ABSTRACT
Brachypodium distachyon has recently emerged as a premier model plant for monocot biology, akin to Arabidopsis thaliana. We previously reported genome-wide transcriptomic and alternative splicing changes occurring in Brachypodium during compatible infections with Panicum mosaic virus (PMV) and its satellite virus (SPMV). Here, we dissected the role of Brachypodium phenylalanine ammonia lyase 1 (PAL1), a key enzyme for phenylpropanoid and salicylic acid (SA) biosynthesis and the induction of plant defenses. Targeted metabolomics profiling of PMV-infected and PMV- plus SPMV-infected (PMV/SPMV) Brachypodium plants revealed enhanced levels of multiple defense-related hormones and metabolites such as cinnamic acid, SA, and fatty acids and lignin precursors during disease progression. The virus-induced accumulation of SA and lignin was significantly suppressed upon knockdown of B. distachyon PAL1 (BdPAL1) using RNA interference (RNAi). The compromised SA accumulation in PMV/SPMV-infected BdPAL1 RNAi plants correlated with weaker induction of multiple SA-related defense gene markers (pathogenesis related 1 [PR-1], PR-3, PR-5, and WRKY75) and enhanced susceptibility to PMV/SPMV compared to that of wild-type (WT) plants. Furthermore, exogenous application of SA alleviated the PMV/SPMV necrotic disease phenotypes and delayed plant death caused by single and mixed infections. Together, our results support an antiviral role for BdPAL1 during compatible host-virus interaction, perhaps as a last resort attempt to rescue the infected plant.
KEYWORDS: plant-virus interactions, grasses, defense hormones, bioenergy, metabolic pathways
INTRODUCTION
Plants, under relentless challenge by pests and pathogens, have evolved a variety of defense mechanisms. The basal host response to pathogen- or microbe-associated molecular patterns (PAMPs or MAMPs, respectively) (1–3) is the activation of specialized transmembrane proteins, pattern recognition receptors (PRRs). This first layer of defense known as P/MAMP-triggered immunity (PTI) is, in turn, suppressed by pathogen-harbored effectors. While a large body of knowledge exists for bacterial pathogen-triggered PTI mechanisms, less is known for virus-triggered PTI responses. A recent study showed that brassinosteroid-insensitive 1-associated kinase 1 (BAK1) or BAK1-like 1 (BKK1) induced immunity in Arabidopsis against Turnip crinkle virus (TCV) infection, and loss-of-function mutations in BAK1 or BKK1 enhanced susceptibility to virus infection (4). Another study demonstrated that Arabidopsis bak1 mutants exhibited increased susceptibility to three RNA viruses (TCV, Tobacco mosaic virus [TMV], and Oilseed rape mosaic virus [ORMV]) during compatible interactions, and crude viral extracts of inoculated leaf tissues induced several PTI markers in a BAK1-dependent manner (5). These studies showed that BAK1-dependent PTI contributed to host antiviral responses.
An effector interferes with various cellular defense pathways to help promote virulence. These effectors are either secreted or, in the case of viruses, expressed within the plant cells. To counter the action of effectors, the host cell responds with a second line of defense, effector-triggered immunity (ETI). Generally, ETI is triggered when a host receptor recognizes an effector, typically a nucleotide binding site-leucine rich repeat (NBS-LRR) protein. NBS-LRR proteins are a type of resistance (R) proteins that prompt a cascade of biochemical reactions that result in a localized hypersensitive response (HR) and/or systemic acquired resistance (SAR) to limit pathogen spread (1, 2). With regard to antiviral defenses, both PTI and ETI can activate downstream defense hormone pathways (e.g., salicylic acid [SA], jasmonic acid [JA], ethylene, or nitric oxide) or mediate changes in the cell wall due to altered composition of lignin, cellulose, and fatty acids; such compounds also act as signaling molecules and precursors to defense hormones and affect host gene expression (6). For instance, earlier studies of an antiviral role of SA in tobacco revealed that SA inhibits replication of Tobacco mosaic virus (TMV) and systemic movement of Cucumber mosaic virus (CMV) in tobacco (7–10). SA application also resulted in reduced accumulation of Potato virus X (PVX) in inoculated tobacco (8). Conversely, unrestricted spread of Potato virus Y (PVY) and enhanced disease symptoms were observed in NahG potato plants that were depleted of SA (11, 12). In bean and cowpea plants, systemic movement of Alfalfa mosaic virus (AMV) was reduced by 90% upon exogenous SA application (13). These and other studies suggest that SA limits virus infection by interfering in multiple steps of viral replication and intercellular spread as well as systemic movement, depending on the virus-host system (14–16).
Recent studies of compatible plant-microbe interactions have certainly advanced our understanding of plant immune responses and strategies used by viruses to target host defenses. Typically, these studies have relied on the dicot model plant Arabidopsis thaliana and Nicotiana spp. However, our understanding of the genetic basis of antiviral defenses in monocots has been at a disadvantage due to the lack of a suitable monocot model plant. Within the past decade, Brachypodium distachyon has been developed as a superb model system for plant research and has opened new avenues for the study of the Poaceae (Gramineae) to include host defense responses to microbes, plant biochemistry, and evolutionary biology (17, 18). The Brachypodium distachyon research community has developed multiple open-access genomic and genetic resources. With the recent expansion of pan-genomics resources for related species in the genus Brachypodium (19), it is now possible to evaluate grass host defense responses to pathogens of significant environmental and economic importance.
For the study of genomic and transcriptomic outcomes of virus infections, Brachypodium has shown its merits. We and others have demonstrated this plant species’ utility in studying Panicum mosaic virus (PMV) and its satellite virus (SPMV), Foxtail mosaic virus (FMV), Barley stripe mosaic virus (BSMV), Brome mosaic virus (BMV), Wheat streak mosaic virus (WSMV), Maize mild mottle virus (MMMV), Sugarcane mosaic virus (SCMV), and Sorghum yellow banding virus (SYBV) (20–28). Brachypodium also supports infections by Sugarcane mosaic virus, Bamboo mosaic virus, and Barley yellow dwarf virus (BYDV-GRV) (29–31). Taken together, Brachypodium is well suited for studies of grass-infecting viruses in several genera.
PMV is the type member of the genus Panicovirus within the family Tombusviridae. The 4,326-nucleotide (nt), single-stranded positive-sense RNA is enveloped in 30-nm icosahedral virions. PMV encodes six open reading frames: two from the genomic RNA to express the replicase-associated proteins (P48 and P112) (32) and four from the subgenomic polycistronic RNA to express a 26-kDa capsid protein (CP) and three nonstructural proteins (P8, P6.6, and P15) that facilitate virus movement (20, 32, 33). PMV is unusual in that it supports replication and movement of two single-stranded RNA satellites: an 826-nt satellite virus (SPMV) and two ∼400-nt satellite RNAs (satRNAs) (27, 28, 34, 35). PMV and its satellites are found in naturally occurring infections of St. Augustinegrass and switchgrass (27, 28, 34–36). SPMV RNA encodes a 17-kDa CP to assemble 16-nm spherical T=1 virions (37, 38). In addition, the SPMV CP elicits a nonhost-hypersensitive response in Nicotiana benthamiana (39). The presence of SPMV in coinfection with PMV exacerbates disease symptoms on systemic leaves in millets and Brachypodium (24, 25). Using PMV, SPMV, and Brachypodium, we have focused our studies on the biology of host-virus interactions.
One early marker of the host response to infection, including to some viruses, is the induction of SA (14). Phenylalanine ammonia lyase (PAL) is a key enzyme for the production of SA and various secondary metabolites—such as monolignols, which are polymerized to form lignin, flavonoids, hydroxycinnamic acid, phenolics, and anthocyanin—which play important roles in plant defense (40–42). Lignin is particularly important, serving as a constituent of secondary cell walls and being involved in defense against several pathogens (43–45). The deposition of lignin strengthens the cell wall and provides a physical barrier to pathogen ingress (44–49).
Our previous work with PMV and PMV plus SPMV (PMV+SPMV) infections of Brachypodium identified genome-wide changes in host gene expression profiles and alternative splicing landscapes (24, 26). Here, we have determined the extent of perturbation in defense-related metabolites during PMV or PMV+SPMV infections (PMV/SPMV) and the function of PAL in antiviral defenses. PAL activity was induced during PMV/SPMV infections, paralleling responses observed for other biotic and abiotic stresses (45, 50–53). PMV/SPMV infections of B. distachyon PAL1 (BdPAL1) RNA interference (RNAi) lines showed lower pathogen-induced SA and lignin production and resulted in exacerbation of disease symptoms compared to that in wild-type (WT) plants. These negative effects were abrogated by exogenous application of SA, thus providing evidence that PAL-mediated SA production in response to PMV/SPMV infection is an important host mechanism for antiviral defenses in grasses.
RESULTS AND DISCUSSION
Identification of primary and secondary metabolic pathways altered by virus infection in Brachypodium.
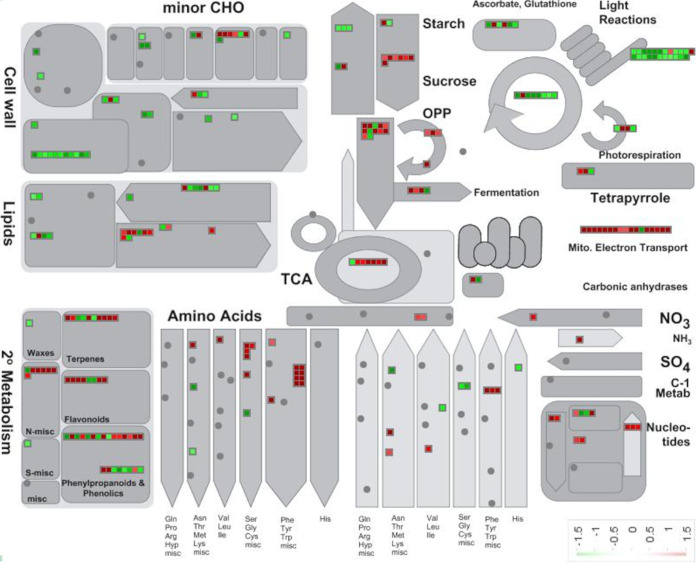
Our previous studies identified widespread transcriptome and splicing changes in response to PMV/SPMV infections in Brachypodium (24, 26). Genes encoding enzymes putatively involved in plant metabolic and biosynthetic pathways affected by PMV infection were identified using the differential gene expression data (stage II, ∼10 to 14 days after infection) and MapMan metabolic pathway analyses (54, 55) (Fig. 1; see also Data Set S1 in the supplemental material). This included gene expression changes predicted to affect production of primary metabolites such as carbohydrates (e.g., glucose and sucrose) that provide cellular energy and function as plant defense signaling molecules (56–58). MapMan analysis of transcripts predicted downregulation of primary metabolites related to light reactions in photosynthesis, which are associated with leaf chlorosis and decreased chlorophyll levels in early infection stages (24, 26). Such breakdown of chloroplast function and the associated leaf chlorosis are known outcomes of plant virus and other plant pathogen infections (59–61).
FIG 1.
MapMan overview of primary and secondary metabolic pathways perturbed during PMV infection in Brachypodium. The square boxes in green and red indicate reduced and induced level of expression (>2 log2 fold-change, FDR < 0.05), respectively. The fold change expression data are provided in Data Set 1 in the supplemental material. Mito, mitochondrial; TCA, tricarboxylic acid; OPP, oxidative pentose phosphate; CHO, carbohydrates.
Gene expression data (>2 log2 fold change, FDR < 0.05) in PMV-infected Brachypodium leaves at stage II. Download Data Set S1, XLSX file, 0.1 MB (52.8KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Metabolic pathways related to cell wall and lipid biosynthesis were also downregulated in PMV-infected plants (Fig. 1). This was associated with an overall decrease in the biomass and the stature of virus-infected plants at later infection stages (24, 26). In contrast, central metabolic pathways associated with respiration, such as the mitochondrial electron transport chain, Krebs cycle, and the oxidative pentose phosphate (OPP) pathway, were induced. These changes may reflect the high demand for energy production in virus-infected plants to sustain cellular activities related to adaptation and defense. Secondary metabolites are also important for host responses to biotic and abiotic stress (62).
In PMV-infected plants, multiple secondary metabolite pathways were substantially induced. These included pathways involved in the production of lignin, flavonoids, phenylpropanoids, phenolics, waxes, and terpenes, all of which play crucial roles in host defense and adaptation to plant stress (Fig. 1). Several nucleotide and amino acid biosynthetic pathways (related to phenylalanine, tyrosine, tryptophan, glycine, serine, and cysteine) were also upregulated, possibly as a consequence of a reprogramming of the cellular transcriptome and proteome. Alternatively, increased amounts of amino acids and nucleotides could be redirected to biosynthesis of secondary metabolites or serve as cofactors for proteins involved in plant defense signal transduction. This bioinformatic analysis of primary and secondary metabolic pathways provided a framework to explore the host mechanisms resulting in phenotypic changes and symptomology induced by virus infection.
PMV and SPMV infection perturb defense hormone and fatty acid profiles.
Salicylic acid (SA) and its derivative methyl-SA (MeSA) are key hormones mediating plant defense responses to pathogens, including viruses (1, 63–65). SA-mediated defenses are induced in both ETI and PTI and during incompatible and compatible interactions. PMV infection modulates the expression profiles of genes in defense hormone pathways such as SA and jasmonic acid (JA) (24, 26). Several pathogenesis-related (PR) genes such as PR-1, PR-3, and PR-5 were found to be upregulated in PMV/SPMV-infected Brachypodium (25).
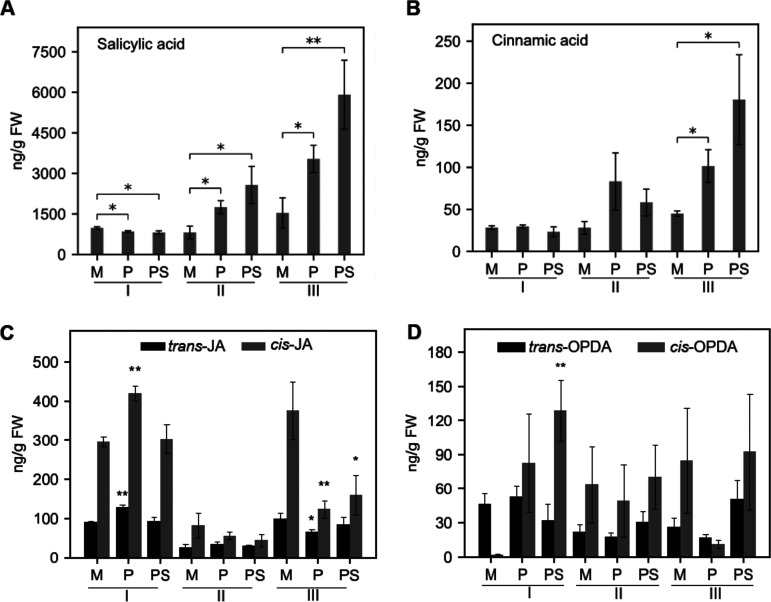
Here, we aimed to investigate if gene expression changes reflected the expected changes in defense-related hormones and their precursors. Brachypodium plants were challenged with PMV or with PMV plus SPMV (PMV+SPMV), followed by targeted metabolic profiling using gas chromatography mass spectrometry (GC-MS). To determine the temporal dynamics of infection, multiple time points were inspected on inoculated leaves: early stage, no visual symptoms (7 days postinfection [dpi], stage I); mid stage, with mild chlorosis symptoms (14 dpi, stage II); and late stage, with severe chlorosis and necrosis symptoms (21 dpi, stage III), as described previously (24). Significant changes in SA accumulation were found in all three stages upon virus infection. The accumulation of cinnamic acid (CA) in PMV- and PMV+SPMV-infected plants was elevated at 14 dpi but not at 7 dpi. The accumulation of SA was significantly higher (P ≤ 0.05) in PMV-infected (1,747 ng/g [fresh weight]) and PMV+SPMV-infected (2,570 ng/g [fresh weight]) plants by stage II than in mock-inoculated plants (SA, 815 ng/g [fresh weight]), and these levels remained elevated through stage III (Fig. 2A). CA concentration was elevated but not significantly affected at stage II upon PMV/SPMV infections; however, it was significantly induced (P ≤ 0.05) at stage III (Fig. 2B). SA and CA concentrations were consistently higher in PMV+SPMV-inoculated plants at stage III than in PMV-inoculated plants. The higher levels of SA and CA in virus-infected plants correspond with the upregulation of several SA signaling genes, including pathogenesis-related (PR) proteins (24, 25). We previously reported that the PMV+SPMV synergism exacerbated disease symptoms and had an additive effect on several defense-related gene expression profiles compared to that from PMV-only infections (24). Such additive affects were also observed in the metabolite profiles, with a higher accumulation of SA and CA in PMV+SPMV-coinfected plants than in plants infected with PMV alone (Fig. 2A and B).
FIG 2.
Quantification of salicylic acid (SA), cinnamic acid (CA), jasmonic acid (JA), and precursors in PMV- and PMV+SPMV-infected Brachypodium. Levels of SA (A), CA (B), trans- and cis-JA (C), and trans- and cis-12-OPDA (D) were determined using GC-MS. The fresh weight (FW) leaf samples were collected at three different stages of disease progression (early stage I, 7 dpi; mid-stage II, 14 dpi; late stage III, 21 dpi), as described previously (24). The y axes represent average contents of respective metabolites from five biological replicates. The error bars represent standard errors of means (n = 3 and n = 4 for early and for mid and late stages, respectively). *, P ≤ 0.05; **, P ≤ 0.01 between mock- and virus-infected samples as determined by two-sample t test (one-tailed). M, mock; P, PMV; PS, PMV+SPMV; OPDA, 12-oxo-phytodienoate.
In a manner similar to that for SA, jasmonic acid (JA) and its derivative methyl-JA (meJA) regulate various physiological and developmental processes as well as responses to biotic stresses, particularly to necrotrophic pathogens, wounding, and herbivores (66–69). Biosynthesis of JA is initiated in the chloroplast with the conversion of linolenic acid to 12-oxo-phytodienoate (12-OPDA), followed by its reduction in the peroxisome by a 12-OPDA reductase through β-oxidation (70, 71). With some exceptions, the JA and SA pathways are antagonistic to each other (25, 72–74). The expression of several JA signaling genes was downregulated in PMV/SPMV-infected Brachypodium, while SA signaling genes were upregulated (24, 25). Metabolite profiling analysis revealed that at stage III, when SA was highest in virus-infected plants (Fig. 2A), cis-JA was significantly lower (P ≤ 0.05) than in mock-infected plants (Fig. 2C), suggesting SA-JA antagonism. Because the cis isoform of JA was highly affected compared to trans, this indicates a downregulation of biosynthesis rather than increased JA conversion or turnover. Interestingly, 12-OPDA, the precursor to JA synthesis, was significantly higher (P ≤ 0.01) in PMV+SPMV-inoculated plants at stage I (Fig. 2D), when both SA and JA were largely unaltered from mock-inoculated plants. This suggests that 12-OPDA or related oxylipins act as signaling molecules, independent of JA, during the early stages of virus infection (75, 76).
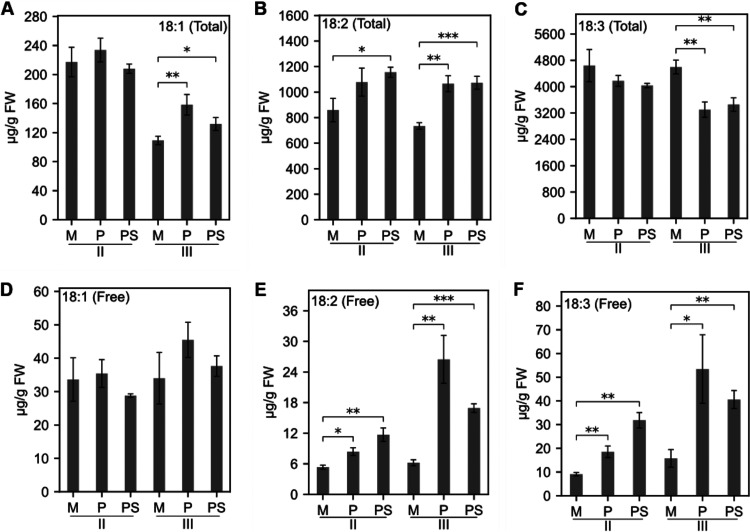
Fatty acids (FAs) are important structural and metabolic constituents of membranes and the cell wall and are involved in intracellular signaling and protein modification processes (77–79). For example, 18-carbon mono- and polyunsaturated FAs such as oleic acid (18:1), linoleic acid (18:2), and linolenic acid (18:3) are precursors to oxylipin-derived defense hormones, with key roles in affecting outcomes in plant-microbe interactions (80–86). We next investigated total and free 18-carbon FA levels in Brachypodium leaves during virus infection at stages II and III (Fig. 3A to F). In stage II, there were no significant changes in levels of total FAs except for linoleic acid (18:2) (Fig. 3A to C). However, at stage III, levels of total oleic acid (18:1) and linoleic acid (18:2) were significantly greater (P ≤ 0.01) in virus-infected plants than in mock-infected plants (Fig. 3A to C). In contrast, total linolenic acid (18:3) levels were significantly lower (P ≤ 0.01) in virus-infected leaves (Fig. 3C).
FIG 3.
Quantification of 18-carbon unsaturated fatty acid levels in PMV- and PMV+SPMV-infected Brachypodium. Levels of free and total 18:1, 18:2, and 18:3 fatty acids in mock-, PMV-, and PMV+SPMV-infected plants at stages II (14 dpi) and III (21 dpi) of infection. The error bars represent standard errors of means (n = 3 and n = 4 for stages II and III, respectively). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001 between mock- and virus-infected samples as determined by two-sample t test (one-tailed). M, mock; P, PMV; PS, PMV+SPMV.
The bottleneck in total 18:3 accumulation corresponds to the decreased expression of FAD7 in PMV/SPMV-infected Brachypodium that we previously observed (24). FAD7 encodes a chloroplast-localized enzyme that desaturates 18:2 to 18:3 (87), and the suppressed expression of FAD7 could result in the accumulation of its substrate, 18:2. Plant membranes, especially chloroplast membranes, are also a repository for 18:3, esterified in glycolipids and phospholipids. Upon cell perturbation or damage, phospholipases are activated, which release free 18:3 from membranes (88). Interestingly, levels of free 18:3 were significantly higher (P ≤ 0.01) in virus-infected plants, as early as stage II (Fig. 3F), suggesting that despite the bottleneck in biosynthesis, during PMV/SPMV infection, there is a greater amount of free 18:3 being released from repositories and/or as breakdown products. Given that plastidic FA-derived lipid signals are critical for modulating plant defense signaling (84, 89, 90), it is possible that the free 18:3 or derived lipid signals are important in host responses to PMV/SPMV infections.
BdPAL1 is required for SA and lignin accumulation during PMV and PMV+SPMV infections.
SA is derived from chorismate or phenylalanine by two key enzymatic pathways involving isochorismate synthase (ICS) or phenylalanine ammonia lyase (PAL), respectively (91–93). Although the ICS pathway is thought to be dominant in Arabidopsis (94), other plant species have various contributions of either ICS or PAL to SA production during infection. For instance, SA is predominantly derived from the phenylpropanoid pathway in tobacco (95). In contrast, in soybean, both ICS and PAL pathways contribute equally to SA biosynthesis upon pathogen infection (96). It is unclear which pathway of SA biosynthesis is dominant in Brachypodium. PAL catalyzes the conversion of phenylalanine to trans-cinnamate by nonoxidative deamination. This is a key rate-limiting step between primary and secondary metabolism (40, 41, 97, 98). The PAL pathway also leads to production of several other critical defense-related metabolites such as lignin and flavonoids. Brachypodium encodes eight putative BdPAL homologs. BdPAL1 (Bradi3g49250) is predominantly expressed in vegetative tissues, and its downregulation by RNA interference (RNAi) resulted in significant changes in defense-related secondary metabolites and enhanced susceptibility to fungal pathogens (50).
We previously showed that PMV/SPMV infection activated multiple SA-related genes (e.g., PR genes) (24, 99). Expression of a few PAL isoforms was also upregulated (data not shown). Because PMV/SPMV infections affected SA biosynthesis (Fig. 2) along with multiple primary and secondary metabolic pathways, including cell wall-related components (Fig. 1), we further examined the role of PAL in PMV/SPMV-induced SA and phenylpropanoid biosynthesis by leveraging the BdPAL1 RNAi plants (50).
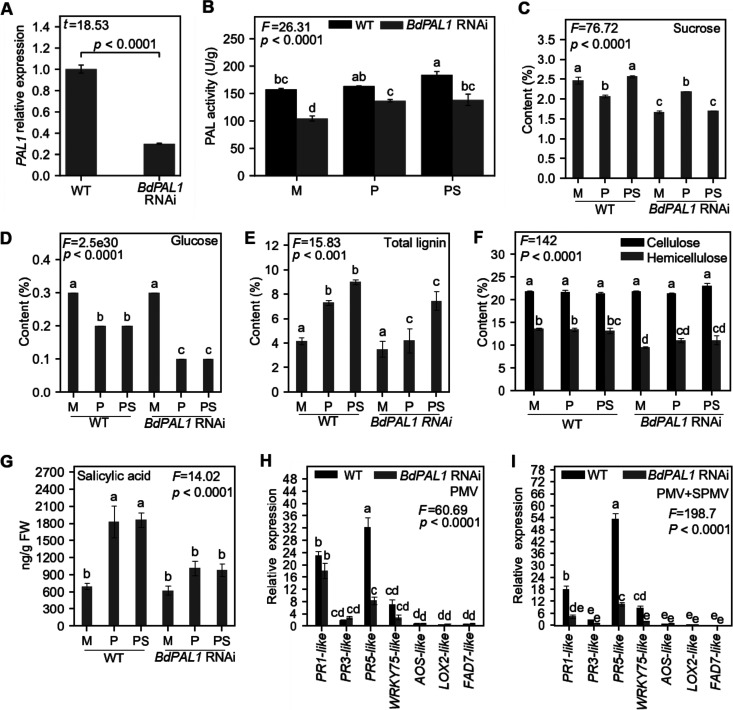
Prior to investigating the host effects of BdPAL1 knockdown on PMV/SPMV infections, we reconfirmed the silencing of BdPAL1 expression and activity in the BdPAL1 RNAi plants compared to those in WT plants (Fig. 4A and B). As anticipated, PMV and PMV+SPMV infections significantly induced PAL activity in WT (P ≤ 0.05) compared to that in mock-inoculated leaves (Fig. 4B). Although PAL activity was induced in PMV- and PMV+SPMV-infected BdPAL1 RNAi plants (P ≤ 0.05), it was consistently lower than in the corresponding WT plants, because of the PAL1 downregulation (Fig. 4B). Next, we quantified defense-related and primary and secondary cell wall-associated compounds in WT and BdPAL1 RNAi plants, in mock-inoculated and PMV/SPMV-infected plants. Among the studied compounds, sucrose levels did not follow any specific pattern of change during virus infection; however, they were lower in healthy BdPAL1 RNAi plants than in WT plants (Fig. 4C). We speculate the decrease in sucrose could be a reflection of PAL function in normal plant growth and development. BdPAL1 RNAi plants do have slightly delayed growth compared to that of the WT (50). Glucose, the end product of photosynthesis, was significantly lower in both WT and BdPAL1 RNAi plants infected with PMV/SPMV than in their healthy counterparts (P ≤ 0.05) (Fig. 4D). The levels of glucose correlated with the decreased level of various enzymes involved in photosynthetic and primary carbohydrate metabolic pathways (Fig. 1) as a result of PMV+SPMV infection, and the observed chlorotic symptomology and stunted growth of the PMV+SPMV-infected plants (Fig. 5C). Although it is difficult to determine if the lowered primary carbohydrates are the cause or consequence of the growth delays and disease phenotypes, they underscore the broader function of PAL1 and their metabolites, including SA, on plant growth, development, and defense processes (50, 100).
FIG 4.
Functional analysis of wild-type (WT) and BdPAL1 RNAi plants during PMV and PMV+SPMV infection. Expression of BdPAL1 (A) and PAL activity (B) in mock-, PMV-, and PMV+SPMV-inoculated WT and BdPAL1 RNAi plants at stage II (14 dpi) of infection. Levels of soluble carbohydrates (sucrose and glucose) (C), structural carbohydrates (cellulose and hemicellulose) and total lignin (D), and salicylic acid (E) at stage II (14 dpi) of infection. M, mock; P, PMV; PS, PMV+SPMV. Relative expression of defense-related genes (SA and JA signaling components) in PMV-infected (F) and PMV+SPMV-infected (G) WT and BdPAL1 RNAi plants. The error bars represent standard errors of means (n = 3 for panels A to D, F, and G and n = 5 for panel E). Statistically significant differences in panel A were determined by two-sample t test (one-tailed) and, in panels B to G, were assessed using one or two-way ANOVA followed by the Tukey’s test. Unlike lowercase letters represent significant differences among the group means (P ≤ 0.05). F and t test statistics of ANOVA and two-sample t tests are indicated.
FIG 5.
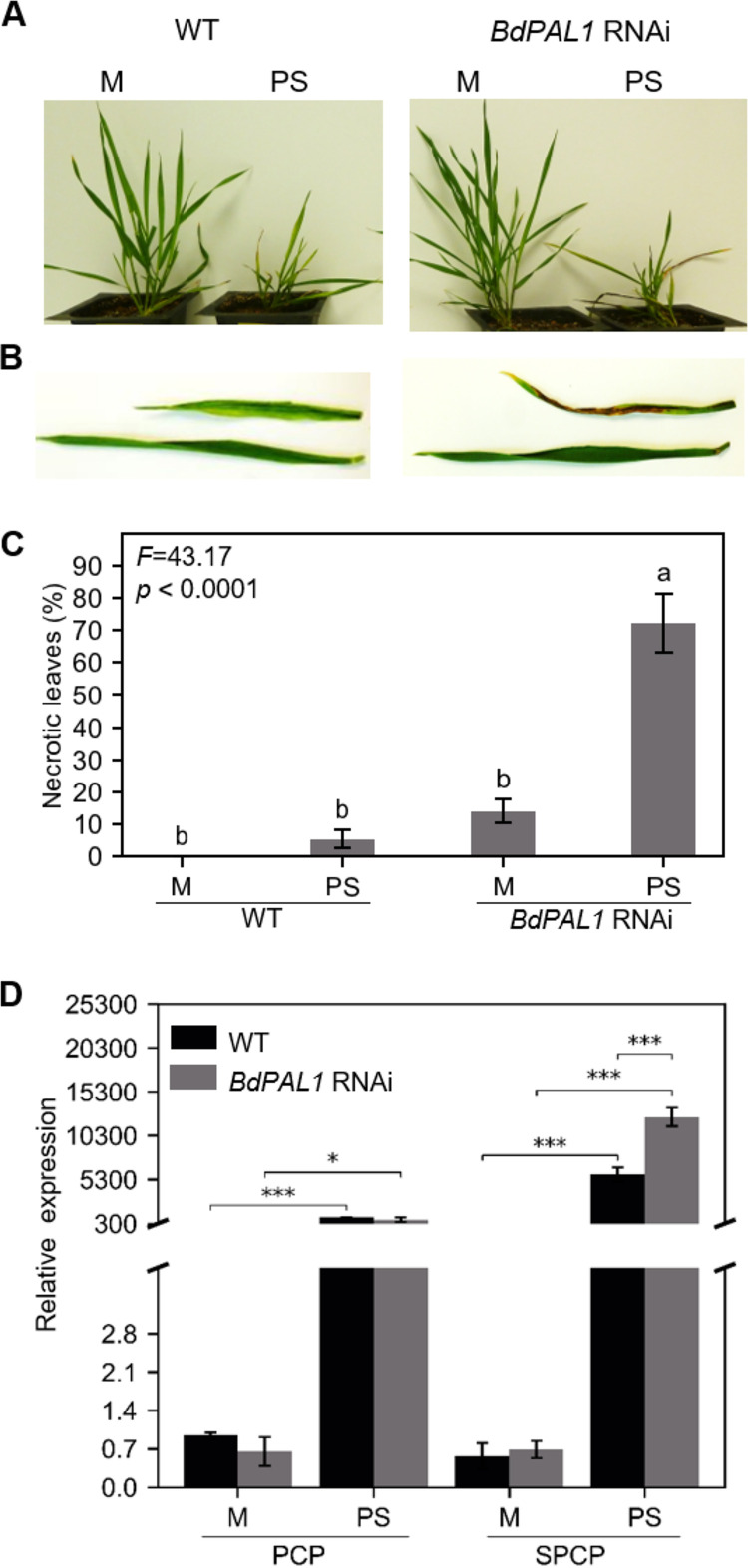
BdPAL1 RNAi plants showed enhanced susceptibility to PMV+SPMV infection. (A) Mock- and PMV+SPMV-infected WT (left) and BdPAL1 RNAi (right) plants at stage III (21 dpi). (B) Closeup of mock- and PMV+SPMV-infected leaves of WT (left) and BdPAL1 RNAi (right) plants at stage III (21 dpi). (C) Percentages of leaves with necrosis of wild-type (WT) and BdPAL1 RNAi plants, mock- and virus-infected plants at stage III (21 dpi). Statistically significant differences were assessed using one-way ANOVA followed by the Tukey’s multiple-comparison test. Unlike lowercase letters represent significant differences among the group means (P ≤ 0.05). F statistics of ANOVA are indicated. (D) RT-qPCR detection of mRNA encoding PMV CP (PCP) and SPMV CP (SPCP) in noninoculated leaves at 14 dpi. M, mock; PS, PMV+SPMV. Statistically significant differences were assessed between mock- and virus-inoculated samples using two-sample t test (one-tailed). *, P ≤ 0.05; ***, P ≤ 0.001. The error bars represent standard errors of the means (n = 3).
Among structural cell wall components, total lignin was significantly induced in WT and BdPAL1 RNAi upon PMV/SPMV infections (P ≤ 0.001), although compared with that in WT, BdPAL1 RNAi plants exhibited low total lignin levels (Fig. 4E). It is possible that the elevated levels of total lignin in WT plants, derived from PAL, may contribute to limiting cell-to-cell movement of PMV and SPMV and/or be a general consequence of tissue stress. Levels of cellulose remained unaltered in healthy and virus-infected WT and BdPAL1 RNAi plants (Fig. 4F). BdPAL1 RNAi lines showed a statistically significant decrease in hemicellulose (P ≤ 0.05), compared to that in WT plants (Fig. 4F). This could be due to the compromised ferulate (FA) production in the BdPAL1 RNAi plants (50). FA is a critical moiety present in grass cell walls, which helps cross-link the cell wall components such as hemicellulose (50). Alternatively, lowered sucrose (Fig. 4C), a carbon source for hemicellulose, may negatively affect its production/levels. There were no significant differences in hemicellulose upon virus infection (Fig. 4F).
Metabolic profiling of SA revealed that virus infection triggered significant increases in SA levels in WT plants (P ≤ 0.05) (Fig. 4G). SA accumulation was slightly higher in virus-infected BdPAL1 RNAi plants than in mock-treated plants but was not statistically significant upon virus infection (Fig. 4G). However, in a manner similar to that for total lignin, virus-triggered SA accumulation was significantly suppressed (P ≤ 0.05) in the BdPAL1 RNAi plants compared to that in WT plants (Fig. 4G). Basal levels of SA in WT and BdPAL1 RNAi plants were largely unaffected (Fig. 4G), suggesting that BdPAL1 primarily contributes to induce SA production upon pathogen infection. The compromised SA levels in PMV+SPMV-infected BdPAL1 RNAi plants correlated with weaker induction of multiple SA-related defense gene markers (PR-1-like, PR-3-like, PR-5-like, and WRKY75-like) (Fig. 4H and I). For instance, PMV/SPMV caused an 18- to 22-fold increase in the PR-1-like gene in WT plants, compared to an ∼2- to 12-fold increase in BdPAL1 RNAi plants. Consistent with previous studies (24, 25, 101–104), we also observed the antagonistic relationship between SA and JA, wherein several JA components (AOS, LOX2, and FAD7) were downregulated in PMV/SPMV-infected WT and BdPAL1 RNAi plants (Fig. 4H to I), which is in agreement with our transcriptomic analyses (24, 25). The suppressed expression of LOX2, AOS, and FAD7 was associated with decreased accumulation of JA in WT plants (Fig. 2C). These findings support the notion that an antagonistic relationship exists between SA and JA during PMV/SPMV infections in Brachypodium.
Inhibition of BdPAL1 results in enhanced susceptibility to PMV and SPMV in Brachypodium.
Next, we determined whether the compromised lignin and SA levels and SA-related defense gene activation in the BdPAL1 RNAi plants influence PMV/SPMV disease outcomes. For this, we assessed the disease symptoms (e.g., chlorotic and necrotic lesions) of WT and BdPAL1 RNAi plants. Typical chlorotic symptoms appeared by 12 to 14 dpi of virus infection in both WT and BdPAL1 RNAi plants, and necrotic symptoms appeared by 21 dpi (Fig. 5A and B). At this time point, the percentage necrosis on the upper noninoculated (systemically infected) leaves was quantified. Severe necrotic lesions were observed on greater than 72% of the leaves of PMV+SPMV-infected BdPAL1 RNAi plants, whereas only ∼5% of the WT leaves displayed necrotic lesions (Fig. 5C). The presence of PMV and SPMV RNA in the respective samples was confirmed by reverse transcription-quantitative PCR (RT-qPCR) (Fig. 5D). Levels of SPMV were significantly higher (P ≤ 0.001) in BdPAL1 RNAi plants (Fig. 5D) than in WT plants and paralleled the severe necrotic symptoms observed on the PMV/SPMV-infected BdPAL1 RNAi plants (Fig. 5B and C). We also observed that 14% of the leaves of nonchallenged BdPAL1 RNAi plants also showed necrotic lesions. These symptoms were reminiscent of spontaneous lesions on lesion mimic mutants (LMMs) which also occur in nonchallenged plants. Several genes are linked to LMMs, many of them involved in plant development, metabolism, and defense signaling (105).
Exogenous SA treatment alleviates the hypersensitive phenotype of BdPAL1 RNAi plants.
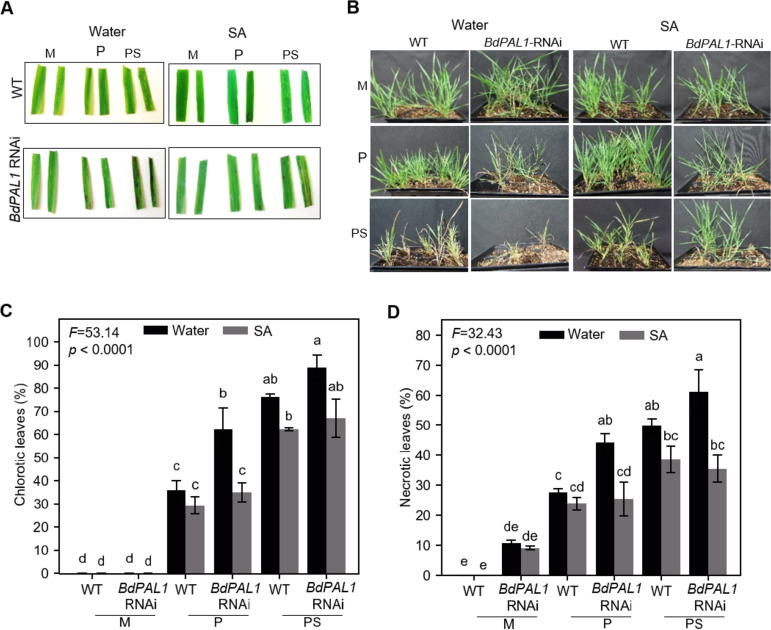
Lastly, we investigated the effects of exogenous supplementation of SA—could it rescue the enhanced PMV/SPMV susceptibility phenotype of BdPAL1 RNAi plants? For this, soil containing WT and BdPAL1 RNAi plants was drenched with 50 ml of water or 100 ppm SA, 24 h prior to inoculation with PMV alone or PMV+SPMV (106). SA was reapplied 24 h postinoculation and then once a week for up to 4 weeks. As expected, on PMV/SPMV-inoculated plants, symptom onset of WT plants, typified by leaf chlorosis, occurred ∼12 to 14 dpi. However, these symptoms were delayed by ∼4 days for SA-treated WT and BdPAL1 RNAi plants. The application of exogenous SA alleviated the necrotic disease phenotype and subsequent plant death at 21 dpi (Fig. 6A) and remained protective through 42 dpi (Fig. 6B). The lower percentage of chlorosis and necrosis on BdPAL1 RNAi plants drenched with SA solution was significant, to levels almost comparable to that for WT plants (P ≤ 0.05) (Fig. 6C and D). It appears the exogenous SA treatment rescued the enhanced susceptibility of BdPAL1 RNAi plants to PMV/SPMV infection. Regarding the possible mechanism of the SA effect on PMV/SPMV, we speculate that the SA applied to the roots, or a mobile SA form (107), could be triggering a systemic acquired resistance (SAR) response. The SAR signal could be perpetuated from roots to distant shoot tissues. Early studies showed that SA not only inhibits virus replication in inoculated tissues but also limits virus cell-to-cell and systemic movement via systemic resistance pathways (7–10, 108, 109). Recent work further bolstered the role of SA-mediated signaling in SAR responses against several biotrophic pathogens, including viruses (11, 12, 107, 110–112). Given that PMV and SPMV accumulate both in shoots and roots (24), it is conceivable that an SA-mediated SAR response originating in the roots could limit PMV/SPMV accumulation on a whole-plant level and attenuate the disease symptoms. Regardless of the exact mechanisms, the results suggest that the induction of the SA pathway in WT plants or supplementation of SA to BdPAL1 knockdown plants can confer tolerance to PMV/SPMV infections.
FIG 6.
Exogenous application of salicylic acid (SA) attenuates PMV and SPMV disease symptoms. Representative chlorosis and necrosis at 21 dpi (A) and overall stunting symptoms at 42 dpi (B) of PMV- and PMV+SPMV-infected plants treated with water or salicylic acid (SA; 100 ppm). Quantification of percent chlorotic (C) and necrotic (D) leaves in wild-type (WT) and BdPAL1 RNAi plants treated either with water or SA (100 ppm) at 21 dpi. Statistically significant differences were assessed using one-way ANOVA followed by the Tukey’s test. Unlike lowercase letters represent significant differences among the group means (P ≤ 0.05). F test statistics of ANOVA are indicated. The error bars represent standard errors of means (n = 3). M, mock; P, PMV; PS, PMV+SPMV.
Conclusion.
We previously established the emerging model grass Brachypodium as an alternate host to study PMV- and SPMV-host interactions and viral synergism (24) and demonstrated by genomic and transcriptomic approaches that several defense-related hormones and signaling pathways are perturbed in PMV/SPMV interactions with Brachypodium (25). Here, we used Brachypodium and PMV/SPMV as a model pathosystem to gain a broader understanding of the defense-related metabolic pathways perturbed during compatible grass virus infections and the role of BdPAL1 in antiviral defenses. We have shown that PMV/SPMV infections triggered changes in various defense-related metabolites, including the defense hormone salicylic acid (SA) and immune-related genes. Specifically, BdPAL1-induced SA is important for tolerating PMV/SPMV. This metabolic, biochemical, and genetic characterization extended our knowledge of PAL-mediated antiviral defenses in a C3 grass and underscores the utility of Brachypodium as a superb model system for the study of grass-microbe interactions. Further translational studies aimed to enhance the PAL pathway or similar conserved immune features of grasses will likely lead to new measures to develop robust disease resistance in the field.
MATERIALS AND METHODS
Plant material, growth conditions, and virus inoculation.
Seeds of Brachypodium wild type (WT, accession Bd21-3) and the BdPAL1 RNAi line were planted in Metro Mix-366 (Sun Gro Horticulture) in 2- by 3-in. pots, stratified at 4°C for 4 days. Information on generation and characterization of BdPAL1 RNAi lines was described previously (50). Plants were maintained in a growth chamber with a photoperiod of 14 h of light (21°C)/10 h of dark (18°C) with light intensity of approximately 250 to 280 mmol/m2s (24). Brachypodium leaves were rub inoculated at the three-leaf stage using in vitro transcribed PMV and SPMV genomic RNA (gRNA), as described previously (24, 113). Briefly, full-length viral cDNA clones of PMV and SPMV in pUC19 were linearized using EcoICR1 and BglII endonucleases, respectively. Linearized infectious viral clones (∼500 ng) were used for in vitro synthesis of gRNA using HiScribe T7 in vitro transcription kit (New England Biolabs). After confirming the size and integrity of RNA by gel electrophoresis, PMV or PMV+SPMV gRNAs were mixed (50% [vol/vol]) in RNA inoculation buffer (50 mM KH2PO4, 50 mM glycine, pH 9.0, 1% bentonite, and 1% Celite) and rub inoculated on the lower-most leaves and cotyledons (∼300 ng of transcript per plant). Throughout this paper, PMV/SPMV indicates results associated with both PMV alone and PMV+SPMV infections. Mock inoculations consisted of RNA inoculation buffer alone. All mock-infected and virus-inoculated plants were covered by a Humi-Dome (Hummert International), incubated in the dark for 24 h, and then moved to the growth chamber.
RNA isolation, RT-qPCR, and molecular diagnostics.
The presence of PMV and SPMV RNA in infected plants was confirmed by quantitative RT-PCR detection of noninoculated (systemically infected) leaves as described previously (24). Briefly, total RNA was isolated from 100 mg of leaf tissue using an RNeasy plant kit (Qiagen) with on-column DNase treatment (Invitrogen). One microgram of RNA was used for cDNA synthesis with a SuperScript IV first-strand cDNA synthesis kit (Invitrogen) and random hexamer primers, followed by PCR amplification to detect PMV and SPMV.
For SA and JA marker gene expression analysis, RNA was isolated from mock- and virus-inoculated leaves at 14 dpi (RNeasy plant kit; Qiagen) according to the manufacturer’s instructions. The RNA concentration and quality were validated using a NanoDrop 1000 and agarose gel electrophoresis, respectively. Two micrograms of RNA was used for cDNA synthesis with a SuperScript IV first-strand cDNA synthesis kit (Invitrogen). Gene expression analysis was performed by RT-qPCR using a CFX384 real-time PCR detection system (Bio-Rad) with iTaq universal SYBR green supermix (Bio-Rad), 1 μl of 2-fold diluted cDNA, and 0.2 μM target specific primers, according to the manufacturer’s instructions. Each assay was performed using three biological replicates (composed of RNA isolated from three different plants) and two technical replicates. The primers were designed using QuantPrime (114) and are listed in Table S1 in the supplemental material. The expression data were normalized to the constitutively expressed ubiquitin 18 (Bradi4g00660) gene (115). The relative quantification of gene expression was performed using the comparative threshold cycle (2−ΔΔCT) method (116) and is presented as relative to mock-inoculated plants, which are set to 1.
Primers used for SA and JA defense marker gene expression analysis. Download Table S1, DOCX file, 0.1 MB (30.2KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
MapMan pathway analysis and GC-MS metabolite profiling.
Brachypodium metabolic pathways affected during PMV infection were determined by MapMan metabolic pathway analysis (54, 55, 117) using the differential gene expression data from a previous transcriptome study (stage II, ∼10 to 14 days after infection [24]). MapMan allows correlating gene expression data placement onto schematic maps or pathways of biological processes. The MapMan mapping units for Brachypodium represent orthologous, nonredundant functional gene ontologies retrieved from similarity searches using BLAST (123) (against Arabidopsis TAIR10, Plant SWISS-Prot, and UniRef90), RPS-BLAST (against NCBI Conserved Domain Database [CDD], EuKaryotic Orthologous Groups [KOG]), and Inter-Pro scan. Only mapping classifications with the highest reliability are retained in these mapping units. Briefly, the normalized expression counts (fragments per kilobase per million [FPKM]) of significantly differentially expressed genes (>2 log2 fold up- or downregulated at a false discovery rate [FDR] of <0.05) were overlaid onto the Brachypodium primary and secondary metabolic maps. A transcript-metabolite correlation map was generated, which provided a bird’s eye view of up- and downregulated pathways, indicated by red and green colored squares, respectively (Fig. 1).
For quantification of metabolites, PMV-, PMV+SPMV-, and mock-inoculated Brachypodium leaves were collected at 7, 14, and 21 dpi. Frozen ground tissue samples (100 mg) were solvent extracted and partitioned into duplicate samples. For phytohormones and free fatty acids, one set of the partitioned samples was methylated, collected on a polymeric adsorbent using vapor-phase extraction (VPE), and analyzed using GC/isobutane chemical ionization MS (CI-MS) as previously described (118). For total fatty acid analysis, the second set of partitioned samples was dried down under liquid nitrogen, and then 1 ml of 1:10 3 M KOH to methanol (MeOH) was added and samples were incubated at 80°C for 1 h. Following incubation, samples were acidified with 50 μl of 6 M HCl, 1 ml of hexane was added, and samples were vortexed. Then, 500 μl was transferred to a 4-ml glass vial and the samples were methylated, collected on a polymeric adsorbent using vapor-phase extraction (VPE), and analyzed using GC/isobutane CI-MS as previously described (118). Metabolite quantification was based d5-JA (C/D/N Isotopes, Canada), or U-13C18:3 (Cambridge Isotope Laboratories) as internal standards.
Primary and secondary cell wall composition analysis.
Cell wall composition analysis, including that of structural carbohydrates (cellulose and hemicellulose), lignin, and nonstructural carbohydrates (glucose and sucrose), of mock- and virus-inoculated Brachypodium leaves was performed at 14 dpi (stage II), as described previously (119, 120). Briefly, leaf samples were collected, freeze-dried, and pulverized in a mortar with a pestle. The dried sample (7 mg) was extracted twice in 2 ml of water with 2 ml of 95% ethanol, sonicated, and centrifuged each time for 20 min. The supernatants were discarded, and the pellets dried overnight at 65°C and subsequently weighed to calculate the percentage of soluble extractives. Next, the dried pellet was hydrolyzed in two steps: (i) by the addition of 70 μl of 72% H2SO4 and incubating for 60 min at 30°C and then (ii) by diluting the acid solution to 3% with ultrapure water and autoclaving for 60 min at 121°C. After autoclaving, the hydrolysis liquor was centrifuged, and 150 μl of the supernatant, diluted to 50%, was used to measure absorbance on a UV-visible spectrophotometer at 240 nm to calculate the percentage of acid soluble lignin. Finally, 1.5 ml of the remaining hydrolyzed liquor was used to analyze structural carbohydrates (percent cellulose and percent hemicellulose) in the sample, as described previously (119, 120). The structural carbohydrate analysis was performed by high-performance liquid chromatography (HPLC) on a system equipped with a refractive index detector using a Shodex sugar SP0810 column after neutralization of the hydrolysis liquor with calcium carbonate. To analyze the acid insoluble lignin, the insoluble residue remaining after hydrolysis was washed with ultrapure water three times to remove the acid, dried overnight at 65°C, and weighed. The percent acid insoluble lignin was estimated by subtracting the percent extractive free ash from the percent acid-insoluble residue. The percent total lignin was the sum of the percent acid-insoluble lignin and percent acid-soluble lignin (119, 120).
Nonstructural carbohydrates were determined as described previously (121, 122), with minor modification. Briefly, 1 ml of 80% ethanol was added to 20 mg of dried sample and incubated in a water bath at 80°C for 20 min. The ethanol in the tubes was decanted, and the process was repeated two times. The decanted ethanol was evaporated by incubating the tube in a water bath at 80°C for 4 h; the remaining volume in the tube was adjusted to 1 ml with ultrapure water, and 40 μl was used to read absorbance at 490 nm. For glucose determination, 180 μl peroxidase/glucose oxidase (PGO) was added, and the sample was incubated at room temperature for 20 min followed by measurement of absorbance at 490 nm. Glucose content was determined by the difference between the background absorbance (before addition of PGO) and the absorbance with PGO (after incubation) and comparing it with a glucose standard curve. For sucrose determination, 40 μl of invertase solution was added to the previous sample and incubated at 35°C for 60 min followed by an absorbance reading at 490 nm. To calculate the sucrose content, the background and glucose absorbances were subtracted from the final absorbance reading using sucrose standard curve.
Measurement of PAL activity.
PAL activity was estimated using a phenylalanine ammonia lyase (PAL) microplate assay kit (Cohesion Biosciences) according to the manufacturer’s instructions. Briefly, 100 mg of leaves from mock- and virus-inoculated WT or BdPAL1 RNAi plants was homogenized with 1 ml of assay buffer in a prechilled mortar and pestle. Homogenized samples were centrifuged at 8,000 × g for 15 min at 4°C. The supernatant was transferred to a new 1.5-ml microcentrifuge tube and kept on ice. Assay samples were prepared in a 96-well plate by adding 10 μl of sample, 130 μl of reaction buffer, and 50 μl of l-phenylalanine (substrate) followed by a 30-min incubation at 30°C. After 30 min, 10 μl of stop solution was added, and PAL activity was quantified by measuring absorption spectra (optical density [OD]) at 290 nm. PAL activity was calculated according to manufacturer’s formula shown below:
Here, one unit (U) is defined as the OD value change of 0.01 per minute, and W is the weight of sample in grams.
Exogenous SA treatment and disease monitoring.
Brachypodium plants were grown to the three-leaf stage under the growth conditions mentioned previously. Then the WT and BdPAL1 RNAi plants were treated with 50 ml of 100 ppm SA (Sigma-Aldrich) or sterile water by soil drenching, followed by PMV or PMV+SPMV inoculation after 24 h (106). The SA treatment was repeated 24 h postinoculation. Then, the SA application was made once every 7 days for up to 4 weeks. Throughout the experiment, we monitored the progression of symptoms on noninoculated leaves (chlorosis and necrosis) and the overall plant habit. At 21 dpi, the noninoculated leaves were scored for percentage of leaves with chlorosis and necrosis symptoms.
Data analysis and statistics.
Data analysis, statistics, and graphs were prepared using Microsoft Excel (version 2009), Python package bioinfokit v1.0.2 (https://github.com/mandadi-lab/bioinfokit), and R package agricolae v1.3-3. Statistically significant differences were analyzed using two-sample t test (one-tailed) or analysis of variance (ANOVA) followed by Tukey’s test for multiple pairwise comparisons.
ACKNOWLEDGMENTS
This project was supported in part by the USDA Agriculture and Food Research Initiative National Institute of Food and Agriculture (NIFA; 2016-67013-24738, HATCH 1023984, and CSP503406 to K.K.M. and K.-B.G.S.) and the US Department of Energy Great Lakes Bioenergy Research Center (Department of Energy, Biological and Environmental Research, Office of Science, grant no. DE-FC02-07ER64494 to J.C.S.).
We thank Brianna Jacques, Gerleene Acuna, and Rolando Mireles (Texas A&M AgriLife Research, Weslaco, TX) for their technical assistance with plant propagation and cell wall composition analysis.
Footnotes
This article is a direct contribution from Karen-Beth G. Scholthof, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Vitaly Citovsky, Stony Brook University, and James Schoelz, University of Missouri.
Citation Pant SR, Irigoyen S, Liu J, Bedre R, Christensen SA, Schmelz EA, Sedbrook JC, Scholthof K-BG, Mandadi KK. 2021. Brachypodium phenylalanine ammonia lyase (PAL) promotes antiviral defenses against Panicum mosaic virus and its satellites. mBio 12:e03518-20. https://doi.org/10.1128/mBio.03518-20.
Contributor Information
Kranthi K. Mandadi, Email: kkmandadi@tamu.edu.
Frederick M. Ausubel, Mass General Hospital
REFERENCES
- 1.Pieterse CM, Leon-Reyes A, Van der Ent S, Van Wees SC. 2009. Networking by small-molecule hormones in plant immunity. Nat Chem Biol 5:308–316. doi: 10.1038/nchembio.164. [DOI] [PubMed] [Google Scholar]
- 2.Jones JD, Dangl JL. 2006. The plant immune system. Nature 444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 3.Mandadi KK, Scholthof K-BG. 2013. Plant immune responses against viruses: how does a virus cause disease? Plant Cell 25:1489–1505. doi: 10.1105/tpc.113.111658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang H, Gou X, He K, Xi D, Du J, Lin H, Li J. 2010. BAK1 and BKK1 in Arabidopsis thaliana confer reduced susceptibility to Turnip crinkle virus. Eur J Plant Pathol 127:149–156. doi: 10.1007/s10658-010-9581-5. [DOI] [Google Scholar]
- 5.Korner CJ, Klauser D, Niehl A, Dominguez-Ferreras A, Chinchilla D, Boller T, Heinlein M, Hann DR. 2013. The immunity regulator BAK1 contributes to resistance against diverse RNA viruses. Mol Plant Microbe Interact 26:1271–1280. doi: 10.1094/MPMI-06-13-0179-R. [DOI] [PubMed] [Google Scholar]
- 6.Macho AP, Zipfel C. 2014. Plant PRRs and the activation of innate immune signaling. Mol Cell 54:263–272. doi: 10.1016/j.molcel.2014.03.028. [DOI] [PubMed] [Google Scholar]
- 7.Chivasa S, Murphy AM, Naylor M, Carr JP. 1997. Salicylic acid interferes with Tobacco mosaic virus replication via a novel salicylhydroxamic acid-sensitive mechanism. Plant Cell 9:547–557. doi: 10.2307/3870506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naylor M, Murphy AM, Berry JO, Carr JP. 1998. Salicylic acid can induce resistance to plant virus movement. Mol Plant Microbe Interact 11:860–868. doi: 10.1094/MPMI.1998.11.9.860. [DOI] [Google Scholar]
- 9.Gaffney T, Friedrich L, Vernooij B, Negrotto D, Nye G, Uknes S, Ward E, Kessmann H, Ryals J. 1993. Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261:754–756. doi: 10.1126/science.261.5122.754. [DOI] [PubMed] [Google Scholar]
- 10.Murphy AM, Carr JP. 2002. Salicylic acid has cell-specific effects on tobacco mosaic virus replication and cell-to-cell movement. Plant Physiol 128:552–563. doi: 10.1104/pp.010688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baebler Š, Witek K, Petek M, Stare K, Tušek-Žnidarič M, Pompe-Novak M, Renaut J, Szajko K, Strzelczyk-Żyta D, Marczewski W, Morgiewicz K, Gruden K, Hennig J. 2014. Salicylic acid is an indispensable component of the Ny-1 resistance-gene-mediated response against Potato virus Y infection in potato. J Exp Bot 65:1095–1109. doi: 10.1093/jxb/ert447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baebler S, Stare K, Kovac M, Blejec A, Prezelj N, Stare T, Kogovsek P, Pompe-Novak M, Rosahl S, Ravnikar M, Gruden K. 2011. Dynamics of responses in compatible potato-Potato virus Y interaction are modulated by salicylic acid. PLoS One 6:e29009. doi: 10.1371/journal.pone.0029009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Huijsduijnen RH, Alblas SW, De Rijk RH, Bol JF. 1986. Induction by salicylic acid of pathogenesis-related proteins and resistance to Alfalfa mosaic virus infection in various plant species. J Gen Virol 67:2135–2143. doi: 10.1099/0022-1317-67-10-2135. [DOI] [Google Scholar]
- 14.Murphy AM, Zhou T, Carr JP. 2020. An update on salicylic acid biosynthesis, its induction and potential exploitation by plant viruses. Curr Opin Virol 42:8–17. doi: 10.1016/j.coviro.2020.02.008. [DOI] [PubMed] [Google Scholar]
- 15.Carr JP, Murphy AM, Tungadi T, Yoon J-Y. 2019. Plant defense signals: players and pawns in plant-virus-vector interactions. Plant Sci 279:87–95. doi: 10.1016/j.plantsci.2018.04.011. [DOI] [PubMed] [Google Scholar]
- 16.Matsuo Y, Novianti F, Takehara M, Fukuhara T, Arie T, Komatsu K. 2019. Acibenzolar-S-methyl restricts infection of Nicotiana benthamiana by Plantago asiatica mosaic virus at two distinct stages. Mol Plant Microbe Interact 32:1475–1486. doi: 10.1094/MPMI-03-19-0087-R. [DOI] [PubMed] [Google Scholar]
- 17.Scholthof K-BG. 2020. Brachypodium and plant viruses: entwined tools for discovery. New Phytol 227:1676–1680. doi: 10.1111/nph.16388. [DOI] [PubMed] [Google Scholar]
- 18.Scholthof K-BG, Irigoyen S, Catalan P, Mandadi KK. 2018. Brachypodium: a monocot grass model genus for plant biology. Plant Cell 30:1673–1694. doi: 10.1105/tpc.18.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gordon SP, Contreras-Moreira B, Woods DP, Des Marais DL, Burgess D, Shu S, Stritt C, Roulin AC, Schackwitz W, Tyler L, Martin J, Lipzen A, Dochy N, Phillips J, Barry K, Geuten K, Budak H, Juenger TE, Amasino R, Caicedo AL, Goodstein D, Davidson P, Mur LAJ, Figueroa M, Freeling M, Catalan P, Vogel JP. 2017. Extensive gene content variation in the Brachypodium distachyon pan-genome correlates with population structure. Nat Commun 8:2184. doi: 10.1038/s41467-017-02292-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turina M, Maruoka M, Monis J, Jackson AO, Scholthof K-BG. 1998. Nucleotide sequence and infectivity of a full-length cDNA clone of panicum mosaic virus. Virology 241:141–155. doi: 10.1006/viro.1997.8939. [DOI] [PubMed] [Google Scholar]
- 21.Cui Y, Lee MY, Huo N, Bragg J, Yan L, Yuan C, Li C, Holditch SJ, Xie J, Luo MC, Li D, Yu J, Martin J, Schackwitz W, Gu YQ, Vogel JP, Jackson AO, Liu Z, Garvin DF. 2012. Fine mapping of the Bsr1 barley stripe mosaic virus resistance gene in the model grass Brachypodium distachyon. PLoS One 7:e38333. doi: 10.1371/journal.pone.0038333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fitzgerald TL, Powell JJ, Schneebeli K, Hsia MM, Gardiner DM, Bragg JN, McIntyre CL, Manners JM, Ayliffe M, Watt M, Vogel JP, Henry RJ, Kazan K. 2015. Brachypodium as an emerging model for cereal-pathogen interactions. Ann Bot 115:717–731. doi: 10.1093/aob/mcv010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee MY, Yan L, Gorter FA, Kim BY, Cui Y, Hu Y, Yuan C, Grindheim J, Ganesan U, Liu Z, Han C, Yu J, Li D, Jackson AO. 2012. Brachypodium distachyon line Bd3-1 resistance is elicited by the Barley stripe mosaic virus triple gene block 1 movement protein. J Gen Virol 93:2729–2739. doi: 10.1099/vir.0.045880-0. [DOI] [PubMed] [Google Scholar]
- 24.Mandadi KK, Scholthof K-B. 2012. Characterization of a viral synergism in the monocot Brachypodium distachyon reveals distinctly altered host molecular processes associated with disease. Plant Physiol 160:1432–1452. doi: 10.1104/pp.112.204362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mandadi KK, Pyle JD, Scholthof K-BG. 2014. Comparative analysis of antiviral responses in Brachypodium distachyon and Setaria viridis reveals conserved and unique outcomes among C3 and C4 plant defenses. Mol Plant Microbe Interact 27:1277–1290. doi: 10.1094/MPMI-05-14-0152-R. [DOI] [PubMed] [Google Scholar]
- 26.Mandadi KK, Scholthof K-BG. 2015. Genomic architecture and functional relationships of intronless, constitutively- and alternatively-spliced genes in Brachypodium distachyon. Plant Signal Behav 10:e1042640. doi: 10.1080/15592324.2015.1042640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pyle JD, Monis J, Scholthof K-BG. 2017. Complete nucleotide sequences and virion particle association of two satellite RNAs of Panicum mosaic virus. Virus Res 240:87–93. doi: 10.1016/j.virusres.2017.06.026. [DOI] [PubMed] [Google Scholar]
- 28.Pyle JD, Scholthof K-BG. 2018. De novo generation of helper virus-satellite chimera RNAs results in disease attenuation and satellite sequence acquisition in a host-dependent manner. Virology 514:182–191. doi: 10.1016/j.virol.2017.11.006. [DOI] [PubMed] [Google Scholar]
- 29.Liou M-R, Huang Y-W, Hu C-C, Lin N-S, Hsu Y-H. 2014. A dual gene-silencing vector system for monocot and dicot plants. Plant Biotechnol J 12:330–343. doi: 10.1111/pbi.12140. [DOI] [PubMed] [Google Scholar]
- 30.Mei Y, Liu G, Zhang C, Hill JH, Whitham SA. 2019. A sugarcane mosaic virus vector for gene expression in maize. Plant Direct 3:e00158. doi: 10.1002/pld3.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tao Y, Nadege SW, Huang C, Zhang P, Song S, Sun L, Wu Y. 2016. Brachypodium distachyon is a suitable host plant for study of Barley yellow dwarf virus. Virus Genes 52:299–302. doi: 10.1007/s11262-016-1297-y. [DOI] [PubMed] [Google Scholar]
- 32.Batten JS, Turina M, Scholthof K-BG. 2006. Panicovirus accumulation is governed by two membrane-associated proteins with a newly identified conserved motif that contributes to pathogenicity. Virol J 3:12. doi: 10.1186/1743-422X-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turina M, Desvoyes B, Scholthof K-BG. 2000. A gene cluster encoded by Panicum mosaic virus is associated with virus movement. Virology 266:120–128. doi: 10.1006/viro.1999.0069. [DOI] [PubMed] [Google Scholar]
- 34.Pyle JD, Mandadi KK, Scholthof K-BG. 2019. Panicum mosaic virus and its satellites acquire RNA modifications associated with host-mediated antiviral degradation. mBio 10:e01900-19. doi: 10.1128/mBio.01900-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pyle JD, Scholthof K-BG. 2017. Biology and pathogenesis of satellite viruses, p 627–636. In Hadidi A, Flores R, Randles JW, Palukaitis P (ed), Viroids and satellites. Academic Press, San Diego, CA. doi: 10.1016/B978-0-12-801498-1.00058-9. [DOI] [Google Scholar]
- 36.Stewart CL, Pyle JD, Jochum CC, Vogel KP, Yuen GY, Scholthof K-BG. 2015. Multi-year pathogen survey of biofuel switchgrass breeding plots reveals high prevalence of infections by Panicum mosaic virus and its satellite virus. Phytopathology 105:1146–1154. doi: 10.1094/PHYTO-03-15-0062-R. [DOI] [PubMed] [Google Scholar]
- 37.Ban N, McPherson A. 1995. The structure of satellite Panicum mosaic virus at 1.9 Å resolution. Nat Struct Biol 2:882–890. doi: 10.1038/nsb1095-882. [DOI] [PubMed] [Google Scholar]
- 38.Desvoyes B, Scholthof K-BG. 2000. RNA:protein interactions associated with satellites of Panicum mosaic virus. FEBS Lett 485:25–28. doi: 10.1016/S0014-5793(00)02177-3. [DOI] [PubMed] [Google Scholar]
- 39.Qiu W, Scholthof K-BG. 2004. Satellite panicum mosaic virus capsid protein elicits symptoms on a nonhost plant and interferes with a suppressor of virus-induced gene silencing. Mol Plant Microbe Interact 17:263–271. doi: 10.1094/MPMI.2004.17.3.263. [DOI] [PubMed] [Google Scholar]
- 40.Dixon RA, Paiva N. 1995. Stress-induced phenylpropanoid metabolism. Plant Cell 7:1085–1097. doi: 10.1105/tpc.7.7.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang J, Gu M, Lai Z, Fan B, Shi K, Zhou YH, Yu JQ, Chen Z. 2010. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol 153:1526–1538. doi: 10.1104/pp.110.157370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qian Y, Lynch JH, Guo L, Rhodes D, Morgan JA, Dudareva N. 2019. Completion of the cytosolic post-chorismate phenylalanine biosynthetic pathway in plants. Nat Commun 10:15. doi: 10.1038/s41467-018-07969-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu L, Zhu L, Tu L, Liu L, Yuan D, Jin L, Long L, Zhang X. 2011. Lignin metabolism has a central role in the resistance of cotton to the wilt fungus Verticillium dahliae as revealed by RNA-Seq-dependent transcriptional analysis and histochemistry. J Exp Bot 62:5607–5621. doi: 10.1093/jxb/err245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gayoso C, Pomar F, Novo-Uzal E, Merino F, de Ilarduya OM. 2010. The Ve-mediated resistance response of the tomato to Verticillium dahliae involves H2O2, peroxidase and lignins and drives PAL gene expression. BMC Plant Biol 10:232. doi: 10.1186/1471-2229-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuan W, Jiang T, Du K, Chen H, Cao Y, Xie J, Li M, Carr JP, Wu B, Fan Z, Zhou T. 2019. Maize phenylalanine ammonia-lyases contribute to resistance to Sugarcane mosaic virus infection, most likely through positive regulation of salicylic acid accumulation. Mol Plant Pathol 20:1365–1378. doi: 10.1111/mpp.12817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Otulak-Kozieł K, Kozieł E, Lockhart B. 2018. Plant cell wall dynamics in compatible and incompatible potato response to infection caused by potato virus Y (PVYNTN). Int J Mol Sci 19:862. doi: 10.3390/ijms19030862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lionetti V, Cervone F, Bellincampi D. 2012. Methyl esterification of pectin plays a role during plant–pathogen interactions and affects plant resistance to diseases. J Plant Physiol 169:1623–1630. doi: 10.1016/j.jplph.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 48.Smit F, Dubery IA. 1997. Cell wall reinforcement in cotton hypocotyls in response to a Verticillium dahliae elicitor. Phytochemistry 44:811–815. doi: 10.1016/S0031-9422(96)00595-X. [DOI] [Google Scholar]
- 49.Bedre R, Rajasekaran K, Mangu VR, Timm LES, Bhatnagar D, Baisakh N. 2015. Genome-wide transcriptome analysis of cotton (Gossypium hirsutum L.) identifies candidate gene signatures in response to aflatoxin producing fungus Aspergillus flavus. PLoS One 10:e0138025. doi: 10.1371/journal.pone.0138025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cass CL, Peraldi A, Dowd PF, Mottiar Y, Santoro N, Karlen SD, Bukhman YV, Foster CE, Thrower N, Bruno LC, Moskvin OV, Johnson ET, Willhoit ME, Phutane M, Ralph J, Mansfield SD, Nicholson P, Sedbrook JC. 2015. Effects of phenylalanine ammonia lyase (PAL) knockdown on cell wall composition, biomass digestibility, and biotic and abiotic stress responses in Brachypodium. J Exp Bot 66:4317–4335. doi: 10.1093/jxb/erv269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pallas JA, Paiva NL, Lamb C, Dixon RA. 1996. Tobacco plants epigenetically suppressed in phenylalanine ammonia‐lyase expression do not develop systemic acquired resistance in response to infection by Tobacco mosaic virus. Plant J 10:281–293. doi: 10.1046/j.1365-313X.1996.10020281.x. [DOI] [Google Scholar]
- 52.He Y, Li X, Zhan F, Xie C, Zu Y, Li Y, Yue M. 2018. Resistance-related physiological response of rice leaves to the compound stress of enhanced UV-B radiation and Magnaporthe oryzae. J Plant Interact 13:321–328. doi: 10.1080/17429145.2018.1478007. [DOI] [Google Scholar]
- 53.Kim DS, Hwang BK. 2014. An important role of the pepper phenylalanine ammonia-lyase gene (PAL1) in salicylic acid-dependent signalling of the defence response to microbial pathogens. J Exp Bot 65:2295–2306. doi: 10.1093/jxb/eru109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Usadel B, Nagel A, Thimm O, Redestig H, Blaesing OE, Palacios-Rojas N, Selbig J, Hannemann J, Piques MC, Steinhauser D, Scheible WR, Gibon Y, Morcuende R, Weicht D, Meyer S, Stitt M. 2005. Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol 138:1195–1204. doi: 10.1104/pp.105.060459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, Kruger P, Selbig J, Muller LA, Rhee SY, Stitt M. 2004. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37:914–939. doi: 10.1111/j.1365-313x.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- 56.Bolton MD. 2009. Primary metabolism and plant defense–fuel for the fire. Mol Plant Microbe Interact 22:487–497. doi: 10.1094/MPMI-22-5-0487. [DOI] [PubMed] [Google Scholar]
- 57.Kangasjarvi S, Neukermans J, Li S, Aro EM, Noctor G. 2012. Photosynthesis, photorespiration, and light signalling in defence responses. J Exp Bot 63:1619–1636. doi: 10.1093/jxb/err402. [DOI] [PubMed] [Google Scholar]
- 58.Kachroo A, Robin GP. 2013. Systemic signaling during plant defense. Curr Opin Plant Biol 16:527–533. doi: 10.1016/j.pbi.2013.06.019. [DOI] [PubMed] [Google Scholar]
- 59.Scholes JD, Rolfe SA. 1996. Photosynthesis in localised regions of oat leaves infected with crown rust (Puccinia coronata): quantitative imaging of chlorophyll fluorescence. Planta 199:573–582. doi: 10.1007/BF00195189. [DOI] [Google Scholar]
- 60.Swarbrick PJ, Schulze-Lefert P, Scholes JD. 2006. Metabolic consequences of susceptibility and resistance (race-specific and broad-spectrum) in barley leaves challenged with powdery mildew. Plant Cell Environ 29:1061–1076. doi: 10.1111/j.1365-3040.2005.01472.x. [DOI] [PubMed] [Google Scholar]
- 61.Bilgin DD, Zavala JA, Zhu J, Clough SJ, Ort DR, DeLucia EH. 2010. Biotic stress globally downregulates photosynthesis genes. Plant Cell Environ 33:1597–1613. doi: 10.1111/j.1365-3040.2010.02167.x. [DOI] [PubMed] [Google Scholar]
- 62.Hendrawati O, Yao Q, Kim HK, Linthorst HJ, Erkelens C, Lefeber AW, Choi YH, Verpoorte R. 2006. Metabolic differentiation of Arabidopsis treated with methyl jasmonate using nuclear magnetic resonance spectroscopy. Plant Sci 170:1118–1124. doi: 10.1016/j.plantsci.2006.01.017. [DOI] [Google Scholar]
- 63.Vlot AC, Dempsey DA, Klessig DF. 2009. Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol 47:177–206. doi: 10.1146/annurev.phyto.050908.135202. [DOI] [PubMed] [Google Scholar]
- 64.Loake G, Grant M. 2007. Salicylic acid in plant defence—the players and protagonists. Curr Opin Plant Biol 10:466–472. doi: 10.1016/j.pbi.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 65.Lukan T, Pompe-Novak M, Baebler S, Tusek-Znidaric M, Kladnik A, Kriznik M, Blejec A, Zagorscak M, Stare K, Dusak B, Coll A, Pollman S, Morgiewicz K, Hennig J, Gruden G. 2020. Precision transcriptomics of viral foci reveals the spatial regulation of immune‐signaling genes and identifies RBOHD as an important player in the incompatible interaction between potato virus Y and potato. Plant J 104:645–661. doi: 10.1111/tpj.14953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Staswick PE, Yuen GY, Lehman CC. 1998. Jasmonate signaling mutants of Arabidopsis are susceptible to the soil fungus Pythium irregulare. Plant J 15:747–754. doi: 10.1046/j.1365-313x.1998.00265.x. [DOI] [PubMed] [Google Scholar]
- 67.Halim VA, Vess A, Scheel D, Rosahl S. 2006. The role of salicylic acid and jasmonic acid in pathogen defence. Plant Biol (Stuttg) 8:307–313. doi: 10.1055/s-2006-924025. [DOI] [PubMed] [Google Scholar]
- 68.Chehab EW, Kaspi R, Savchenko T, Rowe H, Negre-Zakharov F, Kliebenstein D, Dehesh K. 2008. Distinct roles of jasmonates and aldehydes in plant-defense responses. PLoS One 3:e1904. doi: 10.1371/journal.pone.0001904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Glazebrook J. 2005. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- 70.Vick BA, Zimmerman DC. 1984. Biosynthesis of jasmonic acid by several plant-species. Plant Physiol 75:458–461. doi: 10.1104/pp.75.2.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li C, Schilmiller AL, Liu G, Lee GI, Jayanty S, Sageman C, Vrebalov J, Giovannoni JJ, Yagi K, Kobayashi Y, Howe GA. 2005. Role of β-oxidation in jasmonate biosynthesis and systemic wound signaling in tomato. Plant Cell 17:971–986. doi: 10.1105/tpc.104.029108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Harms K, Ramirez I, Peña-Cortés H. 1998. Inhibition of wound-induced accumulation of allene oxide synthase transcripts in flax leaves by aspirin and salicylic acid. Plant Physiol 118:1057–1065. doi: 10.1104/pp.118.3.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.An C, Mou Z. 2011. Salicylic acid and its function in plant immunity. J Integr Plant Biol 53:412–428. doi: 10.1111/j.1744-7909.2011.01043.x. [DOI] [PubMed] [Google Scholar]
- 74.Pieterse CM, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SC. 2012. Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol 28:489–521. doi: 10.1146/annurev-cellbio-092910-154055. [DOI] [PubMed] [Google Scholar]
- 75.Wang K-D, Borrego EJ, Kenerley CM, Kolomiets MV. 2020. Oxylipins other than jasmonic acid are xylem-resident signals regulating systemic resistance induced by Trichoderma virens in maize. Plant Cell 32:166–185. doi: 10.1105/tpc.19.00487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stintzi A, Weber H, Reymond P, Browse J, Farmer EE. 2001. Plant defense in the absence of jasmonic acid: the role of cyclopentenones. Proc Natl Acad Sci U S A 98:12837–12842. doi: 10.1073/pnas.211311098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Savchenko T, Walley JW, Chehab EW, Xiao Y, Kaspi R, Pye MF, Mohamed ME, Lazarus CM, Bostock RM, Dehesh K. 2010. Arachidonic acid: an evolutionarily conserved signaling molecule modulates plant stress signaling networks. Plant Cell 22:3193–3205. doi: 10.1105/tpc.110.073858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jump DB. 2002. Dietary polyunsaturated fatty acids and regulation of gene transcription. Curr Opin Lipidol 13:155–164. doi: 10.1097/00041433-200204000-00007. [DOI] [PubMed] [Google Scholar]
- 79.Duplus E, Glorian M, Forest C. 2000. Fatty acid regulation of gene transcription. J Biol Chem 275:30749–30752. doi: 10.1074/jbc.R000015200. [DOI] [PubMed] [Google Scholar]
- 80.Calvo AM, Hinze LL, Gardner HW, Keller NP. 1999. Sporogenic effect of polyunsaturated fatty acids on development of Aspergillus spp. Appl Environ Microbiol 65:3668–3673. doi: 10.1128/AEM.65.8.3668-3673.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Madi L, Wang XJ, Kobiler A, Lichter A, Prusky D. 2003. Stress on avocado fruits regulates delta(9)-stearoyl ACP desaturase expression, fatty acid composition, antifungal diene level and resistance to Colletotrichum gloeosporioides attack. Physiol Mol Plant Pathol 62:277–283. doi: 10.1016/S0885-5765(03)00076-6. [DOI] [Google Scholar]
- 82.Aliferis KA, Faubert D, Jabaji S. 2014. A metabolic profiling strategy for the dissection of plant defense against fungal pathogens. PLoS One 9:e111930. doi: 10.1371/journal.pone.0111930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Blee E. 2002. Impact of phyto-oxylipins in plant defense. Trends Plant Sci 7:315–321. doi: 10.1016/s1360-1385(02)02290-2. [DOI] [PubMed] [Google Scholar]
- 84.Yaeno T, Matsuda O, Iba K. 2004. Role of chloroplast trienoic fatty acids in plant disease defense responses. Plant J 40:931–941. doi: 10.1111/j.1365-313X.2004.02260.x. [DOI] [PubMed] [Google Scholar]
- 85.Wasternack C. 2007. Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann Bot 100:682–697. doi: 10.1093/aob/mcm079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dave A, Graham IA. 2012. Oxylipin signaling: a distinct role for the jasmonic acid precursor cis-(+)-12-oxo-phytodienoic acid (cis-OPDA). Front Plant Sci 3:42. doi: 10.3389/fpls.2012.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shanklin J, Cahoon EB. 1998. Desaturation and related modifications of fatty acids. Annu Rev Plant Physiol Plant Mol Biol 49:611–641. doi: 10.1146/annurev.arplant.49.1.611. [DOI] [PubMed] [Google Scholar]
- 88.Farmer EE, Ryan CA. 1992. Octadecanoid precursors of jasmonic acid activate the synthesis of wound-inducible proteinase inhibitors. Plant Cell 4:129–134. doi: 10.1105/tpc.4.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kachroo P, Shanklin J, Shah J, Whittle EJ, Klessig DF. 2001. A fatty acid desaturase modulates the activation of defense signaling pathways in plants. Proc Natl Acad Sci U S A 98:9448–9453. doi: 10.1073/pnas.151258398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kachroo A, Lapchyk L, Fukushige H, Hildebrand D, Klessig D, Kachroo P. 2003. Plastidial fatty acid signaling modulates salicylic acid- and jasmonic acid-mediated defense pathways in the Arabidopsis ssi2 mutant. Plant Cell 15:2952–2965. doi: 10.1105/tpc.017301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chaman ME, Copaja SV, Argandona VH. 2003. Relationships between salicylic acid content, phenylalanine ammonia-lyase (PAL) activity, and resistance of barley to aphid infestation. J Agric Food Chem 51:2227–2231. doi: 10.1021/jf020953b. [DOI] [PubMed] [Google Scholar]
- 92.Mauch-Mani B, Slusarenko AJ. 1996. Production of salicylic acid precursors is a major function of phenylalanine ammonia-lyase in the resistance of Arabidopsis to Peronospora parasitica. Plant Cell 8:203–212. doi: 10.1105/tpc.8.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nugroho LH, Verberne MC, Verpoorte R. 2002. Activities of enzymes involved in the phenylpropanoid pathway in constitutively salicylic acid-producing tobacco plants. Plant Physiol Biochem 40:755–760. doi: 10.1016/S0981-9428(02)01437-7. [DOI] [Google Scholar]
- 94.Garcion C, Lohmann A, Lamodiere E, Catinot J, Buchala A, Doermann P, Metraux JP. 2008. Characterization and biological function of the isochorismate synthase2 gene of Arabidopsis. Plant Physiol 147:1279–1287. doi: 10.1104/pp.108.119420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lee H-I, León J, Raskin I. 1995. Biosynthesis and metabolism of salicylic acid. Proc Natl Acad Sci U S A 92:4076–4079. doi: 10.1073/pnas.92.10.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shine M, Yang JW, El‐Habbak M, Nagyabhyru P, Fu DQ, Navarre D, Ghabrial S, Kachroo P, Kachroo A. 2016. Cooperative functioning between phenylalanine ammonia lyase and isochorismate synthase activities contributes to salicylic acid biosynthesis in soybean. New Phytol 212:627–636. doi: 10.1111/nph.14078. [DOI] [PubMed] [Google Scholar]
- 97.Hahlbrock K, Scheel D. 1989. Physiology and molecular biology of phenylpropanoid metabolism. Annu Rev Plant Physiol Plant Mol Biol 40:347–369. doi: 10.1146/annurev.pp.40.060189.002023. [DOI] [Google Scholar]
- 98.Vogt T. 2010. Phenylpropanoid biosynthesis. Mol Plant 3:2–20. doi: 10.1093/mp/ssp106. [DOI] [PubMed] [Google Scholar]
- 99.Mandadi KK, Scholthof K-BG. 2015. Genome-wide analysis of alternative splicing landscapes modulated during plant-virus interactions in Brachypodium distachyon. Plant Cell 27:71–85. doi: 10.1105/tpc.114.133991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rivas-San Vicente M, Plasencia J. 2011. Salicylic acid beyond defence: its role in plant growth and development. J Exp Bot 62:3321–3338. doi: 10.1093/jxb/err031. [DOI] [PubMed] [Google Scholar]
- 101.Glazebrook J, Chen W, Estes B, Chang HS, Nawrath C, Métraux JP, Zhu T, Katagiri F. 2003. Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression phenotyping. Plant J 34:217–228. doi: 10.1046/j.1365-313x.2003.01717.x. [DOI] [PubMed] [Google Scholar]
- 102.De Vos M, Van Oosten VR, Van Poecke RM, Van Pelt JA, Pozo MJ, Mueller MJ, Buchala AJ, Métraux JP, Van Loon LC, Dicke M, Pieterse CM. 2005. Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol Plant Microbe Interact 18:923–937. doi: 10.1094/MPMI-18-0923. [DOI] [PubMed] [Google Scholar]
- 103.Sato M, He S, Tsuda K, Wang L, Coller J, Watanabe Y, Glazebrook J, Katagiri F. 2010. Network modeling reveals prevalent negative regulatory relationships between signaling sectors in Arabidopsis immune signaling. PLoS Pathog 6:e1001011. doi: 10.1371/journal.ppat.1001011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Van der Does D, Leon-Reyes A, Koornneef A, Van Verk MC, Rodenburg N, Pauwels L, Goossens A, Korbes AP, Memelink J, Ritsema T, Van Wees SC, Pieterse CM. 2013. Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1-JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell 25:744–761. doi: 10.1105/tpc.112.108548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bruggeman Q, Raynaud C, Benhamed M, Delarue M. 2015. To die or not to die? Lessons from lesion mimic mutants. Front Plant Sci 6:24. doi: 10.3389/fpls.2015.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.He CY, Wolyn DJ. 2005. Potential role for salicylic acid in induced resistance of asparagus roots to Fusarium oxysporum f.sp. asparagi. Plant Pathol 54:227–232. doi: 10.1111/j.1365-3059.2005.01163.x. [DOI] [Google Scholar]
- 107.Park S-W, Kaimoyo E, Kumar D, Mosher S, Klessig DF. 2007. Methyl salicylate is a critical mobile signal for plant systemic acquired resistance. Science 318:113–116. doi: 10.1126/science.1147113. [DOI] [PubMed] [Google Scholar]
- 108.Mayers CN, Lee KC, Moore CA, Wong SM, Carr JP. 2005. Salicylic acid-induced resistance to Cucumber mosaic virus in squash and Arabidopsis thaliana: contrasting mechanisms of induction and antiviral action. Mol Plant Microbe Interact 18:428–434. doi: 10.1094/MPMI-18-0428. [DOI] [PubMed] [Google Scholar]
- 109.Murphy AM, Chivasa S, Singh DP, Carr JP. 1999. Salicylic acid-induced resistance to viruses and other pathogens: a parting of the ways? Trends Plant Sci 4:155–160. doi: 10.1016/s1360-1385(99)01390-4. [DOI] [PubMed] [Google Scholar]
- 110.Wang W, Withers J, Li H, Zwack PJ, Rusnac D-V, Shi H, Liu L, Yan S, Hinds TR, Guttman M. 2020. Structural basis of salicylic acid perception by Arabidopsis NPR proteins. Nature :1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Klessig DF, Choi HW, Dempsey DA. 2018. Systemic acquired resistance and salicylic acid: past, present, and future. Mol Plant Microbe Interact 31:871–888. doi: 10.1094/MPMI-03-18-0067-CR. [DOI] [PubMed] [Google Scholar]
- 112.Ding Y, Sun T, Ao K, Peng Y, Zhang Y, Li X, Zhang Y. 2018. Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell 173:1454–1467. doi: 10.1016/j.cell.2018.03.044. [DOI] [PubMed] [Google Scholar]
- 113.Qi D, Omarov RT, Scholthof K-BG. 2008. The complex subcellular distribution of satellite panicum mosaic virus capsid protein reflects its multifunctional role during infection. Virology 376:154–164. doi: 10.1016/j.virol.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 114.Arvidsson S, Kwasniewski M, Riano-Pachon DM, Mueller-Roeber B. 2008. QuantPrime—a flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinformatics 9:465. doi: 10.1186/1471-2105-9-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hong SY, Seo PJ, Yang MS, Xiang F, Park CM. 2008. Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real-time PCR. BMC Plant Biol 8:112. doi: 10.1186/1471-2229-8-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 117.Usadel B, Poree F, Nagel A, Lohse M, Czedik-Eysenberg A, Stitt M. 2009. A guide to using MapMan to visualize and compare omics data in plants: a case study in the crop species, Maize. Plant Cell Environ 32:1211–1229. doi: 10.1111/j.1365-3040.2009.01978.x. [DOI] [PubMed] [Google Scholar]
- 118.Schmelz EA, Kaplan F, Huffaker A, Dafoe NJ, Vaughan MM, Ni X, Rocca JR, Alborn HT, Teal PE. 2011. Identity, regulation, and activity of inducible diterpenoid phytoalexins in maize. Proc Natl Acad Sci U S A 108:5455–5460. doi: 10.1073/pnas.1014714108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.DeMartini JD, Studer MH, Wyman CE. 2011. Small-scale and automatable high-throughput compositional analysis of biomass. Biotechnol Bioeng 108:306–312. doi: 10.1002/bit.22937. [DOI] [PubMed] [Google Scholar]
- 120.Sluiter A, Hames B, Ruiz R, Scarlata C, Sluiter J, Templeton D, Crocker D. 2008. Determination of structural carbohydrates and lignin in biomass. Laboratory analytical procedure. https://www.nrel.gov/docs/gen/fy13/42618.pdf.
- 121.Ober ES, Setter TL, Madison JT, Thompson JF, Shapiro PS. 1991. Influence of water deficit on maize endosperm development: enzyme activities and RNA transcripts of starch and zein synthesis, abscisic acid, and cell division. Plant Physiol 97:154–164. doi: 10.1104/pp.97.1.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hendrix DL. 1993. Rapid extraction and analysis of nonstructural carbohydrates in plant tissues. Crop Sci 33:1306–1311. doi: 10.2135/cropsci1993.0011183X003300060037x. [DOI] [Google Scholar]
- 123.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25(17):3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene expression data (>2 log2 fold change, FDR < 0.05) in PMV-infected Brachypodium leaves at stage II. Download Data Set S1, XLSX file, 0.1 MB (52.8KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Primers used for SA and JA defense marker gene expression analysis. Download Table S1, DOCX file, 0.1 MB (30.2KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.