Significance
Sensory nerves innervate bone and are required for normal skeletal development, but the molecular mechanisms through which nerves exert these essential functions have remained obscure. Here, we validate a spatial transcriptomics platform, which allowed us to map changes in the molecular landscape across the developing cranium of mice in which sensory nerve innervation is genetically blocked. We show that sensory, nerve-derived signals, including follistatin-like 1, coordinate cranial bone patterning by regulating the proliferation and differentiation of bone precursor cells within the suture mesenchyme.
Keywords: skeletal innervation, TrkA, calvarial bone, cranial suture, spatial transcriptomics
Abstract
The patterning and ossification of the mammalian skeleton requires the coordinated actions of both intrinsic bone morphogens and extrinsic neurovascular signals, which function in a temporal and spatial fashion to control mesenchymal progenitor cell (MPC) fate. Here, we show the genetic inhibition of tropomyosin receptor kinase A (TrkA) sensory nerve innervation of the developing cranium results in premature calvarial suture closure, associated with a decrease in suture MPC proliferation and increased mineralization. In vitro, axons from peripheral afferent neurons derived from dorsal root ganglions (DRGs) of wild-type mice induce MPC proliferation in a spatially restricted manner via a soluble factor when cocultured in microfluidic chambers. Comparative spatial transcriptomic analysis of the cranial sutures in vivo confirmed a positive association between sensory axons and proliferative MPCs. SpatialTime analysis across the developing suture revealed regional-specific alterations in bone morphogenetic protein (BMP) and TGF-β signaling pathway transcripts in response to TrkA inhibition. RNA sequencing of DRG cell bodies, following direct, axonal coculture with MPCs, confirmed the alterations in BMP/TGF-β signaling pathway transcripts. Among these, the BMP inhibitor follistatin-like 1 (FSTL1) replicated key features of the neural-to-bone influence, including mitogenic and anti-osteogenic effects via the inhibition of BMP/TGF-β signaling. Taken together, our results demonstrate that sensory nerve-derived signals, including FSTL1, function to coordinate cranial bone patterning by regulating MPC proliferation and differentiation in the suture mesenchyme.
The patterning and ossification of the mammalian skeleton requires inductive signals from cells derived from the three primary germ layers, which function in a hierarchical and temporal fashion to control skeletal patterning and bone cell differentiation (1, 2). The flat bones of the cranium are formed through a process termed intramembranous ossification, which does not require a cartilaginous template. During development, cells from the neural crest migrate to positions between the brain and surface ectoderm where they form the cranial suture complex (3). In contrast to endochondral growth plates, which expand through chondrocyte proliferation and hypertrophy, cranial sutures do not have intrinsic growth potential. Instead, they produce new bone at the suture edges in response to both intrinsic, osteogenic stimuli and extrinsic cues emanating from the expanding neurocranium. Thus, for sutures to serve as bone growth sites, they need to remain largely nonossified, with new bone forming only along the edges of the bony plates (4). This dynamic, architectural feature of the cranial suture enables the expansion of the cranial vault to accommodate brain enlargement while also ensuring a stable pool of progenitors.
The current understanding of the molecular mechanisms controlling cranial suture cell dynamics has been facilitated by studies of genetic disorders associated with premature suture closure, collectively called craniosynostosis. Mutational studies in murine models have provided evidence for the involvement of intrinsic signals emanating from the fibroblast growth factor (FGF) (5) and transforming growth factor beta (TGF-β) (6, 7) pathways, as well as osteogenic transcription factors such as Osterix, MSX1/2 (8), and TWIST (9) in the control of suture patency. In addition, recent evidence suggests that blood vessels and peripheral nerves provide additional extrinsic signals which impact skeletal stem cell proliferation and differentiation (10). In this regard, previous studies by our group have demonstrated an essential role for nerve growth factor (NGF)–dependent tropomyosin receptor kinase A (TrkA) signaling by sensory nerves in osteogenesis in the developing long bones (11), in response to mechanical loading (12), and during repair (13, 14). Blockade of sensory nerve ingrowth, either by the inhibition of TrkA signaling or deletion of NGF, retards vascularization and disrupts mineralization.
In this study, we investigated the cellular and molecular mechanisms through which sensory nerves function during cranial bone formation. We find that the blockade of TrkA signaling in vivo reduced sensory nerve innervation of the developing cranium, causing the premature, calvarial suture closure associated with a decrease in suture cell proliferation and increased mineralization along the bony fronts. In vivo spatial transcriptomic analysis revealed that the premature, postnatal suture closure correlated with a spatial misregulation of bone morphogenetic protein (BMP) and TGF-β signaling across the expanse of the developing skull, with subsequent experiments pointing to follistatin-like 1 (FSTL1) as a neural-derived secreted factor through which these effects were exerted.
Results
Innervation of Cranial Bone by TrkA Sensory Nerves Is Required for the Maintenance of Suture Patency.
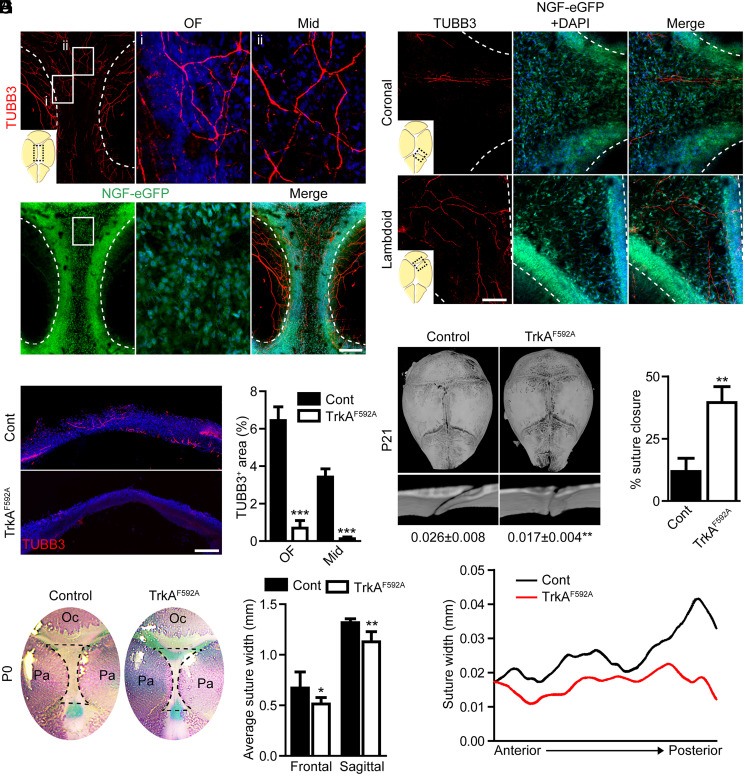
Skeletal nerves were first examined in relation to the developing calvaria in mice (Fig. 1; P0 skulls examined). Whole-mount analysis of the pan-neuronal marker beta III tubulin (TUBB3) demonstrated that skeletal nerve fibers extended toward the midline from the lateral aspects of the developing parietal bones (Fig. 1A). A high density of nerves arborized at the osteogenic front (OF) of the parietal bone (Fig. 1A, i), while a fine basket weave of small nerve fibers crossed over the sagittal suture midline (Fig. 1A, ii). The primary skeletal neurotrophin NGF was assessed using the whole-mount detection of NGF-eGFP reporter activity. Innervation correlated with the abundance of NGF reporter activity, with a robust signal observed throughout the sagittal suture and OF. A similar correlation between skeletal nerve location and NGF reporter activity was observed across the coronal and lambdoid sutures (Fig. 1B). To determine the requirement for sensory nerves in cranial bone development, we analyzed calvaria morphology following the inhibition of TrkA activity using previously validated TrkA knockin transgenic mice (TrkAF592A) (15). TrkAF592A mice harbor a point mutation in the Ntrk1 gene (F592A), rendering the endogenous TrkA kinase sensitive to inhibition by the membrane-permeable, small-molecule 1NMPP1. This chemical–genetic approach has been extensively validated for in vitro and in vivo use (11–14). In our previous studies, we established the dose required to impair the phosphorylation of TrkA in DRGs in vivo and to inhibit the sensory outgrowth in vitro. These studies have also shown that the administration of 1NMPP1 to either wild-type or TrkAF592A heterozygotes have no observable effect on TrkA signaling, skeletal innervation, or skeletal cell behavior (11). Pregnant dams carrying embryos heterozygous and homozygous for the TrkAF592A allele were given 200 µM 1NMPP1 in their drinking water from conception throughout the entire 21 d of gestation. Confocal imaging and histological analysis of sections confirmed the expected reduction in nerve fiber frequency in both the OF and midline of the sagittal suture (Fig. 1 C and D). The examination of cranial bones in newborn offspring showed a significant reduction in the width of the posterior frontal and sagittal sutures in homozygous TrkAF592A mice compared to heterozygote littermate controls (Fig. 1 E and F). No significant changes in suture length were observed between genotypes (posterior frontal suture 3.14 ± 0.17 mm versus 3.30 ± 0.22 mm, P = 0.1836, in control mice; sagittal suture 2.20 ± 0.15 mm versus 2.28 ± 0.13 mm, P = 0.3532, in control mice). To assess the impact of sensory nerves on postnatal, calvarial development, 1NMPP1 was administered by the direct injection to homozygous TrkAF592A and heterozygote littermates, starting at a timepoint after initial suture complex formation (P6), with suture morphology examined at P21. Consistent with embryonic inhibition, microcomputed tomography (micro-CT) analysis of P21 calvariae showed no change in sagittal suture length (2.82 ± 0.13 mm versus 2.76 ± 0.20 mm, P = 0.4891, in control mice) but a reduced average suture width and a high rate of abnormal, flanking parietal bone closure among 1NMPP1-treated TrkAF592A mice (Fig. 1 G and H). Moreover, slice-by-slice, two-dimensional (2D) analyses demonstrated that the posterior sagittal suture was preferentially affected by a loss of sensory innervation, though an overall reduction was observed along the entire anterior–posterior axis (Fig. 1I).
Fig. 1.
TrkA signaling is essential for maintaining cranial suture patency. (A) Whole-mount calvaria images of the sagittal suture within NGF-eGFP reporter animals subjected to TUBB3 immunohistochemical staining. (Scale bar, 500 µm.) (B) Whole-mount calvaria images of the coronal and lambdoid sutures within NGF-eGFP reporter animals subjected to TUBB3 immunohistochemical staining. (Scale bar, 100 µm.) (C) Coronal section of P0 sagittal suture stained for TUBB3. (Scale bar, 100 µm.) (D) Quantification of TUBB3+ nerve area within the OF or midline (Mid) suture mesenchyme. n = 4 to 5 mice/genotype. (E) Calvarial, whole-mount skeletal staining (top-down view) conducted at P0 following treatment with 1NMPP1, initiated at the time of conception. Oc, occipital bone and Pa, parietal bone. (F) Average suture width among posterior frontal and sagittal sutures within control (Cont) and TrkAF592A mice. n = 5 to 9 mice/genotype. (G) Micro-CT reconstructions of P21 calvaria (top-down view) and sagittal suture (coronal cross-section), following treatment with 1NMPP1, initiated at P6. Numbers below indicate the average sagittal suture width. (H) Percent of sagittal suture closure within Cont and TrkAF592A mice. (I) 2D average suture width between flanking parietal bones relative to the anterior–posterior position. n = 7 mice/genotype. Graphs represent average values ± SD, *P < 0.05, and **P < 0.01.
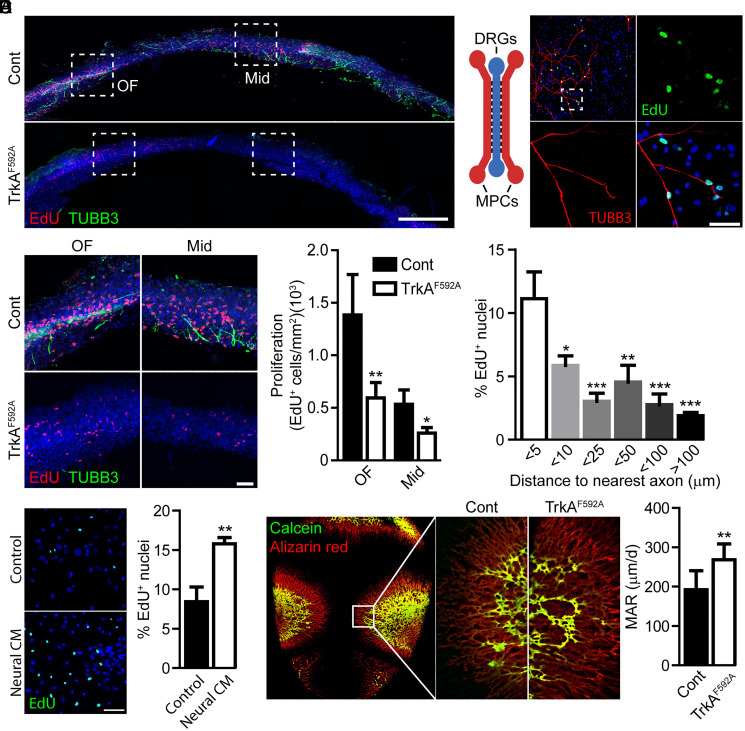
Sutures are maintained by a coordinated proliferative expansion from within the suture mesenchyme and mineralization-based closure occurring along the bony fronts. To directly assess the role of skeletal nerves in cranial suture proliferation, the EdU labeling of TrkAF592A mutant and control littermates was examined in relation to the distribution of TUBB3+ nerve fibers (Fig. 2A). Coronal cross-sections of the P0 sagittal suture and flanking parietal bones were examined. Control animals showed abundant EdU labeling within both the OF and suture midline. In contrast, TrkA mutant calvarial sutures showed a stark reduction in EdU incorporation (Fig. 2B). A similar reduction in proliferation in TrkAF592A calvaria was observed when staining for the proliferative marker Ki67 (SI Appendix, Fig. S1). The reduced proliferative activity in the suture mesenchyme of TrkA mutant mice suggested that sensory nerves might influence suture cell proliferation. To test this idea, we first examined the interaction of peripheral afferent neurons derived from dorsal root ganglia (DRG) with primary calvarial mesenchymal progenitor cells (MPCs) cultured in custom microfluidic chambers (Fig. 2C). In this device, the cell bodies of sensory nerves isolated from the DRG are cultured in a central chamber with their axons traversing through microchannels into adjacent, separate chambers seeded with calvarial MPCs. Following 2 d of cocultures, proliferation, indicated by EdU incorporation, was highest in cells in close proximity to infiltrating axons and decreased with increasing distance from the nearest axon (Fig. 2D). The observed gradient in cell proliferation was hypothesized to be the result of a nerve-released, soluble paracrine factor. The quantification of EdU uptake by MPCs following treatment with neural conditioned media demonstrated a significant increase in proliferation compared to untreated neural media controls (Fig. 2E). In accordance with this finding, qPCR of MPCs treated with neural conditioned media showed significant up-regulation of proliferative markers (Mki67 1.67 ± 0.15, P = 0.0027 and Ccna2 1.53 ± 0.19, P = 0.0122, relative to control).
Fig. 2.
TrkA+ sensory nerves promote the proliferative expansion of the cranial sutures via a spatially restricted, secreted ligand. (A) Proliferation and innervation of the P0 sagittal suture among control (Cont) and TrkAF592A mutant animals, visualized via EdU-labeling and TUBB3 immunohistochemical staining. (Scale bar, 200 µm.) (B) Quantification of EdU incorporation within the OF or midline (Mid) suture mesenchyme among Cont and TrkAF592A mutant animals. (Scale bar, 50 µm.) n = 4. (C) Proliferation of primary MPCs following direct coculture with DRG axons using a microfluidic device, in which chambers are connected via microchannels. (Scale bar, 10 µm.) TUBB3 immunohistochemical staining is used to visualize axons. (D) Spatial distribution of EdU+ MPCs in relation to the nearest DRG axon. n = 4. (E) Quantification of EdU labeling in MPCs following treatment with DRG-derived, neural conditioned media (CM). n = 4. (Scale bar, 25 µm.) (F) MAR of developing parietal bones in Cont and TrkAF592A mutant calvaria. n = 8. Graphs represent average values ± SD, *P < 0.05, **P < 0.01, and ***P < 0.001.
To determine the effects of sensory nerves on mineralization-based suture closure, mineralization rates along the bony fronts were assessed (Fig. 2F). Pregnant dams were injected with calcein and alizarin red at E17.5 and E18.5, respectively, and pups were euthanized at P0. Compared to controls, TrkA mutant mice showed a significant increase in mineral apposition rates (MARs) along the sagittal, suture-flanking regions of the parietal bones. Taken together, these data suggest that TrkA signaling in peripheral afferent neurons are required to maintain MPCs in a proliferative, undifferentiated state.
Comparative Spatial Transcriptomic Analysis of Sutures Reveals Alterations in the TGF-β/BMP Pathways.
To map the molecular landscape within the developing cranial niches in response to the ingrowth of sensory nerves, we employed spatial transcriptomics (16), a method that enables the visualization and quantitation of the transcriptome in tissue sections with high, spatial resolution (see Materials and Methods for details). In brief, slides are coated with an array of poly-T primers, which encode a unique spatial barcode (SI Appendix, Fig. S2A). Hematoxylin and eosin (H&E)–stained sections (SI Appendix, Fig. S2B) undergo enzymatic permeabilization, allowing messenger RNA (mRNA) release and the subsequent capture by the primer-coated slides, visualized through the incorporation of fluorescent nucleotides into the complementary DNA (cDNA) synthesis process (SI Appendix, Fig. S2C). The resolution of the unique spatial barcodes allows matching RNA abundance with their original spatially resolved spots, resulting in RNA-sequencing (RNA-seq) with 2D positional information. To optimize this approach for skeletal tissue, enzymatic permeabilization times on optimization slides were varied in calvarial sections to determine the timing of maximum mRNA recovery and minimal mRNA diffusion on the fluorescently labeled cDNA footprint (SI Appendix, Fig. S2 C and D). Following optimization, spatial gene expression studies were carried out on control and TrkAF592A P0 calvariae. Following sequence alignment and image registration, this spatial technique yielded 1,670 spatial “spots” across the sagittal suture and surrounding parietal bones, periosteum, and dura mater. Each spot contained an average of ∼3,500 unique genes and ∼10,500 unique mRNA molecules, with a slight, though insignificant, bias toward increased transcriptional information derived from TrkAF592A samples (SI Appendix, Fig. S2E).
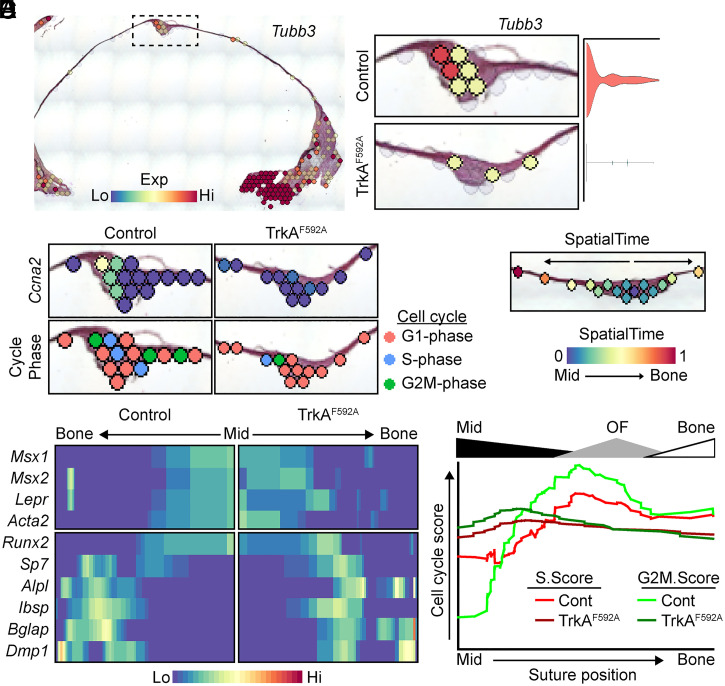
Spatial transcriptomic analyses were conducted on spatial spots mapping to the sagittal suture and immediate, flanking parietal bone. As expected, TrkAF592A calvariae showed significant reductions in suture innervation, indicated by Tubb3 expression (Fig. 3A). In keeping with the previously observed, reduced, and proliferative EdU index, TrkAF592A also showed the reduced expression of the proliferation marker Ccna2 and altered cell-cycle phase assignment using the CellCycleScoring function (17), compared to control mice (Fig. 3B). Of the 12,433 genes expressed in one or more spots within the data pool, only 69 genes were significantly regulated between control and TrkA mutant calvariae, and only seven genes were significantly regulated when adjusted for population-level expression (Dataset S1).
Fig. 3.
In vivo spatial transcriptomics confirms the attenuation of proliferative markers in the sagittal suture of TrkAF592A mice. (A) Spatial feature plot and suture complex quantification of Tubb3 expression. (B) Cell-cycle gene Ccna2 and predictive cell-cycle phase scoring of spots within the sagittal suture of control (Cont) and TrkAF592A mice. (C) Representative calvarial suture with spots analyzed using SpatialTime. (D) Validation of SpatialTime analysis using known suture/progenitor (Top) and osteogenic (Bottom) markers. n = 5 to 7 mice/genotype. (E) Changes in cell-cycle scoring across SpatialTime. Relative SpatialTime regions aligning to the midline (Mid) suture, OF, and flanking parietal bone (Bone) are denoted above graph.
As stated above, calvarial sutures are maintained in spatially defined domains, with mineralization occurring along the bony fronts while simultaneously maintaining MPCs in an undifferentiated state within the suture midline. To determine changes in these spatially distinct domains, we next compared changes in gene expression in TrkAF592A and control mice using a method analogous to the single-cell RNA sequencing (scRNAseq) computational technique of inferring pseudotime (18), referred to herein as “SpatialTime” (SI Appendix, Fig. S3). In this analysis, the distance between each spot within a suture and a manually selected, central anchor point in the midline is measured and normalized to the suture width. Spots from multiple suture samples are then combined to generate a map of the average suture spot gene expression (Fig. 3C). SpatialTime analysis was validated using expression of known suture and osteogenic cell markers (Fig. 3D) and confirmed a notable depression in cell-cycle scoring within the OF (Fig. 3E), consistent with our previous results. Gene expression patterns significantly correlating with SpatialTime are calculated and visualized in heatmaps with the most central midline suture spot positioned on one side and the most distant, flanking bone spot analyzed on the opposite side. This analysis showed a differential expression across SpatialTime of ∼3,500 genes (Dataset S2); as expected, several genes were seen to be regulated across SpatialTime in both control and TrkAF592A calvaria. Transcription factors showing a SpatialTime-dependent expression specific to either control or mutant sutures (SI Appendix, Fig. S4A) were then interrogated using pathway analysis (SI Appendix, Fig. S4B). This data revealed the altered expression of genes involved in stem cell maintenance (control) or differentiation (TrkAF592A). Spatial data confirmed a reduced expression of several factors previously identified to be expressed and/or involved in maintaining suture patency, including Gli1, Twist1, and Noggin, specifically within the midline suture (SI Appendix, Fig S4C). Taken together, these data are compatible with the notion that a loss of suture mesenchyme patency, along with a decline in cell proliferation, contribute to the premature suture closure observed in TrkAF592A mice. In agreement with this idea, qPCR analyses of primary MPCs cocultured in microfluidic devices with DRG axons suggest that nerve-derived signals have the ability to regulate stemness, up-regulating several genes previously linked to stem cell plasticity, pluripotency, and suture maintenance, including Sox2 (3.52 ± 1.71, P < 0.05), Pou5f1 (5.68 ± 2.10, P < 0.05), Klf4 (1.50 ± 0.27, P < 0.05), and Msx2 (2.05 ± 0.55, P < 0.05).
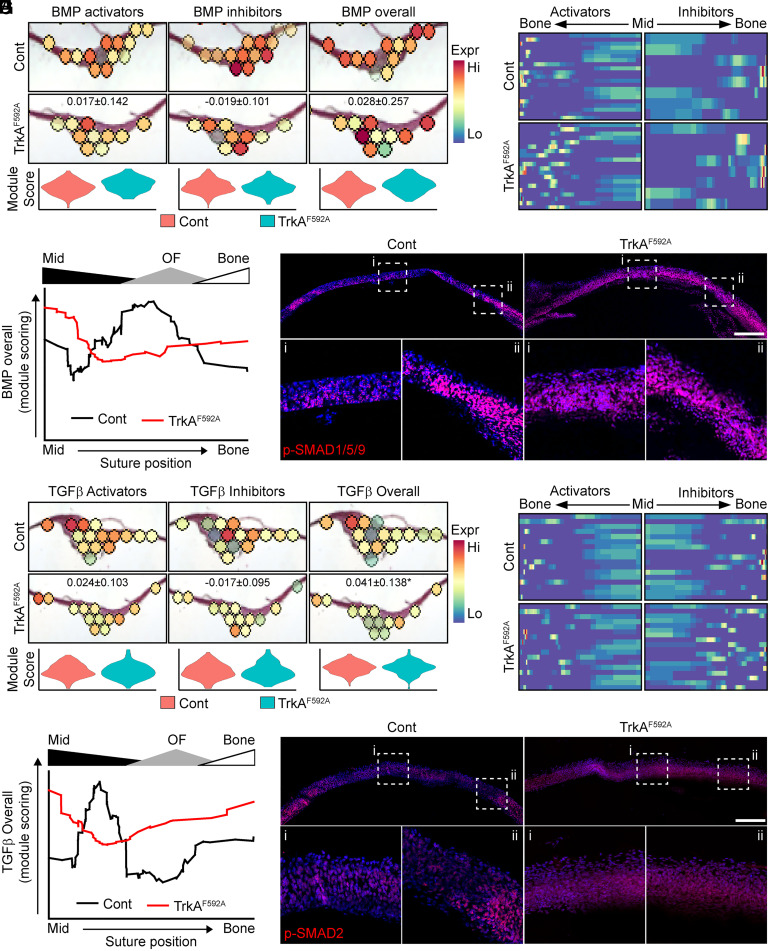
In addition to changes in proliferation and stemness, our pathway analysis also indicated a dysregulation of components of the BMP/TGF-β signaling pathways in TrkAF592A calvariae. In representative control calvariae, modular scoring to assess the expression of BMP activators and inhibitors (see SI Appendix, Table S1 for genes used in module scoring) showed a specific enrichment for BMP activation within the OF, with minimal activation within the suture midline; BMP inhibitory scores showed the inverse spatial localization, with high levels of BMP inhibition within the midline gradually fading within the OF and bone (Fig. 4A). In contrast, this putative BMP signaling gradient was disrupted in the TrkAF592A calvaria. Combined analysis of all sutures using our SpatialTime analysis confirmed a notable disruption in BMP signaling in TrkA mutant calvariae (Fig. 4B). The quantification of overall BMP activation from SpatialTime data confirmed the presence of a robust BMP activation “wave” initiating in the late midline/early OF and extending into the bony fronts (Fig. 4C). This pattern was both blunted and shifted toward the midline suture in TrkAF592A mice. Consistent with these spatial data, immunohistochemistry on calvarial sections showed robust staining for canonical downstream BMP signaling activators p-SMAD1/5/9, primarily localized within the OF and flanking bone (Fig. 4D, ii) in control mice, while TrkA mutant mice showed moderate p-SMAD1/5/9 staining throughout the suture. Analogous results were observed for modular scores of TGF-β signaling; calvaria from TrkAF592A mice showed only modest changes in overall TGF-β activation but exhibited the altered, spatial expression pattern of activators and inhibitors (Fig. 4 E–G). Histological confirmation revealed a near-uniform p-SMAD2 staining throughout the suture of TrkAF592A mice (Fig. 4H).
Fig. 4.
Spatial transcriptomics demonstrate a disruption in BMP/TGF-β signaling in TrkAF592A calvaria. (A) Violin plot and spatial expression data of BMP-signaling module scoring among control (Cont) and TrkAF592A calvaria, including BMP activators, BMP inhibitors, and overall BMP signaling. (B) Heatmap of BMP activator and inhibitor genes expressed across SpatialTime among Cont and TrkAF592A calvaria. (C) Overall BMP activation, based on module scoring across SpatialTime from midline (Mid) suture to flanking parietal bone (Bone) among Cont and TrkAF592A calvaria. Relative SpatialTime regions aligning to the Mid suture, OF, and flanking Bone are denoted above graph. n = 5 to 7 mice/genotype. (D) Canonical BMP signaling, as detected via immunofluorescent staining for p-SMAD1/5/9 using coronal cross-sections of the sagittal suture and Bones. (Scale bar, 200 µm.) (E) Violin plot and spatial expression of TGF-β signaling module scoring among Cont and TrkAF592A calvaria, including TGF-β activators, TGF-β inhibitors, and overall TGF-β signaling. (F) Heatmap of TGF-β activator and inhibitor genes expressed across SpatialTime among Cont and TrkAF592A calvaria. (G) Overall TGF-β activation based on module scoring across SpatialTime, from Mid suture to flanking Bone among Cont and TrkAF592A calvaria. Relative SpatialTime regions aligning to the Mid suture, OF, and flanking Bone are denoted above graph. n = 5 to 7 mice/genotype. (H) Canonical TGF-β signaling, as assessed by immunofluorescent staining for p-SMAD2 using coronal cross-sections of the sagittal suture and bones. (Scale bar, 200 µm.)
In contrast to the spatially restricted pattern of BMP/TGF-β signaling blunted by TrkA inhibition, FGF signaling, another pathway previously implicated in suture maintenance, was found to be uniformly depressed in TrkAF592A mice (SI Appendix, Fig. S5A). However, unlike the previously mentioned pathways, FGF ligands derived from within the suture mesenchyme (SI Appendix, Fig. S5B, red box) showed robust down-regulation. These data suggest that FGF signaling disruption across the suture (SI Appendix, Fig. S5C) may be a secondary effect of sensory denervation, potentially linked to changes in the proliferation described above. Overall, these data suggest that TrkA+ sensory nerves are essential in maintaining signaling gradients within the calvarial suture indispensable for facilitating suture growth and the expansion of the cranial vault, while simultaneously maintaining midline suture patency, thereby preventing premature stem cell depletion and suture closure.
Sensory Nerve-Derived FSTL1 Functions to Maintain MPCs in a Proliferative, Undifferentiated State.
The data described above together suggested that the ingrowth of sensory nerve axons influence cell fate in the cranial suture through soluble signals. To identify candidate molecules, we conducted RNA-seq on DRG cell bodies cultured in the central chamber of our microfluidic devices with or without axonal contact to primary bone marrow stromal cells (BMSCs) in the outer device chambers (SI Appendix, Fig. S6A). A total of 753 up-regulated and 131 down-regulated differentially expressed genes (DEGs) were detected (Dataset S3). Principal component analysis (PCA) and unsupervised, hierarchical clustering showed the clear grouping of sample replicas (SI Appendix, Fig. S6 B and C). Pathway analysis was next conducted on DEGs (SI Appendix, Fig. S6D). DRG axonal–BMSC contact was associated with the up-regulation of genes enriched for the positive regulation of cell migration, proliferation, and epithelial to mesenchymal transition, while down-regulated genes were enriched for the negative regulation of proliferation and pathways related to cell stress and neural apoptosis within the neural cell bodies. In addition, and consistent with our SpatialTime analysis, several pathways related to BMP and TGF-β were observed. Based on our in vivo spatial transcriptomics and these RNA-seq results, we focused on major modulators of the BMP/TGF-β pathway, identifying the up-regulation of the inhibitors Bambi, Ltbp1, Fstl1, Inhba and Fst (SI Appendix, Fig. S6E).
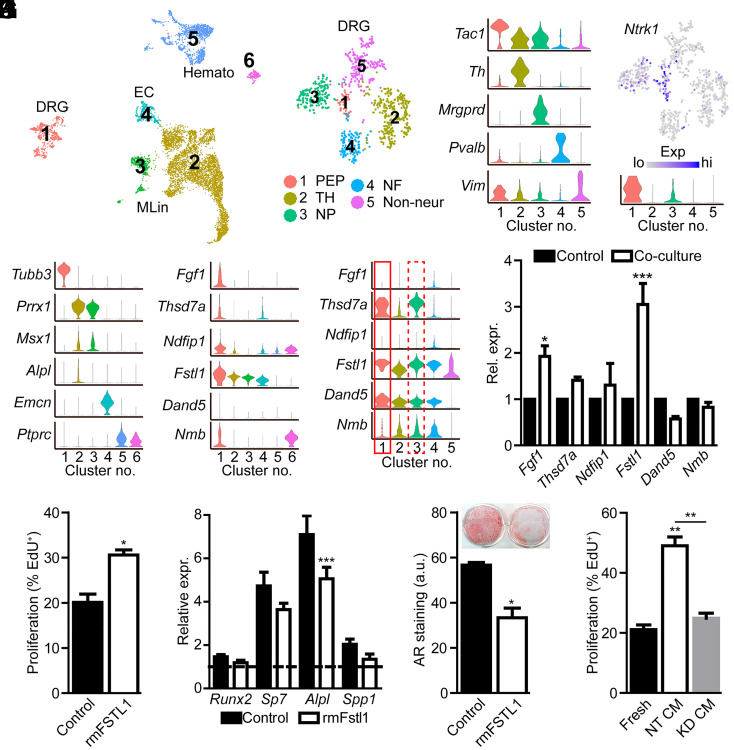
To gain additional support for our experiment findings, we accessed publicly available scRNAseq datasets from long bone and calvaria. DRG resident cells (19) were combined with datasets which comprise cells of the long bone and marrow cavity (20), including endothelial and smooth muscle/pericytes, cells of the mesenchymal lineage, and cells of the hematopoietic lineage (SI Appendix, Fig. S6F). Differential gene expression identified 2,686 genes showing an enriched expression in DRG neurons relative to other cellular components of the bone marrow compartment (Dataset S4). To identify the neural, enriched genes within the context of the calvaria, similar computational analyses were conducted on a recently published scRNAseq dataset derived from the developing, calvarial frontal suture (21). DRG resident cells (19) (cluster 1) were combined with datasets derived from E18.5 calvariae (21), including MPCs (clusters 2 and 3), endothelial cells (cluster 4), and cells of the hematopoietic lineage (clusters 5 and 6) identified through marker gene expression (Fig. 5 A and B). Differential gene expression identified 2,515 genes showing an enriched expression in DRG neurons relative to other cellular components of the calvaria (Dataset S5). Next, gene lists derived from our single-cell long bone and calvaria analyses were filtered for those genes encoding curated secreted proteins (22), identifying 72 candidate genes in the long bone dataset (Dataset S6) and 69 candidate genes from the calvaria dataset (Dataset S7). These lists included known neuropeptides Tac1 (encoding substance P) and Calca (encoding calcitonin gene-related polypeptide). Gene enrichment showed striking overlap in DRG-enriched genes in the context of bone marrow or calvarial cells, both overall, as well as specifically, in those genes encoding secreted proteins (SI Appendix, Fig. S6H). Finally, literature review was used to identify those proteins previously implicated in regulating proliferation, osteo- or chondrogenesis, and maintenance of stemness. Of the genes encoding secreted proteins, violin plots were used to show the relative expression of the six identified candidate genes Fgf1, Thsd7a, Fstl1, Ndfip1, Dand5, and Nmb within our reconstructed calvaria (Fig. 5C) and long bone (SI Appendix, Fig. S6I) environments.
Fig. 5.
FSTL1 is secreted by TrkA+ sensory nerves and regulates MPC proliferation and differentiation. (A) UMAP projection of merged scRNAseq datasets of E18.5 calvarial bone resident cells (21) and lumbar DRG neurons (19). (B) Violin plot of cluster marker genes. (C) Candidate secreted factors showing the enriched expression in DRG neurons relative to other cell populations. (D) Subclustering of DRG cells into distinct neural subtypes. PEP, peptidergic nociceptors; TH, tyrosine hydroxylase containing; NP, nonpeptidergic nociceptors; and NF, neurofilament. (E) Violin plot of DRG subpopulation marker genes. (F) Feature and violin plot showing the expression of TrkA (encoded by Ntrk1) by PEP and NP neural subtypes. (G) Violin plot showing the expression of candidate genes in different neural subtypes. (H) Expression of candidate genes showing the enriched expression in TrkA+ neural subtypes in control DRG cell bodies or following direct, axonal coculture with MPCs. (I) Proliferation of MPCs following treatment with rmFSTL1 (250 ng/mL) for 48 h. (J) Gene expression following early osteogenic induction with or without rmFSTL1. The dotted line denotes values obtained at day 0 of osteogenic induction. (K) Alizarin red (AR) staining and quantification of MPCs following 21 d of osteogenic differentiation with or without rmFSTL1 (250 ng/mL). n = 4. (L) Proliferation of MPCs following treatment with fresh neural media or media condition by nontargeting (NT CM) or Fstl1-targeted (KD CM) siRNA. n = 3. Graphs represent average values ± SD, *P < 0.05, **P < 0.01, and ***P < 0.001. a.u., arbitrary units; EC, endothelial cells.
As DRGs house both TrkA-expressing and nonexpressing neurons, we further refined this list to those secreted factors predominantly produced by TrkA+ neurons. To accomplish this, DRG resident cells were further subclustered to identify neural subpopulations (Fig. 5D), from which peptidergic, and to a lesser extent nonpeptidergic, nociceptive neurons were found to express the TrkA receptor (encoded by Ntrk1) (Fig. 5E). The specific inhibition of TrkA+ neural subtypes was confirmed using predictive scoring, in which gene expression is used to predict how effective each single-cell cluster maps to each spatial spot (SI Appendix, Fig. S7). This analysis confirmed a significant and specific reduction in DRG-predicted spatial spots within the calvarial suture (SI Appendix, Fig. S7A) and further demonstrated that this DRG reduction was specific for TrkA+ peptidergic and nonpeptidergic nociceptors (SI Appendix, Fig. S7B). Violin plots show that only four of the six candidate genes met the criteria of being primarily expressed in TrkA+ neural subtypes (Fig. 5F). Using microfluidic coculture system, qPCR was conducted on DRG cell bodies with or without coculture with MPCs (Fig. 5G).
FSTL1 functions as an antagonistic binding partner of BMP ligands (23) and has previously been implicated in the regulation of cellular proliferation and migration in nonskeletal cells (24) and, as such, was selected as a lead candidate for further investigation. FSTL1 was confirmed to be produced in DRG protein lysates (SI Appendix, Fig. S8A) and localized to within the axons of DRG neurons in vitro (SI Appendix, Fig. S8B). The treatment of calvarial MPCs with rmFSTL1 significantly induced cell proliferation under normal growth conditions (Fig. 5H), which coincided with a reduction in BMP and TGF-β signaling as assessed by p-Smad1/5/9 and p-Smad2 levels, respectively (SI Appendix, Fig. S8C). Under osteogenic induction conditions, treatment with rmFSTL1 was found to modestly inhibit the induction of osteogenic marker genes at day 7 (Fig. 5I), as well as significantly inhibited terminal osteoblast differentiation and mineralization at day 21 of differentiation (Fig. 5J). To confirm FSTL1 as the neural-derived factor through which sensory nerves exert their effects, DRGs were subjected to electroporation and treated with nontargeting small interfering RNA (siRNA) or siRNA targeted to Fstl1. Knockdown was confirmed by qPCR at several time points after electroporation (SI Appendix, Fig. S8D). Conditioned media collected from nontargeted or Fstl1 knockdown cells, as well as fresh neural media, was applied to calvarial MPCs and the proliferation measured by EdU uptake (Fig. 5K). While treatment with nontargeted conditioned media showed a robust increase in MPC proliferation, consistent with our previous results, conditioned media derived from Fstl1-silenced DRGs failed to show any significant, proliferative induction compared to fresh media controls. These data further confirm FSTL1 as the main neural factor through which recipient cell proliferation is modulated.
Discussion
In this study, we report a previously unanticipated requirement for sensory nerve signaling in mouse cranial bone formation (Fig. 6). We show that TrkA+ sensory nerve axons terminate in the calvarial suture mesenchyme, a critical niche for osteogenic precursors, in which they preferentially approximate proliferating suture resident cells. The inhibition of sensory nerve ingrowth by blocking TrkA in vivo reduced proliferative activity of the suture mesenchyme, increased MARs along the flanking parietal bone fronts, and caused premature suture closure. These findings indicate that sensory nerves function to preserve suture patency by delivering soluble factors such as FSTL1 that support the proliferative capacity of the suture mesenchyme and inhibit downstream differentiation cues (e.g., BMP signaling).
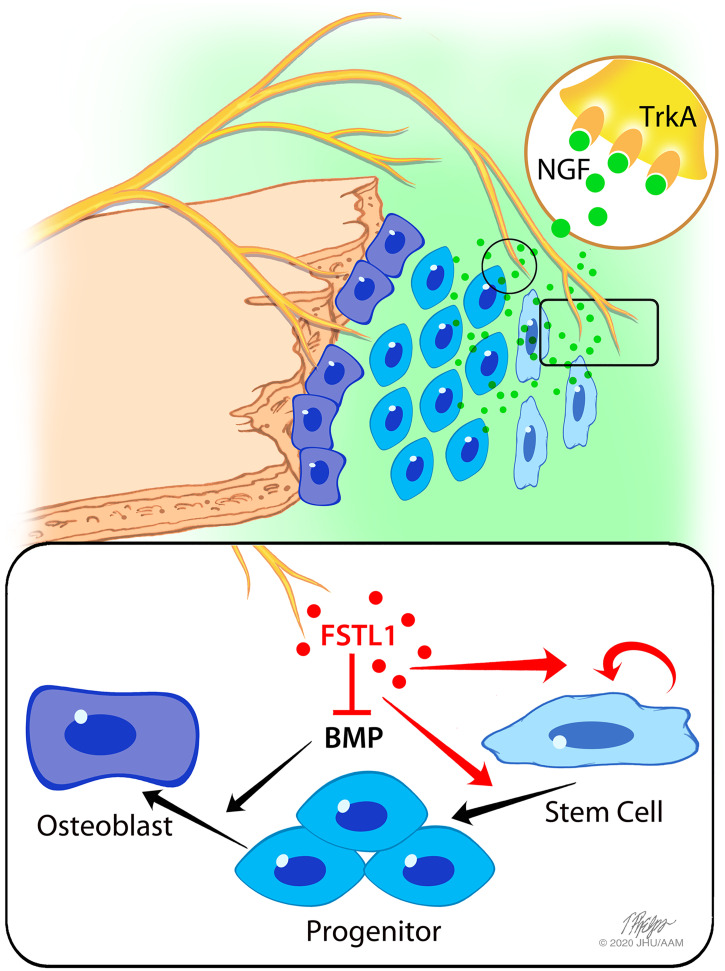
Fig. 6.
Overview of sensory, nerve-mediated regulation of suture plasticity. The suture is maintained by a balance of expansive progenitor cell proliferation and inward closing mineralization along the bony plates. TrkA+ sensory nerves, guided to the cranial suture by NGF, secrete factors such as FSTL1, regulating progenitor activation and expansion and inhibiting BMP-mediated terminal differentiation. TrkA inhibition impairs sensory innervation and reduces suture mesenchyme expansion, resulting in premature suture closure.
As described above, the flat bones that compose the mammalian skull vault develop and expand through a process termed intramembranous ossification. A key anatomical component in this process is the fibrous suture, which serves the dual purposes of permitting cranial bone expansion while simultaneously serving as a reservoir of MPCs. For this process to function correctly, the bulk of the suture mesenchyme must be maintained in a proliferative, undifferentiated state, with active, bone-forming cells restricted to the bony fronts of the cranial bones (4). This dynamic architecture of the developing suture presented a challenge to identify the signaling mechanisms through which sensory nerve signaling impacts suture mesenchymal patency. To overcome these difficulties, we applied spatial transcriptomics, a technique in which spatial barcoding allows transcriptomic data to be mapped to the histological region of origin.
Spatial transcriptomic analysis of calvaria from control mice revealed a strong correlation between sensory innervation and proliferative activity, as well as the expression of progenitor markers (e.g., Gli1, Twist1, etc.), specifically within the midline suture mesenchyme. Furthermore, the expression analysis of MPCs following direct coculture with DRG nerve axons showed the up-regulation of several transcription factors previously linked to the induction of stemness and reprogramming (25). This tight spatial association of these proliferative markers was lost in TrkAF592A calvaria following the inhibition of TrkA signaling, resulting in a sharp decline in suture cell proliferation and culminating in premature suture closure. These findings suggest that sensory nerves preserve suture patency by maintaining mesenchymal cells in a primed, undifferentiated state. The suture mesenchyme consists of a heterogenous cell population. Given the current resolution of spatial transcriptomics, our studies are unable to define whether sensory nerves affect all cellular subtypes within the suture mesenchyme or whether these effects are preferentially disposed to one or more cellular subtypes. Further work is required to determine the cell type–specific effects of sensory innervation. However, our results are compatible with recent findings in the mouse femurs (10), indicating a more generalized role for sensory nerve TrkA signaling in regulating the differentiation status of MPCs.
Another important finding made possible by the spatial transcriptomics approach was the striking, spatially restricted pattern of TGF-β and BMP pathway activation across the developing cranial suture. In this regard, a number of previous studies have implicated TGF-β/BMP signaling in regulating suture patency, closure, and fusion through the use of the altered expression of ligands, receptors, or signaling inhibitors (26). For example, mutations to the TGF-β receptors resulted in premature suture closure (27), while the modulation of the TGF-β ligands had varying effects, with TGFB1 promoting bony fusion (28), TGFB2 promoting suture closure (29), and TGFB3 maintaining suture openness (28, 30). Conversely, BMP signaling is known to promote osteogenic differentiation and suture closure (31), whereas BMP inhibitors, such as Noggin, specifically inhibited BMP signaling within the midline suture (32). Our data using the SpatialTime analysis confirm and advance these previous studies, suggesting the presence of a TGF-β/BMP activation wave which initiates in the late midline/early OF and extends into the bony fronts. The fact that this pattern was both blunted and shifted toward the midline suture in TrkAF592A mice strongly implicates sensory nerve signals as regulators of these TGF-β/BMP signaling patterns. Moreover, our finding that the inhibition of TrkA signaling postnatally also resulted in premature suture closure, combined with our previous observations that sutures, which remain patent into adulthood, are extensively innervated (14) strongly suggests that sensory nerves function throughout life to preserve suture patency.
The further interrogation of downstream components of the TGF-β/BMP signaling pathway using sequencing-based approaches identified Fstl1 as a neural secreted factor involved in the maintenance of suture patency. Fstl1 expression is regulated by TGF-β and has previously been shown to regulate numerous cellular processes, including proliferation, migration, and survival (24). In the context of bone, FSTL1 is a known inhibitor of BMP signaling, directly binding to BMP4 and inhibiting downstream activation (23), similar to the previously described mechanism through which Noggin maintains suture patency. In addition, in vitro studies have demonstrated a link between FSTL1 and osteoclastogenesis (33, 34). Knockout (KO) mice, though lethal early postnatal due to the improper formation of the respiratory system (23), showed skeletal defects, including the shortening and bending of the limbs, curvature of the spine, and displaced head positioning (35). Though no abnormal suture phenotype has been described in these Fstl1 KO mice, it is likely that other gross skeletal abnormalities may have masked any potential alterations in the suture mesenchyme. Our demonstration that FSTL1 is secreted by sensory nerves and effectively mimicked mitotic and anti-osteogenic effects of sensory nerves in cocultured MPCs are compatible with the conclusion that this BMP inhibitor functions as a key, neural-derived regulator of suture plasticity. The fact that the knockdown of Fstl1 in DRGs inhibited these actions further supports this conclusion. However, given the complex nature of suture maintenance and expansion, it is important to emphasize the likely involvement of other factors in this process. Indeed, our SpatialTime analysis revealed the regional-specific expression of several genes previously linked to the regulation of osteogenic differentiation, including FGF ligands (36), members of the semaphorin/neuropilin family (37), FOXO/FOXP (38), Notch target genes (39), SOX transcription factors (40), and the Wnt-regulating Frizzled family (41) within the central suture. While many of the genes did not show a robust change in response to TrkA inhibition, their direct or indirect effects on suture maintenance, patency, and differentiation cannot be discounted. In addition, while this study has focused on the role of sensory nerves in suture patency and closure driven by intramembranous ossification, the role of sensory nerves in suture fusion, mediated by endochondral ossification, remains unknown. Further studies are needed to characterize the normal regulation of suture patency and fusion and how nerves and other surrounding tissue may regulate this patterning.
Materials and Methods
Animals.
All procedures involving mice were approved by the Institutional Animal Care and Use Committee of The Johns Hopkins University (protocol M015M118). NGF-eGFP mice, which express eGFP under the control of the full-length mouse Ngf promoter, were generously donated by the Kawaja Laboratory (42). TrkAF592A mice are commercially available (Jackson Laboratory; stock No. 022362), and dosing with 1NMPP1 (Aurora Analytics LLC) (15) was administered based on previously validated methods (11–14). For the developmental inhibition of TrkA, female heterozygous animals, crossed with TrkAF592A homozygote males, were given water supplemented with 1NMPP1 at a final concentration of 200 µM from the time of conception until birth. Heterozygote littermates were used as controls, and mothers were used for no more than two litters. For early postnatal development, mice were injected intraperitoneally with 5 mM 1NMPP1 at a dose of 17 mg/kg body weight twice weekly from P6 to P21.
Tissue Preparation and Imaging.
Calvariae were collected and fixed for 4 to 6 h in 4% paraformaldehyde at 4 °C then rinsed in phosphate-buffered saline. For thick sections, samples were incubated in 20% sucrose and 2% polyvinylpyrrolidone at 4 °C overnight before embedding in the same solution with an additional 8% gelatin. The 100-µm thick sections were prepared and stained for TUBB3 (Abcam, ab18207, 1:1,000), Ki67 (Abcam, ab16667, 1:100), p-SMAD2 (Cell Signaling, 3108T, 1:200), or p-SMAD1/5/9 (Cell Signaling, 13820T, 1:150). Sections were imaged using a Zeiss LSM 880 laser-scanning confocal microscope (Zeiss) with a z-step of 3 µm and displayed as maximum intensity projections. For in vivo EdU labeling, newborn pups were injected with 1.6 mg/kg EdU (Baseclick) 3 h before euthanasia. Sections were subsequently stained according to manufacturer protocol. In vivo proliferation was quantified as the number of EdU+ puncta/mm2, while TUBB3 area was quantified as the percentage of the suture area labeled with TUBB3 following the thresholding and binarization of a signal using ImageJ. Quantification was conducted on maximum intensity projections. Whole-mount samples were prepared as previously described (43) and imaged using a dissection microscope fitted with a Leica MC 170 HD camera. Individual images were acquired to quantify posterior frontal and sagittal sutures. Suture width was quantified as the total suture area divided by the suture length.
Dynamic Histomorphometry.
For the assessment of dynamic histomorphometry, pregnant females were injected with calcein (10 mg/kg; Sigma-Aldrich C0875) at E17.5 and alizarin red (30 mg/kg; Sigma-Aldrich A3882) at E18.5. At P0, mice were euthanized and whole calvaria dissected out. Calvaria were placed in a Petri dish containing a glass coverslip bottom and imaged using a confocal microscope. To determine MARs, approximate leading-edge boundaries of each label along the parietal bone flanking the sagittal suture were traced, and the average distance was calculated using Bioquant (Bioquant Image Analysis Corporation).
Micro-CT.
Bones were dissected free of soft tissue and evaluated using a SkyScan1275 (Bruker) high-resolution micro-CT imaging system. Each bone was scanned separately at 65 kV and 153 μA with a 0.5-mm aluminum filter to obtain a 10.5-μm voxel size. Scan slices were acquired in the frontal plane by placing the bone parallel to the z-axis of the scanner. NRecon (Bruker) was used to reconstruct images using a beam-hardening correction of 40%, and quantitative analysis was performed using CTAn (Bruker) in accordance with the recommendations of the American Society for Bone and Mineral Research (44). To quantify suture closure, the sagittal suture was isolated on thresholded images, and the number of slices in which flanking parietal bones were in direct contact were quantified as a percent of the total number of slices. Suture width at P21 was calculated by selecting the sagittal suture and flanking bony fronts, applying a 2D shrink wrap region of interest with a stretch factor of 50 pixels, and then calculating a 2D plate model trabecular separation according to its relative position along the anterior–posterior axis.
Cell Culture.
DRG were harvested from mixed sex C57BL/6J mice at embryonic age 13.5 into ice-cold culture medium (neurobasal media supplemented with B27, 1× penicillin/streptomycin, anti-mitotics [20 μM 5-fluoro-2-deoxyuridine 20 μM uridine], 1× Glutamax, and 50 ng/mL NGF) (Thermo Fisher Scientific) (11). Next, DRGs were digested with 1 mg/mL collagenase A (Roche) at 37 °C for 15 min and then with 0.05% trypsin-EDTA at 37 °C for 7 min. Following this, DRGs were washed three times with culture medium and dissociated by trituration with a 1-mL pipette tip. The dissociated neurons were plated into the microfluidic devices at a density of 104 neurons/cm2. Primary mesenchymal cells were isolated by either flushing the central marrow of hind limbs from 8- to 12-wk-old mice (BMSCs used for RNAseq) or through the digestion of early postnatal calvaria, as previously described (45) (MPCs, used for in vitro studies). These cells consist of a heterogenous mesenchymal cell population, a portion of which possesses the ability to form colonies repeatedly after passaging and differentiate, primarily into mineralizing osteoblasts but also down the chondrogenic and adipogenic lineages (SI Appendix, Fig. S9) (4, 46). For simplicity, these cells have been referred to as MPCs, recognizing that not all cells may have these attributes. MPCs were cultured in alpha minimum essential media (αMEM) supplemented with 10% fetal bovine serum (FBS) (Corning). Osteogenic cells were used at passage 1 to 2 for all experiments. Microfluidic platforms, consisting of a poly-dimethylsiloxane device bonded to a glass coverslip, were generated, as previously described (47), and incubated overnight with a 100 μg/mL solution of poly-D-lysine (Sigma-Aldrich) and 10 µg/mL laminin (Gibco). For proliferation assays, calvarial MPCs were seeded at ∼40% confluency, while for osteogenic differentiation, MPCs were seeded at high density and allowed to reach ∼90% confluency. Osteogenic differentiation was conducted by supplementing the media with 10 mM β-glycerol phosphate and 50 μg/mL ascorbic acid (Sigma-Aldrich). To visualize proliferation, cells were treated with 20 µM EdU for 24 h. To calculate EdU–axon distance, TUBB3+ axons were thresholded and dilated to the appropriate distance, followed by mask-based subtraction to identify the number of EdU+ nuclei within each of the stated distances. To generate conditioned media, DRGs were allowed to grow for 2 d in 12-well plates and then cultured in 500 µL of fresh neurobasal media for 24 h. Conditioned media was then sterile filtered at 0.22 µm and frozen at −80 °C until use. For FSTL1 studies, cells were treated with 250 ng/mL recombinant mouse FSTL1 (R&D Biosystems) supplemented into the culture media immediately (proliferation assays) or upon switching to osteogenic inductive media (differentiation assays). For the detection of FSTL1 protein, a rat anti-mouse FSTL1 antibody (R&D Biosystems, MAB17381) was used at 1:100 (immunofluorescence). For fibroblast colony forming units assays (48), primary MPCs were isolated and cultured in regular growth media for 24 h. Next, cells were passaged at density of 500 cells/cm2 and cultured in osteogenic for 10 d. Colonies were then passaged and again reseeded at 500 cells/cm2. Colonies were stained with crystal violet prior to imaging. For lineage induction, primary MPCs were cultured until ∼80% confluent and then grown in either growth or induction conditions: osteogenic media (αMEM, 10% FBS, 50 µg/mL ascorbic acid, and 10 mM β-glycerolphosphate) for 14 d, chondrogenic media (Dulbecco's modified Eagle medium [DMEM], 100 nM dexamethasone, 0.2 mM ascorbic acid, 1 mM sodium pyruvate, 1× insulin, transferrin, and selenous acid supplement, and 10 ng/mL TGF-β1) for 21 d, and adipogenic media (DMEM, 10% FBS, 1 uM dexamethasone, 1ug/mL insulin, and 500 uM IBMX) for 14 d.
RNA Analysis.
DRGs were seeded in the middle chamber of a microfluidic device and allowed to grow to the outer chambers for 4 d. Neural growth media was replaced with mesenchymal growth media in outer chambers only and allowed to acclimatize for 24 h. Next, 105 MPCs (qPCR) or BMSCs (RNAseq) were seeded into the outer chamber and cultured with axons for 48 h. RNA from the DRG cell bodies and mesenchymal cells were collected using the RNeasy mini kit (Qiagen). DRG cell bodies with axons cultured in growth media and MPCs cultured in microfluidic devices without axonal contact were used as controls. For total RNA-seq, only samples with an RNA integrity number value of >8 were used. Following library preparation and Illumina indexing in accordance with manufacturer protocols, samples were subjected to pair-ended sequencing with a read length of 150 bp and a minimum sequencing depth of 20 million reads on an Illumina NovaSeq6000. Sequencing alignment was carried out against the mm10 reference genome following quality controls for error rate distribution, guanine–cytosine content distribution, and removal of low-quality reads. Differential gene expression was analyzed using DESeq2. Significantly regulated genes were identified using a log2-fold change cutoff of 0.25 and a P value <0.05 owing to the relatively low contact between many DRG cell bodies and recipient cell population. PCA was conducted using the 500 genes with the greatest intersample variance and displayed using ClustVis (49). For qPCR, RNA was reverse transcribed using an iScript cDNA Synthesis Kit (Bio-Rad), and cDNA was amplified under standard PCR conditions using iQ SYBR Green Supermix (Bio-Rad). All cDNA samples were run in triplicate, averaged, and normalized to endogenous 18S ribosomal gene expression levels.
Spatial Transcriptomics.
Spatial Transcriptomics was conducted using the Visium Spatial Gene Expression System (10X Genomics). P0 calvaria were harvested and stacked together, simultaneously fresh frozen and embedded in OCT, and then stored at −80 °C. Samples were cut at −30 °C at a thickness of 12 µm and placed directly on the Visium slide. Tissue optimization and gene expression assays were carried out according to manufacturer’s protocol. Briefly, slides were fixed in methanol at −20 °C, dried with isopropanol, and stained using a modified H&E protocol. A tile scan image of all reaction areas was generated using a Leica DM6 B microscope (Leica Microsystems Inc). For optimization, enzymatic permeabilization was conducted for 0 to 40 min, followed by first-strand cDNA synthesis using fluorescent nucleotides and subsequent tissue removal. The whole-tissue optimization slide was again imaged using a Leica microscope and standard Texas red filter cube. To determine optimal permeabilization time, a region of interest was placed around the suture and fluorescent intensity obtained, in which overall total fluorescence was indicative of total mRNA recovery and SD in pixel-level fluorescence throughout the suture indicative of diffusion. Optimal permeabilization time (21 min) was used during library generation using a gene expression slide, with cDNA synthesis conducted with nonfluorescent nucleotides. cDNA is released from the slide and amplified, based on relative cDNA abundance determined by qPCR. Library preparation, clean up, and indexing was conducted using standard procedures. Samples were subjected to pair-ended sequencing using an Illumina HiSeq generating ∼400 M reads. Alignment and demultiplexing was conducted using the SpaceRanger pipeline (raw and processed sequencing data have been deposited in Gene Expression Omnibus [GEO] under accession code GSE174313) with subsequent analyses conducted using Seurat (50) in R. Pathway analysis was conducted using Database for Annotation, Visualization, and Integrated Discovery (51). Pathway scoring was determined using the AddModuleScore function in Seurat using gene lists obtained from validated Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (see SI Appendix, Table S1 for gene lists used for BMP, TGF-β, and FGF signaling). Overall, pathway activation was calculated as the module scoring of activation minus inhibition. For overall suture analysis, each sample was first cropped to include the entire suture complex and ∼2 to 3 spots (∼100 to 200 µm) of flanking parietal bone.
SpatialTime.
For SpatialTime analysis, a reference spatial spot is manually selected from within the central suture based on its histological location. Distances from this central reference spatial spot and all other spots within the cropped suture/flanking bone region are computed using the 2D spatial coordinates and scaled so that the most distant spot has a SpatialTime value of 1. Once the SpatialTime values is calculated for each suture, samples are concatenated to increase statistical power and visualize gene expression across an “average” suture. A slingshot approach is next applied to identify spatially dynamic features (52). This method consists of two steps, the first step is to construct a global lineage through identifying paths using the minimal spanning tree algorithm to connects adjacent clusters. The second step fits the smooth curves using the simultaneous principal curves method, and the SpatialTime are computed by orthogonal projection of the spatial dots. Subsequently, we fit the general additive model using binomial distribution to assess the association between the SpatialTime and the gene expression and identify the spatially dynamic genes.
Protein Analysis.
Microdissected DRGs (L2 to L5) were placed into ice-cold Triton X-100 lysis buffer containing both a protease inhibitor (Sigma-Aldrich, P8340) and a phosphatase inhibitor (Sigma-Aldrich, P2850). DRGs were then minced in ice-cold lysis buffer followed by incubation for 5 min on ice and centrifugation for 10 min at 12,000 rpm for protein extraction. For cells, primary MPCs were seeded in 6-well plates at a density of 50,000 cells/well and cultured in growth media supplemented with or without 250 ng/mL recombinant mouse FSTL1 for 2 d. Cells were then rinsed and subjected to protein extraction. Blots were probed using antibodies against FSTL1 (1:250, MAB17381, R&D Systems), p-SMAD1/5/9 (1:1,000, 13820, Cell Signaling), total SMAD1 (1:1,000, 9743, Cell Signaling), p-SMAD2 (1:1,000, 44–244G, Invitrogen), total SMAD2/3 (1:1,000, 8685, Cell Signaling), or GAPDH (1:1,000, 10494-I-AP, ProteinTech).
siRNA Knockdown Experiments.
Adult mouse DRG neuronal culture and electroporation were performed as previously described (53) with minor adjustments. Lumbar DRGs from 8- to 10-wk-old mixed sex C57BL/6J mice were harvested into ice-cold MEM with 10% FBS and 1× penicillin/streptomycin. The DRGs were then digested with 1 mg/mL type I collagenase (Thermo Fisher Scientific 17018017) and 5 mg/mL dispase II (Thermo Fisher Scientific 17105041) in MEM at 37 °C for 70 min and dissociated into single cells by trituration with a 1-mL pipette tip in the media mentioned above. The dissociated cells were filtered with a 70-μm cell strainer and centrifuged at 500 rcf for 5 min. The pelleted cells were then resuspended with 100 μL electroporation buffer (Lonza VPG-1001) containing 0.2 nmol nontargeting control siRNA (Dharmacon L-001810-10-05) or siRNA against Fstl1 mRNA (Ambion 4390771, ID: s66251), transferred to a 2.0-mm electroporation cuvette and electroporated with Nucleofector II (Lonza) following manufacturer’s manual. After electroporation, the cells were immediately mixed with the desired volume of prewarmed MEM with 5% FBS, 1× penicillin/streptomycin (Thermo Fisher Scientific 15140122), 1× Glutamax (Thermo Fisher Scientific 35050061), and anti-mitotic reagents (20 μM 5-fluoro-2-deoxyuridine and 20 μM uridine, and Sigma-Aldrich F5030 and U3750) and seeded into 12-well plates precoated with 100 μg/mL poly-D-lysine (Sigma-Aldrich P6407) and 10 μg/mL laminin (Thermo Fisher Scientific 23017015). The media was fully changed the following morning.
Statistical Analysis.
Data are expressed as means ± SD. All statistical analyses were conducted using Prism (GraphPad). Comparisons between groups were analyzed by either a Student’s t test (paired or unpaired as stated) or two‐way ANOVA (with repeated measurements or not as appropriate), with a Bonferroni’s posttest as stated. P values less than 0.05 were considered to be statistically significant. SpatialTime graphs were generated by generating a smoothed curve averaged with 30 neighbors using Prism.
Acknowledgments
This work was supported by NIH grant AR068934 and DE027922. T.L.C. receives partial salary support through a Senior Research Career grant from the Veterans Administration. X.-W.W. was supported by K99 EY031742 from the National Eye Institute. We thank Dr. Mei Wan for supplying the p-SMAD2 antibody.
Footnotes
Competing interest statement: 10X Genomics provided supplies and expert consultation for the study. A.W.J. is a paid consultant for Novadip and Lifesprout LLC. This arrangement has been reviewed and approved by Johns Hopkins University in accordance with its conflict-of-interest policies.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2103087118/-/DCSupplemental.
Data Availability
Spatial Transcriptomics data have been deposited in GEO (GSE174313). Previously published data were used for this work GSE59739, FB00001013, and GSE145477 (19–21). All other study data are included in the article and/or supporting information.
References
- 1.Ferguson J. W., Atit R. P., A tale of two cities: The genetic mechanisms governing calvarial bone development. Genesis 57, e23248 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lenton K. A., Nacamuli R. P., Wan D. C., Helms J. A., Longaker M. T., Cranial suture biology. Curr. Top. Dev. Biol. 66, 287–328 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Jiang X., Iseki S., Maxson R. E., Sucov H. M., Morriss-Kay G. M., Tissue origins and interactions in the mammalian skull vault. Dev. Biol. 241, 106–116 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Zhao H., et al., The suture provides a niche for mesenchymal stem cells of craniofacial bones. Nat. Cell Biol. 17, 386–396 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellus G. A., et al., Identical mutations in three different fibroblast growth factor receptor genes in autosomal dominant craniosynostosis syndromes. Nat. Genet. 14, 174–176 (1996). [DOI] [PubMed] [Google Scholar]
- 6.Opperman L. A., Nolen A. A., Ogle R. C., TGF-beta 1, TGF-beta 2, and TGF-beta 3 exhibit distinct patterns of expression during cranial suture formation and obliteration in vivo and in vitro. J. Bone Miner. Res. 12, 301–310 (1997). [DOI] [PubMed] [Google Scholar]
- 7.Roth D. A., et al., Studies in cranial suture biology: Part I. Increased immunoreactivity for TGF-beta isoforms (beta 1, beta 2, and beta 3) during rat cranial suture fusion. J. Bone Miner. Res. 12, 311–321 (1997). [DOI] [PubMed] [Google Scholar]
- 8.Jabs E. W., et al., A mutation in the homeodomain of the human MSX2 gene in a family affected with autosomal dominant craniosynostosis. Cell 75, 443–450 (1993). [DOI] [PubMed] [Google Scholar]
- 9.El Ghouzzi V., et al., Mutations within or upstream of the basic helix-loop-helix domain of the TWIST gene are specific to Saethre-Chotzen syndrome. Eur. J. Hum. Genet. 7, 27–33 (1999). [DOI] [PubMed] [Google Scholar]
- 10.Hu B., et al., Sensory nerves regulate mesenchymal stromal cell lineage commitment by tuning sympathetic tones. J. Clin. Invest. 130, 3483–3498 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomlinson R. E., et al., NGF-TrkA signaling by sensory nerves coordinates the vascularization and ossification of developing endochondral bone. Cell Rep. 16, 2723–2735 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomlinson R. E., et al., NGF-TrkA signaling in sensory nerves is required for skeletal adaptation to mechanical loads in mice. Proc. Natl. Acad. Sci. U.S.A. 114, E3632–E3641 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Z., et al., Fracture repair requires TrkA signaling by skeletal sensory nerves. J. Clin. Invest. 129, 5137–5150 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyers C. A., et al., A neurotrophic mechanism directs sensory nerve transit in cranial bone. Cell Rep. 31, 107696 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen X., et al., A chemical-genetic approach to studying neurotrophin signaling. Neuron 46, 13–21 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Ståhl P. L., et al., Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Nestorowa S., et al., A single-cell resolution map of mouse hematopoietic stem and progenitor cell differentiation. Blood 128, e20–e31 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trapnell C., et al., The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Usoskin D., et al., Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat. Neurosci. 18, 145–153 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Zhong L., et al., Single cell transcriptomics identifies a unique adipose lineage cell population that regulates bone marrow environment. eLife 9, e54695 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holmes G., et al., Integrated transcriptome and network analysis reveals spatiotemporal dynamics of calvarial suturogenesis. Cell Rep. 32, 107871 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meinken J., Walker G., Cooper C. R., Min X. J., MetazSecKB: The human and animal secretome and subcellular proteome knowledgebase. Database (Oxford) 2015, bav077 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geng Y., et al., Follistatin-like 1 (Fstl1) is a bone morphogenetic protein (BMP) 4 signaling antagonist in controlling mouse lung development. Proc. Natl. Acad. Sci. U.S.A. 108, 7058–7063 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mattiotti A., Prakash S., Barnett P., van den Hoff M. J. B., Follistatin-like 1 in development and human diseases. Cell. Mol. Life Sci. 75, 2339–2354 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takahashi K., Yamanaka S., Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Kosty J., Vogel T. W., Insights into the development of molecular therapies for craniosynostosis. Neurosurg. Focus 38, E2 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Loeys B. L., et al., A syndrome of altered cardiovascular, craniofacial, neurocognitive and skeletal development caused by mutations in TGFBR1 or TGFBR2. Nat. Genet. 37, 275–281 (2005). [DOI] [PubMed] [Google Scholar]
- 28.James A. W., Xu Y., Lee J. K., Wang R., Longaker M. T., Differential effects of TGF-beta1 and TGF-beta3 on chondrogenesis in posterofrontal cranial suture-derived mesenchymal cells in vitro. Plast. Reconstr. Surg. 123, 31–43 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lai C.-F., Cheng S.-L., Signal transductions induced by bone morphogenetic protein-2 and transforming growth factor-beta in normal human osteoblastic cells. J. Biol. Chem. 277, 15514–15522 (2002). [DOI] [PubMed] [Google Scholar]
- 30.Opperman L. A., Chhabra A., Cho R. W., Ogle R. C., Cranial suture obliteration is induced by removal of transforming growth factor (TGF)-beta 3 activity and prevented by removal of TGF-beta 2 activity from fetal rat calvaria in vitro. J. Craniofac. Genet. Dev. Biol. 19, 164–173 (1999). [PubMed] [Google Scholar]
- 31.Chen G., et al., BMP signaling in the development and regeneration of cranium bones and maintenance of calvarial stem cells. Front. Cell Dev. Biol. 8, 135 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warren S. M., Brunet L. J., Harland R. M., Economides A. N., Longaker M. T., The BMP antagonist noggin regulates cranial suture fusion. Nature 422, 625–629 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Negri S., et al., Human perivascular stem cells prevent bone graft resorption in osteoporotic contexts by inhibiting osteoclast formation. Stem Cells Transl. Med. 9, 1617–1630 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim H.-J., et al., Follistatin-like 1 promotes osteoclast formation via RANKL-mediated NF-κB activation and M-CSF-induced precursor proliferation. Cell. Signal. 28, 1137–1144 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Sylva M., et al., The BMP antagonist follistatin-like 1 is required for skeletal and lung organogenesis. PLoS One 6, e22616 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ornitz D. M., Marie P. J., Fibroblast growth factors in skeletal development. Curr. Top. Dev. Biol. 133, 195–234 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Fukuda T., et al., Sema3A regulates bone-mass accrual through sensory innervations. Nature 497, 490–493 (2013). Correction in: Nature 500, 612 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Ambrogini E., et al., FoxO-mediated defense against oxidative stress in osteoblasts is indispensable for skeletal homeostasis in mice. Cell Metab. 11, 136–146 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zanotti S., Canalis E., Notch signaling and the skeleton. Endocr. Rev. 37, 223–253 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lefebvre V., Roles and regulation of SOX transcription factors in skeletogenesis. Curr. Top. Dev. Biol. 133, 171–193 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teufel S., Hartmann C., Wnt-signaling in skeletal development. Curr. Top. Dev. Biol. 133, 235–279 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Kawaja M. D., et al., Nerve growth factor promoter activity revealed in mice expressing enhanced green fluorescent protein. J. Comp. Neurol. 519, 2522–2545 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rigueur D., Lyons K. M., Whole-mount skeletal staining. Methods Mol. Biol. 1130, 113–121 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bouxsein M. L., et al., Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. J. Bone Miner. Res. 25, 1468–1486 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Fulzele K., et al., Insulin receptor signaling in osteoblasts regulates postnatal bone acquisition and body composition. Cell 142, 309–319 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maruyama T., Jeong J., Sheu T.-J., Hsu W., Stem cells of the suture mesenchyme in craniofacial bone development, repair and regeneration. Nat. Commun. 7, 10526 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hosmane S., et al., Toll/interleukin-1 receptor domain-containing adapter inducing interferon-β mediates microglial phagocytosis of degenerating axons. J. Neurosci. 32, 7745–7757 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gutierrez G. M., et al., Impaired bone development and increased mesenchymal progenitor cells in calvaria of RB1-/- mice. Proc. Natl. Acad. Sci. U.S.A. 105, 18402–18407 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Metsalu T., Vilo J., ClustVis: A web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Res. 43, W566–W570 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stuart T., et al., Comprehensive integration of single-cell data. Cell 177, 1888-1902.e1821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang W., Sherman B. T., Lempicki R. A., Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 52.Street K., et al., Slingshot: Cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19, 477 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang X.-W., et al., Lin28 signaling supports mammalian PNS and CNS axon regeneration. Cell Rep. 24, 2540–2552.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Spatial Transcriptomics data have been deposited in GEO (GSE174313). Previously published data were used for this work GSE59739, FB00001013, and GSE145477 (19–21). All other study data are included in the article and/or supporting information.