Significance
Many genes in higher organisms are transcribed intermittently rather than continuously. The mechanisms behind this phenomenon, which is often referred to as “noise” in gene expression, are not clear. Building on previous work, we show that this behavior arises because the regulatory regions of eukaryotic genes are usually sequestered within dense chromatin and become accessible to transcription factors only intermittently because of random local decondensation events in chromatin. We constitutively opened the chromatin at the promoters of two genes, which resulted in a reduction in the noise in their transcription. This perturbation reveals that the natural compaction of chromatin is responsible for noisy gene expression and also shows a route to selectively reducing the transcriptional noise from particular genes.
Keywords: single-cell heterogeneity, transcriptional bursting, stochastic mRNA synthesis
Abstract
Many eukaryotic genes are expressed in randomly initiated bursts that are punctuated by periods of quiescence. Here, we show that the intermittent access of the promoters to transcription factors through relatively impervious chromatin contributes to this “noisy” transcription. We tethered a nuclease-deficient Cas9 fused to a histone acetyl transferase at the promoters of two endogenous genes in HeLa cells. An assay for transposase-accessible chromatin using sequencing showed that the activity of the histone acetyl transferase altered the chromatin architecture locally without introducing global changes in the nucleus and rendered the targeted promoters constitutively accessible. We measured the gene expression variability from the gene loci by performing single-molecule fluorescence in situ hybridization against mature messenger RNAs (mRNAs) and by imaging nascent mRNA molecules present at active gene loci in single cells. Because of the increased accessibility of the promoter to transcription factors, the transcription from two genes became less noisy, even when the average levels of expression did not change. In addition to providing evidence for chromatin accessibility as a determinant of the noise in gene expression, our study offers a mechanism for controlling gene expression noise which is otherwise unavoidable.
E ukaryotic cells exhibit remarkable cell-to-cell variability in the levels of messenger RNAs (mRNAs) for many genes, even when the cells are genetically and developmentally identical (1–7). Although variations in global factors such as cell cycle stage, RNA polymerase abundance, and cell size contribute to this heterogeneity, a large fraction of the heterogeneity arises because transcription at the gene locus occurs in randomly initiated bursts that are separated by periods of inactivity (3, 4, 6–9). Why genes are transcribed in such episodic bursts, rather than continuously, is a subject of intense research. Among the ideas that guide current research on the subject are the following: probabilistic assembly of transcription factors (TFs) at the gene promoter (10), abrupt release of paused convoys of RNA polymerases at the end of the gene (11), facilitated transcription reinitiation during bursts (12, 13), stochastic interactions of promoters with distant enhancers (14, 15), and a random chromatin accessibility hypothesis that we are exploring in this report. According to the random chromatin accessibility hypothesis, the regulatory regions of eukaryotic genes (promoters and enhancers) are usually sequestered within chromatin and become accessible to TFs only intermittently because of random local decondensation events (1, 9).
Gene expression heterogeneity is often referred to and analyzed as “noise.” Models designed to explain the observed heterogeneity predict that even under conditions that are permissive for gene expression (such as after induction), gene loci switch randomly between an on state and an off state (two-state model) (9, 16). The bursts of RNA synthesis correspond to the on state, and the intervals between them correspond to the off state. Instances of cells being able to modulate both the frequency and the magnitude of bursts to control mRNA levels have been documented (6, 17, 18).
In early explorations of the role of random chromatin accessibility in stochastic mRNA synthesis, the status of chromatin accessibility was altered in cultured cells using an inhibitor of histone deacetylases (HDACs) (19) and, in a second study in yeast, by deletion of chromatin-remodeling complexes one by one (1). However, since they produce pleotropic genome-wide effects and do not affect all of the cellular HDACs, neither perturbation appreciably altered the noise characteristics of specific genes that were measured (1, 19). We undertook studies in which instead of perturbing the condensation status of the entire genome, chromatin accessibility was increased specifically at the promoters of two particular genes, and the noise characteristics of their expression were studied. To accomplish this task, we tethered a histone acetyl transferase (HAT) to these loci using endonuclease-deficient CRISPR-associated protein (dCas9) constructs. Chromatin accessibility analysis by an assay for transposase-accessible chromatin using sequencing (ATAC-seq) indicated that this HAT tethering increased accessibility in the chromatin near the engineered sites in the promoters. We analyzed the heterogeneity in the number of mature mRNAs and in the number of transcriptionally active gene loci in single cells and found that these perturbations lead to a significant reduction in the noise in the gene expression from the targeted genes. Together with two other recent studies that observed reduction in gene expression noise upon targeted acetylation of gene loci in different biological contexts (20, 21), our results provide strong support for the idea that intermittent accessibility of regulatory regions to TFs is a major determinant of stochastic mRNA synthesis.
Results
Constitutive Tethering of HAT at a Promoter Lowers the Noise in Gene Expression.
To increase the accessibility of a specific gene locus without disturbing other genes in the nucleus, we tethered a HAT to a gene locus via an endonuclease-deficient dCas9, which harbors mutations that render its deoxyribonuclease domain inactive while preserving its ability to bind to the target site (Fig. 1A). We used a previously described construct, dCas9-p300, in which the core HAT domain of the transcriptional coactivator p300 is fused to dCas9 (22). This construct is expected to increase DNA accessibility locally by acetylation of histone H3 on lysine 27 (H3K27ac) (22). This change removes a positive charge from histones in nucleosomes, thereby decreasing their interaction with the negatively charged phosphate groups of DNA, thus relaxing the complex.
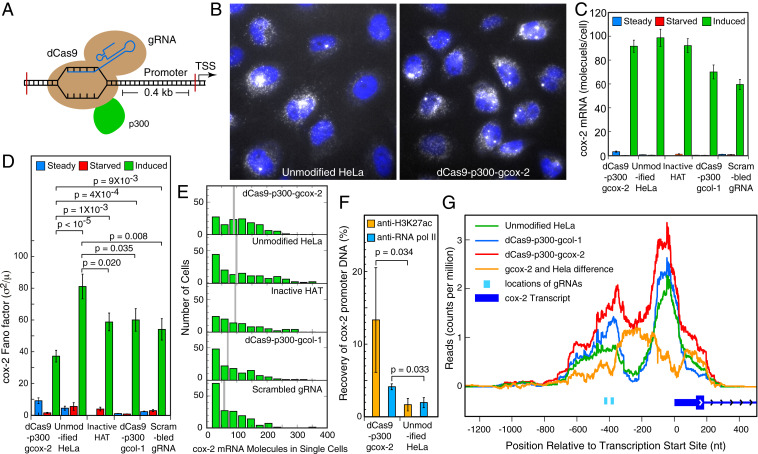
Fig. 1.
dCas9-mediated tethering of a HAT at the promoter of the cox-2 gene (also known as PTGS2) increases accessibility to the locus and decreases noise in expression from it. (A) Schematic depiction of how the dCas9–HAT fusion construct is attached at the promoter of the cox-2 gene. TSS here refers to the transcription start site, and p300 is the HAT used here. (B) Representative smFISH images of cox-2 mRNA molecules (white) in two cell lines 4 h after serum stimulation. The images are merged z-stacks with DAPI-stained nuclei (blue). (C) Average number of cox-2 mRNA molecules in single cells determined by smFISH 4 h after induction of the gene by the addition of serum. (Left to Right) The data are from cell lines expressing dCas9 and cox-2 gRNAs, unmodified HeLa cells, a dCas9 fused to a catalytically inactive HAT and cox-2 specific gRNAs, dCas9 and col-1 gRNAs, and dCas9 and a scrambled gRNA with no target in the cells. (D) Gene expression noise levels in terms of Fano factors (σ2/μ). Error bars represent the standard deviations (SDs) derived by the bootstrapping of single-cell counts obtained from 102 to 169 cells in a single experiment. The calculated probability (P values) are the probabilities that the observed difference in the mean Fano factor values in the pairs indicated by horizontal brackets can arise by chance. (E) Distribution of cox-2 mRNA counts in indicated clones 4 h after induction. Mean values are indicated by gray bars. (F) Percent recovery of cox-2 promoter DNA after ultralow-input native ChIP using antibodies against lysine 27 of histone 3 and the active form of RNA polymerase II after induction of the cox-2 gene for 4 h. Error bars are the SD of the mean from three biological replicates and P values from a single-tailed t test. (G) ATAC-seq analysis of an accessible region of chromatin at the promoter of the cox-2 locus in the indicated cell lines after serum induction. The reads represent counts per million of total reads in the sample (which ranged from 71 to 84 million) that map at each nucleotide. Measures of ATAC-seq quality are presented in SI Appendix, Fig. S4.
We employed two guide RNAs (gRNAs) to attach dCas9 to the promoter region of the gene that encodes cyclooxygenase-2 (cox-2, also known as prostaglandin-endoperoxide synthase 2) on chromosome 1 while avoiding the regions of the promoter where gene specific and general TFs are expected to bind (SI Appendix, Fig. S1). As a control, we also targeted the promoter region of a second gene located on chromosome 11, collagenase-1 (col-1, also known as matrix metalloproteinase-1), with a second set of two gRNAs. As a second control, we utilized a scrambled gRNA, which had no target in the human genome. Finally, as a third control, we used cox-2–specific gRNAs in combination with dCas9-p300-D1399Y in which the HAT domain is catalytically inactive because of a point mutation (22). Since the expression of heterologous proteins and RNAs via transient transfections would result in very high cell-to-cell variation, unrelated to the natural variation we seek to explore, we isolated stable HeLa cell lines that harbor integrated copies of the templates for dCas9-p300 and gRNAs within their genomes. The cell lines were characterized for the expression of dCas9-p300 mRNA and proteins as well as for the presence of gRNA templates within their genome. All cells in each clone expressed the dCas9 constructs with little cell-to-cell variation, as shown by antibodies against FLAG and HA-tags present on the dCas9 constructs, as well as single-molecule fluorescence in situ hybridization (smFISH) for their transcripts (SI Appendix, Fig. S1).
The cox-2 and col-1 genes are induced in serum-starved HeLa cells a few hours after the addition of serum. This occurs because the addition of serum leads to the synthesis of immediate early TFs, such as c-fos and c-jun, that in turn drive the expression of late-response genes, including cox-2 and col-1 (23). The expression of cox-2 and col-1 mRNA is characterized by high cell-to-cell variation (24).
We determined the number of cox-2 mRNA molecules in 102 to 169 single cells of the two cell lines, along with control cell lines and unmodified HeLa cells, by performing smFISH (25), using probes complementary to the coding sequence of cox-2 mRNA (Fig. 1B and SI Appendix, Fig. S2). These measurements indicated that the average number of cox-2 mRNAs after induction in single cells of all five categories were similar to each other (Fig. 1C). None of the cell lines expressed significant levels of cox-2 mRNAs under the conditions of serum starvation or steady state-growth, indicating that the tethering of HAT at the promoter of cox-2 in and of itself is not sufficient for mRNA production; additional gene-specific TFs (c-fos/c-jun heterodimers) that are produced after serum induction must also be present (24).
Cell-to-cell variation, or the noise level in gene expression, is often quantified by the Fano factor (square of the SD [σ] divided by the mean [μ]) that provides a measure of how far a population departs from a Poisson distribution, which would occur if mRNAs were to be produced and degraded steadily with equal rates in different cells (26). The Fano factor for a true Poisson distribution is one. When we determined the Fano factor for the cell populations mentioned above, we found that the cell line dCas9-p300-gcox-2 exhibited a significantly smaller Fano factor than the other cell lines (Fig. 1D). Histograms depicting the mRNA distributions in single cells are presented in Fig. 1E, and evidence of repeatability of the Fano factor measurements is presented in SI Appendix, Fig. S3. This data indicates that coexpression of the HAT construct dCas9-p300, along with the gRNAs designed to tether the HAT construct to the cox-2 gene promoter, leads to a reduction in the level of noise in the expression of the cox-2 gene compared to the controls. However, in this experiment, we also observed that the expression of HAT constructs and control gRNAs lead to some reduction in the Fano factor compared to unmodified HeLa cells, suggesting that the expression of these heterologous constructs may alter the noise characteristics of cox-2 to some extent (see Discussion).
HAT Tethering Increases Acetylation and DNA Accessibility at Targeted Gene Loci.
Since tethering of HAT at the promoter region leads to a decrease in gene expression noise, we asked whether the chromatin in this region of the gene locus had indeed been acetylated and become more accessible to TFs as a result of local HAT activity. Performing these analyses on a bulk cell population rather than on single cells, we first determined if the targeted region of the chromatin showed enhanced H3K27ac. To this end, we performed chromatin immunoprecipitation (ChIP) using an antibody specific to H3K27ac followed by real-time qPCR, amplifying a region of the cox-2 promoter (SI Appendix, Fig. S1A and Dataset S1). H3K27 acetylation is an expected consequence of HAT activity catalyzed by the p300 core that we had employed in our fused protein construct (22). The results indicated that H3K27ac is indeed enriched in the promoter region of the cox-2 gene in cell line dCas9-p300-gcox-2 compared to unmodified HeLa cells in the serum-induced state of the cells (Fig. 1F). We then performed a second ChIP analysis using an antibody against RNA polymerase II, and we found that there were more RNA polymerase II molecules present in the same region of the cox-2 promoter in the dCas9-p300-gcox-2 cell line compared with unmodified HeLa cells (Fig. 1F).
In order to determine whether the promoter had become more accessible as a result of these alterations, we carried out ATAC-seq, which permits very sensitive genome-wide identification of open regions in native chromatin using a hyperactive mutant of Tn5 transposase that inserts sequencing adapters into open regions of the genome (27). Previous studies indicate that the regions near transcription start sites of expressed genes are generally more accessible than the rest of the chromatin (28); accordingly, our ATAC-seq analysis showed focused accessibility peaks near the transcription start sites of many genes, including cox-2 and col-1 (SI Appendix, Fig. S4), as well as genome wide (Dataset S2). A closer analysis on chromatin derived from the cell line dCas9-p300-gcox-2 indicated that the promoter region of the cox-2 gene was more accessible in this cell line compared to unmodified HeLa cells and control cell line dCas9-p300-gcol-1 (Fig. 1G). When the reads mapping to unmodified HeLa cells are subtracted from the reads mapping to dCas9-p300-gcox-2, a differentially more accessible region between the locations of the gRNAs and the transcription start site became apparent (Fig. 1G, orange track). Such changes were not seen at other regions of the chromatin where gRNAs were not targeted (SI Appendix, Fig. S4B).
Utilizing a SunTag Strategy to Increase Accessibility at col-1 Promoter.
In the forgoing experiments in which we studied the impact of HAT tethering on the cox-2 gene, the cell line dCas9-p300-gcol-1 served as a control in which gRNAs were targeted to a gene on a different chromosome. However, when we analyzed the level of noise in col-1 mRNA expression in this cell line, we found that it was only modestly lower than the unaltered HeLa cells (data not presented). This was the case, despite the successful expression of dCas9-p300 and two col-1 promoter-specific gRNAs in the cell line. Col-1 is different from cox-2, as it is expressed in just a few cells in the HeLa cell population after serum induction (Fig. 2B and SI Appendix, Fig S2). In addition, a comparison of ATAC-seq between the two gene loci indicated that while the cox-2 locus exhibits an accessibility peak near the transcription start site both before and after serum induction, the col-1 locus does so only after serum induction (SI Appendix, Fig. S4C). These observations suggest that the col-1 gene locus is highly condensed in HeLa cells.
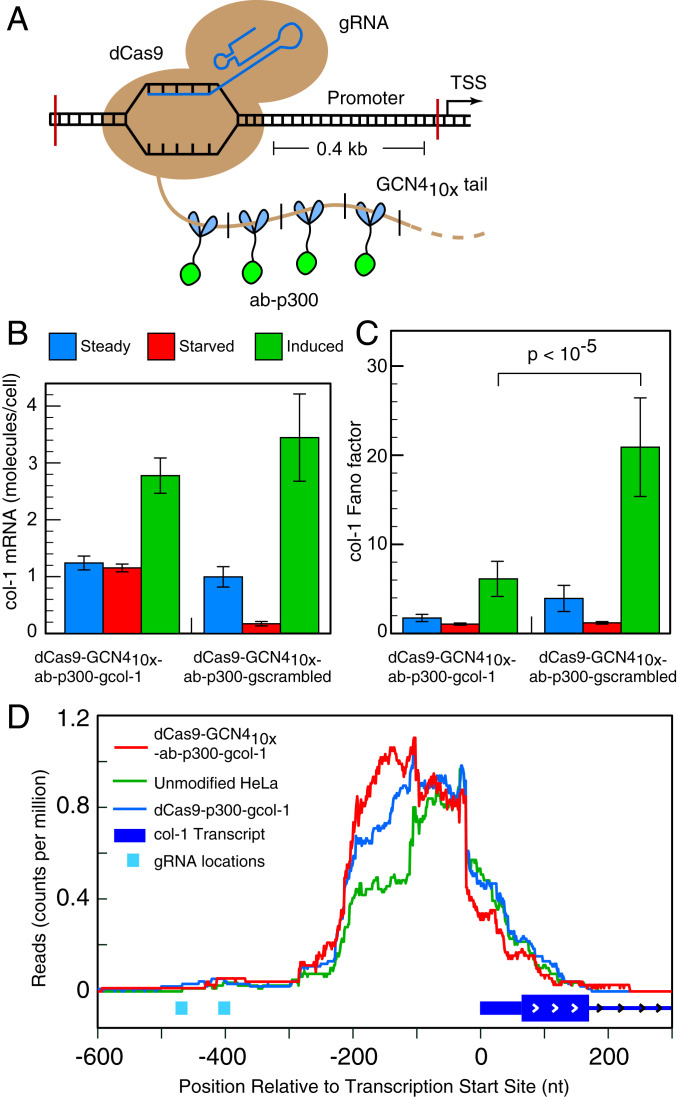
Fig. 2.
Impact of tethering a large number of HAT p300 molecules to the col-1 (also known as MMP1) promoter on the noise in gene expression from that locus. (A) The SunTag strategy for attaching 10 copies of p300 fused to a single-chain antibody against peptide GCN4. (B) Average number of col-1 mRNA molecules in single cells determined by smFISH in cells that are steadily growing, serum starved, or stimulated for 4 h by the addition of serum after starvation. The data are from two different cell lines expressing dCas9-GCN410x, ab-p300, and indicated gRNAs. (C) Fano factor determined from single-cell col-1 mRNA counts. Error bars represent the SDs derived by the bootstrapping of single-cell counts obtained from 122 to 163 cells in a single experiment. The P values are the probabilities that the observed difference in the mean Fano factor values in the pairs indicated by horizontal brackets can arise by chance. (D) ATAC-seq analysis of the accessible region of chromatin at the col-1 promoter of the indicated cell lines after serum induction. The reads represent counts per million of total reads in the sample (which ranged from 69 to 84 million) that map at each nucleotide.
We reasoned that tethering a larger number of HAT constructs would be more effective in decondensing the col-1 locus than a single HAT construct as done above. To this end, we exploited the SunTag strategy (29), which can tether up to 10 effector constructs at the target site using one gRNA. In the SunTag strategy, dCas9 is not fused to the effector protein directly, but it is fused to a multimeric peptide, GCN4 (10 units), in which each GCN4 unit can be bound to a second construct composed of a single-chain variable–fragment antibody against peptide GCN4 fused to the effector protein. Coexpression of the two constructs, along with appropriate gRNAs in the same cell, leads to attachment of the 10 copies of the effector protein to the genomic locus for each gRNA (29).
We fused the p300 core at the C terminus of the single-chain variable–fragment antibody against peptide GCN4 (referred to as ab-p300) (Fig. 2A). This construct, along with dCas9-GCN410x– and col-1–specific gRNAs, was stably integrated into the genome of a HeLa cell line referred to as dCas9-GCN410x-ab-p300-gcol-1. Immunofluorescence imaging showed that the cell line expressed both components of the CRISPR system (SI Appendix, Fig. S1C). We also prepared a control cell line in which the gRNAs were designed against a target that did not exist in the cell (dCas9-GCN410x-ab-p300-gscrambled). We then measured the mean mRNA expression levels and Fano factor for each of the two cell lines growing under steady state, in the serum-starved state, and in the serum-induced state (Fig. 2 B and C). The results showed that the Fano factor is lower for cell line dCas9-GCN410x-ab-p300-gcol-1 than it is for the control cell line, indicating that the tethering of multiple HATs at the col-1 promoter leads to the lowering of gene expression noise from that locus.
To determine whether the attachment of multiple p300 molecules had indeed rendered the locus more open, we examined the accessibility at the locus by ATAC-seq. The results show that a region between the gRNA location and the transcription start site was most accessible in cell line dCas9-GCN410x-ab-p300-gcol-1, less accessible in dCas9-p300-gcol-1, and least accessible in unaltered HeLa cells (Fig. 2D). The results also indicate that the two approaches impact the same region. Overall, we find that HAT tethering at the promoter regions leads to increased accessibility, which lowers the noise in the gene expression from both genes that we examined.
Tethering of HAT to Gene Loci Leads to an Increase in the Number of Transcriptionally Active Loci.
Although statistical measurements such as the Fano factor, particularly with single-cell RNA counts, have been used in many studies, they are a rather indirect measure of dynamic gene activity (the on and off states). When the half-life of mRNA is relatively high, rapid changes in gene activity are reflected in the mRNA population to a lesser extent (9). A more direct visualization of the on and off states of gene loci can be made by imaging the nascent RNA at the gene locus (5, 30–32). During bursts, the rate of mRNA synthesis is higher than its dispersal into the nucleoplasm, and as a consequence, a cluster of nascent mRNA molecules composed of pre-mRNAs in various stages of splicing is often visible at the gene locus (31, 32). Since a majority of introns are spliced cotranscriptionally and are rapidly degraded after splicing, the imaging of these introns using smFISH probes can serve as a tool for the visualization of gene loci that are active at the time of cell fixation (31).
We visualized cox-2 gene loci in their on states using both intron- and exon-specific probes for cox-2 (Fig. 3A). We found that zero to four br ight foci that were specific to the intron and exon probe sets were visible within each HeLa cell nuclei. While exon-specific probes illuminated the bright foci within the nucleus as well as the single-mRNA molecules scattered throughout the cell volume, intron-specific signals were present only at the nuclear foci. These dual-labeled nuclear foci were visible only after serum induction.
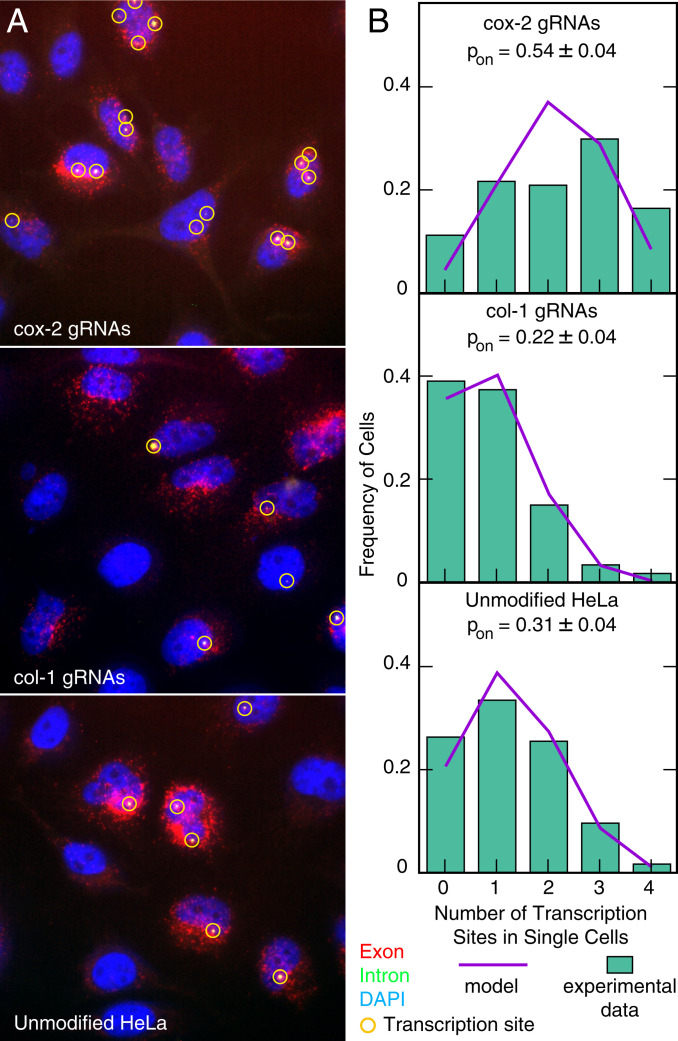
Fig. 3.
Analysis of the number of transcriptionally active cox-2 gene loci (out of four alleles present in HeLa cells) 4 h after serum induction. (A) Representative images of cox-2 mRNA imaged using exon-specific (red) and intron-specific smFISH (green) probe sets from cell lines expressing dCas9 and cox-2 gRNAs, dCas9 and col-1 gRNAs, and unmodified HeLa cells. The images are color-coded, maximum intensity–merged z-stacks. Intron-specific probes in this case reveal only the nascent mRNAs that are yet to be released from the gene loci, whereas the exon-specific probes reveal both the nascent RNAs at the gene locus and the mature mRNAs dispersed throughout the cell. Algorithmically identified transcription sites are marked by yellow circles. (B) The distribution of the number of active transcription sites from 130 to 140 cells (bars) and their best fits to the binomial distribution considering the four copies of the gene are identical and independent (lines). The probabilities of a gene locus being on and their 95% CI are shown.
Using the average intensity of single, well-separated mRNA spots as a “unit intensity,” we could determine the number of mRNA molecules that were present at the gene loci (32). This analysis indicated that, although the number of mRNA molecules present at the bright nuclear foci were highly variable, on average, there were 13 mRNA molecules within the loci (SI Appendix, Fig. S5A).
In order to show that these bright nuclear foci represent nascent mRNAs tethered to the gene loci, we treated HeLa cells with actinomycin D, which rapidly stops transcription. Within 5 min of the addition of actinomycin D, the intensity and the numbers of the foci dropped precipitously (SI Appendix, Fig. S5B). After 60 min of actinomycin D treatment, no nuclear foci were visible. By contrast, mature mRNAs remained unaffected during this period. These observations indicate that the nuclear foci are active transcription sites (TSs). Our observation of up to four TSs is consistent with genome sequencing studies of HeLa cells, which indicate that a large portion of chromosome 1 that surrounds the cox-2 gene is present in four copies within the genome of HeLa cells (33).
To explore whether the tethering of HAT affects the number of active TSs, we imaged the cox-2 TSs in cell lines dCas9-p300-gcox-2, dCas9-p300-gcol-1, and in unmodified HeLa cells 4 h after serum induction. We counted the frequencies of cells exhibiting zero, one, two, three, or four TSs in each cell line (Fig. 3B). The results indicate that the dCas9-p300-gcox-2 cell line possesses more cells with three and four sites and less cells with zero, one, and two sites compared to the dCas9-p300-gcol-1 cell line and unmodified HeLa cells, which display the reverse pattern. Assuming that all four copies of the gene are identical and independent of each other, the number of active TSs was fitted to a binomial distribution. The results show that the probability of a cox-2 gene locus being on is higher in cell line dCas9-p300-gcox-2 in which the gRNAs mediate attachment of HAT at that locus than in cell line dCas9-p300-gcol-1 or in unmodified HeLa cells (Fig. 3B). This observation strongly corroborates the observation above that HAT tethering lowers gene expression variability as measured by the Fano factor of mature mRNAs.
Each Copy of the cox-2 Gene Is Equally Impacted by HAT Tethering.
Since each of the four cox-2 gene loci are virtually identical, as a first approximation, one can assume that the accessibility of each locus to TFs will increase equally in cell line dCas9-p300-gcox-2. However, studies of CRISPR-mediated gene editing of pairs of loci of which one member is in a heterochromatin state and the other in a euchromatin state have revealed that eventually they both get edited, but the heterochromatin allele is slower to be edited (34). Thus, there is a potential for an unequal decompaction of the four loci within the same cells. We therefore set out to develop an e xperimental system to determine whether each of the four loci is equally likely to experience bursts of transcription, or only a subset of them experience that.
In our method, we distinguish different cox-2 gene loci by in situ detection of a naturally occurring single-nucleotide polymorphism (SNP) in nascent RNA clusters at the cox-2 TSs. To identify a distinguishing SNP that is present in the HeLa cell cox-2 gene copies, we interrogated five SNPs in the cox-2 gene that occur in human populations at relatively high frequencies by allele-specific PCR amplification. We found that a cytidine/thymidine SNP exists at 50% frequency within the 3′ untranslated region of HeLa cell cox-2 mRNA at complementary DNA position 2375 (SI Appendix, Fig. S6). This data indicates that two of the four cox-2 alleles in HeLa cells have a C, and the other two alleles have a T at this position. To distinguish the TSs among the expressed alleles, we utilized allele-specific amplified FISH (amp-FISH), which produces distinguishably colored amp-FISH signals for each allele (35). In addition, we simultaneously detected all active gene loci using directly labeled smFISH probe sets against introns (Fig. 4A).
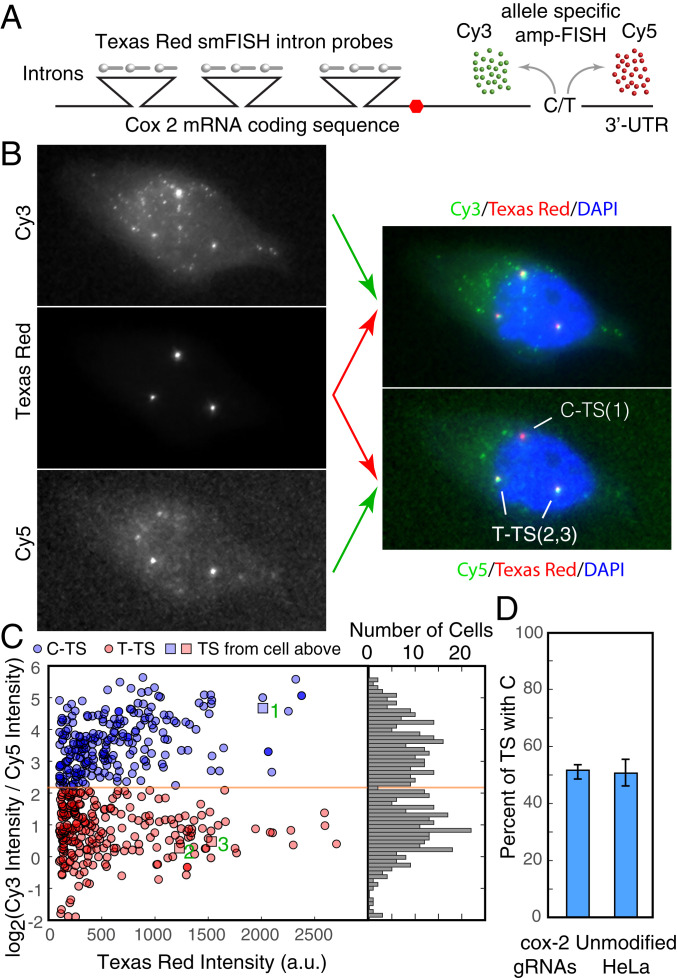
Fig. 4.
Determination of the likelihood with which two variants of the four cox-2 gene loci are turned on. (A) Schematic for using amp-FISH to distinguish two variants of the cox-2 gene that differ by an SNP in the 3′ untranslated region (UTR ) of the gene. One variant has a C at complementary DNA position 2375, and the other variant has a T at the same position. The Texas Red–labeled intron-specific set of smFISH probes label all cox-2 TSs, whereas allele-specific amp-FISH (35) creates a Cy3 spot if C is present and a Cy5 spot if T is present. (B) Analysis of a cell that has three active transcription sites. (Left) Merged z-stacks in each of the three different channels. (Right) Their color-combined versions. (Top Right) The Texas Red image is depicted in red and the Cy3 image in green, whereas in lower right, the Texas Red image is depicted in red and the Cy5 image in green. The determinations of the sites are indicated. (C) Analysis of 493 TSs from 521 cells using intensity measurements (in arbitrary units [a.u.]) at the TSs from the cell of line dCas9-p300-gcox-2 and from unmodified HeLa cells. The population falls into two groups based on the log2 ratio of the Cy3 intensity/Cy5 intensity at the TSs (histogram on Right). The reason that the yellow line does not fall at 0 on the y-axis is because the discriminatory capacities of the two probes are not equal (SI Appendix, Materials and Methods). The intensities of the three TSs depicted in the cell in B are indicated by numbered squares in the scatter diagram. (D) The likelihood of turning on the C sites in cell line dCas9-p300-gcox-2 is compared to the likelihood of turning on C sites in unmodified HeLa cells.
This three-color smFISH/amp-FISH technique can be used to distinguish the two kinds of TSs as illustrated in Fig. 4B, which shows merged z-stacks for the same cell in each of three channels on the left and pairwise color-coded merges of these images on the right. The merged color images show that this cell had three active TSs, one of which had a C genotype, and the other two had a T genotype. After identifying TSs in about 500 cells (in cell line dCas9-p300-gcox-2 and in unmodified HeLa cells) using intron-specific signals, we computed the ratios of C and T SNP-specific amp-FISH signals at the TSs. The log2 of these ratios displayed a bimodal distribution that partitioned the population into two approximately equal halves, with the upper half representing the C genotype and the lower half representing the T genotype (Fig. 4C). While the DNA sequencing data in SI Appendix, Fig. S6 indicates that the C and T alleles are present in two copies each, this data shows that the two alleles are equally likely to be expressed. Furthermore, supporting the same idea, when cells displayed more than two active TSs, the number of active sites with each genotype was always two or less.
In order to determine whether the expression dCas9-p300– and cox-2–specific gRNAs impact the two versions of the cox-2 genes equally or are preferential toward one of the alleles, we measured the frequency of each activated allele in cell line dCas9-p300-gcox-2 and in unmodified HeLa cells (from data in Fig. 4C, which represents both cases) and found that they were indistinguishable from each other (Fig. 4D). This indicates that the probability of turning on each allele is the same in both kinds of cells, even though the dCas9-p300-gcox-2 turns on more gene loci per cell than unmodified HeLa cells. If dCas9-p300 tended to attach to and preferentially decondense one of the alleles, that allele would have been expressed with a higher frequency, altering this balance. This experiment, however, does not address any potential difference that may exist between the two copies of the same allele.
Impact of Increased Accessibility at the Locus on the Kinetic Steps of Transcription.
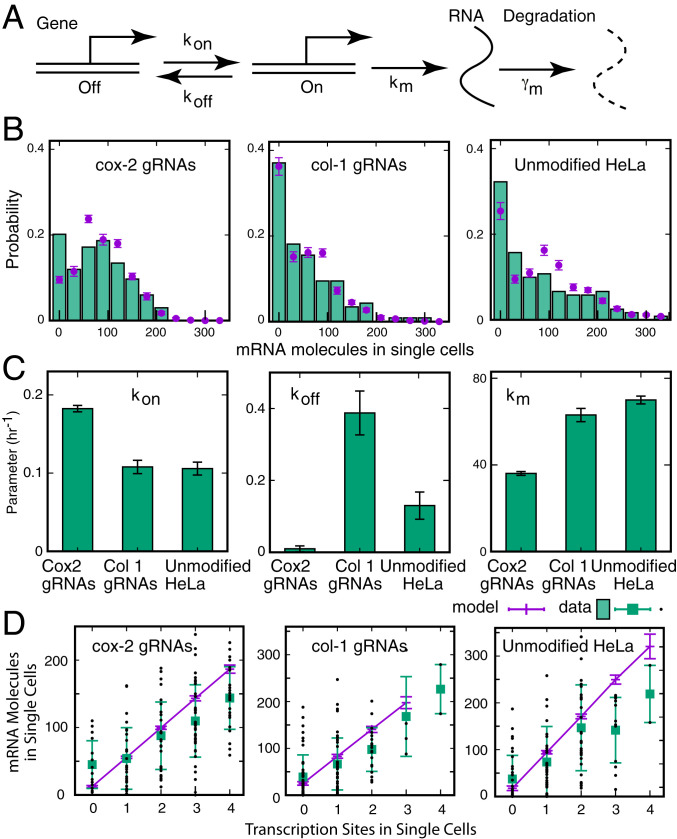
To gain further insights into the mechanisms of burst generation, we analyzed the single-cell cox-2 mRNA distributions and TS data described above using the two-state model of gene expression (Fig. 5A) (9, 16). In our model, we consider four identical copies of the cox-2 gene in the same cells. Each gene copy switches randomly and independently between two states, on and off, with rates kon and koff. While in the on state, mature mRNAs are synthesized with rate km at the gene locus. The degradation rate of cox-2 mRNA is , which is known in HeLa cells (half-life 90 min) (36). We solved our model by assuming that all gene copies are in the off state, and mRNA counts are zero before serum induction (time, t = 0), and then obtained TS and mRNA distributions at 4 h after serum induction (t = 4). The TS distribution is given by a binomial distribution with the probability of the gene in the on state being time dependent (model details provided in SI Appendix). The mRNA count distributions were obtained by running Monte Carlo simulations using the Gillespie algorithm (37). We first estimated the probability of a gene being in the on state by fitting the experimental TS data to the binomial distribution. Then, we inferred the kinetic parameters by fitting the mRNA count data while constraining the model to the estimated on-state probabilities (Fig. 5 B and C). The two-state model not only faithfully captured the measured mRNA count distribution across the three different cell types (Fig. 5A), but it also predicted the correlation between mRNA counts and the number of active TSs in individual cells reasonably well (Fig. 5D). The values of the estimated kinetic parameters indicated that although our system does not reach a steady state in 4 h, the tethering of HAT at the promoter lowers the noise in gene expression throughout the rest of the predicted time course (SI Appendix, Fig. S7).
Fig. 5.
Determination of the kinetic steps in transcription using single-cell mRNA measurements and the two-state model. (A) Each of the four copies of the gene can switch randomly between an on state and an off state, with rates and , producing mRNAs in the on state with rate . After its synthesis, mRNA decays with the rate of γm, which is assumed to be constant. These parameters were inferred from single-cell data using a maximum likelihood estimation method (SI Appendix). (B) Predictions of the two-state model (purple circles) overlaid on the distribution of cox-2–mRNA molecules in single cells. (C) Estimates of the three kinetic parameters from the model. (D) Relationship between the number of active transcription sites and mRNA copy number in the three cell types and the prediction of the model. The error bars correspond to SDs.
Estimates of the three parameters for cox-2 expression in serum-induced cell lines dCas9-p300-gcox-2, dCas9-p300-gcol-1, and HeLa cells at 4 h presented in Fig. 5C indicate that in cell line dCas9-p300-gcox-2 there was an increase in kon and a decrease in koff compared to the two controls. It is expected that kon and koff will depend on the affinity of TF toward the promoter, TF concentration, and the accessibility of the promoter to the TF through chromatin (32, 38). However, since TF affinity and concentration are unlikely to be altered in the dCas9-p300-gcox-2 cell line compared to the controls, we conclude that increased promoter accessibility brought about by HAT tethering increases chromatin accessibility and leads to observed changes in kon and koff. By themselves, these changes would result in an increase in the net amounts of mRNAs per cell, which does not occur (Fig. 1C). However, the system somehow decreases the rate of mRNA synthesis, km, to keep the levels of mRNAs about the same (see Discussion).
A number of studies have identified cell-to-cell variation in the gene extrinsic factors, such as cell cycle stage and size of the cell, as contributing factors in the observed cell-to-cell variability in mRNA copy numbers of some mRNAs (4, 39). However, our system is naturally synchronized with respect to cell cycle because serum starvation arrests cells in the nondividing G0 stage, and after 4 to 6 h of serum induction, most of them will be in the same stage of the cell cycle (23, 24). Furthermore, we found that the copy number of mRNAs is not correlated with cell size in our system (SI Appendix, Fig. S8). Finally, gene extrinsic factors are expected to influence the probability of each of the four cox-2 TSs being on in the same way, suggesting that promoter fluctuations are the main source of variability in our system.
Discussion
Both prokaryotes and eukaryotes exhibit noisy gene expression. In prokaryotes, part of the noise arises because so few of TF molecules are present in each cell that their interactions with the target gene become stochastic (40). This cannot be the case in higher eukaryotes because the TFs are usually present in much larger numbers (10,000 to 300,000 copies per nucleus), which would be thermodynamically sufficient to permit binding of TFs to the DNA most of the time (41). However, even though all four cox-2 loci share the same pool of TFs in the nucleus, only a subset of them is turned on at any given time, suggesting that some of the key determinants of stochasticity lie at the gene locus in eukaryotes.
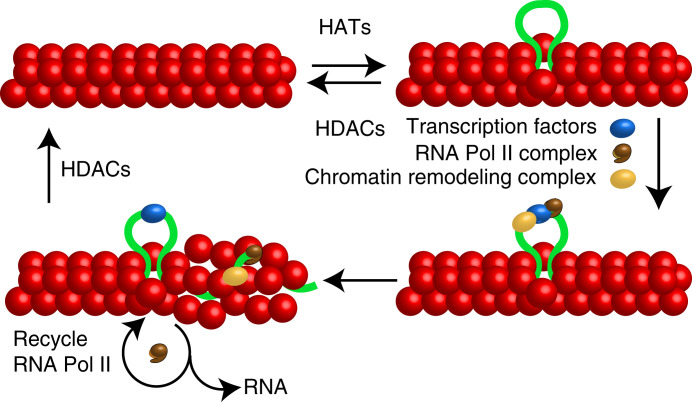
These results support the random chromatin accessibility hypothesis (1, 9, 19) as an explanation of episodic transcriptional bursts in higher eukaryotes. According to this model, gene loci are normally refractory to the binding of otherwise ample gene-specific TFs because they are sequestered within tight chromatin. Random local “breathing” events brought about by encounters with diverse diffusible chromatin decondensation factors, such as HATs, allow the TFs to bind to these loci (Fig. 6). After they bind to the locus, TFs recruit RNA polymerase II and additional chromatin modulating complexes, which, in addition to producing mRNA, decondense the locus in a more sustained manner (42). Convoys of RNA polymerase molecules plow through the gene, one behind the other (11), and are recycled back to the promoter after they complete RNA synthesis (12), thereby producing a burst of mRNA (the on state). RNA polymerase II recycling is facilitated by a reinitiation complex that stays bound to the promoter, while polymerase complexes are copying the gene (43). The burst ends when interactions of the locus with chromatin condensation factors, such as HDACs, render it quiescent again (the off state). In this model, we refer to the intermittent access of TFs to the promoters; however, the same applies to the access of TFs to the enhancers as well as to the interactions between promoters and enhancers. A number of previous studies have provided support for aspects of this model. For example, Dey et al. showed that HIV reporters display higher levels of noise when they are integrated in more dense chromatin regions than in less dense chromatin (17).
Fig. 6.
Schematic depiction of the random chromatin accessibility model for intermittent mRNA synthesis. Chromatin prevents TFs from accessing the regulatory regions of genes. Diffusible chromatin remodelers (HATs, pioneer factors, and HDACs are examples) dynamically decondense and condense chromatin. Occasional exposure of promoters allows gene-specific and general TFs to bind to the gene, which then recruit the RNA polymerase II complex and additional, polymerase-associated, and chromatin-remodeling complexes to the gene locus. The activities of these complexes decondense the gene in a more sustained manner, and RNA polymerase II is recycled repeatedly to produce bursts of mRNA synthesis (the on state). The locus becomes compact again by the stochastic action of chromatin condensing remodelers, and an off state ensues.
Although the dCas9-HAT construct is expected to increase accessibility at the target gene via a gRNA-directed mechanism, in Fig. 1D we observed a decrease in cox-2 Fano factor even when control gRNAs were employed, or when an inactive HAT construct was used, compared to unmodified HeLa cells. This might occur through an unspecified mechanism, or it may represent an experiment-to-experiment variation in HeLa cell Fano factor measurements. However, in light of the repeats of Fano factor measurements presented in SI Appendix, Fig. S3 and previous HeLa cells measurement reported by Shah and Tyagi (24), which indicate that the Fano factor of unmodified HeLa cells is about the same as the controls (ranging from 55 to 70), we prefer the latter explanation.
Although we utilized HATs as the agent of chromatin decondensation, there is a diverse array of other chromatin modulators whose activities include histone methylation, phosphorylation, and ubiquitination, as well as pioneer factors that can access DNA while it is bound to the nucleosome, and they may play a role in providing initial accesses to the TFs (42).
In the present study, the tethering of HAT to the promoter regions of two different gene loci led to increased accessibility for TFs to these genes. This resulted in reduced transcriptional noise from the loci and in the appearance of a larger number of active TSs in single nuclei. An analysis of single-cell counts of mature mRNA molecules and the number of active TSs using the two-state model showed that when HAT is tethered to a gene locus, although the average number of mRNAs produced in single cells does not change appreciably, the rate of turning the gene on, kon, is increased; the rate of turning the gene off, koff, is decreased; and rate of RNA synthesis, km, is decreased. In light of this model, it is reasonable to expect that a constitutively increased TF access to the promoter leads to an increase in kon and a decrease in koff. However, the reason why increased accessibility leads to a reduction in the rate of mRNA synthesis, km, is not clear. A plausible hypothesis is that, although the binding of HAT increases accessibility of RNA polymerase II from the nuclear pool, it adversely affects the recycling of RNA polymerases II back to the promoter after they finish RNA synthesis by changing the local environment at the promoter. As a consequence, fewer RNA molecules are produced in each burst. Support of this hypothesis becomes apparent when we divide the mRNA counts in single cells by the number of active TSs visible in the same cells, which yields the mRNAs produced by an average active TS (accepting the caveat that very nascent TSs do not contribute to the pool of total mRNAs in the cell). This “output” of the average TS was 41 molecules for dCas9-p300-gcox-2, 82 molecules for dCas9-p300-gcol-1, and 75 molecules for unmodified HeLa cells, which is suggestive of the proposed dampening.
Two recent studies that performed similar analyses also support our model. Chen et al. tethered dCas9-p300 to the promoter and the enhancers of the FOS gene in neurons, and Nicolas et al. tethered the same construct to the promoter of the circadian rhythm gene Bmal on a luciferase reporter, discovering that the overall transcriptional noise was reduced by the perturbations (20, 21). However, unlike in our study in which average mRNA levels stayed the same, in these studies they slightly increased (20). There are significant differences in the biological contexts of these studies, including that the FOS gene undergoes negative feedback inhibition (23, 24), and the Bmal gene locus is subjected to circadian rhythmic controls (19, 21). Whether different biological contexts are responsible for the observed differences is presently not clear.
It is interesting to consider whether HAT tethering would impact the noise in the expression of genes situated in the neighborhood of the targeted locus. Our ATAC-seq experiments reveal that the opened chromatin region is restricted to a 100- to 400-nucleotide (nt )-wide region situated 200 to 300 nt downstream of the gRNA locations. Since, in higher eukaryotes, the neighboring genes are often separated by much longer stretches of noncoding DNA, the possibilities of a tethered HAT influencing a neighboring gene seem rather low.
A fruitful avenue of further exploration would be to determine what distinguishes the on state from the off state and how these two states differ from the preinduction off state in terms of the protein factors that are bound to the gene locus.
In addition to revealing random chromatin access as a source of transcriptional noise, our study, together with two previous studies (20, 21), provides a direct way to control noise in gene expression. Although there is a diverse array of natural noise–buffering mechanisms in eukaryotic organisms (24, 44), transcriptional noise is a significant impediment in the construction of artificial gene regulatory pathways and circuits in synthetic biology (45). When constructing these artificial pathways, it will be fruitful to minimize transcriptional noise by HAT tethering. Furthermore, our study provides a tool to explore whether transcriptional noise is indeed the source of the phenomenon, such as the emergence of drug resistance during tumor progression (46), because now we are able to dampen the noise in the expression of any target gene using the dCas9-mediated tethering of HAT.
Materials and Methods
We integrated dCas9-p300 constructs and the gRNA templates into the genome of HeLa cells and isolated stable cell lines expressing both components. The plasmid templates for each component were linearized and cotransfected into HeLa cells followed by a selection of clones using puromycin. The selection marker was present only on the gRNA plasmids. Puromycin-resistant clones were screened for expression of dCas9 by smFISH against dCas9 mRNA to identify clones that express both components. To obtain cell lines expressing the three components of the SunTag strategy, we first transduced with a lentivirus harboring the ab-p300 fusion construct and isolated clones that express it by performing immunofluorescence against the anti-human influenza hemagglutinin tag that is a part of the insert. These clones were further transduced with a lentivirus expressing the dCas9-GCN410x construct, followed by clone selection by performing smFISH against dCas9 mRNA. Finally, gRNA templates were introduced into these clones by transfection, followed by puromycin selection. We used four gRNAs for each gene, but found that only two of them were stably expressed in our clones.
mRNA counts in single cells were obtained by performing smFISH against the coding sequences of the two genes and TS were visualized by smFISH against their intronic sequence. smFISH probe sequences are listed in Dataset S3. To qualify as a transcriptionally active TS, the nuclear foci had to exhibit both intronic and exonic signals.
Details for these methods, cloning of various inserts into plasmid vectors, serum induction, ChiP, image analysis, statistical analysis, and mathematical modeling are provided in SI Appendix. Details of ATAC-seq data analysis are presented in Dataset S4.
Acknowledgments
This research was supported by NIH Grants R01CA227291, R01AI106036, R01GM124446, and R01GM126557. We thank the Office of A dvanced Research Computing at Rutgers University for providing computing resources.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2018640118/-/DCSupplemental.
Data Availability
ATAC-seq data has been deposited in the Gene Expression Omnibus (GSE157399) and freely available Custom Image Processing Software suit is deposited at GitHub (https://github.com/TyagiLab/CountRNASpots). All other study data are included in the article and/or supporting information.
References
- 1.Raser J. M., O’Shea E. K., Control of stochasticity in eukaryotic gene expression. Science 304, 1811–1814 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanchez A., Golding I., Genetic determinants and cellular constraints in noisy gene expression. Science 342, 1188–1193 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Little S. C., Tikhonov M., Gregor T., Precise developmental gene expression arises from globally stochastic transcriptional activity. Cell 154, 789–800 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Padovan-Merhar O., et al., Single mammalian cells compensate for differences in cellular volume and DNA copy number through independent global transcriptional mechanisms. Mol. Cell 58, 339–352 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bahar Halpern K., et al., Bursty gene expression in the intact mammalian liver. Mol. Cell 58, 147–156 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Larsson A. J. M., et al., Genomic encoding of transcriptional burst kinetics. Nature 565, 251–254 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tunnacliffe E., Chubb J. R., What is a transcriptional burst? Trends Genet. 36, 288–297 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Chubb J. R., Trcek T., Shenoy S. M., Singer R. H., Transcriptional pulsing of a developmental gene. Curr. Biol. 16, 1018–1025 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raj A., Peskin C. S., Tranchina D., Vargas D. Y., Tyagi S., Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 4, e309 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Voss T. C., et al., Combinatorial probabilistic chromatin interactions produce transcriptional heterogeneity. J. Cell Sci. 122, 345–356 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tantale K., et al., A single-molecule view of transcription reveals convoys of RNA polymerases and multi-scale bursting. Nat. Commun. 7, 12248 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hebenstreit D., Are gene loops the cause of transcriptional noise? Trends Genet. 29, 333–338 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Bartman C. R., et al., Transcriptional burst initiation and polymerase pause release are key control points of transcriptional regulation. Mol. Cell 73, 519–532.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukaya T., Lim B., Levine M., Enhancer control of transcriptional bursting. Cell 166, 358–368 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartman C. R., Hsu S. C., Hsiung C. C., Raj A., Blobel G. A., Enhancer regulation of transcriptional bursting parameters revealed by forced chromatin looping. Mol. Cell 62, 237–247 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh A., Vargas C. A., Karmakar R., “Stochastic analysis and inference of a two-state genetic promoter model” in Proceedings of the American Control Conference (IEEE, 2013), pp. 4563–4568.
- 17.Dey S. S., Foley J. E., Limsirichai P., Schaffer D. V., Arkin A. P., Orthogonal control of expression mean and variance by epigenetic features at different genomic loci. Mol. Syst. Biol. 11, 806 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dar R. D., et al., Transcriptional burst frequency and burst size are equally modulated across the human genome. Proc. Natl. Acad. Sci. U.S.A. 109, 17454–17459 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suter D. M., et al., Mammalian genes are transcribed with widely different bursting kinetics. Science 332, 472–474 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Chen L. F., et al., Enhancer histone acetylation modulates transcriptional bursting dynamics of neuronal activity-inducible genes. Cell Rep. 26, 1174–1188.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicolas D., Zoller B., Suter D. M., Naef F., Modulation of transcriptional burst frequency by histone acetylation. Proc. Natl. Acad. Sci. U.S.A. 115, 7153–7158 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hilton I. B., et al., Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 33, 510–517 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iyer V. R., et al., The transcriptional program in the response of human fibroblasts to serum. Science 283, 83–87 (1999). [DOI] [PubMed] [Google Scholar]
- 24.Shah K., Tyagi S., Barriers to transmission of transcriptional noise in a c-fos c-jun pathway. Mol. Syst. Biol. 9, 687 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raj A., van den Bogaard P., Rifkin S. A., van Oudenaarden A., Tyagi S., Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods 5, 877–879 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ozbudak E. M., Thattai M., Kurtser I., Grossman A. D., van Oudenaarden A., Regulation of noise in the expression of a single gene. Nat. Genet. 31, 69–73 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Buenrostro J. D., Giresi P. G., Zaba L. C., Chang H. Y., Greenleaf W. J., Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boyle A. P., et al., High-resolution mapping and characterization of open chromatin across the genome. Cell 132, 311–322 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanenbaum M. E., Gilbert L. A., Qi L. S., Weissman J. S., Vale R. D., A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159, 635–646 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vargas D. Y., Raj A., Marras S. A., Kramer F. R., Tyagi S., Mechanism of mRNA transport in the nucleus. Proc. Natl. Acad. Sci. U.S.A. 102, 17008–17013 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vargas D. Y., et al., Single-molecule imaging of transcriptionally coupled and uncoupled splicing. Cell 147, 1054–1065 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Senecal A., et al., Transcription factors modulate c-Fos transcriptional bursts. Cell Rep. 8, 75–83 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Landry J. J., et al., The genomic and transcriptomic landscape of a HeLa cell line. G3 (Bethesda) 3, 1213–1224 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kallimasioti-Pazi E. M., et al., Heterochromatin delays CRISPR-Cas9 mutagenesis but does not influence the outcome of mutagenic DNA repair. PLoS Biol. 16, e2005595 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marras S. A. E., Bushkin Y., Tyagi S., High-fidelity amplified FISH for the detection and allelic discrimination of single mRNA molecules. Proc. Natl. Acad. Sci. U.S.A. 116, 13921–13926 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moore A. E., Young L. E., Dixon D. A., A common single-nucleotide polymorphism in cyclooxygenase-2 disrupts microRNA-mediated regulation. Oncogene 31, 1592–1598 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gillespie D. T., Exact stochastic simulation of coupled chemical reactions. J. Phys. Chem. 81, 2340–2361 (1977). [Google Scholar]
- 38.Xu H., Sepúlveda L. A., Figard L., Sokac A. M., Golding I., Combining protein and mRNA quantification to decipher transcriptional regulation. Nat. Methods 12, 739–742 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Foreman R., Wollman R., Mammalian gene expression variability is explained by underlying cell state. Mol. Syst. Biol. 16, e9146 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elowitz M. B., Levine A. J., Siggia E. D., Swain P. S., Stochastic gene expression in a single cell. Science 297, 1183–1186 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Biggin M. D., Animal transcription networks as highly connected, quantitative continua. Dev. Cell 21, 611–626 (2011). [DOI] [PubMed] [Google Scholar]
- 42.Li B., Carey M., Workman J. L., The role of chromatin during transcription. Cell 128, 707–719 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Yudkovsky N., Ranish J. A., Hahn S., A transcription reinitiation intermediate that is stabilized by activator. Nature 408, 225–229 (2000). [DOI] [PubMed] [Google Scholar]
- 44.Raj A., Rifkin S. A., Andersen E., van Oudenaarden A., Variability in gene expression underlies incomplete penetrance. Nature 463, 913–918 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murphy K. F., Adams R. M., Wang X., Balázsi G., Collins J. J., Tuning and controlling gene expression noise in synthetic gene networks. Nucleic Acids Res. 38, 2712–2726 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shaffer S. M., et al., Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature 546, 431–435 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
ATAC-seq data has been deposited in the Gene Expression Omnibus (GSE157399) and freely available Custom Image Processing Software suit is deposited at GitHub (https://github.com/TyagiLab/CountRNASpots). All other study data are included in the article and/or supporting information.