Significance
During infection, extracellular “labile” heme, released from damaged red blood or parenchymal cells, acts as prototypical alarmin stimulating myeloid cells. A characteristic hallmark of myeloid cell activation is the development of trained immunity, specified as long-lasting adaptations based on transcriptional and epigenetic modifications. In vivo, this is maintained by the rerouting of hematopoiesis. We found that heme is a previously unrecognized trained immunity inducer promoting resistance to bacterial infection in mice. This goes along with extensive long-lasting epigenetic memory in hematopoietic stem cells provoking drastic changes in the transcription factor–binding landscape of myeloid progenitor cells. Given the critical role of heme during infections, we propose that trained immunity is a more general component of innate immunity than previously suggested.
Keywords: heme, trained innate immunity, myelopoiesis, single-nuclei analysis, sepsis
Abstract
Trained immunity defines long-lasting adaptations of innate immunity based on transcriptional and epigenetic modifications of myeloid cells and their bone marrow progenitors [M. Divangahi et al., Nat. Immunol. 22, 2–6 (2021)]. Innate immune cells, however, do not exclusively differentiate between foreign and self but also react to host-derived molecules referred to as alarmins. Extracellular “labile” heme, released during infections, is a bona fide alarmin promoting myeloid cell activation [M. P. Soares, M. T. Bozza, Curr. Opin. Immunol. 38, 94–100 (2016)]. Here, we report that labile heme is a previously unrecognized inducer of trained immunity that confers long-term regulation of lineage specification of hematopoietic stem cells and progenitor cells. In contrast to previous reports on trained immunity, essentially mediated by pathogen-associated molecular patterns, heme training depends on spleen tyrosine kinase signal transduction pathway acting upstream of c-Jun N-terminal kinases. Heme training promotes resistance to sepsis, is associated with the expansion of self-renewing hematopoetic stem cells primed toward myelopoiesis and to the occurrence of a specific myeloid cell population. This is potentially evoked by sustained activity of Nfix, Runx1, and Nfe2l2 and dissociation of the transcriptional repressor Bach2. Previously reported trained immunity inducers are, however, infrequently present in the host, whereas heme abundantly occurs during noninfectious and infectious disease. This difference might explain the vanishing protection exerted by heme training in sepsis over time with sustained long-term myeloid adaptations. Hence, we propose that trained immunity is an integral component of innate immunity with distinct functional differences on infectious disease outcome depending on its induction by pathogenic or endogenous molecules.
The tetrapyrrole heme acts as the prosthetic group of vital hemoproteins such as hemoglobin (1). Following hemolysis or tissue damage, cell-free, “labile” heme can accumulate in plasma (2, 3) and, if not contained, promotes free radical production in an unfettered manner contributing critically to the pathogenesis of severe acute infectious disease as demonstrated for malaria (4) and for bacterial sepsis (5–7). Heme also triggers an inflammatory response, especially on macrophages (Mϕ) through pattern-recognition receptors (PRRs) (8, 9). Hence, heme is perceived as a prototypical damage-associated molecular pattern (DAMP) (10, 11) or alarmin (6).
PRRs mainly evolved to recognize pathogen-associated molecular patterns (PAMPs) expressed by microbes (12). Depending on the origin of the PAMP, PRR-mediated signaling can lead to persistently modulated reactions to subsequent inflammatory cues which manifests as enhanced or dampened ensuing responses. Such responses are referred to as trained immunity if enhanced and as tolerance if dampened (13–16). It is now well established that these long-lasting effects are enabled by specific histone modifications and the expansion of hematopoietic stem and progenitor cells (HSPCs) in vivo and that this can confer an improved host resistance to infection (15, 17–20). Trained immunity is perceived as part of an evolutionarily conserved host defense strategy, acting in a protective manner against different pathogens (13, 20, 21). A similar phenomenon is observed with the sickle cell trait protecting against malaria, which is mediated by chronic and low-dose cellular exposure to “labile” heme exerting protective effects via the induction of the catabolizing enzyme heme oxygenase-1 (HO-1) (22). We here asked whether heme induces long-lasting rewiring of innate immune responses, reminiscent of trained immunity to enhance host resistance to infection.
Results
Heme Induces Trained Immunity.
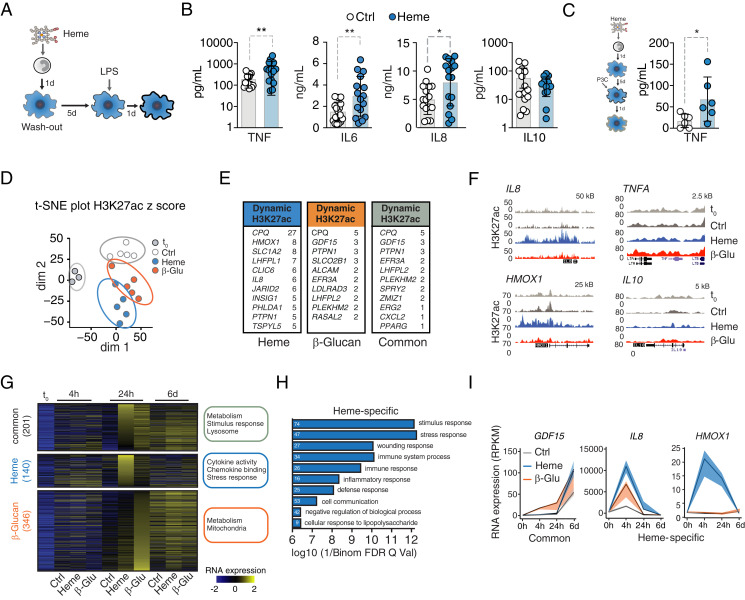
Given the importance of heme for noninfectious and infectious disease, we hypothesized that labile heme might induce innate immune cell training beyond its short-term immunomodulatory properties (9, 23). We first used a well-established in vitro two-hit monocyte-to-macrophage model of trained immunity (Fig. 1A) and assessed whether heme exposure elicits innate immune cell memory, similarly to the established inducer β-glucan (24). Heme was applied to purified primary human monocytes for 24 h at a concentration of 50 μM, a level which can be detected in plasma during severe acute infections (5). After 6 d, the resulting macrophages (Mφ) were exposed to lipopolysaccharide (LPS; 10 ng/mL) as a secondary stimulus. This resulted in increased tumor necrosis factor (TNF) release as compared to nontrained controls, characteristic for trained immunity (Fig. 1B and SI Appendix, Fig. S1A). No remaining heme nor cytokines were detected in the supernatants of the trained cells before restimulation, supporting the notion that cells had returned to steady state prior to secondary challenge, which is consistent with the definition of trained immunity (SI Appendix, Fig. S1 B and C). Having established that heme indeed induces a trained immunity phenotype based on TNF secretion, we further characterized the immune response by measuring different cytokines in the supernatant (SI Appendix, Fig. S1D). The first stimulation with LPS or application of different DAMPs, such as ATP, failed to increase TNF release in response to LPS secondary challenge (SI Appendix, Fig. S1 E and F). Of note, replacing the secondary stimulus with the TLR1/2 agonist Pam3CSK4 resulted in comparable TNF release as observed with LPS (Fig. 1C). This suggests that labile heme induces a distinct signaling cascade that promotes innate immune training.
Fig. 1.
Heme imposes trained immunity and evokes a specific transcriptional signature. (A) Experimental setup. (B) Cytokine release from vehicle- or heme-trained (50 μM) Mφ after LPS restimulation (10 ng/mL) at day 7, measured with LegendPlex. IL-8 measurement by enzyme-linked immunosorbent assay. n = 16 independent donors. (C) TNF release from vehicle- or heme-pretreated Mφ second hit at day 7 with Pam3CSK4. n = 6 independent donors. (D) t-distributed stochastic neighbor embedding (t-SNE) plot of dynamic acetylation peaks in monocytes and vehicle, heme, or β-glucan Mφ 24 h after stimulation. Dim1: differentiation processes. Dim2: variation because of treatment. (E) Genes with highest number of differential H3K27ac peaks within 1 Mb of their TSS. (F) Representative gene loci. (G–I) Transcriptional analysis by RNA-seq. (G) Heatmap of regulated genes at baseline (t0) and at indicated time points after heme or β-glucan exposure. GO terms are listed next to each group of genes. (H) Most abundant pathways based on gene expression analysis 24 h after treatment. White numbers indicate the number of affected genes. (I) Time-resolved expression patterns of genes found with increased H3K27ac. All analyses were performed from n = 5 individual donors. All data mean ± SD unless otherwise stated. Student’s t test, *P ≤ 0.05; **P ≤ 0.01. β-Glu, β-glucan; Ctrl. control; d, days; h, hours; H3K27ac, histone 3 lysine 27 acetylation; HMOX1, heme oxygenase 1; P3C, Pam3CSK4; t0, baseline before stimulation; dim, dimension.
While most of the described biological effects described for heme are linked to its pro-oxidative properties exerted by the central iron atom (1), this appears not to be the case for heme-induced trained immunity. Pre-exposure to protoporphyrin IX (PPIX), which structurally resembles heme but lacks iron, equally increased TNF secretion from human Mφ in response to LPS secondary challenge (SI Appendix, Fig. S1G). Also, several other, but not all, porphyrins could induce trained immunity (SI Appendix, Fig. S1H). In support of this notion, administration of the antioxidant N-Acetyl Cysteine (NAC) did not abolish heme-induced training, despite increased production of reactive oxygen species during induction of trained immunity (SI Appendix, Fig. S1 I–K).
On a molecular level, trained immunity involves histone modifications, most prominently histone 3 lysine 27 acetylation (H3K27ac), and consequently transcriptional alterations (13, 18–20). To confirm whether protein acetylation is involved in the phenotype induced by heme, we applied the unspecific acetylase inhibitor epigallocatechin-3-gallate (EGCG) during heme treatment. This prevented the enhanced TNF release observed after LPS stimulation (SI Appendix, Fig. S1I). Chromatin immunoprecipitation sequencing (ChIP-seq) revealed that heme induces H3K27ac accumulation at distinct genomic regions as well as in common regions with β-glucan, a prototypical inducer of trained immunity (Fig. 1 D–F and SI Appendix, Fig. S2 A–D and Datasets S1 and S2). The vast majority of heme-induced H3K27ac loci were within 500 kb from a gene promoter, with 122 occurring in promoter regions (SI Appendix, Fig. S2E). The second dimension of a t-distributed stochastic neighbor embedding analysis separated heme- and β-glucan–trained and naïve Mφ, indicating treatment-specific H3K27ac patterns (Fig. 1D). Further Gene Ontology (GO) analysis reveals that heme exposure induced H3K27ac at genes involved in wounding and stress. In contrast, H3K27ac levels at genes related to adaptive immune regulation were not induced by heme (SI Appendix, Fig. S2F).
As H3K27ac is associated with transcriptional activation, we performed bulk RNA sequencing (RNA-seq) to compare the transcriptional response of human monocytes and Mφ at different time points after exposure to heme or β-glucan (Fig. 1 G and H and SI Appendix, Fig. S3). The largest number of differentially expressed genes occurred at 24 h after stimulation, with 201 genes commonly induced by heme and β-glucan, 140 induced by heme only, and 346 by β-glucan only (Fig. 1G and SI Appendix, Fig. S3A and Datasets S3–S5). The common signature is related to lysosome maturation and metabolism, critical processes required for increased cytokine production in the context of trained Mφ (25, 26). Pathway analysis using Genomic Regions Enrichment of Annotations Tool demonstrated that genes induced by heme are mainly involved in inflammatory pathways and up-regulated cytokine signaling and stress responses (Fig.1 G and H). However, heme-induced trained immunity was not associated with Mφ polarization into classical M1, M2 (27), or hemorrhage-associated Mφ (M-hem) (28) (SI Appendix, Fig. S3 B–D). The expression of selected genes encoding transcription factors with enriched motifs at gene promoters of heme-induced genes are shown in SI Appendix, Fig. S3E.
Consistent with increased H3K27ac, expression of GDF15, a transcription factor involved in the host response to infection (29), the interleukin (IL) 8 gene, and the heme-degrading gene HMOX1 were increased (Fig. 1I). Heme also induced transcription of heme/iron-related genes FTH1 and HRG1 but not the iron exporter FPN1 (Fig. 1I and SI Appendix, Fig. S3F). Collectively, heme-induced adaptations in human monocytes are associated with a trained innate immune phenotype.
Heme Training Is Mediated via a Spleen Tyrosine Kinase/c-Jun N-Terminal Kinase–Dependent Pathway.
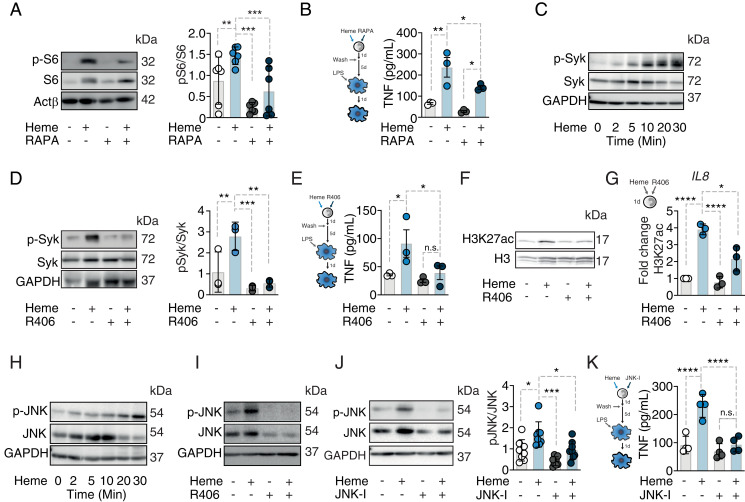
In previous reports, β-glucan–induced trained immunity relied on the activation of the mTOR pathway (30). Although in our settings, heme exposure was also associated with increased phosphorylation of the mTOR target S6 in monocytes (Fig. 2A), mTOR inhibition by rapamycin did not compromise heme training as assessed by TNF secretion (Fig. 2B). Previously, it was shown that heme can signal via a distinct pathway involving the spleen tyrosine kinase (SYK) (8). We can confirm that heme exposure induces SYK phosphorylation in human monocytes (Fig. 2C). Moreover, SYK inhibition by R406 during heme treatment abolished the heme-training phenotype (Fig. 2 D and E) and also reduced the global increase in H3K27ac (Fig. 2F). ChIP-qPCR analysis of the IL8 promoter revealed elevated histone acetylation that was prevented by R406, pointing toward an association of heme-induced SYK activation and H3K27ac (Fig. 2G). In line with the notion that c-Jun N-terminal kinases (JNK) can act downstream of SYK activation (31), we show that heme increased JNK phosphorylation in human monocytes (Fig. 2H), an effect that is abolished by SYK inhibition (Fig. 2I). In addition, application of the JNK inhibitor SP600125 prevented the development of heme training (Fig. 2 J and K), supporting the notion that heme signals via SYK and upstream of JNK to induce trained immunity.
Fig. 2.
Heme training relies on Syk/JNK activation in human monocytes. (A) Western blot of p-S6 24 h after heme ± rapamycin stimulation. (B) TNF production after restimulation with LPS at day 7 of vehicle- or heme-trained Mφ ± rapamycin. The representative Western blots of at least three independent donors are pictured. (C) Western blot of Syk phosphorylation after heme treatment of human monocytes at indicated time points. (D) Syk phosphorylation 20 min after heme stimulation ± the Syk inhibitor R406. (E) TNF production after restimulation with LPS at day 7 of vehicle- or heme-trained Mφ ± R406. (F) Western blot of total H3K27ac 24 h after heme treatment ± R406. (G) ChIP-qPCR analysis of H3K27ac at the IL8 promoter (n = 3 independent donors). (H) Western blot of JNK phosphorylation after heme treatment of human monocytes at indicated time points. (I) Western blot of JNK phosphorylation 20 min after heme stimulation ± R406. (J) Western blot and densitometry of JNK activation 20 min after heme stimulation ± the JNK inhibitor SP600125. (K) TNF production after restimulation with LPS at day 7 of vehicle- or heme-trained Mφ ± SP600125. All enzyme-linked immunosorbent assay results derived ≥3 independent experiment à 2 to 3 donors; Two-way ANOVA with Fisher’s LSD; mean ± SEM, *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; H3, histone 3; H3K27ac, histone 3 lysine 27 acetylation; JNK-I, JNK inhibitor SP600125.
Heme Induces Resistance to Bacterial Sepsis and Promotes Myelopoiesis.
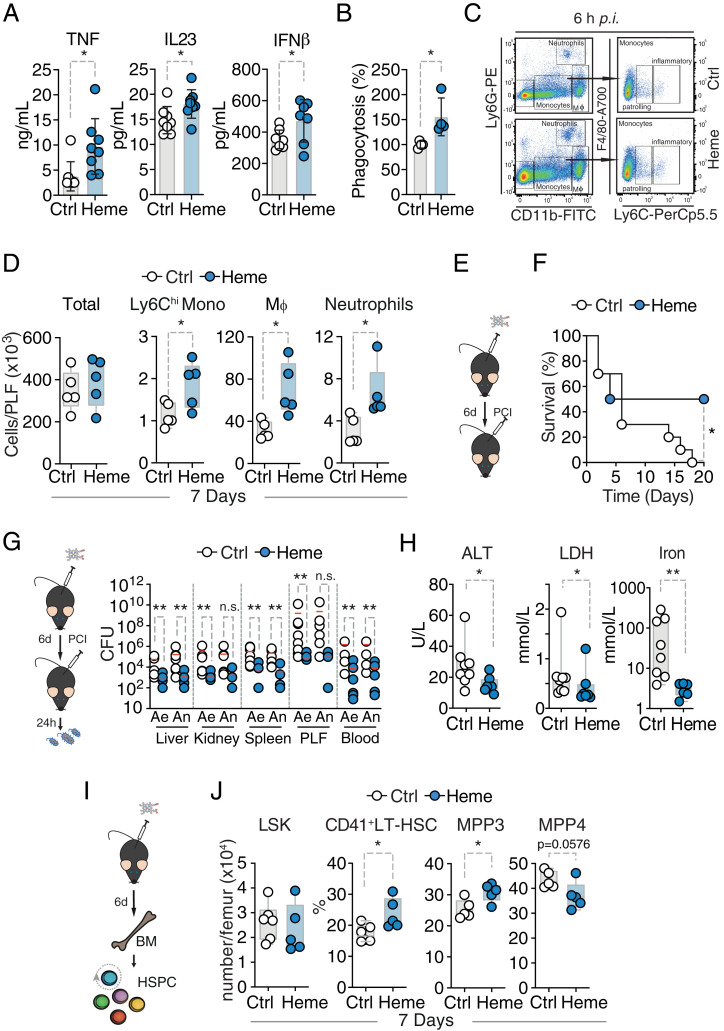
So far, we have provided experimental evidence that heme can induce what is referred to as “peripheral” trained immunity. Long-term effects would be expected only if immune function–altering modifications would also be imprinted in self-renewing cells such as bone marrow HSPCs and potentially affect HSPC expansion and composition (32). In order to investigate whether also “central” trained immunity is induced, C57BL/6 mice were trained by heme or vehicle. An injection of heme at 2 mg/kg did not change the vigor or behavior of the animals (SI Appendix, Fig. S4 A and B). In a first step, bone marrow cells from vehicle- or heme-trained mice were isolated 7 d after heme treatment, differentiated into macrophages and exposed to LPS. Heme-trained cells displayed increased cytokine release after LPS stimulation and enhanced phagocytosis (Fig. 3 A and B). Heme training was also associated with enhanced peritoneal recruitment of phagocytic cells, monocytes, and neutrophils in response to intraperitoneal (i.p.) LPS administration (Fig. 3 C and D) and increased mortality (SI Appendix, Fig. S4C). The deleterious effects of heme training on subsequent LPS-induced systemic inflammation were also observed in Rag2-deficient mice (Rag2−/−) lacking mature B and T cells (SI Appendix, Fig. S4 D and E). This supports the notion that long-term heme-induced effects on immune responses are not mediated by cells of the adaptive immune system. Next, we assessed the host response to infection subsequent to heme training. A total of 7 or 28 d after i.p. injection of heme (2 mg/kg) or vehicle, mice were subjected to polymicrobial sepsis (Fig. 3E) (7). Heme training proved to be protective in polymicrobial sepsis as illustrated by faster weight recovery, thermoregulation maintenance, and lower incidence of mortality (Fig. 3F and SI Appendix, Fig. S4 F and G) when induced 7 d after heme training. Of note, this protective effect was associated with a marked reduction in host–pathogen load (Fig. 3G). Markers of tissue damage as well as serum iron levels were significantly reduced (Fig. 3H). However, when animals were subjected to polymicrobial sepsis 28 d after heme training, no survival benefit was observed. Instead, heme-trained animals had a tendency of increased mortality (SI Appendix, Fig. S4H).
Fig. 3.
Persistent heme effects in vivo. (A) Cytokine release of bone marrow–derived macrophages (BMDM), bone marrow (BM) isolated at day 7 after control (Ctrl; n = 8) or heme (n = 8) injection and 24 h after LPS (10 ng/mL) stimulation. Pool of two independent experiments. (B) Phagocytic capacity of BMDM, BM isolated at day 7 after Ctrl (n = 4) or heme (n = 4) injection and 2 h after pHrodo Red E. coli stimulation. Pool of two independent experiments. (C) Gating of myeloid cells. Neutrophils are CD11b+Ly6G+ and monocytes are CD11b+Ly6G‒ and further distinguished as Ly6Chigh (inflammatory monocytes) and Ly6Clow (patrolling monocytes). Peritoneal Mφ were gated as CD11bhighLy6G‒F4/80+. (D) Cell count and flow cytometry analysis of peritoneal lavage fluid of vehicle- or heme-pretreated mice at day 7 and 6 h after endotoxemia. (E) Experimental setup. (F) Survival of polymicrobial sepsis induced 7 d after vehicle (n = 10) or heme (n = 10) treatment in mice. Pool of two independent experiments. Survival: Fisher’s exact test. (G) Pathogen load and (H) serology of vehicle- (n = 8) or heme- (n = 7) pretreated mice 24 h after sepsis induction. Pool of two independent experiments. Mann–Whitney U test; mean, *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001. (I) Experimental setup. (J) Flow cytometry analysis of indicated HSPC cellular subsets of vehicle- or heme-pretreated mice at day 7. n = 5 to 6 animals per group derived from two independent experiments. Student’s t test; mean ± SD, *P ≤ 0.05; **P ≤ 0.01. Ae, aerob; ALT, alanin aminotransferase; An, anaerob; h, hours; hi, high; IFN, interferon; IL, interleukin; LDH, lactate dehydrogenase; LSK, Lin−Sca1+cKit+; Neutrop, neurophils; n.s., non-significant; PCI, peritoneal contamination and infection; p.i., post injection; PLF, peritoneal lavage fluid.
Our findings suggest that while labile heme can, in principle, promote resistance to infection (short term) as a trade-off, it counteracts the establishment of disease tolerance to bacterial infection presumably by exacerbating the innate immune reaction to bacterial components, such as LPS or as a long-term consequence.
Based on these findings, we postulated that heme elicits alterations in hematopoiesis, similar to previous reports of other trained immunity-inducing agents (17, 33, 34). We analyzed mouse bone marrow 7 and 28 d after heme or vehicle treatment. After 7 d, we did not find a difference in cell number or percentages of LSK cells (Lin−cKit+Sca1+), long-term hematopoetic stem cells (LT-HSCs, CD48−CD150+LSK), or multipotent progenitors (MPP, CD48+CD150−LSK) (Fig. 3 I and J and SI Appendix, Fig. S5 A–C) as compared to vehicle-treated mice. However, we observed an increased frequency of myeloid-biased LT-HSCs (CD41+LT-HSC) (35) as well as of myeloid-biased MPP3 cells (Flt−3CD48+CD150−LSK) at the expense of lymphoid-biased MPP4 cells (Flt3+CD48+CD150−LSK) (36) (Fig. 3 I and J and SI Appendix, Fig. S5 A–C). Myeloid-biased LT-HSCs (CD41+ LT-HSC) and MPP3 expansion was sustained as far as 28 d after heme administration (SI Appendix, Fig. S5D). Furthermore, a significant increase in mature neutrophils and monocyte was observed at this time point in the bone marrow (SI Appendix, Fig. S5E).
Heme Changes the Epigenetic Landscapes of HSPCs.
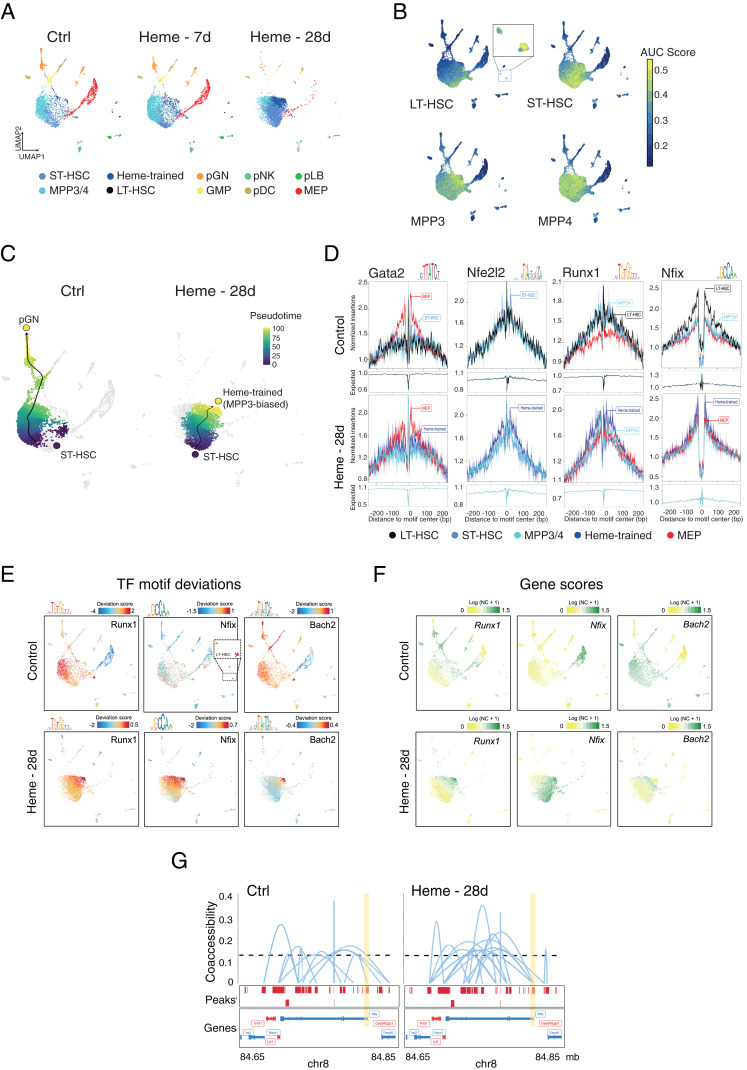
Given the long-term cellular and functional adaptations (Figs. 1–3), we explored whether heme causes changes in the chromatin landscape of HSPCs and assessed genome-wide chromatin accessibility using single nucleus assay for transposase-accessible chromatin using sequencing (snATAC-seq) on fluorescence-activated cell sorting (FACS)-sorted LSK cells 7 or 28 d after a single dose of heme and from control animals (Fig. 4 and SI Appendix, Fig. S6). Using iterative latent semantic indexing (LSI) clustering (37), we identified 10 cell populations, including an HSC/MPP cloud of cells, megakaryocyte–erythroid progenitors (MEP), granulocyte–macrophage progenitors (GMP), and several early committed immune progenitor cells. Strikingly, the clustering revealed changes in chromatin accessibility in the cloud of LSK cells that were subtle at day 7 but became very prominent on day 28 after heme administration as compared to control (Fig. 4A). We classified these shifted HSPC populations as short-term (ST) HSC (STHSC.150-.BM) and the MPP3 biased as “heme-trained” cells (MPP3.48+.BM) (Fig. 4 A and B and SI Appendix, Fig. S6 A and B). Early committed cell subpopulations (e.g., MEP, GMP) were sharply decreased at day 28. Interestingly, the number of long-term HSCs (LTHSC.34-.BM) also diminished after heme administration (Fig. 4A and SI Appendix, Fig. S6A).
Fig. 4.
Single-cell epigenetic landscape of heme-trained HSPCs in mice. (A) UMAP projection of 4,842 (Control), 5,562 (Heme 7d), and 6,052 (Heme 28d) single-nuclei (sn) ATAC profiles from sorted bone marrow–derived LSK cells. Each dot represents an individual cell, and colors represent the identity of the cluster. An annotation of the clusters was performed according to the overlay of epigenetic signatures (ImmGen Database) and known marker genes per cell type. (B) Enrichment of epigenomic ImmGen Database signatures (area under the curve [AUC] scores) of the HSPCs onto UMAP projection of LSK cells. (C) Pseudotime trajectory of pGN differentiation and heme-trained cells from ST-HSCs in control LSK cells and after 28 d of heme training, respectively. (D) TF footprint analysis in the indicated snATAC-seq clusters of non- and heme-treated LSK cells subtracted for Tn5 insertion bias shown below. (E) TF motif scores from bone marrow cells projected onto the UMAP. (F) UMAP projection colored by log-normalized gene scores calculated on the individual LSK cell types. (G) Enhancer–promoter connections (coaccessibility) of the Nfix locus in control- and heme-trained cells. The height of connections indicates the coaccessibility score between the connected peaks. Ctrl, control; pGN, precursor of granulocytes; pDC, precursor of dendritic cells; pNK, precursor of NK cells; pLB, precursor of B lymphocytes; NC, normalized count.
Long-Lasting Alteration of Chromatin Sustains Access for Transcription Factors Promoting Myelopoiesis.
To gain further molecular insight, we arranged the cells on a pseudotime development trajectory (37). In control bone marrow, the myeloid differentiation path (38) from HSC to progranulocytes (indicated by the dark blue and yellow circle, respectively) is evident. Heme application caused a prominent shift in the trajectory, now leading toward the novel heme-trained population (Fig. 4C). To assess factors that direct these different pathways and characterized the epigenetic landscape of de novo heme-trained cells, we performed motif enrichment and transcription factor (TF) footprinting analyses of chromatin-accessible regions (39). The binding sites of Runx1, a TF well known for its role in sustaining the stemness of hematopoietic cells (40), ranked highest, thus validating our approach (Fig. 4 D and E and SI Appendix, Fig. S6C). Next in line is the CCAAT-box, the binding site of nuclear factor I (Nfi) family members that form homo- and heterodimers. Importantly, the relative distribution of the motifs was drastically altered from LT-HSC in control bone marrow to ST-HSCs and heme-treated MPP3-biased cells upon heme treatment correlating with the altered trajectory (Fig. 4E). The motif analysis also included Nfe2l2, a vital regulator of the oxidative stress response and HSC homeostasis (41) (Fig. 4D and SI Appendix, Fig. S6C). Gata2, the master regulator of the MEP lineage, was prominently detected in control and, to a lesser extent, in the bone marrow of heme-treated mice (Fig. 4D). The projection of the TF motif deviation scores onto the Uniform Manifold Approximation and Projection (UMAP) plots visualizes the notable shifts in the abundance and cell-specific occurrences of the TF-binding motifs in control versus heme LSK cells (Fig. 4E). The Runx1 motif was abundant in the MPP3/4 population in control and shifted toward the heme-trained cell population, whereas the Nfi motif shifted from the LT-HSC to the ST-HSC and heme-induced MPP3-biased cells. Thus, the persistence of stemness-associated TFs (Runx, Nfi, Spi, and Elf TF families) characterizes the heme-trained population.
The Runx1 and Nfi-binding motifs do not unequivocally point to the relevant TFs, as several closely related family members bind to the same or very similar motif. Therefore, we inferred the gene expression level (activity score) based on the accessibility of cis-regulatory elements in the proximity of candidate TFs (42) and projected the scores onto the UMAP clustering. The Nuclear Factor One X (Nfix) is the most likely family member to bind to the Nfi motif and regulate the skewed myelopoiesis. At first, the inferred Nfix activity correlates well with the rerouting of the differentiation trajectory from LT-HSC to ST-HSC/heme-trained cluster (Fig. 4F). Secondly, the gene activity score of the Nfib does not follow the altered trajectory, although it seems to play a yet to be discovered role in the HSPC, whereas Nfia and -c appears to play a role in MEPs. Thirdly, Nfix has been implicated in stem cell survival and the regulation of lineage specification toward myeloid cells (43–45) (Fig. 4D and SI Appendix, Fig. S6 C and D).
Heme has recently been reported to inhibit the BTB domain and CNC homolog (Bach) TFs, impeding erythroid commitment and favoring myelopoiesis (46). Our chromatin accessibility analysis shows scattered gene activity as well as motif deviation scores throughout HSCs and early committed myeloid cells but is enriched and more restricted in the heme-trained population compared with control (Fig. 4 E and F). Interestingly, HOMER motif analysis revealed several Bach2-binding sites at the Nfix locus, suggesting a potential regulatory axis between Bach2 and Nfix (Dataset S6). In support of this, heme-trained ST-HSC show time- and treatment-specific accessibility differences at the Mpl and Erg genes (SI Appendix, Fig. S6E), potential downstream regulators of myeloid lineage commitment and survival. Finally, GO enrichment analysis unveiled enrichment in biological processes linked to positive regulation of leukocyte and myeloid cell differentiation and negative regulation of megakaryocyte differentiation in the HSC/heme-trained population, supporting our findings (SI Appendix, Fig. S6F).
Thus, we uncover heme-mediated, long-lasting modifications of the chromatin-accessible landscape that favors myelopoiesis, reminiscent of innate immune training.
Discussion
Trained immunity is a fundamental feature of myeloid cells allowing innate immune cells to respond to rechallenge by the same or an unrelated pathogen in a more efficient manner (18–20, 25, 47). Our data show that extracellular labile heme acts as a potent inducer of trained immunity. In contrast to previous reports, heme training involves a SYK/JNK phosphorylation signal transduction pathway (18–20, 25, 47). How heme induces SYK phosphorylation, independently of TLR4 and JNK signaling–linked histone modifications, remains to be determined. However, it is conceivable that there is no specific heme receptor involved, as SYK activation can occur via autophosphorylation (48), which could be triggered by heme–membrane interaction. It is also unclear to this point how JNK signaling leads to epigenetic modifications and which cofactors are being involved.
Heme possesses a highly redox-active central iron atom, conferring its strong pro-oxidant properties (1). Heme training, however, appears not to rely on this redox activity, as we can show that both scavenging reactive oxygen species (ROS) as well as PPIX and several other metal-containing porphyrins also induces innate immune training in vitro. The reason for this remains to be revealed. It is conceivable that the porphyrin ring itself can induce a yet to be established signal transduction and biological function that acts independently of its iron core. In fact, PPIX activates JNK phosphorylation (49), suggesting that the same pathways are activated as by heme. This notion is supported by our finding that ROS were not increased after heme training and that the application of the antioxidant NAC did not abolish heme-induced training as would be expected if a redox-sensitive mechanism was involved (49). A less likely explanation would be that porphyrins undergo noncatalytic metalation or transmetalation that are at least in principle possible (50, 51), replacing the central metal by iron.
Our data specifically links heme training to IL8 biology, a chemokine especially involved in the recruitment and degranulation of innate immune cells. We report increased IL8 secretion and RNA expression and reveal that IL8 is the gene with the highest number of differential H3K27 acetylation peaks in proximity of the transcription start site. In contrast to the heme-induced IL8 modifications, no histone modification of IL8 was detected after β-glucan exposure. IL8 expression is regulated by histone acetylation (52, 53), which indicates that H3K27ac in the IL8 promotor leads to an increased IL8 chemokine production of heme-pretreated cells in response to LPS. Blocking histone deacetylases increases the transcription of IL8. On the other hand, the suppression of CREB-binding protein, exerting a histone acetyltransferase activity, blocked IL8 expression (53). This was dependent on increased ability of the p65 unit of NFκB to bind to the IL8 promoter in a histone-acetylated environment (53). It is also well known that LPS induces NFκB in a TLR4-dependent manner (54). It follows that histone acetylation in the IL8 promotor region might enhance the binding capacity of the p65 NFκB, activated through the LPS signaling and subsequent enhanced IL8 release.
Despite increased TNF secretion in heme-trained Mφ, we could not find a difference in H3K27ac levels at the TNF promoter after heme treatment when compared to control cells, which is in line with the pan-innate immune training signature observed in both heme- and β-glucan–exposed Mφ that involves the up-regulation of mechanisms resulting in cytokine release but not cytokine gene priming. This is also in agreement with recent findings by other authors who used a model of IFN-γ priming (55). Increased TNF production was also unrelated to H3K27ac in the TNF promotor itself, as IFN-γ–dependent modifications occur at a region 8 kb upstream of the TNF promotor (55). Additionally, trained immunity does not only depend on H3K27ac but also on histone methylation (20, 56). This has been exemplarily shown for activating the activating H3K4me3 mark at promotor regions and the H3K4me1 mark at enhancers in β-glucan–trained Mφ (57). These marks were not part of our investigation.
We also assessed the recently identified heme-responsive motifs (HERM) (58, 59). However, none of the motifs were located in the genes of interest. In addition, analyzing a region of 200 base pairs (bp) upstream and 200 bp downstream of the HERM element, there were only two heme peaks overlapping out of 450 induced by heme. This suggests that HERM motifs might not be relevant for heme-induced trained immunity.
Innate immune training is generally considered a beneficial adaptive long-term phenomenon that improves disease severity and survival of subsequent infections (19, 20, 30, 60). However, evidence for maladaptive responses, that is, nonprotective trained immunity, has been shown in models of chronic sustained inflammation such as atherosclerosis and Mycobacterium tuberculosis infection. When monocytes were exposed to oxidized low-density lipoprotein (oxLDL), these exhibited an increased inflammatory response and enhanced foam cell formation, a hallmark of atherosclerosis progression (61, 62). In vivo, a Western type, high-fat diet given to Ldlr−/− mice, a classical model for atherosclerosis, induced trained immunity (63). The recently published study by Khan et al. provides evidence that in contrast to the vaccine strain Bacillus Calmette–Guérin (bacillus Calmette–Guérin), Mycobacterium tuberculosis does not induce but rather impairs innate immune training (64).
We here provide evidence that heme can induce both protective (i.e., early bacterial sepsis) and deleterious (i.e., endotoxic shock or delayed bacterial sepsis) trained immunity. This is the first demonstration, that the same stimulus has opposing effects depending on the context and timing. In principle, the deleterious innate immune training could manifest at a peripheral level, for example, via enhanced cytokine release and associated tissue damage (65), exceeding the benefits of decreased pathogen loads and at a central level, for example, via maladaptive myelopoiesis, impairing the host response to infection. In beneficial innate immune training, this would manifest at a peripheral level as improved resistance to infection and at the central level as enhanced myelopoiesis supporting pathogen destruction and tissue repair. For heme-induced training, we postulate that maladaptive effects in the LPS model are indeed caused by enhanced tissue damage while no beneficial effects of pathogen destruction can occur since this is a sterile inflammation. Conversely, in sepsis, the protective effect of heme training was directly associated with a significant enhanced resistance to infection, that is, decreased pathogen loads. Whether heme-induced training results in heterologous protective immunity (66) against infections by other classes of pathogens or whether it will exert immunopathology to chronic inflammatory disease needs to be further investigated.
It is widely assumed that innate immune memory is encoded in the epigenome. However, the proof in vivo has been scarce so far (57). We document striking, long-term changes in cellular, transcriptional, and chromatin accessibility 28 d following a one-time exposure to heme, which did not affect the animals' vigor. This prominent response to heme manifests as the expansion of ST-HSC–like cells and the appearance of a heme-trained cell population. Chromatin footprinting and motif analysis revealed drastic changes in the binding sites of TF Nfix along with the well-known Runx1, Nfe2l2, and Bach2. A plausible scenario linking the reported down-regulation of Bach1/2 repressor TFs (46) by heme with our observed long-term alterations of the chromatin-accessible regions of heme-trained HSPCs favors persistence of myeloid differentiation and stemness-related genes. Our data also suggest that hosts exposed to labile heme, as it would occur in malaria, bacterial sepsis, or other form of hemolysis, will likely prime hematopoietic stem cells toward adapted myelopoiesis. Whether this striking long-term modulation of the hematopoiesis is responsible for the maladaptive response to sepsis 28 d after heme training remains to be established.
Generally, several open questions remain and will be tested experimentally in the future, for example, how long does heme training last? Can the training signature be reversed by heme training as has been shown for β-glucan (26)? Does heme training confer protection to other, also noncommunicable disease, and if so, how would this be explained evolutionarily? Is heme the training mediator, or does heme application result in the release of cytokines or other molecules that induce innate immune training, and if so, is this a universal feature or trainer specific?
In conclusion, we here reveal that heme, a bona fide alarmin, induces peripheral as well as central innate immune memory regulating host response to infection. Previously reported trained immunity inducers are, however, infrequently present in the host, whereas heme abundantly occurs during noninfectious and infectious disease. Hence, we propose that trained immunity is an integral component of innate immunity to confer long-term host protection.
Materials and Methods
Preparation of Pooled Human Serum.
All procedures were approved by the Ethics Committee of the University Hospital Jena, license reference 4899–07/16. Blood was collected from healthy volunteers into a S-Monovette (Sarstedt), and serum was collected according to manufactures instructions. Serum was pooled and centrifuged, sterile filtrated, and stored at −80 °C until usage.
Isolation of Peripheral Blood Mononuclear Cells and Human Monocytes.
Peripheral blood mononuclear cells were isolated from buffy coats that were obtained from the Institute of Transfusion Medicine, Jena University Hospital, or from blood freshly donated by volunteers (ethical approval of the Ethics Committee of the Jena University Hospital, license reference 4899–07/16). Isolation was performed as previously described (24).
Isolation of Murine Bone Marrow Cells.
Bone marrow was isolated from femora and tibiae of male 8- to 12-wk-old C57BL/6J mice. Bones were flushed with Roswell Park Memorial Institute medium (RPMI) containing 10% fetal calf serum (FCS) and 1% Pen/Strep and stable glutamine. Cells were centrifuged at 4 °C (7 min; 350 g). Erythrocytes were lysed by resuspending the pellet in red blood cell lysis buffer (150 mM NH4Cl; 10 mM KHCO3; 130 μM ethylenediaminetetraacetic acid (EDTA)). The reaction was stopped by adding RPMI and cells were again centrifuged thereafter. Cells were used directly or stored at −150 °C as cryogenic stock.
Differentiation of Murine Bone Marrow–Derived Macrophages.
Cells from murine bone marrow were cultured in RPMI containing 10% FCS and 1% Pen/Strep. Differentiation was induced by adding 20 ng/mL granulocyte–macrophage colony-stimulating factor (GM-CSF; Peprotech). After 1 wk, cells were harvested and seeded on well plates and incubated for 1 d with GM-CSF (20 ng/mL; Peprotech). Training experiments were performed with these cells according to the monocyte training protocol. During this time, 0.2 ng/mL GM-CSF was always added to the medium. For the determination of cytokine levels released from bone marrow macrophages of vehicle- or heme-trained mice, cells were restimulated with LPS. Cytokines were measured 24 h after stimulation.
Monocyte Training.
Monocyte training was adapted from Bekkering et al. (24). To initiate training, cells were either incubated with culture medium (RPMI 1640 Dutch modification supplemented with 10% pooled human serum, 2 mM GlutaMAX, and 1 mM pyruvate and 10 μg/mL gentamicin) as a negative control or with the respective stimuli for 24 h. For inhibition experiments, cells were incubated with the respective inhibitor 1 h prior to stimulation and throughout the 24 h of stimulation. After 24 h, cells were washed, and new culture medium with serum was added to the cells. On day 6, cells were stimulated with 10 ng/mL LPS in culture medium. Supernatants were collected 24 h later, and cytokine levels were determined. Cells were either lysed by radioimmunoprecipitation assay buffer (protein analysis) or in Qiazol (transcriptional analysis).
Pharmacological Approaches.
Stimulation was performed with 50 μM heme or 50 μM PPIX (Sigma-Aldrich or Frontier Scientific) and 50 μM of other porphyrins as indicated, unless otherwise stated. All were prepared as described in ref. 2. For β-glucan training, cells were stimulated with 1 μg/mL β−1,3-(D)-glucan. Acetyltransferases were inhibited with EGCG (15 μM; Sigma-Aldrich) (16). R406 (1 μM; InvivoGen) was used for inhibiting spleen tyrosine kinase activation. The JNK pathway was blocked by SP600125 (10 μM; InvivoGen). Rapamycin (10 nM; InvivoGen) was used for blocking mTOR-mediated signal transduction. ROS were scavenged by N-acetylcystein (10 mM; Fluka). ROS measurements were performed by incubating the cells with H2DCFDA and flow cytometry analysis. Ultrapure LPS from Escherichia coli O55:B5 (Cat#: tlrl-pb5lps) and ATP was purchased from InvivoGen.
Animal Experiments.
Experimental procedures were ethically reviewed and approved by the Ethics Committee of the Instituto Gulbenkian de Ciência (license reference: A009/2011) by Direção Geral de Alimentação e Veterinária (license reference: 0420/000/000/2012) or for animal experiments at the Jena University Hospital by the regional animal welfare committee (registration no.: 02–044/16, Thuringian State Office for Consumer Protection and Food Safety). All experiments conducted on animals followed the Portuguese (Decreto-Lei no. 113/2013) or the German legislation on the protection of animals and the European legislations (Directive 2010/63/EU) concerning housing, husbandry, and animal welfare. C57BL/6 mice were bred and maintained under specific pathogen-free conditions at the Instituto Gulbenkian de Ciência or at the Jena University Hospital (10/14 h day/night; fed ad libitum). Rag2−/− mice were bred and maintained at the Instituto Gulbenkian de Ciência, Oeiras, Portugal. Mice received 2 mg/kg body weight (BW) heme or vehicle (NaOH; pH 7.4) via i.p. injection. For endotoxic shock survival experiments and flow cytometric analysis of peritoneal lavage fluid cells, mice received 15 mg/kg LPS (E. coli O55:B5; Sigma-Aldrich) 1 wk after heme treatment.
Polymicrobial sepsis was induced using a well-characterized, standardized human stool suspension (peritoneal contamination and infection) (7). Animals received one i.p. injection of 0.85 μL feces/g BW and 25 μL/g BW sterile 0.9% NaCl solution. Temperature, weight, and clinical severity were monitored using an established score system (7).
Quantification and Statistical Analysis.
Survival plots were analyzed as indicated. One-way ANOVA with Fisher’s least significant difference (LSD) was used to analyze more than two groups. Two-way ANOVA with Fisher’s LSD was used when analyzing more than one group and time point. Student’s t test was used for pairwise comparisons. For the analysis of the pathogen load, the Mann–Whitney U test was used due to the high variance of SDs. Floy cytometry and FACS data analysis was performed using FlowJo (Tree Star) software. Mean and SD were used whenever single values were compared. Mean and SEM were used to compare means of distinct experiments with replicates within the same experiment and setting.
RNA-seq analysis.
To determine gene expression levels, RNA-seq reads were aligned to the Ensembl v68 human transcriptome using Bowtie 1. The quantification of gene expression was performed using Many-against-Many sequence searching (MMSEQ). Differential expression was determined using DESeq. Differentially expressed genes were determined using pairwise comparison between treatments, with a fold change > 2.5, adjusted P value of < 0.05, and Reads per kilobase of transcript per Million mapped reads (RPKM) ≥ 5.
ChIP-seq analysis.
Sequencing reads were aligned to human genome assembly hg19 (National Center for Biotechnology Information [NCBI] version 37) using bwa. Duplicate and low-quality reads were removed after alignment using Samtools and Bamtools. For peak calling, the BAM files were first filtered to remove the reads with a mapping quality less than 15 followed by fragment size modeling. MACS2 was used to call the peaks. H3K27ac peaks were called using the default (narrow) setting. Reads/peak tables were merged using Bedtools. Data were normalized using the R package DESeq1, and then pairwise comparisons were performed (fold change > 1.5; P value < 0.05 and mean reads/peak ≥ 20 in any condition) to determine the differential peaks.
snATAC-seq data preprocessing and analysis.
Binary base call (BCL) files were demultiplexed by Cell Ranger ATAC (version 1.1.0) using “cellranger-atac mkfastq.” FASTQ files were aligned to the mm10 reference genome using “cellranger-atac count” with default parameters. This pipeline also marks duplicated reads and identifies transposase cut sites. Cells were filtered, keeping those with a transcription start site (TSS) enrichment of at least eight and more than 1,000 unique fragments. This approach allows the detection of TSSs without having a defined peak set. Since the peak calling was performed separately, peaks do not overlap perfectly and might be treated as entirely different features. Using the Granges package, we find overlaps of the chromosome coordinates among samples as previously described (37). The obtained count matrix was used to generate a union peak set, first binarizing the top 20,000 most accessible sites followed by term frequency–inverse document frequency transformation. We then used singular value decomposition implementation and created a Seurat (67) (version 3.1.5) object for nonlinear dimension reduction. LSI scores were then used for clustering, and a visual representation of the clusters was performed using a UMAP projection. To annotate the cell clusters and perform GO enrichment analysis, we used the union peak set and looked for the enrichment of epigenomic signatures, as well as linking regions to genes, and determined GO terms that are enriched using CisTopic (version 0.3.0) (68). Bulk ATAC profiles of purified cell types from the mouse immune system (ImmGen Database) were used as a reference (38). Additionally, each accessible region was linked to the nearest gene based on CHIPseeker annotations, and the probability of each region to a defined set of marker genes (gene activity score) was depicted as the log of normalized counts + 1. To infer the pseudotime trajectory, we aligned and ordered in the two-dimensional UMAP subspace the single cells across an expected differentiation path. The order's significance is obtained after permuting the trajectory 5,000 times and computing the average rank of the ordering for the permuted and input trajectory as described elsewhere (37). Enhancer–promoter connections were detected using Cicero (version 1.3.4.9) using default parameters (42). Motif enrichment and TF footprinting analyses and the aggregation of cells from each condition to create a bulk track per cluster for peak calling and visualization were carried out using ArchR (version 0.9.5) (39). Details of the analyses can be checked at https://www.archrproject.com. All plots and analyses were performed using R (version 4.0).
For further details, see SI Appendix, Material and Methods.
Acknowledgments
We thank Dr. David L. Williams, Ph.D. from the Quillen College of Medicine East Tennessee State University for providing the β-glucan used in this study. We also thank Maziar Divangahi, Ph.D. from the McGill University Montreal, Canada, for his insightful comments on the manuscript. J.G., M. Bauer, and S.W. were supported by the Integrated Research and Treatment Center–Center for Sepsis Control and Care (CSCC) at the Jena University Hospital. The CSCC is funded by the German Ministry of Education and Research (BMBF No. 01EO1502). M. Bauer and E.J. were supported by Deutsche Forschungsgemeinschaft (DFG) Grant GRK 1715/2. E.J., M.P.S., M. Bauer, and S.W. are currently funded by the DFG under Germanýs Excellence Strategy—EXC 2051—Project Identification 390713860. S.W. is currently funded by DFG Project No. WE 4971/6-1 and BMBF Project No. 01EN2001. T.C. was supported by a grant from the DFG (SFB/TRR 127, Project A3). B.N. is supported by an National Health and Medical Research Council (NHMRC) (Australia) Investigator Grant (No. 1173314). M. Bauer was supported by the DFG FOR 1738. M.G.N. is supported by a European Research Council Advanced Grant (No. 833247) and a Spinoza Grant of the Netherlands Organization for Scientific Research. W.L.M. is supported by the Italian National Operational Programme on Research 2014 to 2020 (PON AIM 1859703-2): PON Ricerca e Innovazione 2014 to 2020—Azione I.2—D.D. n.407, 27.02.2018 “Attraction and International Mobility”—Line 2 (Researchers Attraction). C.R.-M. is supported by the European Union’s Horizon 2020 Skłodowska-Curie Actions (Project AiPBAND) under Grant No. 76428, and H.G.S. is supported by the Princess Maxima Center and a ZonMW Grant No. 91216061. We are part of the International Trained Immunity (INTRIM) consortium.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2102698118/-/DCSupplemental.
Data Availability
RNA sequencing and snATAC-seq data have been deposited in NCBI Gene Expression Omnibus (GSE111003).
References
- 1.Bozza M. T., Jeney V., Pro-inflammatory actions of heme and other hemoglobin-derived DAMPs. Front. Immunol. 11, 1323 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gouveia Z., et al., Characterization of plasma labile heme in hemolytic conditions. FEBS J. 284, 3278–3301 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Englert F. A., et al., Labile heme impairs hepatic microcirculation and promotes hepatic injury. Arch. Biochem. Biophys. 672, 108075 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Ferreira A., Balla J., Jeney V., Balla G., Soares M. P., A central role for free heme in the pathogenesis of severe malaria: The missing link? J. Mol. Med. (Berl.) 86, 1097–1111 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Larsen R., et al., A central role for free heme in the pathogenesis of severe sepsis. Sci. Transl. Med. 2, 51ra71 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Soares M. P., Bozza M. T., Red alert: Labile heme is an alarmin. Curr. Opin. Immunol. 38, 94–100 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Weis S., et al., Metabolic adaptation establishes disease tolerance to sepsis. Cell 169, 1263–1275.e14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Figueiredo R. T., et al., Characterization of heme as activator of toll-like receptor 4. J. Biol. Chem. 282, 20221–20229 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Dutra F. F., et al., Hemolysis-induced lethality involves inflammasome activation by heme. Proc. Natl. Acad. Sci. U.S.A. 111, E4110–E4118 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matzinger P., The danger model: A renewed sense of self. Science 296, 301–305 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Chen G. Y., Nuñez G., Sterile inflammation: Sensing and reacting to damage. Nat. Rev. Immunol. 10, 826–837 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janeway C. A. J. Jr., Medzhitov R., Innate immune recognition. Annu. Rev. Immunol. 20, 197–216 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Netea M. G., et al., Trained immunity: A program of innate immune memory in health and disease. Science 352, aaf1098 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beeson P. B., Tolerance to bacterial pyrogens: I. Factors influencing its development. J. Exp. Med. 86, 29–38 (1947). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foster S. L., Hargreaves D. C., Medzhitov R., Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature 447, 972–978 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Ifrim D. C., et al., Trained immunity or tolerance: Opposing functional programs induced in human monocytes after engagement of various pattern recognition receptors. Clin. Vaccine Immunol. 21, 534–545 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitroulis I., et al., Modulation of myelopoiesis progenitors is an integral component of trained immunity. Cell 172, 147–161.e12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kleinnijenhuis J., et al., Bacille calmette-guerin induces NOD2-dependent nonspecific protection from reinfection via epigenetic reprogramming of monocytes. Proc. Natl. Acad. Sci. U.S.A. 109, 17537–17542 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quintin J., et al., Candida albicans infection affords protection against reinfection via functional reprogramming of monocytes. Cell Host Microbe 12, 223–232 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saeed S., et al., Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science 345, 1251086 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Penkov S., Mitroulis I., Hajishengallis G., Chavakis T., Immunometabolic crosstalk: An ancestral principle of trained immunity? Trends Immunol. 40, 1–11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferreira A., et al., Sickle hemoglobin confers tolerance to plasmodium infection. Cell 145, 398–409 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Fernandez P. L., et al., Heme amplifies the innate immune response to microbial molecules through spleen tyrosine kinase (Syk)-dependent reactive oxygen species generation. J. Biol. Chem. 285, 32844–32851 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bekkering S., et al., In vitro experimental model of trained innate immunity in human primary monocytes. Clin. Vaccine Immunol. 23, 926–933 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arts R. J., et al., Glutaminolysis and fumarate accumulation integrate immunometabolic and epigenetic programs in trained immunity. Cell Metab. 24, 807–819 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novakovic B., et al., β-glucan reverses the epigenetic state of LPS-induced immunological tolerance. Cell 167, 1354–1368.e14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bouhlel M. A., et al., PPARgamma activation primes human monocytes into alternative M2 macrophages with anti-inflammatory properties. Cell Metab. 6, 137–143 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Finn A. V., et al., Hemoglobin directs macrophage differentiation and prevents foam cell formation in human atherosclerotic plaques. J. Am. Coll. Cardiol. 59, 166–177 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santos I., et al., CXCL5-mediated recruitment of neutrophils into the peritoneal cavity of Gdf15-deficient mice protects against abdominal sepsis. Proc. Natl. Acad. Sci. U.S.A. 117, 12281–12287 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng S. C., et al., mTOR- and HIF-1α-mediated aerobic glycolysis as metabolic basis for trained immunity. Science 345, 1250684 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dutra F. F., Bozza M. T., Heme on innate immunity and inflammation. Front. Pharmacol. 5, 115 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Divangahi M., et al., Trained immunity, tolerance, priming and differentiation: Distinct immunological processes. Nat. Immunol. 22, 2–6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaufmann E., et al., BCG educates hematopoietic stem cells to generate protective innate immunity against tuberculosis. Cell 172, 176–190.e19 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Chavakis T., Mitroulis I., Hajishengallis G., Hematopoietic progenitor cells as integrative hubs for adaptation to and fine-tuning of inflammation. Nat. Immunol. 20, 802–811 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gekas C., Graf T., CD41 expression marks myeloid-biased adult hematopoietic stem cells and increases with age. Blood 121, 4463–4472 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Pietras E. M., et al., Functionally distinct subsets of lineage-biased multipotent progenitors control blood production in normal and regenerative conditions. Cell Stem Cell 17, 35–46 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Satpathy A. T., et al., Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat. Biotechnol. 37, 925–936 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoshida H., et al.; Immunological Genome Project, The cis-regulatory atlas of the mouse immune system. Cell 176, 897–912.e20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.J. M. Granjaet al., . ArchR is a scalable software package for integrative single-cell chromatin accessibility analysis. Nat. Genet. 53, 403–411 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tober J., Yzaguirre A. D., Piwarzyk E., Speck N. A., Distinct temporal requirements for Runx1 in hematopoietic progenitors and stem cells. Development 140, 3765–3776 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsai J. J., et al., Nrf2 regulates haematopoietic stem cell function. Nat. Cell Biol. 15, 309–316 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pliner H. A., et al., Cicero predicts cis-regulatory DNA interactions from single-cell chromatin accessibility data. Mol. Cell 71, 858–871.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hall T., et al., Nfix promotes survival of immature hematopoietic cells via regulation of c-Mpl. Stem Cells 36, 943–950 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holmfeldt P., et al., Nfix is a novel regulator of murine hematopoietic stem and progenitor cell survival. Blood 122, 2987–2996 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Connor C., et al., Nfix expression critically modulates early B lymphopoiesis and myelopoiesis. PLoS One 10, e0120102 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kato H., et al., Infection perturbs Bach2- and Bach1-dependent erythroid lineage ‘choice’ to cause anemia. Nat. Immunol. 19, 1059–1070 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Domínguez-Andrés J., et al., The itaconate pathway is a central regulatory node linking innate immune tolerance and trained immunity. Cell Metab. 29, 211–220.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Tsang E., et al., Molecular mechanism of the Syk activation switch. J. Biol. Chem. 283, 32650–32659 (2008). [DOI] [PubMed] [Google Scholar]
- 49.Xu H., et al., Protoporphyrin IX induces a necrotic cell death in human THP-1 macrophages through activation of reactive oxygen species/c-Jun N-terminal protein kinase pathway and opening of mitochondrial permeability transition pore. Cell. Physiol. Biochem. 34, 1835–1848 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Shipovskov S., et al., Metallation of the transition-state inhibitor N-methyl mesoporphyrin by ferrochelatase: Implications for the catalytic reaction mechanism. J. Mol. Biol. 352, 1081–1090 (2005). [DOI] [PubMed] [Google Scholar]
- 51.Kawamura-Konishi Y., et al., Kinetic characterization of antibody-catalyzed insertion of a metal ion into porphyrin. J. Biochem. 119, 857–862 (1996). [DOI] [PubMed] [Google Scholar]
- 52.Gilmour P. S., Rahman I., Donaldson K., MacNee W., Histone acetylation regulates epithelial IL-8 release mediated by oxidative stress from environmental particles. Am. J. Physiol. Lung Cell. Mol. Physiol. 284, L533–L540 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Gatla H. R., Zou Y., Uddin M. M., Vancurova I., Epigenetic regulation of interleukin-8 expression by class I HDAC and CBP in ovarian cancer cells. Oncotarget 8, 70798–70810 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Medzhitov R., Janeway C. Jr., Innate immune recognition: Mechanisms and pathways. Immunol. Rev. 173, 89–97 (2000). [DOI] [PubMed] [Google Scholar]
- 55.Chow N. A., Jasenosky L. D., Goldfeld A. E., A distal locus element mediates IFN-γ priming of lipopolysaccharide-stimulated TNF gene expression. Cell Rep. 9, 1718–1728 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fanucchi S., et al., Immune genes are primed for robust transcription by proximal long noncoding RNAs located in nuclear compartments. Nat. Genet. 51, 138–150 (2019). [DOI] [PubMed] [Google Scholar]
- 57.Cirovic B., et al., BCG vaccination in humans elicits trained immunity via the hematopoietic progenitor compartment. Cell Host Microbe 28, 322–334.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liao R., et al., Discovering how heme controls genome function through heme-omics. Cell Rep. 31, 107832 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tanimura N., et al., GATA/heme multi-omics reveals a trace metal-dependent cellular differentiation mechanism. Dev. Cell 46, 581–594.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kleinnijenhuis J., et al., BCG-induced trained immunity in NK cells: Role for non-specific protection to infection. Clin. Immunol. 155, 213–219 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bekkering S., et al., Oxidized low-density lipoprotein induces long-term proinflammatory cytokine production and foam cell formation via epigenetic reprogramming of monocytes. Arterioscler. Thromb. Vasc. Biol. 34, 1731–1738 (2014). [DOI] [PubMed] [Google Scholar]
- 62.Sohrabi Y., et al., OxLDL-mediated immunologic memory in endothelial cells. J. Mol. Cell. Cardiol. 146, 121–132 (2020). [DOI] [PubMed] [Google Scholar]
- 63.Christ A., et al., Western diet triggers NLRP3-dependent innate immune reprogramming. Cell 172, 162–175.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Khan N., et al., M. tuberculosis reprograms hematopoietic stem cells to limit myelopoiesis and impair trained immunity. Cell 183, 752–770.e22 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mandal P., et al., Caspase-8 collaborates with caspase-11 to drive tissue damage and execution of endotoxic shock. Immunity 49, 42–55.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Benn C. S., Netea M. G., Selin L. K., Aaby P., A small jab - A big effect: Nonspecific immunomodulation by vaccines. Trends Immunol. 34, 431–439 (2013). [DOI] [PubMed] [Google Scholar]
- 67.Stuart T., et al., Comprehensive integration of single-cell data. Cell 177, 1888–1902 e1821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bravo González-Blas C., et al., cisTopic: Cis-regulatory topic modeling on single-cell ATAC-seq data. Nat. Methods 16, 397–400 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
RNA sequencing and snATAC-seq data have been deposited in NCBI Gene Expression Omnibus (GSE111003).