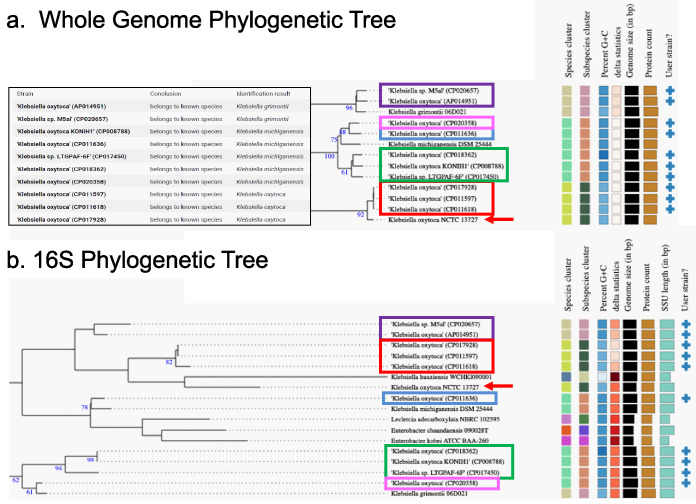
FIG 7.
Whole-genome and 16S phylogenetic analyses. Ten K. oxytoca strains represented in the Athena database were compared with 11,300 strains in the TYGS database to identify closely related strains. The partial trees shown include all branches needed to place the 10 “user strains” selected from the Athena database. Colored boxes are used to indicate groups of genomes that sorted together. (a) Whole-genome phylogeny. The inset box indicates how the input Athena strains were identified by TYGS. The tree with detailed relationships was inferred with FastME 2.1.6.1 (37) from GBDP distances calculated from genome sequences. The numbers above branches are GBDP pseudobootstrap support values of >60% from 100 replications, with an average branch support of 85.5%. (b) 16S phylogeny tree inferred with FastME 2.1.6.1 from GBDP distances calculated from 16S rRNA gene sequences. The numbers above branches are GBDP pseudobootstrap support values of >60% from 100 replications, with an average branch support of 53.3%. GenBank accession numbers are in parentheses. SSU, small subunit.