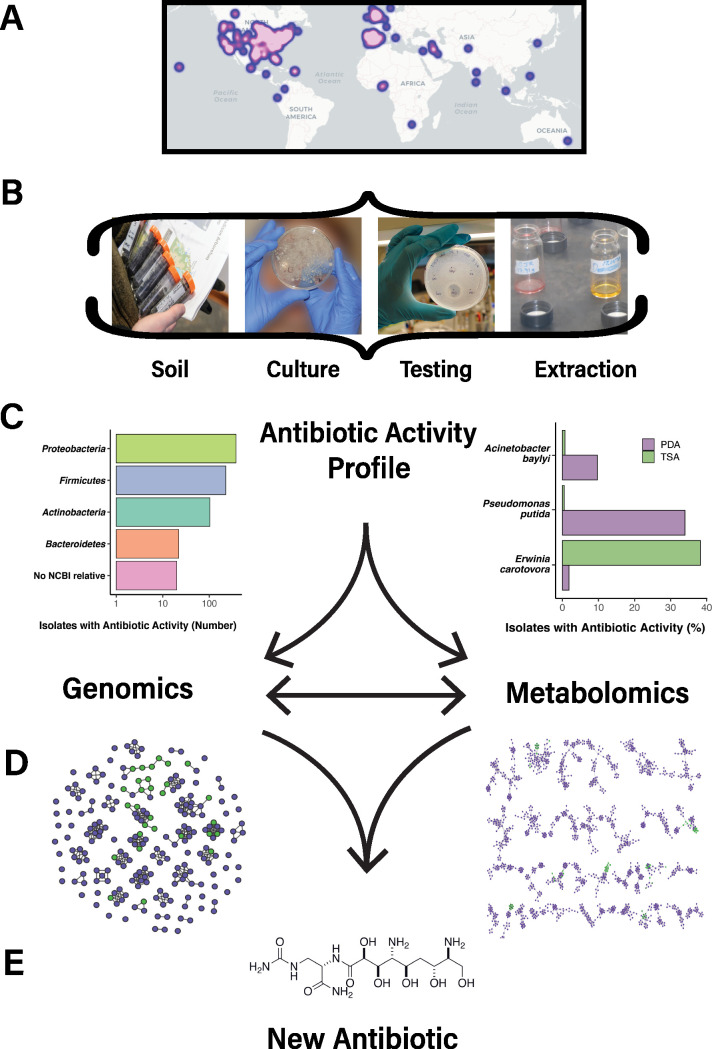
FIG 1.
Tiny Earth Chemistry Hub pipeline. (A) Heat map of global Tiny Earth partner institutions. (B) Strains are catalogued and stored at the Chemistry Hub where each strain is then tested against the safe relatives of ESKAPE pathogens to verify activity profile. Molecules from active strains are extracted in organic solvents. (C, left) Phylum affiliations of strains submitted to the Chemistry Hub as determined by 16S rRNA gene sequencing. NCBI (National Center for Biotechnology Information) catalogues all known sequenced organisms. (C, right) Growth medium differentially affects discovery rate of antibiotic-producing bacteria based on target bacteria. Tryptic soy agar (TSA) enhances isolation of bacteria that inhibit Erwinia carotovora, while potato dextrose agar (PDA) favors inhibition of Acinetobacter baylyi and Pseudomonas putida. Results shown for the three target bacteria are from three separate soils, suggesting soil origin influences media specificity. (D, left) DNA is extracted for whole-genome sequencing and chemical compounds are harvested for mass determination to search for novel patterns. Putative biosynthetic pathways for potential antibiotics are organized in clusters by similarity, with green circles representing similarity to pathways of known function and purple indicating those of unknown function. (D, right) Families of molecules produced by sequenced strains with the same color legend as the left panel. (E) Using both genomic and metabolomic data, strains with potentially novel chemistry are prioritized to purify molecules and determine the structure and activity of a new antibiotic; the structure of soil-derived zwittermicin (37) is shown as an example.