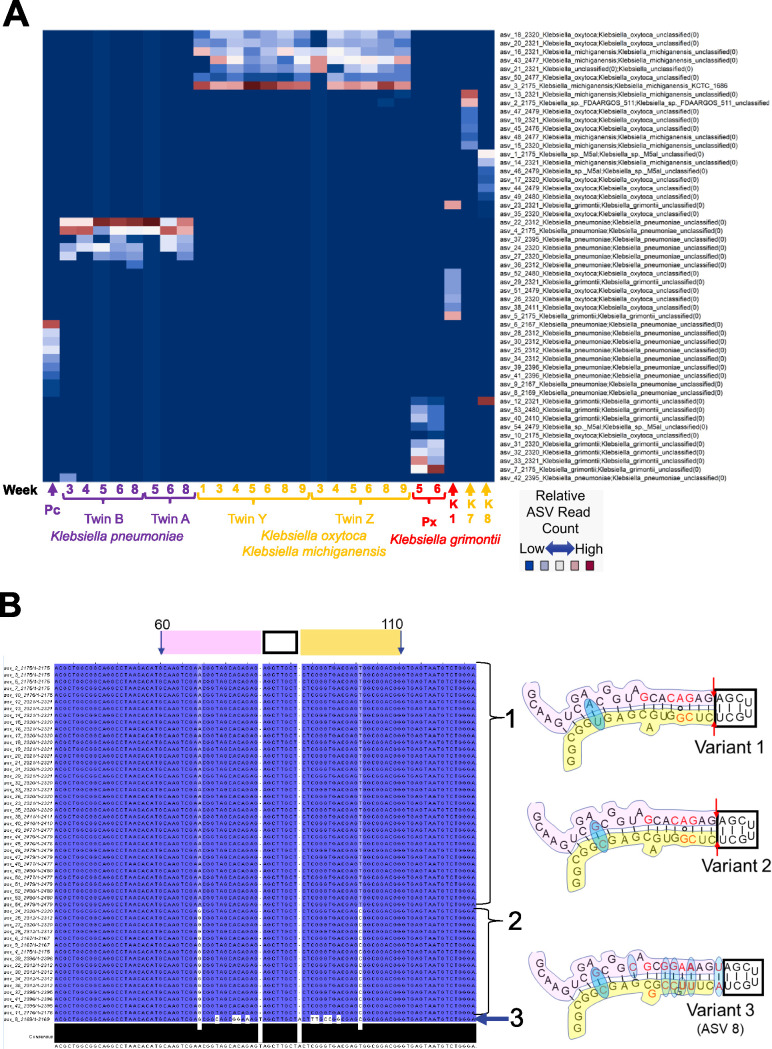
FIG 3.
Klebsiella amplicon sequence variant fingerprints. (A) Relative-abundance heat map of ∼2,100- to 2,500-base StrainID amplicon sequence variants (ASVs). Samples sharing Klebsiella strains have similar ASV fingerprint patterns. On the x axis, sample IDs are indicated by brackets, and numbers indicate the week since birth when the sample was obtained. Individual samples are indicated by arrows. “Pc” and “Px” indicate fecal sample ASVs from unrelated preterm infants, classified as K. pneumoniae and K. grimontii, respectively. K1, K7, and K8 represent ASVs from Klebsiella isolates from unrelated preterm infants who developed necrotizing enterocolitis. Reads classified by SBanalyzer as Klebsiella were analyzed using DADA2, resulting in the 54 Klebsiella ASVs listed on the y axis. The ASV number is followed by the four-digit ASV length and the taxonomy assigned for that ASV by SBanalyzer. Each coordinate representing an ASV in a sample was colored according to the relative number of reads. Possible species-level taxonomic assignments for the fingerprint in each sample based on the best mapping results on the y axis are indicated by the label colors on the x axis for K. pneumoniae, K. oxytoca/K. michiganensis, and K. grimontii. (B) Klebsiella ASVs in V1 maintain a conserved stem-loop structure. All 54 Klebsiella ASVs were aligned in Jalview (40), with each base numbered according to the alignment and shaded by percent identity. Dark blue variant bases match the most abundant variations, and white indicates a rare variant base. Bases 60 to 110 are indicated at the top of the alignment, part of the 16S V1 region containing a conserved stem-loop based on the E. coli 2-D structure (41). The 5′ stem, loop, and 3′ stem are marked above the alignment by pink, black, and yellow boxes, respectively. Only three variant structures were identified at the V1 loop, shown at the right. Brackets 1 and 2 indicate the ASVs with variants 1 and 2, respectively. Arrow 3 indicates ASV 8, the only ASV with a variant 3 sequence. The pink, black, and yellow regions of the variant structures match the sequence in the alignment window. The red bases in the structures indicate Klebsiella-specific sequences common to these variants that differ from the E. coli structure. The red arrows in variants 1 and 2 indicate a deletion from the E. coli and variant 3 structures. The blue ovals indicate base variations between the aligned Klebsiella ASVs.