Abstract
Induction of GCN4 translation in amino acid-starved cells involves the inhibition of initiator tRNAMet binding to eukaryotic translation initiation factor 2 (eIF2) in response to eIF2 phosphorylation by protein kinase GCN2. It was shown previously that GCN4 translation could be induced independently of GCN2 by overexpressing a mutant tRNAAACVal (tRNAVal*) or the RNA component of RNase MRP encoded by NME1. Here we show that overexpression of the tRNA pseudouridine 55 synthase encoded by PUS4 also leads to translational derepression of GCN4 (Gcd− phenotype) independently of eIF2 phosphorylation. Surprisingly, the Gcd− phenotype of high-copy-number PUS4 (hcPUS4) did not require PUS4 enzymatic activity, and several lines of evidence indicate that PUS4 overexpression did not diminish functional initiator tRNAMet levels. The presence of hcPUS4 or hcNME1 led to the accumulation of certain tRNA precursors, and their Gcd− phenotypes were reversed by overexpressing the RNA component of RNase P (RPR1), responsible for 5′-end processing of all tRNAs. Consistently, overexpression of a mutant pre-tRNATyr that cannot be processed by RNase P had a Gcd− phenotype. Interestingly, the Gcd− phenotype of hcPUS4 also was reversed by overexpressing LOS1, required for efficient nuclear export of tRNA, and los1Δ cells have a Gcd− phenotype. Overproduced PUS4 appears to impede 5′-end processing or export of certain tRNAs in the nucleus in a manner remedied by increased expression of RNase P or LOS1, respectively. The mutant tRNAVal* showed nuclear accumulation in otherwise wild-type cells, suggesting a defect in export to the cytoplasm. We propose that yeast contains a nuclear surveillance system that perceives defects in processing or export of tRNA and evokes a reduction in translation initiation at the step of initiator tRNAMet binding to the ribosome.
Starvation of yeast cells for amino acids or purines leads to increased expression of GCN4, a transcriptional activator of amino acid biosynthetic enzymes (general amino acid control). GCN4 expression is stimulated at the translational level by a mechanism involving four short upstream open reading frames (uORFs) in its mRNA leader. During growth on amino acid-replete medium, scanning ribosomes translate the first uORF (uORF1) and reinitiate downstream at uORF2, uORF3, or uORF4 but cannot reinitiate again at the GCN4 start codon. In amino acid-starved cells, eukaryotic translation initiation factor 2 (eIF2) is phosphorylated on its α subunit by protein kinase GCN2, and the phosphorylated eIF2 inhibits the guanine nucleotide exchange factor for eIF2, known as eIF2B. Consequently, formation of the ternary complex containing eIF2, GTP, and initiator methionyl-tRNA (Met-tRNAiMet) is reduced, impairing delivery of tRNAiMet to the ribosome. In GCN4 mRNA, the ensuing delay in rebinding of ternary complex to 40S ribosomes which have translated uORF1 allows them to scan past uORF2 to uORF4 and reinitiate downstream at the GCN4 start codon instead (25, 26). Thus, GCN4 translation is induced under conditions of diminished ternary-complex formation.
It is thought that GCN2 is activated in amino acid-starved cells by uncharged tRNAs (34, 42, 43, 54) which accumulate under these conditions and bind to a regulatory domain in GCN2 homologous to histidyl-tRNA synthetases (55, 57, 58). Because starvation for any of several amino acids elicits activation of GCN2 and attendant derepression of GCN4 (34, 57), it is probable that most uncharged tRNAs can bind to GCN2 and activate its kinase function. Two positive regulators of GCN2, encoded by GCN1 and GCN20 (41, 53), show sequence similarity to translation elongation factor eEF3 and have ribosome binding activities (40). It has been proposed that the GCN1-GCN20 complex functions at the ribosome in promoting activation of GCN2 by uncharged tRNAs which have entered the decoding site (40).
There are several instances where GCN4 translation is stimulated in a manner dependent on the uORFs but independent of GCN2 and eIF2 phosphorylation. Mutations in subunits of eIF2 or eIF2B appear to reduce the functions of these two factors and mimic the effects of eIF2 phosphorylation in restricting ternary-complex formation. These mutations constitutively derepress GCN4 translation and the amino acid biosynthetic enzymes subject to general amino acid control (Gcd− phenotype). The same phenotype is observed for deletions that reduce the number of IMT genes encoding tRNAiMet (14) and thereby decrease the steady-state level of this component of the ternary complex. Mutations in GCD10 (18, 22) and GCD14 (10, 13), whose products are required for methylation of adenosine-58 in tRNAiMet (1), also have GCN2-independent Gcd− phenotypes. Lack of m1A58 specifically impairs 5′-end processing and stability of tRNAiMet. In these instances, the Gcd− phenotype can be explained by a reduction in the ternary-complex level independently of eIF2 phosphorylation by GCN2.
Previously, we observed GCN2-independent derepression of GCN4 translation in cells overexpressing tRNAs under conditions where it was presumed that the excess tRNA was not aminoacylated efficiently. This occurred most notably with a mutant form of tRNAAACVal harboring an A-to-G substitution in the 3′-terminal nucleotide (tRNAVal*), which is expected to impair aminoacylation by valyl-tRNA synthetase. Overexpression of tRNAVal* did not lead to eIF2 phosphorylation in strains containing GCN2; however, it exacerbated the growth defect of a GCN2c mutant (expressing a constitutively active kinase) in which eIF2 is hyperphosphorylated and thus impaired in general translation initiation. The latter findings suggested that excess tRNAVal* leads to reduced eIF2 function by a mechanism independent of eIF2 phosphorylation (54). To explain these findings, we proposed that yeast cells have a second sensor of uncharged tRNA besides GCN2 that also constrains eIF2 activity. Moreover, because tRNAVal* overexpression did not activate GCN2, it seemed possible that this defective tRNA was physically sequestered from GCN2 (54).
GCN2-independent derepression of GCN4 translation also was elicited by overexpression of NME1 (51), encoding the RNA component of RNase MRP (48). RNase MRP is involved in processing rRNA, and it was suggested that defects in ribosome biogenesis caused by NME1 overexpression could impair GCN4 translational control. Partial derepression of GCN4 translation additionally occurred during growth on rich medium in mutant strains with constitutively high levels of protein kinase A (PKA) function (RAS2Val19 and bcy1Δ mutants) (16). It is unknown how elevated PKA function impairs translational control of GCN4.
In this report we show that overexpression of the tRNA pseudouridine 55 synthase encoded by PUS4 (7) stimulates GCN4 translation independently of GCN2 and its phosphorylation site on eIF2. We present several lines of evidence that PUS4 overexpression does not reduce the amount of functional tRNAiMet as the means of limiting ternary-complex formation or its utilization in translation initiation. Instead, it appears that excess PUS4 impedes the 5′-end processing and export of certain tRNAs in the nucleus. The same mechanism seems to apply to overexpressed NME1; moreover, overproduction of a mutant pre-tRNA that cannot be processed by RNase P elicits a GCN2-independent Gcd− phenotype. These and other findings strongly suggest that yeast contains a surveillance system that perceives defects in tRNA processing or transport in the nucleus and reduces the efficiency of translation initiation in the cytoplasm in response.
MATERIALS AND METHODS
Identification of PUS4 as a high-copy-number suppressor of gcn2-1.
Plasmid pAH14 was isolated previously as a high-copy-number suppressor of a gcn2-1 mutant (27). Sequencing the ends of the DNA insert and comparison with the complete yeast genome sequence showed that the insert is ∼5 kb and contains three genes from chromosome XIV: RFC3 (36), MID1 (30), and PUS4 (7). Subclones pHQ536 carrying MID1 and PUS4, pHQ537 carrying PUS4, and pHQ538 carrying RFC3 and MID1 were constructed from pAH14 (see below) and introduced into gcn2-1 mutant H113, and the resulting transformants were tested for growth on medium containing 3-aminotriazole (3-AT). Only plasmids pHQ536 and pHQ537 conferred 3-AT resistance, indicating that PUS4 is a high-copy-number suppressor of gcn2-1. To confirm this conclusion, 5′ and 3′ deletions (pHQ546 and pHQ545, respectively) and an internal frameshift mutation (pHQ575) in PUS4 were constructed (see below). None of these plasmids conferred 3-AT resistance in strain H113, and neither did the single-copy-number PUS4 plasmid pHQ543 that we constructed.
Yeast strains and plasmid construction.
All yeast strains except HQY316 used in this study were described previously and are listed in Table 1. HQY316 was constructed from H1895 by replacing LOS1 with the los1Δ::hisG::URA3::hisG allele using a ∼4.8-kb EcoRI-BglII fragment from plasmid pHQ871. The resulting los1Δ strain was identified by PCR and further confirmed by complementation of its Gcd− phenotype by LOS1 plasmid pHQ860. The plasmids used in this work were constructed as follows. Plasmid pHQ536 containing MID1 (30) and PUS4 was constructed by inserting an ∼4-kb SalI fragment from pAH14 into YEplac181 (20) at the SalI site. An ∼2.0-kb BglII-Asp718 fragment containing PUS4 (7) from pHQ536 was inserted into YEplac181 between the BamHI and Asp718 sites to produce plasmid pHQ537. An ∼3.8-kb NheI-XbaI fragment containing RFC3 (36) and MID1 from pAH14 was inserted into YEplac181 at the XbaI site to produce pHQ538. To construct the C-terminal deletion of PUS4, an ∼1.5-kb XbaI fragment from pHQ537 was subcloned into YEplac181 at the XbaI site to produce pHQ545, encoding PUS4 amino acids 1 to 342. The N-terminal deletion of PUS4 was constructed by removing the BamHI fragment from pHQ537 to produce plasmid pHQ546, in which the 5′ noncoding region and first 74 codons of PUS4 were deleted. A frameshift mutation in PUS4 was constructed by digesting pAH14 with BamHI, filling in the ends, and religating to produce pHQ575. Single-copy-number plasmid pHQ543 bearing PUS4 was constructed by inserting an ∼1.8-kb BglII-NaeI fragment from pHQ536 into YCplac111 (20). High-copy-number PUS4 plasmid pHQ547 was constructed by inserting the BglII-SphI fragment containing PUS4 from pAH14 into YEp24 between the BamHI and SphI sites. The single-copy-number GCN2 plasmid pHQ548 was created by inserting the XbaI-SalI fragment from p722 (56) into YCplac111. To add the hemagglutinin (HA) epitope to PUS4, NruI and MluI sites were first introduced into pHQ537 immediately 5′ to the PUS4 stop codon by site-directed mutagenesis, using the Quik-Change site-directed mutagenesis kit (Stratagene), producing plasmid pHQ732. An ∼100-bp PCR fragment encoding three copies of HA with Ecl136II and MluI ends was then inserted between the NruI-MluI sites of pHQ732 to produce plasmid pHQ753 encoding PUS4-HA. An ∼2-kb SalI fragment encoding PUS4-HA from pHQ753 was inserted into YCplac111 and pHQ583 to produce single-copy-number and high-copy-number plasmids pHQ771 and pHQ839, respectively. pHQ583 is a derivative of YEplac181 in which the polycloning sites SacI to BamHI were deleted by filling in the EcoRI and XbaI sites and religating. pHQ853 and pHQ857, encoding pus4-1-HA and pus4-2-HA, respectively, were derived from pHQ839 by site-directed mutagenesis using the Quik-Change site-directed mutagenesis kit.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| H113 | MATα his1-29 ura3-52 leu2-3 leu2-112 gcn2-1 [HIS4-lacZ, URA3] | 27 |
| H1816 | MATa ura3-52 leu2-3 leu2-112 trp1-Δ63 gcn2Δ sui2Δ [GCN4-lacZ, TRP1] [SUI2 LEU2] | 13a |
| H1817 | MATa ura3-52 leu2-3 leu2-112 trp1-Δ63 gcn2Δ sui2Δ [GCN4-lacZ, TRP1] [SUI2-S51A LEU2] | 13a |
| H1894 | MATa ura3-52 leu2-3 leu2-112 trp1-Δ63 gcn2Δ | T. Dever |
| H1895 | MATa ura3-52 leu2-3 leu2-112 trp1-Δ63 gcn2Δ [GCN4-lacZ, TRP1] | T. Dever |
| H1897 | MATa ura3-52 leu2-3 leu2-112 trp1-Δ63 GCN2 sui2Δ [GCN4-lacZ, TRP1] [SUI2-S51A LEU2] | 13a |
| H1937 | MATα gcn2::LEU2 ura3-52 leu2-3 leu2-112 [HIS4-lacZ, ura3-52] | 54 |
| HQY316 | MATa ura3-52 leu2-3 leu2-112 trp1-Δ63 gcn2Δ los1Δ::hisG::URA3::hisG [GCN4-lacZ, TRP1] | This work |
Plasmid pHQ731, used for in vitro synthesis of tRNAAsp mut#2 (8), was constructed by inserting a 52-bp double-stranded oligonucleotide encoding the T7 promoter and tRNAAsp mut#2 into pUC18 at the SmaI site. To construct hcNME1 and hcNME1/RPR1 plasmids, an EcoRI linker was first added to the filled-in BamHI site of pDK45, an NME1-bearing plasmid obtained from Lasse Lindahl, to produce pHQ859. The ∼0.7-kb EcoRI fragment containing NME1 from pHQ859 was then inserted into YEplac181 and pHQ682 at their respective EcoRI sites to produce high-copy-number plasmids pHQ862 and pHQ863 containing NME1 and NME1/RPR1, respectively. pHQ682 was constructed by inserting the ∼1.3-kb EcoRI-HindIII fragment containing RPR1 from pDK42, an RPR1-bearing plasmid obtained from Lasse Lindahl, into YEplac181 between the EcoRI and HindIII sites. An EcoRI linker was also added to the filled-in HindIII site of pDK42, so that an ∼1.3-kb EcoRI fragment bearing RPR1 could be isolated from the resulting plasmid (pHQ858) and inserted into pHQ839 at the EcoRI site, producing high-copy-number plasmid pHQ864 bearing PUS4-HA and RPR1.
LOS1-bearing plasmids YEpLOS1 and YCpLOS1 were described previously (29). To construct the los1Δ plasmid, a BamHI linker was first added at the PvuII site of YCpLOS1 to produce plasmid pHQ868 and a BamHI fragment containing hisG::URA3::hisG was inserted at the BamHI site to produce plasmid pHQ871. hcLOS1 plasmid pHQ860 was constructed by inserting a 5.3-kb SphI fragment from YCpLOS1 into the SphI site of YEplac181. Plasmids pHQ982 and pHQ985 encoding wild-type and mutant pre-tRNAGUATyr, respectively, were constructed by inserting PCR-synthesized genomic DNA fragments with EcoRI and BamHI sites at the 5′ and 3′ ends, respectively, between the corresponding sites in high-copy-number plasmid YEplac181. The genomic DNA fragments containing 170 and 18 bp of 5′ and 3′ noncoding DNA, respectively, were synthesized using the following oligonucleotide primers: 5′-CCGGAATTCCTGTATTAGTCGATATACCACC-3′ (forward primer), 5′-CGCGGATCCGCAAGATTTAAAAAAATATCTCCCGGGGGCGA-3′ (reverse primer for the wild-type 3′ trailer), and 5′-CGCGGATCCGCAAGATTTAAAAAAATACGACTCCCGGGGGCGA-3′ (reverse primer for the mutant 3′ trailer).
Assay of HIS4-lacZ and GCN4-lacZ fusions.
Assays were conducted using cell extracts prepared from cultures grown in SD medium containing only the required supplements as described previously (37). For repression conditions, saturated cultures were diluted 1:50 into fresh medium and harvested in mid-logarithmic phase after 6 h of growth. For derepression conditions, cultures were grown for 2 h under repression conditions and then for 6 h after adding 3-AT to 10 mM, 5-methyltryptophan (5-MT) to 2 mM, or sulfometuron methyl (SM) to 0.5 μg/ml.
Assay of yeast tRNA pseudouridine 55 synthase.
Synthesis of pseudouridine is accompanied by the release of a proton from carbon 5 in the pyrimidine ring of the uridine base (12); therefore, release of tritium from [5-3H]uridine-labeled tRNA can be used as a measure of pseudouridine 55 synthase activity (46). PUS4, the S. cerevisiae enzyme, can catalyze the formation of pseudouridine 55 in a model substrate corresponding to the acceptor stem and TψC stem-loop of tRNAAsp (mut#2 minihelix) (8). Accordingly, we assayed PUS4 activity in cell extracts by measuring the release of tritium (46) from mut#2 minihelix RNA synthesized in vitro in the presence of [5-3H]UTP. tRNAAsp mut#2 RNA labeled with [5-3H]uridine was synthesized in vitro as previously described (46). Briefly, 10 μg of MvaI-digested pHQ731 was mixed with 100 μCi of [5-3H]UTP (14.5 Ci/mmol; Amersham), dried under vacuum, and resuspended in 100 μl of a reaction mixture containing 40 mM Tris-HCl (pH 7.9), 6 mM MgCl2, 2 mM spermidine, 10 mM NaCl, 10 mM dithiothreitol, 10 mM GMP, 1 mM each GTP, CTP, and ATP, 250 μM UTP, and 100 U of RNasin (Promega). The reaction was initiated by adding 100 U of T7 RNA polymerase (Promega), and the mixture was incubated at 37°C for 2 h. Afterwards, the mixture was extracted once with phenol-chloroform (1:1) and the [3H]RNA was ethanol precipitated and resuspended in water pretreated with diethylpyrocarbonate.
The tritium release assay for pseudouridine synthase was conducted as described previously (46). Briefly, the reaction mixture contained 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 10 mM dithiothreitol, 0.2 mg of bovine serum albumin per ml, 80 U of RNasin (Promega), and 3H-labeled tRNAAsp mut#2 (2.5 × 106 cpm). The reaction was started by adding S100 yeast whole-cell extract to a total volume of 100 μl, and the mixture was incubated at 30°C for 30 min. The reaction was terminated by adding 0.3 ml of a suspension of Norit A (12% in 0.1 N HCl). After 2 min at room temperature, the mixture was centrifuged and the radioactivity in the supernatant was determined. The activity of pseudouridine 55 synthase in the whole-cell extract was expressed as cpm of 3H released per microgram of protein. S100 yeast whole-cell extracts were prepared as described previously (4).
Analysis of tRNA modification and aminoacylation in vivo.
The primer extension assay used for mapping pseudouridine residues was conducted as described previously (5–7). In this assay, RNA is treated with 1-cyclohexyl-3-(2-morpholinoethyl)-carbodiimide metho-p-toluenesulfonate (CMCT), a chemical that reacts with both pseudouridine and uridine residues. The presence of CMCT-coupled nucleosides in tRNA impedes reverse transcription primed by an oligonucleotide annealed 3′ to the CMCT-coupled base. Because CMCT-pseudouridine is more resistant than CMCT-uridine to alkali treatment, the locations of pseudouridine residues in a tRNA molecule can be deduced from the presence of alkali-insensitive blocks to reverse transcription (5, 6). Chromatography of aminoacylated tRNAiMet and elongator tRNAMet (tRNAeMet) on RPC-5 resin was carried out as described previously (1). For Northern analysis of in vivo-aminoacylated tRNAs, total RNA was prepared under acidic conditions and resolved by electrophoresis on acid-urea gels as described previously (52). The following oligonucleotides were used to probe the Northern blots: 5′-TGGTAGCGCCGCTCGGTTTCGAATCC-3′ (tRNAiMet), 5′-TGCTCCAGGGGAGGTTCGAACTCTCGACC-3′ (tRNAeMet), 5′-CACTCACGATGGGGGTCGAA-3′ (tRNAUCUArg), 5′-TGCTCGAGGTGGGGA/TTTGAACCCACGACGG-3′ (tRNAUAUIle), 5′-GATTGCAGCACCTGAGTTTCGCGTTATGG-3′ (5S rRNA), and 5′-GGTGGGAGACTTTCAACCCAAAGC-3′ (NME1).
Fluorescence in situ hybridization.
The fluorescence in situ hybridization procedure was conducted as described previously (47), except that transformants carrying plasmids were grown at 30°C to log phase. The oligonucleotides used were probe 04 (47), to detect tRNAUAUIle, and 5′-CGCCCAGGATCGAACTG GGGACGTTCTGCGTGTTAAGCAGATGCCATAACCGACTAGACC-3′, to detect tRNAAACVal.
RESULTS
Overexpression of PUS4, encoding tRNA pseudouridine 55 synthase, derepresses GCN4 translation in the absence of eIF2 kinase GCN2.
In an effort to identify a novel regulator of GCN2, we analyzed a previously described high-copy-number plasmid, pAH14, which suppresses the 3-AT-sensitive (3-ATs) phenotype of a gcn2-1 mutant (27). 3-AT is a competitive inhibitor of the histidine biosynthetic enzyme encoded by HIS3, and GCN4-mediated derepression of HIS3 transcription is required for growth in the presence of this inhibitor. Accordingly, gcn2 mutants are 3-ATs because they fail to derepress GCN4 translation in response to histidine starvation. Suppression of the 3-ATs phenotype of gcn2-1 by pAH14 suggested that HIS3 derepression had been restored independently of GCN2. Sequencing the ends of the genomic DNA insert in pAH14 revealed that it contains three genes from chromosome XIV: RFC3 (36), MID1 (30), and PUS4, of which the last encodes tRNA pseudouridine 55 synthase (7). By analyzing subclones of pAH14, we determined that high-copy-number PUS4 was sufficient for suppression of gcn2-1 and that deletions removing the 5′ or 3′ end of the PUS4 ORF or introduction of a frameshift mutation in PUS4 abolished suppression (see Materials and Methods). Moreover, PUS4 on a low-copy-number plasmid failed to suppress the gcn2-1 allele (data not shown). Thus, we concluded that PUS4 is a high-copy-number suppressor of gcn2-1.
We found that high-copy-number PUS4 (hcPUS4) suppressed the 3-ATs phenotypes of a gcn2Δ mutant and a strain containing an Ala substitution in the GCN2 phosphorylation site in eIF2α, Ser-51 (the SUI2-S51A allele) (Fig. 1). Thus, it appeared that hcPUS4 derepresses HIS3 expression independently of eIF2α phosphorylation by GCN2, an event required in wild-type cells for increased translation of GCN4 mRNA. Analysis of a HIS4-lacZ fusion showed that expression of HIS4, another target of GCN4, was derepressed ca. threefold in gcn2Δ transformants bearing hcPUS4 (Table 2). Similar degrees of HIS4-lacZ derepression were observed in gcn2Δ cells bearing hcPUS4 in the presence or absence of inhibitors of histidine (3-AT), tryptophan (5-MT), or isoleucine-valine (SM) biosynthesis. These findings suggested that GCN4 expression was constitutively derepressed by hcPUS4 independently of both amino acid starvation and eIF2 phosphorylation by GCN2. Supporting this conclusion, expression of a GCN4-lacZ fusion was derepressed five- to sixfold in gcn2Δ cells bearing hcPUS4 in the presence or absence of 3-AT (Table 3). In contrast, expression of a GCN4-lacZ fusion lacking all four uORFs required for translational control was unaffected by hcPUS4 (Table 3). These last results indicate that hcPUS4 stimulates GCN4 expression at the translational level. In agreement with this conclusion, the presence of hcPUS4 had no effect on steady-state GCN4 mRNA levels (data not shown).
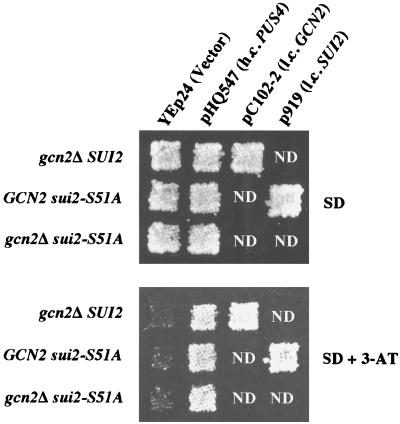
FIG. 1.
High-copy-number plasmid encoding PUS4 derepresses histidine biosynthetic genes in the absence of GCN2 and Ser-51 of eIF2α. Isogenic strains H1816 (gcn2Δ SUI2), H1897 (GCN2 sui2-S51A), and H1817 (gcn2Δ sui2-S51A) were transformed with the indicated plasmids, replica-plated to SD medium or to SD medium containing 30 mM 3-AT, and incubated for 3 days at 30°C. pHQ547 is a high-copy-number (h.c.) plasmid containing PUS4; pC102-2 is a low-copy-number (l.c.) plasmid containing GCN2, and p919 is a low-copy-number (l.c.) plasmid containing SUI2.
TABLE 2.
Effect of hcPUS4 on HIS4-lacZ expression in a gcn2Δ strain starved for different amino acids
| Plasmid | Genea | β-Galactosidase activity (U)b
|
|||
|---|---|---|---|---|---|
| R | 3-AT | 5-MT | SM | ||
| YEp24 | None | 120 | 120 | 110 | 120 |
| p585 | GCN2 (l.c.) | 210 | 650 | 640 | 920 |
| pHQ547 | PUS4 (h.c.) | 330 | 370 | 290 | 450 |
| p1362 | tRNAVal* (h.c.) | 360 | 530 | 280 | 510 |
l.c., low copy number; h.c., high copy number.
Transformants of strain H1937 (gcn2Δ) carrying the indicated plasmid were grown under repressing (R) nonstarvation condition or under derepressing conditions generated by adding 3-AT for histidine starvation, 5-MT for tryptophan starvation, or SM for leucine, isoleucine, and valine starvation. Extracts were prepared and assayed for enzyme activity, and the results shown are means from three transformants. The β-galactosidase activity is expressed as nanomoles of o-nitrophenyl-β-d-galactopyranoside hydrolyzed per minute per milligram of protein. The standard errors are less than 20%.
TABLE 3.
Derepression of GCN4-lacZ expression by hcPUS4 in a gcn2Δ strain requires uORFs in GCN4 mRNA
| Plasmid (gene) | β-Galactosidase activity (U)a
|
|||
|---|---|---|---|---|
| p180
|
p227
|
|||
| R | DR | R | DR | |
| YEplac181 (vector) | 5 | 8 | 1,800 | 1,900 |
| pHQ537 (hcPUS4) | 25 | 48 | 1,900 | 1,900 |
| pHQ548 (lcGCN2) | 14 | 158 | 1,900 | 1,800 |
The gcn2Δ strain H1894 was transformed with empty vector, high-copy-number plasmid pHQ537 carrying PUS4, or low-copy-number GCN2 (lcGCN2) plasmid pHQ548 and cotransformed with p180 or p227 harboring GCN4-lacZ fusions containing all four uORFs or no uORFs, respectively, in the GCN4 mRNA leader. Transformants were grown under repressing (R) conditions or under derepressing (DR) conditions imposed by adding 3-AT to elicit histidine starvation. Extracts of the transformants were assayed for enzyme activity, and the results are means from three transformants. The β-galactosidase activity is expressed as nanomoles of o-nitrophenyl-β-d-galactopyranoside hydrolyzed per minute per milligram of protein. The standard errors are less than 20%.
A high-copy-number plasmid encoding the mutant tRNAAACVal described above (hctRNAVal*) (54) led to slightly higher levels of GCN4-lacZ expression in a gcn2Δ strain than did hcPUS4; however, the presence of both plasmids in the same transformants did not increase GCN4 expression in an additive fashion (Table 4). This nonadditivity suggests that overexpression of PUS4 or tRNAVal* leads to derepression of GCN4 expression by a common mechanism.
TABLE 4.
Nonadditive effects of high-copy-number plasmids carrying tRNAVal* and PUS4 on derepression of GCN4-lacZ expression in a gcn2Δ strain
| Plasmid 1 | Plasmid 2 | β-Galactosidase activity (U)a
|
|
|---|---|---|---|
| R | DR | ||
| YEp24 (vector) | YEplac181 (vector) | 4 | 5 |
| YEp24 | pHQ537 (hcPUS4) | 16 | 35 |
| p1362 (hctRNAVal*) | YEplac181 | 23 | 45 |
| p1362 | pHQ537 | 22 | 55 |
Transformants of strain H1895 (gcn2Δ) bearing the indicated plasmids were grown under repressing (R) nonstarvation conditions or under derepressing (DR) conditions produced by adding 3-AT for histidine starvation. Extracts were assayed for enzyme activity, and the results shown are means from three or more transformants. The standard errors are less than 20%.
hcPUS4 leads to elevated pseudouridine 55 synthase activity in vivo.
To show that cells bearing hcPUS4 contain increased amounts of PUS4 protein, we assayed pseudouridine 55 synthase activity in cell extracts (see Materials and Methods). As shown in Table 5, extracts of transformants containing hcPUS4 or a functional HA-tagged form of hcPUS4 contained 15 to 16 times as much pseudouridine 55 synthase activity than did the corresponding extract from the vector transformant. This increase in enzyme activity was similar in magnitude to the increase in PUS4 protein levels measured in extracts from transformants bearing the HA-tagged PUS4 allele (PUS4-HA) on high-copy-number versus single-copy-number plasmids, as judged by immunoblot analysis with anti-HA antibodies (Table 5). Based on these results, we conclude that hcPUS4 leads to a large increase in the level of PUS4 enzyme activity in vivo.
TABLE 5.
Phenotypes and level of pseudouridine 55 synthase activity conferred by PUS4 allelesa
| Plasmid | Genotype | Growth on 3-AT | Relative ψ55 synthase activity | Relative protein level |
|---|---|---|---|---|
| Vector | PUS4 | − | 1 | NAb |
| pHQ537 | hcPUS4 | + | 16 | NA |
| pHQ771 | lcPUS4-HA | − | NDc | 6 |
| pHQ839 | hcPUS4-HA | + | 15 | 100 |
| pHQ853 | hcpus4-1-HA | + | 1 | 66 |
| pHQ857 | hcpus4-2-HA | − | 1 | 14 |
Transformants of strain H1895 (gcn2Δ) bearing the indicated plasmids were tested for growth on medium containing 3-AT (30 mM). Extracts were assayed for pseudouridine 55 (ψ55) synthase activity and for levels of PUS4-HA by Western blot analysis. PUS4-HA proteins were detected using anti-HA antibodies and an enhanced chemiluminescence system (Amersham) to visualize immune complexes. The Western blot signals were quantified with a scanner (Silverscanner III) and NIH Image software (version 1.61).
NA, not applicable.
ND, not determined.
Increased pseudouridine 55 synthase activity is not required for suppression of gcn2 mutations by hcPUS4.
The observations that overexpressing the mutant tRNAVal* derepressed GCN4 translation independently of GCN2 (54), that hctRNAVal* and hcPUS4 had nonadditive effects on GCN4 expression, and that PUS4 is a tRNA modification enzyme led us to consider that overexpression of PUS4 might lead to aberrant pseudouridine formation in tRNAs and impede aminoacylation by their cognate aminoacyl-tRNA synthetases. If this occurred with tRNAiMet, the only known tRNA in Saccharomyces cerevisiae that normally lacks this modification (49), it would lower ternary-complex levels and thereby derepress GCN4 translation in gcn2Δ cells.
Several observations preclude the possibility of aberrant pseudouridine formation in tRNAiMet or in any other tRNAs in hcPUS4 transformants. First, we found no evidence for increased pseudouridine levels in total tRNA prepared from gcn2Δ transformants carrying hcPUS4 versus vector alone. When total tRNA isolated from these transformants was digested to nucleosides and resolved by high-pressure liquid chromatography (19), there was no significant difference in the amount of pseudouridine relative to other nucleosides, conventional or modified, between the two tRNA samples (data not shown). For example, the ratios of pseudouridine to t6A (N6-threonylcarbamoyladenosine) in the vector and hcPUS4 transformants were 6.65 and 6.82, respectively; the corresponding ratios of pseudouridine to m22G (N2,N2-dimethylguanosine; a modification at guanosine-26) were 2.61 and 2.68.
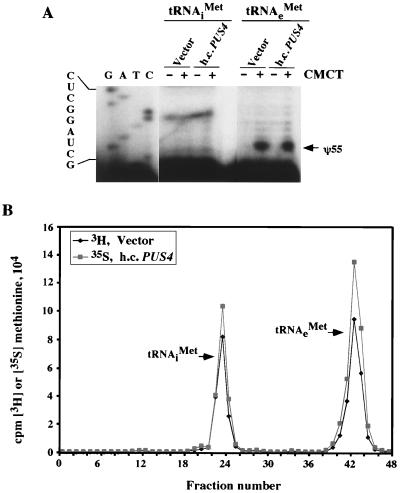
To determine whether overexpression of PUS4 leads specifically to formation of pseudouridine-55 in tRNAiMet, total tRNA was isolated from gcn2Δ transformants containing hcPUS4 or vector alone and subjected to a primer extension assay for mapping pseudouridine residues (see Materials and Methods). By applying this technique with primers that anneal 3′ to position 55 in tRNAiMet or tRNAeMet, we observed the expected block to reverse transcription at pseudouridine 55 in tRNAeMet from transformants containing hcPUS4 or vector alone. In contrast, we observed no block at this location in tRNAiMet that was enhanced by the presence of hcPUS4 (Fig. 2A). Similar results were obtained using total tRNAs prepared from cultures starved for histidine by 3-AT treatment (data not shown). Thus, hcPUS4 does not lead to detectable amounts of pseudouridine 55 in tRNAiMet or diminish this modification in tRNAeMet.
FIG. 2.
High-copy-number PUS4 does not alter base modification of tRNAiMet. (A) Samples of total tRNA (10 μg) prepared from gcn2Δ strain H1894 carrying empty vector (YEplac181) or hcPUS4 plasmid (pHQ537) were treated (+) or not treated (−) with CMCT, and 1 μg was reverse transcribed using end-labeled primers complementary to tRNAiMet or tRNAeMet (nucleotides 60 to 76). Reverse transcription products were resolved in an 8% sequencing gel. The strong stops in reverse transcription of CMCT-treated tRNA correspond to pseudouridine-55 (ψ55), as indicated by the arrow. On the left of the gel is a sequence ladder of initiator tRNAMet. The strong stops at position 52 for tRNAiMet observed independently of CMCT presumably arise from strong secondary structure. (B) The same tRNA samples as in panel A were aminoacylated with [3H]methionine or [35S]methionine, and ca. 500,000 cpm was resolved on an RPC-5 column. Radioactivity in each fraction (2 ml) was measured by liquid scintillation and plotted against the fraction number. The elution positions of the methionine-accepting tRNAs are indicated at the appropriate positions.
In accordance with the above findings, we obtained strong evidence that pseudouridine 55 synthase activity is not required for the suppressor activity of hcPUS4. Using site-directed mutagenesis, we altered PUS4 residues 283 to 286 from TYIR to AAAA (producing pus4-1-HA) or altered residues 74 to 77 from LDPL to AAAA (pus4-2-HA) and introduced the mutant alleles into a gcn2Δ strain on high-copy-number plasmids. The residues altered by these mutations are conserved in pseudouridine 55 synthases among bacteria and yeast (32). As shown in Table 5, neither high-copy-number mutant allele led to pseudouridine 55 synthase activity in extracts above the background level produced by chromosomal PUS4. Immunoblot analysis showed that expression of pus4-1-HA was only slightly reduced, whereas pus4-2-HA expression was greatly decreased, compared to wild-type PUS4-HA (Table 5). Surprisingly, hcpus4-1-HA was indistinguishable from hcPUS4-HA in suppressing the 3-ATs phenotype of the gcn2Δ mutant (Table 5), suggesting that hcPUS4 suppressor activity does not require elevated pseudouridine 55 synthase activity. The fact that hcpus4-2-HA was inactive as a dosage suppressor can be explained by the fact that it was not highly expressed (Table 5).
Evidence that hcPUS4 elicits derepression of GCN4 partly by interfering with 5′-end processing of tRNA by RNase P.
Although the pseudouridine 55 synthase activity of PUS4 is not required for its suppressor activity, it was possible that increased binding of overexpressed PUS4 to one or more tRNAs would restrict the access of other enzymes involved in modification or processing of these tRNAs. As indicated above, we were particularly interested in possible differences in the structure or function of tRNAiMet that could reduce ternary-complex formation. To investigate this last possibility, we first aminoacylated total tRNA from transformants containing hcPUS4 or vector alone with [35S]methionine or [3H]methionine, respectively, and resolved the labeled tRNAs by RPC-5 column chromatography (31). tRNAs that differ by only a single methyl group can be resolved by RPC-5 chromatography (15). The results in Fig. 2B show that the elution positions of [35S]methionine-charged tRNAiMet and tRNAeMet were identical between gcn2Δ transformants bearing hcPUS4 and vector alone. These results suggest that mature methionine-accepting tRNAs are modified identically in cells overexpressing PUS4 and wild-type cells; however, it is possible that certain modifications would not alter the behavior of methionyl-tRNAs on RPC-5 chromatography.
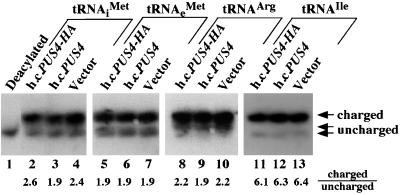
In a second approach, we investigated whether hcPUS4 led to reductions in the efficiency of tRNAiMet aminoacylation in vivo which might arise from a defect in one or more steps in the production of tRNAiMet. The degree of aminoacylation of a tRNA in vivo can be measured by isolating total tRNA at pH 4.5 to preserve the aminoacyl-tRNA linkage and resolving the aminoacylated and deacylated forms by gel electrophoresis followed by Northern blot hybridization (52). When this technique was carried out with tRNA isolated from transformants bearing hcPUS4 versus vector alone, we observed no significant differences in the charged-to-uncharged ratios for tRNAiMet, tRNAeMet, tRNAUCUArg, tRNAUAUIle, and tRNACAALeu (Fig. 3 and data not shown). These results suggest that tRNAiMet, as well as four other tRNAs analyzed by this technique, are aminoacylated with similar efficiencies in cells overexpressing PUS4 and in wild-type cells.
FIG. 3.
Evidence that hcPUS4 does not reduce in vivo aminoacylation of various tRNAs. Total RNAs prepared under acidic conditions (52) from strain H1894 (gcn2Δ) carrying empty vector (YEplac181) or high-copy-number plasmids carrying PUS4 (pHQ537) or PUS4-HA (pHQ839) were resolved by electrophoresis on an acid-urea polyacrylamide gel and subjected to Northern blot analysis. The same blot was probed with radiolabeled oligonucleotides that specifically hybridized to the indicated tRNAs by stripping one probe from the blot before using the next. An aliquot of tRNAiMet was deacylated in 2 M Tris-HCl (pH 8.0) and loaded in lane 1. The intensities of the hybridization signals corresponding to charged and uncharged tRNAs were quantified by phosphorimaging analysis, and the ratios of charged to uncharged tRNA signals are listed below each lane.
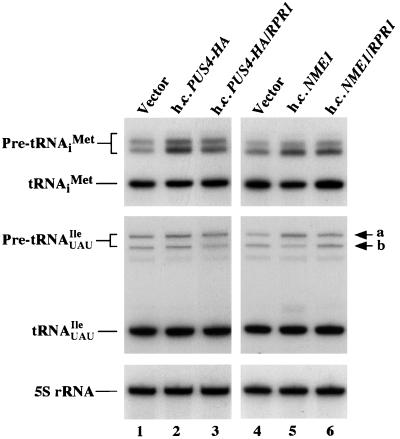
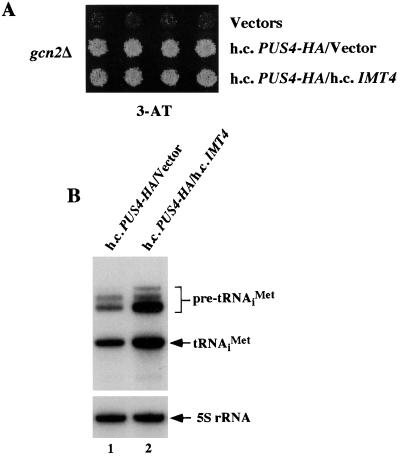
We also measured the total steady-state levels of tRNAiMet, tRNAUAUIle, and tRNACCATrp by Northern analysis and observed no large differences for the mature forms of these tRNAs in gcn2Δ strains bearing hcPUS4-HA versus vector alone (Fig. 4, lanes 1 and 2, and data not shown). Interestingly, the hcPUS4-HA transformants showed significant accumulation of the various precursors of tRNAiMet containing both 5′ and 3′ extensions that are transcribed from different IMT genes (Fig. 4, lanes 1 and 2), leading to a precursor/mature tRNAiMet ratio ca. twofold greater than that of the vector transformant (Table 6). After normalizing for the amounts of 5S RNA in the samples, we calculated that the hcPUS4-HA transformants contained 93% of the wild-type level of mature tRNAiMet. Thus, it appears that the hcPUS4-HA transformants process tRNAiMet precursors more slowly than does the wild type but this defect does not substantially reduce the steady-state level of mature tRNAiMet. The hcPUS4-HA transformants also showed slight accumulation of the larger tRNAUAUIle precursor (Fig. 4), which corresponds to the primary transcript containing 5′ and 3′ extensions plus the intron (44); again, little or no reduction in the level of mature tRNAUAUIle was evident (Table 6). The presence of hcPUS4 had no detectable effect on the levels of precursor or mature tRNACCATrp precursor (data not shown). These findings suggest that PUS4 overexpression decreases the rate at which 5′ and 3′ extensions are removed from a subset of tRNAs.
FIG. 4.
Overexpressing PUS4 or NME1 leads to accumulation of untrimmed tRNA precursors. Total RNA (9 μg) prepared from gcn2Δ strains carrying empty vector YEplac181 or high-copy-number plasmids pHQ839 (h.c.PUS4-HA), pHQ864 (h.c.PUS4-HA/RPR1), (Vector) pHQ862 (h.c.NME1), and pHQ863 (h.c.NME1/RPR1) were subjected to Northern blot analysis and probed with a radiolabeled oligonucleotide complementary to tRNAiMet. The same blots were stripped and reprobed with radiolabeled oligonucleotides specific for tRNAUAUIle or 5S rRNA (see Materials and Methods). The positions of pre-tRNAiMet, mature tRNAiMet, pre-tRNAUAUIle, mature tRNAUAUIle, and 5S rRNA are indicated on the left. The primary transcript (upper band) and the 5′- and 3′-end-processed intron-containing pre-tRNAUAUIle (lower band) are indicated on the right by the letters a and b, respectively.
TABLE 6.
Effect of overexpression of NME1, PUS4, and RPR1 on 5′ processing of tRNAa
| Plasmid | tRNAiMetb
|
tRNAUAUIleb
|
||||
|---|---|---|---|---|---|---|
| Normalized amount
|
p/m ratio | Normalized amount
|
p/m ratio | |||
| p | m | p | m | |||
| Group 1 | ||||||
| Vector | 100 | 100 | 0.50 | 100 | 100 | 0.13 |
| hcPUS4-HA | 194 | 93 | 1.05 | 138 | 97 | 0.19 |
| hcPUS4-HA/RPR1 | 164 | 108 | 0.77 | 112 | 101 | 0.14 |
| Group 2 | ||||||
| Vector | 100 | 100 | 0.47 | 100 | 100 | 0.07 |
| hcNME1 | 136 | 70 | 0.92 | 219 | 99 | 0.16 |
| hcNME1/RPR1 | 127 | 105 | 0.57 | 150 | 105 | 0.10 |
The intensities of the hybridization signals in the autoradiograms were quantified by phosphorimager analysis and normalized to the 5S rRNA signals. The data from Fig. 4 and from a replicate experiment not shown were averaged.
The values for the tRNAiMet precursors or the primary tRNAUAUIle precursor (p) in the transformants harboring hcPUS4-HA or hcNME1 are expressed relative to the corresponding precursor levels in the vector transformants, which are assigned a value of 100. The levels of mature (m) tRNAs are similarly normalized to the levels observed in the vector transformants. p/m is the ratio of the absolute amounts of precursor and mature tRNAs (normalized for 5S rRNA content) for a given tRNA in each transformant.
Although we observed only a small reduction in the steady-state level of mature tRNAiMet in hcPUS4-HA transformants (Table 6), it was important to determine whether this defect was responsible for the suppressor phenotype of hcPUS4. We showed previously that a high-copy-number plasmid bearing IMT4, encoding tRNAiMet, overcame the Gcd− phenotype of gcd10 mutations that reduce steady-state levels of mature tRNAiMet (1). In contrast, we saw little or no effect of hcIMT4 on the phenotype of hcPUS4 (Fig. 5A) even though it produced ca. fourfold-higher levels of mature tRNAiMet (Fig. 5B). We conclude that the Gcd− phenotype of hcPUS4 does not arise from a reduction in the steady-state level of mature tRNAiMet.
FIG. 5.
Overexpression of initiator tRNAMet does not suppress the Gcd− phenotype of hcPUS4. (A) Transformants of strain H1894 (gcn2Δ) bearing high-copy-number plasmids YEplac181 and YEp24 (Vectors), pHQ839 and YEp24 (h.c.PUS4-HA/vector), or pHQ839 and pC50 (h.c.PUS4-HA/h.c.IMT4) were replica-plated to SD medium containing 30 mM 3-AT and incubated for 3 days at 30°C. (B) Total RNA (6 μg) isolated from strains carrying the indicated high-copy-number plasmids were subjected to Northern blot analysis and probed with an oligonucleotide specific for initiator tRNAMet.
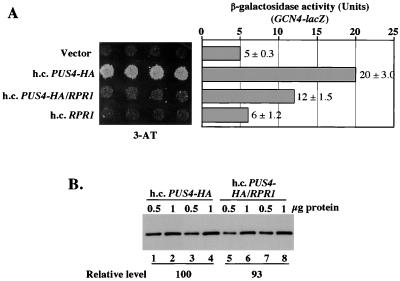
The observation that hcPUS4 leads to accumulation of untrimmed tRNAiMet precursors suggested that an overabundance of these molecules in the nucleus might be a signal for activating GCN4 translation by a GCN2-independent pathway. Because removal of the 5′ leader by RNase P appears to be required for subsequent removal of the 3′ trailer (44), we asked whether overexpression of the RNA component of RNase P, encoded by RPR1, would suppress the Gcd− phenotype of hcPUS4 in gcn2Δ cells. As shown in Fig. 6A, the presence of RPR1 in the same high-copy-number plasmid bearing PUS4-HA overcame the 3-ATr phenotype and partially suppressed the derepression of GCN4 expression, conferred by hcPUS4-HA. Immunoblot analysis indicated that hcRPR1 did not significantly affect PUS4-HA expression in these cells (Fig. 6B). Thus, overexpression of RPR1 overrides the suppressor function of hcPUS4 and does not simply reduce the extent of PUS4 overproduction. Northern blot analysis showed that the presence of RPR1 with PUS4-HA in the same high-copy-number plasmid decreased the precursor/mature ratios for tRNAiMet and tRNAUAUIle from 1.05 to 0.77 and 0.19 to 0.14, respectively (Fig. 4 and Table 6). These results are in agreement with the idea that hcPUS4 elicits derepression of GCN4, at least in part, by interfering with 5′-end processing of certain tRNAs by RNase P.
FIG. 6.
Overexpression of RPR1 reduces the Gcd− phenotype of hcPUS4. (A) Transformants of strain H1895 (gcn2Δ) bearing high-copy-number plasmids YEplac181 (Vector), pHQ839 (h.c.PUS4-HA), pHQ864 (h.c.PUS4-HA/RPR1), or pHQ682 (h.c.RPR1) were replica-plated to SC medium containing 30 mM 3-AT and incubated for 3 days at 30°C (left panel). Extracts from the same transformants grown under repressing (nonstarvation) conditions were assayed for β-galactosidase activity, and the results shown in the right panel are the means and standard deviations from three individual transformants. (B) Expression of PUS4-HA in transformants carrying high-copy-number plasmids pHQ839 (h.c.PUS4-HA) or pHQ864 (h.c.PUS4-HA/RPR1) measured by Western blot analysis. PUS4-HA was detected by anti-HA antibody and visualized by enhanced chemiluminescence. The intensities of bands were calculated with a scanner (Silverscanner III) and NIH image software (version 1.61). The relative levels were calculated by averaging the band intensities from two independent extract preparations for each transformant (lanes 1 to 4, pHQ839 transformant; lanes 5 to 8, pHQ864 transformant).
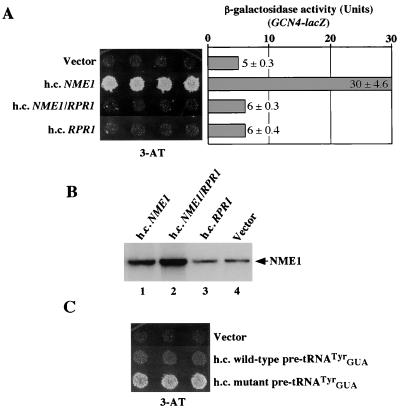
Other evidence that accumulation of unprocessed pre-tRNAs stimulates GCN4 translation by a GCN2-independent pathway relates to the previous observation that high-copy-number NME1 also triggers this response (51). NME1 encodes the RNA component of ribonuclease MRP, involved in pre-rRNA processing (48). Because RNases MRP and P are ribonucleoprotein complexes which share numerous protein subunits (11, 39), we considered that NME1 overexpression might titrate protein subunits away from RPR1 RNA. The ensuing reduction in RNase P levels would impair the processing of one or more pre-tRNAs, and the unprocessed precursors would trigger GCN2-independent derepression of GCN4 translation. According to this hypothesis, simultaneous overexpression of RPR1 and NME1 should reverse the titration of subunits from RNase P and reduce the concentration of tRNA precursors, thereby restoring the repression of GCN4 translation. As shown in Fig. 7A, the presence of hcNME1 suppressed the 3-ATs phenotype of a gcn2Δ mutant (Gcd− phenotype) and the presence of RPR1 on the same high-copy-number plasmid eliminated the suppressor activity of hcNME1. The antagonistic effect of hcRPR1 on hcNME1 suppressor activity did not involve a reduction in NME1 expression (Fig. 7B).
FIG. 7.
Overexpression of RPR1 reduces the Gcd− phenotype of hcNME1. (A) Transformants of strain H1895 (gcn2Δ) bearing high-copy-number plasmids YEplac181 (Vector), pHQ862 (h.c.NME1), pHQ863 (h.c.NME1/RPR1), or pHQ682 (h.c.RPR1) were replica-plated to SC medium containing 30 mM 3-AT and incubated for 3 days at 30°C (left panel). Extracts from the same transformants grown under repressing (nonstarvation) conditions were assayed for β-galactosidase activity, and the results shown in the right panel are the means and standard deviations of activities from three individual transformants. (B) Expression of NME1 in transformants carrying high-copy-number plasmids YEplac181 (Vector), pHQ862 (h.c.NME1), pHQ863 (h.c.NME1/RPR1), and pHQ682 (h.c.RPR1) was measured by Northern blot analysis using radiolabeled oligonucleotide specific to NME1. (C) Transformants of strain H1895 (gcn2Δ) bearing high-copy-number plasmids YEplac181 (Vector), pHQ982 (h.c. wild-type pre-tRNATyrGUA) and pHQ985 (h.c. mutant pre-tRNATyrGUA) were replica-plated to SC medium containing 30 mM 3-AT and incubated for 3 days at 30°C.
Northern analysis revealed that the hcNME1 transformants had increased amounts of precursors and decreased levels of mature tRNAiMet compared to the vector transformants (Fig. 4, lanes 4 and 5), with a twofold increase in the precursor/mature ratio (0.92 versus 0.47 [Table 6]) for this tRNA. A twofold increase in the amount of unprocessed primary transcript for tRNAUAUIle also was observed in the hcNME1 transformants with respect to the vector transformants (Fig. 4 and Table 6). The presence of hcRPR1 together with hcNME1 decreased the precursor/mature ratio from 0.92 to 0.57 for tRNAiMet and from 0.16 to 0.10 for pre-tRNAUAUIle in the hcNME1 transformant (Table 6). These data are consistent with the idea that hcNME1 leads to derepression of GCN4 by interfering with 5′-end processing of tRNAs by RNase P. Because introduction of hcIMT4 did not reverse the Gcd− phenotype of hcNME1 (data not shown), it most probably results from accumulation of unprocessed pre-tRNAs rather than depletion of mature tRNAiMet.
To provide more direct evidence that untrimmed pre-tRNA elicits derepression of GCN4 translation, we examined the consequences of overexpressing a mutant form of pre-tRNAGUATyr that cannot be processed by yeast RNase P in vitro. Three base changes were introduced into wild-type pre-tRNAGUATyr to extend the length of uninterrupted helix in the aminoacyl stem (35). In accordance with our hypothesis, the gene encoding the stem extension mutant of pre-tRNAGUATyr on a high-copy-number plasmid conferred a Gcd− phenotype in the gcn2Δ strain whereas the corresponding plasmid encoding wild-type pre-tRNAGUATyr did not (Fig. 7C). The results of Northern analysis confirmed that the stem extension mutation impaired processing of the pre-tRNAGUATyr in vivo (data not shown).
Evidence that PUS4 overexpression elicits derepression of GCN4 partly by interfering with nuclear export of tRNAs.
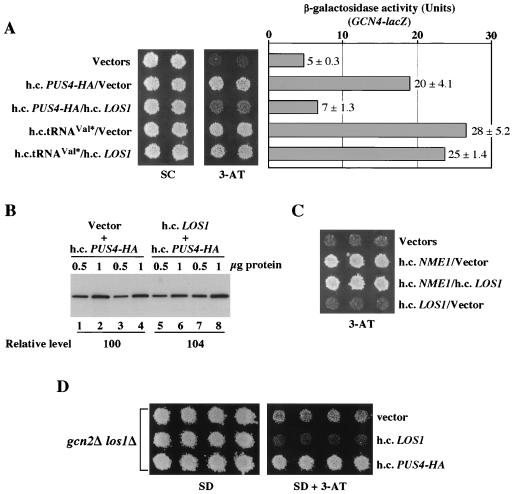
It is thought that tRNA export in mammalian cells requires exportin-t (Xpo-t), which binds tRNA directly with high affinity (33). It also requires the GTP-bound form of Ran (RanGTP), which forms a complex with Xpo-t and tRNA (2, 33) involving extensive interactions with the backbone of the TψC and acceptor arms of the tRNA (3). LOS1 is a yeast homolog of Xpo-t (2, 33), and the nuclear accumulation of tRNA observed in a los1Δ mutant (47) plus the ability of LOS1 to interact with Ran-GTP in a tRNA-dependent fashion (23) have implicated LOS1 in tRNA export from the yeast nucleus. If LOS1 resembles Xpo-t in binding to the TψC and acceptor arms of tRNA, overexpressed PUS4 might compete with LOS1 for tRNA binding and interfere with tRNA export. This possibility is consistent with the findings that PUS4 can form stable complexes with tRNA in vitro (45), that a minimal substrate for enzymatic formation of pseudouridine 55 by PUS4 is a TψC stem-loop structure (8), and that pseudouridine 55 synthase in Escherichia coli requires the TψC stem-loop to catalyze pseudouridine formation (21). The accumulation of mature tRNA in the nucleus resulting from inhibition of LOS1 function by PUS4 might be a signal for derepression of GCN4 translation. According to this hypothesis, overexpression of LOS1 should reduce the derepression of GCN4 elicited by hcPUS4.
In agreement with this prediction, LOS1 on a high-copy-number plasmid partially overcame the ability of hcPUS4-HA to confer 3-ATr and derepression of GCN4-lacZ translation in gcn2Δ cells without reducing the expression of PUS4-HA (Fig. 8A and B). In contrast, hcLOS1 had little effect on these same phenotypes when conferred by hctRNAVal* (Fig. 8A) or hcNME1 (Fig. 8C). This result is consistent with the idea that hcPUS4 elicits derepression of GCN4 in part by interfering with LOS1 function and producing the accumulation of mature tRNA in the nucleus. The hctRNAVal* and hcNME1 suppressors, by contrast, would derepress GCN4 by producing defective or unprocessed tRNAs, respectively, without directly interfering with tRNA export. As expected, Northern analysis showed that hcLOS1 did not reduce the accumulation of tRNA precursors in cells bearing hcPUS4 (data not shown).
FIG. 8.
Overexpression of LOS1 reduces the Gcd− phenotype of hcPUS4. (A) Transformants of strain H1895 (gcn2Δ) bearing high-copy-number plasmids YEplac181/YEp24 (Vectors), pHQ839/YEp24 (h.c.PUS4-HA/Vector), pHQ839/YEpLOS1 (h.c.PUS4-HA/h.c.LOS1), p856/YEp24 (h.c.tRNAVal*/Vector), and p856/YEpLOS1 (h.c.tRNAVal*/h.c.LOS1) were replica-plated to SC medium containing 30 mM 3-AT and incubated for 3 days at 30°C (left panel). Extracts from the same transformants grown under repressing (nonstarvation) conditions were assayed for β-galactosidase activity, and the results shown in the right panel are the means and standard deviations from three individual transformants. (B) Expression of PUS4-HA in transformants carrying high-copy-number plasmids pHQ839/Vector (Vector + h.c.PUS4-HA) or pHQ839/YEpLOS1 (h.c.LOS1 + h.c.PUS4-HA) measured by Western blot analysis. PUS4-HA was detected with an anti-HA antibody and visualized by enhanced chemiluminescence. Intensities of bands were calculated with a scanner (Silverscanner III) and NIH image software (version 1.61). The relative levels were calculated by averaging the band intensities from two independent extract preparations for each transformant (lanes 1 to 4, pHQ839/Vector transformant; lanes 5 to 8, pHQ839/YEpLOS1 transformant). (C) Transformants of strain H1895 (gcn2Δ) bearing high-copy-number plasmids YEplac181/YEp24 (Vectors), pHQ862/YEp24 (h.c.NME1/Vector), pHQ862/YEpLOS1 (h.c.NME1/h.c.LOS1), or YEplac181/YEpLOS1 (Vector/h.c.LOS1) were replica-plated to SC medium containing 30 mM 3-AT and incubated for 3 days at 30°C. (D) Deletion of LOS1 has a Gcd− phenotype. Transformants of strain HQY316 (gcn2Δlos1Δ) bearing plasmids YEplac181 (vector), pHQ860 (h.c.LOS1), and pHQ839 (h.c.PUS4-HA) were replica-plated to SD medium containing the required supplements and 30 mM 3-AT and incubated for 3 days at 30°C.
If nuclear accumulation of mature tRNA elicits GCN2-independent derepression of GCN4, inactivation of LOS1 should increase GCN4 expression in a gcn2Δ mutant. In agreement with this prediction, deletion of LOS1 partially suppressed the 3-ATs phenotype of the gcn2Δ strain (Fig. 8D). We also found that introduction of hcPUS4 into the los1Δ gcn2Δ double mutant led to even greater 3-ATr (Fig. 8D). This last observation can be explained by proposing that inhibiting LOS1 function in tRNA export is only one component of the derepression signal generated by hcPUS4. As indicated above, the fact that hcRPR1 partially reversed the Gcd− phenotype of hcPUS4 also points to a defect in tRNA 5′-end processing elicited by PUS4 overexpression.
In an effort to provide independent evidence that hcPUS4 derepresses GCN4 translation partly by interfering with LOS1-mediated tRNA export, we carried out fluorescence in situ hybridization to visualize the cellular distributions of various tRNAs in cells overexpressing PUS4. For tRNAUAUIle, we consistently observed nuclear accumulation in most cells bearing hcPUS4 versus vector alone (Fig. 9A and B). In addition, the presence of hcLOS1 reduced the extent and frequency of tRNAUAUIle nuclear accumulation compared to the situation with hcPUS4 alone (Fig. 9B and C). These results support the idea that PUS4 overexpression impedes nuclear export of tRNAUAUIle in a manner that can be overcome by increased expression of LOS1. Similar results were observed for tRNAAACVal, although the extent of nuclear accumulation conferred by hcPUS4 was less pronounced. No significant nuclear accumulation was detected for tRNAiMet, tRNAAAUIle, tRNAGUATyr, and tRNACAALeu. Thus, it appears that PUS4 overexpression interferes with nuclear export of a subset of tRNAs. (The fact that hcPUS4 did not produce detectable nuclear accumulation of tRNAiMet despite accumulation of its untrimmed precursors in this strain [Fig. 4] may be explained by the fact that tRNAiMet exhibits a more intense nuclear signal than the other tRNAs we examined in wild-type cells, presumably indicating a relatively large nuclear pool of mature tRNAiMet under normal conditions.)
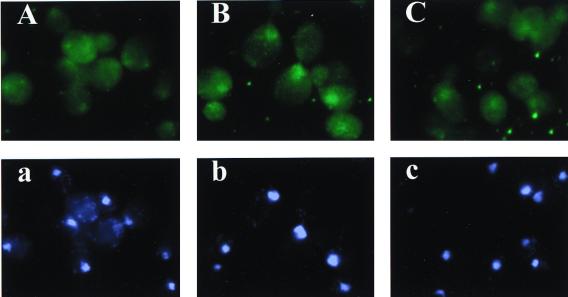
FIG. 9.
hcPUS4-HA leads to nuclear accumulation of tRNAUAUIle detected by fluorescence in situ hybridization. Cells of transformants of strain H1895 (gcn2Δ) bearing high-copy-number plasmids YEplac181/YEp24 (vectors) (A and a), pHQ839/YEp24 (h.c.PUS4-HA/vector) (B and b), or pHQ839/YEpLOS1 (h.c.PUS4-HA/h.c.LOS1) (C and c) were subjected to fluorescence in situ hybridization using a probe specific for tRNAUAUIle (panels A, B, and C) or stained with 4′,6-diamidino-2-phenylindole (DAPI) to visualize nuclei (panels a, b, and c).
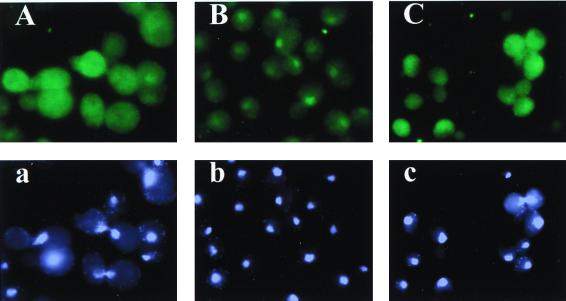
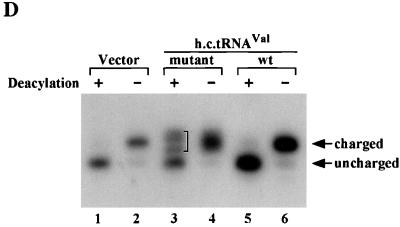
Interestingly, we observed significant nuclear accumulation of tRNAVal* in strains overexpressing this mutant tRNA versus the corresponding wild-type tRNAAACVal species (Fig. 10B and C). We previously proposed that the mutation in the 3′-terminal nucleotide of tRNAVal* would impede aminoacylation in vivo because the same substitution reduced charging in vitro of a yeast tRNAAACVal model substrate (minihelix) (17) and of E. coli tRNAAACVal (50). To test this prediction, we used Northern analysis under acidic conditions to analyze the relative amounts of deacylated tRNAAACVal in cells overexpressing tRNAVal* and in those overexpressing wild-type tRNAAACVal. The results in Fig. 10D showed that essentially all of the overexpressed wild-type tRNAAACVal was acylated in vivo, since the vast majority of this sample (lane 6) comigrated with the acylated form of native tRNAAACVal rather than with the faster-migrating deacylated tRNA (lanes 2 and 1, respectively). Unexpectedly, it appeared that the mutant tRNAVal* molecules in the deacylated sample (lane 3) migrated more slowly and were more heterogeneous than the deacylated wild-type tRNAAACVal (lane 1), suggesting a defect in processing the mutant tRNA. The overexpressed mutant tRNAVal* in the acylated sample (lane 4) was only slightly more heterogeneous than the acylated wild-type tRNAAACVal (lane 6). This last observation, plus the fact that the aberrant species in deacylated tRNAVal* roughly comigrated with acylated wild-type tRNAAACVal, led us to propose that the tRNAVal* molecules are aberrantly processed and aminoacylated inefficiently in vivo.
FIG. 10.
Evidence that mutant tRNAVal* is defective for aminoacylation and processing and is retained in the nucleus. (A to C) Cells of transformants of strain H1937 (gcn2Δ) bearing empty vector YEp24 (A and a), p1362 (tRNAVal*) (B and b), or p1308 (wild-type tRNAVal) (C and c) were subjected to fluorescence in situ hybridization using a probe specific for tRNAAACVal (A, B, and C) or stained with DAPI to visualize nuclei (a, b, and c). (D) Total RNAs prepared under acidic conditions from the strains analyzed in panels A to C were resolved by electrophoresis on an acid-urea polyacrylamide gel and subjected to Northern blot analysis using a probe specific for tRNAAACVal. In lanes 1, 3, and 5, the RNA samples were deacylated in 2 M Tris-HCl (pH 8.0) prior to electrophoresis.
Recent findings indicate that tRNAs are aminoacylated in the nucleus and that this reaction stimulates their export to the cytoplasm both in mammalian cells (38) and in yeast (47a). This might explain our finding that mutant tRNAVal* accumulated in the nucleus (Fig. 10B). However, it is possible that the mutation in tRNAVal* also weakens its interaction with LOS1 (3), either directly or because of incomplete processing of the acceptor stem (Fig. 10D). In any case, the nuclear retention of tRNAVal* provides strong support for the idea that defects in the maturation of tRNA in the nucleus or in its export to the cytoplasm can trigger derepression of GCN4 translation. Moreover, it can explain why hctRNAVal* failed to stimulate eIF2α phosphorylation by GCN2, which are both presumably restricted to the cytoplasm (54).
DISCUSSION
Evidence that unprocessed tRNAs in the nucleus elicit derepression of GCN4 translation independently of eIF2α phosphorylation.
GCN4 translation can be stimulated independently of GCN2 in mutants with lesions in subunits of eIF2 or eIF2B, in the genes encoding tRNAiMet, or in the GCD10- or GCD14-encoded proteins required for methylation of adenosine-58 in tRNAiMet. It is thought that all of these mutations mimic the effects of GCN2-mediated eIF2α phosphorylation by lowering the concentration of ternary complexes in the cytoplasm. It was suggested that a defect in ribosome biogenesis was responsible for derepressing GCN4 translation in gcn2 cells overexpressing NME1 (51). Our finding that the Gcd− phenotype of hcNME1 was suppressed by overexpressing RPR1 points to a reduction in RNase P levels and diminished tRNA 5′-end processing as the cause of derepression. We propose that overexpression of NME1 reduces RNase P levels by titrating from RPR1 one or more protein subunits shared between RNases MRP and P. Consistent with this hypothesis, we detected an ca. twofold increase in the precursor/mature ratio for tRNAiMet in the hcNME1 strain versus the wild type, and this phenotype was partially reversed by hcRPR1. The small reduction in mature initiator tRNAMet abundance caused by hcNME1 cannot account for its Gcd− phenotype, because it was not suppressed by hcIMT4. Instead, we propose that an increase in the levels of unprocessed tRNAs in the nucleus activates a regulatory mechanism that down-regulates ternary complex binding to 40S ribosomes by an amount sufficient to derepress GCN4 translation. We cannot exclude the possibility that a defect in ribosome biogenesis also contributes to the derepression of GCN4 conferred by hcNME1.
A more direct demonstration that unprocessed tRNAs trigger GCN2-independent derepression of GCN4 was provided by our finding that an overexpressed mutant pre-tRNAGUATyr that cannot be processed by RNase P also elicits a Gcd− phenotype in gcn2Δ cells. Because unprocessed pre-tRNAs are not exported (9, 38, 47), this result provides strong evidence that the accumulated pre-tRNAs are recognized in the nucleus and send a signal to the cytoplasm, which leads to increased translation of GCN4 mRNA. This signalling mechanism may additionally account for the Gcd− phenotype of hctRNAVal*, because this mutant tRNA appeared to be processed aberrantly and was retained in the nucleus.
Evidence that overexpression of PUS4 elicits GCN2-independent derepression of GCN4 by impeding nuclear export and 5′-end processing of tRNAs.
The derepression of GCN4 translation in cells overexpressing PUS4 also seems to be triggered partly by the accumulation of pre-tRNAs in the nucleus. The Gcd− phenotype of hcPUS4 was partially reversed by hcRPR1, suggesting that overexpressed PUS4 interferes with 5′-end processing by RNase P. Consistent with this model, we observed a ca. twofold increase in the precursor/mature ratio for tRNAiMet in the hcPUS4 transformant, which was reversed by cooverexpressing RPR1. The postulated interference with RNase P exerted by overexpressed PUS4 could involve direct competition between these two enzymes for binding to a subset of tRNA precursors. This idea is ostensibly at odds with the fact that 5′-end processing of pre-tRNAiMet was impaired by hcPUS4 even though this tRNA is not a substrate for PUS4. When overexpressed 15-fold, however, PUS4 may bind tightly to pre-tRNAiMet and block access of RNase P even though it fails to synthesize pseudouridine-55. Alternatively, PUS4 and RNase P may interact with a common tRNA chaperone that facilitates the activities of both enzymes, and overexpression of PUS4 could reduce the availability of this hypothetical chaperone for 5′-end processing of pre-tRNAiMet by RNase P.
Unlike the situation with hcNME1, the Gcd− phenotype of hcPUS4 was partially suppressed by hcLOS1 in addition to hcRPR1. Because LOS1 appears to be the yeast homologue of mammalian exportin-t, the suppression by hcLOS1 could indicate that overexpression of PUS4 impedes tRNA export and that increased nuclear accumulation of one or more fully processed tRNAs contributes to the derepression of GCN4 translation. Consistent with this interpretation, a los1Δ mutant had a Gcd− phenotype, albeit weaker than that of hcPUS4, and we observed nuclear accumulation of tRNAUAUIle in strains bearing hcPUS4 that was reversed by cooverexpressing LOS1. At the same time, we did not observe convincing nuclear accumulation of several other tRNAs examined in strains harboring hcPUS4. Thus, overexpressed PUS4 seems to inhibit LOS1-dependent nuclear export of only a subset of tRNAs. This inhibition might involve competition between LOS1 and PUS4 for binding to the affected tRNAs. Presumably, the selective nuclear retention of tRNAs is sufficient to trigger derepression of GCN4 only when combined with the accumulation of certain pre-tRNAs which results from inhibition of RNase P by overexpressed PUS4.
Considering that removal of introns from pre-tRNAs is defective in los1 mutants (28), it is conceivable that the Gcd− phenotype of los1Δ cells results from accumulation of unspliced pre-tRNAs in the nucleus rather than from nuclear retention of fully processed tRNAs. Similarly, it could be argued that the tRNAUAUIle species retained in the nucleus of hcPUS4 transformants (Fig. 9) are incompletely processed molecules rather than fully matured tRNAs. However, the latter possibility seems inconsistent with the fact that hcPUS4 produces only a small increase in the relative abundance of pre-tRNAUAUIle, which is a minor fraction of the combined pool of precursor and mature forms of this tRNA (Fig. 4). Thus, the increase in pre-tRNAUAUIle abundance seems insufficient to account for its considerable nuclear retention in hcPUS4 transformants. Assuming that LOS1 is the tRNA exportin of yeast, it may be simpler to propose that hcLOS1 overcomes the nuclear retention of mature tRNAUAUIle in cells overexpressing PUS4 rather than suggesting that it corrects a processing defect. Accordingly, we consider it likely that an overabundance of mature tRNA in the nucleus, as well as accumulation of unprocessed pre-tRNAs, can trigger derepression of GCN4 by the GCN2-independent pathway.
It is thought that GCN2 is stimulated in the cytoplasm of amino acid-starved cells by uncharged tRNAs that interact with translating ribosomes. In view of recent findings that tRNAs are aminoacylated in the nucleus (38), we considered the possibility that uncharged tRNA in the nucleus could be a signal for GCN2-independent derepression of GCN4. Consistent with this model, we obtained evidence that tRNAVal* is retained in the nucleus and is aminoacylated inefficiently, possibly because of a processing defect. Either impeding 5′-end processing by overexpressing NME1 or PUS4, or overproducing a mutant tRNA that cannot be processed by RNase P, should also produce an excess of pre-tRNAs in the nucleus that cannot be charged. Increased binding of overexpressed PUS4 to mature tRNAs in the nucleus might block their interaction with aminoacyl-tRNA synthetases or, by impeding export, generate increased nuclear pools of tRNA which outstrip the enzymatic capacity of synthetases in the nucleus. (The fact that hcPUS4 did not perceptibly increase the proportion of total cellular tRNA that was uncharged could be explained by stipulating that only a small fraction of the mature tRNA is located in the nucleus.) Finally, this model could account for the GCN2-independent derepression of GCN4 that accompanies overproduction of wild-type tRNAs under conditions of reduced aminoacylation (54). The idea that GCN4 translation can be induced by uncharged tRNA in the nucleus is attractive; however, it seems equally possible that an excess of unprocessed or untransported tRNA in the nucleus, regardless of its aminoacylation status, is the primary signal for this derepression mechanism.
Under adverse environmental conditions where processing, modification or transport of tRNA is impaired, it could be advantageous to decrease the rate of protein synthesis. The inhibition of ternary-complex formation by phosphorylation of eIF2 is a widely employed mechanism to down-regulate translation under conditions of starvation or stress (24). Our results indicate that ternary-complex formation or utilization is reduced by a mechanism other than eIF2α phosphorylation in response to malfunctions in tRNA biogenesis. This may provide a useful strategy for coupling the rate of translation initiation in the cytoplasm with nuclear events involved in producing functional tRNA molecules that can participate in protein synthesis.
ACKNOWLEDGMENTS
We thank Lasse Lindahl for the NME1 and RPR1 plasmids and David Engelke for advice and gifts of plasmids. We thank Bobbie Felix for help in preparation of the manuscript and members of the Hinnebusch and Dever laboratories for discussion.
G.R.B. was supported by grants from the National Science Research Council (BU-2930) and the Swedish Cancer Society (project 680), and A.K.H. was supported by NIH grant GM27930.
REFERENCES
- 1.Anderson J, Phan L, Cuesta R, Carlson B A, Pak M, Asano K, Bjork G R, Tamame M, Hinnebusch A G. The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev. 1998;12:3650–3662. doi: 10.1101/gad.12.23.3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arts G J, Fornerod M, Mattaj I J. Identification of a nuclear export receptor for tRNA. Curr Biol. 1998;8:305–314. doi: 10.1016/s0960-9822(98)70130-7. [DOI] [PubMed] [Google Scholar]
- 3.Arts G J, Kuersten S, Romby P, Ehresmann B, Mattaj I W. The role of exportin-t in selective nuclear export of mature tRNAs. EMBO J. 1998;17:7430–7441. doi: 10.1093/emboj/17.24.7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Auxillien S, Crain P F, Trewyn R W, Grosjean H. Mechanism, specificity and general properties of the yeast enzyme catalysing the formation of inosine 34 in the anticodon of transfer RNA. J Mol Biol. 1996;262:437–458. doi: 10.1006/jmbi.1996.0527. [DOI] [PubMed] [Google Scholar]
- 5.Bakin A, Ofengand J. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: analysis by the application of a new sequencing technique. Biochemistry. 1993;32:9754–9762. doi: 10.1021/bi00088a030. [DOI] [PubMed] [Google Scholar]
- 6.Bakin A, Ofengand J. Mapping of the 13 pseudouridine residues in Saccharomyces cerevisiae small subunit ribosomal RNA to nucleotide resolution. Nucleic Acids Res. 1995;23:3290–3294. doi: 10.1093/nar/23.16.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becker H F, Motorin Y, Planta R J, Grosjean H. The yeast gene YNL292w encodes a pseudouridine synthase (Pus4) catalyzing the formation of ψ55 in both mitochondrial and cytoplasmic tRNAs. Nucleic Acids Res. 1997;25:4493–4499. doi: 10.1093/nar/25.22.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Becker H F, Motorin Y, Sissler M, Florentz C, Grosjean H. Major identity determinants for enzymatic formation of ribothymidine and pseudouridine in the Tψ-loop of yeast tRNAs. J Mol Biol. 1997;274:505–518. doi: 10.1006/jmbi.1997.1417. [DOI] [PubMed] [Google Scholar]
- 9.Bertrand E, Houser-Scott F, Kendall A, Singer R H, Engelke D R. Nucleolar localization of early tRNA processing. Genes Dev. 1998;12:2463–2468. doi: 10.1101/gad.12.16.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calvo O, Cuesta R, Anderson J, Gutierrez N, Garcia-Barrio M T, Hinnebusch A G, Tamame M. GCD14p, a repressor of GCN4 translation, cooperates with Gcd10p and Lhp1p in the maturation of initiator methionyl-tRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:4167–4181. doi: 10.1128/mcb.19.6.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chamberlain J R, Lee Y, Lane W S, Engelke D R. Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes Dev. 1998;12:1678–1690. doi: 10.1101/gad.12.11.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cortese R, Kammen H O, Spengler S J, Ames B N. Biosynthesis of pseudouridine in transfer ribonucleic acid. J Biol Chem. 1974;249:1103–1108. [PubMed] [Google Scholar]
- 13.Cuesta R, Hinnebusch A G, Tamame M. Identification of GCD14 and GCD15, novel genes required for translational repression of GCN4 mRNA in Saccharomyces cerevisiae. Genetics. 1998;148:1007–1020. doi: 10.1093/genetics/148.3.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13a.Dever T E, Feng L, Wek R C, Cigan A M, Donahue T D, Hinnebusch A G. Phosphorylation of initiation factor 2α by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell. 1992;68:585–596. doi: 10.1016/0092-8674(92)90193-g. [DOI] [PubMed] [Google Scholar]
- 14.Dever T E, Yang W, Åström S, Byström A S, Hinnebusch A G. Modulation of tRNAiMet, eIF-2 and eIF-2B expression shows that GCN4 translation is inversely coupled to the level of eIF-2 · GTP · Met-tRNAiMet ternary complexes. Mol Cell Biol. 1995;15:6351–6363. doi: 10.1128/mcb.15.11.6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diamond A M, Choi I S, Crain P F, Hashizume T, Pomerantz S C, Cruz R, Steer C J, Hill K E, Burk R F, McCloskey J A, Hatfield D L. Dietary selenium affects methylation of the wobble nucleoside in the anticodon of selenocysteine tRNA[Ser]Sec*. J Biol Chem. 1993;268:14215–14223. [PubMed] [Google Scholar]
- 16.Engelberg D, Klein C, Martinetto H, Struhl K, Karin M. The UV response involving the ras signaling pathway and AP-1 transcription factors is conserved between yeast and mammals. Cell. 1994;77:381–390. doi: 10.1016/0092-8674(94)90153-8. [DOI] [PubMed] [Google Scholar]
- 17.Frugier M, Florentz C, Giege R. Anticodon-independent aminoacylation of an RNA minihelix with valine. Proc Natl Acad Sci USA. 1992;89:3990–3994. doi: 10.1073/pnas.89.9.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Barrio M T, Naranda T, Cuesta R, Hinnebusch A G, Hershey J W B, Tamame M. GCD10, a translational repressor of GCN4, is the RNA-binding subunit of eukaryotic translation initiation factor-3. Genes Dev. 1995;9:1781–1796. doi: 10.1101/gad.9.14.1781. [DOI] [PubMed] [Google Scholar]
- 19.Gehrke C W, Kuo K C. Ribonucleoside analysis by reversed-phase high-performance liquid chromatography. In: Gehrke C W, Kuo K C, editors. Chromatography and modification of nucleosides. Amsterdam, The Netherlands: Elsevier; 1990. pp. A3–A71. [DOI] [PubMed] [Google Scholar]
- 20.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 21.Gu X, Yu M, Ivanetich K M, Santi D V. Molecular recognition of tRNA by tRNA pseudouridine 55 synthase. Biochemistry. 1998;37:339–343. doi: 10.1021/bi971590p. [DOI] [PubMed] [Google Scholar]
- 22.Harashima S, Hinnebusch A G. Multiple GCD genes required for repression of GCN4, a transcriptional activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:3990–3998. doi: 10.1128/mcb.6.11.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hellmuth K, Lau D M, Bischoff F R, Kunzler M, Hurt E, Simos G. Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA. Mol Cell Biol. 1998;18:6374–6386. doi: 10.1128/mcb.18.11.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hinnebusch A G. The eIF-2α kinases: regulators of protein synthesis in starvation and stress. Semin Cell Biol. 1994;5:417–426. doi: 10.1006/scel.1994.1049. [DOI] [PubMed] [Google Scholar]
- 25.Hinnebusch A G. Translational control of GCN4: gene-specific regulation by phosphorylation of eIF2. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 199–244. [Google Scholar]
- 26.Hinnebusch A G. Translational regulation of yeast GCN4: a window on factors that control initiator-tRNA binding to the ribosome. J Biol Chem. 1997;272:21661–21664. doi: 10.1074/jbc.272.35.21661. [DOI] [PubMed] [Google Scholar]
- 27.Hinnebusch A G, Fink G R. Positive regulation in the general amino acid control of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1983;80:5374–5378. doi: 10.1073/pnas.80.17.5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hopper A K, Schultz L D, Shapiro R A. Processing of intervening sequences: a new yeast mutant which fails to excise intervening sequences from precursor tRNAs. Cell. 1980;19:741–751. doi: 10.1016/s0092-8674(80)80050-x. [DOI] [PubMed] [Google Scholar]
- 29.Hurt D J, Wang S S, Lin Y H, Hopper A K. Cloning and characterization of LOS1, a Saccharomyces cerevisiae gene that affects tRNA splicing. Mol Cell Biol. 1987;7:1208–1216. doi: 10.1128/mcb.7.3.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iida H, Nakamura H, Ono T, Okumura M S, Anraku Y. MID1, a novel Saccharomyces cerevisiae gene encoding a plasma membrane protein, is required for Ca2+ influx and mating. Mol Cell Biol. 1994;14:8259–8271. doi: 10.1128/mcb.14.12.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelmers A D, Heatherly D E. Columns for rapid chromatographic separation of small amounts of tracer-labeled transfer ribonucleic acids. Anal Biochem. 1971;44:486–495. doi: 10.1016/0003-2697(71)90236-3. [DOI] [PubMed] [Google Scholar]
- 32.Koonin E V. Pseudouridine synthases: four families of enzymes containing a putative uridine-binding motif also conserved in dUTPases and dCTP deaminases. Nucleic Acids Res. 1996;24:2411–2415. doi: 10.1093/nar/24.12.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kutay U, Lipowsky G, Izaurraide E, Bischoff F R, Schwartzmaier P, Hartmann E, Gorlich D. Identification of a tRNA-specific nuclear export receptor. Mol Cell. 1998;1:359–369. doi: 10.1016/s1097-2765(00)80036-2. [DOI] [PubMed] [Google Scholar]
- 34.Lanker S, Bushman J L, Hinnebusch A G, Trachsel H, Mueller P P. Autoregulation of the yeast lysyl-tRNA synthetase gene GCD5/KRS1 by translational and transcriptional control mechanisms. Cell. 1992;70:647–657. doi: 10.1016/0092-8674(92)90433-d. [DOI] [PubMed] [Google Scholar]
- 35.Lee Y, Kindelberger D W, Lee J Y, McClennen S, Chamberlain J, Engelke D R. Nuclear pre-tRNA terminal structure and RNase P recognition. RNA. 1997;3:175–185. [PMC free article] [PubMed] [Google Scholar]
- 36.Li X, Burgers P M. Molecular cloning and expression of the Saccharomyces cerevisiae RFC3 gene, an essential component of replication factor C. Proc Natl Acad Sci USA. 1994;91:868–872. doi: 10.1073/pnas.91.3.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lucchini G, Hinnebusch A G, Chen C, Fink G R. Positive regulatory interactions of the HIS4 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:1326–1333. doi: 10.1128/mcb.4.7.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lund E, Dahlberg J E. Proofreading and aminoacylation of tRNAs before export from the nucleus. Science. 1998;282:2082–2085. doi: 10.1126/science.282.5396.2082. [DOI] [PubMed] [Google Scholar]
- 39.Lygerou Z, Mitchell P, Petfalski E, Seraphin B, Tollervey D. The POP1 gene encodes protein component common to the RNase MRP and RNase P ribonucleoproteins. Genes Dev. 1994;8:1423–1433. doi: 10.1101/gad.8.12.1423. [DOI] [PubMed] [Google Scholar]
- 40.Marton M J, Vazquez de Aldana C R, Qiu H, Chakraburtty K, Hinnebusch A G. Evidence that GCN1 and GCN20, translational regulators of GCN4, function on enlongating ribosomes in activation of the eIF2α kinase GCN2. Mol Cell Biol. 1997;17:4474–4489. doi: 10.1128/mcb.17.8.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marton M J, Crouch D, Hinnebusch A G. GCN1, a translational activator of GCN4 in S. cerevisiae, is required for phosphorylation of eukaryotic translation initiation factor 2 by protein kinase GCN2. Mol Cell Biol. 1993;13:3541–3556. doi: 10.1128/mcb.13.6.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Messenguy F, Delforge J. Role of transfer ribonucleic acids in the regulation of several biosyntheses in Saccharomyces cerevisiae. Eur J Biochem. 1976;67:335–339. doi: 10.1111/j.1432-1033.1976.tb10696.x. [DOI] [PubMed] [Google Scholar]
- 43.Niederberger P, Aebi M, Huetter R. Influence of the general control of amino acid biosynthesis on cell growth and cell viability in Saccharomyces cerevisiae. J Gen Microbiol. 1983;129:2571–2583. [Google Scholar]
- 44.O'Connor J P, Peebles C L. In vivo pre-tRNA processing in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:425–439. doi: 10.1128/mcb.11.1.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samuelsson T. Interactions of transfer RNA pseudouridine synthases with RNAs substituted with fluorouracil. Nucleic Acids Res. 1991;19:6139–6144. doi: 10.1093/nar/19.22.6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Samuelsson T, Olsson M. Transfer RNA pseudouridine synthases in Saccharomyces cerevisiae. J Biol Chem. 1990;265:8782–8787. [PubMed] [Google Scholar]
- 47.Sarkar S, Hopper A K. tRNA nuclear export in Saccharomyces cerevisiae: in situ hybridization analysis. Mol Biol Cell. 1998;9:3041–3055. doi: 10.1091/mbc.9.11.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47a.Sarkar S, Azad A K, Hopper A K. Nuclear tRNA aminoacylation and its role in nuclear export of endogenous tRNAs in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1999;96:14366–14371. doi: 10.1073/pnas.96.25.14366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmitt M E, Clayton D A. Yeast site-specific ribonucleoprotein endoribonuclease MRP contains an RNA component homologous to mammalian RNAse MRP RNA and essential for cell viability. Genes Dev. 1992;6:1975–1985. doi: 10.1101/gad.6.10.1975. [DOI] [PubMed] [Google Scholar]
- 49.Sprinzl M, Hartmann T, Meissner F, Moll J, Vorderwulbecke T. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1987;15:r53–r188. doi: 10.1093/nar/15.suppl.r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tamura K, Himeno H, Asahara H, Hasegawa T, Shimizu M. Identity determinants of E. coli tRNAVal. Biochem Biophys Res Commun. 1991;177:619–623. doi: 10.1016/0006-291x(91)91833-x. [DOI] [PubMed] [Google Scholar]
- 51.Tavernarakis N, Alexandraki D, Liodis P, Tzamarias D, Thireos G. Gene overexpression reveals alternative mechanisms that induce GCN4 mRNA translation. Gene. 1996;179:271–277. doi: 10.1016/s0378-1119(96)00379-4. [DOI] [PubMed] [Google Scholar]
- 52.Varshney U, Lee C P, RajBhandary U L. Direct analysis of aminoacylation levels of tRNA as in vivo. J Biol Chem. 1991;266:24712–24718. [PubMed] [Google Scholar]
- 53.Vazquez de Aldana C R, Marton M J, Hinnebusch A G. GCN20, a novel ATP binding cassette protein, and GCN1 reside in a complex that mediates activation of the eIF-2α kinase GCN2 in amino acid-starved cells. EMBO J. 1995;14:3184–3199. doi: 10.1002/j.1460-2075.1995.tb07321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vazquez de Aldana C R, Wek R C, San Segundo P, Truesdell A G, Hinnebusch A G. Multicopy tRNA genes functionally suppress mutations in yeast eIF-2α kinase GCN2: evidence for separate pathways coupling GCN4 expression to uncharged tRNA. Mol Cell Biol. 1994;14:7920–7932. doi: 10.1128/mcb.14.12.7920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wek R C, Jackson B M, Hinnebusch A G. Juxtaposition of domains homologous to protein kinases and histidyl-tRNA synthetases in GCN2 protein suggests a mechanism for coupling GCN4 expression to amino acid availability. Proc Natl Acad Sci USA. 1989;86:4579–4583. doi: 10.1073/pnas.86.12.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wek R C, Ramirez M, Jackson B M, Hinnebusch A G. Identification of positive-acting domains in GCN2 protein kinase required for translational activation of GCN4 expression. Mol Cell Biol. 1990;10:2820–2831. doi: 10.1128/mcb.10.6.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wek S A, Zhu S, Wek R C. The histidyl-tRNA synthetase-related sequence in the eIF-2α protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol Cell Biol. 1995;15:4497–4506. doi: 10.1128/mcb.15.8.4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu S, Sobolev A Y, Wek R C. Histidyl-tRNA synthetase-related sequences in GCN2 protein kinase regulate in vitro phosphorylation of eIF-2. J Biol Chem. 1996;271:24989–24994. doi: 10.1074/jbc.271.40.24989. [DOI] [PubMed] [Google Scholar]