Abstract
Mice homozygous for the s1Acrg deletion at the Ednrb locus arrest at embryonic day 8.5. To determine the molecular basis of this defect, we initiated positional cloning of the s1Acrg minimal region. The mouse Uch-L3 (ubiquitin C-terminal hydrolase L3) gene was mapped within the s1Acrg minimal region. Because Uch-L3 transcripts were present in embryonic structures relevant to the s1Acrg phenotype, we created a targeted mutation in Uch-L3 to address its role during development and its possible contribution to the s1Acrg phenotype. Mice homozygous for the mutation Uch-L3Δ3-7 were viable, with no obvious developmental or histological abnormalities. Although high levels of Uch-L3 RNA were detected in testes and thymus, Uch-L3Δ3-7 homozygotes were fertile, and no defect in intrathymic T-cell differentiation was detected. We conclude that the s1Acrg phenotype is either complex and multigenic or due to the loss of another gene within the region. We propose that Uch-L3 may be functionally redundant with its homologue Uch-L1.
The analysis of induced mutations has proven to be a powerful method for identifying genes involved in development in many species. The first genetic screen for induced mutations in the mouse was the specific locus test (SLT) (22; for a review, see reference 19). With a variety of mutagens, the SLT generated new alleles over seven tester loci chosen for their easily scored mutant phenotypes. The molecular lesions ranged in size from single base changes to large deletions spanning multiple centimorgans. These alleles have been useful in the positional cloning of the genes underlying the tester loci themselves, such as short ear/Bmp5 (11). In addition, large deletion alleles have also been used to assign biological function to chromosomal regions flanking the specific loci. For the albino-linked deletion region required for embryonic ectoderm development (eed), the corresponding gene was identified through positional cloning (27).
The piebald (s) locus was one of the specific loci used in the SLT. piebald encodes the endothelin B receptor (EDNRB), a G protein-coupled seven-transmembrane receptor required for the migration of two neural crest derivatives, melanocytes and enteric ganglia (8, 26). Mice homozygous for a null allele of Ednrb are amelanocytic and develop megacolon, resulting in juvenile lethality (12, 15). Many Ednrb alleles generated in the SLT are deletions that exhibit a more severe phenotype than the loss of Ednrb alone, most likely due to the loss of linked essential genes (16). Through phenotypic analysis of individual deletions combined with molecular mapping of deletion breakpoints and complementation analysis of deletion alleles, chromosomal regions associated with distinct developmental defects were defined (17). These include embryonic lethality, neonatal lethality, and skeletal and central nervous system defects.
The s1Acrg deletion results in embryonic lethality; based on complementation analysis, the portion of the deletion associated with this defect was defined as the s1Acrg minimal region (17). Embryos homozygous for s1Acrg arrest at embryonic day 8.5 and display a complex phenotype that includes cranial neural tube defects, altered somite and notochord morphology, and a failure to complete embryonic turning and heart looping morphogenesis (T. P. O'Brien, personal communication). Based on histological and molecular marker analyses, this phenotype results from defects that are already evident in the primitive streak and node. Although the s1Acrg deletion phenotype may be multigenic, several single-gene mutations lead to arrest at embryonic day 8.5 with a similar phenotype (for a review, see reference 3). Therefore, the s1Acrg phenotype could result from the loss of a single gene that is essential during development.
To determine the molecular basis of the s1Acrg phenotype, we initiated an analysis of the genes within the minimal region. For this purpose, a 1.4-Mb contig of P1, bacterial artificial chromosome (BAC), and yeast artificial chromosome clones was constructed (L. J. Kurihara, E. Semenova, D. L. Metallinos, X.-J. Guan, R. S. Ingram, A. Goddard, and S. M. Tilghman, unpublished data). Based on the low CpG content of syntenic human chromosome 13 (6) and the small (compared to other chromosomes) number of human expressed sequence tags (ESTs) mapping to chromosome 13 (24), we predicted that the s1Acrg region is gene poor. Indeed, no CpG islands were identified within the contig. However three ESTs were mapped by sequence analysis of a single CpG-rich BAC clone. In addition, two human genes that map proximal to EDNRB cross-hybridized to the mouse s1Acrg contig (Kurihara et al., unpublished data). One of these genes is human UCH-L3, which encodes ubiquitin C-terminal hydrolase L3.
The ubiquitin pathway is constitutive and essential for the turnover of many short-lived regulatory proteins as well as damaged proteins (for a review, see reference 18). However, mutations within ubiquitin pathway enzymes have revealed distinct phenotypes due to either their substrate specificity or particular spatial or temporal expression patterns. Moreover, certain mutations have indicated a role for the ubiquitin pathway during development. For example, loss of the mouse UbcM4 ubiquitin-conjugating enzyme leads to embryonic lethality (7), the Caenorhabditis elegans let-70/ubc-2 ubiquitin-conjugating enzyme is essential for larval development (32), and mutation of the Drosophila fat facets deubiquitinating enzyme leads to defects in eye cell fate determination (9).
Here we report the characterization of the mouse Uch-L3 gene and show that its expression pattern during embryogenesis makes it a candidate for a gene underlying the s1Acrg defect. To directly test whether the absence of Uch-L3 alone leads to embryonic lethality, we generated a targeted mutation in this gene.
MATERIALS AND METHODS
Isolation of Uch-L3.
A human EST corresponding to a UCH-L3 cDNA was shown by low-stringency hybridization to map to a BAC contig of the s1Acrg minimal region. The human UCH-L3 probe was used to isolate mouse cDNAs from an embryonic day 17.5 λgt11 library (Clontech). Phage inserts from purified clones were amplified by PCR, cloned into the TA vector (Invitrogen), and sequenced with an ABI Prism labeling kit using an ABI 373 sequencer. Two partial cDNAs (mUCH4 and mUCH12) and one full-length cDNA (mUCH14) were isolated.
Expression analysis.
Whole-mount in situ hybridization to embryos was performed as described by Wilkinson and Nieto (30). Digoxigenin-labeled RNA probes were synthesized with T7 polymerase. The antisense Uch-L3 probe included exons 3 to 10 from mUCH14 linearized with StuI. The sense control probe included exons 1 to 8 from mUCH4 linearized with BglII.
Total RNA was extracted from mouse tissues with Trizol (GIBCO/BRL). Fifteen micrograms of RNA was separated in 1% agarose gels containing morpholinepropanesulfonic acid (MOPS)-formaldehyde and transferred to Hybond N+ membranes (Amersham). Blots were hybridized in Church buffer (2) at 65°C and washed in 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate at 23 and 65°C. Radiolabeled probes were synthesized from fragments of Uch-L3 (wild-type mUCH4 and mutant Δ3-7 [Uch- L3Δ3-7]) and β-actin cDNA clones.
Reverse transcription (RT)-PCR was performed with a cDNA cycle kit (Invitrogen). Primers used to amplify Uch-L3Δ3-7 RNA were 5′-ATGGAGGGTCAACGCTGGCT-3′ and 5′-GGTGTTTCTGTCAAGATGCTAT-3′. PCR products were cloned with a TOPO-TA kit (Invitrogen) and sequenced with the ABI Prism labeling kit using an ABI 373 sequencer.
Generation and analysis of Uch-L3Δ3-7 mutant mice.
To delete the 9.5-kb region encoding exons 3 to 7, two flanking genomic DNA fragments were subcloned into the targeting vector pLOX-PNT, which contains the neomycin resistance gene driven by the phosphoglycerol kinase 2 promoter (PGK-NEO) and herpes simplex virus thymidine kinase (25). Targeting arms were subcloned from a BAC containing Uch-L3 into the Bluescript vector, where polylinker restriction sites and HindIII/KpnI adapters were utilized for subsequent cloning into pLOX-PNT. The targeting arms included a 3.25-kb SpeI fragment upstream of exon 3 at the 5′ end and a 4-kb HindIII fragment downstream of exon 7 at the 3′ end.
The Uch-L3Δ3-7 targeting construct was linearized at a unique NotI site and electroporated into CJ7 embryonic stem (ES) cells (28), followed by selection with 125 μg of active G418 (Sigma) per ml and 1 μM ganciclovir (Roche). Following colony purification, ES cell DNA was extracted and digested with either HindIII (5′ arm) or PstI (3′ arm), separated in 1% agarose–Tris-borate-EDTA (TBE) gels, and transferred to Hybond N+ membranes. Radiolabeled probes were PCR products generated from genomic DNA flanking each targeting arm, denoted 5′ and 3′ probes. Correctly targeted ES cell clones were obtained at a frequency of one in nine G418-selected colonies.
Three independent ES cell clones (A2, F3, and C11) were injected into C57BL/6 blastocysts and implanted into pseudopregnant mice. Chimeras were bred to C57BL/6 mice, and their agouti progeny were genotyped. PCR genotyping was performed on tail DNA with a common forward primer from the genomic sequence flanking the deletion (5′-GGAACTACTGAGCCATATGTGC-3′). This primer was used with either a reverse primer derived from endogenous DNA within the deletion for detecting the wild-type allele (5′-CCGACTTACTCCATCACTTCAC-3′) or a reverse PGK primer from the NEO cassette for detecting the targeted allele (5′-CTTGTGTAGCGCCAAGTGC-3′). PCR conditions were 35 cycles at 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min. Fluorescence-activated cell sorter (FACS) analysis was performed on thymus and spleen cells as described by Beavis and Pennline (1).
RESULTS AND DISCUSSION
Isolation of mouse Uch-L3.
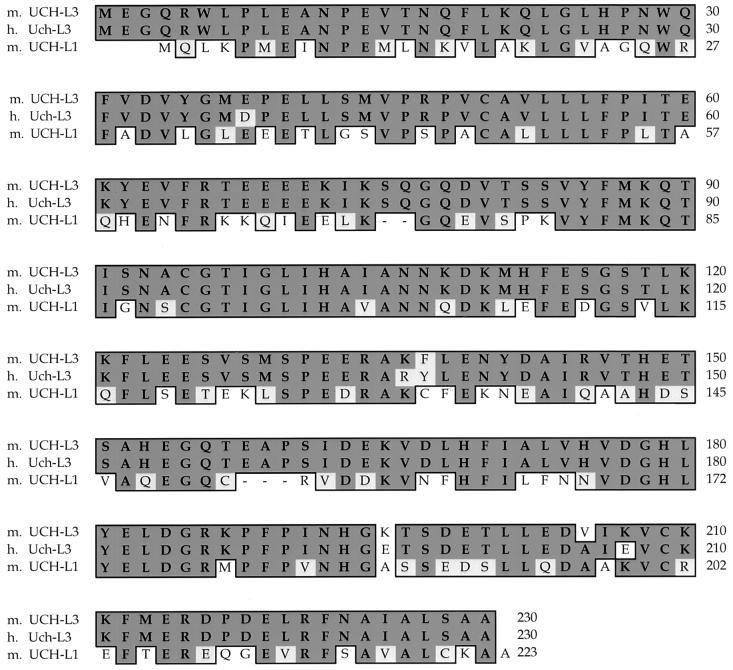
Ednrb maps at 51 cM on mouse chromosome 14, a region that is syntenic with human chromosome 13q22. As the s1Acrg region maps immediately proximal to Ednrb, we searched the National Center for Biotechnology Information human gene map for genes linked to human EDNRB and identified UCH-L3. We then found that the human UCH-L3 cDNA cross-hybridized to the BAC contig over the s1Acrg minimal region (Kurihara et al., unpublished data). To isolate the mouse Uch-L3 gene, the human UCH-L3 probe was used to screen a mouse cDNA library. Sequence analysis of mouse Uch-L3 cDNAs revealed an ∼900-nucleotide transcript with a predicted open reading frame encoding 230 amino acids (Fig. 1). The predicted mouse UCH-L3 protein displays 96% identity to its human orthologue Uch-L3 and 52% identity to its mouse paralogue UCH-L1 (Fig. 2).
FIG. 1.
Uch-L3 gene sequence and structure. Exon (Ex) boundaries are denoted above the nucleotide sequence. The exon 4-exon 5 boundary was ambiguous, as indicated. The start and stop codons and residues deleted in Uch-L3Δ3-7 are underlined; the caret denotes conserved cysteine 95.
FIG. 2.
Alignment of the mouse (m.) UCH-L3 amino acid sequence with human (h.) Uch-L3 and mouse UCH-L1. Identical residues are boxed and darkly shaded, and conserved changes are boxed and lightly shaded. Dashes indicate gaps relative to mUCH-L3.
The UCH family of deubiquitinating enzymes consists of two members, UCH-L1 and UCH-L3. This small conserved family differs from the larger and highly diverse UBP family of deubiquitinating enzymes (for reviews, see references 4 and 31). Although enzymatic activity has been confirmed for members of both families in vitro, the in vivo substrate specificity and function of the majority of these enzymes remain unknown. Recently, a mutation in UCH-L1 was linked to Parkinson's disease in humans (14) and to the gracile axonal dystrophy (gad) mutation in mice (23). Because the loss of Uch-L1 results in the accumulation of protein aggregates, leading to neurodegeneration, the likely in vivo function of Uch-L1 is to stimulate protein degradation in neurons where it is primarily expressed.
Expression of Uch-L3.
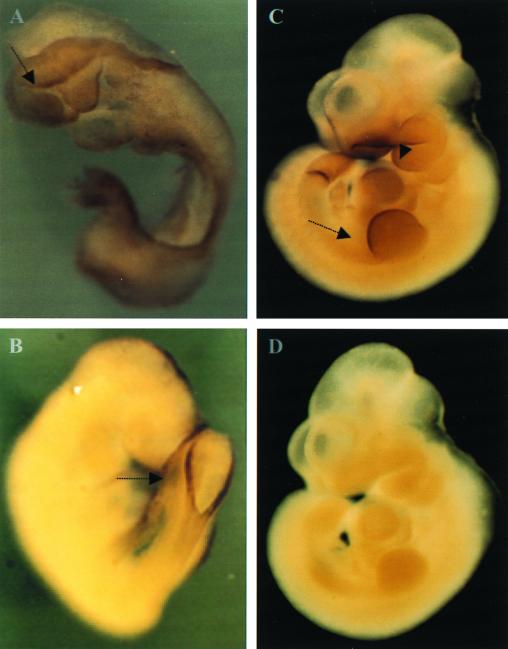
To consider Uch-L3 as a candidate gene for s1Acrg-dependent embryonic lethality, the expression of Uch-L3 at embryonic day 8.5 was verified by RT-PCR (data not shown). In addition, Uch-L3 transcripts were found within structures relevant to the s1Acrg phenotype by whole-mount in situ hybridization (Fig. 3). These include the edges of the open neural folds, which fail to close in s1Acrg mice, and the somites, which are disorganized. Uch-L3 transcripts were also present in structures that form after embryonic day 8.5, including the rim of the posterior neuropore, the apical ectodermal ridge of the limb buds, the branchial arches, the somites, and the tail bud. Combined with its location in s1Acrg, this expression pattern is consistent with a role for Uch-L3 during embryogenesis.
FIG. 3.
Analysis of Uch-L3 transcripts by in situ hybridization. (A) An embryonic day 8.5 (e8.5) embryo is stained at the open edge of the anterior and posterior neural folds (arrow). Staining throughout the embryo was also detected. (B) An e9.5 embryo shows staining at the rim of the posterior neuropore (arrow). (C) An e10.5 embryo is stained at the branchial arches (arrowhead), apical ectodermal ridge (arrow), somites, and tail bud. (D) An e10.5 embryo hybridized with the control sense Uch-L3 probe shows no staining.
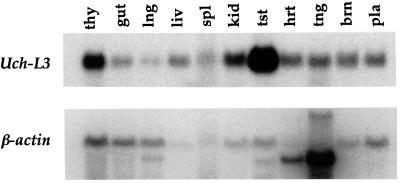
To characterize the expression of Uch-L3 in adult mice, Northern analysis of RNAs isolated from multiple organs was performed (Fig. 4). The Uch-L3 transcript was ∼900 nucleotides long, as predicted by the cDNA sequence. Although Uch-L3 RNA was detected in all tissues analyzed, particularly high levels were present in the testes and to a lesser degree in the thymus. This result suggests that Uch-L3 may also have a role in adult mice, particularly during spermatogenesis or intrathymic T-cell differentiation, both of which are dependent on the ubiquitin pathway.
FIG. 4.
Uch-L3 expression in adult tissues. Total RNAs from the tissues indicated were hybridized to both Uch-L3 and β-actin probes. The order of the lanes, from left to right, is thymus, gut, lung, liver, spleen, kidney, testis, heart, tongue, brain, and placenta. Based on ethidium bromide staining of rRNA bands, relatively equivalent amounts of RNA were loaded in each lane (data not shown).
Generation and analysis of Uch-L3Δ3-7 mice.
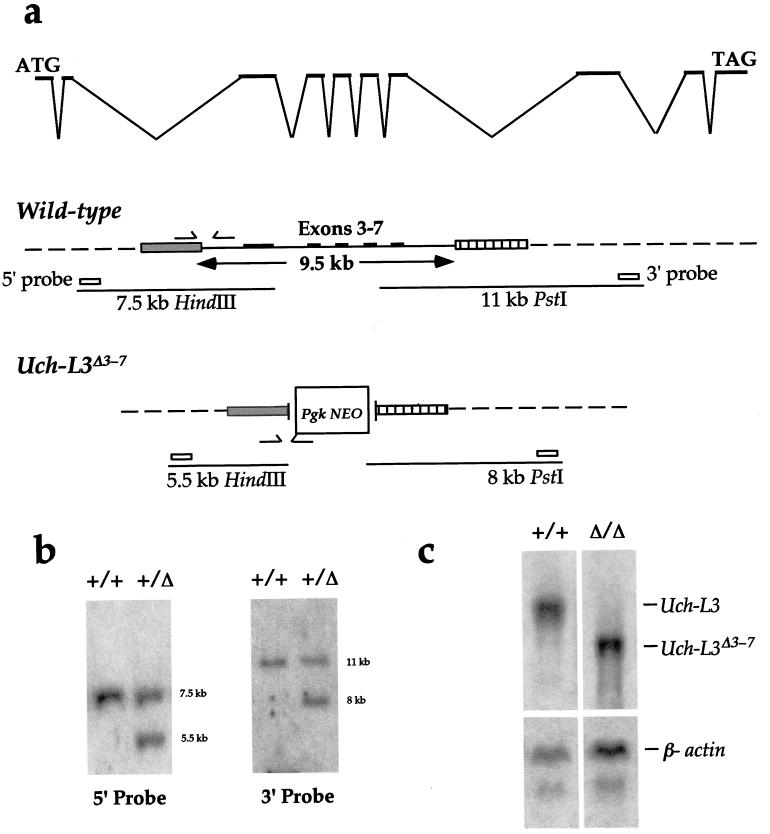
To directly address the role of Uch-L3 during development, mice with a targeted mutation were generated. To design this allele, we first determined the genomic structure of Uch-L3 by alignment of the cDNA sequence with corresponding fragments of the BAC genomic sequence (Fig. 1 and 5a). Restriction mapping of BAC clones was also used to estimate the size of the Uch-L3 locus at 47 to 60 kb. Exons 1 and 2 are ∼100 bp apart; up to 15 kb downstream lie exons 3 to 7, which are clustered within 9.5 kb; and exons 8 to 10 lie at least 15 kb further downstream.
FIG. 5.
Construction of a targeted Uch-L3Δ3-7 allele. (a) The genomic structure of Uch-L3 is indicated in the top line. The wild-type allele depicts the SpeI (gray box) and HindIII (hatched box) fragments used as targeting arms flanking exons 3 to 7. The wild-type allele was detected as a 7.5-kb HindIII fragment with the 5′ probe and as an 11-kb PstI fragment with the 3′ probe. The arrows indicate the primers used to detect the wild-type allele by PCR. The Uch-L3Δ3-7 allele depicts the replacement of exons 3 to 7 with PGK-NEO following targeting. The targeted allele was detected as a 5.5-kb HindIII fragment with the 5′ probe and as an 8-kb PstI fragment with the 3′ probe. The arrows indicate the primers used to detect the targeted allele by PCR. (b) Southern blot hybridization of the 5′ and 3′ probes to wild-type (+/+) and heterozygous (+/Δ) mouse DNAs digested with HindIII (5′) or PstI (3′) to detect the wild-type and targeted restriction fragments. (c) Total RNAs from wild-type (+/+) and Uch-L3Δ3-7 (Δ/Δ) testes were hybridized to Uch-L3 and β-actin probes.
Since only a portion of Uch-L3 could be targeted due to its large size, it was most critical to remove residue 95, the catalytic cysteine that is essential for hydrolase activity in vitro (13). Because the crystal structure of human Uch-L3 predicts a small, single-domain hydrolase (10), it is unlikely that Uch-L3 possesses any other enzymatic activity. Therefore, we created a deletion of clustered exons 3 to 7 which removed up to 90% of the protein, including C95 (Fig. 5a). If exon 2 spliced over PGK-NEO in frame to exon 8, the resulting 90-amino-acid protein would still lack C95 and hydrolase activity.
Because the s1Acrg phenotype is recessive, Uch-L3Δ3-7 heterozygotes were bred to homozygosity. Mice homozygous for Uch-L3Δ3-7 were obtained at weaning at the expected Mendelian frequency. To assess the transcripts from Uch-L3Δ3-7, Northern analysis and RT-PCR were performed. As shown in Fig. 5c, a truncated transcript was present in homozygotes at a level equivalent to that of the wild-type transcript. RT-PCR revealed that the Uch-L3Δ3-7 RNA was composed of two products. One, which included exons 1 and 2 spliced in frame to exons 8 to 10, would be capable of encoding a 90-amino-acid fusion protein. The other, which included exons 1 and 2 spliced out of frame to exons 9 and 10, would encode only the first 18 amino acids of the protein. These results confirm that mice lacking functional Uch-L3 are viable. Furthermore, we generated Uch-L3Δ3-7/s1Acrg compound heterozygotes to determine whether the loss of Uch-L3, together with a haploid copy of s1Acrg, would be deleterious. However, the offspring were viable and fertile. While we cannot rule out the possibility that the loss of Uch-L3 contributes to the s1Acrg phenotype, its loss alone cannot account for embryonic lethality. Thus, the s1Acrg phenotype is either complex and multigenic or due to another gene within the minimal region.
Mice homozygous for Uch-L3Δ3-7 developed to maturity with no obvious abnormalities. Although Uch-L3 is expressed in embryonic structures required for skeletal patterning, no abnormalities were identified in specimens of Uch-L3Δ3-7 neonates that were stained with alcian blue-alizarin red and cleared to view cartilage and bone (data not shown). Particular attention was paid to the axial skeleton, limbs, and craniofacial structures, which are derived from Uch-L3-expressing embryonic tissues. Similarly, although Uch-L3 is expressed in many adult tissues, no histological defects were observed in hematoxylin-eosin-stained sections of mutant kidney, spleen, thymus, lymph node, intestine, liver, lung, adrenal gland, testis, ovary, brain, or heart (data not shown). Because high levels of Uch-L3 RNA were detected in wild-type testes and to a lesser degree in thymus, we determined whether the functions of these organs were affected in Uch-L3Δ3-7 homozygotes.
Within the testes, the ubiquitin pathway is required during spermatogenesis, as shown by the male sterility that is associated with the loss of the mouse HR6B ubiquitin-conjugating enzyme (21). Based on the mutant phenotype, HR6B appears to be required during postmeiotic chromatin condensation, when histones are replaced by transition proteins and protamines. However, fertility and sperm morphology were unaffected in Uch-L3Δ3-7 homozygous mice (data not shown).
Within the thymus, differentiation of CD4− CD8+ T lymphocytes is dependent on the generation of major histocompatibility complex (MHC) I peptide antigens by the ubiquitin pathway (for a review, see reference 20). For example, a mutation of the ubiquitin proteasome component LMP2 leads to a 49% reduction in CD4− CD8+ T lymphocytes within the thymus (29). A mutation of the proteasome component LMP7 leads to a 25 to 45% decrease in MHC I cell surface staining (5), another event that is dependent on MHC I peptide antigens. However, in Uch-L3Δ3-7 mice, no significant reduction in CD4− CD8+ T lymphocytes within the thymus or spleen was observed by FACS analysis with CD4, CD8, and T-cell receptor αβ antisera (Table 1). In addition, MHC I cell surface staining was not significantly reduced in Uch-L3Δ3-7 mice when assayed by FACS analysis with H-2Kb antiserum (Table 1).
TABLE 1.
T-cell differentiation and MHC I expression in Uch-L3 mutant mice
| Cell type | % of CD4− CD8+ cellsa | Surface MHC I staining (arbitrary units)b |
|---|---|---|
| Thymus | ||
| +/+ | 7.98 ± 2.28 | 48.31 ± 2.5 |
| −/− | 8.89 ± 3.19 | 46.09 ± 1.15 |
| Spleen | ||
| +/+ | 7.85 ± 1.20 | 47.45 ± 2.7 |
| −/− | 6.50 ± 1.43 | 49.71 ± 4.26 |
Compared to the wild-type results, the percentage of CD4− CD8+ cells was increased by 10% (P, 0.30) in mutant thymus and reduced by 17% (P, 0.07) in mutant spleen (n = 5).
Relative surface MHC I staining (arbitrary units) was decreased by 5% (P, 0.03) in mutant thymus and increased by 5% (P, 0.17) in mutant spleen (n = 5).
The absence of either an embryonic or an adult phenotype in Uch-L3Δ3-7 mice implies either that Uch-L3 performs an undetected nonessential function or that Uch-L3 is functionally redundant with Uch-L1. Uch-L3 and Uch-L1 display 52% identity, and their expression patterns overlap in several tissues, including the brain and testes, where Uch-L3 is present at high levels. Loss of Uch-L1 leads to distinct neurological defects, but it is possible that the simultaneous loss of both Uch-L1 and Uch-L3 would exacerbate these defects and result in additional defects in other organs. Where overlapping expression patterns have not been demonstrated, such as during embryogenesis, Uch-L3 and Uch-L1 function would be either dispensable or possibly redundant with that of members of the UBP family of deubiquitinating enzymes, which do not share sequence conservation with the UCH family. However, given the degree of sequence divergence between UCH and UBP deubiquitinating enzymes, it is expected that they possess distinct substrate specificities. Experiments are under way to test these premises.
ACKNOWLEDGMENTS
We are grateful to Robert S. Ingram for sequencing of cDNA clones and to Audrey Goddard at Genentech, Inc., for genomic DNA sequencing. We also thank Se-Ho Park, Albert Bendelac, and Andrew Beavis for antisera and FACS analysis. The histopathologic analysis of mutant mice was performed at the University of California Davis Histo-Pathology Laboratory.
L.J.K. was supported by an NRSA award from the National Institutes of Health. S.M.T. is an investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Beavis A J, Pennline K J. Simultaneous measurement of five cell surface antigens by five-colour immunofluorescence. Cytometry. 1994;15:371–376. doi: 10.1002/cyto.990150413. [DOI] [PubMed] [Google Scholar]
- 2.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Copp A J. Death before birth: clues from gene knockouts and mutations. Trends Genet. 1995;11:87–93. doi: 10.1016/S0168-9525(00)89008-3. [DOI] [PubMed] [Google Scholar]
- 4.D'Andrea A, Pellman D. Deubiquitinating enzymes: a new class of biological regulators. Crit Rev Biochem Mol Biol. 1998;33:337–352. doi: 10.1080/10409239891204251. [DOI] [PubMed] [Google Scholar]
- 5.Fehling H J, Swat W, Laplace C, Kuhn R, Rajewsky K, Muller U, von Boehmer H. MHC class I expression in mice lacking the proteasome subunit LMP-7. Science. 1994;265:1234–1237. doi: 10.1126/science.8066463. [DOI] [PubMed] [Google Scholar]
- 6.Gardiner K. Base composition and gene distribution: critical patterns in mammalian genome organization. Trends Genet. 1996;12:519–524. doi: 10.1016/s0168-9525(97)81400-x. [DOI] [PubMed] [Google Scholar]
- 7.Harbers K, Muller U, Grams A, Li E, Jaenisch R, Franz T. Provirus integration into a gene encoding a ubiquitin-conjugating enzyme results in a placental defect and embryonic lethality. Proc Natl Acad Sci USA. 1996;93:12412–12417. doi: 10.1073/pnas.93.22.12412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hosoda K, Hammer R E, Richardson J A, Baynash A G, Cheung J C, Giaid A, Yanagisawa M. Targeted and natural (piebald-lethal) mutations of endothelin-B receptor gene produce megacolon associated with spotted coat color in mice. Cell. 1994;79:1267–1276. doi: 10.1016/0092-8674(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 9.Huang Y, Baker R T, Fischer-Vize J A. Control of cell fate by a deubiquitinating enzyme encoded by the fat facets gene. Science. 1995;270:1828–1831. doi: 10.1126/science.270.5243.1828. [DOI] [PubMed] [Google Scholar]
- 10.Johnston S C, Larsen C N, Cook W J, Wilkinson K D, Hill C P. Crystal structure of a deubiquitinating enzyme (human UCH-L3) at 1.8 A resolution. EMBO J. 1997;16:3787–3796. doi: 10.1093/emboj/16.13.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kingsley D M, Bland A E, Grubber J M, Marker P C, Russell L B, Copeland N G, Jenkins N A. The mouse short ear skeletal morphogenesis locus is associated with defects in a bone morphogenetic member of the TGF beta superfamily. Cell. 1992;71:399–410. doi: 10.1016/0092-8674(92)90510-j. [DOI] [PubMed] [Google Scholar]
- 12.Lane P W. Association of megacolon with two recessive spotting genes in the mouse. J Hered. 1966;57:29–31. doi: 10.1093/oxfordjournals.jhered.a107457. [DOI] [PubMed] [Google Scholar]
- 13.Larsen C N, Price J S, Wilkinson K D. Substrate binding and catalysis by ubiquitin C-terminal hydrolases: identification of two active site residues. Biochemistry. 1996;35:6735–6744. doi: 10.1021/bi960099f. [DOI] [PubMed] [Google Scholar]
- 14.Leroy E, Boyer R, Auburger G, Leube B, Ulm G, Mezey E, Harta G, Brownstein M J, Jonnalagada S, Chernova T, Dehejia A, Lavedan C, Gasser T, Steinbach P J, Wilkinson K D, Polymeropoulos M H. The ubiquitin pathway in Parkinson's disease. Nature. 1998;395:451–452. doi: 10.1038/26652. [DOI] [PubMed] [Google Scholar]
- 15.Mayer T C. The development of piebald spotting in mice. Dev Biol. 1965;11:319–334. doi: 10.1016/0012-1606(65)90042-4. [DOI] [PubMed] [Google Scholar]
- 16.Metallinos D L, Oppenheimer A J, Rinchik E M, Russell L B, Dietrich W, Tilghman S M. Fine structure mapping and deletion analysis of the murine piebald locus. Genetics. 1994;136:217–223. doi: 10.1093/genetics/136.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Brien T P, Metallinos D L, Chen H, Shin M K, Tilghman S M. Complementation mapping of skeletal and central nervous system abnormalities in mice of the piebald deletion complex. Genetics. 1996;143:447–461. doi: 10.1093/genetics/143.1.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peters J-M, Harris J R, Finley D. Ubiquitin and the biology of the cell. New York, N.Y: Plenum Press; 1998. [Google Scholar]
- 19.Rinchick E M, Russell L B. Germ-line deletion mutations in the mouse: tools for intensive functional and physical mapping of regions of the mammalian genome. In: Davies K E, Tilghman S M, editors. Genome analysis. 1. Genetic and physical mapping. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. pp. 121–158. [Google Scholar]
- 20.Rock K L, Goldberg A L. Degradation of cell proteins and the generation of MHC class I presented peptides. Annu Rev Immunol. 1999;17:739–779. doi: 10.1146/annurev.immunol.17.1.739. [DOI] [PubMed] [Google Scholar]
- 21.Roest H P, van Klaveren J, de Wit J, van Gurp C G, Koken M H, Vermey M, van Roijen J H, Hoogerbrugge J W, Vreeburg J T, Baarends W M, Bootsma D, Grootegoed J A, Hoeijmakers J H. Inactivation of the HR6B ubiquitin-conjugating DNA repair enzyme in mice causes male sterility associated with chromatin modification. Cell. 1996;86:799–810. doi: 10.1016/s0092-8674(00)80154-3. [DOI] [PubMed] [Google Scholar]
- 22.Russell W L. X-ray induced mutations in mice. Cold Spring Harbor Symp Quant Biol. 1951;16:327–336. doi: 10.1101/sqb.1951.016.01.024. [DOI] [PubMed] [Google Scholar]
- 23.Saigoh K, Wang Y L, Suh J G, Yamanishi T, Sakai Y, Kiyosawa H, Harada T, Ichihara N, Wakana S, Kikuchi T, Wada K. Intragenic deletion in the gene encoding ubiquitin carboxy-terminal hydrolase in gad mice. Nat Genet. 1999;23:47–51. doi: 10.1038/12647. [DOI] [PubMed] [Google Scholar]
- 24.Schuler G D, Boguski M S, Stewart E A, Stein L D, Gyapay G, Rice K, White R E, Rodriguez-Tome P, Aggarwal A, Bajorek E, Bentolila S, Birren B B, Butler A, Castle A B, Chiannilkulchai N, Chu A, Clee C, Cowles S, Day P J, Dibling T, Drouot N, Dunham I, Duprat S, East C, Hudson T J, et al. A gene map of the human genome. Science. 1996;274:540–546. [PubMed] [Google Scholar]
- 25.Shalaby F, Rossant J, Yamaguchi T P, Gertsenstein M, Wu X F, Breitman M L, Schuh A C. Failure of blood-island formation and vasculogenesis in Flk-1-deficient mice. Nature. 1995;376:62–66. doi: 10.1038/376062a0. [DOI] [PubMed] [Google Scholar]
- 26.Shin M K, Levorse J M, Ingram R S, Tilghman S M. The temporal requirement for endothelin receptor-B signalling during neural crest development. Nature. 1999;402:496–501. doi: 10.1038/990040. [DOI] [PubMed] [Google Scholar]
- 27.Shumacher A, Faust C, Magnuson T. Positional cloning of a global regulator of anterior-posterior patterning in mice. Nature. 1996;383:250–253. doi: 10.1038/383250a0. [DOI] [PubMed] [Google Scholar]
- 28.Swiatek P J, Gridley T. Perinatal lethality and defects in hindbrain development in mice homozygous for a targeted mutation of the zinc finger gene Krox20. Genes Dev. 1993;7:2071–2084. doi: 10.1101/gad.7.11.2071. [DOI] [PubMed] [Google Scholar]
- 29.Van Kaer L, Ashton-Rickardt P G, Eichelberger M, Gaczynska M, Nagashima K, Rock K L, Goldberg A L, Doherty P C, Tonegawa S. Altered peptidase and viral-specific T cell response in LMP2 mutant mice. Immunity. 1994;1:533–541. doi: 10.1016/1074-7613(94)90043-4. [DOI] [PubMed] [Google Scholar]
- 30.Wilkinson D G, Nieto M A. Detection of messenger RNA by in situ hybridization to tissue sections and whole mounts. Methods Enzymol. 1993;225:361–373. doi: 10.1016/0076-6879(93)25025-w. [DOI] [PubMed] [Google Scholar]
- 31.Wilkinson K D. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J. 1997;11:1245–1256. doi: 10.1096/fasebj.11.14.9409543. [DOI] [PubMed] [Google Scholar]
- 32.Zhen M, Schein J E, Baillie D L, Candido E P. An essential ubiquitin-conjugating enzyme with tissue and developmental specificity in the nematode Caenorhabditis elegans. EMBO J. 1996;15:3229–3237. [PMC free article] [PubMed] [Google Scholar]