Significance
Single amino acid changes causative of neurologic disease often map to the cross-β forming regions of low-complexity (LC) domains. All such mutations studied to date lead to enhanced avidity of cross-β interactions. The LC domain of the fused in sarcoma (FUS) RNA binding protein contains three different regions that are capable of forming labile cross-β interactions. Here we describe the perplexing effect of amyotrophic lateral sclerosis (ALS)-causing mutations localized to the LC domain of FUS to substantially weaken its ability to form one of its three cross-β interactions. An understanding of how these mutations abet uncontrolled polymerization of the FUS LC domain may represent an important clue as to how LC domains achieve their proper biological function.
Keywords: FUS, low-complexity sequence, cross-beta polymer, ALS mutation, neurodegenerative disease
Abstract
The low-complexity (LC) domain of the fused in sarcoma (FUS) RNA binding protein self-associates in a manner causing phase separation from an aqueous environment. Incubation of the FUS LC domain under physiologically normal conditions of salt and pH leads to rapid formation of liquid-like droplets that mature into a gel-like state. Both examples of phase separation have enabled reductionist biochemical assays allowing discovery of an N-terminal region of 57 residues that assembles into a labile, cross-β structure. Here we provide evidence of a nonoverlapping, C-terminal region of the FUS LC domain that also forms specific cross-β interactions. We propose that biologic function of the FUS LC domain may operate via the mutually exclusive use of these N- and C-terminal cross-β cores. Neurodegenerative disease–causing mutations in the FUS LC domain are shown to imbalance the two cross-β cores, offering an unanticipated concept of LC domain function and dysfunction.
Upwards of 20% of the proteomes of eukaryotic cells consist of polypeptide sequences that are of low complexity (LC) (1, 2). These atypical polypeptide domains are composed of only a subset of the 20 amino acids normally required for proteins to assume their distinct, three-dimensional shapes. As studied in the monomeric state in biochemical preparations outside of cells, LC sequences remain unfolded. As such, they have been described as intrinsically disordered regions (IDRs) of the proteome.
Unbiased proteomic studies employing thermal perturbation as a probe for structural order within cells have shown that a surprisingly large fraction of protein domains thought to be intrinsically disordered may instead function in states of labile, structural order (3). Numerous reports published over the past several years have also given evidence that LC domains can undergo phase separation upon purification and incubation in test tube assays (4, 5). Purified LC domains can self-associate in a manner causing them to partition out of aqueous solution by forming liquid-like droplets. Following incubation, these droplets solidify into a gel-like state. If the interactions driving phase separation of LC domains properly reflect their biological function, this line of research may help reveal how these unusual protein domains actually work in living cells.
Studies of the fused in sarcoma (FUS) RNA binding protein offered an early example of the unusual behavior of phase separation by an LC domain (6, 7). Incubation of the purified FUS LC domain initially triggers the formation of an opalescent suspension composed of liquid-like droplets. Sustained incubation of the same preparation leads to a more stable, gel-like state. Electron microscopy and X-ray diffraction analysis revealed FUS hydrogels to be composed of uniform, amyloid-like polymers (7). Unlike pathogenic, prion-like amyloids, FUS polymers are labile to disassembly upon dilution. A molecular structure of FUS polymers has been resolved by the use of solid-state NMR (ssNMR) spectroscopy (8). Among the 214 residues constituting the FUS LC domain, polymers were found to form via the organization of in-register, cross-β interactions localized between residues 39 and 95.
Structural studies of FUS polymers revealed two differences from pathogenic amyloids, such as α-synuclein or polymers formed from the Aβ fragment long understood to form hyperstable aggregates in Alzheimer’s disease patients. First, FUS polymers reproducibly form the same, monomorphic structure via an N-terminally localized cross-β core. By contrast, pathogenic fibrils formed from α-synuclein or Aβ can adopt any of a number of different, inordinately stable structures (9, 10). Second, the subunit interface holding α-synuclein or Aβ polymers together are replete with hydrophobic amino acids believed to contribute to extreme polymer stability. The molecular structure of FUS polymers revealed but a single hydrophobic amino acid, proline residue 72, within the 57 residues constituting the subunit interface (8). The paucity of hydrophobic residues at the subunit interface of FUS polymers may, at least in part, explain polymer lability.
Studies of the LC domain of the hnRNPA2 protein have yielded similar findings. The LC domain of hnRNPA2 becomes phase separated into liquid-like droplets that, with time, also mature into a gel-like state (11, 12). Cross-β interactions formed by a region 40 to 50 residues in length also define hnRNPA2 LC domain polymers (11, 13, 14). Human genetic studies of patients suffering from various forms of neurological disease have identified mutations in the genes encoding hnRNPA1, hnRNPA2, and hnRNPDL (15, 16). These recurrent, disease-causing mutations commonly alter a conserved aspartic acid residue located within the cross-β core known to hold hnRNPA2 LC domain polymers together (11, 13, 17).
A simplistic interpretation of these observations offers that evolution has favored the presence of an aspartic acid residue within the cross-β cores formed from hnRNP LC domains as a means of tuning the balance of polymer stability/lability. Proximal disposition of this aspartic acid residue, as dictated by the in-register organization of the polymer interface, has been hypothesized to impart instability resulting from repulsive charge:charge interactions (13). Mutations removing these destabilizing interactions, especially when replacing the conserved aspartic acid residue with valine, may enhance polymer stability and lead to disease pathophysiology.
The relatively straightforward understanding of disease-causing mutations in the LC domains of three different hnRNP proteins does not translate to human genetic studies of FUS. Two of the most prominent amyotrophic lateral sclerosis (ALS)-causing mutations within the FUS LC domain include a missense mutation, changing glycine residue 156 to glutamic acid, and the deletion of glycine residues 174 and 175 (18–20). Neither of these mutations is anywhere close to the cross-β core of FUS that is located between residues 39 to 95.
The inability of our current understanding of the FUS cross-β core to explain substantive human genetic studies prompted reexamination of the pathway by which soluble monomers are captured by hydrogel samples composed of the intact LC domain of FUS. As will be described, these studies reveal surprising and unanticipated observations. The baroque pathway of subunit recruitment into existing FUS polymers offers an unconventional perspective as to how disease-causing mutations in the FUS LC domain manifest their pathophysiology at a molecular level. These studies further offer a means of understanding how the FUS LC domain is prevented from runaway polymerization via either of its cross-β cores.
Results
Identification of a Region of the FUS LC Domain Critical for Hydrogel Binding.
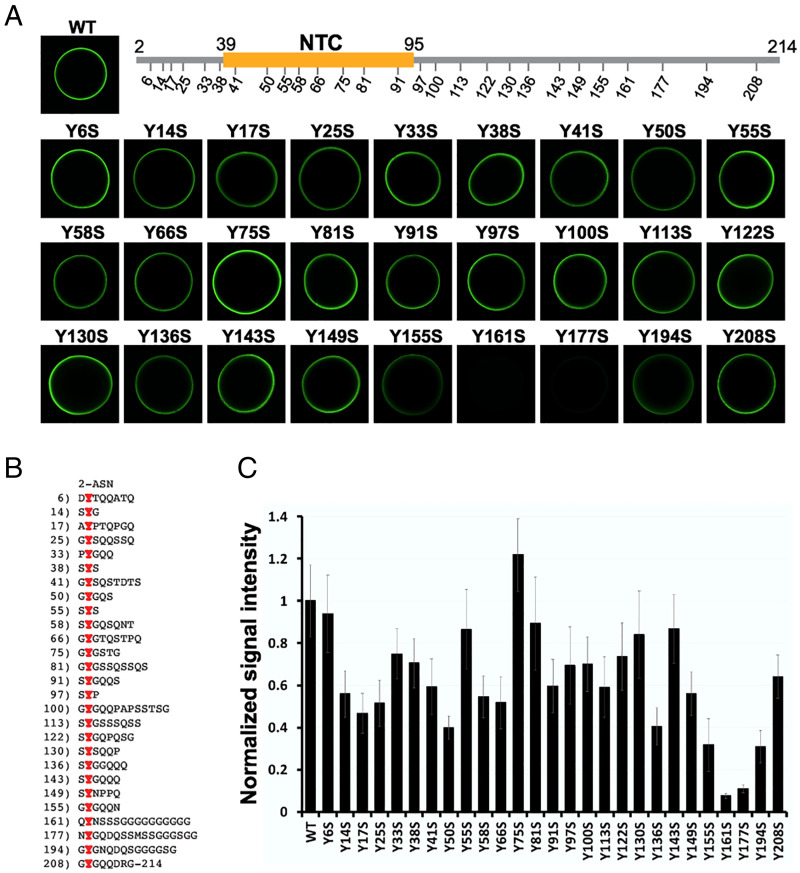
The sequence of the FUS LC domain is quasirepetitive, containing 27 repeats of the tripeptide sequence G/S-Y-G/S (Fig. 1B). Early mutational studies gave evidence that the tyrosine residues of these repeats are correlatively important for the binding of FUS to hydrogel preparations formed from the LC domain of FUS itself, as well as its recruitment to RNA granules in living cells (7). In these experiments, hydrogels were prepared from a fusion protein linking mCherry to the native LC domain of FUS, and test proteins were GFP fusions to either the native LC domain of FUS or mutated variants thereof. We have repeated these same experiments using 27 variants of the FUS LC domain bearing single tyrosine-to-serine mutations. Little or small effect on hydrogel binding was observed for 24 of the 27 variants. Three variants, wherein tyrosine residues 155, 161, or 177 were individually changed to serine, bound to mCherry:FUS hydrogels considerably less well than the native protein (Fig. 1 A and C). It is notable that these mutation-sensitive tyrosine residues localize far distal to the cross-β core housed between residues 39 and 95.
Fig. 1.
Identification of a C-terminal region of the FUS LC domain required for hydrogel binding. (A) Twenty-seven individual tyrosine-to-serine mutations were made across the FUS low-complexity domain. The yellow region of the schematic diagram of the FUS LC domain corresponds to the location of the NTC determined by ssNMR (8). Native protein (wild type [WT]) and individual point mutants were linked to GFP, expressed in bacteria, purified, and tested in binding assays using hydrogel droplets formed from mCherry linked to the intact LC domain of FUS. Hydrogel binding activity was reduced most significantly by the Y161S and Y177S mutants. (B) Amino acid sequence of the FUS LC domain with tyrosine residues shown in red. Residue numbers for the individual tyrosines are shown at Left. (C) Quantitation of hydrogel binding assays shown in A. Intensities are an average of three measurements.
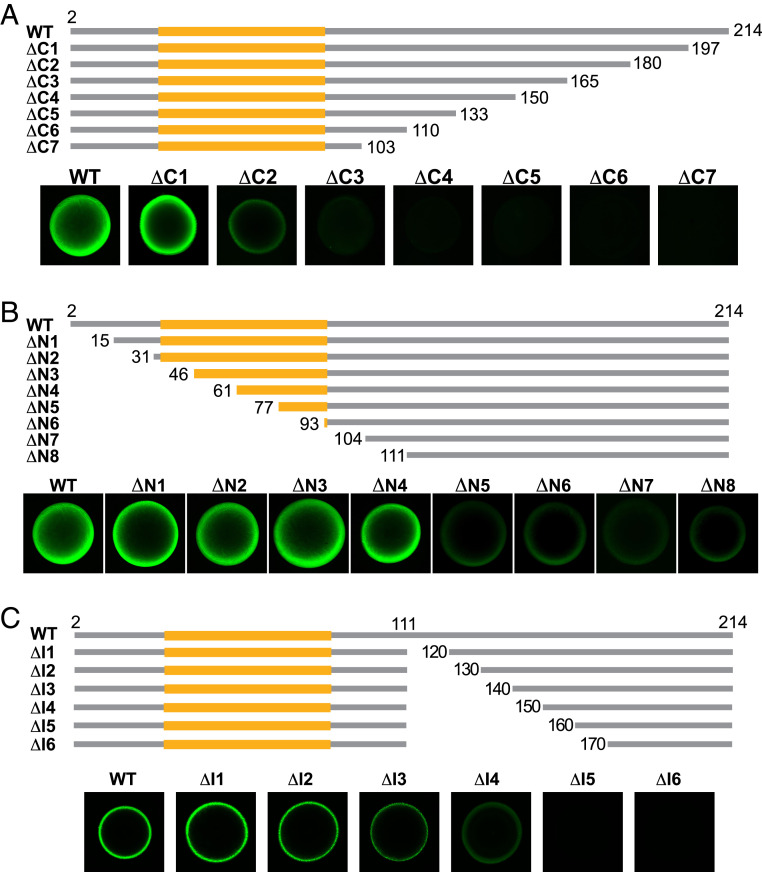
Three sets of deletion mutants were prepared to further investigate regions of the FUS LC domain important for hydrogel binding. One set of deletions systematically truncated the LC domain from its C terminus. As shown in Fig. 2A, removal of 17 residues yielded a protein (ΔC1) that retained strong hydrogel binding activity. The next variant missing 34 residues (ΔC2) retained residual binding, yet all remaining deletion mutants were unable to bind hydrogels formed by the intact LC domain of FUS (Fig. 2A). From these studies we define a boundary around residue 190 that marks the C-terminal location of a region required for monomeric GFP test proteins to bind mCherry hydrogels formed by the intact FUS LC domain.
Fig. 2.
Hydrogel binding analysis of three sets of deletion variants of the FUS LC domain. (A) Seven deletion mutants progressing from the C terminus of the FUS LC domain were linked to GFP, expressed in bacteria, purified, and tested in hydrogel binding assays as described in Fig. 1. One deletion mutant (ΔC1) displayed robust hydrogel binding activity, another (ΔC2) retained residual binding activity, and all other deletion mutants lost hydrogel binding activity. (B) Eight deletion mutants progressing from the N terminus of the FUS LC domain were linked to GFP, expressed in bacteria, purified, and assayed for hydrogel binding. Four deletion mutants (ΔN1–ΔN4) displayed robust hydrogel binding activity, and four deletion mutants (ΔN5–ΔN8) retained attenuated hydrogel binding activity. (C) Six internal deletion mutants progressing in 10 amino acid increments from residue 111 were linked to GFP, expressed in bacteria, purified, and assayed for hydrogel binding. Four internal deletion mutants (ΔI1–ΔI4) displayed hydrogel binding activity and two did not (ΔI5 and ΔI6). Yellow regions in schematic diagrams designate the location of the NTC of the FUS LC domain as defined by its atomic structure spanning residues 39 to 95 (8).
Analysis of N-terminal truncations of the FUS LC domain yielded the surprising observation that upwards of half of the region specifying the cross-β core, spanning residues 39 to 95, could be deleted without significantly affecting hydrogel binding (Fig. 2B). The ΔN4 variant displayed strong hydrogel binding despite lacking 22 residues of the cross-β core. It was likewise surprising that four deletion mutants missing even greater amounts of the FUS LC domain, two of which eliminated the entire cross-β core (ΔN7 and ΔN8), retained attenuated but readily detectible hydrogel binding activity. In combination, these systematic mutagenesis experiments give evidence that a region of the FUS LC domain located far distal to the cross-β core is required for an unstructured test protein to bind hydrogel preparations of the FUS LC domain.
In a third set of deletion mutations, we initiated truncation downstream of the N-terminal cross-β core starting at residue 111. Deletions were extended from this point and enlarged at 10 amino acid increments, internally removing as few as 9 residues and extending up to as much as 59 residues. Each variant was expressed as a GFP fusion, purified and tested for binding to mCherry hydrogel samples formed from the intact FUS LC domain. The internal deletion missing 29 amino acids (ΔI3), and all variants missing fewer residues of the FUS LC domain, exhibited hydrogel binding. By contrast, the variant missing 39 internal residues (ΔI4) exhibited reduced hydrogel binding, and those missing 49 or 59 residues (ΔI5 or ΔI6) revealed no binding (Fig. 2C). In combination with studies of C-terminal truncations (Fig. 2A), analysis of these internal deletion mutants define a region of roughly 40 amino acids, located between residues 150 and 190 of the FUS LC domain, required for monomeric test protein to bind mCherry:FUS hydrogels.
Interpretation of these experiments assumes that the mCherry:FUS hydrogel droplets are composed of polymers assembled via the same cross-β core whose structure was resolved at the atomic level by solid-state NMR spectroscopy (8). Such studies were performed on the isolated LC domain of FUS not appended to mCherry or GFP. It was formally possible that polymers formed from the mCherry:FUS fusion protein used in this study might be different from polymers formed from the isolated FUS LC domain. To test this possibility we employed intein chemistry to ligate unlabeled GFP to a uniformly, 13C,15N-labeled form of the FUS LC domain. The latter protein was allowed to polymerize and be evaluated by solid-state NMR spectroscopy. As shown in SI Appendix, Fig. S1, polymers made from the segmentally labeled GFP:FUS chimeric protein yielded NMR spectra indistinguishable from polymers prepared from the isolated, uniformly labeled LC domain of FUS.
Identification of a Cross-β Core within the C-Terminal Half of the FUS LC Domain.
The ΔN8 deletion variant contains residues 111 to 214 of the FUS LC domain. From here forward we designate this as the C-terminal half of the FUS LC domain. This protein fragment of 104 residues was expressed in bacterial cells, purified, and incubated under conditions receptive to phase transition (SI Appendix, Materials and Methods). Upon incubation at neutral pH and physiological concentration of monovalent salt (SI Appendix, Materials and Methods), this C-terminal half of the FUS LC domain became phase separated into a gel-like state. When observed by transmission electron microscopy, the hydrogel was found to be composed of uniform, unbranched polymers (SI Appendix, Fig. S2A). X-ray diffraction analysis of hydrogels formed from the C-terminal half of the FUS LC domain revealed diffraction rings at 4.7 and 10 Å (SI Appendix, Fig. S2B). Finally, when analyzed by semidenaturing agarose gel electrophoresis (SDD-AGE), the observed polymers were labile to disassembly (SI Appendix, Fig. S2C).
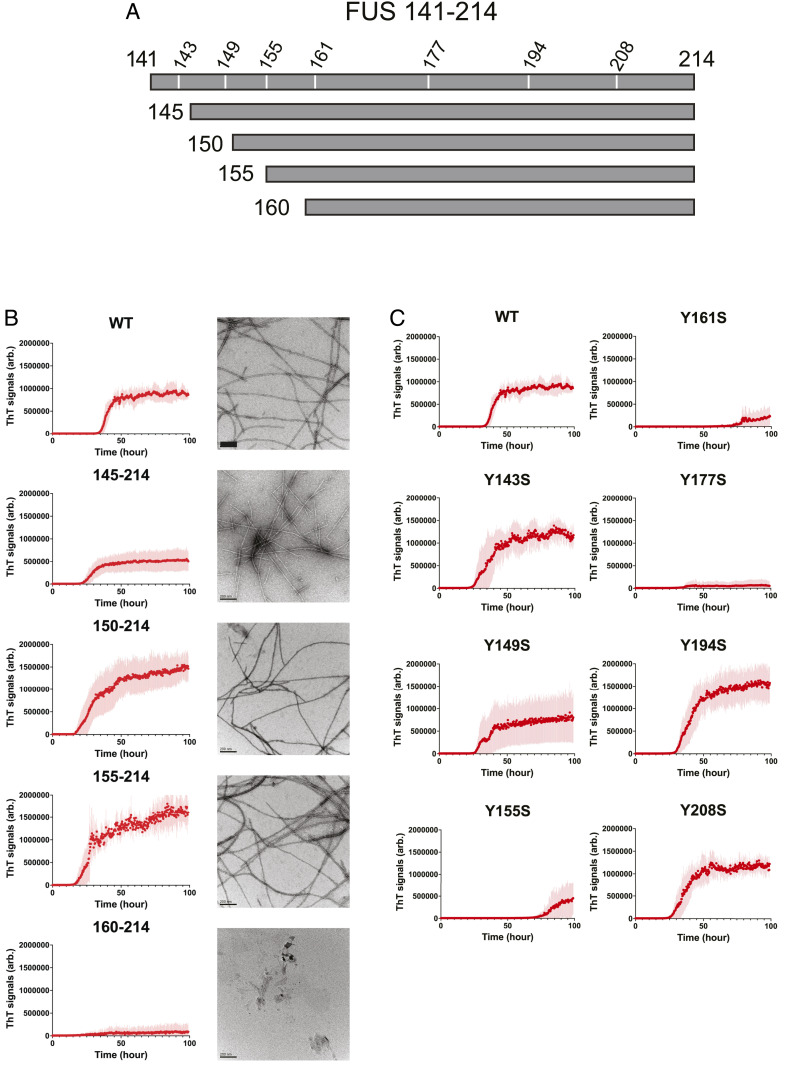
Truncations of the C-terminal half of the FUS LC domain were prepared having N termini at residues 141, 145, 150, 155, and 160, together with a common C terminus at residue 214 (Fig. 3A). Following incubation under conditions receptive to phase transition, each variant was evaluated for its capacity to polymerize both by time-dependent acquisition of thioflavin-T fluorescence and transmission electron microscopy (Fig. 3B). All variants were observed to form homogenous polymers, save for the most truncated fragment bearing an N terminus at residue 160. We thus conclude that the region of the FUS LC domain located between residues 155 and 190 specifies a secondary cross-β core distinct from that of the N-terminal half of the FUS LC domain characterized extensively in previous studies (8).
Fig. 3.
Polymerization capacity of a fragment of the FUS LC domain spanning residues 141 to 214. (A) Schematic diagram of a C-terminal region of the FUS LC domain having the capacity to form labile cross-β polymers. Locations of seven tyrosine residues are designated numerically (143, 149, 155, 161, 177, 194, and 208). Truncations incrementally removing 4, 10, 15, or 20 residues from the N terminus are shown below the parental 141 to 214 fragment of the FUS LC domain. (B) Parental and N-terminal truncations were incubated under conditions receptive to polymerization (SI Appendix, Materials and Methods). Assays for time-dependent acquisition of thioflavin-T fluorescence (Left) and electron microscopy (Right) were used to monitor formation of amyloid-like polymers. (Scale bar, 200 μm.) (C) Acquisition of thioflavin-T fluorescence at a function of time was compared for fragments of the FUS LC domain spanning residues 141 to 214 bearing either the native sequence or that of variants carrying a single tyrosine-to-serine mutation. Each graph displays fluorescence increase (y axis) relative to time of incubation (x axis). Four mutants, including Y143S, Y149S, Y194S, and Y208S, revealed evidence of polymerization similar to the wild-type protein. Three mutants, including Y155S, Y161S, and Y177S, revealed substantially impeded capacity for polymerization. arb.: arbitrary unit.
The laboratory of R. Tycko has recently described the structure of cross-β polymers formed from the C-terminal half of the FUS LC domain (21). The structural core of the Tycko polymers, whose atomic fold was resolved by cryoelectron microscopy, is specified by residues 112 to 150 of the FUS LC domain. This region of 39 amino acids is not required for soluble test protein to bind to mCherry hydrogels formed from the intact LC domain of FUS (Fig. 2C). It is likewise clear that the cross-β core described by R. Tycko and colleagues is entirely distinct from the C-terminal cross-β core described herein. Truncated variants of the C-terminal half of the FUS LC domain completely lacking residues 111 to 150 are capable of polymerization (Fig. 3B).
To compare cross-β polymers corresponding to those described by the R. Tycko laboratory (21) with those described in this study, fragments of the FUS LC domain spanning either residues 111 to 160 or 141 to 214 were purified and allowed to polymerize under conditions receptive to phase separation. Both fragments readily formed cross-β polymers that were evaluated by electron microscopy, X-ray diffraction, semidenaturing agarose gel electrophoresis, and solid-state NMR spectroscopy. Both samples yielded homogeneous, unbranched polymers as viewed by electron microscopy (SI Appendix, Fig. S3A). Both samples revealed X-ray diffraction images consistent with cross-β amyloid-like polymers (SI Appendix, Fig. S3B). Both samples of polymers were labile to disassembly when subjected to SDD-AGE (SI Appendix, Fig. S3C). Finally, the two samples revealed different spectra as evaluated by solid-state NMR spectroscopy (SI Appendix, Fig. S3D). For reasons we do not yet understand, the region specifying the cross-β core characterized by R. Tycko and colleagues (residue 112 to 150) is not required for hydrogel binding by the intact FUS LC domain (Figs. 1 and 2). By contrast, the region of the FUS LC domain specifying the C-terminal cross-β core described in this report (155 to 190) is vitally required for hydrogel binding.
From here forward we will refer to the region spanning residues 39 to 95 of the FUS LC domain as specifying the boundaries of the predominating, N-terminal cross-β core (NTC) of the FUS LC domain (8). We will further term the region spanning residues 155 to 190 as specifying the boundaries of the secondary, C-terminal cross-β core (CTC) of the FUS LC domain that has been characterized in the present study.
In order to investigate the possible relationship between this secondary, C-terminal cross-β core and the capacity of soluble FUS LC domain monomers to bind mCherry hydrogels formed from the intact LC domain of FUS (Figs. 1 and 2), we investigated the effects of single tyrosine-to-serine (Y-to-S) mutations upon polymerization of the fragment of the FUS LC domain spanning residues 141 to 214 (Fig. 3A). As shown in Fig. 3C, four of the single Y-to-S mutants (Y143S, Y149S, Y194S, and Y208S) polymerized in a manner indistinguishable from the wild-type fragment, whereas three of the single Y-to-S mutants (Y155S, Y161S, and Y177S) were severely compromised in polymerization, exactly corresponding to the inability of the same mutants in the intact LC domain to incorporate into hydrogels (see Fig. 1C). The exactly correspondent pattern of polymerization by the C-terminal 141 to 214 segment with hydrogel capture of the intact LC domain thus indicates that the C-terminal region of the LC domain alone can recapture aspects of cross-β behavior in the intact LC domain.
Evidence of Molecular Specificity in Elongation of Both NTC and CTC Cross-β Polymers.
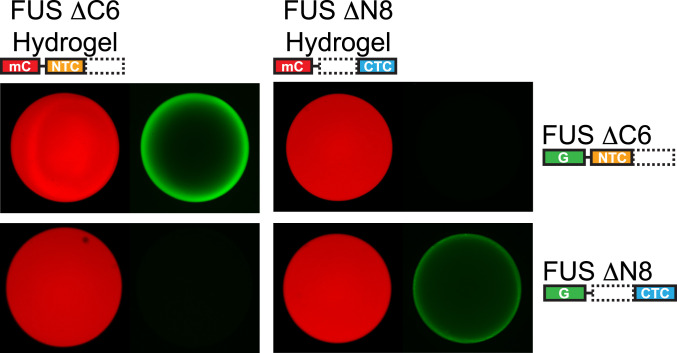
In efforts to complete a more thorough analysis of the two regions allowing for self-association of the FUS LC domain, we prepared hydrogels composed of either the NTC (residues 2 to 110) or the CTC (residues 111 to 214) and tested their respective abilities to bind two GFP-tagged test proteins. Both hydrogels were made from mCherry fusion proteins. The two GFP test proteins used to interrogate the two hydrogel samples included one composed of the N-terminal half of the LC domain (residues 2 to 110) or a second composed of the C-terminal half of the LC domain (residues 141 to 214). The latter protein was purposely truncated at its N terminus to remove the region shown recently by R. Tycko and colleagues to be capable of forming cross-β polymers (21).
As shown in Fig. 4, the GFP fusion protein linked to the N-terminal half of the LC domain (residues 2 to 110) bound only the hydrogel made from the NTC itself. The GFP fusion protein linked to the CTC (residues 141 to 214) bound only the hydrogel made from the C-terminal half of the LC domain. Having observed that the GFP fusion protein containing the NTC does not bind to mCherry:CTC hydrogels, and that the GFP fusion protein containing the CTC does not bind to mCherry:NTC hydrogels, we conclude that these binding reactions offer evidence of molecular specificity.
Fig. 4.
Specificity of binding of N- and C-terminal halves of the FUS low-complexity domain to hydrogel polymers formed from the same regions. mCherry fusion proteins linked to either the N- terminal half (residues 2 to 110) or C-terminal half (residues 111 to 214) of the FUS LC domain were challenged with soluble GFP fusion proteins corresponding to the same two halves of the protein. mCherry hydrogels composed of the N-terminal half of the FUS LC domain bound the GFP fusion protein linked to the same N-terminal half of the protein, but not the C-terminal half (Left). mCherry hydrogels composed of the C-terminal half of the FUS LC domain bound the GFP fusion protein linked to the same C-terminal half of the protein, but not the N-terminal half (Right).
Kinetic Formation Rates and Stabilities of the N- and C-Terminal Cross-β Cores of the FUS LC Domain.
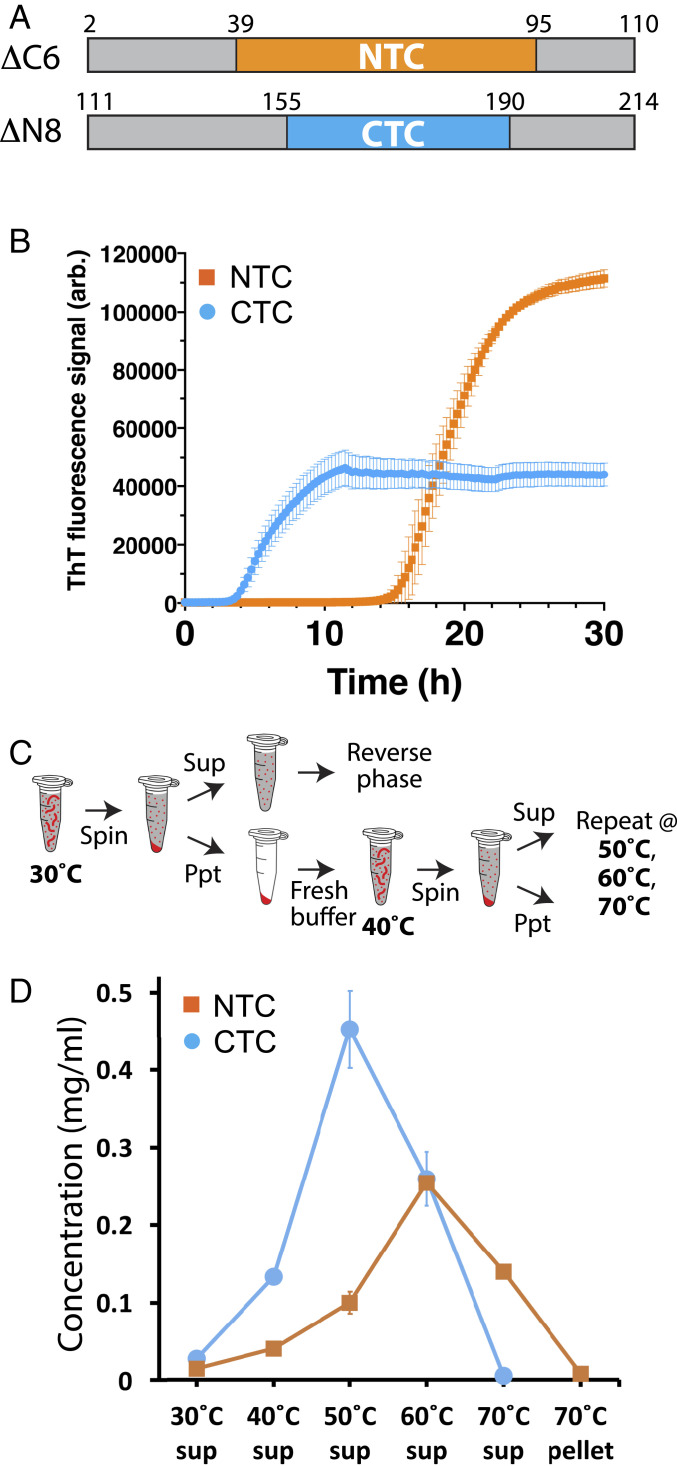
Purified, tag-free fragments corresponding to the N- and C-terminal halves of the FUS LC domain (Fig. 5A) were diluted out of denaturant and monitored for polymerization by acquisition of thioflavin-T fluorescence. As shown in Fig. 5B, the fragment corresponding to the C-terminal half of the FUS LC domain polymerized more rapidly than the fragment corresponding to the N-terminal half. In order to measure the stabilities of the NTC and CTC polymers, assembled polymers were exposed to graded increases in temperature. Release of soluble monomers was monitored by reverse-phase column chromatography (Fig. 5C). Roughly, half of the CTC polymers were soluble at 50 °C, and all of the sample was converted to monomers at 60 °C. By contrast, the NTC polymers required 10 °C higher temperature to achieve, respectively, half-maximal or full disassembly.
Fig. 5.
Measurements of polymerization rates and stabilities of polymers formed from the N- and C-terminal halves of the FUS low-complexity domain. (A) Schematic diagram of protein fragments corresponding to N- and C-terminal halves of the FUS low-complexity domain. Boxed region shown in yellow corresponds to structural boundaries of the NTC of the FUS LC domain (residues 39 to 95). The boxed region shown in cyan corresponds to functional boundaries of the CTC-forming region of the FUS LC domain (residues 155 to 190). (B) Protein fragments shown in A were expressed in bacteria, purified, and incubated under conditions of neutral pH and physiological monovalent salt for 30 h. Thioflavin-T fluorescence (y axis) was measured as a function of time (x axis). (C) Schematic diagram depicting methods used to monitor the release of monomer subunits from existing polymers as a function of temperature. Existing polymers were incubated at 30 °C and isolated from solution by ultracentrifugation. The supernatant was analyzed by reverse-phase chromatography to quantify soluble monomeric proteins. Pelleted polymers were resuspended in fresh buffer, incubated at 40 °C, and subjected to the same process of centrifugation and supernatant analysis. This process was repeated at 50 °C, 60 °C, and 70 °C. (D) Polymerized samples of NTC and CTC polymers were incubated at varying temperatures (x axis) and monitored for the release of soluble monomers by reverse-phase column chromatography (y axis) as described in C. Little monomeric protein was observed to be released from polymers incubated at 30 °C. CTC polymers released more monomers than NTC polymers at 40 °C and 50 °C and were completely solubilized at 60 °C. NTC polymers required incubation at 70 °C to effect complete solubilization. sup: supernatant.
The properties of the FUS LC domain, as well as N- and C-terminal fragments thereof, may well be altered when studies in the context of the intact FUS protein. It has been noted that an arginine:glycine (RG)-rich domain exists within a C-terminal region of FUS and proposed that cation:π interactions may facilitate intermolecular interactions between tyrosine residues of the N-terminal LC domain of FUS and arginine residues within its C-terminal RG domain (22, 23). Perhaps importantly, the LC domain of the FUS, Ewing sarcoma (EWS) and TAF15 proteins are all understood to function in isolation of remaining parts of their respective polypeptides as oncogenic fusion proteins (24). We cautiously contend, as such, that reductionist studies as exemplified in Fig. 5 may be valid.
ALS-Causing Variants Destabilize the C-Terminal Cross-β Core of the FUS Low-Complexity Domain.
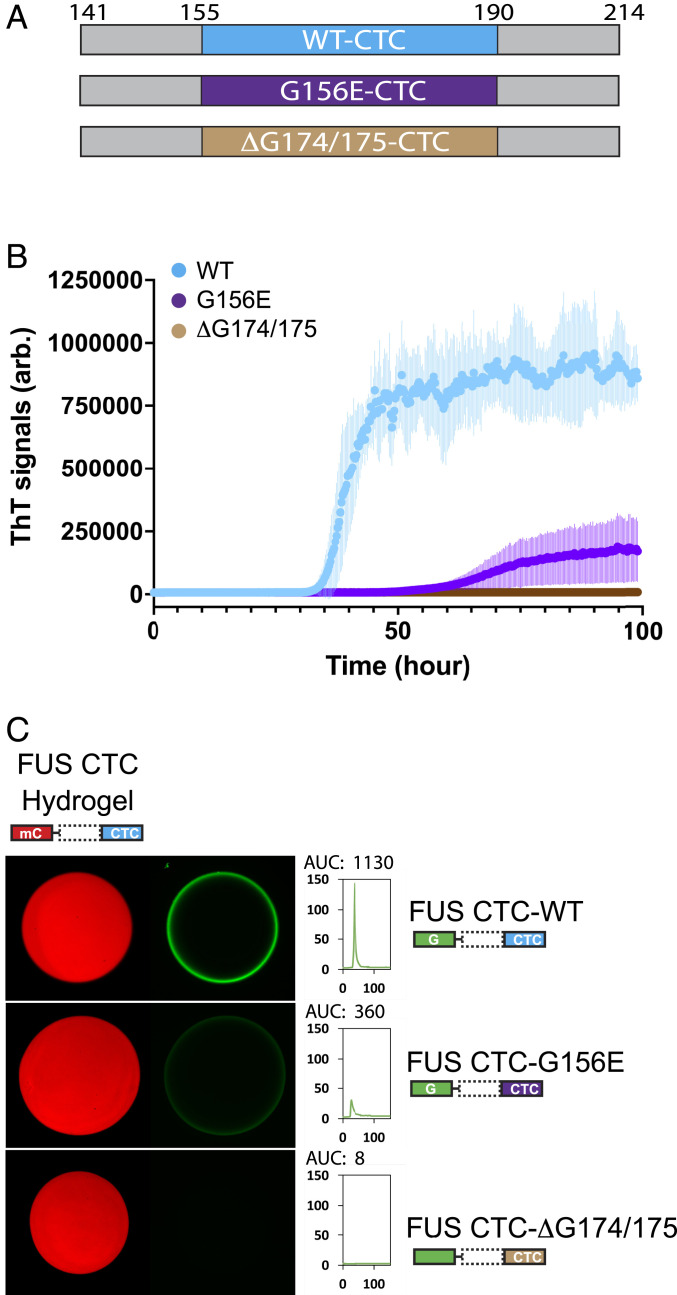
Variants of the FUS LC domain reported to predispose patients to ALS include a glycine-to-glutamic acid missense mutation of residue 156 (G156E) (18) and the deletion of glycine residues 174 and 175 (ΔG174/G175) (19, 20, 25). Recognizing that these variants map within the C-terminal cross-β core of the FUS LC domain, we initially expressed and purified both variants as GFP fusion proteins in the context of the isolated C-terminal core. Upon incubation under conditions normally leading to phase transition, we were surprised to observe that neither variant was able to form cross-β polymers within a time frame in which the native protein readily polymerized.
In order to investigate these observations more carefully, purified, tag-free monomeric protein was tested for polymerization via assays of thioflavin-T fluorescence. Fig. 6B shows polymerization assays for the truncated C-terminal core (FUS 141 to 214) in its native sequence configuration as compared with variants carrying either the G156E missense mutation or G174/G175 deletion (ΔG174/G175) (Fig. 6A). Although anticipating that the ALS-disposing variants might prompt the formation of aberrantly stable or more rapidly forming cross-β polymers, we observed no detectable polymerization for the ΔG174/G175 variant and significantly delayed polymerization for the G156E variant (Fig. 6B). When tested in binding assays to hydrogels formed from the CTC alone, weak binding (G156E) or no binding (ΔG174/G175) was observed for these ALS-disposing mutants (Fig. 6C).
Fig. 6.
Measurements of polymerization and hydrogel binding of ALS-causing mutations as assayed within the isolated C-terminal cross-β core of the FUS low-complexity domain. (A) Schematic diagram of protein fragments used to express native FUS LC domain (cyan) and ALS-causing variants (G156E, purple; ΔG174/175, tan). (B) The region spanning residues 141 to 214 of the FUS LC domain was expressed in its native form as well as when carrying the G156E or ΔG174/G175 ALS-disposing variants. Purified protein was incubated under conditions of neutral pH and physiological monovalent salt for 100 h. Thioflavin-T fluorescence (y axis) was measured as a function of time (x axis), giving evidence of polymerization by the native CTC fragment (blue line), delayed polymerization for the G156E variant (purple line), and no polymerization for the ΔG174/G175 variant (tan line). (C) The region spanning residues 141 to 214 of the FUS LC domain containing the C-terminal cross-β core was expressed as a GFP fusion in its native form as well as when carrying the G156E or ΔG174/175 ALS-disposing variants. Purified GFP-tagged proteins were incubated with hydrogels formed from mCherry linked to the C-terminal half of the FUS LC domain. Hydrogel binding activity evident for the GFP fusion linked to the native C-terminal half of the FUS LC domain was attenuated for the G156E variant and absent for the ΔG174/G175 variant. AUC: area under the curve.
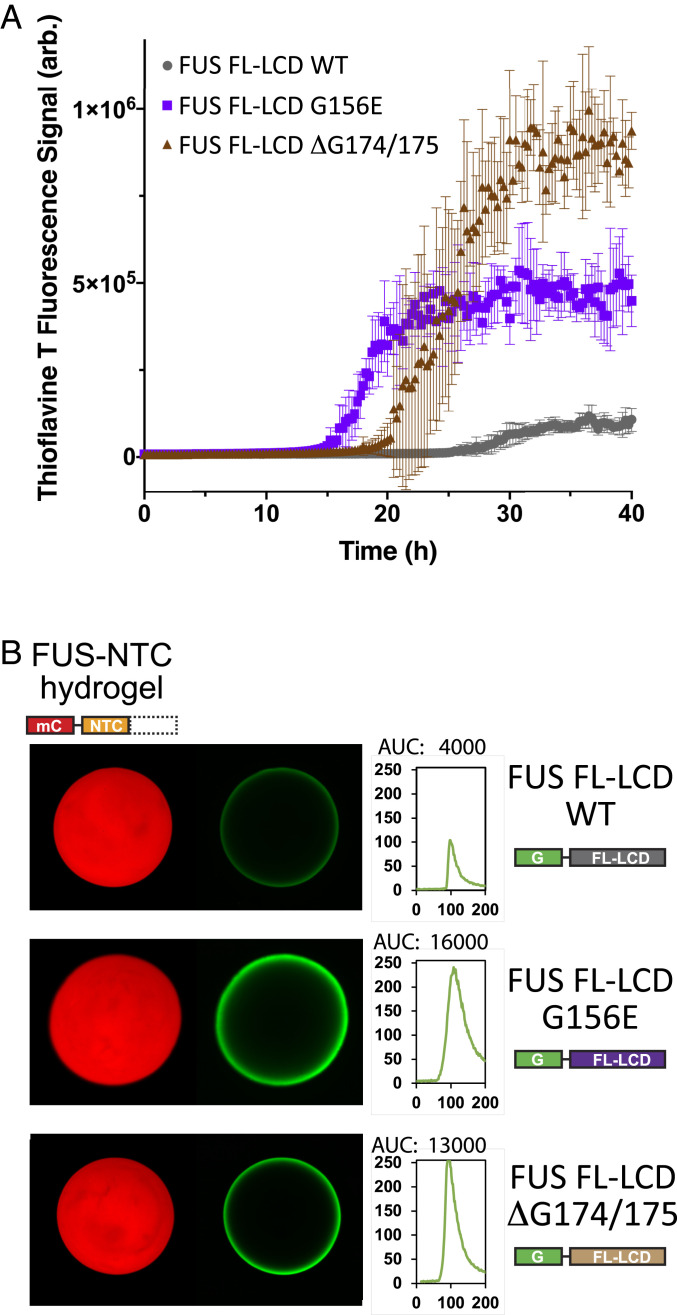
The G156E or ΔG174/G175 ALS-disposing variants were further analyzed in the context of the full-length LC domain of FUS by two assays: 1) Acquisition of thioflavin-T fluorescence and 2) hydrogel binding. For the former assay, tag-free proteins corresponding to the native LC domain of FUS, and both ALS-disposing variants, were incubated under conditions of neutral pH and physiological monovalent salt in the presence of thioflavin T. As shown in Fig. 7A, both ALS-disposing variants acquired thioflavin-T fluorescence considerably more rapidly than the native FUS LC domain. This result, which is consistent with published studies of the G156E ALS-disposing variant of the FUS LC domain (26, 27), was notable in revealing the opposite pattern of polymerization from that observed when the three proteins were studied in the context of the isolated, C-terminal half of the FUS LC domain (Fig. 6B).
Fig. 7.
Measurements of polymerization and hydrogel binding of ALS-causing mutations as assayed within the intact low-complexity domain of FUS. (A) Full-length derivatives of the FUS low-complexity domain bearing the native amino acid sequence (WT) or that carrying either of two ALS-disposing variants, were expressed as 6His-tagged proteins in bacteria, purified, and incubated under conditions of neutral pH and physiological monovalent salt in the presence of thioflavin-T. The y axis presents thioflavin-T fluorescence; the x axis presents time of incubation. (B) The same three segments of the FUS low-complexity domain were expressed as GFP-tagged proteins in bacteria, purified, and incubated under conditions of neutral pH and physiological monovalent salt with hydrogels formed from mCherry linked to the isolated N-terminal cross-β core of the FUS low-complexity domain. Scans depicted to the Right of hydrogel images present quantitation of GFP signal intensity as measured at hydrogel perimeters. The G156E and ΔG174/G175 variants of the FUS low-complexity domain (Bottom two rows) displayed between three- and fourfold greater binding intensities than that observed for the native FUS protein (Top row).
For hydrogel binding assays, each of the three proteins was linked to GFP, expressed in bacteria, purified, and incubated with mCherry hydrogels formed from the N-terminal half of the FUS LC domain. To our surprise, both ALS-disposing variants bound more prominently to mCherry hydrogels formed from the NTC alone (Fig. 7B). Why do variants carrying mutations that inactivate the C-terminal cross-β core bind NTC hydrogels more strongly than the intact LC domain of FUS? This observation, we propose, may offer an unanticipated clue as to how low-complexity domains function in living cells.
Discussion
In initiating the experiments described herein, we anticipated a simple pathway for the binding of soluble FUS monomers to precast hydrogels. Given the presence of the structured NTC as the defining feature of mCherry hydrogels formed by the intact LC domain of FUS, our expectation was that the soluble, unstructured test protein would be capable of sampling the contours of the folded NTC located at polymer termini and simply slot itself into the existing protein fold. If so, the amino acid region specifying the NTC would have been the most important region of the FUS LC domain required for a soluble test protein to bind to hydrogel droplets. This expectation was not met. We instead observed that a distinct, C-terminal region of the FUS LC domain was considerably more important for hydrogel binding. These unexpected observations led to characterization of a different and nonoverlapping cross-β forming region that we now designate as the CTC of the FUS LC domain.
What are we to make of these unexpected observations? What follows are hypothetical thoughts supported only in part by experimental observations. We offer these thoughts as correlates that may be helpful in considering how the FUS LC domain might achieve its biological function in a manner avoiding runaway polymerization from either the NTC or CTC.
Correlate 1: Cross-β Formation Is Achieved More Readily by Two Unstructured Regions than if One Region Already Exists in a Cross-β Conformation.
The sole way in which the FUS LC domain can exist in the unique cross-β structure that has been resolved by solid-state NMR spectroscopy is if at least two molecules have coalesced. In other words, the cross-β conformation cannot be assumed by a monomer on its own. Once two molecules have assembled into the cross-β state, conformational freedom of the amino acids localized within the structured region is restricted. If one copy of the FUS LC domain exists in the structurally ordered state as the terminal cap of an existing cross-β polymer, and another is free and unstructured, we hypothesize the latter to have difficulty in finding the former.
It has been extensively postulated that π:π stacking of aromatic amino acid side chains may be important for self-associative interactions between unstructured LC domains (28). The FUS LC domain contains 27 tyrosine residues that are important for self-association of the protein (Fig. 1) (7). Once assembled into the cross-β structural state, a subset of these tyrosine residues become conformationally restricted (8). We hypothesize that conformational restriction may impede the ability of an unstructured monomer to participate in tyrosine:tyrosine-mediated π:π stacking interactions within the region imposed to exist in the structurally ordered state.
If correct, this correlate may explain why the C-terminal region of FUS monomers is more important than the N-terminal region for a soluble test protein to bind hydrogels composed of NTC polymers. The LC domain of the soluble, GFP-tagged monomers whose binding is being assayed exists in an unstructured state. Likewise, the C-terminal region of FUS subunits already existing in hydrogel polymers is also unstructured so long as the polymers are assembled by the NTC (8). Since the C-terminal regions of both the hydrogel recipient of binding and test monomers are unstructured, we predict that the probability of forming a CTC cross-β interaction should exceed that of an NTC interaction.
Correlate 2: Formation of Both NTC and CTC Cross-β Structures within a Single Polypeptide Is Sufficiently Unfavorable to Impart Mutual Exclusion.
Hydrogels formed by the intact LC domain of FUS contain uniform polymers thousands of subunits in length. These FUS polymers are held together by cross-β interactions specified by the NTC (8). The unstructured C-terminal region of the FUS LC domains extends laterally from polymers in bottle brush fashion, such that each polymer displays thousands of free and unstructured C-terminal regions. Here we show that when challenged with a soluble, unstructured test monomer, binding by the test protein is considerably more dependent upon the sequences of its C-terminal region than its N-terminal region.
Each polymer within a hydrogel has but two termini exposing the molded conformation of the structured, NTC cross-β assembly. By contrast, the same polymers display thousands of laterally extending C-terminal regions that are unstructured. Given this fact, coupled with correlate 1 as described above, it comes as no surprise that binding of the unstructured test protein is inordinately reliant upon the integrity of its C-terminal domain (Figs. 1 and 2).
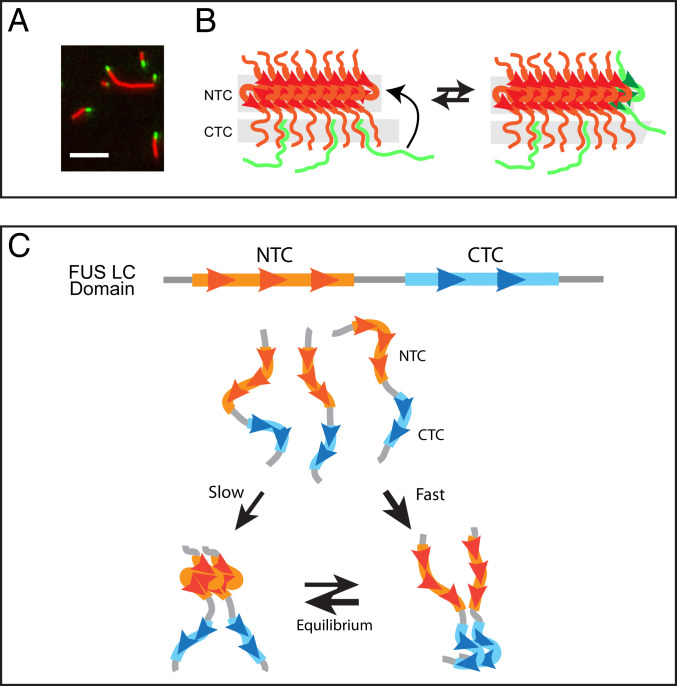
These findings are at odds, however, with simple microscopic observations of the process of polymer growth (7). Following coincubation of mCherry:FUS polymers with soluble GFP:FUS test monomers, we used total internal reflection fluorescence (TIRF) microscopy to image polymer growth as a function of time. As shown in Fig. 8A, we observed no evidence of GFP binding to the lateral sides of preassembled mCherry:FUS polymers. Instead, time-dependent polymer growth revealed GFP-labeled termini. From such studies we offer that C-terminal interactions between the soluble test protein and the lateral sides of existing polymers must be unstable. It is possible that transient π:π stacking interactions via tyrosine side chains may enable these unstable, CTC:CTC interactions.
Fig. 8.
Hypothetical pathway of FUS low-complexity domain binding to hydrogel polymers and conceptual pathway for initial self-association of soluble monomers. (A) TIRF microscopic imaging of mCherry:FUS polymers incubated with GFP:FUS monomers. Copolymerization of GFP test protein into existing mCherry:FUS polymers can be seen as green tips extending from red fibrils. (B) Schematic conceptualization of the process of copolymerization. Existing FUS polymer (red) is interpreted to initially bind soluble FUS monomers (green) via interactions wherein the CTC of test proteins attempts to form cross-β interaction with lateral surfaces of existing polymers (Left). Initial, unstable interaction is interpreted to be in equilibrium with copolymerization onto the N-terminal cross-β core (Right). (C) Soluble, unstructured FUS low-complexity domain monomers (Top) are interpreted to preferentially self-associate via C-terminal cross-β core interactions (Right). Slower yet more stable N-terminal cross-β self-association (Left) is proposed to isomerize from dimers held together by the less stable, C-terminal cross-β core. Simultaneous formation of N- and C-terminal cross-β interactions within a single polypeptide is interpreted to be impermissible.
We speculatively attribute these discordant observations to the fact that, owing to the correlate of mutual exclusion, a stable CTC cross-β structure cannot assemble on subunits of a polymer formed by the NTC cross-β structure. We instead imagine—as schematized in Fig. 8B—that transient attempts toward CTC formation weakly adhere the soluble GFP-labeled test protein to mCherry-labeled hydrogel polymers. Stable hydrogel binding is limited to the event wherein the test protein copolymerizes onto termini of existing polymers. In other words, C-terminal interactions between test protein and hydrogel may weakly retain the test protein in a position sufficiently proximal to polymer termini to enable execution of an unfavorable molecular event. Even though, according to correlate 1, binding of an unstructured N-terminal region to the cross-β structure at polymer termini is disfavored, it eventually transpires.
A more obvious indication of mutual exclusivity derives from structural studies of hydrogel polymers formed from the intact LC domain of FUS. Solid-state NMR studies of these polymers have resolved the monomorphic structure of the N-terminal cross-β core (8). The in-register conformation of protomers held together by the NTC cause the unstructured, C-terminal domain of FUS to protrude laterally from the polymer core in a specified geometry. These extending, C-terminal regions of the polymer are flexibly positioned 4.7 Å apart. That the aligned, C-terminal regions do not adopt the cross-β conformation readily observed when the isolated CTC is incubated at high concentration (Figs. 3 and 4 and SI Appendix, Fig. S2) bolsters the concept that individual molecules of the FUS LC domain cannot simultaneously form both N- and C-terminal cross-β cores.
Finally, the concept of mutual exclusion may explain the perplexing data shown in Fig. 7B. mCherry hydrogel droplets composed solely of the NTC of FUS were challenged with GFP-tagged protein corresponding to the full-length LC domain bearing its native sequence, or that of either ALS variant. We offer that the enhanced binding of the latter proteins reflects the fact that, unlike the native LC domain, they cannot form CTC cross-β structures. According to correlate 2, mutual exclusion predicts an impediment to copolymerization with NTC polymers of the hydrogel if the test protein can itself form CTC cross-β interactions. Since the native FUS LC domain can form CTC interactions, but the ALS variants cannot, the latter test proteins are able to bind NTC-only hydrogels more readily than the former.
Implications of Correlates 1 and 2.
Correlates 1 and 2 represent simplified interpretations of the data included in this and earlier studies of the FUS LC domain. Despite their hypothetical nature, these correlates may be useful in thinking about questions of both narrow focus and potentially broad significance.
On the more narrow side, these correlates allow us to consider how certain ALS-causing mutations within the FUS LC domain might lead to aberrant protein aggregation. We have found that the G156E and ΔG174/G175 mutations destabilize the CTC (Fig. 6B). In contrast to destabilizing the isolated CTC, these very same ALS-disposing mutations significantly enhance the propensity of the intact LC domain of FUS to form runaway polymers (Fig. 7A). Following the mutual exclusion teaching of correlate 2, variants of the FUS LC domain lacking a functional CTC are unable to deploy CTC-specified cross-β interactions to interfere with NTC interactions. As such, runaway NTC polymerization takes place (Fig. 7A). We make note of the fact that this behavior of the G156E mutation within the FUS LC domain has already been documented by S. Alberti and colleagues and P. St George-Hyslop and colleagues (26, 27).
We offer the schematic diagram shown in Fig. 8C as an illustration of the concept of mutual exclusion. Kinetic parameters may favor initial self-association of the FUS LC domain to form the CTC cross-β structure (Fig. 5B). The resulting proximity of two unstructured NTC regions is predicted to facilitate a process of isomerization that begets formation of the slightly more stable NTC cross-β structure. Important to the thesis articulated herein, the concept of mutual exclusion demands that the initial CTC cross-β structure be disassembled in order for the NTC structure to form.
Once equilibrium has been reached, we offer two reasons to account for impediments to further growth of NTC polymers. First, if a third FUS LC domain were to use its CTC to invade the free CTC of the dimer held together by NTC cross-β interactions, mutual exclusivity would demand that NTC interactions dissolve (correlate 2). The same idea would guard against the ability of a third FUS LC domain to invade the free and unstructured NTC of a dimer held together by CTC cross-β interactions. Should runaway polymerization by either cross-β core take place, the correlate of mutual exclusion may no longer apply. If a series of NTCs are coassembled into a long polymer, we propose that an incoming monomer is unable to use its CTC to productively coalesce with any of the unstructured CTC domains extending laterally from the polymer. We offer that this option is voided because each NTC internal to the polymer is bordered on both sides by another structured NTC. Whereas the forces of mutual exclusion are proposed to be sufficient to force disassembly of a pair of molecules organized in the cross-β conformation (Fig. 8C), they may be inadequate to force dissolution of a cross-β assembly bearing the forces of structural order from both sides.
Our second reason for hypothesizing orderly limitation of FUS polymer growth derives from correlate 1. Upon encountering either of the dimers shown in Fig. 8C, an unstructured FUS LC domain would prefer interaction with the region of FUS not already existing in the cross-β structural state. If the dimer were held together by NTC interactions, the incoming protein is predicted to use its CTC in an attempt to form cross-β interactions with the unstructured CTC of the dimer. Reciprocally, if the dimer were held together by CTC interactions, the incoming protein would be expected to use its NTC to attempt self-association with the unstructured NTC of the dimer. These preferences, as specified in correlate 1, are attributed to conformal restriction. If the bias of conformational restriction is strong, and if mutual exclusivity is likewise strong, the combination of these limitations should prevent the FUS LC domain from polymerizing any further than the dimeric state.
We close with emphasis on the multitude of unknowns that cloud our simplistic ideas. The FUS LC domain is subject to many forms of posttranslational modification (PTM) that may influence behavior of the NTC and CTC domains. Differential regulatory effects of PTMs might allow the FUS LC domain to expand either NTC- or CTC-mediated polymerization on demand. The FUS protein likewise shuttles from the nucleus to cytoplasm and back and uses its LC domain for any of a number of heterotypic interactions with other proteins. These variables can be understood to open the opportunity for the FUS LC domain to deviate from the dimeric ground state imposed by correlates 1 and 2. Despite the complexities of this science, we have faith in the value of the reductionist approach exemplified by the experiments described herein.
Materials and Methods
See SI Appendix, Materials and Methods for detailed materials and methods that describe 1) cloning of expression plasmids; 2) protein expression and purification; 3) hydrogel binding assays; 4) polymer extension assays; 5) polymer formation; 6) X-ray diffraction; 7) SDD-AGE; 8) thioflavin-T assays; 9) thermal stability assays; and 10) sample preparation for ssNMR.
Acknowledgments
We thank Robert Tycko and Myungwoon Lee for help in recording ssNMR spectra. We thank Deepak Nijhawan and Glen Liszczak for thoughtful discussions regarding the research described herein. We also thank Lillian Sutherland, Lily Sumrow, and Leeju Wu for plasmid cloning. S.L.M. was supported by National Institute of General Medical Sciences Grant 5R35GM130358 and National Cancer Institute Grant 1U54CA231649, as well as unrestricted funding from an anonymous donor.
Footnotes
Reviewers: A.L.H., Yale University School of Medicine; and P.S.G.-H., University of Cambridge.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2114412118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Radó-Trilla N., Albà M., Dissecting the role of low-complexity regions in the evolution of vertebrate proteins. BMC Evol. Biol. 12, 155 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Toll-Riera M., Radó-Trilla N., Martys F., Albà M. M., Role of low-complexity sequences in the formation of novel protein coding sequences. Mol. Biol. Evol. 29, 883–886 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Leuenberger P., et al., Cell-wide analysis of protein thermal unfolding reveals determinants of thermostability. Science 355, eaai7825 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Banani S. F., Lee H. O., Hyman A. A., Rosen M. K., Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shin Y., Brangwynne C. P., Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Han T. W., et al., Cell-free formation of RNA granules: Bound RNAs identify features and components of cellular assemblies. Cell 149, 768–779 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Kato M., et al., Cell-free formation of RNA granules: Low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murray D. T., et al., Structure of FUS protein fibrils and its relevance to self-assembly and phase separation of low-complexity domains. Cell 171, 615–627.e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tycko R., Solid-state NMR studies of amyloid fibril structure. Annu. Rev. Phys. Chem. 62, 279–299 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fändrich M., Meinhardt J., Grigorieff N., Structural polymorphism of Alzheimer Abeta and other amyloid fibrils. Prion 3, 89–93 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiang S., et al., The LC domain of hnRNPA2 adopts similar conformations in hydrogel polymers, liquid-like droplets, and nuclei. Cell 163, 829–839 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ryan V. H., et al., Mechanistic view of hnRNPA2 low-complexity domain structure, interactions, and phase separation altered by mutation and arginine methylation. Mol. Cell 69, 465–479.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murray D. T., et al., Structural characterization of the D290V mutation site in hnRNPA2 low-complexity-domain polymers. Proc. Natl. Acad. Sci. U.S.A. 115, E9782–E9791 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu J., et al., CryoEM structure of the low-complexity domain of hnRNPA2 and its conversion to pathogenic amyloid. Nat. Commun. 11, 4090 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim H. J., et al., Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 495, 467–473 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vieira N. M., et al., A defect in the RNA-processing protein HNRPDL causes limb-girdle muscular dystrophy 1G (LGMD1G). Hum. Mol. Genet. 23, 4103–4110 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Lin Y., et al., Toxic PR poly-dipeptides encoded by the C9orf72 repeat expansion target LC domain polymers. Cell 167, 789–802.e12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ticozzi N., et al., Analysis of FUS gene mutation in familial amyotrophic lateral sclerosis within an Italian cohort. Neurology 73, 1180–1185 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.T. J. Kwiatkowski, Jr, et al., Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 323, 1205–1208 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Rademakers R., et al., Fus gene mutations in familial and sporadic amyotrophic lateral sclerosis. Muscle Nerve 42, 170–176 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee M., Ghosh U., Thurber K. R., Kato M., Tycko R., Molecular structure and interactions within amyloid-like fibrils formed by a low-complexity protein sequence from FUS. Nat. Commun. 11, 5735 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qamar S., et al., FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation-π interactions. Cell 173, 720–734.e15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J., et al., A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell 174, 688–699.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kovar H., Dr. Jekyll and Mr. Hyde: The two faces of the FUS/EWS/TAF15 protein family. Sarcoma 2011, 837474 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan J., et al., Frameshift and novel mutations in FUS in familial amyotrophic lateral sclerosis and ALS/dementia. Neurology 75, 807–814 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patel A., et al., A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Murakami T., et al., ALS/FTD mutation-induced phase transition of FUS liquid droplets and reversible hydrogels into irreversible hydrogels impairs RNP granule function. Neuron 88, 678–690 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vernon R. M., et al., Pi-Pi contacts are an overlooked protein feature relevant to phase separation. eLife 7, e31486 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All study data are included in the article and/or supporting information.