Significance
Plants’ adaptations to and divergence in arid deserts have long fascinated scientists and the general public. Here, we present a genomic analysis of two congeneric desert plant species that clarifies their evolutionary history and shows that their common ancestor arose from a hybrid polyploidization, which provided genomic foundations for their survival in deserts. The whole-genome duplication was followed by translocation-based rearrangements of the ancestral chromosomes. Rapid evolution of genes in these reshuffled chromosomes contributed greatly to the divergences of the two species in desert microhabitats during which gene flow was continuous. Our results provide insights into plant adaptation in the arid deserts and highlight the significance of polyploidy-driven chromosomal structural variations in species divergence.
Keywords: desert plants, polyploidization, chromosomal structural variation, microhabitat divergence
Abstract
Deserts exert strong selection pressures on plants, but the underlying genomic drivers of ecological adaptation and subsequent speciation remain largely unknown. Here, we generated de novo genome assemblies and conducted population genomic analyses of the psammophytic genus Pugionium (Brassicaceae). Our results indicated that this bispecific genus had undergone an allopolyploid event, and the two parental genomes were derived from two ancestral lineages with different chromosome numbers and structures. The postpolyploid expansion of gene families related to abiotic stress responses and lignin biosynthesis facilitated environmental adaptations of the genus to desert habitats. Population genomic analyses of both species further revealed their recent divergence with continuous gene flow, and the most divergent regions were found to be centered on three highly structurally reshuffled chromosomes. Genes under selection in these regions, which were mainly located in one of the two subgenomes, contributed greatly to the interspecific divergence in microhabitat adaptation.
As individuals of each plant species cannot simply move to avoid stresses, biologists since Darwin’s era have been fascinated by their adaptations to environments with harsh climatic conditions such as deserts (1, 2). Several mechanisms generate novel genetic variation that enables plants to adapt to their environment, such as point mutation, gene duplication, and polyploidization (3–5). As one of these mechanisms, whole-genome duplication (WGD) or polyploidization, has frequently occurred throughout the evolutionary history of plants (6–8), and many polyploidization events have putative associations with environmental changes and subsequent adaptation to new niches (9, 10). Two types of polyploidy are recognized in plants and other organisms: autopolyploidy (involving duplication of a single species’ genome) and allopolyploidy (involving combination of different species’ genomes) (7, 11). The relative proportion of autopolyploids and allopolyploids are comparable, but allopolyploidy is generally expected to provide higher adaptive potential (12, 13). This is because it not only allows the pairing of chromosomes from each parent, with diploid-like meiotic behavior and disomic inheritance, but also leads to extensive chromosomal structural variations, morphological innovations, novel genic interactions, and hybrid vigor (7, 14). Such evolutionary consequences have been repeatedly confirmed in allopolyploid model species, for example, Arabidopsis, and widely cultivated allopolyploid crops, including cotton, wheat, and oilseed rape (15–19).
In general, polyploid plants exhibit higher drought and salt tolerance than their diploid relatives (12–15); however, little is known about how subsequent speciation and diversification occur in such polyploid genomes. To improve such understanding, we examined the contribution of polyploidization (if any) to adaptive evolution and speciation in the bispecific genus Pugionium, endemic to the Kubuqi Desert and Mu Us Desert in northwestern China (20). These inland deserts arose from rapid climate transformation since the early Miocene (21, 22), with consequent changes in vegetation, forest retreats, and the emergence of aridity-adapted species (23). The genus Pugionium belongs to the family Brassicaceae, which includes more than 3,700 species distributed around the world (24, 25). Many species of Brassicaceae are economically important crops that are cultivated as vegetables, condiments, fodder, and oilseeds (18, 24, 26). In addition, allopolyploidy—usually associated with chromosomal structural variations (fusion, shuffling, or translocation) and changes in chromosome base number—is prevalent in the family (24, 27–29). Brassicaceae are divided into five major lineages (24), and Pugionium belongs to Expanded Lineage II with isolated position (30). Although young leaves and shoots of Pugionium are consumed as vegetables by local communities (23), both species produce highly lignified roots, stems, and silicles, which have a clear adaptive value in dry and salty deserts. Pugionium cornutum has long roots and an erect stem that can be more than 1.5 m tall, while Pugionium dolabratum produces short roots and numerous basal “bushy” branches (Fig. 1A). Most populations of the two species have no overlapped distributions, but they do occur rarely in the same site with distinct microhabitat divergence (23) (SI Appendix, Results). Furthermore, P. cornutum and P. dolabratum are confined to the mobile and fixed dunes, respectively (23), and also display differences in leaf and silicle morphology, including the sizes of silicle valves and wings (31). In this study, we first sequenced and assembled genomes of the two Pugionium species and then assessed the genomic changes that had taken place during the ancestral adaptation of the genus to the desert environment. Next, we examined the genomic divergence of both species at the population level to investigate how speciation might occur in the desert.
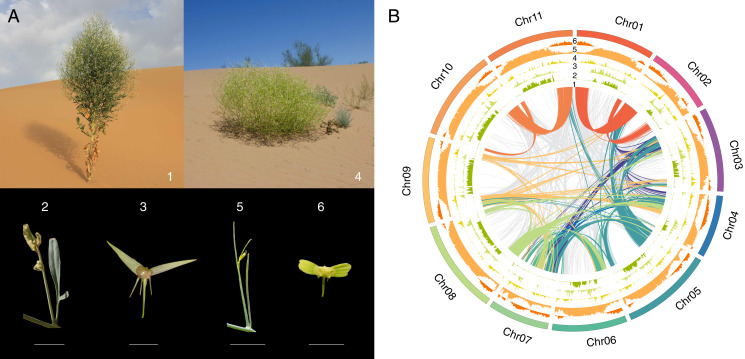
Fig. 1.
The contrasted habit and morphology of the two Pugionium species and genomic structure of P. cornutum. (A) Morphological and habitat divergence of the two species (1, 2, and 3 for P. cornutum and 4, 5, and 6 for P. dolabratum) on the basal branching and stem height, leaf (lobe width), silique morphology (valve and wing length and angle ), and habitat (mobile and fixed dunes). (Scale bar: 1 cm.) (B) Collinearity within the P. cornutum genome. Color-coded lines in the middle (1) show gene synteny between chromosomes. Histograms from inside to outside show frequencies of tandem repeats (2), LTR/Gypsy retrotransposons (3), LTR/Copia retrotransposons (4), overall repetitive contents (5), and densities of genes (6), respectively.
Results
De Novo Genome Assemblies of the Two Species.
Our examination of DAPI-stained mitotic chromosome spreads revealed 11 chromosome pairs (2n = 22) in both Pugionium species (SI Appendix, Fig. S1). The genome sizes were estimated to be 570 and 606 Mb for P. cornutum and P. dolabratum, respectively (SI Appendix, Figs. S2 and S3). A high-quality reference genome of P. cornutum was obtained with 81.3 Gb (143x) Nanopore long reads and 44.7 Gb (78x) short reads (SI Appendix, Table S1). With the aid of the chromosome conformation capture technique (SI Appendix, Fig. S4), the genome of P. cornutum was further assembled into 11 chromosomes (Fig. 1B and SI Appendix, Fig. S5). The resulting assembly of P. cornutum was 550 Mb, with a scaffold N50 of 37.1 Mb and a contig N50 of 311.7 kb (SI Appendix, Table S2). For P. dolabratum, 211 Gb (356x) short reads and 10.7 Gb (18x) Pacbio long reads were used to de novo assemble the genome into large scaffolds, with scaffold N50 being 357.8 kb and contig N50 being 68.4 kb (SI Appendix, Tables S3 and S4). We assessed the quality of genome assemblies using RNA sequencing (RNA-seq ) data obtained from roots, stems, leaves, and flowers (SI Appendix, Table S5). The results showed that most coding regions were well represented in the assemblies (SI Appendix, Table S6). Moreover, 97.9 and 97.4% of the 2,326 eudicot-specific BUSCO genes were identified in the genome assemblies of P. cornutum and P. dolabratum, respectively (SI Appendix, Table S7).
In total, 72.8 and 65.0% of the genome sequences were identified as repetitive elements for P. cornutum and P. dolabratum, respectively (Fig. 1B and SI Appendix, Tables S8 and S9), and the vast majority of repeats were classified as tandem repeats and long terminal repeat (LTR ) retrotransposons. An analysis of LTR retrotransposons indicated an increase in the activity during the last three million years (SI Appendix, Fig. S6). A total of 31,412 and 30,614 protein-coding genes were predicted for P. cornutum and P. dolabratum, respectively (SI Appendix, Table S10), and 27,982 (89.1%) of these genes were distributed on the assembled chromosomes of P. cornutum. In addition, most of these genes were successfully annotated by at least one public database (SI Appendix, Table S11), with complete BUSCO scores of 95.1 and 94.2% for P. cornutum and P. dolabratum, respectively (SI Appendix, Table S12), indicating near completion of both the assemblies and annotations.
WGD by Allotetraploidy.
Our comparative chromosome painting analyses based on cross-species hybridization, using BAC contigs specific to the chromosomes of Arabidopsis thaliana, suggested two copies of genomic blocks (GBs) in the Pugionium pachytene chromosome complements (SI Appendix, Fig. S7). This pointed to a potential WGD (tetraploidization) that had occurred during the origin of Pugionium. This WGD was further confirmed by genome collinearity and synonymous divergences of paralogous gene pairs within collinear blocks (Fig. 1B and SI Appendix, Figs. S8–S11). Based on the divergence of paralogous gene pairs, this WGD was estimated to have occurred ∼18 Mya (SI Appendix, Fig. S9) when Lineages I and II diverged (30, 32) and was more recent than the family-specific At-α WGD (∼43 Mya) (33). Phylogenetic analyses of different datasets were then performed to examine whether the tetraploidy arose from autopolyploidization or allopolyploidization. We first constructed gene trees using six species, that is, P. cornutum, Arabidopsis lyrata, Capsella rubella, Eutrema salsugineum, Schrenkiella parvula, and Aethionema arabicum, and assessed the pattern of gene tree topologies. For the 5,461 genes that were single copy in each of the six genomes, the placement of P. cornutum as sister to Lineage II was supported by 42.0% (bootstrap supports ≥70% ) of gene trees, while 17.8% (bootstrap supports ≥70%) placed P. cornutum sister to Lineage I plus II (SI Appendix, Fig. S12), suggesting a likely hybrid origin because of the high inconsistent tree topologies. Then, we carried out phylogenetic analyses of the two duplicate paralogs from the At-α polyploidization and the possible homologs in Pugionium and Eutrema. Most duplicated homologs in Pugionium did not cluster into one monophyletic group as expected for autopolyploidization (SI Appendix, Fig. S12). Finally, 8,268 gene trees constructed based on homolog groups that contain one gene from A. arabicum and at least one homolog in all other genomes were used to perform multilabeled trees (MUL-trees) analysis, and the optimal MUL-tree also supported an allopolyploid origin of P. cornutum (SI Appendix, Fig. S13).
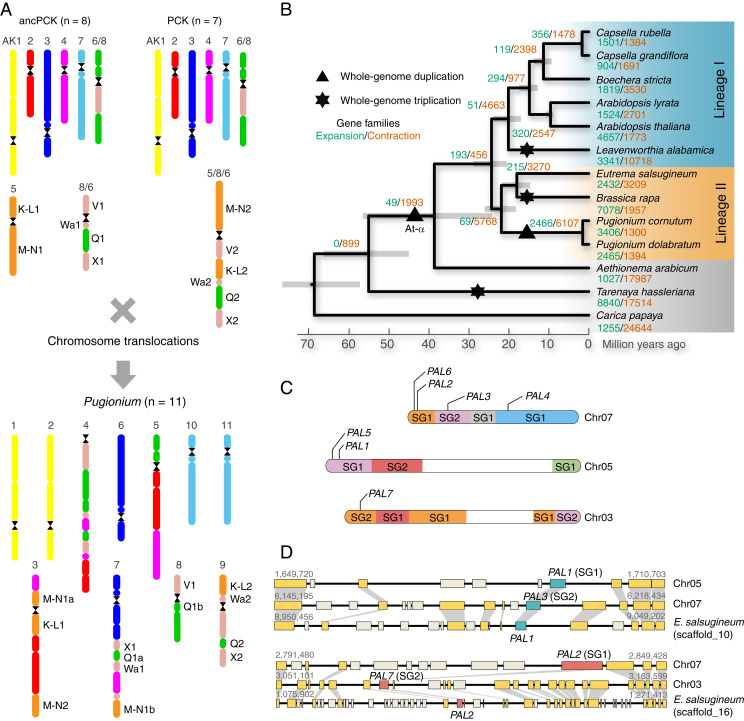
In order to further confirm allopolyploidization and uncover the origin of the two parental Pugionium (sub)genomes, the genome of P. cornutum was used to examine the association of GBs specific to previously defined ancestral Brassicaceae genomes—ancestral Proto-Calepineae Karyotype (ancPCK, n = 8) (29) and Proto-Calepineae Karyotype (PCK, n = 7) (27). The conserved association of blocks K-L and M-N on Pugionium chromosome 3 indicated that one parental (sub)genome was ancPCK-like (denoted as SG1, Fig. 2A and SI Appendix, Fig. S14). In contrast, the association K-L+Wa on Pugionium chromosome 9 pointed to a PCK-like (sub)genome (denoted as SG2). Despite the extensive postpolyploidization shuffling, these comparative analyses have collectively shown that the ancestral Pugionium genome originated through an allotetraploid WGD based on hybridization between ancPCK- and PCK-like genomes (Fig. 2A). This ancestral allopolyploid genome experienced an extensive postpolyploid diploidization, reducing the chromosome number from n = 15 to n = 11 (Fig. 2A). Among the 11 chromosomes in the Pugionium genome, five chromosomes remained conserved (chromosomes 1, 2, 6, 10 and 11), whereas the remaining six chromosomes were greatly reshuffled by translocations and inversions (Fig. 2A). Three chromosomes (3, 4, 7) showed high chromosomal structural variations as compared to the ancestral genomes.
Fig. 2.
The origin and postpolyploid evolution of the allotetraploid Pugionium genome. (A) The ancestral Pugionium genome presumably originated from hybridization between an ancPCK-like genome (n = 8, subgenome SG1) and a PCK-like genome (n = 7, subgenome SG2). Capital letters denote GBs and their associations important for inferring the ancestral (sub)genomes. (B) Phylogenetic tree for Pugionium and 11 other plants. A WGD was identified in Pugionium, paralleling independent mesohexaploidy events in Leavenworthia and Brassica. More changes in the numbers of gene family members apparently occurred in the ancestor of P. cornutum and P. dolabratum because of polyploidization than in the ancestor of E. salsugineum and Brassica rapa. (C) The seven phenylalanine ammonia-lyase (PAL) genes on three Pugionium chromosomes derived from allopolyploidization. (D) Collinear gene blocks between Pugionium and E. salsugineum indicate that genes of the PAL family from both ancestral parents were retained in Pugionium; other collinear genes are shown in yellow, and genes not in collinear blocks are displayed in creamy white.
Biased Gene Fractionation and Gene Family Expansion.
According to their associated gene tree topologies, duplicated GBs in the genome of P. cornutum were partitioned into subgenomes SG1 and SG2 (SI Appendix, Figs. S15 and S16 and Table S13). Based on the modeled postpolyploidization interchromosomal rearrangements and loss of chromosomal segments (SI Appendix, Fig. S14), we identified a total of 10,985 and 14,936 protein-coding genes in the subgenome SG1 and SG2, respectively. Here, biased fractionation resulted in the preferential retention of genes from one parental genome (SI Appendix, Table S14). Based on these 14,936 genes, which were phylogenetically closer to Lineage II, the divergence time between subgenome SG2 and E. salsugineum was estimated to be ∼12 Mya. This suggests that the allotetraploid WGD recovered for Pugionium or with its related but unsampled genera should have occurred around this age or later. We then examined expression levels of the homeologous gene pairs in order to investigate the presence or absence of the subgenome dominance. Using RNA-seq data from different tissues, we found biased gene expressions between the two subgenomes with genes located in the subgenome SG2 having significantly higher expression than those from SG1 (SI Appendix, Figs. S17 and S18). In addition, around 42.0% of the homoeologous gene pairs were estimated to show at least twofold differentiated expressions between the two subgenomes (SI Appendix, Fig. S19).
We next determined gene families experienced expansion and contraction in the Pugionium genus based on annotated genomes of the two Pugionium species and other species from Brassicaceae (Fig. 2B and SI Appendix, Table S15). Out of the 2,466 gene families specifically expanded in Pugionium, 2,143 contained duplicated genes derived from WGD as determined by the presence of collinear blocks. Gene families expanded via WGD were enriched in multiple Gene Ontology categories related to organ developments and stress responses, including “leaf development,” “root development,” “seed development,” “cellular response to salt stress,” and “response to light stimulus,” while those expanded via tandem duplications were overrepresented in functional categories associated with “root meristem growth,” “secondary metabolite biosynthetic process,” and “DNA (cytosine-5-)-methyltransferase activity” (Datasets S1 and S2). We found that 40 out of 58 transcription factor gene families had expanded in Pugionium (SI Appendix, Table S16). Most of them are involved in responses to abiotic stress. For example, members of RAV and GRAS gene families were reported to respond to salty and cold stresses. In addition, we found that gene families related to ionic and osmotic equilibrium (CIPK and CDPK), drought tolerance (ABF and DREBs), and lignin biosynthetic pathway (PAL and MSBP) were also expanded within Pugionium (Fig. 2 C and D and SI Appendix, Figs. S20–S22 and Tables S17–S19). Expansions of these gene families should have supplied genetic foundations for this genus to adapt to the challenging habitats. In addition, we also found that genes located in the subgenome SG2 showed higher expression levels than those in SG1 in these gene families, which further confirmed that the biased gene expression played a likely role for plant adaptation during diploidization after allopolyploidization (SI Appendix, Table S18).
Interspecific Divergence of the Allopolyploid Genome.
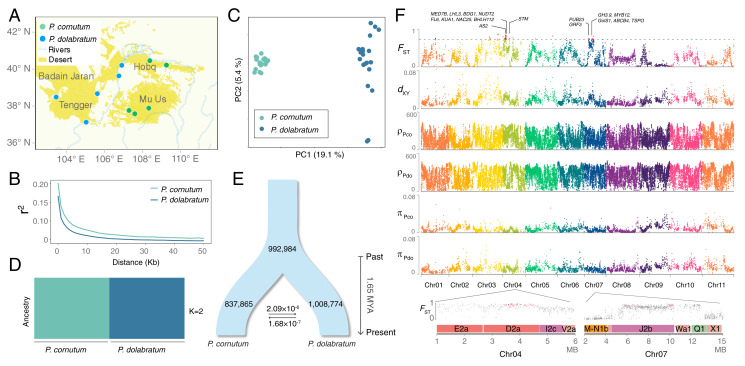
In addition to morphological differentiation (Fig. 1A), two Pugionium species appear to show local adaptation to different microhabitats (SI Appendix, Figs. S23–S26 and Tables S20–S23) as found for other closely related desert plants (34). To explore the genetic basis of the divergence, we conducted whole-genome resequencing of five populations (a total of 20 individuals) for each species (Fig. 3A and SI Appendix, Table S24). The linkage disequilibrium of both species decayed to half maximum within 5 kb (Fig. 3B). The principal component (PC) analysis distinguished the two species along PC1 (variance explained 19.1%, Tracy–Widom P = 3.0 × 10−13; Fig. 3C), and our population structure analysis similarly revealed two distinct genetic clusters (Fig. 3D and SI Appendix, Fig. S27). We then evaluated four models of speciation, that is, strict isolation, isolation with migration, isolation after migration, and secondary contact (SI Appendix, Fig. S28 and Table S25), using a composite likelihood approach. The best-fit model suggested that two Pugionium species diverged with a continuous gene flow (SI Appendix, Table S26) around 1.65 Mya (Fig. 3E), suggesting sympatric or parapatric speciation through microhabitat selections.
Fig. 3.
Population structure and interspecies divergence of Pugionium. (A) Locations of 10 sampled populations. (B) Linkage disequilibrium decay based on the squared correlation coefficient between SNPs in P. cornutum and P. dolabratum populations. (C) Results of the PC analyses of SNPs within the two species. (D) Population structure of all sampled individuals of the two species (with K = 2 as the best inferred value). (E) The best-fit demographic divergence of the two species modeled by fastsimcoal2. Effective population sizes, divergence time and estimates of gene flow between species are displayed on the schematic plot. MYA, million years ago. (F) Manhattan plots of FST, dXY, ρ, and π between and within the two Pugionium species using a 50-kb nonoverlapping window. Genomic regions of high divergence in 11 chromosomes (Top) and genes under selection in those regions on chromosome 4 and 7 (Bottom) are highlighted in red. Pco, P. cornutum; Pdo, P. dolabratum.
We identified a total of 42 genomic regions (50 kb in size) in assembled chromosomes of P. cornutum that exhibited high divergence between P. cornutum and P. dolabratum (i.e., upper 1% of the empirical FST distribution) (SI Appendix, Table S27), which also had significantly elevated dXY compared to other genomic regions (P = 2.1 × 10−13, Mann–Whitney U test; Fig. 3F). Out of these 42 regions, 27 and 15 were identified in subgenome SG1 and SG2, respectively, without biased distributions (P = 0.06) (Fig. 3F and SI Appendix, Table S27). However, 86% of these regions were found to be located on chromosomes 3, 4, and 7 (Fig. 3F), which were formed by recombination among multiple ancestral chromosomes (SI Appendix, Fig. S14). Genome-wide FST and dXY were positively correlated, especially for the three chromosomes (SI Appendix, Fig. S29). Furthermore, nucleotide diversity was found to be significantly lower in these regions for both P. cornutum (P = 5.3 × 10−15) and P. dolabratum (P < 2.2 × 10−16; Fig. 3F), suggesting that selection might have acted on these regions. A total of 236 genes were identified from these highly divergent regions, and the vast majority of these genes were found to be located on chromosome 4 (68.6%) and 7 (28.8%). The expression of some of these genes in four different tissues showed contrasting difference between the two species (SI Appendix, Figs. S30 and S31). Using a Hudson–Kreitman–Aguadé test, 197 of these genes were inferred to be under selection, and most of them were located in subgenome SG2 (SI Appendix, Table S28). Homologs of these genes were identified to be involved in root development (BDG1, KUA1, ABCB4, GH3.9), leaf morphogenesis (AS2, KUA1, FL6, GRF3), xylem differentiation (LHL3), seed germination and seedling development (NAC25, MED7B, STM), salt tolerance (BHLH112, GolS1, TSPO), drought resistance (BDG1, PUB23), oxidative stress response (NUDT2), and flavonoid biosynthesis (MYB12) (Fig. 3F). The two Pugionium species have distinct differences in morphology and habitat, with P. cornutum only occurring on mobile dunes, whereas P. dolabratum is distributed in fixed or semifixed deserts (Fig. 1A). They displayed contrasting patterns in seed germination speed and growth rate in response to salinity stress and desert burials (SI Appendix, Figs. S23–S26 and Table S23). Therefore, the divergence selection of those genes might be responsible for morphological differentiation of root, shoot, and leaf and further contributed to local adaptation of the two Pugionium species to different microhabitats.
To further test whether copy number variations of specific gene families between the two species contributed to speciation, we used the two de novo genomes to identify the genes of three amino acid loop extension (TALE) and histidine kinases (HKs) gene families, members of which were revealed to have crucial functions in regulating various development processes and responses to abiotic stress in plants (SI Appendix). Compared to other Brassicaceae species, these families were expanded in both Pugionium species but with interspecific copy number variations between them (SI Appendix, Figs. S32 and S33 and Table S29). In the TALE gene family, we found that P. dolabratum contained more copies for BLH11, KNAT2, and KNAT6 compared with P. cornutum. The BLH11 ortholog from Medicago truncatula (PINNA1) was identified as a determinacy factor during leaf morphogenesis (35). In Arabidopsis, KNAT2 and KNAT6 were also confirmed to play essential roles in regulating proximal–distal development of leaves by the repression from AS2 (36), which was also found to have experienced positive selection in the two Pugionium species (SI Appendix, Table S28). For the HKs gene family, more homologic copies were detected in P. cornutum for AHK2 and AHK4, which encode two cytokinin receptors involved in shoot and root development, as well as tolerance to salt and drought stress (37–39). In addition, expression divergence of these gene copies was also detected between the two species (SI Appendix, Figs. S32 and S33). Thus, the copy number variations in these gene families may also have contributed to the morphological divergences between the two species as well as the respective adaptations to mobile and stable desert dunes.
Discussion
Based on comparative chromosome painting analyses (SI Appendix, Fig. S7) and divergence distributions of the paired paralogs, we inferred the occurrence of a WGD presumably specific to the genus Pugionium and clear postpolyploid chromosomal structural variation. Further analyses suggested that this WGD probably involved allopolyploidization rather than autopolyploidization and occurred around 12 Mya or later, postdating the divergence of two ancestral parental lineages (n = 8 and 7, respectively; Fig. 2A) ∼18 Mya. Similar allopolyploidizations, involving ancPCK- and PCK-like parental genomes, were previously reported in the genus Ricotia (n = 13 and 14) (40) and Lunaria (n = 14) (41). However, the ancestral allopolyploid Pugionium genome experienced more extensive descending dysploidy (from n = 15 to n = 11) during its postpolyploid diploidization, associated with the origin of three highly rearranged chromosomes (Fig. 2A). The allopolyploid origin of Pugionium seems to have facilitated its survival through adaptation to the changing environments of northwest China during their desertification since the early Miocene (21, 42). Inter alia, gene families involved in drought tolerance, ionic and osmotic equilibrium, and lignin biosynthesis expanded in the Pugionium genomes significantly (SI Appendix, Fig. S20 and Table S17).
Genomic evidence indicates that the two species started to diverge around 1.65 Mya, during the Quaternary, when a global increase in aridity (20, 43, 44) might have led to the development of contrasting desert microhabitats, mobile and fixed dunes, thereby promoting the initial divergence of the two species through microhabitat adaptation with parapatric or sympatric distribution. This hypothesis is corroborated by our speciation modeling of joint site frequency spectra across the total genome, which suggests the occurrence of continuous and strong gene flow through their evolutionary divergence history. We further found that the high-divergence regions in the Pugionium allopolyploid genome were mainly distributed on three chromosomes with most structural variations generated by translocation-based reshuffling during postpolyploidization diploidization. In addition to copy number variations of gene families, genes with positive selection signals in these regions are highly involved in root development, leaf morphogenesis, and microhabitat adaptation (seed germination and dry/salt tolerance), corresponding well with interspecific divergences in these respects (SI Appendix, Tables S21 and S23). Therefore, our results suggest that polyploidy-driven chromosomal structural variation may have played an important role in subsequent speciation and further extensive diversification (45) in addition to well-known rapid differentiations of the duplicated genes and novel genic interactions (46).
Materials and Methods
Mitotic chromosome spreads were used primarily for chromosome counting and pachytene spreads for comparative chromosome painting analysis. Long reads were generated using GridION and PacBio RS II. Paired-end and mate-pair short reads were generated using the MGISeq 2000 and Illumina HiSeq platforms. Genomes were assembled using MaSuRCA. Transposable elements were identified using Tandem Repeats Finder, RepeatMasker, RepeatModeler, and LTR_Finder. Genes were predicted using AUGUSTUS, GlimmerHMM, PASA, Exonerate, and EVidenceModeler. Collinear gene blocks were identified with MCscanX. Synonymous substitution rates were calculated using PAML. Following genome alignments and chaining by LASTZ, GRAMPA was used to determine the likeliest mode of polyploidy. Gene expression levels were estimated using Salmon and DESeq. Clean reads from population data were mapped to the P. cornutum genome using the bwa-men algorithm. Genome-wide single nucleotide polymorphisms (SNPs) were called by GATK. ADMIXTURE and Eigensoft were used for population structure analysis. Coalescence-based simulation of speciation patterns was performed in fastsimcoal2. The interspecific reproductive isolation within the genus, and differences between the two species in microhabitat adaptation, were experimentally confirmed at desert sites. Detailed information on all the experimental and analytical procedures is available in SI Appendix.
Acknowledgments
This work was equally supported by the Second Tibetan Plateau Scientific Expedition and Research program (2019QZKK0502), the National Natural Science Foundation of China (91731301, 91331102, and 41771055), and also the Fundamental Research Funds for the Central Universities (SCU2021D006 and 2020SCUNL207). T.M. and M.A.L. were supported by the Central European Institute of Technology 2020 project (LQ1601).
Footnotes
Reviewers: M.A.B., The University of Arizona; and L.L., Fudan University.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2025711118/-/DCSupplemental.
Data Availability
The whole-genome sequencing data, transcriptome sequencing data, and genome assemblies have been deposited in the National Center for Biotechnology Information Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) under accession numbers PRJNA685118 and PRJNA760666.
Change History
November 2, 2021: The Supporting Information has been updated.
References
- 1.Dassanayake M., et al., The genome of the extremophile crucifer Thellungiella parvula. Nat. Genet. 43, 913–918 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyer J. S., Plant productivity and environment. Science 218, 443–448 (1982). [DOI] [PubMed] [Google Scholar]
- 3.Anderson J. T., Willis J. H., Mitchell-Olds T., Evolutionary genetics of plant adaptation. Trends Genet. 27, 258–266 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turner T. L., Bourne E. C., Von Wettberg E. J., Hu T. T., Nuzhdin S. V., Population resequencing reveals local adaptation of Arabidopsis lyrata to serpentine soils. Nat. Genet. 42, 260–263 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Li L. F., Li Y. L., Jia Y., Caicedo A. L., Olsen K. M., Signatures of adaptation in the weedy rice genome. Nat. Genet. 49, 811–814 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Jiao Y., et al., Ancestral polyploidy in seed plants and angiosperms. Nature 473, 97–100 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Soltis P. S., Soltis D. E., Ancient WGD events as drivers of key innovations in angiosperms. Curr. Opin. Plant Biol. 30, 159–165 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Marburger S., et al., Interspecific introgression mediates adaptation to whole genome duplication. Nat. Commun. 10, 5218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu S., Han B., Jiao Y., Genetic contribution of paleopolyploidy to adaptive evolution in angiosperms. Mol. Plant 13, 59–71 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Beilstein M. A., et al., Evolution of the telomere-associated protein POT1a in Arabidopsis thaliana is characterized by positive selection to reinforce protein–protein interaction. Mol. Biol. Evol. 32, 1329–1341 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stebbins G. L. Jr., Types of polyploids; their classification and significance. Adv. Genet. 1, 403–429 (1947). [DOI] [PubMed] [Google Scholar]
- 12.Barker M. S., Arrigo N., Baniaga A. E., Li Z., Levin D. A., On the relative abundance of autopolyploids and allopolyploids. New Phytol. 210, 391–398 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Estep M. C., et al., Allopolyploidy, diversification, and the Miocene grassland expansion. Proc. Natl. Acad. Sci. U.S.A. 111, 15149–15154 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor S. A., Larson E. L., Insights from genomes into the evolutionary importance and prevalence of hybridization in nature. Nat. Ecol. Evol. 3, 170–177 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Ding M., Chen Z. J., Epigenetic perspectives on the evolution and domestication of polyploid plant and crops. Curr. Opin. Plant Biol. 42, 37–48 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang G., et al., Genome sequence of Gossypium herbaceum and genome updates of Gossypium arboreum and Gossypium hirsutum provide insights into cotton A-genome evolution. Nat. Genet. 52, 516–524 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marcussen T., et al., Ancient hybridizations among the ancestral genomes of bread wheat. Science 345, 1250092 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Chalhoub B., et al., Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950–953 (2014). [DOI] [PubMed] [Google Scholar]
- 19.del Pozo J. C., Ramirez-Parra E., Whole genome duplications in plants: an overview from Arabidopsis. J. Exp. Bot. 66, 6991–7003 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Yang X., Desert research in northwestern China–A brief review. Géomorphologie Reli. Process. Environ. 4, 275–284 (2006). [Google Scholar]
- 21.Guo Z. T., et al., A major reorganization of Asian climate by the early Miocene. Clim. Past 4, 153–174 (2008). [Google Scholar]
- 22.Guo Z. T., et al., Onset of Asian desertification by 22 Myr ago inferred from loess deposits in China. Nature 416, 159–163 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Yu Q., Wang Q., Wang A., Wu G., Liu J., Interspecific delimitation and phylogenetic origin of Pugionium (Brassicaceae). J. Syst. Evol. 48, 195–206 (2010). [Google Scholar]
- 24.Nikolov L. A., et al., Resolving the backbone of the Brassicaceae phylogeny for investigating trait diversity. New Phytol. 222, 1638–1651 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Al-Shehbaz I. A., Beilstein M. A., Kellogg E. A., Systematics and phylogeny of the Brassicaceae (Cruciferae): An overview. Plant Syst. Evol. 259, 89–120 (2006). [Google Scholar]
- 26.Schmidt R., Bancroft I., Eds., Genetics and Genomics of the Brassicaceae (Springer, 2011). [Google Scholar]
- 27.Mandáková T., Lysak M. A., Chromosomal phylogeny and karyotype evolution in x=7 crucifer species (Brassicaceae). Plant Cell 20, 2559–2570 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kagale S., et al., Polyploid evolution of the Brassicaceae during the Cenozoic era. Plant Cell 26, 2777–2791 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geiser C., Mandáková T., Arrigo N., Lysak M. A., Parisod C., Repeated whole-genome duplication, karyotype reshuffling, and biased retention of stress-responding genes in Buckler mustard. Plant Cell 28, 17–27 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo X., et al., Plastome phylogeny and early diversification of Brassicaceae. BMC Genomics 18, 176 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Q., Abbott R. J., Yu Q. , Lin K., Liu J. , Pleistocene climate change and the origin of two desert plant species, Pugionium cornutum and Pugionium dolabratum (Brassicaceae), in northwest China. New Phytol. 199, 277–287 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Huang C. , et al., Resolution of Brassicaceae phylogeny using nuclear genes uncovers nested radiations and supports convergent morphological evolution. Mol. Biol. Evol. 33, 394–412 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kiefer C., et al., Interspecies association mapping links reduced CG to TG substitution rates to the loss of gene-body methylation. Nat. Plants 5, 846–855 (2019). [DOI] [PubMed] [Google Scholar]
- 34.DiVittorio C. T., et al., Natural selection maintains species despite frequent hybridization in the desert shrub Encelia. Proc. Natl. Acad. Sci. U.S.A. 117, 33373–33383 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He L., et al., A molecular framework underlying the compound leaf pattern of Medicago truncatula. Nat. Plants 6, 511–521 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Machida C., Nakagawa A., Kojima S., Takahashi H., Machida Y., The complex of ASYMMETRIC LEAVES (AS) proteins plays a central role in antagonistic interactions of genes for leaf polarity specification in Arabidopsis. Wiley Interdiscip. Rev. Dev. Biol. 4, 655–671 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishiyama R., et al., Arabidopsis AHP2, AHP3, and AHP5 histidine phosphotransfer proteins function as redundant negative regulators of drought stress response. Proc. Natl. Acad. Sci. U.S.A. 110, 4840–4845 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tran L. S., et al., Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 20623–20628 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishimura C., et al., Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 16, 1365–1377 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mandáková T., Guo X., Özüdoğru B., Mummenhoff K., Lysak M. A., Hybridization-facilitated genome merger and repeated chromosome fusion after 8 million years. Plant J. 96, 748–760 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Guo X., et al., Linked by ancestral bonds: Multiple whole-genome duplications and reticulate evolution in a Brassicaceae tribe. Mol. Biol. Evol. 38, 1695–1714 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu X., et al., Where were the monsoon regions and arid zones in Asia prior to the Tibetan Plateau uplift? Natl. Sci. Rev. 2, 403–416 (2015). [Google Scholar]
- 43.Hövermann J., Süssenberger H., Zur Klimageschichte Hoch- und Ostasiens. Berl. Geogr. Stud. 20, 173–186 (1986). [Google Scholar]
- 44.Huang J., Yu H., Guan X., Wang G., Guo R., Accelerated dryland expansion under climate change. Nat. Clim. Chang. 6, 166–171 (2016). [Google Scholar]
- 45.Walden N., et al., Nested whole-genome duplications coincide with diversification and high morphological disparity in Brassicaceae. Nat. Commun. 11, 3795 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van de Peer Y., Ashman T. L., Soltis P. S., Soltis D. E., Polyploidy: An evolutionary and ecological force in stressful times. Plant Cell 33, 11–26 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The whole-genome sequencing data, transcriptome sequencing data, and genome assemblies have been deposited in the National Center for Biotechnology Information Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) under accession numbers PRJNA685118 and PRJNA760666.