Abstract
Currently, Coronavirus disease (COVID-19), one of the most infectious diseases in the 21st century, is diagnosed using RT-PCR testing, CT scans and/or Chest X-Ray (CXR) images. CT (Computed Tomography) scanners and RT-PCR testing are not available in most medical centers and hence in many cases CXR images become the most time/cost effective tool for assisting clinicians in making decisions. Deep learning neural networks have a great potential for building COVID-19 triage systems and detecting COVID-19 patients, especially patients with low severity. Unfortunately, current databases do not allow building such systems as they are highly heterogeneous and biased towards severe cases. This article is three-fold: (i) we demystify the high sensitivities achieved by most recent COVID-19 classification models, (ii) under a close collaboration with Hospital Universitario Clínico San Cecilio, Granada, Spain, we built COVIDGR-1.0, a homogeneous and balanced database that includes all levels of severity, from normal with Positive RT-PCR, Mild, Moderate to Severe. COVIDGR-1.0 contains 426 positive and 426 negative PA (PosteroAnterior) CXR views and (iii) we propose COVID Smart Data based Network (COVID-SDNet) methodology for improving the generalization capacity of COVID-classification models. Our approach reaches good and stable results with an accuracy of 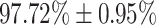 ,
, 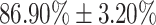 ,
, 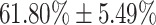 in severe, moderate and mild COVID-19 severity levels. Our approach could help in the early detection of COVID-19. COVIDGR-1.0 along with the severity level labels are available to the scientific community through this link https://dasci.es/es/transferencia/open-data/covidgr/.
in severe, moderate and mild COVID-19 severity levels. Our approach could help in the early detection of COVID-19. COVIDGR-1.0 along with the severity level labels are available to the scientific community through this link https://dasci.es/es/transferencia/open-data/covidgr/.
Keywords: COVID-19, convolutional neural networks, smart data
I. Introduction
In the last months, the world has been witnessing how COVID-19 pandemic is increasingly infecting a large mass of people very fast everywhere in the world. The trends are not clear yet but some research confirm that this problem may persist until 2024 [1]. Besides, prevalence studies conducted in several countries reveal that a tiny proportion of the population have developed antibodies after exposure to the virus, e.g., 5% in Spain.1 This means that frequently a large number of patients will need to be assessed in small time intervals by few number of clinicians and with very few resources.
In general, COVID-19 diagnosis is carried out using at least one of these three tests.
-
•
Computed Tomography (CT) scans-based assessment: it consists in analyzing 3D radiographic images from different angles. The needed equipment for this assessment is not available in most hospitals and it takes more than 15 minutes per patient in addition to the time required for CT decontamination.2
-
•
Reverse Transcription Polymerase Chain Reaction (RT-PCR) test: it detects the viral RNA from sputum or nasopharyngeal swab [2]. It requires specific material and equipment, which are not easily accessible and it takes at least 12 hours, which is not desirable as positive COVID-19 patients should be identified and tracked as soon as possible. Some studies found that RT-PCR results from several tests at different points from the same patients were variable during the course of the illness producing a high false-negative rate [3]. The authors suggested that RT-PCR test should be combined with other clinical tests such as CT.
-
•
Chest X-Ray (CXR): The required equipment for this assessment are less cumbersome and can be lightweight and transportable. In general, this type of resources is more available than the required for RT-PCR and CT-scan tests. In addition, CXR test takes about 15 seconds per patient [2], which makes CXR one of the most time/cost effective assessment tools.
Few recent studies provide estimates on expert radiologists sensitivity in the diagnosis of COVID-19 based on CT scans, RT-PCR and CXR. A study on a set of 51 patients with chest CT and RT-PCR essay performed within 3 days, reported a sensitivity in CT of 98% compared with RT-PCR sensitivity of 71% [4]. A different study on 64 patients (26 men, mean age 56 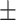 19 years) reported a sensitivity of 69% for CXR compared with 91% for initial RT-PCR [2]. According to an analysis of 636 ambulatory patients [5], most patients presenting to urgent care centers with confirmed coronavirus disease 2019 have normal or mildly abnormal findings on CXR. Only 58.3% of these patients are correctly diagnosed by the expert eye.
19 years) reported a sensitivity of 69% for CXR compared with 91% for initial RT-PCR [2]. According to an analysis of 636 ambulatory patients [5], most patients presenting to urgent care centers with confirmed coronavirus disease 2019 have normal or mildly abnormal findings on CXR. Only 58.3% of these patients are correctly diagnosed by the expert eye.
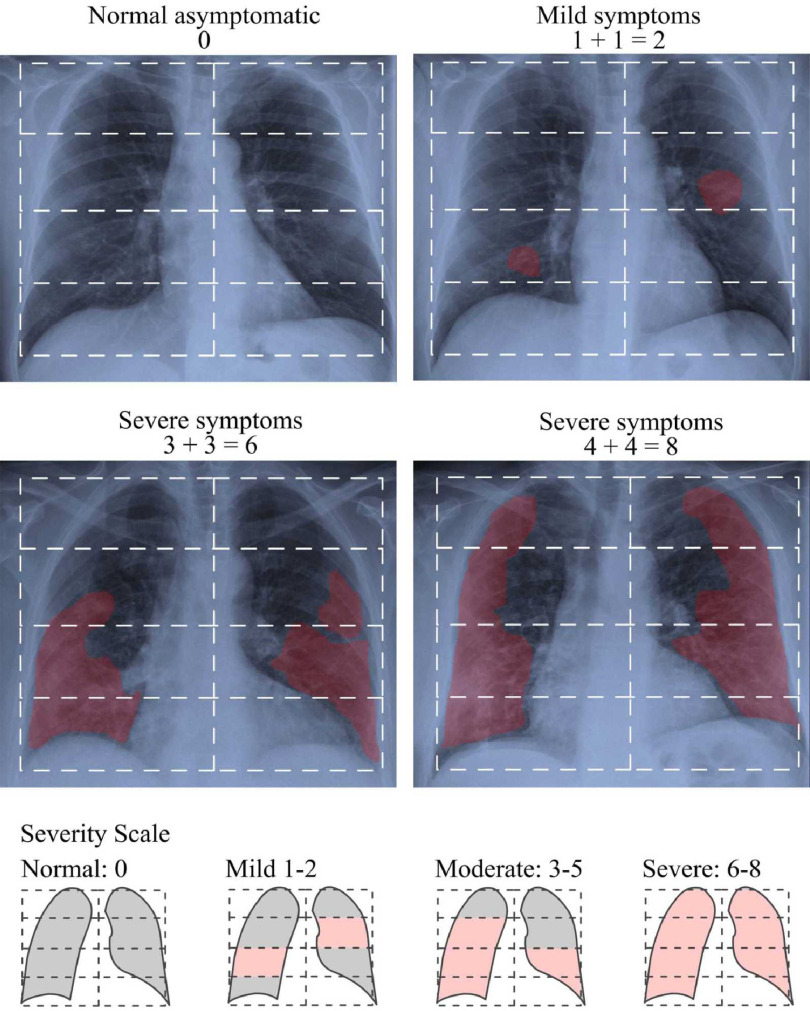
In a recent study [2], authors proposed simplifying the quantification of the level of severity by adapting a previously defined Radiographic Assessment of Lung Edema (RALE) score [6] to COVID-19. This new score is calculated by assigning a value between 0-4 to each lung depending on the extent of visual features such as, consolidation and ground glass opacities, in the four parts of each lung as depicted in Fig. 1. Based on this score, experts can identify the level of severity of the infection among four severity stages, Normal 0, Mild 1-2, Moderate 3-5 and Severe 6-8. In practice, a patient classified by expert radiologist as Normal can have positive RT-PCR. We refer to these cases as Normal-PCR+. Expert annotation adopted in this work is based in this score.
Fig. 1.
The stratification of radiological severity of COVID-19. Examples of how RALE index is calculated.
Automated image analysis via Deep learning (DL) models have a great potential to optimize the role of CXR images for a fast diagnosis of COVID-19. A robust and accurate DL model could serve as a triage method and as a support for medical decision making. An increasing number of recent works claim achieving impressive sensitivities 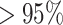 , far higher than expert radiologists. These high sensitivities are due to the bias in the most used COVID-19 dataset, COVID-19 Image Data Collection
[7]. This dataset includes a very small number of COVID-19 positive cases, coming from highly heterogeneous sources (at least 15 countries) and most cases are severe patients, an issue that drastically reduces its clinical value. To populate Non-COVID and Healthy classes, AI researchers are using CXR images from diverse pulmonary disease repositories. The obtained models will have no clinical value as well since they will be unable to detect patients with low and moderate severity, which are the target of a clinical triage system. In view of this situation, there is still a huge need for higher quality datasets built under the same clinical protocol and under a close collaboration with expert radiologists.
, far higher than expert radiologists. These high sensitivities are due to the bias in the most used COVID-19 dataset, COVID-19 Image Data Collection
[7]. This dataset includes a very small number of COVID-19 positive cases, coming from highly heterogeneous sources (at least 15 countries) and most cases are severe patients, an issue that drastically reduces its clinical value. To populate Non-COVID and Healthy classes, AI researchers are using CXR images from diverse pulmonary disease repositories. The obtained models will have no clinical value as well since they will be unable to detect patients with low and moderate severity, which are the target of a clinical triage system. In view of this situation, there is still a huge need for higher quality datasets built under the same clinical protocol and under a close collaboration with expert radiologists.
Multiple studies have proven that higher quality data ensures higher quality models. The concept of Smart Data refers to the process of converting raw data into higher quality data with higher concentration of useful information [8]. Smart data includes all pre-processing methods that improve value and veracity of data. Examples of these methods include noise elimination, data-augmentation [9] and data transformation [10] among other techniques.
In this work, we designed a high clinical quality dataset, named COVIDGR-1.0 that includes four levels of severity, Normal-PCR+, Mild, Moderate and Severe. We identified these four severity levels from a recent COVID-19 radiological study [2]. We also propose COVID Smart Data based Network (COVID-SDNet) methodology. It combines segmentation, data-augmentation and data transformations together with an appropriate Convolutional Neural Network (CNN) for inference.
The contributions of this paper can be summarized as follows:
-
•
We analyze reliability, potential and limitations of the most used COVID-19 CXR datasets and models.
-
•
From a data perspective, we provide the first public dataset, called COVIDGR-1.0, that quantifies COVID-19 in terms of severity levels, normal, mild, moderate and severe, with the aim of building triage systems with high clinical value.
-
•
From a pre-processing perspective, we combined several methods. To eliminate irrelevant information from the input CXR images, we used a new pre-processing method called segmentation-based cropping. To increase discrimination capacity of the classification model, we used a class-inherent transformation method inspired by GANs.
-
•
From a post-processing perspective, we proposed a new inference process that fuses the predictions of the four transformed classes obtained by the class-inherent transformation method to calculate the final prediction.
-
•
From a global perspective, we designed a novel methodology, named COVID-SDNet, with a high generalization capacity for COVID-19 classification based on CXR images. COVID-SDNet combines segmentation, data-transformation, data-augmentation, and a suitable CNN model together with an inference approach to get the final prediction.
Experiments demonstrate that our approach reaches good and stable results especially in moderate and severe levels, with 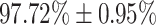 and
and 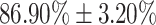 respectively. Lower accuracies were obtained in mild and normal-PCR+ severity levels with
respectively. Lower accuracies were obtained in mild and normal-PCR+ severity levels with 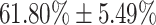 and
and 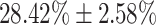 , respectively.
, respectively.
This article is organized as follows: A review of the most used datasets and COVID-19 classification approaches is provided in Section II. Section III describes how COVIDGR-1.0 is built and organized. Our approach is presented in Section IV. Experiments, comparisons and results are provided in Section V. The inspection of the model's decision using heatmaps is provided in Section VI and the conclusions are pointed out in Section VII.
II. Related Works
The last months have known an increasing number of works exploring the potential of deep learning models for automating COVID-19 diagnosis based on CXR images. The results are promising but still too much work needs to be done at the level of data and models design. Given the potential bias in this type of problems, several studies include explication methods to their models. This section analyzes the advantages and limitations of current datasets an models for building automatic COVID-19 diagnosis systems with and without decision explication.
A. Datasets
There does not exist yet a high quality collection of CXR images for building COVID-19 diagnosis systems of high clinical value. Currently, the main source for COVID-19 class is COVID-19 Image Data Collection [7]. It contains 76 positive and 26 negative PA views. These images were obtained from highly heterogeneous equipment from all around the world. Another example of COVID-19 dataset is Figure-1-COVID-19 Chest X-ray Dataset Initiative [11]. To build Non-COVID classes, most studies are using CXR from one or multiple public pulmonary disease data-sets. Examples of these repositories are:
-
•
RSNA Pneumonia CXR challenge dataset on Kaggle [12].
-
•
ChestX-ray8 dataset [13].
-
•
MIMIC-CXR dataset [14].
-
•
PadChest dataset [15].
For instance, COVIDx 1.0 [16] was built by combining three public datasets: (i) COVID-19 Image Data Collection [7], (ii) Figure-1-COVID-19 Chest X-ray Dataset Initiative [11] and (iii) RSNA Pneumonia Detection Challenge dataset [12]. COVIDx 2.0 was built by re-organizing COVIDx 1.0 into three classes, Normal (healthy), Pneumonia and COVID-19, using 201 CXR images for COVID class, including PA(PosteroAnterior) and AP(AnteroPosterior) views (see Table I). Notice that for a correct learning front view (PA) and back view (AP) cannot be mixed in the same class.
TABLE I. A Brief Description of COVIDx Dataset [7] (Only PA Views are Counted).
| Version | Normal(healthy) | Pneumonia | COVID-19 |
|---|---|---|---|
| 1.0 | 1,583 | 4,273 (Bacterial+viral) | 76 |
| 2.0 | 8,066 | 8,614 | 190 |
Although the value of these datasets is unquestionable as they are being useful for carrying out first studies and reformulations, they do not guarantee useful triage systems for the next reasons. It is not clear what annotation protocol has been followed for constructing the positive class in COVID-19 Image Data Collection. The included data is highly heterogeneous and hence DL-models can rely on other aspects than COVID visual features to differentiate between the involved classes. This dataset does not provide a representative spectrum of COVID-19 severity levels, most positive cases are of severe patients [17]. In addition, an interesting critical analysis of these datasets has shown that CNN models obtain similar results with and without eliminating most of the lungs in the input X-Ray images [18], which confirms again that there is a huge need of COVID-19 datasets with high clinical value.
Our claim is that the design of a high quality dataset must be done under a close collaboration between expert radiologists and AI experts. The annotations must follow the same protocol and representative numbers of all levels of severity, especially Mild and Moderate levels, must be included.
B. DL Classification Models
Existing related works are not directly comparable as they consider different combinations of public data-sets and different experimental setup. A brief summary of these works is provided in Table II.
TABLE II. Summary of Related Works That Analyze Variations of COVIDx With CNN.
| Ref. | Classes | Datasets | Model | Partition | Sens. | Acc. |
|---|---|---|---|---|---|---|
| [16] | Normal, Pneumonia, COVID | COVIDx 1.0 | COVIDNet | 98% - 2% | 87.1% | 92.6% |
| [19] | Normal, COVID | COVIDx 1.0 | COVID-CAPS | 98% - 2% | 90% | 95.7% |
| ([20]) | No-Findings, COVID | [7] + [13] | DarkCovidNet | 5-FCV | 90.65% | 98.08% |
| No-Findings, Pneumonia, COVID | 5-FCV | 97.9% | 87.02% | |||
| [21] | Normal, Pneumonia, COVID | COVIDx 2.0+[12] | VGG-19 + DenseNet-161 | 70% - 30% | 93% | 96.77% |
| [22] | Normal, Bacterial, Viral, COVID | [7]+[12] | Bayesian ResNet50V2 | 80% - 20% | 85.71% | 89.82% |
| ([23]) | Normal, Pneumonia, COVID | [7] + [12]+ other sources | MobileNet | 10-FCV | 98.66% | 96.78% |
The most related studies to ours as they proposed different models to the typical ones are [16] and [19]. In [16], the authors designed a deep network, called COVIDNet. They affirmed that COVIDNet reaches an overall accuracy of 92.6%, with 97.0% sensitivity in Normal class, 90.0% in Non-COVID-19 and 87.1% in COVID-19. The authors of a smaller network, called COVID-CAPS [19], also claim that their model achieved an accuracy of 98.7%, sensitivity of 90%, and specificity of 95.8%. These results look too impressive when compared to expert radiologist sensitivity, 69%. This can be explained by the fact that the used dataset is biased to severe COVID cases [17]. In addition, the performed experiments in both cited works are not statistically reliable as they were evaluated on one single partition. The stability of these models, in terms of standard deviation, has not been reported.
C. DL Classification Models With Explanation Approaches
Several interesting explanations were proposed to help inspect the predictions of DL-models [21], [22] although all their classification models were trained and validated on variations of COVIDx. The authors in [21] first use an ensemble of two CNN networks to predict the class of the input image, as Normal, Pneumonia or COVID. Then highlight class-discriminating regions in the input CXR image using gradient-guided class activation maps (Grad-CAM++) and layer-wise relevance propagation (LRP). In [22], the authors proposed explaining the decision of the classification model to radiologists using different saliency map types together with uncertainty estimations (i.e., how certain is the model in the prediction).
III. COVIDGR-1.0: Data Acquisition, Annotation and Organization
Instead of starting with an extremely large and noisy dataset, one can build a small and smart dataset then augment it in a way it increases the performance of the model. This approach has proven effective in multiple studies. This is particularly true in the medical field, where access to data is heavily protected due to privacy concerns and costly expert annotation.
Under a close collaboration with four highly trained radiologists from Hospital Universitario Clínico San Cecilio, Granada, Spain, we first established a protocol on how CXR images are selected and annotated to be included in the dataset. A CXR image is annotated as COVID-19 positive if both RT-PCR test and expert radiologist confirm that decision within less than 24 hours. CXR with positive PCR that were annotated by expert radiologists as Normal are labeled as Normal-PCR+. The involved radiologists annotated the level of severity of positive cases based on RALE score as: Normal-PCR+, Mild, Moderate and Severe.
COVIDGR-1.0 is organized into two classes, positive and negative. It contains 852 images distributed into 426 positive and 426 negative cases, more details are provided in Table III. All the images were obtained from the same equipment and under the same X-ray regime. Only PosteriorAnterior (PA) view is considered. COVIDGR-1.0 along with the severity level labels are available to the scientific community through this link: https://dasci.es/es/transferencia/open-data/covidgr/.
TABLE III. A Brief Summary of COVIDGR-1.0 Dataset. All Samples in COVIDGR 1.0 are Segmented CXR Images Considering Only PA View.
| Dataset | Class | #images | women | men | #img. per severity level |
|---|---|---|---|---|---|
| COVIDGR-1.0 | Negative | 426 | 239 | 187 | |
| COVID-19 | 426 | 190 | 236 | Normal-PCR+: 76 | |
| Mild: 100 | |||||
| Moderate: 171 | |||||
| Severe: 79 |
IV. COVID-SDNet Methodology
In this section, we describe COVID-SDNet methodology in detail, covering pre-processing to produce smart data, including segmentation and data transformation for increasing discrimination between positive and negative classes, combined with a deep CNN for classification.
One of the pieces of COVID-SDNet is the CNN-based classifier. We have selected Resnet-50 initialized with ImageNet weights for a transfer learning approach. To adapt this CNN to our problem, we have removed the last layer of the net and added a 512 neurons layer with ReLU activation and a two or four neurons layer (according to the considered number of classes) with softmax activation.
Let 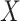 be the set of
be the set of 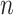 images and
images and 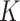 the total number of classes. Each image
the total number of classes. Each image 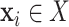 has a true label
has a true label 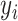 with
with 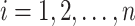 . The softmax function computes the probability that an image belongs to class
. The softmax function computes the probability that an image belongs to class 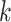 with
with 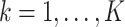 . Let
. Let 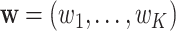 be the output of the last fully connected layer before the softmax activation is applied. Then, this function is defined as:
be the output of the last fully connected layer before the softmax activation is applied. Then, this function is defined as: 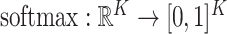 ,
,
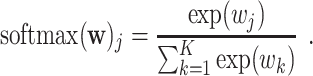 |
Let 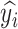 be the class prediction of the network for the image
be the class prediction of the network for the image 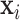 , then
, then 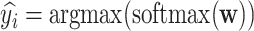 , where
, where 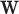 is the output vector of the last layer before softmax is applied for the input
is the output vector of the last layer before softmax is applied for the input 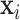 .
.
All the layers of the network were fine-tuned. We used a batch size of 16 and SGD as optimizer.
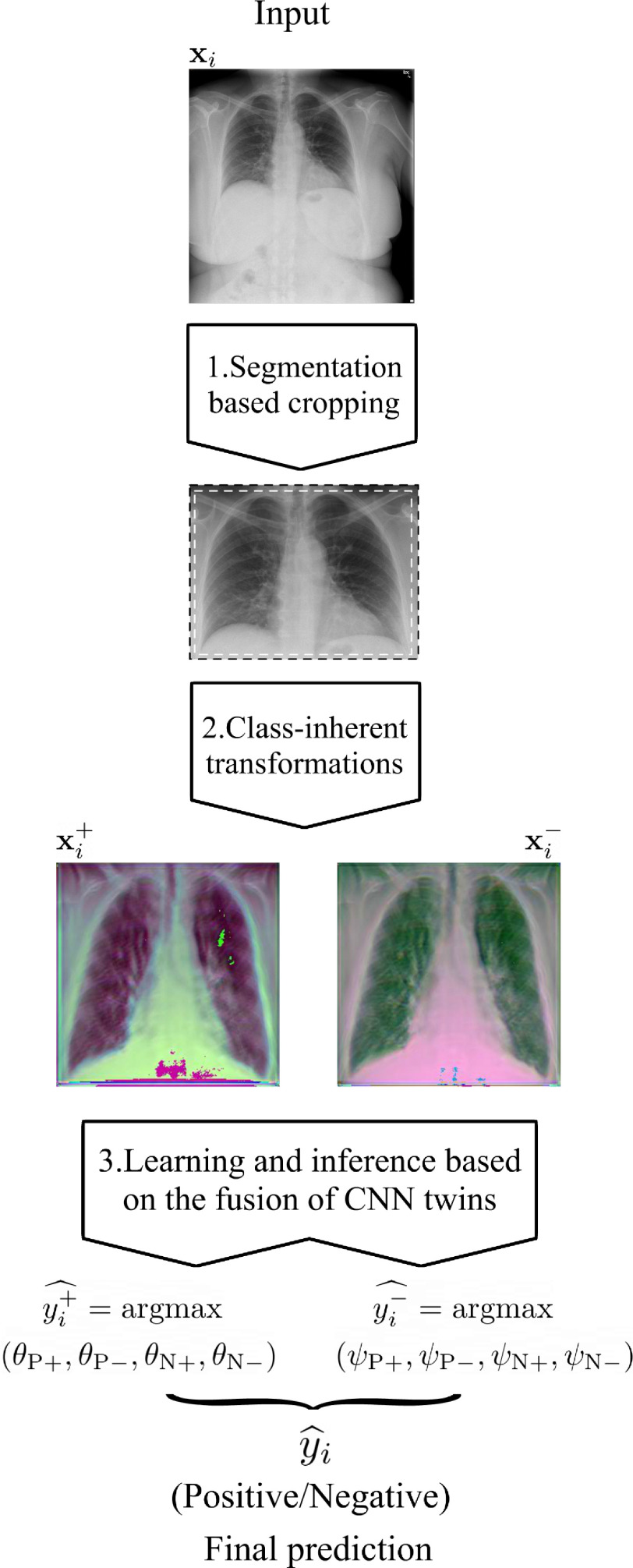
The main stages of COVID-SDNet are three, two associated to pre-processing for producing quality data (smart data stages) and the learning and inference process. A flowchart of COVID-SDNet is depicted in Fig. 2.
Fig. 2.
Flowchart of the proposed COVID-SDNet methodology.
1) Segmentation-Based Cropping: Unnecessary Information Elimination: Different CXR equipment brands include different extra information about the patient in the sides and contour of CXR images. The position and size of the patient may also imply the inclusion of more parts of the body, e.g., arms, neck, stomach. As this information may alter the learning of the classification model, first, we segment the lungs using the U-Net segmentation model provided in [24], pre-trained on Tuberculosis Chest X-ray Image datasets [25] and RSNA Pneumonia CXR challenge dataset [12]. Then, we calculate the smallest rectangle that delimits the left and right segmented-lungs. Finally, to avoid eliminating useful information, we add 2.5% of pixels to the left, right, up and down sides of the rectangle. The resulting rectangle is cropped. An illustration with example of this pre-processing is shown in Fig. 3.
Fig. 3.
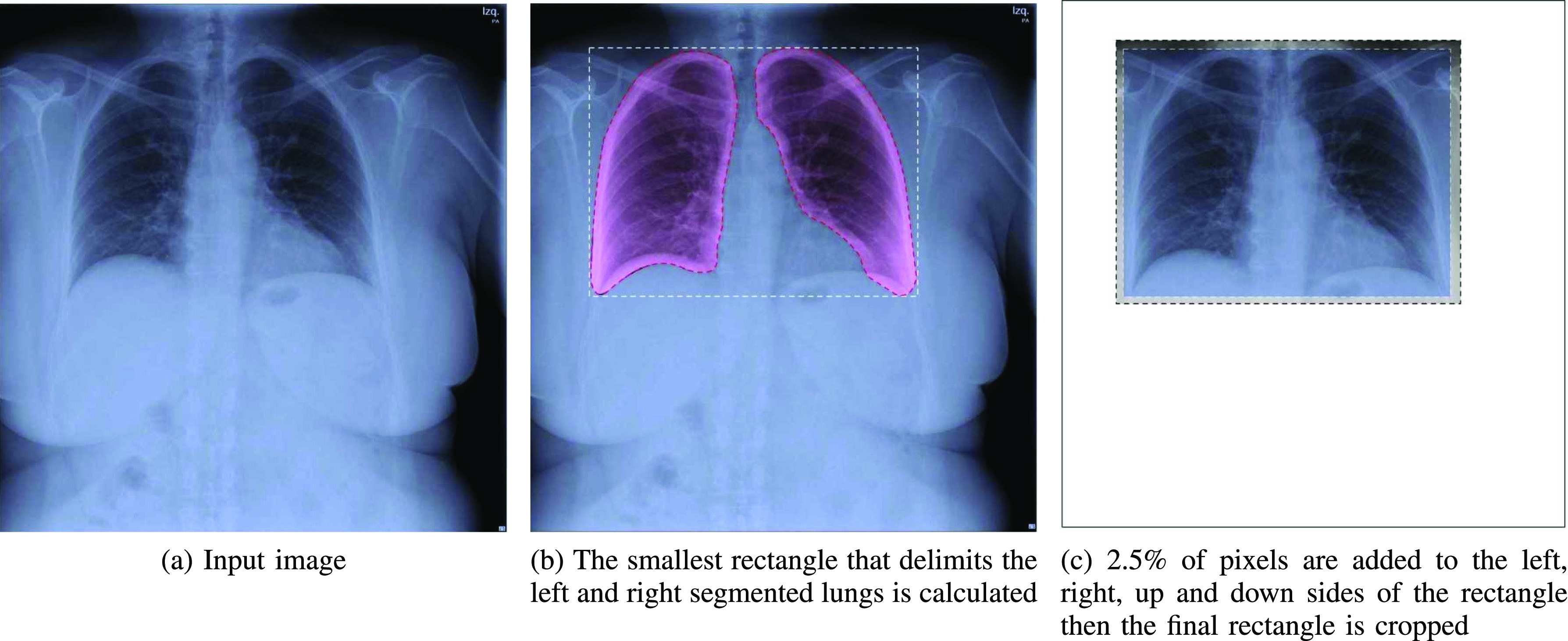
The segmentation-based cropping pre-processing applied to the input X-ray image.
2) Class-Inherent Transformations Network: To increase the discrimination capacity of the classification model, we used, FuCiTNet [10], a Class-inherent transformations (CiT) Network inspired by GANs (Generative Adversarial Networks). This transformation method is actually an array of two generators 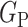 and
and 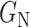 , where
, where 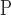 refers to the positive class and
refers to the positive class and 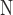 refers to the negative class.
refers to the negative class. 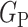 learns the inherent-class transformations of the positive class
learns the inherent-class transformations of the positive class 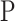 and
and 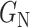 learns the inherent-class transformations of the negative class
learns the inherent-class transformations of the negative class 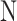 . In other words,
. In other words, 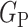 learns the transformations that bring an input image from its own
learns the transformations that bring an input image from its own 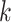 domain, with
domain, with 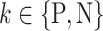 , to the
, to the 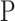 class domain. Similarly,
class domain. Similarly, 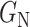 learns the transformations that bring the input image from its
learns the transformations that bring the input image from its 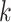 space, with
space, with 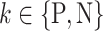 , to the
, to the 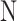 class space. The classification loss is introduced in the generators to drive the learning of each specific
class space. The classification loss is introduced in the generators to drive the learning of each specific 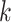 -class transformations. That is, each generator is optimized based on the following loss function:
-class transformations. That is, each generator is optimized based on the following loss function:
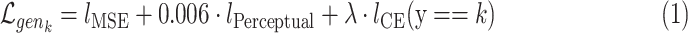 |
Where 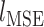 is a pixel-wise Mean Square Error,
is a pixel-wise Mean Square Error, 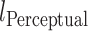 is a perception Mean Square Error and
is a perception Mean Square Error and 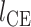 is the classifier loss. The weighted factor
is the classifier loss. The weighted factor 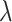 indicates how much the generator must change its outcome to suit the classifier. More details about these transformation networks can be found in [10].
indicates how much the generator must change its outcome to suit the classifier. More details about these transformation networks can be found in [10].
The architecture of the generators consists of 5 identical residual blocks. Each block has two convolutional layers with 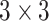 kernels and 64 feature maps followed by batch-normalization layers and Parametric ReLU as activation function. The last residual block is followed by a final convolutional layer which reduces the output image channels to 3 to match the input's dimensions. The classifier is a ResNet-18 which consists of an initial convolutional layer with
kernels and 64 feature maps followed by batch-normalization layers and Parametric ReLU as activation function. The last residual block is followed by a final convolutional layer which reduces the output image channels to 3 to match the input's dimensions. The classifier is a ResNet-18 which consists of an initial convolutional layer with 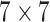 kernels and 64 feature maps followed by a
kernels and 64 feature maps followed by a 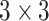 max pool layer. Then, 4 blocks of two convolutional layers with
max pool layer. Then, 4 blocks of two convolutional layers with 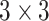 kernels with 64, 128, 256 and 512 feature maps respectively followed by a
kernels with 64, 128, 256 and 512 feature maps respectively followed by a 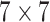 average pooling and one fully connected layer which outputs a vector of
average pooling and one fully connected layer which outputs a vector of 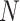 elements. ReLU is used as activation function.
elements. ReLU is used as activation function.
Once the generators learn the corresponding transformations, the dataset is processed using 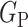 and
and 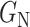 . Two pair of images
. Two pair of images 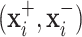 will be obtained from each input image
will be obtained from each input image 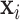 ,
, 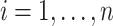 , where
, where 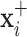 and
and 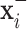 are respectively the positively and negatively transformed images of
are respectively the positively and negatively transformed images of 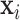 . Note that, once the entire dataset is processed, we have four classes (
. Note that, once the entire dataset is processed, we have four classes (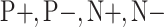 ) instead the original
) instead the original 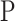 and
and 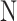 classes. Let
classes. Let 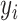 be the class of
be the class of 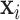 ,
, 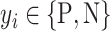 . If
. If 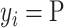 ,
,  and
and 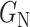 will produce the positive transformation
will produce the positive transformation 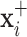 with
with 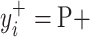 and the negative transformation
and the negative transformation 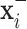 with
with 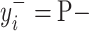 , respectively. If
, respectively. If 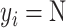 ,
, 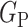 and
and 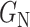 will produce the positive transformation
will produce the positive transformation 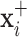 with
with 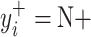 and the negative transformation
and the negative transformation 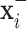 with
with 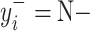 , respectively. Fig. 4 illustrates with example the transformations applied by
, respectively. Fig. 4 illustrates with example the transformations applied by 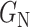 and
and 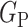 . Notice that these transformations are not meant to be interpretable by the human eye but rather help the classification model better distinguish between the different classes.
. Notice that these transformations are not meant to be interpretable by the human eye but rather help the classification model better distinguish between the different classes.
Fig. 4.
Class-inherent transformations applied to a negative sample. a) Original negative sample; b) Negative transformation; c) Positive transformation.
3) Learning and Inference Based on the Fusion of CNN Twins: The CNN classification model described above in this section (Resnet-50) is trained to predict the new four classes: 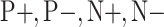 . The output of the network (after softmax is applied) for each transformed image associated to the original one is a vector
. The output of the network (after softmax is applied) for each transformed image associated to the original one is a vector 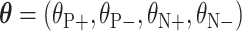 , where
, where 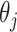 is the probability of the transformed image to belong to class
is the probability of the transformed image to belong to class 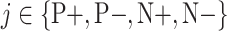 . Herein, we propose an inference process to fuse the output of the two transformed images
. Herein, we propose an inference process to fuse the output of the two transformed images 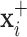 and
and 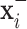 to predict the label of the original image
to predict the label of the original image 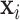 . In this way, for each pair
. In this way, for each pair 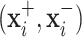 , the prediction of the original image
, the prediction of the original image 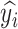 will be either
will be either 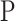 or
or 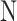 . Let
. Let 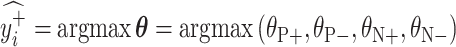 and
and 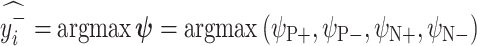 be the ResNet-50 predictions for
be the ResNet-50 predictions for 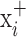 and
and 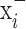 respectively. Then:
respectively. Then:
-
1)
If
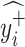 =
=  and
and 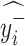 =
= 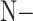 , then
, then 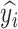 = N.
= N. -
2)
If
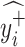 =
= 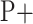 and
and 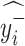 =
= 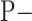 , then
, then 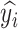 = P.
= P. -
3)If none of the above applies, then
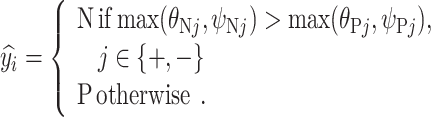
Experimentally, we used a batch size of 16 and SGD as optimizer.
V. Experiments and Results
In this section we (1) provide all the information about the used experimental setup, (2) evaluate two state-of-the-art COVID classification models and FuCiTNet alone [10] on our dataset then, analyze (3) the impact of data pre-processing and (4) Normal-PCR+ severity level on our approach.
A. Experimental Setup
Due to the high variations between different executions, we performed 5 different 5 fold cross validations in all the experiments. Each experiment uses 80% of COVIDGR-1.0 for training and the remaining 20% for testing. To choose when to stop the training process, we used a random 10% of each training set for validation. In each experiment, a proper set of data-augmentation techniques is carefully selected. All results, in terms of sensitivity, specificity, precision, F1 and accuracy, are presented using the average values and the standard deviation of the 25 executions. The used metrics are calculated as follows:
 |
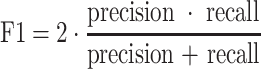 |
TP and TN refers respectively to the number of true positives and true negatives.
B. Analysis of COVIDNet and COVID-CAPS
We compare our approach with the two most related approaches to ours, COVIDNet [16] and COVID-CAPS [19].
-
•
COVIDNet: Currently, the authors of this network provide three versions, namely A, B and C, available at [26]. A has the largest number of trainable parameters, followed by B and C. We performed two evaluations of each network in such a way that the results will be comparable to ours.
-
•
First, we tested COVIDNet-A, COVIDNet-B and COVIDNet-C, pre-trained on COVIDx, directly on our dataset by considering only two classes: Normal (negative), and COVID-19 (positive). The whole dataset (426 positive images and 426 negative images) is evaluated. We report in Table IV recall and precision results for Normal and COVID-19 classes.
-
•
Second, we retrained COVIDNet on our dataset. It is important to note that as only a checkpoint of each model is available, we could not remove the last layer of these networks, which has three neurons. We used 5 different 5 fold cross validations. In order to be able to retrain COVIDNet models, we had to add a third Pneumonia class into our dataset. We randomly selected 426 images from the Pneumonia class in COVIDx dataset. We used the same hyper-parameters as the ones indicated in their training script, that is, 10 epochs, a batch size of 8 and a learning rate of 0.0002. We changed covid_weight to 1 and covid_percent to 0.33 since we had the same number of images in all the classes. Similarly, we report in Table IV recall and precision of our two classes, Normal and COVID-19, and omit recall and precision of Pneumonia class. The accuracy reported in the same table only takes into account the images from our two classes. As with our models, we report here the mean and standard deviation of all metrics.
Although we analyzed all three A, B and C variations of COVIDNet, for simplicity we only report the results of the best one.
COVID-CAPS: This is a capsule network-based model proposed in [19]. Its architecture is notably smaller than COVIDNet, which implies a dramatically lower number of trainable parameters. Since the authors also provide a checkpoint with weights trained in the COVIDx dataset, we were able to follow a similar procedure than with COVIDNet:
-
•
First, we tested the pretrained weights using COVIDx on COVIDGR-1.0 dataset. COVID-CAPS is designed to predict two classes, so we reused the same architecture with the new dataset and compute the evaluation metrics shown in Table IV.
-
•
Second, COVID-CAPS architecture was retrained over the COVIDGR-1.0 dataset. This process finetunes the weights to improve class separation. The retraining process is performed using the same setup and hyper-parameters reported by the authors. Adam optimizer is used across 100 epochs with a batch size of 16. Class weights were omitted as with COVIDNet, since this dataset contains balanced classes in training as well as in test. Evaluation metrics are computed for five sets of 5-fold cross-validation test subsets and summarized in Table IV.
-
•
TABLE IV. COVIDNet and COVID-CAPS Results on Our Dataset.
| Class | Negative | Positive (COVID-19) | Accuracy | ||
|---|---|---|---|---|---|
| Metric | Specificity | Precision | Sensitivity | Precision | |
| COVIDNet-CXR A [16] | 0.23 | 16.00 | 99.29 | 33.54 | 49.76 |
| Retrained COVIDNet-CXR A |
88.82 0.90 0.90
|
3.36 6.15 6.15 |
46.82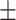 17.59 17.59 |
81.65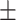 6.02 6.02
|
67.82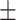 6.11 6.11
|
| COVID-CAPS [19] | 26.30 | 45.81 | 69.01 | 48.36 | 47.66 |
| Retrained COVID-CAPS | 65.74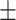 9.93 9.93 |
65.62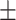 3.98 3.98
|
64.93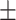 9.71 9.71 |
66.07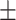 4.49 4.49 |
65.34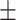 3.26 3.26 |
The results from Table IV show that COVIDNet and COVID-CAPS trained on COVIDx overestimate COVID-19 class in our dataset, i.e., most images are classified as positive, resulting in very high sensitivities but at the cost of low positive predictive value. However, when COVIDNet and COVID-CAPS are re-trained on COVIDGR-1.0 they achieve slightly better overall accuracy and a higher balance between sensitivity and specificity, although they seem to acquire a bias favoring the negative class. In general, none of these models perform adequately for the detection of the disease from CXR images in our dataset.
C. Results and Analysis of COVID Prediction
The results of the baseline COVID classification model considering all the levels of severity, with and without segmentation, FuCiTNet [10], and COVID-SDNet are shown in Table V.
TABLE V. Results of COVID-19 Prediction Using Retrained COVIDNet-CXR A, Retrained COVID-CAPS, ResNet-50 With and Without Segmentation, FuCiTNet and COVID-SDNet. All Four Levels of Severity in the Positive Class are Taken Into Account.
| Class | N | P | Accuracy | |||||
|---|---|---|---|---|---|---|---|---|
| Metric | Specificity | Precision | F1 | Sensitivity | Precision | F1 | ||
| COVIDNet-CXR | 88.82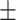 0.90 0.90 |
3.36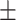 6.15 6.15 |
73.31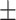 3.79 3.79 |
46.82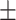 17.59 17.59 |
81.65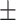 6.02 6.02 |
56.94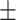 15.05 15.05 |
67.82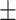 6.11 6.11 |
|
| COVID-CAPS | 65.74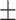 9.93 9.93 |
65.62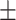 3.98 3.98 |
65.15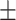 5.02 5.02 |
64.93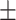 9.71 9.71 |
66.07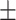 4.49 4.49 |
64.87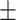 4.92 4.92 |
65.34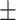 3.26 3.26 |
|
| Without seg. | 79.87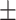 8.91 8.91 |
71.91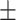 3.12 3.12 |
75.40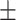 4.91 4.91 |
68.63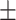 6.08 6.08 |
78.75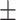 6.31 6.31 |
72.689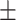 3.45 3.45 |
74.25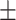 3.61 3.61 |
|
| With seg. | 78.41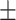 7.09 7.09 |
73.36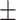 4.66 4.66 |
75.46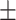 2.97 2.97 |
70.80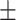 8.26 8.26 |
77.17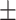 4.79 4.79 |
73.40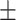 4.01 4.01 |
74.60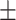 2.93 2.93 |
|
| FuCiTNet |
80.79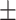 6.98 6.98
|
72.00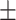 4.48 4.48 |
75.84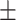 3.18 3.18 |
67.90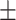 8.58 8.58 |
78.48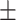 4.99 4.99 |
72.35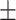 4.76 4.76 |
74.35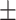 3.34 3.34 |
|
| COVID-SDNet | ||||||||
79.76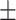 6.19 6.19 |
74.74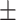 3.89 3.89
|
76.94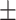 2.82 2.82
|
72.59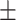 6.77 6.77
|
78.67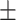 4.70 4.70
|
75.71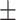 3.35 3.35
|
76.18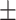 2.70 2.70
|
||
In general, COVID-SDNet achieves better and more stable results than the rest of approaches. In particular, COVID-SDNet achieved the highest balance between specificity and sensitivity with 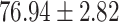 F1 in the negative class and
F1 in the negative class and 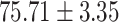 F1 in the positive class. Most importantly, COVID-SDNet achieved the best sensitivity
F1 in the positive class. Most importantly, COVID-SDNet achieved the best sensitivity 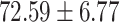 and accuracy with
and accuracy with 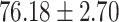 . FuCiTNet provides in general good but lower and less stable results than COVID-SDNet. When comparing the results of the baseline classification model with and without segmentation, we can observe that the use of segmentation improves substantially the sensitivity, which is the most important criteria for a triage system. This can be explained by the fact that segmentation allows the model to focus on most important parts of the CXR image.
. FuCiTNet provides in general good but lower and less stable results than COVID-SDNet. When comparing the results of the baseline classification model with and without segmentation, we can observe that the use of segmentation improves substantially the sensitivity, which is the most important criteria for a triage system. This can be explained by the fact that segmentation allows the model to focus on most important parts of the CXR image.
Analysis Per Severity Level
To determine which levels are the hardest to distinguish by the best approach, we have analyzed the accuracy per severity level (S), with 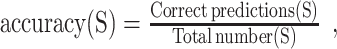 where
where 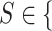 Normal-PCR+, Mild, Moderate, Severe
Normal-PCR+, Mild, Moderate, Severe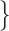 . The results are shown in Table VI.
. The results are shown in Table VI.
TABLE VI. Results of COVID-SDNet Per Severity Level.
| S (Severity level) | accuracy (S)(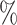 ) ) |
|---|---|
| Normal-PCR+ | 28.42 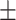 2.58 2.58 |
| Mild | 61.80 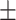 5.49 5.49 |
| Moderate | 86.90 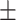 3.20 3.20 |
| Severe | 97.72 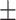 0.95 0.95 |
As it can be seen from these results, COVID-SDNet correctly distinguish Moderate and Severe levels with an accuracy of 86.90% and 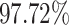 , respectively. This is due to the fact that Moderate and Severe CRX images contain more important visual features than Mild and Normal-PCR+ which ease the classification task. Normal-PCR+ and Mild cases are much more difficult to identify as they contain few or none visual features. These results are coherent with the clinical studies provided in [5] and [2] which report that expert sensitivity is very low in Normal-PCR+ and Mild infection levels. Recall that the expert eye does not see any visual signs in Normal-PCR+ although the PCR is positive. Those cases are actually considered as asymptomatic patients.
, respectively. This is due to the fact that Moderate and Severe CRX images contain more important visual features than Mild and Normal-PCR+ which ease the classification task. Normal-PCR+ and Mild cases are much more difficult to identify as they contain few or none visual features. These results are coherent with the clinical studies provided in [5] and [2] which report that expert sensitivity is very low in Normal-PCR+ and Mild infection levels. Recall that the expert eye does not see any visual signs in Normal-PCR+ although the PCR is positive. Those cases are actually considered as asymptomatic patients.
D. Analysis of the Impact of Normal-PCR+
To analyze the impact of Normal-PCR+ class on COVID-19 classification, we trained and evaluated the baseline model, FuciTNet, COVID-SDNet classification stage, COVIDNet-CXR-A and COVID-CAPS, on COVIDGR-1.0 by eliminating Normal-PCR+. The results are summarized in Table VII.
TABLE VII. Results of the Baseline Classification Model With Segmentation, COVID-SDNet, Retrained COVIDNet-CXR-A and Retrained COVID-CAPS. Only Three Levels of Severity are Considered, Mild, Moderate and Severe.
| Class | N | P | Accuracy | ||||
|---|---|---|---|---|---|---|---|
| Metric | Specificity | Precision | F1 | Sensitivity | Precision | F1 | |
| COVIDNet-CXR | 83.42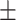 15.39 15.39 |
69.73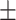 10.34 10.34 |
74.45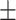 8.86 8.86 |
61.82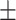 22.44 22.44 |
79.50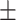 11.47 11.47 |
65.64 15.90 15.90 |
72.62 7.6 7.6 |
| COVID-CAPS | 65.09 10.51 10.51 |
71.72 5.57 5.57 |
67.52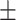 5.29 5.29 |
73.31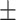 9.74 9.74 |
68.40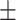 5.13 5.13 |
70.20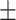 4.31 4.31 |
69.20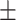 3.61 3.61 |
| With seg. | 80.57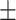 8.72 8.72 |
78.68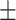 6.57 6.57 |
78.97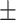 3.20 3.20 |
76.80 10.15 10.15 |
80.70 5.56 5.56 |
78.01 4.29 4.29 |
78.69 3.00 3.00 |
| FuCiTNet | 82.63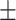 6.61 6.61 |
79.94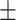 4.28 4.28
|
81.05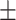 3.44 3.44 |
78.91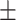 5.88 5.88
|
82.43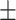 5.43 5.43 |
80.37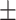 3.16 3.16 |
80.77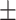 3.15 3.15 |
| COVID-SDNet |
85.20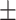 5.38 5.38
|
78.88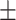 3.89 3.89 |
81.75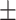 2.74 2.74
|
76.80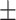 6.30 6.30 |
84.23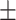 4.59 4.59
|
80.07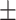 0.04 0.04
|
81.00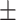 2.87 2.87
|
Overall, all the approaches systematically provide better results when eliminating Normal-PCR+ from the training and test processes, including COVIDNet-CXR-A and COVID-CAPS. In particular, COVID-SDNet still represents the best and most stable approach.
E. Analysis Per Severity Level
A further analysis of the accuracy at the level of each severity degree (see Table VIII) demonstrates that eliminating Normal-PCR+ decreases the accuracy in Mild and Moderate severity levels by 15.8% and 1.52% respectively.
TABLE VIII. Results of COVID-SDNet by Severity Level Without Considering Normal-PCR+.
| S (Severity level) | accuracy (S)(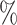 ) ) |
|---|---|
| Mild | 46.00 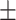 7.10 7.10 |
| Moderate | 85.38 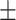 1.85 1.85 |
| Severe | 97.22 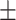 1.86 1.86 |
These results show that although Normal-PCR+ is the hardest level to predict, its presence improves the accuracy of lower severity levels, especially Mild level.
VI. Inspection of Model's Decision
Automatic DL diagnosis systems alone are not mature yet to replace expert radiologists. To help clinician making decisions, these tools must be interpretable so that clinicians can decide whether to trust the model or not [27]. We inspect what led our model make a decision by showing the regions of the input image that triggered that decision along with its counterfactual explanation by showing the parts that explain the opposite class. We adapted Grad-CAM method [28] to explain the decision of the negative and positive class.
Figs. 5, 6, and 7 show (a) the original CXR image, (b) visual explanation by means of a heat-map that highlights the regions/pixels which led the model to output the actual prediction and (c) its counterfactual explanation using a heat-map that highlights the regions/pixels which had the highest impact on predicting the opposite class. Higher intensity in the heat-map indicates higher importance of the corresponding pixel in the decision. The larger higher intensity areas in the heat-map determine the final class. However, Fig. 8(b) represents first the counterfactual explanation and Fig. 8(c) represents the explanation of the actual decision.
Fig. 5.
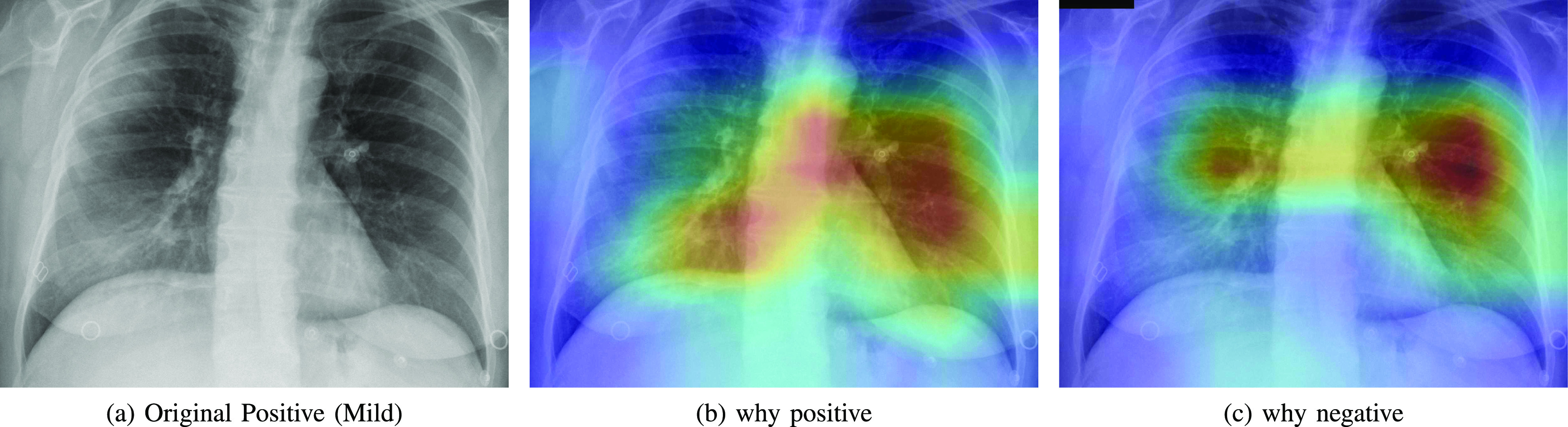
Heatmap showing the parts of the input image that triggered the positive prediction (b) and counterfactual explanation (c).
Fig. 6.
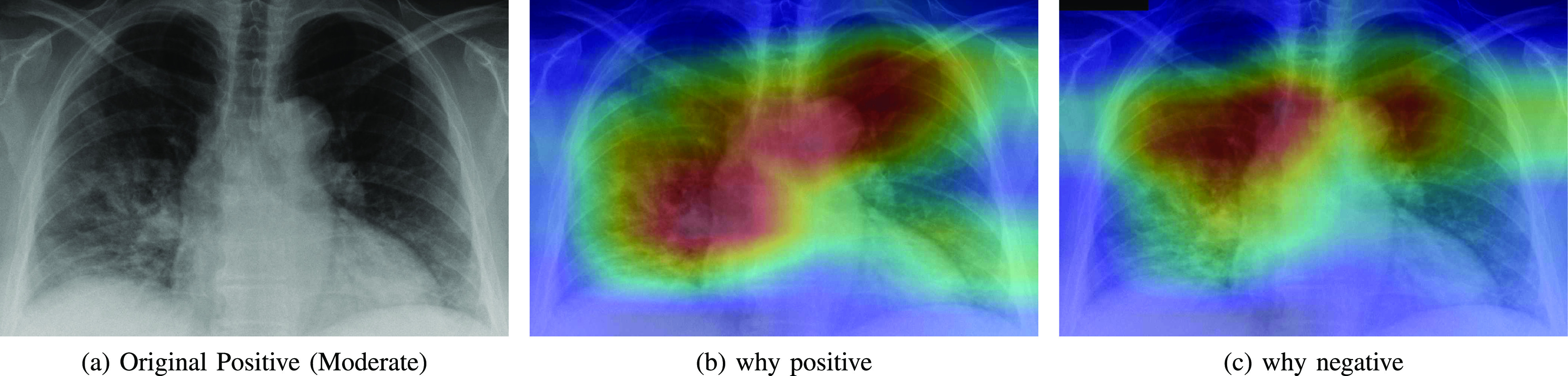
Heatmap showing the parts of the input image that triggered the positive prediction (b) and counterfactual explanation (c).
Fig. 7.
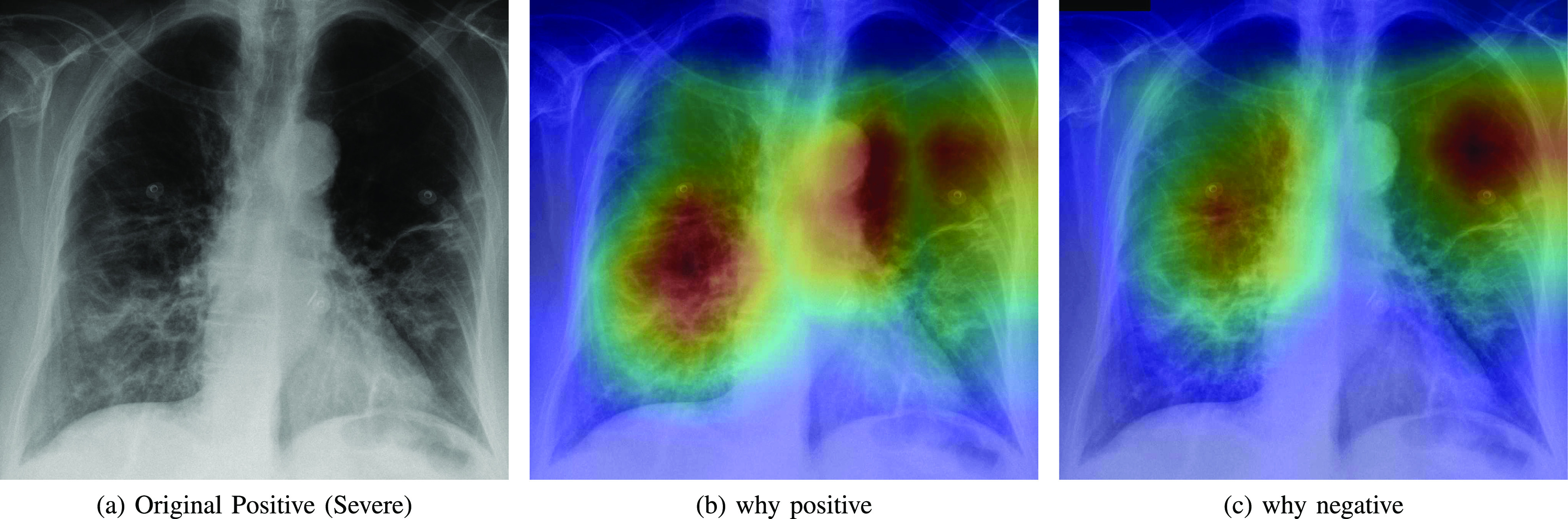
Heatmap showing the parts of the input image that triggered the positive prediction (b) and counterfactual explanation (c).
Fig. 8.
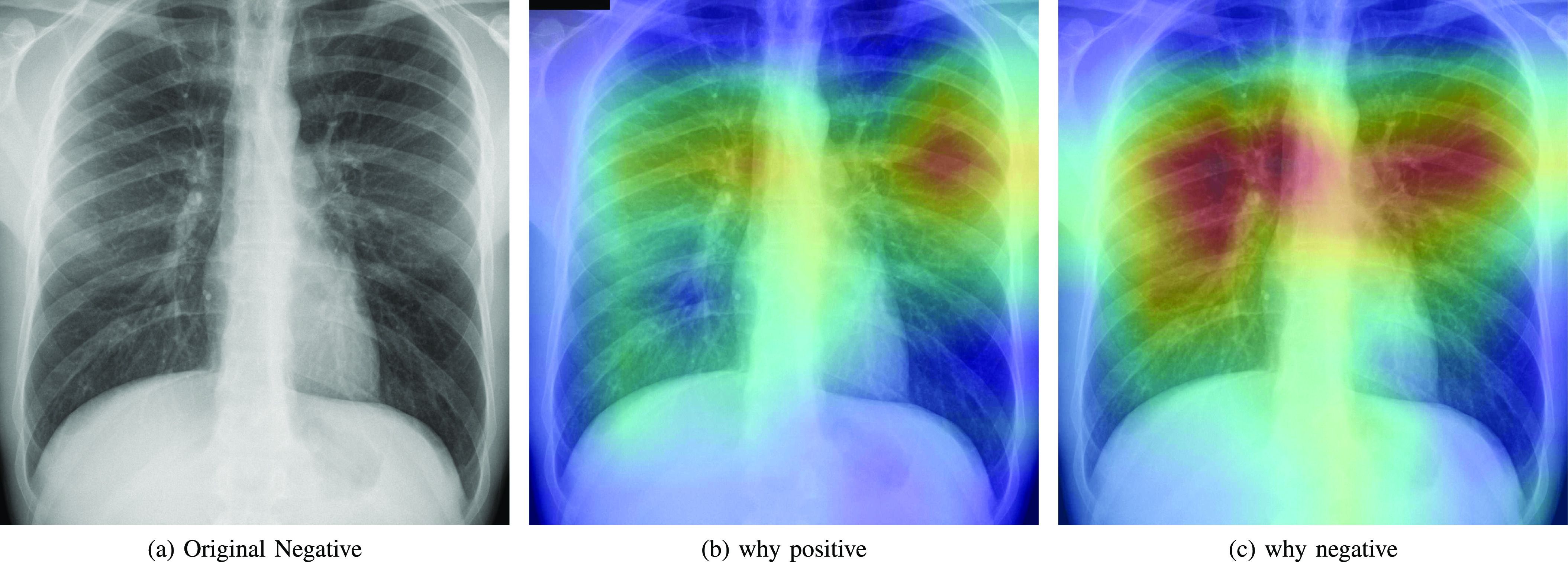
Heatmap that explains the parts of the input image that triggered the counterfactual explanation (b) and the negative actual prediction (c).
As expected, negative and positive interpretations are complementary, i.e, areas which triggered the correct decision are opposite, in most cases, to the areas that triggered the decision towards negative. In CXR images with different severity levels, the heat-maps correctly point out opaque regions due to different levels of infiltrates, consolidations and also to osteoarthritis.
In particular, in Fig. 5(b), the red areas in the right lung points out a region with infiltrates and also osteoarthritis in the spine region. Fig. 6(b) correctly shows moderate infiltrates in the right lower and lower-middle lung fields in addition to a dilation of ascending aorta and aortic arch (red color in the center). Fig. 5(c) shows normal upper-middle fields of both lungs (less important on the left due to aortic dilation). Fig. 7(b) indicates an important bilateral pulmonary involvement with consolidations.
As it can be observed in Fig. 8(c), the explanation of the negative class correctly highlights a symmetric bilateral pattern that occupies a larger lung volume especially in regions with high density. In fact, a very similar pattern is shown in the counterfactual explanation of the positive class in Fig. 5(c), 6(c) and 7(c).
VII. Conclusion
This article introduced a dataset, named COVIDGR-1.0, with high clinical value. COVIDGR-1.0 includes the four main COVID severity levels identified by a recent radiological study [2]. We proposed a methodology, called COVID-SDNet, that combines segmentation, data-augmentation and data transformation. The obtained results show the high generalization capacity of COVID-SDNet, specially on severe and moderate levels as they include important visual features. The existence of few or none visual features in Mild and Normal-PCR+ reduces the opportunities for improvement.
As main conclusions, we must highlight that COVID-SDNet can be used in a triage system to detect especially moderate and severe patients. Finally, we must also mention that more robust and accurate triage system can be built by fusing our approach with other approaches such as the one proposed in [29].
As future work, we are working on enriching COVIDGR-1.0 with more CXR images coming from different hospitals. We are planning to explore the use of additional clinical information along with CXR images to improve the prediction performance.
Funding Statement
This work was supported by the project DeepSCOP-Ayudas Fundación BBVA a Equipos de Investigación Científica en Big Data 2018, COVID19_RX-Ayudas Fundación BBVA a Equipos de Investigación Científica SARS-CoV-2 y COVID-19 2020, and the Spanish Ministry of Science and Technology under the project TIN2017-89517-P. S. Tabik was supported by the Ramon y Cajal Programme (RYC-2015-18136). A. Gómez-Ríos was supported by the FPU Programme FPU16/04765. D. Charte was supported by the FPU Programme FPU17/04069. J. Suárez was supported by the FPU Programme FPU18/05989. E.G was supported by the European Research Council (ERC Grant agreement 647038 [BIODESERT]). This project is approved by the Provincial Research Ethics Committee of Granada.
Footnotes
[Online]. Available: https://english.elpais.com/society/2020-05-14/antibody-study-shows-just-5-of-spaniards-have-contracted-the-coronavirus.html.
[Online]. Available: //www.acr.org/Advocacy-and-Economics/ACR-Position-Statements/Recommendations-for-Chest-Radiography-and-CT-for-Suspected-COVID19-Infection.
Contributor Information
S. Tabik, Email: siham@ugr.es.
A. Gómez-Ríos, Email: anabelgrios@decsai.ugr.es.
J. L. Martín-Rodríguez, Email: joseluismartin.rx@hotmail.com.
I. Sevillano-García, Email: isega24ivan@gmail.com.
M. Rey-Area, Email: mreyarea@gmail.com.
D. Charte, Email: fdavidcl@ugr.es.
E. Guirado, Email: geesecillo@gmail.com.
J. L. Suárez, Email: jlsuarezdiaz@ugr.es.
J. Luengo, Email: julianlm@decsai.ugr.es.
M. A. Valero-González, Email: valerogonzalez@yahoo.es.
P. García-Villanova, Email: pgvillanova@gmail.com.
E. Olmedo-Sánchez, Email: euolm@yahoo.es.
F. Herrera, Email: herrera@decsai.ugr.es.
References
- [1].Kissler S. M. et al. , “Projecting the transmission dynamics of SARS-CoV-2 through the postpandemic period,” Science, vol. 368, no. 6493, pp. 860–868, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wong H. et al. , “Frequency and distribution of chest radiographic findings in COVID-19 positive patients,” Radiology, 2020, Art. no. 201160. [DOI] [PMC free article] [PubMed]
- [3].Li Y. et al. , “Stability issues of RT-PCR testing of SARS-CoV-2 for hospitalized patients clinically diagnosed with COVID-19,” J. Med. Virol., vol. 92, no. 7 pp. 903–908, Jul. 2020, doi: 10.1002/jmv.25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Fang Y. et al. , “Sensitivity of chest CT for COVID-19: Comparison to RT-PCR,” Radiology, 2020, Art. no. 200432. [DOI] [PMC free article] [PubMed]
- [5].Weinstock M. et al. , “Chest X-ray findings in 636 ambulatory patients with COVID-19 presenting to an urgent care center: A normal chest X-ray is no guarantee,” J. Urgent Care Med., vol. 14, no. 7, pp. 13–18, 2020. [Google Scholar]
- [6].Warren M. A. et al. , “Severity scoring of lung oedema on the chest radiograph is associated with clinical outcomes in ards,” Thorax, vol. 73, no. 9, pp. 840–846, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Cohen J. P. et al. , “COVID-19 image data collection,” 2020, arXiv 2003.11597.
- [8].Luengo J. et al. , Big Data Preprocessing - Enabling Smart Data. Springer, 2020. [Google Scholar]
- [9].Tabik S. et al. , “A snapshot of image pre-processing for convolutional neural networks: Case study of MNIST,” Int. J. Comput. Intell. Syst., vol. 10, no. 1, pp. 555–568, 2017. [Google Scholar]
- [10].Rey-Area M., Guirado E., Tabik S., and Ruiz-Hidalgo J., “FuCiTNet: Improving the generalization of deep learning networks by the fusion of learned class-inherent transformations,” Inf. Fusion, vol. 63, pp. 188–195, 2020. [Google Scholar]
- [11].Chung et al. , “Figure 1 COVID-19 chest X-ray dataset initiative,” 2020. [Online]. Available: https://github.com/agchung/Figure1-COVID-chestxray-dataset
- [12].“Radiological Society of North America. RSNA pneumonia detection challenge,” 2019. [Online]. Available: https://www.kaggle.com/c/rsnapneumonia-detection-challenge/data [DOI] [PubMed]
- [13].Wang X. et al. , “Chest X-ray8: Hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit., 2017, pp. 2097–2106. [Google Scholar]
- [14].Johnson A. E. et al. , “MIMIC-CXR: A large publicly available database of labeled chest radiographs,” 2019, arXiv:1901.07042. [DOI] [PMC free article] [PubMed]
- [15].Bustos A. et al. , “PadChest: A large chest X-ray image dataset with multi-label annotated reports,” 2019, arXiv:1901.07441. [DOI] [PubMed]
- [16].Wang L. et al. , “COVID-Net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest radiography images,” 2020. [DOI] [PMC free article] [PubMed]
- [17].Kundu S. et al. , “How might AI and chest imaging help unravel COVID-19's mysteries?,” Radiol. Artif. Intell., vol. 2, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Maguolo G. and Nanni L., “A critic evaluation of methods for COVID-19 automatic detection from x-ray images,” 2020, arXiv:2004.12823. [DOI] [PMC free article] [PubMed]
- [19].Afshar P. et al. , “COVID-CAPS: A capsule network-based framework for identification of COVID-19 cases from x-ray images,” 2020, arXiv:2004.02696. [DOI] [PMC free article] [PubMed]
- [20].Ozturk T. et al. , “Automated detection of COVID-19 cases using deep neural networks with X-ray images,” Comput. Biol. Med., 2020, Art. no. 103792. [DOI] [PMC free article] [PubMed]
- [21].Karim M. et al. , “DeepCOVIDExplainer: Explainable COVID-19 predictions based on chest X-ray images,” 2020, arXiv:2004.04582.
- [22].Ghoshal B. and Tucker A., “Estimating uncertainty and interpretability in deep learning for coronavirus (COVID-19) detection,” 2020, arXiv:2003.10769.
- [23].Apostolopoulos I. and Mpesiana T., “COVID-19: Automatic detection from X-ray images utilizing transfer learning with convolutional neural networks,” Phys. Eng. Sci. Med., vol. 43, no. 2, pp. 635–640, Jun. 2020, doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Mineo E., “U-Net lung segmentation,” [Online]. Available: https://www.kaggle.com/eduardomineo/u-net-lung-segmentation-montgomery-shenzhen, 2020.
- [25].Jaeger S., Candemir S., Antani S., Wáng Y.-X. J., Lu P.-X., and Thoma G., “Two public chest X-ray datasets for computer-aided screening of pulmonary diseases,” Quantitative Imag. Med. Surg., vol. 4, no. 6, p. 475, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Warren M. A. et al. , “COVIDNet,” 2020. [Online]. Available: https://github.com/lindawangg/COVID-Net
- [27].Arrieta A. et al. , “Explainable artificial intelligence (XAI): Concepts, taxonomies, opportunities and challenges toward responsible AI,” Inf. Fusion, vol. 58, pp. 82–115, 2020. [Google Scholar]
- [28].Selvaraju R. et al. , “Grad-CAM: Visual explanations from deep networks via gradient-based localization,” in Proc. IEEE Int. Conf. Comput. Vis., 2017, pp. 618–626. [Google Scholar]
- [29].Cohen J. P. et al. , “Predicting COVID-19 pneumonia severity on chest X-ray with deep learning,” 2020, arXiv:2005.11856. [DOI] [PMC free article] [PubMed] [Google Scholar]