DBOs (14).
Stereo figure showing superposition of noncrystallographic molecules A and B of P. aeruginosa PBP2 active site complexed with WCK 5153. Two PBP2 molecules were observed in the asymmetric unit. Molecule A and molecule B are shown with orange and green carbon atoms, respectively. Hydrogen bonds between WCK 5153 and either PBP2 molecule A or molecule B are shown as black or gray dashed lines, respectively. A crystallographically observed water in the active site of molecule B is shown (“wat”). WCK 5153 is shown in ball-and-stick form with its different moieties labeled as in Fig. 2B. Download FIG S2, TIF file, 2.5 MB (2.5MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Within hydrogen bonding distance of the first motif is the second conserved active site motif, S-X-N/D (S384-N385-D386), located between the α13 and α14 helices. D386 hydrogen bonds with the nitrogen(s) of the diacylhydrazide moiety of WCK 5153 in both molecules A and B (Fig. 2B and Fig. S2). In molecule B, the nitrogen atom of the diacylhydrazide moiety closest to the piperidine ring is slightly rotated and makes a water-mediated interaction (Fig. S2); this water molecule also hydrogen bonds with the backbone carbonyl oxygen of I450. The third PBP motif, K-T/S-G-T (K538-S539-G540-T541), is in the β18 strand; the side chain of S539 forms a hydrogen bond with the sulfate of WCK 5153, and T541 interacts with the carbonyl oxygen.
Additional active site residues include W367, which interacts with the hydrophobic face of the DBO piperidine ring of the WCK 5153 scaffold. Also, the N-terminal end of the α21 helix, via the backbone nitrogen of S592 as well as its side chain, forms hydrogen bonds with the sulfate of WCK 5153 (Fig. 2B and Fig. S2). This α21 helix provides a helix dipole-sulfate interaction; such interactions are also observed in unrelated proteins (18, 19).
On the other side of the active site, I450 forms a 4.1-Å van der Waals interaction with the carbon atom of the diacylhydrazide moiety that is closest to the R1-pyrrolidine ring. Additionally, the nitrogen of the Q452 side chain interacts with that same end of the diacylhydrazide moiety, but with its oxygen atom. Overall, the diacylhydrazide moiety of WCK 5153 is significantly involved in active-site interactions; this likely explains the preference of WCK 5153 (and similar zidebactam) over WCK 4234 and avibactam to inhibit P. aeruginosa PBP2, culminating in their more potent antibacterial properties.
The terminal R1-group pyrrolidine ring of WCK 5153 makes relatively few interactions; one of its carbon atoms establishes a 3.9-Å hydrophobic van der Waals interaction with the main chain C carbon atom of N366 (Fig. 2B and Fig. S2). The tertiary carbon of this pyrrolidine ring forms a 3.7-Å van der Waals interaction with the hydroxyl oxygen atom of Y390. This apolar-polar interaction would typically constitute an unfavorable interaction of burying a hydrogen bond donor (i.e., the Y390 hydroxyl). However, this is likely not an unfavorable interaction as the Y390 hydroxyl group already forms a weak 3.4- to 3.5-Å hydrogen-bond interaction with the backbone oxygen of R365. Finally, R369 makes a weak 3.5-Å interaction with WCK 5153 (only in molecule A; Fig. 2B and Fig. S2).
Comparison with avibactam-bound E. coli PBP2 structure.
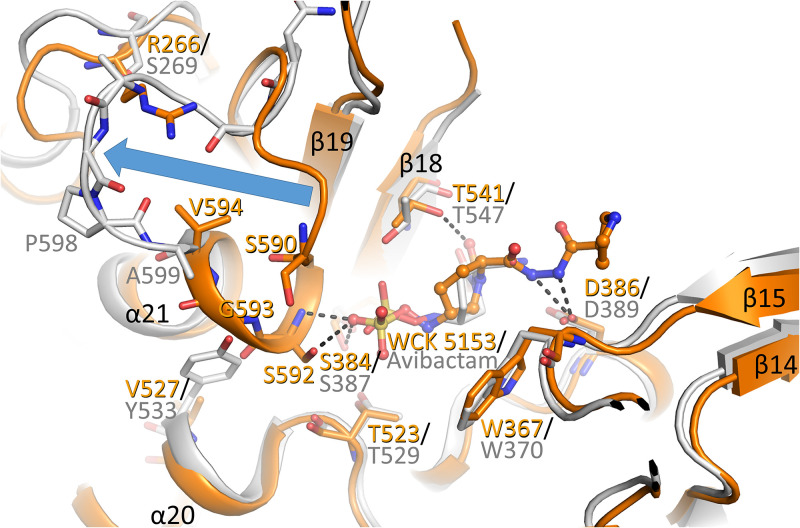
The P. aeruginosa PBP2 structure is similar to the E. coli PBP2 complex (Fig. 3). Also, avibactam bound to E. coli PBP2 makes similar active site interactions as WCK 5153 with P. aeruginosa PBP2. These ligand interaction similarities include the covalent bond with the catalytic serine, and the sulfate of avibactam hydrogen bonding with S545, which is similar to WCK 5153 interacting with the equivalent S539 in P. aeruginosa PBP2 (Fig. 2B). The sulfate moiety in avibactam also makes a hydrogen bond with E. coli PBP2 T547; this interaction is not observed with the equivalent T541 in the WCK 5153 P. aeruginosa complex as the latter side chain is oriented differently. Besides this minor difference, there is one other significant conformational difference near the sulfate binding region which involves the N terminus of helix α21 (Fig. 3). This first turn of the P. aeruginosa PBP2 N-terminal α-helix is “unwound” in the E. coli PBP2 structure and is repositioned away from the active-site region (Fig. 3). This particular α-helical turn (comprising P. aeruginosa PBP2 residues 591 to 594) provides interactions with the sulfate moiety of WCK 5153 via the hydrogen bond(s) involving residue S592 (Fig. 2 and 3 and Fig. S2). These interactions are not present with the sulfate moiety of avibactam when bound to E. coli PBP2 due to this conformational difference (Fig. 3). There are steric and other reasons for this conformational difference in E. coli PBP2 (P. aeruginosa residue numbering in parentheses): the larger Y533 (V527), the smaller S269 (R266), helix-breaking proline P598 (G593), and this helix-loop region also being one residue shorter in E. coli PBP2 (Fig. 3 and Fig. S1).
FIG 3.
Superpositioning of P. aeruginosa PBP2:WCK 5153 complex with the E. coli PBP2:avibactam complex structure. The P. aeruginosa PBP2 structure (orange) and the E. coli PBP2 structure (gray, PDB ID 6G9F) are shown with their respective ligands shown in ball-and-stick representation. The large conformational difference of the N-terminal part of the helix α21 and connecting loop in the P. aeruginosa structure from that in the E. coli PBP2 structure is depicted by the blue arrow.
This DBO sulfate binding region of PBPs is also the region that recognizes the carboxyl moiety in β-lactam antibiotics. The above-noted conformational differences between E. coli PBP2 and P. aeruginosa PBP2 could be, in part, the basis for why the latter PBP is more difficult to inhibit by β-lactam antibiotics: the IC50 values for ceftobiprole, ceftazidime, cefepime, and imipenem are between 8 and 15 times greater for P. aeruginosa PBP2 than for E. coli PBP2 (20). Other β-lactams have similar IC50 differences (21).
Comparison with WCK 5153 bound KPC-2 β-lactamase structure.
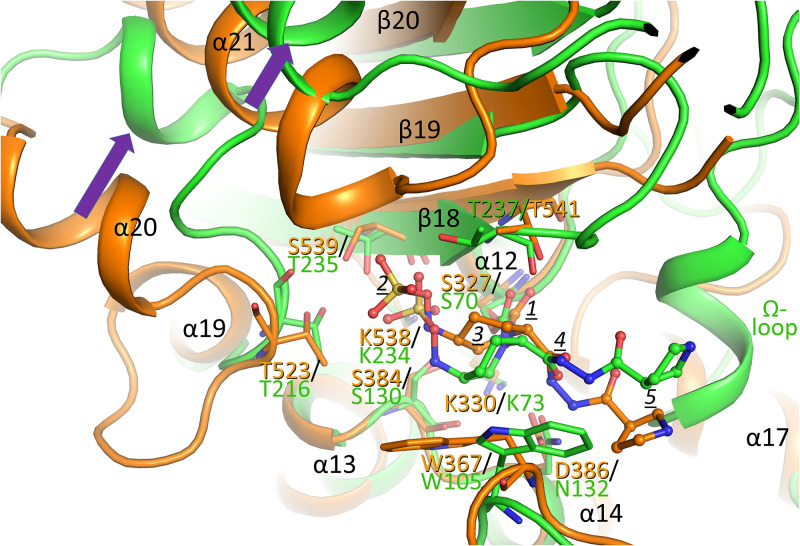
With the WCK 5153 bound PBP2 structure solved, a comparison is made to the same DBO when bound to KPC-2 β-lactamase (11). Superpositioning of the WCK 5153 bound structures of P. aeruginosa PBP2 and KPC-2 reveals several secondary-structure elements that align well. These elements include PBP2 β-strands β18, β19, and β20, the catalytic S327-containing α12 helix, the α13 and α14 helices, and the loop that connects these last two helices (Fig. 4). KPC-2 contains the same three conserved motifs as described above for PBPs. Residues in these motifs include the catalytic serine (S70) and the conserved active site lysine residues (K234 and K73 in KPC-2, which are analogous to K538 and K330, respectively, in PBP2). The active sites even contain a conserved tryptophan that provides hydrophobic interactions with the DBO, although it is rotated in a different position in KPC-2 (residues W367 in PBP2 and W105 in KPC-2; Fig. 4).
FIG 4.
Superpositioning of WCK 5153 inhibited complexes of PBP2 and KPC-2 β-lactamase. PBP2 is depicted in orange and KPC-2 in green. The WCK 5153 molecules are shown in a ball-and-stick model; equivalent key active-site residues of PBP2 and KPC-2 are labeled and shown in stick model. The Ω-loop present in KPC-2 is indicated; this loop is not present in PBPs. Conformational shifts between the α20 and α21 helices in PBP2 and their respective corresponding helices in KPC-2 are indicated by purple arrows. The moieties of WCK 5153 are labeled as in Fig. 2B. The active site Cα atoms of PBP2 residues 325 to 334 (α12 helix region containing catalytic S327), 381 to 389 (α13 region), 535 to 540 (β18), and 566 to 570 (β19) were superpositioned onto their equivalent atoms of KPC-2 β-lactamase (residues 68 to 77, 127 to 135, 231 to 236, and 244 to 248, respectively), yielding an RMSD of 0.87 Å for 30 Cα atoms.
Although KPC-2 also contains the conserved motif S-X-N/D, KPC-2 differs in that it possesses an asparagine at position 132, whereas PBP2 contains an aspartate at the structurally equivalent position 386 (Fig. 4). This difference is very relevant to WCK 5153 binding; in PBP2, the nitrogens of the diacylhydrazide donate hydrogen bonds to D386, whereas this interaction is not possible in KPC-2 since it has N132 at this position (the hydrogen bond accepting oxygen is now a hydrogen bond donating amide NH). This results in a key conformational difference in WCK 5153, which involves the diacylhydrazide oxygen near the label 4 in Fig. 4. In PBP2, the dihedral of the bond involving this diacylhydrazide oxygen atom orients the adjacent nitrogen to hydrogen bond with D386. In KPC-2 this equivalent residue is N132, which causes this diacylhydrazide moiety to reorient such that now the adjacent oxygen of the diacylhydrazide moiety interacts with N132. This conformational change suggests that WCK 5153 demonstrates structural plasticity in that the diacylhydrazide moiety can flip conformation to accommodate binding to both N132-containing KPC-2 and the D386-containing P. aeruginosa PBP2. The rest of WCK 5153 adopts a somewhat similar position and conformation in the active site in KPC-2, although the piperidine ring and attached sulfate moiety are shifted. This shift is perhaps due to the different orientations in the tryptophan (W105 in KPC-2; note that this residue can also adopt different conformations in KPC-2 [22]). Additionally, the end of the β18 strand also harbors a shift when comparing PBP2 and KPC-2 (KPC-2 residue T237, which is equivalent to T541 in PBP2). These residues are also close to the piperidine ring of the DBO. Overall, these DBO shifts cause the sulfate moiety in KPC-2 to make hydrogen bonds with S130, T235, and T237.
In contrast, the sulfate moiety of WCK 5153 in the PBP2 structure is shifted (more outward) toward the end of the α21 helix, forming two hydrogen-bond interactions with the N terminus of this α21 helix and one hydrogen bond with S539 (equivalent to T235 in KPC-2; Fig. 2B and Fig. 4). Both the PBP2 α20 and α21 helices also have corresponding helices in KPC-2, but those helices are shifted (Fig. 4, purple arrows). Remarkably, WCK 5153 is still able to make different yet substantial active site interactions involving its sulfate moiety despite these noted active site differences with KPC-2. As in PBP2, the pyrrolidine moiety of WCK 5153 is making few interactions in KPC-2 (Fig. 2B and Fig. 4) (11).
DSF analysis of WCK 5153 and zidebactam binding to PBP2.
To probe the effect of WCK 5153 and zidebactam binding on the thermal stability of P. aeruginosa PBP2, we carried out differential scanning fluorimetry (DSF)/thermal shift assay experiments. Both DBOs significantly increased the melting temperature (Tm) of PBP2, which increased from 46.1 ± 0.1°C, when in the absence of ligands, to 52.1 ± 0.1°C and 51.5 ± 0.1°C in the presence of 600 μM WCK 5153 or zidebactam, respectively (Fig. 5A). These values are almost identical at much lower DBO concentrations, 52.2 ± 0.0°C and 51.4 ± 0.1°C in the presence of 9.4 μM WCK 5153 or zidebactam, respectively, indicating their high affinity. These thermal shifts indicate that the binding of either of these DBOs stabilizes PBP2, contributing to their effective inhibition, which translates into their standalone antibacterial activity. These two DBO compounds differ only in the R1-group, where WCK 5153 contains a five-membered pyrrolidine ring and zidebactam a six-membered piperidine ring (Fig. 1A). The previously reported IC50 values of WCK 5153 and zidebactam for PBP2 are 0.14 and 0.26 μg/ml, respectively (13); these values follow the same trend in that WCK 5153 binding leads to a stronger PBP2 stabilization than for zidebactam binding. Nevertheless, it is important to note there is no clear correlation between change in protein stability and affinity of a ligand; the latter is dependent on the rates of the inhibition reaction steps whereas the former is dependent on the combined enthalpic and entropic effects when affecting the thermal stability of the covalent protein-inhibitor complex (23).
FIG 5.
Differential scanning fluorimetry (DSF) measurement of WCK 5153 and zidebactam binding to PBP2 and modeling of zidebactam. (A) DSF thermal shift assay of WCK 5153 and zidebactam binding to PBP2. The derivative of the change in fluorescence is plotted versus temperature. Experiments were performed in duplicate (a representative curve is depicted). (B) Modeling of zidebactam in P. aeruginosa PBP2 active site. The coordinates of zidebactam were obtained by transplanting most of the atom coordinates from the similar WCK 5153 when bound to PBP2, yet with the pyrrolidine ring being replaced by the piperidine ring of zidebactam using the zidebactam piperidine conformation when complexed to KPC-2 (11). This modeling and superpositioning were done using COOT. The electrostatic potential map of the PBP2 active site is shown as generated using APBS in PyMOL. Zidebactam in shown in ball-and-stick representation, and the individual moieties are labeled as in Fig. 2B with the noted change that 5 now represents the (larger) piperidine ring of the R1-group.
To complement the thermal shift data, we modeled zidebactam in the PBP2 active site using the WCK 5153 complex (Fig. 5B) as a starting model. The zidebactam modeling demonstrated a very similar binding mode as WCK 5153, which is not surprising due to their close chemical similarity. The R1-group piperidine ring is also quite solvent exposed, like the corresponding pyrrolidine ring in WCK 5153. This observation perhaps could be a potential reason for WCK 5153’s slightly higher PBP2 affinity (13) and increased PBP2 stability (Fig. 5B): this ring is overall relatively hydrophobic, except for the secondary amine, is larger, and exists in chair conformation in zidebactam versus the envelope form of pyrrolidine ring in WCK 5153. A larger, more hydrophobic solvent-exposed ring could be energetically less favorable for PBP2 binding and stabilization for zidebactam. Note that compared with zidebactam, WCK 5153 also yields lower MIC values against P. aeruginosa, both with and without a partnered β-lactam (11).
Probing DBO binding to P. aeruginosa PBP3.
Zidebactam and WCK 5153 do not inhibit P. aeruginosa PBP3 activity, probably because of their weak affinities (IC50 values for zidebactam and WCK 5153 are >4 μg/ml for PBP3 as determined using Bocillin-based competition assays with PBP-containing membrane preparations [13]). Comparison of previously reported P. aeruginosa PBP3 structures (24–28) and PBP2 structure (resolved in this study) revealed similarities (the active sites of P. aeruginosa PBP2 and PBP3 have 6/13 active site residues that are identical); hence, we hypothesized that DBO molecules can also bind to PBP3. Such insights could be useful for optimizing future DBOs that can drive activity by inhibiting multiple PBPs. Hence, we first grew P. aeruginosa apo-PBP3 crystals and subsequently incubated three DBOs (zidebactam, avibactam, and WCK 4234) with these apo-PBP3 crystals, and their structures were solved.
PBP3 shares a similar catalytic TP domain with PBP2, yet with a few differences. In PBP2, helices α10 and α11 in the TP domain are replaced in PBP3 by a short loop. Also, PBP3 has a somewhat smaller linker domain compared with PBP2 as the secondary structure elements α5 and η4 present in PBP2 are deleted and replaced with a short loop. P. aeruginosa PBP3 structures with three DBO compounds, avibactam, WCK 4234, and zidebactam, are resolved at 2.39, 2.08, and 2.50 Å, respectively. A representative PBP3 DBO complex with WCK 4234 structure is the highest resolution, and the best-resolved structure of the three is shown (Fig. 6). The overall structures of the DBO complexes are all very similar: the zidebactam and avibactam complexes superimpose onto the WCK 4234 PBP3 complex with RMSD values of 0.55 and 0.49 Å, respectively.
FIG 6.
Crystal structure of P. aeruginosa PBP3 in complex with WCK 4234. The noncatalytic N-terminal domain and catalytic TP domain are colored cyan and gray, respectively. Bound WCK 4234 is shown in spheres with green carbon atoms. Secondary structure elements near the active site are labeled (numbering as in reference 24).
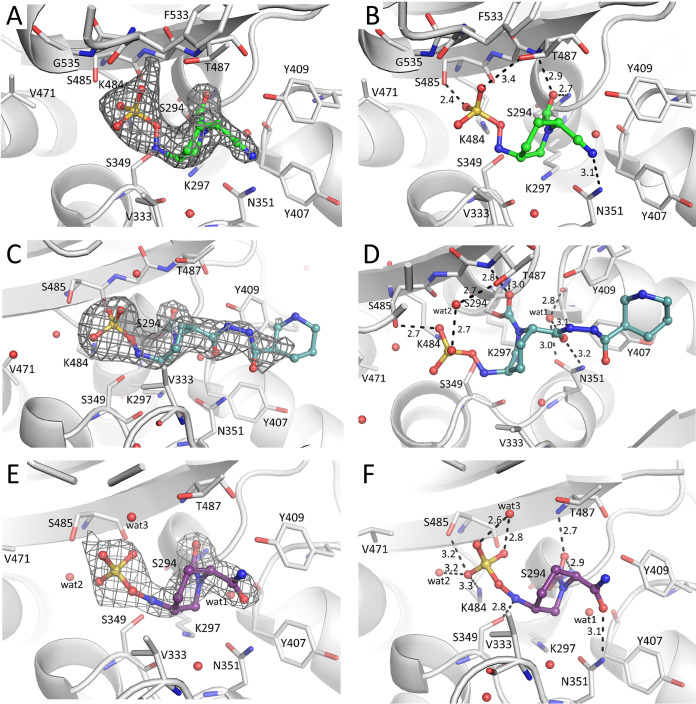
The PBP3 active site S-X-X-K motif (S294-T295-V296-K297) is located at the end of the α2 helix. The S-X-N/D motif (S349-S350-N351) is located between helices α4 and α5, and the third motif, K-T/S-G-T (K484-S485-G486-T487), is part of the β3 strand (Fig. 6 and 7). In all these three structures, several flexible loop segments (residue ranges 197 to 207, 490 to 500, and 528 to 534) could not be modeled; in particular, the last loop located at the beginning of α11 helix (containing residues Y532 and F533) is known to undergo various conformational changes during ligand binding and catalysis.
FIG 7.
Crystal structures of WCK 4234, zidebactam, and avibactam bound to P. aeruginosa PBP3. (A, C, and E) Electron difference density for WCK 4234, zidebactam, and avibactam, respectively, contoured at 2.75 σ level. (B, D, and F) Interactions of DBO ligands in the active site of PBP3 for WCK 4234, zidebactam, and avibactam, respectively.
DBO interactions in PBP3 active sites.
Zidebactam, WCK 4234, and avibactam are covalently bound to the catalytic S294 (Fig. 7). The carbonyl oxygen of each of the DBOs is situated in the oxyanion hole formed by the backbone nitrogens of S294 and T487 (Fig. 7). The sulfate moiety hydrogen bonds with S485 and, depending on the DBO, also with either S349, K484, or T487 (Fig. 7). The R1-group moieties of the DBOs were well defined: for WCK 4234, its nitrile moiety, and for avibactam, its amide moiety. Both these moieties form hydrogen bonds with N351 (Fig. 7B and F). In the zidebactam complex, N351 interacts with the diacylhydrazide oxygen atom closest to the piperidine scaffold of the DBO (Fig. 7D). This zidebactam oxygen also makes indirect active site interactions via hydrogen bonding to a water molecule (wat1). For zidebactam, electron density was present for the diacylhydrazide moiety and the terminal piperidine moiety of the R1-group of zidebactam, but mainly for the tertiary carbon atom (Fig. 7C). This tertiary carbon atom of the R1-group piperidine establishes hydrophobic interactions with Y409 (3.8-Å distance) and is at a larger distance from Y407 (4.8 Å). The carbon atom of the diacylhydrazide moiety connected to this tertiary carbon atom of the R1-group piperidine also makes hydrophobic interactions with Y409 (4.3 Å). Overall, the hydrogen bond interaction with N351 is conserved in all three DBO complexes, yet the moiety making this interaction varies depending on the R1-group of the DBO.
Comparison of PBP3:zidebactam complex with PBP2:WCK 5153 complex.
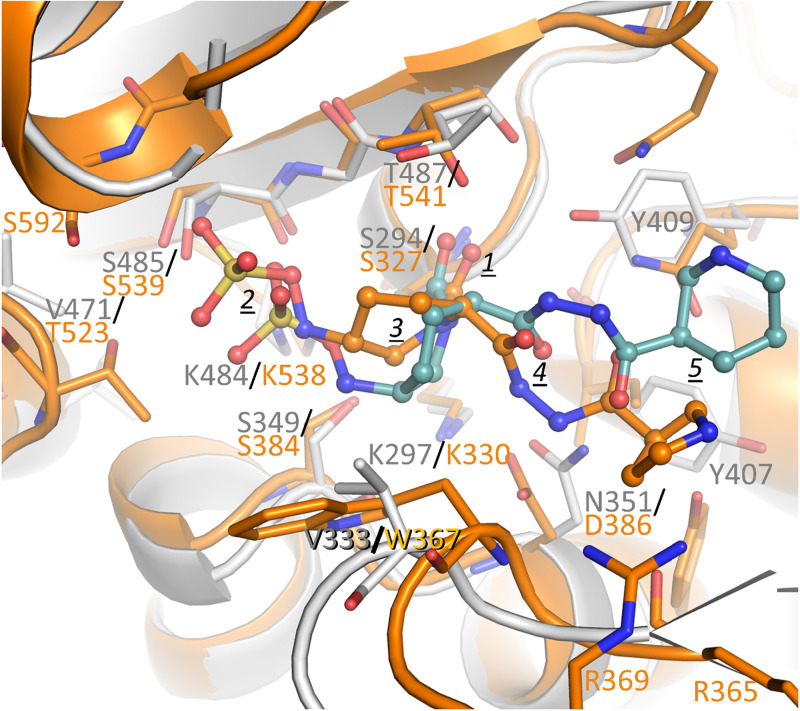
The zidebactam PBP3 structure was compared against WCK 5153 complexed to PBP2 (Fig. 8). Their carbonyl oxygen atoms are both situated in their respective PBP oxyanion holes. In contrast, the main DBO piperidine ring and attached sulfate moiety are shifted such that the sulfate moiety in the PBP2 complex is positioned 1.9 Å further away from the catalytic serine. These shifts are perhaps due to the sulfate of WCK 5153 needing to reach the N-terminal part of the α21 helix in PBP2 to make hydrogen bonding and helix-dipole interactions (Fig. 2B and Fig. 8). Additionally, the larger W367 residue in PBP2, which is the smaller V333 in PBP3, might contribute to this difference.
FIG 8.
Superposition of WCK 5153 inhibited complex of PBP2 with zidebactam complexed to PBP3. PBP2 and bound WCK 5153 are depicted in orange, and PBP3 is in gray with its bound zidebactam ligand in teal carbon atoms. The moieties of WCK 5153 and zidebactam are labeled as in Fig. 2B. The active site Cα atoms of PBP2 residues 325 to 333 (α12 helix region containing catalytic S327), 364 to 367 (includes β15), 378 to 387 (α13 region), 447 to 452 (α17), and 537 to 542 (β18) were superpositioned onto their equivalent atoms in PBP3 (residues 292 to 300, 330 to 333, 343 to 352, 404 to 409, and 483 to 488, respectively), yielding an RMSD of 0.81 Å for 35 Cα atoms.
The PBP2 and PBP3 active sites contain some notable differences. In the S-X-D/N motif in PBP3, this last residue is N351, whereas in PBP2 this residue is D386. Despite this difference, these residues both interact with the diacylhydrazide of the DBOs. The DBOs can accomplish this by reorienting the diacylhydrazide moiety in PBP3 analogous to when bound to the N132-containing KPC-2 (see above). A second important active site difference is that PBP2 contains a large hydrophobic W367 that interacts with the hydrophobic face of the main piperidine ring of WCK 5153, whereas this residue is V333 in PBP3. This interaction could, in part, explain the WCK 5153 (and similar zidebactam) higher affinity for PBP2 than for PBP3. The terminal 5- or 6-membered ring of the R1-groups of WCK 5153 and zidebactam, respectively, makes only limited interactions as described above.
Comparison of PBP3:DBO with KPC-2:DBO complexes.
Similar to the PBP2:WCK 5153 complex above, we superimposed the zidebactam-bound complexes of PBP3 and KPC-2 β-lactamase, a DBO complex structure determined previously (11) (Fig. S3). The superposition shows that the main DBO piperidine ring and attached sulfate moiety superimpose quite well (sulfate moieties are at a distance of 0.6 Å). Both PBP3 and KPC-2 share an asparagine at the S-X-N/D motif. The diacylhydrazide moiety of zidebactam is therefore oriented in a similar fashion in both structures with the oxygen interacting with the nitrogen atom of N351 in PBP2 and N132 in KPC-2 (Fig. S3). An active site difference involves KPC-2 having a large aromatic tryptophan at position 105 corresponding to the equivalent, yet smaller, V333 in PBP3; this makes the KPC-2 active site somewhat like PBP2 although the conformations of their respective tryptophans are different (Fig. 4 and 8 and Fig. S3).
Superposition of zidebactam bound complexes of P. aeruginosa PBP3 and KPC-2 β-lactamase. PBP3 is shown in gray, and its bound zidebactam ligand is in teal carbon atoms; KPC-2 and its bound zidebactam are colored green. The active site Cα atoms of PBP3 residues 294 to 301, 346 to 353, 481 to 486, and 505 to 509 were superpositioned onto their equivalent atoms in KPC-2 (residues 70 to 77, 127 to 134, 231 to 236, and 244 to 248), yielding an RMSD of 0.99 Å for 27 Cα atoms. Download FIG S3, TIF file, 1.6 MB (1.6MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
An additional difference compared to the active site of KPC-2 is in the β-strand that contains the K-T/S-G-T motif. The last residue of this motif (T487 in PBP3 and T237 in KPC-2) is bent more inward toward the active site crevice for KPC-2 compared with PBP3 (Fig. S3). The terminal piperidine of the R1-group of zidebactam in both structures makes only minimal interactions. An additional difference encompasses the Ω-loop in KPC-2, which is absent in the PBP3 and other PBPs. This Ω-loop contains E166 and N170 needed for deacylation of bound ligands, which allows the β-lactamase to hydrolyze β-lactam substrates. Finally, the two helices that shifted when comparing P. aeruginosa PBP2 and E. coli PBP2 (Fig. 4) are also repositioned in KPC-2 compared with P. aeruginosa PBP3 (Fig. S3, purple arrows).
Regarding the other DBOs, the hydrogen bonds of the nitrile and amide moiety of WCK 4234 and avibactam, respectively, with N351 when bound to PBP3 are conserved in KPC-2 via their interaction with N132 (11).
Comparison with other PBP3 inhibitor complexes.
Several conformational changes often observed in β-lactam binding to P. aeruginosa PBP3 (24–28) are not observed in the DBO complexes. In most previously determined PBP3 complexes, an aromatic wall is formed with residues Y532, F533, and also Y503 moving close to the hydrophobic parts of the ligand (24). Figure S4A shows this for the carbenicillin PBP3 complex. The F533 side chain often approaches the hydrophobic V333 residue on the other side of the active site to form a hydrophobic bridge (24). The phenyl moiety of carbenicillin (b) and its dimethyl groups (c) attract these aromatic wall residues. In the ceftazidime complex, the dimethyl groups of the 2-carboxypropan-2-yl moiety (b) likely also contribute to the hydrophobic attraction of forming the aromatic wall (Fig. S4B). Note that Y532 and F533 could be resolved only in the higher-resolution WCK 4234 PBP3 complex; these residues are too disordered to be modeled in the zidebactam and avibactam PBP3 complexes. A possible explanation for why the DBO inhibitor complexes do not show this aromatic wall conformation is as follows. The DBO inhibitors have only two hydrophobic carbon atoms that are part of the main DBO piperidine ring that are solvent exposed; this is likely not enough for the aromatic wall to make these hydrophobic interactions as seen in other PBP3:inhibitor complexes.
Active site views of superpositions of PBP3:WCK 4234 complex with β-lactam complexes of PBP3 and apo-PBP3. (A) Superposition with the PBP3:carbenicillin complex (PDB ID 3OCL [25]; shown in salmon color). Important secondary structure elements β3, β4, and α11 and relevant active site residues are labeled. All residues labeled are shown in black labels except for Y532 and F533 in the PBP3:WCK 4234 complex, which are in gray as this region adopts a different conformation. Three moieties of carbenicillin are labeled: amide (a), phenyl (b), and dimethyl substituents of the thiazolidine ring (c). (B) Superposition with the PBP3:ceftazidime complex (PDB ID 3PBO [24]). Four moieties of ceftazidime are labeled: amide (a), dimethyl groups of 2-carboxypropan-2-yl group (b), methyl substituent of thiazine ring (c) (methyl group is left after pyridine elimination, see reference 29), and aminothiazole ring (d). (C) Superposition with the apo-PBP3 (PDB ID 6HZR [26]). The view is enlarged from panels A and B as the residue F533 position in the apo-PBP3 structure is more distant from the active site. (D) Superposition with the PBP3:meropenem complex (PDB ID 3PBR [24]). One atom of meropenem is labeled, that of the sulfur atom attached to the pyrroline ring of meropenem (c). Download FIG S4, TIF file, 2.5 MB (2.5MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
A second conformational change often observed in PBP3:inhibitor complexes involves the bending of the β-strands β3 and β4 toward the active site. These β-strand shifts allow hydrogen bonding interactions with the amide moiety (a) of ligands like carbenicillin and ceftazidime (24, 25) (Fig. S4A and B). In the latter case, these shifts also allow the formation of hydrogen bonding interactions with aminothiazole moieties in inhibitors such as ceftazidime (d in Fig. S4B). This amide moiety is also a key moiety in the d-Ala-d-Ala end of the peptidoglycan substrate of PBPs. Such a β-lactam-induced conformational change is also observed in N. gonorrhoeae PBP2 (16).
Third, PBP inhibitors that have an aminothiazole ring, such as ceftazidime, are accommodated in the active site by having Y409 swing out to create a pocket and also to provide stacking interactions with this aminothiazole moiety (d in Fig. S4B) (24, 25).
The PBP3:DBO protein conformation is structurally most similar to that of the apo-PBP3 structure (Fig. S4C) as well as the meropenem bound PBP3 structure (Fig. S4D). The β3 and β4 strands are in the same position in all these structures. This similarity is likely a consequence of either an absence of an amide moiety or the situation that, when present in WCK 4234, its amide is not correctly positioned for such a β3 strand interaction. Despite this similarity, the meropenem bound structure does show a partial formation of the aromatic wall via the shift of only F533 interacting with the hydrophobic parts of the sulfur-containing substituent of meropenem (c) (Fig. S4D). Note that even though the tested DBOs bind to P. aeruginosa PBP3, the binding is not of sufficiently high affinity to be able to contribute in antibacterial activity. Avibactam and WCK 4234 also do not possess activity against P. aeruginosa.
DSF experiments of DBO binding to PBP3.
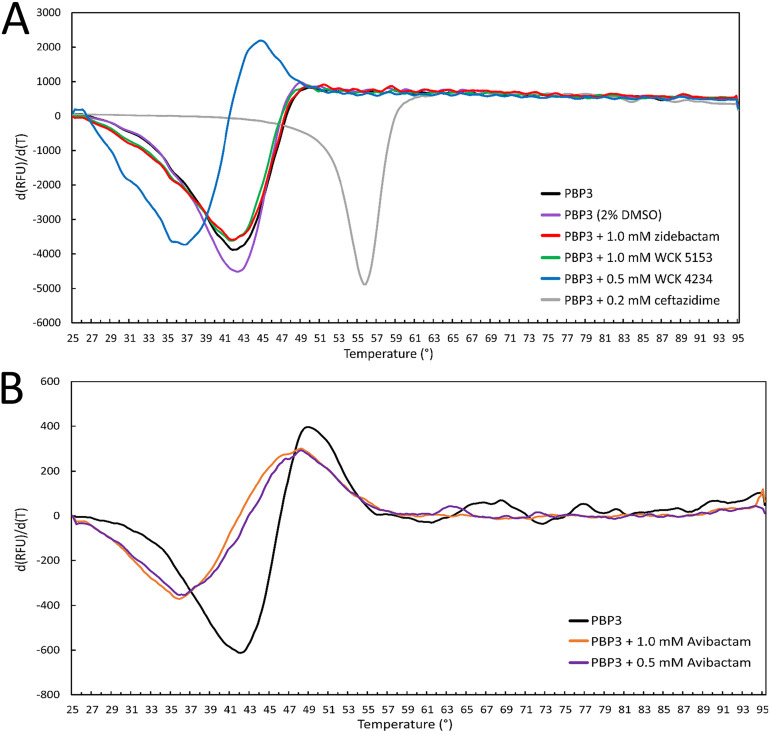
In contrast to the increase in thermal stability observed with PBP2, zidebactam and WCK 5153, each at 1 mM, lowered the apo-PBP3 Tm of 42.8 ± 0.3°C to 41.9 ± 0.1°C (Fig. 9). WCK 4234 binding decreased the Tm even more significantly, to 37.0°C. Avibactam at 1.0 and 0.5 mM also decreased the Tm of PBP3 to 35.5 ± 0.4 and 35.9 ± 0.1°C, respectively (Fig. 9). As a positive control for comparison, ceftazidime binding to PBP3 led to a large increase in Tm, as observed previously (Fig. 9) (24, 25, 29).
FIG 9.
DSF thermal shift measurement of DBOs binding to PBP3. (A) Zidebactam, WCK 5153, and WCK 4234 binding to PBP3 at the indicated concentrations. The compounds were dissolved in DMSO with a final DMSO concentration of 2% in the DSF experiment; a PBP3 2% DMSO control experiment is therefore also included. For comparison, ceftazidime is a positive control since it showed a large increase in Tm as previously published (29). Experiments were done in duplicate, and a representative curve is shown for each experiment. (B) Avibactam binding to PBP3. Data are plotted similarly as in Fig. 5. Avibactam was dissolved in water. Experiments were done in triplicate, and a representative curve is shown for each experiment.
The decreasing effects on the Tm of PBP3 by DBO binding are likely due to the absence of hydrophobic moieties on the piperidine ring DBO scaffolds or R1-groups that attract the aromatic wall to interact with the ligand. For example, carbenicillin binding also leads to a substantial increase in the Tm of PBP3 by 13.2°C (25). The decrease in Tm of avibactam and WCK 4234 binding to PBP3 is similar to that obtained for meropenem and imipenem with a reduction in Tm of 4 and 6°C, respectively (24). Meropenem also does not induce the full aromatic wall interactions (Fig. S4D), and imipenem has a very different conformation of this Y532/F533 region (24). That zidebactam/WCK 5153 binding led to a minor negative effect on Tm compared with the more destabilizing effects of WCK 4234 and avibactam can be explained by analyzing the similarities and differences of their binding modes. First, the similarities entail that all three DBOs form a hydrogen bond with N351, and their DBO sulfate active site interactions are also quite similar (Fig. 7). The one key difference that could explain the Tm differences is that zidebactam makes additional stabilizing hydrophobic interactions via its diacylhydrazide moiety and adjacent tertiary carbon atom of the piperidine moiety with mainly Y409 (Fig. 7). We anticipate that the corresponding similar pyrrolidine ring in WCK 5153 is making similar interactions. The much smaller DBOs WCK 4234 and avibactam cannot make such stabilizing hydrophobic interactions. Note that, as mentioned above, there is no clear correlation between change in protein stability and affinity of a ligand. For example, meropenem and ceftazidime have very different, even opposite effects on the Tm of PBP3, yet their 50% effective concentration (EC50) values of PBP3 binding are nearly identical (24).
The DSF results aid in understanding important differences of DBO binding to PBP2 and PBP3. For P. aeruginosa PBP2 binding of either WCK 5153 or zidebactam caused an upward Tm shift of 5 to 6°C with DBO concentrations as low as 9.4 μM, indicating effective inhibition of PBP2. However, these DBOs did not show a stabilizing effect when binding to PBP3. Although it is not a necessity to have a stabilizing effect for efficient PBP3 binding, most potent PBP3 inhibitors do achieve that.
Conclusion.
In summary, we present co-crystallized and modeled structures of WCK 5153 and zidebactam with P. aeruginosa PBP2, respectively. The PBP2 structure represents the first atomic-level analysis for this P. aeruginosa PBP and provides possible explanations for this PBP’s decreased susceptibility to be inhibited by β-lactam antibiotics compared to its E. coli equivalent PBP. For comparison, we also determined crystal structures of DBOs zidebactam, WCK 4234, and avibactam in complex with P. aeruginosa PBP3 to explain the DBOs’ weaker affinity for this PBP. We show that in addition to the covalent bond, WCK 5153 binds PBP2 via (a) interactions of the carbonyl oxygen with the oxyanion hole, (b) interactions with S539 and the C-terminal end of helix α21 by the sulfate moiety forming a helix-dipole interaction, (c) interactions by the diacylhydrazide moiety with D386 and Q452, (d) hydrophobic interactions of the pyrrolidine ring, and (e) interactions of the hydrophobic part of the piperidine ring with W367. Upon binding, the DBOs stabilize PBP2, yet destabilize PBP3, as measured via thermal shift assays. Structural comparisons indicate that a strong contributor to WCK 5153 and zidebactam preferentially binding to PBP2 and KPC-2 is the presence of a tryptophan residue to provide hydrophobic interactions; this residue is nonaromatic in PBP3. For zidebactam and WCK 5153, our results show that the diacylhydrazide moiety can adopt different orientations to interact as either a hydrogen acceptor with asparagine (in PBP3 and KPC-2) or a donor with aspartate (in PBP2) in the conserved S-X-N/D motif. This diacylhydrazide moiety of zidebactam and WCK 5153 is relatively more involved in active site interactions than DBOs with smaller side chains like nitrile in WCK 4234 and amide in avibactam. These diacylhydrazide interactions combined with the modest hydrophobic interactions of the R1-group piperidine/pyrrolidine likely explain the preference of zidebactam and WCK 5153 over WCK 4234 and avibactam to inhibit P. aeruginosa PBP2 effectively and exhibit potent antibacterial properties.
These molecular insights and the ability of DBOs to be “dual-target” inhibitors could be utilized for optimization efforts to develop novel potent β-lactamase-resistant non-β-lactam DBOs with the ability to bind and functionally inhibit multiple PBPs.
MATERIALS AND METHODS
Protein expression, purification, and crystallization. (i) PBP2.
P. aeruginosa PBP2 (39 to 646 aa) lacking the N-terminal membrane anchoring region (residues 1 to 38) was subcloned into the NdeI cleavage site of the pET28a vector to add an N-terminal histidine tag. The PBP2 plasmid was transformed into LOBSTR RIL competent cells (Kerafast). Six- to 8-liter cultures were grown in terrific broth medium with kanamycin 50 μg/ml and induced at 0.6 to 0.8 OD600 using 50 μM IPTG while shaking for 14 to 16 h at 18°C. After pelleting by centrifugation, the cells were suspended in lysis buffer (50 mM Tris, pH 8.0, 300 mM NaCl, 10% glycerol, and 10 mM imidazole). The lysis buffer was supplemented with one cOmplete EDTA-free protease inhibitor tablet (Sigma-Aldrich) and 1,000 units of Benzonase (ACROBiosystems) and 1 mM MgCl2. Cells were lysed using an Avestin Emulsiflex B-15 apparatus at 4°C, and the cell suspension was centrifuged at 30,597 × g to remove cell debris. The supernatant was incubated with freshly washed Ni-NTA beads and rocked for 2 h at 4°C. The Ni-NTA beads were washed with 50 mM Tris, pH 8.0, 300 mM NaCl, 10% glycerol, 25 mM imidazole buffer and then eluted with the same buffer containing 250 mM imidazole. The elution fractions were dialyzed overnight against 3 liters of 50 mM Tris, pH 8.0, 300 mM NaCl, and 10% glycerol buffer. After centrifugation at 13,000 rpm to remove possible precipitation, a second round of His-tag purification was carried out using a 5-ml Ni-NTA HisTrap column (GE Life Sciences). After loading the PBP2 protein, the HisTrap column was washed with 50 mM Tris, pH 8.0, 300 mM NaCl, 10% glycerol, and 20 mM imidazole buffer. The protein was eluted using an imidazole gradient of 20 to 250 mM. The purity of the fractions was checked using SDS-PAGE gel analysis, and the corresponding fractions were pooled and dialyzed against the above-described dialysis buffer. WCK 5153 was subsequently added at a 1:50 protein/compound molar ratio, and the inhibited PBP2 was concentrated to 2.9 mg/ml prior to cocrystallization. The cocrystallization trials with DBO compounds resulted in crystallization hits in MCSG crystallization screens (Anatrace) using the sitting drop method at 20°C. The best crystals were grown in 0.1 M HEPES, pH 7.5, 0.2 M ammonium acetate, and 25% polyethylene glycol (PEG) 3350. The crystal was cryoprotected using perfluoropolyether (Hampton Research) prior to freezing the crystal in liquid nitrogen for data collection.
(ii) PBP3.
The P. aeruginosa PBP3 protein expression and purification were carried out as described earlier (24, 29, 30). PBP3 crystals were grown via the sitting drop method using previously published conditions: 30% polyethylene glycol 4000, 0.2 M MgCl2, 0.1 M Tris, pH 8.5 (24). Apo-PBP3 crystals were used for soaking the individual DBO inhibitors in mother liquor as follows. Crystals were soaked with 2.5 mM zidebactam for 23 h, or with 3 mM either WCK 4234 or avibactam for 1 h prior to freezing the crystals in liquid nitrogen.
Data collection and refinement. (i) PBP2.
A needle-shaped crystal of the cocrystallized PBP2 and WCK 5153 complex was used for data collection at the microfocus FMX 17-ID-2 beamline at NSLS-II, and diffraction data were processed using XDS (31) to 2.87-Å resolution (Table 1). The PBP2 structure was solved via molecular replacement using PHASER (32) with the TP domain of E. coli PBP2 structure (14) (PDB ID 6G9F) as a search model. The solution yielded two molecules in the asymmetric unit. The crystallographic refinement was carried out using REFMAC (33) and PHENIX (34), and model building was done using COOT (35). Noncrystallographic symmetry restraints were applied during refinement in PHENIX. Refinement parameter files for WCK 5153 were generated using eLBOW (36). The final structure yielded an R-factor/Rfree of 25.9/31.7% and contained two molecules in the asymmetric unit, each containing a covalently bound WCK 5153. In addition, there are 20 water molecules and one chloride ion. PBP2 chain A contains residues 60 to 87, 162 to 222, 238 to 544, and 561 to 622; chain B contains residues 63 to 86, 164 to 183, 189 to 219, 240 to 544, and 562 to 621. This refined PBP2 structure encompasses most of the construct and includes the anchor, linker, and TP domains. However, the head domain (residues 88 to 161) is not included as it could not be modeled confidently for either monomer due to poor electron density likely due to disorder. The two non-crystallographically related PBP2 molecules are very similar to each other (0.69-Å root mean square deviation for 440 Cα atoms); the 2 monomers in the asymmetric unit do not represent a dimer as no relevant interfaces were observed using PISA (37).
TABLE 1.
Data collection and refinement statistics for crystal structures of PBP2 and PBP3 DBO complexes
| Data collection | PBP2 WCK 5153 | PBP3 WCK 4234 | PBP3 avibactam | PBP3 zidebactam |
|---|---|---|---|---|
| Wavelength (Å) | 0.979331 | 0.97928 | 0.97928 | 0.97946 |
| Resolution range (Å)a | 2.87–29.67 (2.87–3.00) | 2.09–29.25 (2.09–2.14) | 2.39–29.58 (2.39–2.45) | 2.50–25.0 (2.50–2.59) |
| Space group | P21 | P212121 | P212121 | P212121 |
| Unit cell (Å,°) | 82.0, 75.9, 97.3, 90, 106.7, 90 | 66.9, 78.9, 87.2, 90, 90, 90 | 67.9, 80.1, 87.8, 90, 90, 90 | 68.4, 80.0, 88.1, 90, 90, 90 |
| Completeness (%) | 90.8 (86.1) | 99.4 (94.3) | 99.3 (93.4) | 99.9 (100.0) |
| No. of unique reflections | 23,998 (3,298) | 27,862 (1,915) | 19,459 (1,323) | 17,225 (1,711) |
| Total no. of observations | 81,726 | 183,701 | 122,435 | 207,103 |
| Multiplicity | 3.4 (3.3) | 6.6 (6.2) | 6.3 (6.2) | 12.0 (12.8) |
| Mean I/σ(I) | 5.2 (2.0) | 8.2 (1.7) | 10.9 (2.3) | 22.3 (4.2) |
| Mean CC(1/2) (%) | 97.8 (76.5) | 99.4 (65.8) | 99.3 (73.3) | 99.7 (94.2) |
| R merge | 0.158 (0.514) | 0.134 (0.920) | 0.196 (0.885) | 0.182 (1.21) |
| Refinement | ||||
| Resolution range refinement (Å) | 2.87–29.67 | 2.09–29.25 | 2.39–29.58 | 2.50–24.94 |
| R work | 0.2580 | 0.1951 | 0.1930 | 0.1996 |
| R free | 0.3165 | 0.2421 | 0.2577 | 0.2719 |
| Ligands | 2 ligands, 1 chloride ion | 1 ligand | 1 ligand | 1 ligand |
| No. of water molecules | 20 | 84 | 114 | 71 |
| RMSD bond lengths (Å) | 0.003 | 0.007 | 0.008 | 0.007 |
| RMSD Bond angles (°) | 0.724 | 0.896 | 1.004 | 1.372 |
| B-factors protein (Å2) | 42.8 | 37.9 | 35.0 | 50.2 |
| B-factors ligands (Å2) | 44.6 | 39.1 | 40.2 | 74.8 |
| Ramachandran favored (%) | 95.6 | 96.8 | 96.0 | 95.4 |
| Ramachandran outliers (%) | 0.57 | 0.43 | 0.0 | 0.22 |
Values in parentheses are for the highest-resolution shell.
(ii) PBP3.
Avibactam- and WCK 4234-soaked PBP3 crystals were used for data collection at the AMX 17-ID-1 beamline at NSLS-II and processed using XDS (Table 1). Data for the avibactam and WCK 4234 complexes were diffracted to 2.39- and 2.09-Å resolution, respectively. Data for the zidebactam-soaked PBP3 crystal were collected at SSRL beamline 12-2, and the data were processed to 2.5-Å resolution using HKL-3000 (38) (Table 1). The three DBO PBP3 complex structures were solved by molecular replacement using PHASER with the PBP3-imipenem complex protein coordinates as the search model (PDB ID 3PBQ) (24). COOT was used for model building, and REFMAC and PHENIX were used for crystallographic refinement. The WCK 4234 complex contains PBP3 residues 56 to 196, 204 to 272, 276 to 467, 471 to 490, 503 to 527, and 532 to 559. The avibactam complex contains PBP3 residues 57 to 190, 213 to 489, 502 to 527, and 536 to 560. The zidebactam complex contains PBP3 residues 57 to 191, 211 to 489, 501 to 527, and 535 to 559. The refinement parameter files for the DBO ligands were generated using eLBOW. The refined R-factor/Rfree values are listed in Table 1. The refined structures yielded >94% of the residues in the favorable region of the Ramachandran plot as calculated using MolProbity (39) (Table 1).
Molecular figures were generated using PyMOL (www.pymol.org). Coordinates and structure factors of the P. aeruginosa PBP2 WCK 5153 complex and the PBP3 complexes with WCK 4234, avibactam, and zidebactam were deposited in PDB.
DSF experiments.
The DSF thermal shift assay was conducted on a CFX96-C1000-Touch thermocycler (Bio-Rad) similarly to what we previously described (29, 30). The reactions were carried out in the buffer containing 50 mM HEPES, pH 8.0, 300 mM NaCl, 10% glycerol at a sample volume of 30 μl, with 1.4 μM PBP2 either with or without WCK 5153 and zidebactam compounds dissolved in dimethyl sulfoxide (DMSO). Equivalent DMSO controls were also included, and reactions were run in duplicates. All the reaction mixtures included SYPRO orange dye (Fisher Scientific) at 10× concentration, and the thermal scan was conducted between 25 and 70°C with 0.2°C/min intervals.
For PBP3 (3 μM), the DSF experiments were conducted for all four DBO compounds (zidebactam, WCK 5153, WCK 4234, and avibactam) at the indicated concentrations. The first three DBOs were dissolved in DMSO, and DSF experiments were carried out in duplicate. Avibactam was dissolved in water, and DSF experiments with avibactam-containing mixtures were carried out in triplicate. Ceftazidime (at 200 μM) was used as a positive control similarly to what was previously described (29, 30). The reaction buffer contained 14 mM Tris, pH 8.0, 280 mM NaCl, 7% glycerol, and 10× SYPRO orange.
Data availability.
Coordinates and structure factors of the P. aeruginosa PBP2 WCK 5153 complex and the PBP3 complexes with WCK 4234, avibactam, and zidebactam were deposited in the Worldwide Protein Data Bank (wwPDB) with accession numbers 7KIS, 7KIT, 7KIV, and 7KIW, respectively.
ACKNOWLEDGMENTS
This study was funded by a grant from Wockhardt (to R.A.B. and F.V.D.A.). Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (NIH) to R.A.B. under award numbers R01AI100560, R01AI063517, and R01AI072219 and has been funded in whole or part with federal funds from the National Institute of Allergy and Infectious Diseases of the NIH, Department of Health and Human Services, under award number U19AI110819. This study was also supported in part by funds and/or facilities provided by the Cleveland Department of Veterans Affairs, award number 1I01BX001974 to R.A.B. from the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development, and the Geriatric Research Education and Clinical Center VISN 10. This research used the FMX and AMX beamlines of the National Synchrotron Light Source II, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Brookhaven National Laboratory under contract no. DE-SC0012704. The Center for BioMolecular Structure (CBMS) is primarily supported by the National Institutes of Health, National Institute of General Medical Sciences (NIGMS), through a Center Core P30 Grant (P30GM133893), and by the DOE Office of Biological and Environmental Research (KP1605010). Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under contract no. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (P41GM103393).
The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH or the Department of Veterans Affairs.
We thank Dr. Vivien Yee for data collection at NSLS as well as thank beamline support at NSLS and SSRL for their help with data collection. Zidebactam, WCK 5153, and WCK 4234 compounds were obtained from Wockhardt.
Footnotes
Citation Rajavel M, Kumar V, Nguyen H, Wyatt J, Marshall SH, Papp-Wallace KM, Deshpande P, Bhavsar S, Yeole R, Bhagwat S, Patel M, Bonomo RA, van den Akker F. 2021. Structural characterization of diazabicyclooctane β-lactam “enhancers” in complex with penicillin-binding proteins PBP2 and PBP3 of Pseudomonas aeruginosa. mBio 12:e03058-20. https://doi.org/10.1128/mBio.03058-20.
Contributor Information
Focco van den Akker, Email: focco.vandenakker@case.edu.
Gerard D. Wright, McMaster University
REFERENCES
- 1.Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, Ouellette M, Outterson K, Patel J, Cavaleri M, Cox EM, Houchens CR, Grayson ML, Hansen P, Singh N, Theuretzbacher U, Magrini N, WHO Pathogens Priority List Working Group. 2018. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 2.Mullane EM, Avery LM, Nicolau DP. 2019. Comparative evaluation of the in vitro activities of WCK 5222 (cefepime-zidebactam) and combination antibiotic therapies against carbapenem-resistant Pseudomonas aeruginosa. Antimicrob Agents Chemother 64:e01669-20. doi: 10.1128/AAC.01669-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Livermore DM, Mushtaq S, Warner M, Vickers A, Woodford N. 2017. In vitro activity of cefepime/zidebactam (WCK 5222) against Gram-negative bacteria. J Antimicrob Chemother 72:1373–1385. doi: 10.1093/jac/dkw593. [DOI] [PubMed] [Google Scholar]
- 4.Sader HS, Castanheira M, Huband M, Jones RN, Flamm RK. 2017. WCK 5222 (cefepime-zidebactam) antimicrobial activity against clinical isolates of Gram-negative bacteria collected worldwide in 2015. Antimicrob Agents Chemother 61:e00072-17. doi: 10.1128/AAC.00072-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sader HS, Rhomberg PR, Flamm RK, Jones RN, Castanheira M. 2017. WCK 5222 (cefepime/zidebactam) antimicrobial activity tested against Gram-negative organisms producing clinically relevant beta-lactamases. J Antimicrob Chemother 72:1696–1703. doi: 10.1093/jac/dkx050. [DOI] [PubMed] [Google Scholar]
- 6.Moya B, Bhagwat S, Cabot G, Bou G, Patel M, Oliver A. 2020. Effective inhibition of PBPs by cefepime and zidebactam in the presence of VIM-1 drives potent bactericidal activity against MBL-expressing Pseudomonas aeruginosa. J Antimicrob Chemother 75:1474–1478. doi: 10.1093/jac/dkaa036. [DOI] [PubMed] [Google Scholar]
- 7.Monogue ML, Tabor-Rennie J, Abdelraouf K, Nicolau DP. 2019. In vivo efficacy of WCK 5222 (cefepime-zidebactam) against multidrug-resistant Pseudomonas aeruginosa in the neutropenic murine thigh infection model. Antimicrob Agents Chemother 63:e00233-19. doi: 10.1128/AAC.00233-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kidd JM, Abdelraouf K, Nicolau DP. 2020. Efficacy of human-simulated bronchopulmonary exposures of cefepime, zidebactam and the combination (WCK 5222) against MDR Pseudomonas aeruginosa in a neutropenic murine pneumonia model. J Antimicrob Chemother 75:149–155. doi: 10.1093/jac/dkz414. [DOI] [PubMed] [Google Scholar]
- 9.den Blaauwen T, de Pedro MA, Nguyen-Disteche M, Ayala JA. 2008. Morphogenesis of rod-shaped sacculi. FEMS Microbiol Rev 32:321–344. doi: 10.1111/j.1574-6976.2007.00090.x. [DOI] [PubMed] [Google Scholar]
- 10.Papp-Wallace KM, Bonomo RA. 2016. New beta-lactamase inhibitors in the clinic. Infect Dis Clin North Am 30:441–464. doi: 10.1016/j.idc.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papp-Wallace KM, Nguyen NQ, Jacobs MR, Bethel CR, Barnes MD, Kumar V, Bajaksouzian S, Rudin SD, Rather PN, Bhavsar S, Ravikumar T, Deshpande PK, Patil V, Yeole R, Bhagwat SS, Patel MV, van den Akker F, Bonomo RA. 2018. Strategic approaches to overcome resistance against gram-negative pathogens using beta-lactamase inhibitors and beta-lactam enhancers: activity of three novel diazabicyclooctanes WCK 5153, zidebactam (WCK 5107), and WCK 4234. J Med Chem 61:4067–4086. doi: 10.1021/acs.jmedchem.8b00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asli A, Brouillette E, Krause KM, Nichols WW, Malouin F. 2016. Distinctive binding of avibactam to penicillin-binding proteins of gram-negative and gram-positive bacteria. Antimicrob Agents Chemother 60:752–756. doi: 10.1128/AAC.02102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moya B, Barcelo IM, Bhagwat S, Patel M, Bou G, Papp-Wallace KM, Bonomo RA, Oliver A. 2017. WCK 5107 (zidebactam) and WCK 5153 are novel inhibitors of PBP2 showing potent "beta-lactam enhancer" activity against Pseudomonas aeruginosa, including multidrug-resistant metallo-beta-lactamase-producing high-risk clones. Antimicrob Agents Chemother 61:e02529-16. doi: 10.1128/AAC.02529-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levy N, Bruneau JM, Le Rouzic E, Bonnard D, Le Strat F, Caravano A, Chevreuil F, Barbion J, Chasset S, Ledoussal B, Moreau F, Ruff M. 2019. Structural basis for E. coli penicillin binding protein (PBP) 2 inhibition, a platform for drug design. J Med Chem 62:4742–4754. doi: 10.1021/acs.jmedchem.9b00338. [DOI] [PubMed] [Google Scholar]
- 15.Contreras-Martel C, Martins A, Ecobichon C, Trindade DM, Mattei PJ, Hicham S, Hardouin P, Ghachi ME, Boneca IG, Dessen A. 2017. Molecular architecture of the PBP2-MreC core bacterial cell wall synthesis complex. Nat Commun 8:776. doi: 10.1038/s41467-017-00783-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh A, Tomberg J, Nicholas RA, Davies C. 2019. Recognition of the beta-lactam carboxylate triggers acylation of Neisseria gonorrhoeae penicillin-binding protein 2. J Biol Chem 294:14020–14032. doi: 10.1074/jbc.RA119.009942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tomberg J, Fedarovich A, Vincent LR, Jerse AE, Unemo M, Davies C, Nicholas RA. 2017. Alanine 501 mutations in penicillin-binding protein 2 from Neisseria gonorrhoeae: structure, mechanism, and effects on cephalosporin resistance and biological fitness. Biochemistry 56:1140–1150. doi: 10.1021/acs.biochem.6b01030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aqvist J, Luecke H, Quiocho FA, Warshel A. 1991. Dipoles localized at helix termini of proteins stabilize charges. Proc Natl Acad Sci U S A 88:2026–2030. doi: 10.1073/pnas.88.5.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vijayaraghavan J, Kumar V, Krishnan NP, Kaufhold RT, Zeng X, Lin J, van den Akker F. 2018. Structural studies and molecular dynamics simulations suggest a processive mechanism of exolytic lytic transglycosylase from Campylobacter jejuni. PLoS One 13:e0197136. doi: 10.1371/journal.pone.0197136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davies TA, Page MG, Shang W, Andrew T, Kania M, Bush K. 2007. Binding of ceftobiprole and comparators to the penicillin-binding proteins of Escherichia coli, Pseudomonas aeruginosa, Staphylococcus aureus, and Streptococcus pneumoniae. Antimicrob Agents Chemother 51:2621–2624. doi: 10.1128/AAC.00029-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davies TA, Shang W, Bush K, Flamm RK. 2008. Affinity of doripenem and comparators to penicillin-binding proteins in Escherichia coli and Pseudomonas aeruginosa. Antimicrob Agents Chemother 52:1510–1512. doi: 10.1128/AAC.01529-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen NQ, Krishnan NP, Rojas LJ, Prati F, Caselli E, Romagnoli C, Bonomo RA, van den Akker F. 2016. Crystal structures of KPC-2 and SHV-1 beta-Lactamases in complex with the boronic acid transition state analog S02030. Antimicrob Agents Chemother 60:1760–1766. doi: 10.1128/AAC.02643-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bergsdorf C, Wright SK. 2018. A guide to run affinity screens using differential scanning fluorimetry and surface plasmon resonance assays. Methods Enzymol 610:135–165. doi: 10.1016/bs.mie.2018.09.015. [DOI] [PubMed] [Google Scholar]
- 24.Han S, Zaniewski RP, Marr ES, Lacey BM, Tomaras AP, Evdokimov A, Miller JR, Shanmugasundaram V. 2010. Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 107:22002–22007. doi: 10.1073/pnas.1013092107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sainsbury S, Bird L, Rao V, Shepherd SM, Stuart DI, Hunter WN, Owens RJ, Ren J. 2011. Crystal structures of penicillin-binding protein 3 from Pseudomonas aeruginosa: comparison of native and antibiotic-bound forms. J Mol Biol 405:173–184. doi: 10.1016/j.jmb.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bellini D, Koekemoer L, Newman H, Dowson CG. 2019. Novel and improved crystal structures of H. influenzae, E. coli and P aeruginosa penicillin-binding protein 3 (PBP3) and N. gonorrhoeae PBP2: toward a better understanding of beta-lactam target-mediated resistance. J Mol Biol 431:3501–3519. doi: 10.1016/j.jmb.2019.07.010. [DOI] [PubMed] [Google Scholar]
- 27.Sacco MD, Kroeck KG, Kemp MT, Zhang X, Andrews LD, Chen Y. 2019. Influence of the alpha-methoxy group on the reaction of temocillin with Pseudomonas aeruginosa PBP3 and CTX-M-14 beta-lactamase. Antimicrob Agents Chemother 64:e01473-19. doi: 10.1128/AAC.01473-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ren J, Nettleship JE, Males A, Stuart DI, Owens RJ. 2016. Crystal structures of penicillin-binding protein 3 in complexes with azlocillin and cefoperazone in both acylated and deacylated forms. FEBS Lett 590:288–297. doi: 10.1002/1873-3468.12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar V, Tang C, Bethel CR, Papp-Wallace KM, Wyatt J, Desarbre E, Bonomo RA, van den Akker F. 2020. Structural insights into ceftobiprole inhibition of Pseudomonas aeruginosa penicillin-binding protein 3. Antimicrob Agents Chemother 64:e00106-20. doi: 10.1128/AAC.00106-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldberg J, Nguyen H, Kumar V, Spencer E, Hoyer D, Marshall E, Cmolik A, O’Shea M, Marshall S, Hujer A, Hujer K, Rudin SD, Domitrovic TN, Bethel C, Papp-Wallace KM, Logan L, Perez F, Jacobs M, van Duin D, Kreiswirth B, Bonomo RA, Plummer M, van den Akker F. 2020. A gamma-lactam siderophore antibiotic effective against multidrug-resistant gram-negative bacilli. J Med Chem 63:5990–6002. doi: 10.1021/acs.jmedchem.0c00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kabsch W. 2010. XDS. Acta Crystallogr D Biol Crystallogr 66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. 2011. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr 67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 36.Moriarty NW, Grosse-Kunstleve RW, Adams PD. 2009. electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr D Biol Crystallogr 65:1074–1080. doi: 10.1107/S0907444909029436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krissinel E, Henrick K. 2007. Inference of macromolecular assemblies from crystalline state. J Mol Biol 372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 38.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. 2006. HKL-3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr 62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 39.Chen VB, Arendall WB, III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Structure-based sequence alignment of PBP2 sequences from P. aeruginosa, E. coli, H. pylori, and N. gonorrhoeae. Assigned secondary elements in P. aeruginosa PBP2 are indicated above the sequences (α and β; 310 helices are labeled as “η”). Residues not included in the crystallized construct or too disordered to be modeled are indicated by a gray interrupted line above the sequence. Residues either conserved or similar to the respective residue in P. aeruginosa are shown as red and orange, respectively. Fully conserved residues in all four sequences are indicated by an asterisk below the sequence. Active site residues are indicated by green boxes. Download FIG S1, TIF file, 2.6 MB (2.6MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Stereo figure showing superposition of noncrystallographic molecules A and B of P. aeruginosa PBP2 active site complexed with WCK 5153. Two PBP2 molecules were observed in the asymmetric unit. Molecule A and molecule B are shown with orange and green carbon atoms, respectively. Hydrogen bonds between WCK 5153 and either PBP2 molecule A or molecule B are shown as black or gray dashed lines, respectively. A crystallographically observed water in the active site of molecule B is shown (“wat”). WCK 5153 is shown in ball-and-stick form with its different moieties labeled as in Fig. 2B. Download FIG S2, TIF file, 2.5 MB (2.5MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Superposition of zidebactam bound complexes of P. aeruginosa PBP3 and KPC-2 β-lactamase. PBP3 is shown in gray, and its bound zidebactam ligand is in teal carbon atoms; KPC-2 and its bound zidebactam are colored green. The active site Cα atoms of PBP3 residues 294 to 301, 346 to 353, 481 to 486, and 505 to 509 were superpositioned onto their equivalent atoms in KPC-2 (residues 70 to 77, 127 to 134, 231 to 236, and 244 to 248), yielding an RMSD of 0.99 Å for 27 Cα atoms. Download FIG S3, TIF file, 1.6 MB (1.6MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Active site views of superpositions of PBP3:WCK 4234 complex with β-lactam complexes of PBP3 and apo-PBP3. (A) Superposition with the PBP3:carbenicillin complex (PDB ID 3OCL [25]; shown in salmon color). Important secondary structure elements β3, β4, and α11 and relevant active site residues are labeled. All residues labeled are shown in black labels except for Y532 and F533 in the PBP3:WCK 4234 complex, which are in gray as this region adopts a different conformation. Three moieties of carbenicillin are labeled: amide (a), phenyl (b), and dimethyl substituents of the thiazolidine ring (c). (B) Superposition with the PBP3:ceftazidime complex (PDB ID 3PBO [24]). Four moieties of ceftazidime are labeled: amide (a), dimethyl groups of 2-carboxypropan-2-yl group (b), methyl substituent of thiazine ring (c) (methyl group is left after pyridine elimination, see reference 29), and aminothiazole ring (d). (C) Superposition with the apo-PBP3 (PDB ID 6HZR [26]). The view is enlarged from panels A and B as the residue F533 position in the apo-PBP3 structure is more distant from the active site. (D) Superposition with the PBP3:meropenem complex (PDB ID 3PBR [24]). One atom of meropenem is labeled, that of the sulfur atom attached to the pyrroline ring of meropenem (c). Download FIG S4, TIF file, 2.5 MB (2.5MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data Availability Statement
Coordinates and structure factors of the P. aeruginosa PBP2 WCK 5153 complex and the PBP3 complexes with WCK 4234, avibactam, and zidebactam were deposited in the Worldwide Protein Data Bank (wwPDB) with accession numbers 7KIS, 7KIT, 7KIV, and 7KIW, respectively.