Abstract
Evidence indicates that common variants found in genome-wide association studies increase risk of disease through gene regulation via expression Quantitative Trait Loci. Using multiple genome-wide methods, we examined if Single Nucleotide Polymorphisms increase risk of Amyotrophic Lateral Sclerosis through expression Quantitative Trait Loci, and whether expression Quantitative Trait Loci expression is consistent across people who had Amyotrophic Lateral Sclerosis and those who did not. In combining public expression Quantitative Trait Loci data with Amyotrophic Lateral Sclerosis genome-wide association studies, we used Summary-data-based Mendelian Randomization to confirm that SCFD1 was the only gene that was genome-wide significant in mediating Amyotrophic Lateral Sclerosis risk via expression Quantitative Trait Loci (Summary-data-based Mendelian Randomization beta = 0.20, standard error = 0.04, P-value = 4.29 × 10−6). Using post-mortem motor cortex, we tested whether expression Quantitative Trait Loci showed significant differences in expression between Amyotrophic Lateral Sclerosis (n = 76) and controls (n = 25), genome-wide. Of 20 757 genes analysed, the two most significant expression Quantitative Trait Loci to show differential in expression between Amyotrophic Lateral Sclerosis and controls involve two known Amyotrophic Lateral Sclerosis genes (SCFD1 and VCP). Cis-acting SCFD1 expression Quantitative Trait Loci downstream of the gene showed significant differences in expression between Amyotrophic Lateral Sclerosis and controls (top expression Quantitative Trait Loci beta = 0.34, standard error = 0.063, P-value = 4.54 × 10−7). These SCFD1 expression Quantitative Trait Loci also significantly modified Amyotrophic Lateral Sclerosis survival (number of samples = 4265, hazard ratio = 1.11, 95% confidence interval = 1.05–1.17, P-value = 2.06 × 10−4) and act as an Amyotrophic Lateral Sclerosis trans-expression Quantitative Trait Loci hotspot for a wider network of genes enriched for SCFD1 function and Amyotrophic Lateral Sclerosis pathways. Using gene-set analyses, we found the genes that correlate with this trans-expression Quantitative Trait Loci hotspot significantly increase risk of Amyotrophic Lateral Sclerosis (beta = 0.247, standard deviation = 0.017, P = 0.001) and schizophrenia (beta = 0.263, standard deviation = 0.008, P-value = 1.18 × 10−5), a disease that genetically correlates with Amyotrophic Lateral Sclerosis. In summary, SCFD1 expression Quantitative Trait Loci are a major factor in Amyotrophic Lateral Sclerosis, not only influencing disease risk but are differentially expressed in post-mortem Amyotrophic Lateral Sclerosis. SCFD1 expression Quantitative Trait Loci show distinct expression profiles in Amyotrophic Lateral Sclerosis that correlate with a wider network of genes that also confer risk of the disease and modify the disease’s duration.
Keywords: amyotrophic lateral sclerosis, expression quantitative trait loci, GWAS, Mendelian randomization, vesicle-mediated transport
Previous research indicates DNA variants that increase risk of Amyotrophic Lateral Sclerosis (ALS) correlate with SCFD1 RNA activity. Iacoangeli et al. report that SCFD1 RNA and DNA variants are differentially active in ALS, which not only increases risk of the disease but modifies its duration, confirming their involvement in ALS.
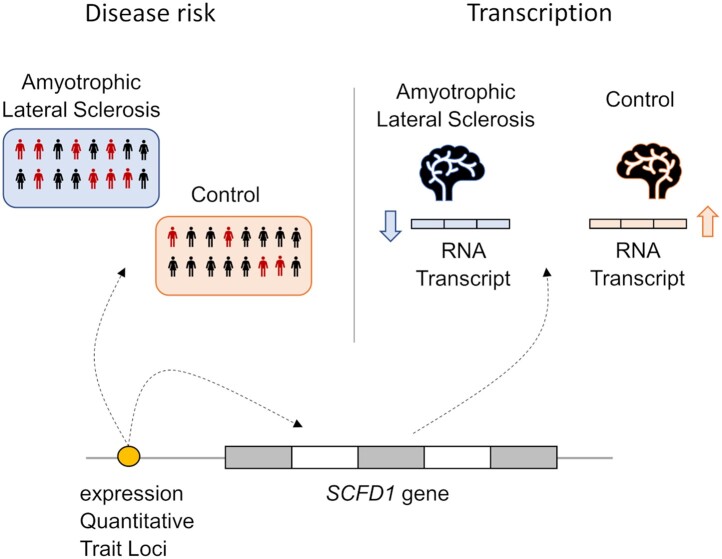
Graphical Abstract
Graphical Abstract.
Introduction
Amyotrophic lateral sclerosis (ALS) is characterized by the progressive degeneration of upper and lower motor neurons, which leads to muscle weakness and loss of voluntary control of the limbs. More than 50% of people with ALS have extra-motor symptoms including cognitive and behavioural impairments. Multiple genetic, environment and lifestyle factors coincide to cause the disease. Most of our understanding of what causes ALS is derived from genetic, proteomic, functional and epidemiological research. Two disease-modifying treatments are licensed for ALS, Riluzole1 and Edaravone,2 but their exact molecular effects on the disease mechanism remain unclear.
Our genetic understanding of ALS is derived from rare variants that co-segregate with disease in families and variants common in the population [single nucleotide polymorphism (SNPs)] that associate with disease risk. Rare variants often provide a link between genes and protein dysfunction. Genes implicated in ALS that have rare deleterious mutations, such as TARDBP,3–5FUS,6,7SOD1,8OPTN,9ANXA11,10UBQLN2,11 suggest protein dysfunction in RNA metabolism, protein homeostasis, oxidative stress and vesicle transport.
SNPs that associate with disease risk, typically identified through genome-wide association studies (GWAS), can have a less obvious relationship with disease mechanism. SNPs often implicate multiple genes within a genomic region and can be very common in the general population where the increase in disease risk is marginal. For ALS there is also little reconciliation between with pathways identified through rare, large effect variants and genes identified using GWAS.
There is increasing evidence that most common SNPs that associate with disease regulate gene expression.12 One method in identifying regulatory SNPs is to analyse whether changes in genotype correlate with changes in gene expression; referred to as expression Quantitative Trait Loci (eQTL). eQTLs can help clarify which gene(s) in a genomic region are modifying disease risk, can provide insights into how, and also provide information on which tissues and cells are affected.13 eQTLs have been associated with increased risk of schizophrenia,14 Alzheimer’s disease15 and Parkinson’s disease,16 as examples.
Using public GWAS and eQTL data, we confirmed the most significant gene to increase risk of ALS via eQTLs is SCFD1, which concurs with previous studies using different methods.17,18 In an independent analysis, we find SCFD1 eQTL expression is significantly different in post-mortem ALS compared to controls. And that SCFD1 eQTLs are hotspot in ALS that significantly correlate with major ALS pathways and ALS survival. Our findings indicate that while SCFD1 eQTLs increase risk of ALS, the expression profile of these eQTLs is different in ALS compared to controls, and these changes correlate with disease duration and pathways known to be compromised in the disease, namely vesicle-mediated transport, RNA metabolism and transmembrane trafficking.
Materials and methods
Samples and cohorts
Genomic datasets
We used the largest ALS GWAS19 for which summary statistics were publicly available, which included 80 610 individuals (20 806 cases and 59 804 controls) of European ancestry and 10 031 417 SNPs. In this study, the authors compared allelic frequency between ALS cases and controls from the UK, USA, Italy, France and Belgium (8229 cases and 36 329 controls in total) by logistic regression and a fixed-effects meta-analysis, integrating our antecedent European ALS GWAS (12 577 cases and 23 475 controls).20
For the survival analysis, we used the largest ALS survival GWAS available.21 In this study, the authors analysed if alleles associated with changes in ALS survival (from date of diagnosis to date of death) for 4256 ALS patients and 7 174 392 SNPs, using genome-wide Cox proportional hazards regression and controlling for age of onset, sex, and four principal components.
Publicly available eQTL datasets
eQTL data were downloaded from the Genotype-Tissue Expression (GTEx) portal,13 for frontal cortex (BA9), cortex, cerebellum and cerebellar hemisphere. We used the GTEx Analysis V8 (dbGaP Accession phs000424. v8. p2 accessed 10/05/2020), the latest release at the time. For consistency with genotype datasets used in this study, we mapped the hg38 genomic coordinates of the GTEx V8 data onto hg19 using the liftover tool in UCSC Genome Browser.
Post-mortem samples
Post-mortem motor cortex was provided by the MRC London Neurodegenerative Diseases Brain Bank based at the Institute of Psychiatry, Psychology & Neuroscience, King’s College London. Ethical approval was granted via local committee at King’s College London and MRC London Neurodegenerative Diseases Brain Bank. Tissue was flash frozen and stored at −80°C. One hundred milligrams of tissue blocks were excised. RNA and DNA were isolated from the same tissue block.
Samples were selected based on availability. The cohort was age-sex matched manually (n = 108) and sex, age and post-mortem delay were analysed for statistically significant differences using a Student’s t-test (Supplementary Results). Five samples were removed as they were outliers in the genomic principal components’ analyses (indicating non-European ancestry) and two samples removed showing significant shared identity by descent (indicating familial relations); see Supplementary Methods 3 and Results 2. The final cohort consisted of 76 ALS and ALS-FTD donors, and 25 non-ALS age-sex matched controls with no known neurological disease (pathology below hp-τ and BNE\Braak stages 2; Supplementary Table 1). Owing to clinical data collected at the time of donation, we are unable to estimate the proportion of pure ALS compared to ALS patients who had FTD involvement. The ALS cohort was screened for ALS mutations, where 5 donors had the C9orf72 repeat mutation, one donor had an ANXA11 D40G mutation, and one donor had a PFN1 E117G mutation.
Statistical analyses
Genome-wide eQTL analyses using genomic summary-statistics and GTEx
See Supplementary Methods 1 for gene-level genome-wide association study.
Mendelian Randomization analyses
To analyse the effects that eQTLs have on ALS risk we performed summary-data-based Mendelian Randomization (SMR) with Heidi tests, using the SMR tool.22 We used eQTL summary statistics from the GTEx Portal for cortical and cerebellar tissues as exposure, and the ALS GWAS disease status as an outcome. We selected frontal cortex and cerebellar eQTLs from GTEx with a P-value < 5 × 10−8 and cortex eQTLs with P-value < 5 × 10−7. SNPs extracted from the ALS GWAS that were not present in the GTEx eQTL dataset were removed.
Direction of effects between exposure and SNP allele were harmonized. A total of 11 728 eQTL gene-targets were tested and 11 198 passed the heterogeneity filter (PHEIDI > 0.05, with number of SNPs > 2). We set the significant threshold at P = 0.05/11,198 (4.5 × 10−6) to correct for multiple-testing. To confirm these results, we used an eQTL dataset from Braineac16 (see Supplementary Methods and Supplementary Results).
Genome-wide QTL analyses in post-mortem motor cortex
See Supplementary Methods for RNA-sequencing, genotyping and imputation methods.
Statistically significant eQTLs, with a nominal P-value = 1 × 10−5, identified in GTEx were downloaded for cortex, frontal lobe (BA9), cerebellum and cerebellar hemisphere. These eQTLs were extracted from our post-mortem genotype imputation dataset based on their reference SNP identifier, position and effect allele. The resulting file was a post-mortem genotype dataset consisting of GTEx brain-specific eQTLs only.
Post-mortem motor cortex RNA-sequence data were normalized using variance stabilizing transformation (VST) at the gene-level (Ensembl GRCh37). Genes with a read-count less than 10, normalized expression for genes at a minimal value, and genes showing variance within the lower quartile genome-wide, were removed, resulting in 20 757 testable Ensembl gene IDs. For distribution, minimum, quartile and maximum normalized expression estimates see Supplementary Fig. 1A.
Matrix eQTL23 was used to import and analyse eQTLs genome-wide. Genotype data, gene expression data, SNP maps and gene locations were standardized to GRCh37 (hg19). Matrix eQTL uses an additive linear least squares model to test for association between SNPs and gene-level expression. We removed SNPs with a minor allele count less than five in cases and controls separately, to allow sufficient statistical power to detect variation in gene expression that correlated with genotypes. We also removed outlier samples and SNPs (see Supplementary Methods). See previous paragraph on removing genes with insufficient variation after normalization.
Given that we were interested in identifying eQTLs that had differential expression in ALS donors compared to controls, our model tested for an interaction between genotype and disease status on the gene expression, genome-wide. This model incorporated covariates: sex, age of death, post-mortem delay and surrogate variables (see Supplementary Fig. 1B). Surrogate variable analysis was calculated using SVA24 and SVASeq.25 We also implemented a model with covariates RNA integrity number (RIN) and flow-cell (n = 2), and these covariates did not show significant impact on gene expression. As our library preparation protocols accounted for RNA fragmentation profiles and surrogate variable analysis controls for RNA degradation effects, we omitted RIN from the final analysis. Reported fold-changes, beta values, standard error and P-values are from the interaction term in additive linear model. P-values were adjusted for multiple testing using false discovery rate (FDR).
Evidence indicates that the correlation between SNPs and gene expression beyond 850 kb becomes marginal,12 therefore, cis-acting eQTLs were categorized as being within an 850 kb region from SNP to gene boundary (determined by Ensembl gene GRCh37coordinates). eQTLs were categorized as trans-acting if greater than this region.
Analyses above were implemented using DESeq226 and MatrixEQTL.23 eQTL and Manhattan plots were created using ggplots in R,27 LocusZoom28 and rMVP (https://github.com/xiaolei-lab/rMVP Accessed 13 October 2021).
Post-mortem cell deconvolution analysis
BRETIGEA was used to estimate cell composition per sample using BRETIGEA’s cell marker dataset (50 markers).29 To analysis differences in cell composition by type across samples, we used a linear regression model to compare cell-type estimates between ALS and controls controlling for variables age of death, post-mortem delay, RIN, and surrogate variables from the previous analysis. We report beta, standard error and non-adjusted P-values from these analysis in the Supplementary Results.
ALS survival genotype and gene-level analysis
To analyse the effect of eQTLs on ALS survival, we used Cox proportional hazards GWAS summary statistics21; see Genomic Datasets.
For gene-level and gene-set analyses, we used MAGMA30 incorporating a flanking region of 40 kb downstream and upstream of the genes, to cover the SCFD1 eQTL haplotype. The ALS survival GWAS summary results were used to assess change in survival as a function of eQTLs and raw genotype data used for Kaplan–Meier plots (using R) to show changes in survival as a function of SCFD1 eQTL genotypes.
Co-expression analyses
To identify genes that co-express with SCFD1 and correlate with SCFD1 eQTLs, we incorporated a Pearson’s r correlation analysis using normalized expression values from post-mortem ALS RNA-sequence data. This created a one-to-many correlation matrix, where SCFD1 r and P-values were calculated for each trans-acting gene that associated with post-mortem ALS via SCFD1 downstream eQTLs. Because we are interested in a wider network of genes, we report non-adjusted P-values unless stated others. Correlation matrices were plotted using PerformanceAnalytics and ggplots in R.
Gene function enrichment and over-representation analyses
We implemented three gene function enrichment analyses. The first was a rank-ordered analysis using g: profiler (https://biit.cs.ut.ee/gprofiler/gost). Categories with a g: profiler adjusted P-value < 0.05 were deemed statistically significant. Generic Enrichment Map (GEM) and Gene Matrix Transposed (GMT) files from this analysis were used for the Gene Set Enrichment Analyses (GSEA) analyses.31 For the second enrichment analysis, we used Enrichr (https://amp.pharm.mssm.edu/Enrichr/), where categories with rank-based adjusted P-values < 0.05 were regarding as statistically significant unless stated otherwise. The third function enrichment analysis was performed on SCFD1 positively and negatively co-expressed genes, using 5000 permutations and GMT files generated from g: profiler. Gene ontology (GO) categories with an FDR corrected q-value < 0.05 were deemed statistically significant, unless stated otherwise. To test if genes significantly overlapped between two gene-sets we implemented an over-representation adapted from: http://nemates.org/MA/progs/overlap_stats.html (Supplementary Methods).
Data availability
Datasets are available, on reasonable request. The post-mortem genetic and RNA-sequence datasets are available from the corresponding author, the GWAS survival datasets are available through Dr Isabella Fogh, and the Braineac eQTL datasets are available through the UK Brain Expression Consortium.
Results
ALS GWAS and eQTLSCFD1 eQTLs increase risk of ALS
To confirm and contextualize which SNPs increase risk of ALS via eQTLs, we performed genome-wide summary-data-based Mendelian Randomization (SMR) using eQTLs derived from cortex, cerebellum and blood as instrumental variables, gene expression as exposure, and ALS disease status as outcome. Here, SMR is testing the likelihood that SNPs that increase risk of ALS do so through modifying gene expression. Filtering for genes with PHEIDI > 0.05, we found SCFD1 was the only gene to show genome-wide significant association with ALS in cerebellum [SMR beta = 0.20, SMR standard error (SE) = 0.043, SMR P-value = 4.29 × 10−6]. In addition, SCFD1 eQTLs showed association with ALS with the lowest genome-wide P-values in frontal cortex (SMR beta = 0.27, SMR SE = 0.072, SMR P-value = 1.34 × 10−4). In GTEx, there are no cortical SCFD1 eQTLs with a P-value < 5 × 10−8 so we ran the SMR analysis using a GTEx P-value threshold = 5 × 10−7, where SCFD1 had the second lowest association P-value (SMR beta = 0.30, SMR SE = 0.085, SMR P-value = 3.81 × 10−4); see Table 1 and Supplementary Fig. 2, and see Supplementary Table 2 for the genome-wide gene-level association analysis.
Table 1.
Results from genome wide SMR analysis using GTEx eQTLs from Cortex, Cerebellum, Frontal Cortex, and Blood, and ALS GWAS SNPs, where SMR P-value < 1 × 10−3
| Cerebellum | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | Chr. | Top SNP BP | GWAS P-value | eQTL P-value | SMR Beta | SMR SE | SMR P-value | Heidi P-value | Heidi N SNPs |
| SCFD1 | 14 | 30622112 | 2.64 × 10–7 | 3.14 × 10–24 | 0.197 | 0.043 | 4.29 × 10–6a | 0.695 | 20 |
| TRIM65 | 17 | 75880335 | 6.36 × 10–5 | 1.76 × 10–30 | −0.083 | 0.022 | 1.62 × 10–4 | 0.869 | 20 |
| TRIP11 | 14 | 91965991 | 8.08 × 10–6 | 2.45 × 10–8 | 0.265 | 0.072 | 2.16 × 10–4 | 0.765 | 18 |
| MESDC2 | 15 | 80946289 | 4.51 × 10–4 | 1.04 × 10–13 | 0.109 | 0.034 | 1.50 × 10–3 | 0.634 | 20 |
|
| |||||||||
| Cortex | |||||||||
|
| |||||||||
| Gene | Chr. | Top SNP BP | GWAS P-value | eQTL P-value | SMR Beta | SMR SE | SMR P-value | Heidi P-value | Heidi N SNPs |
|
| |||||||||
| FBF1 | 17 | 75909574 | 8.54 × 10–5 | 4.41 × 10–22 | −0.104 | 0.028 | 2.63 × 10–4 | 0.686 | 20 |
| SCFD1 | 14 | 30622112 | 1.09 × 10–6 | 2.11 × 10–7 | 0.301 | 0.085 | 3.81 × 10–4 | 0.513 | 13 |
| ZNF391 | 6 | 27374615 | 1.63 × 10–4 | 4.39 × 10–11 | −0.159 | 0.049 | 1.07 × 10–3 | 0.495 | 20 |
| PLEKHG5 | 1 | 6466196 | 1.36 × 10–4 | 1.16 × 10–7 | 0.271 | 0.087 | 1.93 × 10–3 | 0.297 | 7 |
|
| |||||||||
| Frontal cortex | |||||||||
|
| |||||||||
| Gene | Chr. | Top SNP BP | GWAS P-value | eQTL P-value | SMR Beta | SMR SE | SMR P-value | Heidi P-value | Heidi N SNPs |
|
| |||||||||
| SCFD1 | 14 | 30622112 | 2.24 × 10–7 | 1.41 × 10–8 | 0.273 | 0.072 | 1.34 × 10–4 | 0.945 | 20 |
| DENND6B | 22 | 50309030 | 1.17 × 10–4 | 7.99 × 10–16 | 0.182 | 0.052 | 5.25 × 10–4 | 0.731 | 20 |
| GOLGA6L10 | 15 | 82339998 | 2.34 × 10–4 | 3.16 × 10–18 | 0.064 | 0.019 | 6.80 × 10–4 | 0.753 | 20 |
| TESC | 12 | 117038923 | 1.96 × 10–4 | 1.69 × 10–10 | −0.171 | 0.053 | 1.29 × 10–3 | 0.123 | 20 |
|
| |||||||||
| Blood | |||||||||
|
| |||||||||
| Gene | Chr. | Top SNP BP | GWAS P-value | eQTL P-value | SMR Beta | SMR SE | SMR P-value | Heidi P-value | Heidi N SNPs |
|
| |||||||||
| SCFD1 | 14 | 30622112 | 3.87 × 10–7 | 1.88 × 10–56 | −0.258 | 0.053 | 1.38 × 10–6a | 0.634 | 20 |
| GGNBP2 | 17 | 36544888 | 3.27 × 10–6 | 1.83 × 10–27 | 0.434 | 0.102 | 1.90 × 10–5 | 0.279 | 20 |
| TNIP1 | 5 | 151029945 | 7.14 × 10–7 | 1.52 × 10–15 | −0.45 | 0.107 | 2.56 × 10–5 | 0.538 | 7 |
| PLXNB2 | 22 | 50274979 | 7.45 × 10–6 | 3.72 × 10–12 | −0.547 | 0.146 | 1.71 × 10–4 | 0.236 | 20 |
For Cerebellum, Frontal Cortex and Blood, SCFD1 eQTLs showed the most significant association with ALS genome wide. For Cortex SCFD1 eQTLs were second most significant to associate ALS genome-wide. Genes with PHEIDI < 0.05 (Heidi P-value) were removed. Genes organized by their GWAS P-value association. Chr: Chromosome. Top SNP BP: Genomic base-pair location of most significantly associated SNP with ALS.
aGenome-wide significant.
SCFD1 eQTLs that increase risk of ALS do not correlate with SCFD1 expression in ALS motor cortex
The most significant SCFD1 SNP to associate with ALS is rs10139154.20 It is a significant eQTL in GTEx (V8) for cerebellum [Normalized Effect Size (NES) = 0.330, SE = 0.033, P-value = 7.54 × 10−19], cerebellar hemisphere (NES = 0.314, SE = 0.036, P-value = 1.33 × 10−14), frontal cortex (NES = 0.233, SE = 0.046, P-value = 1.7 × 10−6) and cortex (NES = 0.18, SE = 0.041, P-value = 2.2 × 10−5). In GTEx, for each rs10139154 T allele there is an increase in SCFD1 expression. Using our post-mortem control data, we confirmed this correlation (beta = 0.16, SE = 0.047, P-value = 0.03). However, the rs10139154 eQTL in our post-mortem ALS cohort we did not find significant correlation with SCFD1 expression (beta = 0.02, P-value = 0.12) (Supplementary Fig. 3). This finding indicates SCFD1 eQTLs that associate with disease risk may have a different relationship between genotype and expression in post-mortem ALS compared to controls.
Post-mortem differential eQTL expression
SCFD1 eQTLs show significant differences in expression in post-mortem ALS
Given eQTLs that increase risk of ALS showed differences in the relationship between genotype and expression in patients compared to controls, we performed a post-mortem eQTL differential expression analysis modelling the effect of disease status by genotype. We ran this analysis genome-wide for cis-acting eQTLs in post-mortem motor cortex comparing age and sex matched ALS (n = 76 and) non-ALS control donors (n = 25) (see Supplementary Result and Supplementary Table 1).
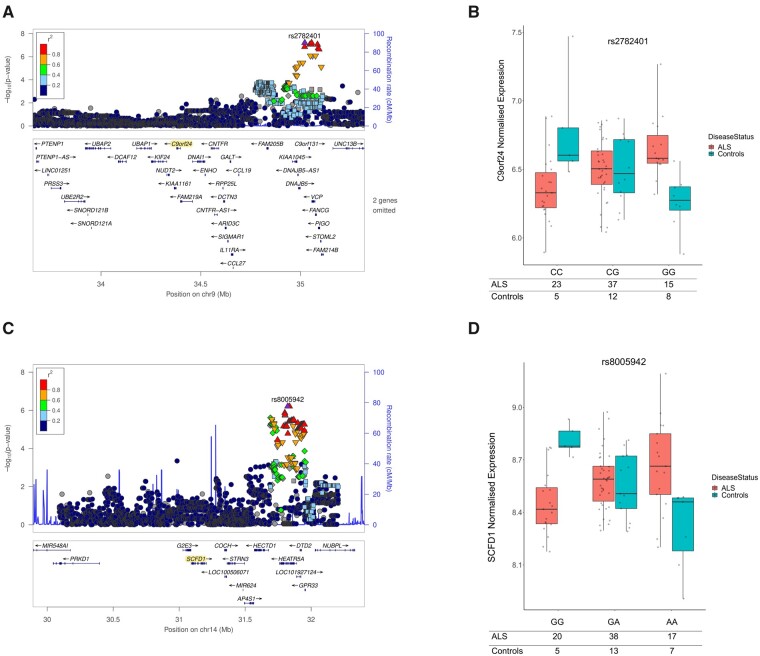
The most statistically significant cis-acting eQTL to show differential expression was for gene C9orf24. The eQTL locus was upstream of C9orf24 and formed a haplotype over ALS genes VCP and DNAJB5. For the most significant eQTL rs2782401, the addition of each G allele correlated with an increase in C9orf24 expression in ALS donors, in contrast to controls where G alleles correlated with a decrease in C9orf24 expression (top eQTL = rs2782401, beta = 0.44, SE = 0.076, P-value = 6.30 × 10−8); see Fig. 1A and B. rs2782401 is a significant GTEx eQTL for VCP (NES = 0.10, SE = 0.018, P-value = 3.57 × 10−9) and DNAJB5 (NES = −0.186, SE = 0.037, P-value = 7.07 × 10−7).
Figure 1.
The most significant eQTL loci differentially expressed in post-mortem ALS compared to controls. (A) Locus plot of C9orf24 eQTLs, where P-values are derived from differences in C9orf24 eQTL expression between ALS cases and controls. Points are C9orf24 eQTLs, point-colour represent r2 LD with the most significant C9orf24 eQTL (rs2782401) to associate with ALS, the x-axis is the genomic location of the eQTLs, and the y-axis is the −log10(P-value) of the eQTL association test. Upward-triangles LD r2 > 0.8, downward-triangles LD r2 between 0.8 and 0.6, rhombi LD r2 between 0.6 and 0.4, and squares LD r2 between 0.4 and 0.2. (B) Jitter-boxplot of C9orf24 eQTL rs2782401 expression by genotype (beta = 0.44, P-value = 6.30 × 10−8). Statistics taken from the linear additive model testing eQTL association with ALS modelling genotype by disease status. X-axis: rs2782401 genotypes; y-axis: normalized C9orf24 expression; Points: C9orf24 expression estimates by sample, differentiated into controls (red) and ALS cases (blue). (C) Locus plot of SCFD1 eQTLs, where P-values are derived from differences in SCFD1 eQTL expression between ALS cases and controls. Dots are SCFD1 eQTLs, dot-colour represent r2 LD with the most significant SCFD1 eQTL (rs8005942) to associate with ALS, the x-axis is the genomic location of the eQTLs, and the y-axis is the −log10(P-value) of the eQTL association test. Upward-triangles LD r2 > 0.8, downward-triangles LD r2 between 0.8 and 0.6, rhombi LD r2 between 0.6 and 0.4, and squares LD r2 between 0.4 and 0.2. (D) Jitter-boxplot of SCFD1 eQTL rs8005942 expression by genotype (beta = 0.34, P-value = 4.45 × 10−7). Statistics taken from the linear additive model testing eQTL association with ALS modelling genotype by disease status. X-axis: rs8005942 genotypes; y-axis: normalized SCFD1 expression; Points: SCFD1 expression estimates by sample, differentiated into controls (red) and ALS cases (blue). In summary, this figure. shows that there are eQTLs that are differentially expressed in ALS compared to controls. The top two most significant loci both implicate ALS genes, with the first loci (A) located at ALS gene VCP and the second loci (C) showing differential eQTL expression of ALS gene SCFD1.
The second most significant cis-acting eQTL to show differential expression in ALS was for SCFD1. These eQTLs were approximately 500 kb downstream of SCFD1 and were largely concentrated across genes HEATR5A, DTD2, HECTD1 and NUBPL (top eQTL = rs8005942, beta = 0.34, SE = 0.063, P-value = 4.54 × 10−7); see Fig. 1C and D. While a decrease in SCFD1 gene expression significantly correlated with each additional in rs8005942 A allele in controls, the opposite was found for ALS samples, where an increase in SCFD1 expression was found with each additional A allele (see Supplementary Table 3).
Post-mortem cell deconvolution analysis
In our post-mortem differential eQTL expression analysis, we have used surrogate variable analysis to control for differences in cell heterogeneity across samples and sample groups. To further examine if the above findings could be explained by differences in cell composition, we performed cell deconvolution analysis to estimate the presence of cell types across all samples. In comparing ALS samples with controls, we found that disease status in our model did not predict differences in cell estimates for any cell type (see Supplementary Table 4). Furthermore, there were no significant differences in RIN estimates between ALS and controls (beta = 0.299, SE = 0.230, P-value = 0.190).
Differentially expressed SCFD1 eQTLs significantly modify ALS survival
In our first analysis (SMR using GTEx-derived eQTLs), we confirmed eQTLs either within or immediately adjacent to SCFD1 increased risk of ALS. In our second post-mortem analysis, testing for differences in eQTL expression between ALS and control donors, we identified an additional locus of SCFD1 eQTLs downstream of the gene that associated with disease status. These two SCFD1 eQTL loci are not in linkage disequilibrium. Given that the SCFD1 downstream eQTLs do not necessarily increase risk of ALS we examined their influence on survival.
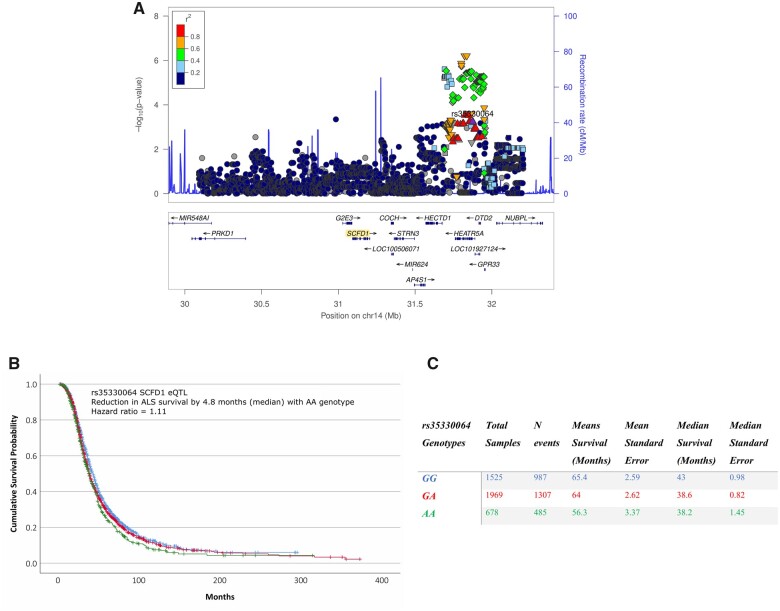
We used summary statistics from a 2016 ALS GWAS21 that estimated the effect of genotypes on ALS survival. We filtered SNPs within a 1 Mb flanking region of SCFD1. The most significant SNP to modify ALS survival in the region was rs35330064, which is linkage disequilibrium with eQTLs that show differences in expression post-mortem. Homozygosity for the rs35330064 minor allele (AA) reduced survival in ALS by a median of 4.8 months (Hazard ratio = 1.11, 95% CI 1.05–1.17, P-value = 2.06 × 10−4); see Fig. 2. rs35330064 was in linkage disequilibrium with the eQTLs that showed differential expression. For example, rs35330064 homozygosity (AA) showed significant linkage disequilibrium with rs8005942 (AA) (r2 = 0.687, D′ = 1, Chi-Square = 125.10, P-value < 1 × 10−4), the most significant eQTL to show differential expression in ALS in this region (see Supplementary Fig. 4).
Figure 2.
The relationship between SCFD1 eQTLs and SNPs that modify ALS survival. (A) Locus plot of SCFD1 eQTLs, where P-values are derived from differences in SCFD1 eQTL expression between ALS cases and controls. Points are SCFD1 eQTLs, point-colour represent r2 LD with the most significant SCFD1 eQTL (rs35330064) to associate with ALS survival, the x-axis is the genomic location of the eQTLs, and the y-axis is the −log10(P-value) of the eQTL association test. Upward-triangles LD r2 > 0.8, downward-triangles LD r2 between 0.8 and 0.6, rhombi LD r2 between 0.6 and 0.4, and squares LD r2 between 0.4 and 0.2. (B) Kaplan–Meier plot and table results showing the effect of rs35330064 genotypes on ALS survival. Kaplan–Meier x-axis show ALS survival in months, y-axis is the cumulative survival probability, and legend table displays in the effect of rs35330064 genotypes on ALS survival. In summary, this figure. shows that SCFD1 eQTLs that are differentially expressed in ALS significantly influence the disease duration of ALS.
rs35330064 and rs8005942 eQTLs are a part of a haplotype spanning genes HEATR5A, DTD2, NUBPL AND GPR33 (see Fig. 2A). To understand if the region was influencing ALS survival, we performed a gene-set analysis of SNPs in these four genes using summary statistics from the 2016 ALS survival GWAS. Collectively, these four genes significantly influenced survival, where beta = 2.20, SE = 0.032, P-value = 3.45 × 10−5. The most significant gene to show association with changes in ALS survival was for HEATR5A (Z Statistic = 2.38, P-value = 0.007), where most of the eQTLs that show differential expression in post-mortem donors are concentrated. In general, the directionality of genotypes indicates that increased SCFD1 expression may reduce ALS survival.
SCFD1 eQTLs form a trans-QTL hotspot that associates with post-mortem ALS
We used the post-mortem motor cortex dataset to test for trans-acting eQTLs that show significant differences in expression between ALS (n = 76) and non-ALS donors (n = 25). The most significant trans-acting eQTLs that was differentially expressed in ALS, after genome-wide FDR correction, was for gene PPP1R8 (top eQTL = rs56314035_C, beta = −0.371, SE = 0.053, P-value = 3.55 × 10−10); Supplementary Fig. 5. Notably, the base pair locations and genotypes for these PPP1R8 trans-acting eQTLs were the same as the SCFD1 eQTLs (from the post-mortem cis-acting eQTL analyses); Fig. 1A. SCFD1 significantly co-expressed with PPP1R8 in our post-mortem ALS samples, where Pearson’s r = 0.386 and P-value = 6.18 × 10−4.
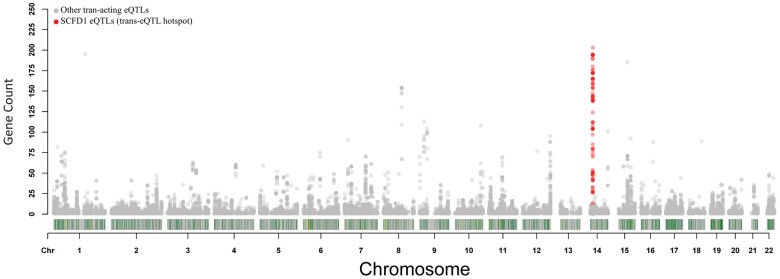
Similar to PPP1R8, we found that many of the trans-acting eQTLs that show differences in expression in post-mortem ALS, significantly overlapped with SCFD1 eQTLs. To explore this, we expanded our search for trans-acting eQTLs that were differentially expressed in ALS, by increasing the P-value to P < 5 × 10−5. We then mapped each of these trans-acting eQTLs by their genomic location and calculated the number genes the eQTLs correlated with. We found that the number of genes (n = 382) that were differentially expressed via trans-acting eQTLs in ALS, and which were located in SCFD1 eQTL locus downstream of the gene, to be the highest in the genome compared to other loci (see Fig. 3). These 382 genes also show significant levels of co-expression with SCFD1 (see Supplementary Results; see Supplementary Fig. 6).
Figure 3.
Count of genes that show association with ALS via trans-acting eQTLs by genomic location. Dots represent trans-acting eQTLs that show significant differences in expression comparing post-mortem ALS to controls. The x-axis is the genomic locations of these ALS-associated trans-acting eQTLs. The y-axis displays the number of genes that correlate with these ALS-associated trans-acting eQTLs. The highlighted dots (red) are the cis-acting SCFD1 eQTLs that associated with post-mortem ALS, from the post-mortem genome-wide cis-eQTL analysis. In summary, this figure displays the genomic locations of trans-acting eQTLs that show significant differences in expression between ALS and controls. Trans-acting eQTLs that correlate with the expression of multiple genes are called trans-eQTL hotspots. We found that SCFD1 eQTLs (red dots) formed a trans-eQTL hotspot that showed significant differences in expression in ALS, for a large number of genes throughout the genome. This indicates that SCFD1 eQTLs that associate with ALS are impacting the transcription of other genes at a distance, and that these changes in expression significantly associate with ALS disease status.
In summary, SCFD1 cis-acting eQTLs that show significant differences in expression between ALS and controls are not just correlated with SCFD1, but correlate with a wider network of genes that show expression changes in post-mortem ALS. And the number of genes that associate with post-mortem ALS via trans-acting eQTLs are most concentrated in this SCFD1 locus, more than anywhere else on the genome. These results suggest that SCFD1 eQTLs are having a wider impact on transcriptional pathways, that are specific to ALS, which we will explore next. We will refer to this SCFD1 eQTL locus as the SCFD1 trans-eQTL hotspot.
Genes that associate with the SCFD1 trans-eQTL hotspot are enriched for ALS pathways
To assess the wider effects of the SCFD1 trans-eQTL hotspot in ALS, we selected genes with differentially expressed trans-acting eQTLs where P < 5 × 10−5. We also included an additional 8 genes which are proximal to the SCFD1 trans-eQTL hotspot and show differences in expression in ALS compared to controls: SCFD1, G2E3, HEATR5A, HECTD1, NUBPL, COCH and DTD2.
Using a final list of 389 genes, we performed gene function enrichment analyses (see Table 2A and Supplementary Table 5). We found genes that associate with post-mortem ALS via the SCFD1 trans-eQTL hotspot were significantly enriched for SCFD1 function and ALS pathways. SCFD1 is involved in retrograde vesicle-mediated protein transport from the ER-to-Golgi, and the regulation of synaptic vesicle docking and exocytosis. These results indicate that SCFD1 eQTLs do not only correlate with SCFD1 expression but with a wider functional pathway that is differentially expressed in post-mortem ALS.
Table 2.
(A–D) Gene function enrichment results from various analyses
| A. Functional categories enriched for genes that are differentially expressed in ALS via the SCFD1 trans-eQTL hotspot | |||
|---|---|---|---|
| Term Name | Term ID | Adj. P-value | Tool |
| Regulation of vesicle mediated transporta | GO:0060627 | 2.24 × 10–5 | g:profiler |
| Cellular, protein and organelle localisationa | GO:0051641 | 7.08 × 10–7 | g:profiler |
| Cytoplasmic vesiclesa | GO:0031410 | 0.004 | g:profiler |
| Vesicle mediated transport in synapse | GO:0099003 | 7.58 × 10–7 | g:profiler |
| Vesicle docking involved in exocytosis\exocytic vesicles | GO:0070382 | 3.30 × 10–5 | g:profiler |
| Calcium/calmodulin signalling | KEGG:04020 | 0.006 | g:profiler |
| The glutamatergic synapse | GO:0098978 | 4.20 × 10–6 | g:profiler |
| mRNA splicing via spliceosome | GO:0048024 | 0.001b | g:profiler |
| Microtubule genes | GO:0005874 | 0.002b | g:profiler |
| Schizophrenia | NA | 3.99 × 10–4 | Enrichr: DisGeNET |
| Motor neuron | NA | 2.78 × 10–14 | Enrichr: ARCHS4 |
| Brain | NA | 3.56 × 10–43 | Enrichr: ARCHS4 |
| Cerebral cortex | NA | 1.35 × 10–34 | Enrichr: ARCHS4 |
| Spinal cord | NA | 7.86 × 10–20 | Enrichr: ARCHS4 |
| Prefrontal cortex | NA | 4.10 × 10–9 | Enrichr: ARCHS4 |
| Abnormal CNS synaptic transmission | NA | 4.05 × 10–3 | Enrichr: MGI |
| Impaired motor coordination | NA | 0.064 | Enrichr: MGI |
|
| |||
| B. Functional categories enriched for trans-acting genes that negatively co-express with SCFD1 | |||
|
| |||
| Term name | Term ID |
FDR
q-value |
Tool |
|
| |||
| Regulation of vesicle mediated transporta | GO:0060627 | 0.004 | GSEA |
| Regulation of localizationa | GO:0032879 | 0.03 | GSEA |
| Integral component of membranea | GO:0016021 | 2.262 × 10–5 | GSEA |
| Secretiona | GO:0046903 | 0.004 | GSEA |
| Regulation of trans-synaptic signalling | GO:0099177 | 3.926 × 10–4 | GSEA |
| Exocytic vesicles | GO:0070382 | 0.001 | GSEA |
| Glutamatergic synapse | GO:0098978 | 0.01 | GSEA |
|
| |||
| C. Functional categories enriched for trans-acting genes that positively co-express with SCFD1 | |||
|
| |||
| Term name | Term ID |
FDR
q-value |
Tool |
|
| |||
| RNA-binding | GO:0003723 | <1 × 10–5 | GSEA |
| Ribonucleoprotein complex | GO:1990904 | <1 × 10–5 | GSEA |
| Nuclear lumen | GO:0031981 | 9.617 × 10–5 | GSEA |
| Negative regulation of gene expression | GO:0010629 | 0.001 | GSEA |
| RNA splicing | GO:0008380 | 0.002 | GSEA |
|
| |||
| D. Functional categories enriched for genes that co-express with SCFD1 and modify schizophrenia risk | |||
|
| |||
| Term name | Term ID |
FDR
q-value |
Tool |
|
| |||
| Voltage-gated potassium channel activity | GO:0022843 | 2.21 × 10–5 | g:profiler |
| Potassium ion transmembrane transporter activity | GO:0015079 | 3.46 × 10–5 | g:profiler |
| Synaptic signalling | GO:0099536 | 6.95 × 10–8 | g:profiler |
| Regulation of vesicle mediated transporta | GO:0060627 | 0.002 | g:profiler |
| Cholinergic Synapse | KEGG:04725 | 6.36 × 10–4 | g:profiler |
| Negative regulation of NMDA receptor-mediated neuronal transmission | REAC | 0.007 | g:profiler |
| Unblocking of NMDA receptors, glutamate binding and activation | REAC | 0.007 | g:profiler |
aFunctional category of which SCFD1 is a member.
bRaw P-value reported.
Genes that associate with the ALS SCFD1 trans-eQTL hotspot are enriched for disease risk
Our previous analysis of SCFD1 trans-eQTL genes (n = 389) identified significant enrichment of genes involved in schizophrenia risk (Table 2A). Genetic correlation between ALS and schizophrenia risk has been previously shown, where SCFD1 was identified as a candidate gene driving risk of both diseases.32 Performing gene-set analysis on the latest publicly available schizophrenia GWAS,33 we found that the 389 genes that associated with post-mortem ALS via the SCFD1 trans-eQTL hotspot were enriched for SNPs that modify schizophrenia risk [beta = 0.263, standard deviation (SD) = 0.036, P-value = 1.18 × 10−5]. Thirteen genes from this set had a statistically significant association below the standard GWAS threshold of P ≤ 5 × 10−8, 38 genes below the MAGMA genome-wide threshold of P ≤ 2.7 × 10−5, and 148 genes below P < 0.05.
Using results from the 2018 ALS GWAS,19 we performed gene-set analysis on the 387 genes that associated with post-mortem ALS status via the SCFD1 trans-eQTL hotspot (we removed SCFD1 and G2E3 from the gene-set, due to their significant association with the disease). This analysis revealed a marginal increase in ALS risk, where beta = 0.05, SD = 0.008, P = 0.094. However, we found that the top 100 genes from this list (ranked by their association with post-mortem ALS) were significantly enriched for SNPs that increased ALS risk, where beta = 0.247, SD = 0.017, P = 0.001.
Discussion
In our first analysis, we confirm that SCFD1 was the only gene that is genome-wide significant in mediating ALS risk via eQTLs. While we were able to confirm a correlation between ALS risk eQTLs and SCFD1 expression in our post-mortem controls, in our ALS donors the correlation was lost, indicating that SCFD1 eQTL expression may be influenced by other factors related to the disease.
In our second analysis using motor cortex, we tested if cis-acting eQTLs showed differences in expression between ALS and control donors, genome-wide. The top two most significant eQTL loci that show differential expression were for SCFD1 and C9orf24. Differentially expressed SCFD1 eQTLs significantly associated with ALS survival, as well as a number of differentially expressed trans-acting eQTLs across the genome. The genes that correlate with trans-acting eQTL ALS-specific changes were highly enriched for SCFD1 function, ALS pathways, and risk of schizophrenia and ALS. ALS and schizophrenia genetically correlate, where SCFD1 is a candidate overlapping gene.32
In addition to the gene-targets, the genomic location of the top two eQTLs to show differential expression in ALS are relevant to the disease as well. For C9orf24, the eQTLs are localized nearby in genes VCP and DNAJB5. VCP is an ALS gene first implicated in the disease in 2010 through the identification of an autosomal dominant point mutation in familial ALS (Johnson et al., 2010). Genomic variants in DNAJB5 have shown association with neuromuscular diseases, including Charcot-Marie-Tooth disease.34 The genomic location of differentially expressed SCFD1 eQTLs, span a number of nearby genes including NUBPL, which itself has shown association with ALS via a TDP-43 conditional FDR analysis.35
Factors influencing differential eQTL expression
SCFD1 alleles that increase ALS risk via GWAS, increase SCFD1 expression in brain in non-ALS controls. We confirmed this using two sets of post-mortem controls: GTEx and our non-ALS cohort. When we examine ALS donors in our post-mortem data, the correlation between ALS SCFD1 risk alleles and SCFD1 expression is not present. In addition, we show cis-acting eQTLs downstream of SCFD1 are differentially expressed in ALS. These results indicate the genomic regulation of SCFD1 eQTLs, in ALS specifically, may be influenced by additional factors.
There are a number of factors that can influence eQTL expression, which show complex relationships between SNPs and gene expression similar to our findings. These include epistatic interactions,36,37 environmental factors,36 methylation,38 chromatin interactions,39 and drug-eQTL interactions.40 An additional factor may involve pre-disease processes influencing the genomic regulation of SCFD1 in ALS, as has been found with TDP-43 depletion and decreased expression of ALS GWAS gene UNC13A.41
Our finding that differentially expressed SCFD1 eQTLs influence ALS survival suggests that the genomic regulation of SCFD1 expression is important to the disease process. Through the analysis of trans-acting eQTLs in the SCFD1 locus, our results indicate differences the regulation of SCFD1 and SCFD1 eQTLs are to likely involve functional pathways known to ALS pathology. Research, explored below, shows modifying SCFD1 expression can be both protective and toxic to cells under stress, and it is plausible that how this regulatory response occurs depends on a person’s genotypes proximal to SCFD1, thereby modifying disease vulnerability and duration.
Changes in SCFD1 expression are functionally relevant to ALS
SCFD1 is a key component in Endoplasmic Reticulum (ER) to Golgi retrograde vesicle transport,42,43 vesicle docking in exocytosis,44 collagen/procollagen ER export45 and autophagy. These pathways have been a major focus in ALS research. Mutations found in genes that play a major role in these pathways cause ALS. The most recent was for gene ANXA11,10 a protein essential for stabilization of SEC31A at ER exit sites,46 a process critical in ER-Golgi vesicle-mediated trafficking.
Previous research into SCFD1 function found that knockdown in zebrafish embryos significantly disrupts ER-to-Golgi transport, leading to intracellular protein build-up and an unfolded protein response.47 In a study using SH-SY5Y cells, SCFD1 expression significantly responds to neurotoxin-induced oxidative stress, where increased expression exhibited anti-apoptotic effects by supressing morphological changes to the ER, where SCFD1-antisense transfected cells accelerated apoptosis.48SCFD1 also colocalizes with SEC31 at ER exit sites, similarly to ANXA11, although SCFD1 knockdown in HeLa cells did not disrupt SEC31 ER localization.45
In our insilico study, vesicle-mediated transport genes involved in cytoplasmic vesicles, localization, and exocytosis, significantly correlated with SCFD1 eQTL expression. Given that SCFD1 eQTLs increase ALS risk and were differentially expressed, these pathways may be differentially regulated depending a person’s genotypes, which confers risk of the disease.
We also found SCFD1 eQTLs significantly correlated with pathways involved in ALS but not attributed to SCFD1 function itself, including glutamatergic and GABAergic synapses, calcium signalling, microtubule genes, RNA-binding and mRNA splicing. The relationship between synaptic glutamate and Ca2+ signalling is believed to selectively predispose motor neurons to excitotoxicity in ALS and is the putative mechanism of the ALS drug Riluzole.1 We found SCFD1 expression negatively correlated with genes involved in synaptic glutamate, GABA and calcium signalling. This included ADCY1, the gene to most significantly negatively co-express with SCFD1. ADCY1 is almost exclusively expressed in brain, regulated by calcium, and is involved in regulation of synaptic vesicle exocytosis, and calcium-responsive adenylate cyclase binding. Similarly, we found ALS gene UNC13A49 negatively correlated with SCFD1 expression, which is also involved in glutamatergic-mediated synapses and synaptic vesicles.
Schizophrenia, ALS and SCFD1
Genes that associated with ALS in our post-mortem analysis via SCFD1 eQTLs, significantly overlap with genes that increase schizophrenia risk. Higher incidences of schizophrenia in families with a history of ALS have been shown since 2013.50 Genetic correlation between ALS and schizophrenia risk was also identified in 2017,32 where SCFD1 was one of 31 candidate genes that overlapped between the diseases. Why this correlation exists is not understood.
Our results support that SCFD1 has role in both ALS and schizophrenia and provides insights into the pathways that links them. We found (i) genes that correlate with SCFD1 eQTLs increase risk of both diseases and (ii) these genes were significantly enriched for pathways known to be involved in the pathology of both diseases, specifically cholinergic synapses, synaptic signalling, potassium transmembrane activity, NMDA-receptor activity and glutamate-binding, and vesicle-mediated transport. Our results indicate that the genetic correlation between ALS and schizophrenia may be driven by shared genomically regulated pathways, which, given the higher incidence of the schizophrenia in ALS families, is heritable risk factor. Further clarification of regulatory loci shared across these diseases could help prioritize drug therapies used in schizophrenia as candidate targets for ALS.
Limitations
A concern in using post-mortem whole-tissue samples is that differences in cell heterogeneity between ALS and control cohorts may lead to confounding results. We have used two methods to control and analyse differences in cell composition between samples and sample groups. We have integrated surrogate variables into our model when testing for differences in eQTL expression between ALS and controls. SVA controls for differences in heterogeneity caused by unavailable confounding factors.24 We performed cell deconvolution analysis to estimate cell composition for each sample and found no significant differences in cell-type between ALS and control groups in our model. If SCFD1 was a marker for motor neuron loss then we would expect gene-level differential expression between ALS and control cohorts, regardless of patient genotypes, which we did not. It is also worth noting that there is significant variability in motor neuron loss across ALS patients.51
In our post-mortem analysis, we identified significant differences in SCFD1 expression between ALS and control donors for both homozygotes of the rarer genotype and the common genotype. Using the post-mortem analysis only, it is unclear how increased or decreased expression of SCFD1 associates with increased ALS risk. Our survival analysis showed increased SCFD1 expression correlated with a quicker disease duration, which given alleles that increase risk of ALS via GWAS also increase SCFD1 expression, may initially indicate that overexpression contributes to the disease onset and process. However, this does not necessarily concur with in vivo and in vitro functional analyses of SCFD1 that indicate increased expression can act as a protective factor against oxidative stress and apoptosis, while a decrease in SCFD1 can lead to cell stress vulnerability. Therefore, an alternative explanation is that increased SCFD1 expression could be reflecting a response to another unknown factor which is accelerating the disease process.
Summary
We confirmed and expanded SCFD1’s involvement in ALS. We confirm that SCFD1 eQTLs significantly increase ALS risk but are differentially expressed in post-mortem ALS, which correlates with the disease’s duration. SCFD1 is a key component in a large transcriptional network that correlates with ALS pathways and genes enriched for schizophrenia risk, a disease known to genetically correlate with ALS.
Supplementary material
Supplementary material is available at Brain Communications online.
Supplementary Material
Acknowledgements
We would like to thank and acknowledge the MND Association for funding of the post-mortem RNA-sequence and genotype data, as a part of fellowship grant Jones/Oct15/958-799. We would like to thank and acknowledge the MRC London Neurodegenerative Diseases Brain Bank at the Institute of Psychiatry, Psychology and Neuroscience, King's College London. With special thanks to the people who have donated tissue to the advancement of clinical sciences. We would like to acknowledge the Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data used for the analyses described in this manuscript were obtained from the GTEx Portal on 07/01/2019. We would like to thank UK Brain Expression Consortium (UKBEC) for their post-mortem brain eQTL data. This project has received funding from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme (grant agreement n° 772376—EScORIAL. We would like to acknowledge Science Foundation Ireland for their contribution of genetic data to this study. This work was in part supported by the Knut and Alice Wallenberg Foundation, the Swedish Brain Foundation, the Ulla-Carin Lindquist Foundation and the Research Council. This work was in part supported by the Italian Ministry of Health (Ministero della Salute, Ricerca Sanitaria Finalizzata, grant RF-2016-02362405), the European Commission’s Health Seventh Framework Programme (FP7/2007-2013 under grant agreement 259867), the Italian Ministry of Education, University and Research (Progetti di Ricerca di Rilevante Interesse Nazionale, PRIN, grant 2017SNW5MB), the Joint Programme—Neurodegenerative Disease Research (Strength and Brain-Mend projects), granted by Italian Ministry of Education, University and Research. This study was performed under the Department of Excellence grant of the Italian Ministry of Education, University and Research to the ‘Rita Levi Montalcini’ Department of Neuroscience, University of Torino, Italy. This is an EU Joint Programme—Neurodegenerative Disease Research (JPND) project. The project is supported through the following funding organizations under the aegis of JPND—www.jpnd.eu (UK), Medical Research Council (MR/L501529/1; MR/R024804/1) and Economic and Social Research Council (ES/L008238/1)) and through the Motor Neurone Disease Association. This study represents independent research part funded by the National Institute for Health Research (NIHR) Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London. Samples used in this research were in part obtained from the UK National DNA Bank for MND Research, funded by the MND Association and the Wellcome Trust. We would like to thank people with MND and their families for their participation in this project. We acknowledge sample management undertaken by Biobanking Solutions funded by the Medical Research Council at the Centre for Integrated Genomic Medical Research, University of Manchester.
Funding
This work was funded by the Motor Neurone Disease Association (grant: Jones/Oct15/958–799). AAC is an NIHR Senior Investigator. This is an EU Joint Programme—Neurodegenerative Disease Research (JPND) project. The project is supported through the following funding organizations under the aegis of JPND—www.jpnd.eu (UK), Medical Research Council (MR/L501529/1; MR/R024804/1) and Economic and Social Research Council (ES/L008238/1)) and through the Motor Neurone Disease Association. This study represents independent research part funded by the National Institute for Health Research (NIHR) Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London.
Competing interests
A.A.-C. reports grants from Motor Neurone Disease Association, grants from Betty Messenger Foundation, grants from Anonymous Family Trust, grants from Joint Programme on Neurodegeneration via MRC, grants from NIHR during the conduct of the study, other from Mitsubishi Tanabe Pharma, other from OrionPharma, other from Cytokinetics, other from Chronos Therapeutics, outside the submitted work.
V.S. is in the Editorial Board of Amyotrophic Lateral Sclerosis, European Neurology, American Journal of Neurodegenerative Diseases, Frontiers in Neurology; received compensation for consulting services and/or speaking activities from AveXis, Cytokinetics, Italfarmaco, and Zambon; and receives or has received research supports from the Italian Ministry of Health (Grant RF-201302355764), Fondazione Italiana di Ricerca per la SLA—AriSLA (Grants Exomefals and Novals), Fondazione Regionale per la Ricerca Biomedica Regione Lombardia (Project nr. 2015–0023), and E-RARE JTC 2018 (Project Repetomics).
P.M.A. serves on scientific advisory boards for Biogen, Genentech-Roche and Orphazyme on matters regarding genetics in ALS clinical trials and have received financial compensation for these services.
A.C. serves on scientific advisory boards for Mitsubishi Tanabe, Roche, and Cytokinetics, and has received a research grant from Italfarmaco.
P.V.D. holds a senior clinical investigatorship of FWO-Vlaanderen and is supported by E. von Behring Chair for Neuromuscular and Neurodegenerative Disorders, the ALS Liga België and the KU Leuven funds “Een Hart voor ALS”, “Laeversfonds voor ALS Onderzoek” and the “Valéry Perrier Race against ALS Fund”. Several authors of this publication are member of the European Reference Network for Rare Neuromuscular Diseases (ERN-NMD).
Appendix
We would like to thank and acknowledge the contribution of brain eQTL data via the UK Brain Expression Consortium. We would like to personally acknowledge:
Professor John Hardy1, Michael E Weale2, Mina Ryten1,3,4, Daniah Trabzuni1,5, Adaikalavan Ramasamy1,6, Colin Smith7, Manuel Sebastian Guelfi1, Karishma D’sa1,2, Paola Forabosco8
1Institute of Neurology, University College London (UCL), London WC1N 3BG, UK
2Department of Medical and Molecular Genetics, King’s College London, 8th Floor, Tower Wing, Guy’s Hospital, London SE1 9RT, UK
3Genetics and Genomic Medicine, Great Ormond Street Institute of Child Health, University College London, London WC1E 6BT, UK
4NIHR Great Ormond Street Hospital Biomedical Research Centre, University College London, London, UK
5Department of Genetics, King Faisal Specialist Hospital and Research Centre, PO Box 3354, Riyadh 11211, Saudi Arabia
6Singapore Institute for Clinical Sciences, Brenner Centre for Molecular Medicine, Singapore, Singapore
7Department of Neuropathology, MRC Sudden Death Brain Bank Project, University of Edinburgh, Wilkie Building, Teviot Place, Edinburgh EH8 9AG
8Istituto di Ricerca Genetica e Biomedica, CNR, Cagliari
Glossary
- ALS
amyotrophic lateral sclerosis
- GWAS
genome-wide association study
- CI
confidence Interval
- D′
D-prime
- eQTL
expression Quantitative Trait Loci
- ER
endoplasmic reticulum
- FDR
false discovery rate
- FTD
frontotemporal dementia
- GO
gene ontology
- GRCh37
Genome Reference Consortium human build 37
- GTEx
genotype-tissue expression project
- HG19
human genome build 19
- hp-τ
hyperphosphorylated tau
- KB
kilobase
- NES
normalized effect size
- r 2
R-squared
- RIN
RNA integrity number
- rsID
reference SNP identifier
- SD
standard deviation
- SE
standard error
- SMR
summary-data-based Mendelian Randomization
- SNP
single nucleotide polymorphism
- VST
variance stabilizing transformation
Contributor Information
UK Brain Expression Consortium:
John Wim Hardy, Michael E Adriano Weale, Mina Richard J Ryten, Daniah Orla Trabzuni, Adaikalavan Christopher E Ramasamy, Colin Leonard H Smith, Manuel Sebastian Peter M Guelfi, Karishma Bradley N D’sa, and Paola Vincenzo Forabosco
References
- 1. Bensimon G, Lacomblez L, Meininger V, Group TAS.. A controlled trial of riluzole in amyotrophic lateral sclerosis. N Engl J Med. 1994;330(9):585–591. [DOI] [PubMed] [Google Scholar]
- 2. Abe K, Aoki M, Tsuji S, et al. Safety and efficacy of edaravone in well defined patients with amyotrophic lateral sclerosis: A randomised, double-blind, placebo-controlled trial. Lancet Neurol. 2017;16(7):505–512. [DOI] [PubMed] [Google Scholar]
- 3. Sreedharan J, Blair IP, Tripathi VB, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319(5870):1668–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kabashi E, Valdmanis PN, Dion P, et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet. 2008;40(5):572–574. [DOI] [PubMed] [Google Scholar]
- 5. Van Deerlin VM, Leverenz JB, Bekris LM, et al. TARDBP mutations in amyotrophic lateral sclerosis with TDP-43 neuropathology: A genetic and histopathological analysis. Lancet Neurol. 2008;7(5):409–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vance C, Rogelj B, Hortobagyi T, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323(5918):1208–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kwiatkowski TJ Jr, Bosco DA, LeClerc AL, Tamrazian E, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323(5918):1205–1208. [DOI] [PubMed] [Google Scholar]
- 8. Rosen DR, Siddique T, Patterson D, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362(6415):59–62. [DOI] [PubMed] [Google Scholar]
- 9. Maruyama H, Morino H, Ito H, et al. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 2010;465(7295):223–226. [DOI] [PubMed] [Google Scholar]
- 10. Smith BN, Topp SD, Fallini C, et al. Mutations in the vesicular trafficking protein annexin A11 are associated with amyotrophic lateral sclerosis. Sci Transl Med. 2017;9(388):eaad9157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Deng HX, Chen W, Hong ST, et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477(7363):211–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Aguet F, Brown AA, Castel SE, et al. ; eQTL Manuscript Working Group. Genetic effects on gene expression across human tissues. Nature. 2017;550(7675):204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lonsdale J, Thomas J, Salvatore M, et al. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45(6):580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bettella F, Brown AA, Smeland OB, et al. Cross-tissue eQTL enrichment of associations in schizophrenia. PLoS One. 2018;13(9):e0202812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Karch CM, Ezerskiy LA, Bertelsen S, Goate AM; Alzheimer’s Disease Genetics Consortium (ADGC). Alzheimer’s disease risk polymorphisms regulate gene expression in the ZCWPW1 and the CELF1 loci. PLoS One. 2016;11(2):e0148717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ramasamy A, Trabzuni D, Guelfi S, et al. ; North American Brain Expression Consortium. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci. 2014;17(10):1418–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Iacoangeli A, Lin T, Al Khleifat A, et al. Genome-wide meta-analysis finds the ACSL5-ZDHHC6 locus is associated with ALS and links weight loss to the disease genetics. Cell Rep. 2020;33(4):108323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Saez-Atienzar S, Bandres-Ciga S, Langston RG, et al. Genetic analysis of amyotrophic lateral sclerosis identifies contributing pathways and cell types. Sci Adv. 2021;7:eabd9036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nicolas A, Kenna K, Renton AE, et al. Genome-wide analyses identify KIF5A as a novel ALS gene. Neuron. 2018;97:1268–1283.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. van Rheenen W, Shatunov A, Dekker AM, et al. ; NNIPPS Study Group. Genome-wide association analyses identify new risk variants and the genetic architecture of amyotrophic lateral sclerosis. Nat Genet. 2016;48(9):1043–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fogh I, Lin K, Tiloca C, et al. Association of a locus in the CAMTA1 gene with survival in patients with sporadic amyotrophic lateral sclerosis. JAMA Neurol. 2016;73(7):812–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhu Z, Zhang F, Hu H, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016;48(5):481–487. [DOI] [PubMed] [Google Scholar]
- 23. Shabalin AA. Matrix eQTL: Ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28(10):1353–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Leek JT, Storey JD.. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007;3(9):1724–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Leek JT. svaseq: Removing batch effects and other unwanted noise from sequencing data. Nucleic Acids Res. 2014;42(21):e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Love MI, Huber W, Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wickham H. ggplot2: Elegant graphics for data analysis. Springer-Verlag New York; 2009. https://ggplot2.tidyverse.or. [Google Scholar]
- 28. Pruim RJ, Welch RP, Sanna S, Teslovich TM, et al. LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McKenzie AT, Wang M, Hauberg ME, et al. Brain cell type specific gene expression and co-expression network architectures. Sci Rep. 2018;8(1):1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. de Leeuw CA, Mooij JM, Heskes T, Posthuma D.. MAGMA: Generalized gene-set analysis of GWAS data. PLOS Comput Biol. 2015;11(4):e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McLaughlin RL, Schijven D, Van Rheenen W, et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium. Genetic correlation between amyotrophic lateral sclerosis and schizophrenia. Nat Commun. 2017;8(1):14774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pardiñas AF, Holmans P, Pocklington AJ, et al. ; CRESTAR Consortium. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet. 2018;50(3):381–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gonzaga-Jauregui C, Harel T, Gambin T, et al. ; Baylor-Hopkins Center for Mendelian Genomics. Exome sequence analysis suggests that genetic burden contributes to phenotypic variability and complex neuropathy. Cell Rep. 2015;12(7):1169–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Karch CM, Wen N, Fan CC, et al. ; International Frontotemporal Dementia (FTD)–Genomics Consortium, International Collaboration for Frontotemporal Dementia, Progressive Supranuclear Palsy (PSP) Genetics Consortium, and International Parkinson’s Disease Genomics Consortium. Selective genetic overlap between amyotrophic lateral sclerosis and diseases of the frontotemporal dementia spectrum. JAMA Neurol. 2018;75(7):860–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Brown AA, Buil A, Viñuela A, et al. Genetic interactions affecting human gene expression identified by variance association mapping. Elife. 2014;3:e01381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lareau CA, White BC, Oberg AL, Kennedy RB, Poland GA, McKinney BA.. An interaction quantitative trait loci tool implicates epistatic functional variants in an apoptosis pathway in smallpox vaccine eQTL data. Genes Immun. 2016;17(4):244–250. [DOI] [PubMed] [Google Scholar]
- 38. Ek WE, Rask-Andersen M, Karlsson T, Enroth S, Gyllensten U, Johansson Å.. Genetic variants influencing phenotypic variance heterogeneity. Hum Mol Genet. 2018;27(5):799–810. [DOI] [PubMed] [Google Scholar]
- 39. Fadason T, Schierding W, Lumley T, O’Sullivan JM.. Chromatin interactions and expression quantitative trait loci reveal genetic drivers of multimorbidities. Nat Commun. 2018;9(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Davenport EE, Amariuta T, Gutierrez-Arcelus M, et al. Discovering in vivo cytokine-eQTL interactions from a lupus clinical trial. Genome Biol. 2018;19(1):168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brown A-L, Wilkins OG, Keuss MJ, et al. Common ALS/FTD risk variants in UNC13A exacerbate its cryptic splicing 1 and loss upon TDP-43 mislocalization. bioRxiv2021. doi: 10.1101/2021.04.02.438170. [DOI]
- 42. Dascher C, Ossig R, Gallwitz D, Schmitt HD.. Identification and structure of four yeast genes (SLY) that are able to suppress the functional loss of YPT1, a member of the RAS superfamily. Mol Cell Biol. 1991;11(2):872–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ossig R, Dascher C, Trepte HH, Schmitt HD, Gallwitz D.. The yeast SLY gene products, suppressors of defects in the essential GTP-binding Ypt1 protein, may act in endoplasmic reticulum-to-Golgi transport. Mol Cell Biol. 1991;11(6):2980–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Halachmi N, Lev Z.. The Sec1 family: A novel family of proteins involved in synaptic transmission and general secretion. J Neurochem. 1996;66(3):889–897. [DOI] [PubMed] [Google Scholar]
- 45. Nogueira C, Erlmann P, Villeneuve J, et al. SLY1 and syntaxin 18 specify a distinct pathway for procollagen VII export from the endoplasmic reticulum. Elife. 2014;2014:e02784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shibata H, Kanadome T, Sugiura H, et al. A new role for annexin A11 in the early secretory pathway via stabilizing Sec31A protein at the endoplasmic reticulum exit sites (ERES). J Biol Chem. 2015;290(8):4981–4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hou N, Yang Y, Scott IC, Lou X.. The Sec domain protein Scfd1 facilitates trafficking of ECM components during chondrogenesis. Dev Biol. 2017;421(1):8–15. [DOI] [PubMed] [Google Scholar]
- 48. Bando Y, Katayama T, Taniguchi M, et al. RA410/Sly1 suppresses MPP+ and 6-hydroxydopamine-induced cell death in SH-SY5Y cells. Neurobiol Dis. 2005;18(1):143–151. [DOI] [PubMed] [Google Scholar]
- 49. van Es MA, Veldink JH, Saris CGJ, et al. Genome-wide association study identifies 19p13.3 (UNC13A) and 9p21.2 as susceptibility loci for sporadic amyotrophic lateral sclerosis. Nat Genet. 2009;41(10):1083–1087. [DOI] [PubMed] [Google Scholar]
- 50. Byrne S, Heverin M, Elamin M, et al. Aggregation of neurologic and neuropsychiatric disease in amyotrophic lateral sclerosis kindreds: A population-based case-control cohort study of familial and sporadic amyotrophic lateral sclerosis. Ann Neurol. 2013;74(5):699–708. [DOI] [PubMed] [Google Scholar]
- 51. Ravits J, Laurie P, Fan Y, Moore DH.. Implications of ALS focality: Rostral–caudal distribution of lower motor neuron loss postmortem. Neurology. 2007;68(19):1576–1582. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Datasets are available, on reasonable request. The post-mortem genetic and RNA-sequence datasets are available from the corresponding author, the GWAS survival datasets are available through Dr Isabella Fogh, and the Braineac eQTL datasets are available through the UK Brain Expression Consortium.