Abstract
Since its outbreak in December 2019, the persistent coronavirus disease (COVID-19) became a global health emergency. It is imperative to develop a prognostic tool to identify high-risk patients and assist in the formulation of treatment plans. We retrospectively collected 366 severe or critical COVID-19 patients from four centers, including 70 patients who died within 14 days (labeled as high-risk patients) since their initial CT scan and 296 who survived more than 14 days or were cured (labeled as low-risk patients). We developed a 3D densely connected convolutional neural network (termed De-COVID19-Net) to predict the probability of COVID-19 patients belonging to the high-risk or low-risk group, combining CT and clinical information. The area under the curve (AUC) and other evaluation techniques were used to assess our model. The De-COVID19-Net yielded an AUC of 0.952 (95% confidence interval, 0.928-0.977) on the training set and 0.943 (0.904-0.981) on the test set. The stratified analyses indicated that our model's performance is independent of age, sex, and with/without chronic diseases. The Kaplan-Meier analysis revealed that our model could significantly categorize patients into high-risk and low-risk groups (p < 0.001). In conclusion, De-COVID19-Net can non-invasively predict whether a patient will die shortly based on the patient's initial CT scan with an impressive performance, which indicated that it could be used as a potential prognosis tool to alert high-risk patients and intervene in advance.
Keywords: Coronavirus disease 2019 (COVID-19), prognosis, computed tomography, deep learning, artificial intelligence
I. Introduction
Since its outbreak in December 2019, the persistent coronavirus disease (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has become a global health emergency [1]–[3]. On January 30th, 2020, the World Health Organization (WHO) confirmed the outbreak as a Public Health Emergency of International Concern (PHEIC) [4]. Up until October 19th, 2020, there have been more than 39,596,000 people among over 200 countries/territories/areas reported to be infected, and more than 1,107,000 of them died [5]. Due to the rapid spread rate and considerable mortality, the epidemic areas are suffering an unprecedented lack of medical resources [1], [6]–[10]. On account of the high false-negative rates and the lack of Reverse Transcription-Polymerase Chain Reaction (RT-PCR) kits early in the COVID-19 outbreak, the readily available medical imaging, such as chest computed tomography (CT) and X-ray, have been recommended as an alternative method to identifying COVID-19 patients [11]–[14]. Although some imaging characteristics which are non-specific in CT images may indicate COVID-19 pneumonia, many types of viral pneumonia are highly similar in CT appearance, and even cannot be identified by a radiologist. Besides, some confirmed COVID-19 patients experienced a rapid deterioration process (such as shock, acute respiratory distress syndrome, and multiple organ failure) due to an unknown progress mechanism [8]. Therefore, the prognostic tools, which could be used to detect high-risk patients with a malignant prognosis, are urgently needed in clinical practice.
Artificial intelligence (AI), especially deep learning, is an emerging methodology in the medical field [15]–[17] and is making a significant contribution to fight COVID-19 [18]. Several AI-based published studies for diagnosing COVID-19 showed acceptable performance. Wang et al. proposed a residual learning network combined with the prior-attention mechanism to screen COVID-19 patients from CT images [19]. Li et al. [20] used ResNet-50 as the backbone to extract the feature of every 2D CT slice and integrated them using max-pooling to discriminate COVID-19 with community-acquired pneumonia and non-pneumonia patients. Huang et al. [21] used an automatic deep learning method to evaluate lung burden changes in COVID-19 patients via serial CT scans. Bai et al. [22] conducted research and concluded that AI assistance could help radiologists in distinguishing COVID-19 pneumonia from non-COVID-19 pneumonia based on chest CT. However, whether the AI-energized prognostic tools can help identify high-risk patients from their medical images still needs to be discussed.
Therefore, in this study, we aimed to develop a deep learning-based prognostic tool that can non-invasively predict whether a patient will die shortly (within 14 days) based on the patient's initial CT scan and clinical information. Specifically, we enrolled 366 four-center severe or critical COVID-19 patients, based on which we develop a 3D convolutional neural network (3D-CNN) termed as De-COVID19-Net to predict the probability of patients’ death within two weeks. The model merged image features and clinical information with a customized training strategy. Compared with the clinical model, radiomics-based model, and pure CNN model, De-COVID19-Net showed the best performance.
II. Materials and Methods
The statistical analyses were conducted using Python programming language (version 3.7.6; http://python.org). The De-COVID19-Net was constructed using the PyTorch package (version 1.4.0; http://pytorch.org).
Fig. 1 shows the workflow of this study.
Fig. 1.

The flowchart of this study. We segmented the CT images and obtained the lung volumes after pre-processing (A). Used the lung volumes and clinical information as input, and we trained the constructed De-COVID19-Net (B). We used multiple evaluation techniques to demonstrate the performance of our model (C).
A. Patients and Follow-up
In this study, we retrospectively recruited 366 patients who were confirmed with severe or critical COVID-19 pneumonia. The patients were from four centers, including Renmin Hospital of Wuhan University (RHWU, n = 317), Huangshi Central Hospital (HCH, n = 28), the Second Affiliated Hospital of Harbin Medical University (SAHHMU, n = 13), and Henan Provincial People's Hospital (HPPH, n = 8). The patients were enlisted between December 12th, 2019, and March 18th, 2020. The institutional review boards of the four hospitals approved this study and waived the requirement for informed consent. We assessed the severity grade of the patients according to the definition in the diagnosis and Treatment Protocol for Novel Coronavirus Pneumonia (Trial Version 7) released by the National Health Commission & State Administration of Traditional Chinese Medicine, People's Republic of China [23].
The inclusion criteria were as follows: 1) the patient had etiologically confirmed COVID-19 pneumonia by RT-PCR tests; 2) the patient was diagnosed as having severe or critical COVID-19 pneumonia according to the protocol; 3) the patient was cured; or 4) the patient died within 14 days; or 5) the patient had regular follow-up for at least 14 days since his/her first CT scan date. Eight patients were excluded because his/her CT image had severe artifacts. Finally, we enrolled 366 patients’ initial CT images and clinical information since admission. We integrated the four-center data and divided them into the training and test sets in a ratio of 2:1 according to a computerized random number generator.
The CT protocols are detailed in Supplementary Section A. The clinical characteristics, including sex, age, the severity grade of the disease (severe or critical), with or without chronic diseases, follow-up duration, and outcome (cured, died, or hospitalized), were retrieved from patients’ electronic medical records. The chronic diseases included hypertension, diabetes, cancer, chronic pulmonary disease, chronic renal failure, cardiovascular or cerebrovascular disease, hepatitis, and liver cirrhosis. Some patients suffered from multiple chronic diseases. From the date of the CT scan, patients who were cured or survived more than 14 days (termed low-risk patients) were labeled as 0, whereas those who died within 14 days (termed high-risk patients) were labeled as 1.
We used the Mann-Whitney U test to measure distribution differences between the continuous variables of the training and test sets, and the Chi-square test to measure categorical variables. A two-side P-value < 0.05 indicated statistical significance.
B. CT Image Segmentation and Pre-processing
All enrolled patients underwent the non-contrast enhanced chest CT scan in the supine position from the level of the upper thoracic inlet to the posterior level of the costophrenic angle.
The lung region was mainly analyzed in this study. We used a threshold-based segmentation method to obtain the lung region automatically. Specifically, we binarized the CT image by the threshold of the Hounsfield Unit = −300 to get the trunk of a human body. Then, given the seed nodes automatically by computer, the flood fill algorithm would seek all voxels connected to the seed nodes in the 3-dimensional space and return the connected domains. We used the closed operation to denoise and retain the largest connected domain as the lung region, blocking the non-lung area such as the background and the trachea. Finally, the 3D lung volumes were derived and resampled to the size of 200 × 240 × 360 using B-spline interpolation [23].
To train a robust and convincing model, we applied several pre-processing methods. First, we windowed the intensities of the derived lung volumes to [−1024, −100] to erase other non-lung tissues and organs (e.g., heart, spine, bones). Then, we standardized the volumes with z-score normalization. During the training, we augmented the data by randomly scaling the volumes to 0.9–1.1, and then randomly cropping it to the size of 150 × 200 × 320 as input. We provided an example of the derived lung region and corresponding lung mask in Fig. 2.
Fig. 2.
An example of a lung mask and the corresponding derived lung region is shown in three perspectives (axial, coronal, sagittal). The first row shows the original CT. The second row illustrates the segmented binary mask using our segmentation pipeline. The third row indicates the step of cropping and resizing. With intensity normalization and data augmentation, we obtained the last row as our model's input.
C. Model Construction and Training
Rather than the traditional 2D-CNN, which was widely applied in natural images, we proposed a 121-layer 3D-CNN architecture termed De-COVID19-Net as our prediction model. The use of 3D convolution can properly synthesize the spatial information of the CT image, which is intuitive and consistent with the way doctors diagnose. The De-COVID19-Net was inspired by DenseNet [24]. Specifically, the model is comprised of four dense blocks, each of which is stacked with multiple convolutional units. A convolutional unit consists of a batch normalization layer, a ReLU activation layer, and a 3D convolutional layer. Inside each dense block, all convolutional units are densely connected in a feedforward style. In this way, the multi-layer image features will be appropriately synthesized. We integrated clinical information (sex, age, severity grade, and with/without chronic disease) and image features in the fully connected layer which would then deduce the prediction. We used a dropout rate of 0.5 in the fully connected layers to mitigate over-fitting. The detailed topological structure of De-COVID19-Net is illustrated in Supplementary Section B.
To tackle the problem of data imbalance, we adopted focal loss [25] as the model's criterion, which can balance the contribution of learning from high-risk and low-risk samples. The formula is
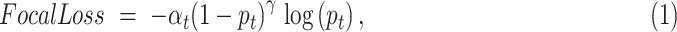 |
where 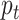 is the predicted probability that the sample is high-risk;
is the predicted probability that the sample is high-risk; 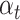 controls the weight of high-risk and low-risk samples; and
controls the weight of high-risk and low-risk samples; and  would push the model to put more concentration on the hard-to-classify samples. Following the instruction of [25], we used
would push the model to put more concentration on the hard-to-classify samples. Following the instruction of [25], we used 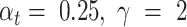 in our model training.
in our model training.
To better merge the CT image features and clinical information, we have customized the training strategy:
-
1)
We only used CT images to train the model for 50 epochs with a learning rate of 1e-5;
-
2)
We froze the weights of the convolutional blocks and involved the clinical information with CT images, followed by further training of the fully connected layers for 20 epochs with a learning rate of 5e-6;
-
3)
Finally, we fine-tuned the whole model for 10 epochs with a learning rate of 5e-6.
During the training, the Adam optimizer was applied to update the model's weights. The batch size was 8, and the weight-decay was 1e-2. We trained De-COVID19-Net on the training set using an NVIDIA Titan RTX Graphics Card.
D. Model Evaluation
We used the ROC curve and the area under the curve (AUC) to quantify the De-COVID19-Net's performance of predicting patients’ death in both the training and test sets.
Using a threshold, the model can classify if a patient belongs to the high-risk or low-risk groups. The threshold was determined by the weighted Youden's index. Youden's index is a statistic that depicts the performance of a dichotomous diagnostic model. Researchers often use the index to determine the threshold of the model's continuous output for binary classification, when false negatives and false positives are considered equally harmful. However, the sensitivity of the model is worth more attention than its specificity, because the missed diagnosis will bring more significant harm in a COVID-19 situation. Therefore, we introduced weighted Youden's index [26], [27]:
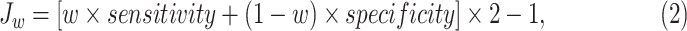 |
where 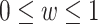 . The
. The  and
and 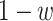 reflect the relative importance regarding false negatives or false positives, respectively. When
reflect the relative importance regarding false negatives or false positives, respectively. When  is equal to 0.5, the formula degenerates to the original Youden's index. According to
is equal to 0.5, the formula degenerates to the original Youden's index. According to 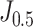 and
and 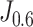 on the training set, we derived two thresholds, by which the accuracy, sensitivity, and specificity were measured respectively.
on the training set, we derived two thresholds, by which the accuracy, sensitivity, and specificity were measured respectively.
We performed the decision curve analysis [28], [29] to discuss the benefit of the model when considering different threshold probabilities as the cut-off.
Moreover, Kaplan-Meier analysis [30] and the log-rank test were performed to assess the prognosis performance of the De-COVID19-Net.
We performed stratified analyses in terms of age, sex, and chronic diseases to verify the reliability of the model. Furthermore, Delong's test [31] was used to compare the ROC curves of subgroups in the stratified analysis.
To demonstrate that the model has learned the abstract mappings between image features and clinical outcomes, we visualized the gradient-weighted class activation maps (Grad-CAM) [32] of the model's last convolutional filter.
E. Model Comparison
In order to verify the performance of our De-COVID19-Net pipeline (3D DenseNet + segmentation + clinical information), we compared it with other prognosis methods, which are detailed below:
-
1)
Clinical model: We constructed a logistic regression model based on clinical information (sex, age, severity grade, and with/without chronic disease) to predict if a patient is at high risk of death.
-
2)
Radiomics-based model: Radiomics, an emerging tool for medical image analysis, was used for COVID-19's diagnosis and prognosis [33]–[35]. Inspired by the works, we extracted radiomic features of the lung volumes and built a prognosis model. The construction of the radiomics-based model is detailed in Supplementary Section C.
-
3)
Pure DenseNet: The model removed the segmentation step and clinical information input from the De-COVID19-Net pipeline. We randomly resized the CT volumes to [150, 200, 320] as input without segmentation. This model was trained for 50 epochs using the learning rate of 1e-5.
-
4)
DenseNet + segmentation: The model removed the clinical information input from the De-COVID19-Net pipeline, while it kept the segmentation procedure. This model is obtained after the first stage of our training strategy.
III. Experiments
A. Patient Characteristics
A total of 366 severe or critical COVID-19 patients (average age = 62.0 years, standard deviation = 16.0 years) from four centers were enrolled in this study and were divided into the training set (n = 246) and test set (n = 120) with a ratio of 2:1. The data from each center roughly maintained a 2:1 ratio between the training set and the test set. We analyzed the distribution of clinical characteristics in the training set and test set by the Chi-square test for categorical variables and the Mann-Whitney U test for continuous variables, and found no statistically significant difference. The detailed information is shown in Table I. In Supplementary Section D, we also provided the distribution of patients’ survival time who finally died.
TABLE I. Clinical Characteristics of Patients.
|
Characteristics |
Training set | Test set | P value |
|---|---|---|---|
| Center, No. (%) | 0.682 | ||
| RHWU | 213 (86.6%) | 104 (86.6%) | |
| HCH | 20 (8.1%) | 8 (6.7%) | |
| SAHHMU | 7 (2.9%) | 6 (5.0%) | |
| HPPH | 6 (2.4%) | 2 (1.7%) | |
| Age, average ± SD, years | 61.46 ± 16.06 | 63.13 ± 15.76 | 0.170 |
| Sex, No. (%) | 0.746 | ||
| Female | 131 (53.3%) | 61 (50.8%) | |
| Male | 115 (46.7%) | 59 (49.2%) | |
| With Chronic diseases, No. (%) | 0.288 | ||
| Yes | 77 (31.3%) | 45 (37.5%) | |
| No | 169 (68.7%) | 75 (62.5%) | |
| Severity grade, No. (%) | 0.404 | ||
| Severe illness | 176 (71.5%) | 80 (66.7%) | |
| Critical illness | 70 (28.5%) | 40 (33.3%) | |
| Follow-up duration, average ± SD, days | 19.25 ± 12.84 | 17.33 ± 10.39 | 0.097 |
| Follow-up outcome *, No. (%) | 0.767 | ||
| Died within 14 days | 46 (18.7%) | 24 (20.0%) | |
| Survived more than 14 days | 200 (81.3%) | 96 (80.0%) |
The distribution of clinical characteristics in the training set and the test set were assessed by a Chi-square test for discrete variables or Mann-Whitney U test for continuous variables.
*The predicted targets: died within 14 days (high-risk group, labeled as 1); survived more than 14 days (low-risk group, labeled as 0).
B. Model Evaluation and Comparison
De-COVID19-Net was trained with a customized strategy based on the processed lung volumes and clinical information. We compared four other models (clinical model, radiomic model, pure DenseNet, and DenseNet + Segmentation) with the proposed De-COVID-Net. De-COVID19-Net yielded an AUC of 0.952 (95% confidence interval [CI], 0.928-0.977) for the training set, and 0.943 (95% CI, 0.904-0.981) for the test set, which indicated the best discrimination performance among other models (Fig. 3). Besides, De-COVID19-Net showed comparable performance among the two datasets (Delong test, p = 0.674). Other indicators, including accuracy, sensitivity, and specificity, were calculated based on 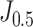 and
and 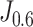 .
.
Fig. 3.
The receiver operating characteristic (ROC) curves of models’ prediction in the training (a) and test sets (b). The proposed De-COVID19-Net workflow had the highest AUC compared with other methods.
Table II shows that De-COVID19-Net achieved the best performance in all metrics. Compared with pure DenseNet, the model had a significant gain from the segmentation step. After incorporating the clinical model, the final model has also been improved. In comparison, the clinical model and radiomic model were far from desirable, especially in sensitivity which is important in the urgent situation.
TABLE II. Performances of Different Models.
| Evaluation Metric | Clinical model | Radiomic model | Pure DenseNet | DenseNet + Seg | De-COVID19-Net | |
|---|---|---|---|---|---|---|
| Training Set |
AUC | 0.740 [0.649, 0.829] | 0.865 [0.807, 0.922] | 0.874 [0.824, 0.923] | 0.922 [0.887, 0.957] | 0.952 [0.928, 0.977] |
| Accuracy J0.5 | 0.752 | 0.821 | 0.780 | 0.793 | 0.882 | |
| Sensitivity J0.5 | 0.630 | 0.783 | 0.848 | 0.935 | 0.957 | |
| Specificity J0.5 | 0.780 | 0.830 | 0.765 | 0.760 | 0.865 | |
| Accuracy J0.6 | 0.626 | 0,772 | 0.756 | 0.744 | 0.850 | |
| Sensitivity J0.6 | 0.804 | 0.848 | 0.848 | 1.000 | 1.000 | |
| Specificity J0.6 | 0.585 | 0.755 | 0.735 | 0.685 | 0.815 | |
| Test Set |
AUC | 0.733 [0.608, 0.855] | 0.850 [0.771, 0.927] | 0.870 [0.802, 0.937] | 0.906 [0.852, 0.959] | 0.943 [0.904, 0.981] |
| Accuracy J0.5 | 0.717 | 0.783 | 0.742 | 0.783 | 0.875 | |
| Sensitivity J0.5 | 0.542 | 0.708 | 0.792 | 0.917 | 0.917 | |
| Specificity J0.5 | 0.760 | 0.802 | 0.729 | 0.750 | 0.864 | |
| Accuracy J0.6 | 0.633 | 0.775 | 0.717 | 0.725 | 0.842 | |
| Sensitivity J0.6 | 0.708 | 0.792 | 0.833 | 1.000 | 1.00 | |
| Specificity J0.6 | 0.615 | 0.s771 | 0.688 | 0.656 | 0.802 | |
The thresholds to calculate accuracy, sensitivity, and specificity were obtained by the weighted Youden's Index 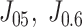 . The AUC confidence intervals were obtained by a 2000-time bootstrap. Our proposed De-COVID19-Net achieved the best performance in all metrics (in bold). The construction of the models is illustrated in Section II. E.
. The AUC confidence intervals were obtained by a 2000-time bootstrap. Our proposed De-COVID19-Net achieved the best performance in all metrics (in bold). The construction of the models is illustrated in Section II. E.
To prove the superiority of the model when considering different threshold probabilities, we draw decision curves. According to Fig. 4, patients would benefit more from the prediction of De-COVID19-Net in most ranges (threshold probability > 5%) compared with other schemes. An interpretation of the decision curve is provided in Supplementary Section E.
Fig. 4.
The decision curves. We compared De-COVID19-Net with five other schemes. The results show that our model has a greater or equal net benefit in most ranges.
C. Follow-up Analysis
The enrolled patients’ follow-up duration was between 0 (died on the day of the CT scan) and 62 days (average = 18.62 days, standard deviation = 12.13 days). To demonstrate the model's prognosis performance, we respectively partitioned the patients into the predicted-high-risk group and predicted-low-risk group according to the threshold decided by the weighted Youden's Index 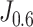 . Then, we performed a Kaplan-Meier analysis on the two groups. According to Fig. 5, the model can distinguish the two groups (log-rank test, p < 0.001).
. Then, we performed a Kaplan-Meier analysis on the two groups. According to Fig. 5, the model can distinguish the two groups (log-rank test, p < 0.001).
Fig. 5.
The Kaplan-Meier curves. We partitioned the patients into the predicted-high-risk group and predicted-low-risk group according to the threshold determined by the weighted Youden's index 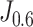 , and plotted the curves. The shaded areas represent the confidence interval.
, and plotted the curves. The shaded areas represent the confidence interval.
D. Stratified Analyses
We conducted four stratified analysis experiments according to patients’ age, sex, and chronic diseases. The results are shown in Fig. 6.
Fig. 6.

Stratified analyses according to age, sex, and with/without chronic diseases. In Kaplan-Meier curves, we partitioned the patients into the predicted-high-risk group and predicted-low-risk group according to the threshold determined by the weighted Youden's index  . As for age, we partitioned the patients into the younger group and elderly group by their median age (64).
. As for age, we partitioned the patients into the younger group and elderly group by their median age (64).
In terms of age, we partitioned the patients into the younger group and elderly group by their median age (64). The results of stratified analyses indicated that the effects of our model were independent of patients’ age or sex.
Clinical experiences suggested that chronic diseases are often a high-risk factor for death. Therefore, we also conducted a stratified analysis comparing patient subgroups with or without chronic diseases. According to Fig. 6, our model demonstrated equally impressive discriminating ability in both subgroups (Delong test, p = 0.905).
We also performed a Kaplan-Meier analysis on each subgroup. For each subgroup, we divided the patients into the predicted-high-risk group and predicted-low-risk group using the threshold derived from the weighted Youden's index 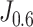 . According to Fig. 6, the Kaplan-Meier curves suggested that our model can successfully stratify every clinical subgroup into two groups, which has significantly different follow-up durations and outcomes (log-rank test, p < 0.001 for every subgroup).
. According to Fig. 6, the Kaplan-Meier curves suggested that our model can successfully stratify every clinical subgroup into two groups, which has significantly different follow-up durations and outcomes (log-rank test, p < 0.001 for every subgroup).
E. Deep Learning Feature Visualization
We selected two cases: a 61-year-old male who died 8 days after his CT scan (the high-risk patient), and a 70-year-old male who was discharged 17 days after his CT scan (the low-risk patient). The two cases were classified correctly by our model. Using gradient-weighted class activation mapping (Grad-CAM), we visualized two patients’ feature maps, which were synthesized by the model's last convolutional layer. Fig. 7 suggested that the model was highly sensitive to areas of pneumonia in the high-risk patient but not in the low-risk patient.
Fig. 7.
A) A 70-year-old male who was discharged 17 days after his CT scan (the low-risk patient); B) A 61-year-old male who died 8 days after his CT scan (the high-risk patient). Both of them were diagnosed as critical patients, showing ground-glass opacities (black arrow), consolidation (green arrow), and interlobular septal thickening (red arrow). The activation map indicated that our model was sensitive to the imaging textures of the high-risk patient, while it had a low response in the low-risk patient.
IV. Discussion
In this study, we collected 366 severe or critical COVID-19 patients from four centers, based on which we explored a deep learning-based method to predict the probability of the patients’ death within 14 days from the CT images. Our proposed model, which was termed as De-COVID19-Net, showed promising performance in the training set and test set. The model showed excellent ability to differentiate high-risk (died within 14 days) and low-risk (survived more than 14 days) COVID-19 patients according to the CT images, which suggested that the model could be considered as a powerful tool to assess the risk of patients’ poor outcome. Those who were rated as high-risk patients require more elaborate intensive care and treatment interventions. The workflow takes about one minute to segment and inference an input CT image. We have already provided the model on the website http://www.radiomics.net.cn/platform.html for open access.
The CT symptom of patchy consolidation might be a high-risk factor associated with dying [36]. This symptom can be observed in both dead and surviving patients [14], [37], may not be observed in patients upon admission, and can hardly be used for prognosis. Based on the impressive performance of our model, we argue that CT images imply information that can prompt patients' prognosis, and our deep learning-based model can mine and make full use of them to alert high-risk patients. In the case of tight medical resources, it is necessary to leverage the readily available CT images to classify high-risk patients further.
Some studies suggested that older age and chronic diseases are the potential risk factors in prompting in-patients’ poor prognosis [36], [37]. Radiomics was also proved to be a potential method for COVID-19 diagnosis [33], [34]. Inspired by these works, we constructed a clinical model and a radiomics-based model based on our dataset. The evaluation metrics (Table II, Fig. 3) and decision curves (Fig. 4) showed that our proposed model achieved the best performance in all metrics. Low sensitivities of the clinical model and radiomics model are the fatal flaw when consider deploying it in the clinic. Table II also indicates that the De-COVID19-Net model gained performance from the segmentation procedure and by incorporating imaging features and clinical information.
Several measures, including the ROC curves, decision curve analysis, Kaplan-Meier analysis, and Grad-CAM, have validated the performance of our model. Furthermore, stratified analyses were conducted to demonstrate the robustness and stability of the model among different subgroups. The stratified analyses proved that our model performed equally well in different clinical subgroups and was independent of patients’ age, sex, and whether the patient suffered from chronic diseases.
Lung region segmentation is an essential procedure in AI-based medical image analysis, including diagnosis and prognosis. In our study, to better explore the lung image information, we used an automatic lung segmentation algorithm based on threshold segmentation following morphological optimization and data augmentations, by which we avoided tedious manual annotation and gained good results. With the low-cost threshold segmentation and pre-processing procedures, we can block out most bones, non-lung organs and tissues. In this way, we forced the model to pay attention to the lung regions. With random cropping, we can make extensive use of the dataset and avoid early overfitting, meanwhile reducing the training burden.
In many medical prognosis studies, researchers often used survival analysis models, such as the Cox proportional hazards regression model or Fine and Gray competing risk regression model. We did not use the survival analysis model because instead of regressing out survival time after a CT scan, it might make more sense to regress out survival time after symptom onset. Besides, it may be futile to predict survival time. Patients often go through a rapid time from admission to the clinical outcome (death or cure), which are affected by many complex factors. Consequently, we binary classified the high-risk and low-risk patients according to the scores outputted by the model's fully connected layer, rather than constructing a survival model which is not appropriate to the actual situation.
The activation maps (Fig. 7) suggest that the sensitive areas which caused the high response of our model are consistent with the suspicious areas in a clinical diagnosis. In terms of a high-risk patient, the model was attracted by the COVID-19 typical imaging characteristics, such as ground-glass opacities, consolidation, centrilobular nodules, and interlobular septal thickening. In terms of low-risk patients, even though the patients exhibited similar imaging characteristics with high-risk patients, our model is less sensitive to them and gives a lower score. The subtle difference might be noticed by experienced radiologists observing these characteristics closely. However, it takes much patience and time, which is expensive in a busy working environment with heavy work pressure. Therefore, our model has the potential to help clinicians quickly identify critically ill COVID-19 cases, provide non-invasive and personalized prognostic means, and propose recommendations for patients’ surveillance and management. Although our model can handle most cases, it is still likely to make mistakes, mainly due to the noise from clinical factors or unusual lung manifestation, which could be alleviated by incorporating more data. We also provided a false-positive example and a false-negative example in Supplementary Section F.
This study still has some limitations. Firstly, we lack an independent external validation set. The reason is that the proportion of data from Wuhan and non-Wuhan areas varies greatly, and it is not enough to constitute another data set. We will validate and improve our model with more data in the future. Secondly, we used the binary classification model to deal with patients’ prognosis problem, because the situation does not lend itself to the hypothesis of popular survival analysis models. Therefore, a survival analysis model, which can predict if and when a patient will die or be cured, might be needed. Thirdly, as a more accessible modality, chest X-rays may also have the potential to help detect high-risk patients. Generally, patients did not perform both CT and chest X-ray scans in the same period. It is difficult to obtain paired data for comparative studies. We will try to explore the effect of chest X-rays in detecting high-risk patients in future studies.
V. Conclusion
We proposed a deep learning-based prognosis model based on initial CT images. The model has the potential to non-invasively predict the death risk of critical COVID-19 patients within the next two weeks. The model achieved impressive performance on the four-center dataset, which indicated that a CT scan combined with an AI method could be used as a powerful prognosis tool to alert high-risk patients.
Acknowledgment
We would like to dedicate this manuscript to the people who made selfless contributions in fighting this disaster. We thanked Prof. Qingguo Xie and Peng Xiao from Huazhong Science and Technology University for their help about the data collection in Wuhan.
Funding Statement
This work was supported by the National Key R&D Program of China (2017YFC1309100, 2017YFA0205200), Novel Coronavirus Pneumonia Emergency Key Project of Science and Technology of Hubei Province (2020FCA015), National Natural Science Foundation of China under Grants 82022036, 91959130, 81971776, 81771924, 6202790004, 81930053, 81871332, Natural Science Foundation of Beijing Municipality (L182061), Strategic Priority Research Program of the Chinese Academy of Sciences (XDB 38040200), the Project of High-Level Talents Team Introduction in Zhuhai City (Zhuhai HLHPTP201703), and Youth Innovation Promotion Association CAS (2017175).
Contributor Information
Lingwei Meng, Email: menglingwei2018@ia.ac.cn.
Di Dong, Email: di.dong@ia.ac.cn.
Liang Li, Email: liliang_082@163.com.
Meng Niu, Email: niumeng@cmu.edu.cn.
Yan Bai, Email: resonance2010@126.com.
Meiyun Wang, Email: mywang@ha.edu.cn.
Xiaoming Qiu, Email: qxm2020cov@163.com.
Yunfei Zha, Email: zhayunfei999@126.com.
Jie Tian, Email: tian@ieee.org.
References
- [1].Fisher D. and Heymann D., “Q&A: The novel coronavirus outbreak causing COVID-19,” BMC Med, vol. 18, no. 1, pp. 18–20, 2020.32000763 [Google Scholar]
- [2].Han R., Huang L., Jiang H., Dong J., Peng H., and Zhang D., “Early clinical and CT manifestations of coronavirus disease 2019 (COVID-19) pneumonia,” Amer. J. Roentgenol., vol. 215, no. 2, pp. 338–343, Aug. 2020, doi: 10.2214/AJR.20.22961. [DOI] [PubMed] [Google Scholar]
- [3].Zhu N. et al. , “A novel coronavirus from patients with pneumonia in china, 2019,” New. Engl. J. Med., vol. 382, no. 8, pp. 727–733, Feb. 2020, doi: 10.1056/nejmoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sohrabi C. et al. , “World health organization declares global emergency: A review of the 2019 novel coronavirus (COVID-19),” Int. J. Surg., vol. 76, pp. 71–76, Apr. 2020, doi: 10.1016/j.ijsu.2020.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].World Health Organization, “Coronavirus disease (COVID-19) pandemic,” 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019
- [6].Zhou S., Wang Y., Zhu T., and Xia L., “CT features of coronavirus disease 2019 (COVID-19) pneumonia in 62 patients in wuhan, china,” Amer. J. Roentgenol., vol. 214, no. 6, pp. 1287–1294, Jun. 2020, doi: 10.2214/AJR.20.22975. [DOI] [PubMed] [Google Scholar]
- [7].Guan W. et al. , “Clinical characteristics of coronavirus disease 2019 in china,” N. Engl. J. Med., vol. 382, no. 18, pp. 1708–1720, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Chen N. et al. , “Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in wuhan, china: A descriptive study,” Lancet, vol. 395, no. 10223, pp. 507–513, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ji Y., Ma Z., Peppelenbosch M. P., and Pan Q., “Potential association between COVID-19 mortality and health-care resource availability,” Lancet Glob. Heal., vol. 8, no. 4, 2020, Art. no. e480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Jarvis C. I. et al. , “Quantifying the impact of physical distance measures on the transmission of COVID-19 in the U.K.,” BMC Med., vol. 18, no. 1, p. 124, May 2020, doi: 10.1186/s12916-020-01597-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Xie X., Zhong Z., Zhao W., Zheng C., Wang F., and Liu J., “Chest CT for typical 2019-nCoV pneumonia: Relationship to negative RT-PCR testing,” Radiology, vol. 296, no. 2, pp. E41–E45, 2020, doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Bernheim A. et al. , “Chest CT findings in coronavirus disease-19 (COVID-19): Relationship to duration of infection,” Radiology, vol. 295, no. 3, pp. 685–691, Jun. 2020, doi: 10.1148/radiol.2020200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kanne J. P., “Chest CT findings in 2019 novel coronavirus (2019-nCoV) infections from wuhan, china: Key points for the radiologist,” Radiol., vol. 295, no. 1, pp. 16–17, Apr. 2020, doi: 10.1148/radiol.2020200241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Dong D. et al. , “The role of imaging in the detection and management of COVID-19: A review,” IEEE Rev. Biomed. Eng., vol. 3333, no. c, 2020, doi: 10.1109/RBME.2020.2990959. [DOI] [PubMed] [Google Scholar]
- [15].Bi W. L. et al. , “Artificial intelligence in cancer imaging: Clinical challenges and applications,” cA. Cancer J. Clin., vol. 69, no. 2, pp. 127–157, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Dong D. et al. , “Development and validation of an individualized nomogram to identify occult peritoneal metastasis in patients with advanced gastric cancer,” Ann. Oncol., vol. 30, no. 3, pp. 431–438, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Dong D. et al. , “Deep learning radiomic nomogram can predict the number of lymph node metastasis in locally advanced gastric cancer: An international multicenter study,” Ann. Oncol., vol. 31, no. 7, pp. 912–920, 2020. [DOI] [PubMed] [Google Scholar]
- [18].Shi F. et al. , “Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for covid-19,” IEEE Rev. Biomed. Eng., to be published, doi: 10.1109/RBME.2020.2987975. [DOI] [PubMed]
- [19].Wang J. et al. , “Prior-attention residual learning for more discriminative COVID-19 screening in CT images,” vol. 39, no. 8, pp. 2572–2583, Aug. 2020, doi: 10.1109/TMI.2020.2994908. [DOI] [PubMed] [Google Scholar]
- [20].Li L. et al. , “Using artificial intelligence to detect COVID-19 and community-acquired pneumonia based on pulmonary ct: Evaluation of the diagnostic accuracy,” Radiology, vol. 296, no. 2, pp. E65–E71, 2020, doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Huang L. et al. , “Serial quantitative chest CT assessment of COVID-19: Deep-learning approach,” Radiol. Cardiothorac. Imag., vol. 2, no. 2, 2020, Art. no. e200075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bai H. X. et al. , “AI augmentation of radiologist performance in distinguishing COVID-19 from pneumonia of other etiology on chest cT,” Radiology, vol. 296, no. 3, pp. E156–E165, Sep. 2020, doi: 10.1148/radiol.2020201491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lehmann T. M., Gonner C., and Spitzer K., “Addendum: B-spline interpolation in medical image processing,” IEEE Trans. Med. Imaging, vol. 20, no. 7, pp. 660–665, Jul. 2001. [DOI] [PubMed] [Google Scholar]
- [24].Huang G., Liu Z., Van Der Maaten L., and Weinberger K. Q., “Densely connected convolutional networks,” in Proc. - 30th IEEE Conf. Comput Vision Pattern Recognit., CVPR 2017, vol. 2017-Janua, pp. 2261–2269, 2017. [Google Scholar]
- [25].Lin T.-Y., Goyal P., Girshick R., He K., and Dollár P., “Focal loss for dense object detection,” in Proc. IEEE Int. Conf. Comput. Vision, 2017, pp. 2980–2988. [DOI] [PubMed] [Google Scholar]
- [26].Li D., Shen F., Yin Y., Peng J., and Chen P., “Weighted youden index and its two-independent-sample comparison based on weighted sensitivity and specificity,” Chin. Med. J. (Engl)., vol. 126, no. 6, pp. 1150–1154, 2013. [PubMed] [Google Scholar]
- [27].Rücker G. and Schumacher M., “Summary ROC curve based on a weighted youden index for selecting an optimal cutpoint in meta-analysis of diagnostic accuracy,” Stat. Med., vol. 29, no. 30, pp. 3069–3078, 2010. [DOI] [PubMed] [Google Scholar]
- [28].Kerr K. F., Brown M. D., Zhu K., and Janes H., “Assessing the clinical impact of risk prediction models with decision curves: Guidance for correct interpretation and appropriate use,” J. Clin. Oncol., vol. 34, no. 21, 2016, Art. no. 2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Vickers A. J. and Elkin E. B., “Decision curve analysis: A novel method for evaluating prediction models,” Med. Decis. Mak., vol. 26, no. 6, pp. 565–574, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kaplan E. L. and Meier P., “Nonparametric estimation from incomplete observations,” J. Am. Stat. Assoc., vol. 53, no. 282, pp. 457–481, 1958. [Google Scholar]
- [31].DeLong E. R., DeLong D. M., and Clarke-Pearson D. L., “Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach,” Biometrics, vol. 44, no. 3, p. 837, Sep. 1988, doi: 10.2307/2531595. [DOI] [PubMed] [Google Scholar]
- [32].Selvaraju R. R., Cogswell M., Das A., Vedantam R., Parikh D., and Batra D., “Grad-cam: Visual explanations from deep networks via gradient-based localization,” in Proc. IEEE Int. Conf. Comput. Vision, 2017, pp. 618–626. [Google Scholar]
- [33].Fang M. et al. , “CT radiomics can help screen the coronavirus disease 2019 (COVID-19): A preliminary study,” Sci. China Inf. Sci., vol. 63, no. 7, pp. 1–8, 2020. [Google Scholar]
- [34].Wu Q. et al. , “Radiomics analysis of computed tomography helps predict poor prognostic outcome in COVID-19,” Theranostics, vol. 10, no. 16, pp. 7231–7244, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Meng L. et al. , “2D and 3D CT radiomic features performance comparison in characterization of gastric cancer: A multi-center study,” IEEE J. Biomed. Heal. Inform., vol. 2194, no. c, pp. 1–1, 2020. [DOI] [PubMed] [Google Scholar]
- [36].Zhang L. et al. , “Clinical characteristics of COVID-19-infected cancer patients: A retrospective case study in three hospitals within wuhan, china,” Ann. Oncol., 2020, doi: 10.1016/j.annonc.2020.03.296. [DOI] [PMC free article] [PubMed]
- [37].Zhou F. et al. , “Clinical course and risk factors for mortality of adult inpatients with COVID-19 in wuhan, china: A retrospective cohort study,” Lancet, vol. 395, no. 10229, pp. 1054–1062, Mar. 2020, doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]







