Abstract
The main protease of SARS-CoV-2 is a critical target for the design and development of antiviral drugs. 2.5 M compounds were used in this study to train an LSTM generative network via transfer learning in order to identify the four best candidates capable of inhibiting the main proteases in SARS-CoV-2. The network was fine-tuned over ten generations, with each generation resulting in higher binding affinity scores. The binding affinities and interactions between the selected candidates and the SARS-CoV-2 main protease are predicted using a molecular docking simulation using AutoDock Vina. The compounds selected have a strong interaction with the key MET 165 and Cys145 residues. Molecular dynamics (MD) simulations were run for 150ns to validate the docking results on the top four ligands. Additionally, root-mean-square deviation (RMSD), root-mean-square fluctuation (RMSF), and hydrogen bond analysis strongly support these findings. Furthermore, the MM-PBSA free energy calculations revealed that these chemical molecules have stable and favorable energies, resulting in a strong binding with Mpro's binding site. This study's extensive computational and statistical analyses indicate that the selected candidates may be used as potential inhibitors against the SARS-CoV-2 in-silico environment. However, additional in-vitro, in-vivo, and clinical trials are required to demonstrate their true efficacy.
Keywords: SARS-CoV-2, Deep learning, Main protease, Molecular docking, Molecular dynamic simulation
1. Introduction
The recent COVID-19 pandemic, caused by the Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2), has posed numerous challenges to global health and other facets of human life. COVID-19 appears to be our generation's worst infectious disease pandemic, based on the number of infections, fatalities, and unprecedented demand for healthcare services [1]. Fever, cough, pneumonia, myalgia or fatigue, and complex dyspnea were among the most frequently reported symptoms in the study cases; however, headache, diarrhea, runny nose, hemoptysis, and phlegm-producing cough were reported as less frequently occurring symptoms [2,3].
Coronavirus genomes are translated into two types of proteins within the host cell: structural proteins such as Spike (S), Envelope (E), Matrix (M), and Nucleocapsid (N), and nonstructural proteins such as 3-C like protease (3CLpro, nsp5), and RNA Dependent RNA Polymerase (RdRp, nsp12) [4]. Due to its critical role in the viral life cycle and post-translational processing of replicas polyproteins, the main protease (Mpro) of SARS-CoV-2 has received considerable attention [5,6]. Mpro and other nonstructural proteins (papain-like protease, helicase, RNA-dependent RNA polymerase) and the spike glycoprotein are required for virus-host cell receptor interactions during viral entry [7]. The main protease (also known as 3C-like protease) is one of the most attractive antiviral drug targets, particularly in the design and development of SARS drugs [8,9]. As a result, inhibiting the activity of this enzyme would prevent the transcription of the virus inside infected cells [10].
The majority of researchers concentrate on repurposing existing antivirals and drugs to combat SARS-CoV-2 [[11], [12], [13], [14], [15], [16]]. Although some of these repurposed drugs, such as Kaletra, eliminated COVID-19 symptoms in early clinical trials, they demonstrated no benefit in patients with severe symptoms [17]. Thus, it is necessary to develop more effective and capable new chemical entities (NCEs) to target the 3CL protease in the virus specifically. Thanks to the most recent technological advancement in AI, scientists can now extract existing knowledge and use it to investigate the virtually limitless chemical space and create new small molecules with the necessary biological and physicochemical properties to treat various diseases [18,19]. It is worth noting that artificial intelligence-based methods have recently been used to synthesize novel molecules with antibacterial properties [20].
Despite these efforts, additional validation techniques such as molecular dynamics simulation, combined with AI and stochastic-based methods such as docking simulation, are required to increase the efficiency of currently used pipelines. This study proposed a de novo pipeline to generate novel drug-like chemical compounds to accomplish this goal. This was accomplished by training and fine-tuning an LSTM network over several generations to generate ligands with increasing binding energy with each generation. Molecular docking was used to determine the binding affinities and molecular interactions of various ligands with the SARS-CoV2-Mpro. Furthermore, molecular dynamic simulations followed by MM-PBSA analysis were performed on the top-ranking representative structures identified through cluster analysis of docking simulations to validate the proposed candidates' interaction and binding affinity. The resulting compounds can be submitted to biological evaluations as lead inhibitors for the treatment of SARS-CoV-2.
2. Methods and computational details
2.1. Data preparation
The datasets used to train the deep neural network models were obtained from ChEMBL and ZINC. The SMILE format collected 2.1 million compound structures from the ChEMBL dataset [21] and 1.9 million molecular structures from Moses [22], a subset of the ZINC dataset. Following that, the SMILEs with lengths ranging from 34 to 128 were extracted. After removing stereochemistry, salts, and molecules containing undesirable atoms or groups, a total of 2.5 million SMILEs was used to train the neural network.
2.2. Training the network with deep learning algorithms
The LSTM_Chem network was used to generate drugs, with its weights trained on ChEMBL. This method is used for de novo molecular design using generative recurrent neural networks (RNNs) referred to as long short-term memory (LSTM) cells. This computational model accurately captures the syntax of molecular representations in terms of SMILEs strings and generates molecules similar to the training data. Later, to adapt the model to the proposed method, the model parameters were changed to allow for the generation of new chemicals using the data described in the previous section. Transfer learning is the process of adapting a trained model to a new dataset.
A batch size of 512 SMILEs was specified in the network configuration, thereby enabling the network to be used during the training session. The network configuration was set to 30 iterations, but validation loss remained constant (<0.0005) from iteration 21 onwards, increasing rapidly in later iterations. As a result, the network was stopped during this iteration to avoid overfitting.
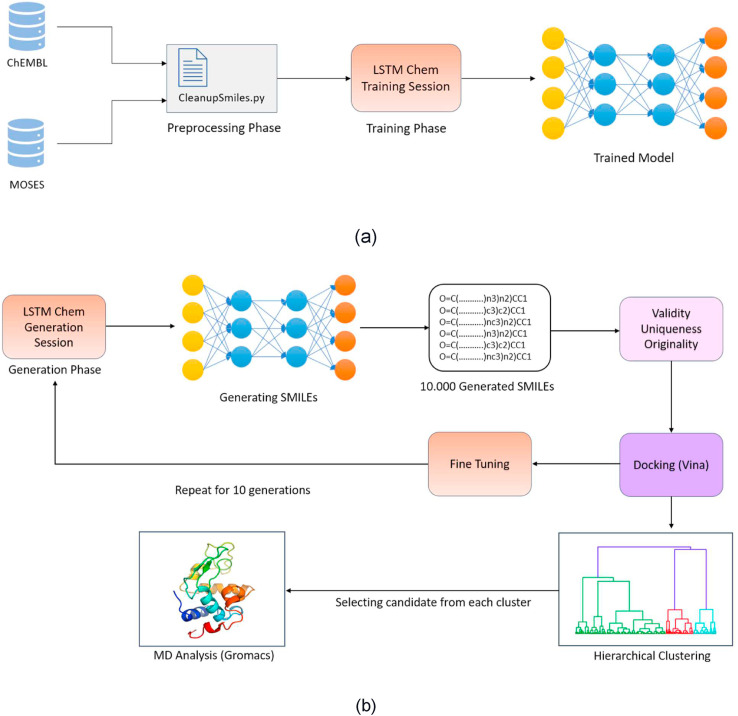
Following the training phase, which took nearly five days on an Nvidia GeForce RTX 2080Ti, the weights obtained were saved in an hdf5 file. The training phase is summarized in Fig. 1 (a).
Fig. 1.
(a). A block diagram illustrating the network's training phase. (b) A block diagram depicting the generation, fine-tuning, and evaluation sessions.
2.3. De novo structure generation
The modified LSTM_Chem network generated new molecular compounds similar to those in this study dataset. A total of 10,000 new molecules were created during the first generation. The generated SMILEs were checked for uniqueness, validity, and originality using the RDKit library.
‘Validity’ determines whether the deep learning system's generated SMILEs are indeed valid SMILE candidates for a molecule. ‘Uniqueness' determines the compound's uniqueness within the dataset. In other words, the compound is not being a duplicate. ‘Originality’ determines whether the generated SMILEs are unique or do not appear in any datasets. Finally, SMILEs that satisfy all three criteria are retained as eligible molecules (Supplementary Table 2).
Afterward, 30 SMILEs were randomly selected from the validated set, and their Tanimoto similarity to other validated SMILEs was calculated. The threshold was primarily set to 0.05. If there were similar insufficient compounds, the threshold was increased incrementally until 1000 SMILE candidates were found. Additionally, a new list of HIV Inhibitor SMILEs was added to the existing list. Ultimately, the list was docked with the 6LU7 protein.
2.4. Molecular docking
The RCSB Protein Data Bank (PDB ID: 6LU7) was accessed to download the crystal structure of the SARS-CoV-2 main protease (SARS-CoV-2 Mpro) in complex with the inhibitor N3 [10]. AutoDockTools-1.5.6 was used to prepare the protein by removing water atoms and the native ligand from the active site, adding polar hydrogen atoms and charges, and converting the protein and ligand PDB files to a PDBQT format. A molecular docking study was performed using the AutoDock Vina virtual screening program (version 1.1.2) to determine the protein's interacting residues with specific ligands [23]. Re-docking 6LU7 with its crystallographic inhibitor was performed to validate docking studies.
The grid box's center points and dimensions were set to target the active site of the main protease protein [24], with the center at X: −11.493, Y: 10.857, Z: 62.522 (Å), and the grid box's dimensions set to X: 30, Y: 30, and Z: 30 (Å). The binding affinities of the compounds were calculated and ranked according to their highest negative values, which corresponded to their best binding affinities [25]. Docking and re-docking results at each docking position were analyzed to confirm the interactions of compounds with the Mpro active site residues. 3D and 2D representations of protein-ligand complexes were visualized using Chimera and Discovery Studio Client 2017 software, respectively.
2.5. Evaluation and tuning
At this stage, a genetic algorithm was used to fine-tune the network. After docking, the SMILEs were sorted according to the Binding Affinity criterion. A total of 35 initial SMILEs were chosen and added to a new list. Following that, these 35 SMILEs were compared to the rest using the Tanimoto similarity criterion. Furthermore, the five SMILEs that possess the slightest resemblance to the new list were added. Moreover, the weight of each molecule was determined. A weight adjustment score was defined in order to prioritize the molecules with the smallest mass.
Consequently, the weight-adjusted score for each SMILE was calculated to prioritize the molecules with lighter compounds. The obtained list was sorted by Weight Adjustment Score; the first five entries were selected and added to the new list. Inspired by the fundamental genetic algorithms for reinforcing random mutations, another five SMILEs were randomly selected and added to the list from the basic generation (generation 0). In total, 50 SMILEs were chosen based on these criteria.
The 50 SMILEs were used to fine-tune the LSTM network. Over ten iterations, the network was trained using this list. After updating the weights, 10,000 brand-new SMILEs were generated for the subsequent generation, validity, uniqueness, and originality were calculated, and the docking and fine-tuning steps were repeated for a total of ten generations, as illustrated in Fig. 1(b).
All ten generations were combined and sorted based on their binding affinity score in this step. Given the total number of generated ligands, 20,866, only those with a binding affinity score of less than ten were selected. Following that, the list was expanded to include HIV inhibitors and Remdesivir. Then, all of the SMILEs and their associated properties were saved in a file, along with the ligands used to calculate binding affinity. AutoDock Vina was used for performing the binding operation. After calculating the output binding affinity scores, the average score and the score with the lowest value were chosen. The degree of similarity between all ligands and Remdesivir and HIV inhibitors was then determined.
2.6. Hierarchical cluster analysis
Due to the time required to simulate each ligand using the Gromacs software [26], the clustering operation is typically performed to examine the representative of each cluster. As a result, a fingerprint was extracted for each ligand in this phase, and then Tanimoto similarities between all the fingerprints were computed, resulting in a 4158*4158 upper triangular matrix. The similarity between the two fingerprints was calculated as:
| (1) |
denotes the Tanimoto coefficient for two fingerprints A and B. and denote the bits set in each fingerprint, respectively. represents the bits set that are common in both fingerprints. The average linkage method was then used to calculate the distance between the object pairs, where each pair included an object from each cluster. The distance of a pair was computed as:
| (2) |
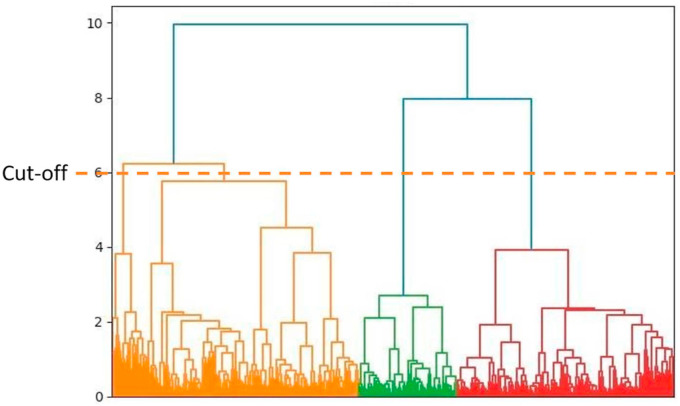
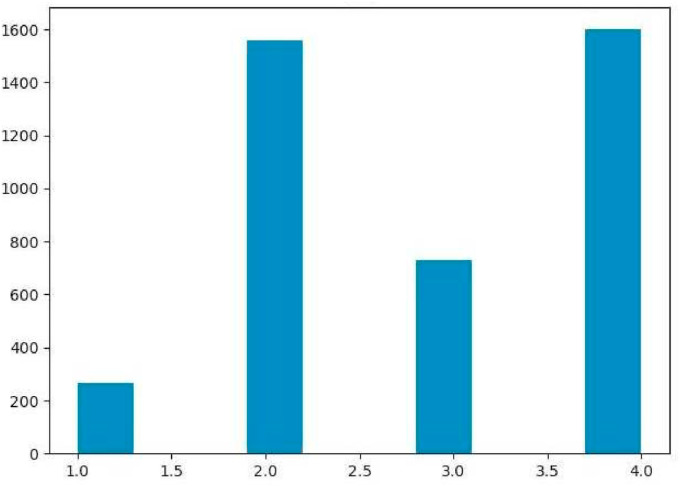
Where denotes the sum of all object pairs between two clusters, h, and k. and represent the total number of objects in each cluster. The clusters with minimum distance were merged during each step, forming a new cluster. The hierarchical clustering process was continued until all clusters were merged as one. According to Fig. 2 , a dendrogram chart was created for the clustering process. A cut-off at a distance of six was used to bring the process to a halt on four clusters. The total number of fingerprints in each cluster is depicted in Fig. 3 . The top-ranked potential compounds in each cluster were chosen for further analysis based on their sorted binding affinities. The optimal pose of protein-ligand complexes determined through docking simulations was used as the initial coordinates for MD simulations.
Fig. 2.
The outcome of the hierarchical clustering process. By setting the cut-off distance to six, similar ligands were divided into four clusters.
Fig. 3.
The total number of fingerprints in each cluster.
2.7. Molecular dynamics simulation
The molecular dynamics simulation of the best docking pose of four top-ranked compounds in complex with Mpro was accomplished using the GROMACS simulation package (version 5.1.2) running on a Linux GPU server. Amber99sb force field was utilized to generate force field parameters and define the atom types. Using an ACPYPE web server, the topology and coordinate files for the selected ligands and native ligands of the main protease (N3) and Remdesivir were created [27,28].
The protein complexes were solvated in the cubic box with TIP3P water molecules [29] and then neutralized by adding 0.15 mol/L Na/Cl-ions. The system was minimized using the steepest descent algorithm. The systems were then equilibrated using NVT and NPT with 100 ps steps, respectively, using a V-rescale Berendsen thermostat and a Parrinello-Rahman. The heating of the systems was gradually increased from 0 to 300 K, and the pressure of the systems was set to 1 atm for the NVT and NPT ensembles, respectively.
The SHAKE algorithm was used to constrain all systems' covalently bonded hydrogen atoms during the process [30]. Periodic boundary simulations used the particle mesh Ewald (PME) method, and long-range interactions used PME. The final step of molecular dynamics simulation took 150 ns, with each step lasting 2fs in the periodic boundary conditions. Following that, trajectories were saved, and the data analyzed. The root mean square deviations (RMSD) and residue root mean square fluctuation (RMSF) of each snapshot were calculated to determine the equilibrium time and stability of the protein backbone atoms. Finally, the trajectories results were subjected to VMD to analyze the ligand's binding mode during simulation [31]. The Pymol graphical software (version 2.3.2) was applied to generate the ligand-protein conformational analysis [32].
2.8. Binding free energy and per-residue decomposition calculations
One way to validate the intermolecular strength of interactions is to calculate the binding free energies of protein-ligand complexes, which provide insight into the relative importance of various chemical energies contributing to overall stability. The Molecular Mechanics Poisson–Boltzmann Surface Area (MM-PBSA) method is a relatively simple technique for quantifying the binding free energies of ligands docked to a receptor [33]. The thermal mmgbsa.py python script was used to run MM-PBSA over a 150-ns period, and the average computed binding energy was reported along with the standard deviation. In order to estimate the non-polar energy contributions, the solvent-accessible surface area (SASA) was calculated.
In general, the following equations can be used to calculate the binding free energy of a protein with a ligand in a solvent:
| (3) |
Where Gprotein and Gligand denote total free energies of the isolated protein and ligand in the solvent and Gcomplex denotes the total free energy of the protein−ligand complex, respectively. Furthermore, the free energy for each entity can be expressed as:
| (4) |
Where x denotes the protein or ligand or protein−ligand complex. The average molecular mechanics' potential energy in a vacuum is defined as ⟨EMM⟩. TS represents the entropic contribution to the free energy in a vacuum, where T and S denote the temperature and entropy, respectively. ⟨Gsolvation⟩ denotes the free energy of solvation [34,35].
2.8.1. Molecular mechanics potential energy
EMM is the vacuum potential energy that includes both bonded and unbonded interactions. It is calculated as the following term using the molecular mechanics (MM) force-field parameters:
| (5) |
Where Ebonded denotes the bonded interactions comprising dihedral, angle, bond, and improper interactions. The nonbonded interactions (Enonbonded) consist of both electrostatic (Eelec) and van der Waals (EvdW) interactions and are modeled by a Coulomb and Lennard-Jones (LJ) potential function, respectively. Where ΔEbonded is always considered as zero [36].
2.9. Free energy of solvation and contribution of residues to the binding energy
The free energy of solvation is the energy required to transfer a solute from a vacuum to a solvent. In the MM-PBSA approach, the free energy of solvation is calculated using an implicit solvent model via the following:
| (6) |
Where Gpolar and Gnonpolar denote the electrostatic and non-electrostatic contributions to the solvation free energy, respectively. The electrostatic term, Gpolar, is calculated by solving the Poisson−Boltzmann (PB) equation [37].
Additionally, the energetic contribution of the amino acids involved in inhibitor binding was calculated using per-residue decomposition analysis. The binding free energy decomposition and overall binding energy of the protein-ligand complex were calculated using the g_mmpbsa tool [34]. For MM-PBSA calculations and individual amino acid contributions, the Python scripts “MmPbSaStat.py” and “MmPbSaDecomp.py” were used.
2.9.1. The radius of gyration (Rg)
The radius of gyration was investigated as a function of the protein structure's compression and the protein's changes over the simulation time [38].
The Rg is calculated using the following formula:
| (7) |
Where N denotes the number of protein atoms, ri and rj denote the coordinates of an atom i and the center of mass, respectively.
Moreover, the energetic contribution of the amino acids involved in inhibitor binding was determined using per-residue decomposition analysis. Binding free energy decomposition was performed using the g_mmpbsa tool. This tool decomposed the total binding energy of the protein-ligand complex. The Python scripts “MmPbSaStat.py” and “MmPbSaDecomp.py” were used to calculate the MM-PBSA and the contribution of individual amino acids [34].
2.10. Principal component analysis (PCA) on MD simulation data
The “gmx covar” was used to generate the eigenvalues and eigenvectors by computing and diagonalizing the covariance matrix. Then, the corresponding eigenvectors were analyzed using GROMACS' “gmx anaeig” tool [39]. The protein fluctuations' covariance matrix was diagonalized to obtain the eigenvalues. The cosine content of the first three principal components was determined to calculate the statistical significance of the trajectories' convergence. When the first principal component is similar to a cosine with a half-period, it has been demonstrated that the sampling is far from converging [40]. In this regard, the probability of conformational sampling convergence increases as the cosine similarity decreases.
3. Discussion
3.1. Deep learning approaches in drug design
Due to the severity and impact of the COVID-19 pandemic, various approaches based on artificial intelligence (AI) and deep neural networks (DNN) have been developed to combat the disease [[11], [12], [13], [14], [15], [16]]. However, no definitive cure for the disease has been discovered to date. As a result, the current study focused on the de novo design of novel antiviral agents for SARS-CoV-2. Deep learning methods such as ConvLSTM and CNN were used to predict binding affinities, and Drug-Target Interaction (DTI) values between existing compounds and the SARS-CoV-2 Mpro protein [41,42]. The KIBA dataset, which contains 52,498 chemical entities, was used as a repository for these studies.
Since deep learning methods require a large amount of data for practical training and updating model weights, these studies lack sufficient data. Through ChEMBL, a reinforcement learning method was proposed to generate potential inhibitors of the SARS-CoV-2 3CL protease [43]. Another study used an LSTM network to generate inhibitors of SARS-CoV-2 Mpro and a classifier to predict bioactivity [44]. Virtual screening was used exclusively in these reports to evaluate the predicted compounds in the protein's binding site.
Deep learning techniques are excellent at detecting and learning similarities, and deep models can also learn to generate similarities to what they have learned. Because this study aimed to identify a series of compounds capable of treating COVID-19, a deep neural network was trained using a large dataset of chemical compounds. The fully trained network was used to generate new compounds similar to those found in the dataset. Valid, drug-like compounds were chosen for each generation.
LSTMs are typically used to predict sequences. LSTM_Chem derives its knowledge from relatively large databases containing molecular structures in the form of SMILE and generates a series of similar samples with a high degree of variation. A total of 10000 of these compounds are generated, with 1000 being chemically valid and unique, even though SMILE is a sequence and the machine is unaware of any chemical rules. A concerted effort has been made to repurpose this tool for use in the treatment of COVID-19. To this end, following validation, each SMILE was docked with the SARS-CoV-2 main protease to identify those with the highest binding energy, which were then used to improve the network by fine-tuning it after each generation.
A recent study chose the binding affinity score between the novel generated ligands and the SARS-CoV-2 3CL protease as the primary measurement criterion. Additionally, a previous study on novel generated ligands achieved a maximum binding affinity score of −38.07 kJ/mol, whereas selected ligands' maximum binding affinity score was −54 kJ/mol (the complete table is available in supplementary table l).
3.2. In-silico studies
Computer-aided drug design (CADD) has emerged as a valuable approach in modern drug discovery due to its ability to reduce the cost and labor associated with the process significantly. Therefore, due to the reliability of their predictions, molecular docking, molecular dynamics, QSAR, and ADMET tools have become critical components of the CADD [45]. Molecular docking predicts the predominant binding modes between a ligand and a protein by hypothesizing how ligands interact in a protein's active site and thus rank candidate ligands [46]. The Mpro of SARS-CoV-2 was docked against selected generated hit compounds in this study. Four candidates were selected from among them based on their high binding affinity and clustering analysis.
3.3. Molecular docking simulation
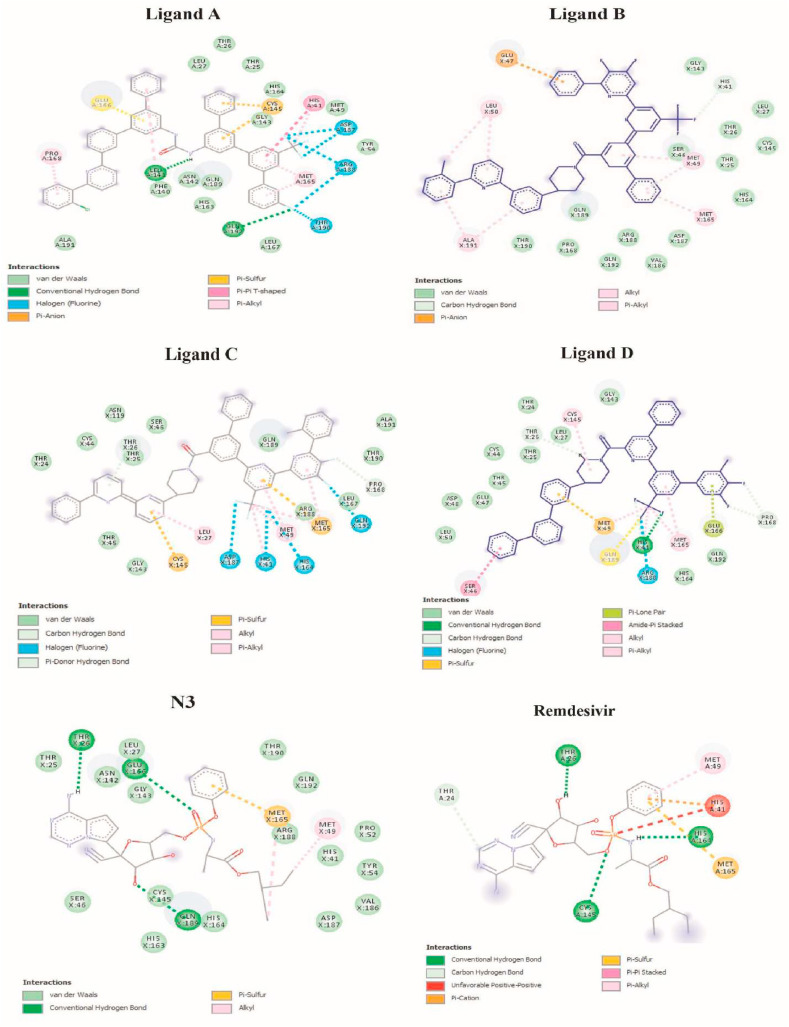
The molecular docking simulation has been widely used to predict bioactive compounds and repurpose drugs against various disease- and infection-specific drug targetable proteins [46]. This study used this method to predict the potential inhibitory effect of selected generated bioactive compounds on SARS-CoV-2 infection via Mpro inhibition. The compounds' binding affinities were calculated and ranked according to their higher negative values. All compounds were classified into four clusters following cluster analysis, and the top candidate in each cluster was chosen for further analysis. When compared to the N3 inhibitor and Remdesivir drug as references, the four top compounds demonstrated significant docking conformations with binding affinity energies > -31.8 kJ/mol in the active pocket of Mpro. Table 1 summarizes the comparative docking results, including the binding energy and intermolecular interaction profile in the active site of Mpro between the selected compounds and the native ligand N3. Fig. 4 depicts the occupancy of compounds in the active site of 6LU7.
Table 1.
Docking results for the top four compounds and native ligand (N3), as well as Remdesivir, within the binding pocket of the SARS-CoV-2 main protease (PDB ID: 6LU7).
| No | Compound | Entry Name | Docking score (KJ/mol) | Interacting residues |
|---|---|---|---|---|
| 1 | Ligand A* | 1-(3-(6-(2-chlorophenyl)pyridin-2-yl)-[1,1':3′,1″-terphenyl]-5′-yl)-3-(3‴-fluoro-5''-(trifluoromethyl)-[1,1':3′,1'':3″,1‴-quaterphenyl]-5′-yl)urea | −46.024 | – |
| 2 | Ligand B** | (4-([1,1':3′,1″-terphenyl]-3-yl)piperidin-1-yl)(3-(4′-chloro-5′-fluoro-6′-phenyl-4-(trifluoromethyl)-[2,2′-bipyridin]-6-yl)-5-(pyridin-2-yl)phenyl)methanone | −50.208 | SER 46/H-donor |
| 3 | Ligand C*** | (4-(3-(6-(2-chlorophenyl)pyridin-2-yl)phenyl)piperidin-1-yl)(6-(3″-fluoro-5-(trifluoromethyl)-[1,1':3′,1″-terphenyl]-3-yl)-4-phenylpyridin-2-yl)methanone | −50.6264 | MET 165/H-donor GLY 143/H-acceptor CYS 145/H-acceptor CYS 145/H-acceptor |
| 4 | Ligand D**** | (4-(3-(6-(2-chlorophenyl)pyridin-2-yl)phenyl)piperidin-1-yl)(6-(3″-fluoro-5-(trifluoromethyl)-[1,1':3′,1″-terphenyl]-3-yl)-4-phenylpyridin-2-yl)methanone | −51.8816 | MET 165/H-donor CYS 145/H-acceptor CYS 145/H-acceptor THR 26/pi-H ASN 142/pi-H GLU 166/pi-H |
| Inhibitor from crystal structures | ||||
| 5 | N3 | n-[(5-methylisoxazol-3-yl)carbonyl]alanyl-l-valyl-ñ1∼-((1r,2z)-4-(benzyloxy)-4-oxo-1-{[(3r)-2-oxopyrrolidin-3-yl]methyl}but-2-enyl)-l-leucinamide | −31.7984 | HIS 164/H-donor GLN 189/H-donor MET 165/H-donor ASN 142/H-donor THR 26/H-acceptor ASN 142/H-acceptor |
| 6 | Remdesivir | 2-ethylbutyl (2S)-2-[[[(2R,3S,4R,5R)-5-(4-aminopyrrolo[2,1-f][1,2,4]triazin-7-yl)-5-cyano-3,4-dihydroxyoxolan-2-yl]methoxy-phenoxyphosphoryl]amino]propanoate | −27.196 | GLN 189/H-donor MET 49/H-donor GLN 189/pi-H |
* cluster1-compound 1.
** cluster2-compound 1.
*** cluster3-compound1.
**** cluster4-compound1.
Fig. 4.
The 2D binding mode and molecular interaction of selected ligands in Mpro's active site (PDB ID: 6LU7).
Investigating the docking results revealed that four of the top selected molecules could be used as potential Mpro inhibitors. These molecules had a higher affinity for binding to the active site of Mpro than the native ligand (N3). Notably, certain compounds docked with SARS-CoV-2 Mpro exhibited one or more types of interaction with the catalytic site, including H-bond acceptor, H-bond donor, and pi-H interaction; thus, they can be proposed as inhibitors of the main protease. Shafi Mahmud et al. (2020) investigated plant-derived compounds as potential inhibitors of the SARS-CoV-2 main protease using virtual screening and molecular dynamics simulations [47]. In comparison to the study results, the docking analysis of this study revealed higher bonding energy.
3.4. Molecular dynamics simulation and analysis of MD trajectories
Molecular dynamics simulations can be used to investigate the docked complexes and crystal protease complexes with the inhibitors N3 and Remdesivir as additional positive references to determine their stability and intermolecular interaction profiles over time [48].
RMSD, RMSF, the number of hydrogen bonds, the radius of gyration (Rg), and contribution energy of the residue are obtained from MD simulations using the GROMACS routines. The graphs generated by all of these analyses are frequently used to predict the affinity of selected compounds for the active site of the Mpro protein.
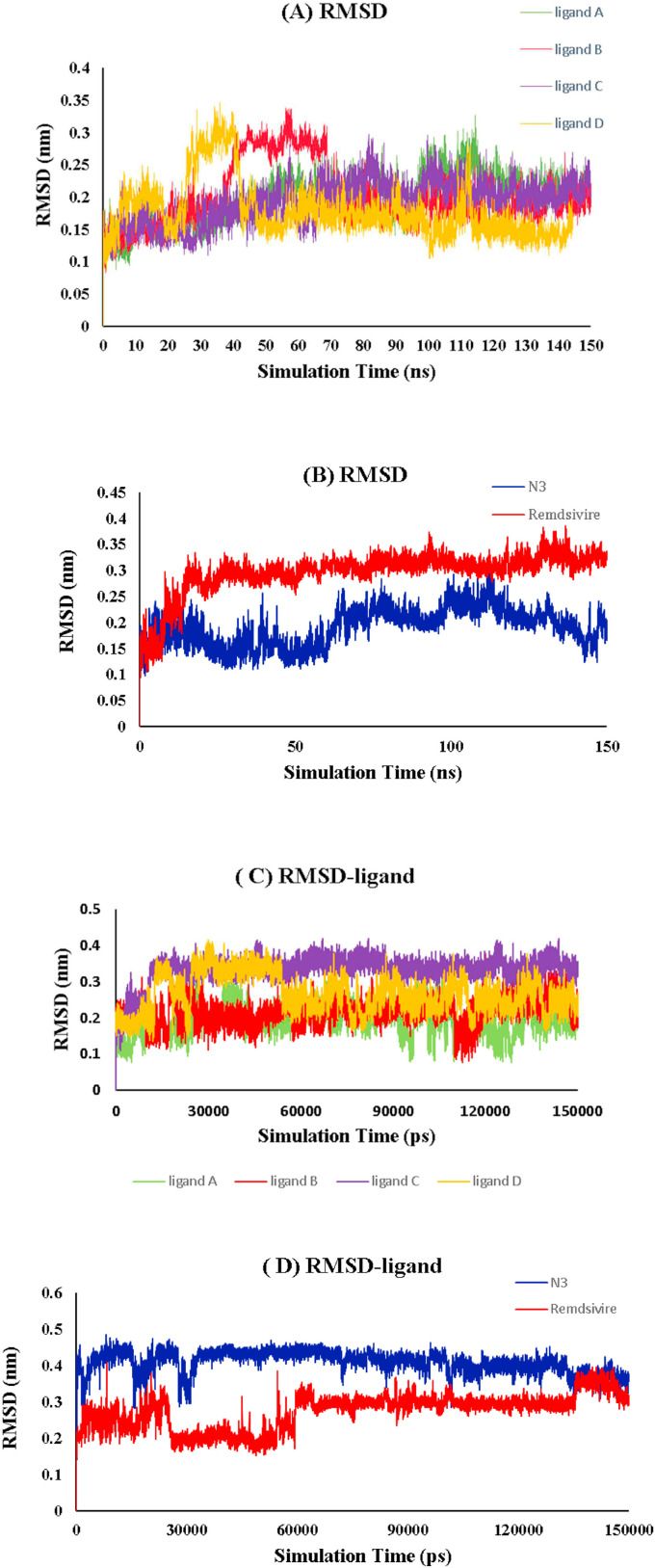
The present study used molecular dynamics (MD) simulation to predict the intermolecular interactions between the atoms of four selected compounds located within the Mpro active site residues. The root mean square deviation (RMSD) between the backbone of protein atoms and the primary confirmation was calculated for each complex to determine the system's stability and quantify the degree of conformational change in the docked complexes' ligands and protein. The plot of the RMSD for all simulation systems is shown in Fig. 5 . The RMSD of all systems was between 0.13 and 0.3 nm. As illustrated in Fig. 5 (A), after nearly 60ns from the start of the simulation, the RMSD of all the systems reached a plateau with minimal fluctuation, indicating that the selected compounds were stabilized and fit into the protein's active site. The RMSD of Mpro complexes docked with ligands is significantly more stable than the native co-crystal ligand Inhibitor N3 and Remdesivir.
Fig. 5.
(A) RMSD of the protein backbone in complex with four ligands. (B) RMSD of the protein backbone in complex with N3 and Remdesivir at 150 ns MD simulations. (C) RMSD of the four ligands. (D) RMSD of N3 and Remdesivir in complex with Mpro.
Fig. 5 (A) illustrates that the ligand D complex has the lowest RMSD of all the complexes. The fluctuation of ligand D was observed between 30 and 40ns and remained stable at 0.15 nm until the simulation time expired. On a 150ns time scale, ligand B exhibited fluctuations at two distinct intervals. The first stable conformation lasted between 6 and 40ns, while the second lasted between 70 and 150ns. This value eventually decreases, indicating that ligand B may alter the conformation of Mpro in the binding region. The RMSD of ligand A varied between 0.12 and 0.23 nm throughout the simulation and remained constant at 0.23 nm, but there was a limited fluctuation between 100 and 120ns (RMSD >0.25 nm).
During simulation analysis, similar ligand A and ligand C conformations exhibited different behavior with protein binding regions. This observation was confirmed by the RMSF plot of their local changes in residue levels. According to Fig. 5 (B), the Remdesivir-protein complex's RMSD reached 0.3 nm from 0 to 13 ns; however, this value significantly increased after 150ns, reaching 0.29 nm. When the RMSD of the N3 complex was determined, a steady increase in the RMSD after 50ns was observed (average RMSD >0.18 nm).
The RMSD of each ligand snapshot relative to the initial structure was calculated to monitor ligand stability in all studied systems throughout the simulation. As illustrated in Fig. 5 (C), the average RMSD of four randomly chosen ligands (A, B, C, and D) in complex with Mpro was 0.19, 0.21, 0.33, and 0.26 nm during simulation times, respectively, with deviations from average structures ranging from 0.15 to 0.35 nm. The RMSD of the selected ligands in all systems approached a plateau, indicating that the ligands were relatively stable in the protein's active site. As illustrated in Fig. 5 (C), ligand C had the highest RMSD of all complexes, indicating that ligand C was less stable in complex with Mpro. This measure of reference flexibility was analyzed using RMSD and is depicted in Fig. 5 (D). The RMSD of N3 and Remdesivir tend to converge after 30 and 60ns, respectively, indicating that the ligands were stable and exhibited slight fluctuation. N3 and Remdesivir had an average RMSD of 0.40 and 0.27 nm, respectively. The results indicated that Remdesivir possessed a high degree of stability in the active packet.
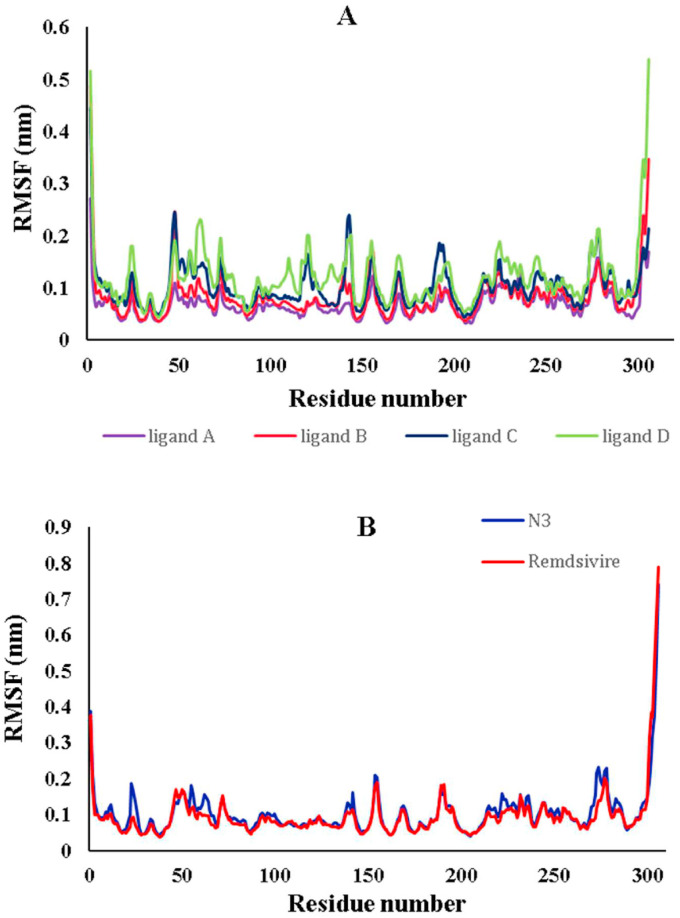
The RMSF of Mpro's backbone residue was calculated to account for amino acid residue fluctuation. Each residue's flexibility was examined to understand better the extent to which ligand binding affects protein flexibility during the equilibrium simulation time. The lowest RMSF values for amino acid residues indicated the receptor's stability, rigidity, and compactness. The RMSF values for several selected complexes have been calculated and are depicted in Fig. 6 (A) . The results indicated that the protein backbone with selected ligands did not affect the behavior of protein structures during the equilibrium simulation time. The RMSF plot established that no changes in the structure of Mpro occurred due to the binding of the selected compounds. No amino acid residue had an RMSF value greater than 0.2 nm, based on the insignificant changes observed for involved residues in the active site. The average RMSF values for Mpro amino acid residues were 0.065, 0.076, 0.093, 0.111, 0.104, and 0.098 nm for selected ligand A, ligand B, ligand C, ligand D, native ligand Inhibitor N3, and Remdesivir, respectively. The results indicated that selected ligands had lower RMSF values than the native ligands Inhibitor N3 and Remdesivir. As illustrated in Fig. 6 (A), the binding of ligand A results in the protein being the most flexible in all directions compared to the protein and the other complexes. Fig. 6 (B) indicates that the primary protease formed stable complexes with reference compounds [47].
Fig. 6.
The RMSF values of the protein's backbone throughout the simulations, where the ordinate is the RMSF (nm), and the abscissa is the residue number.
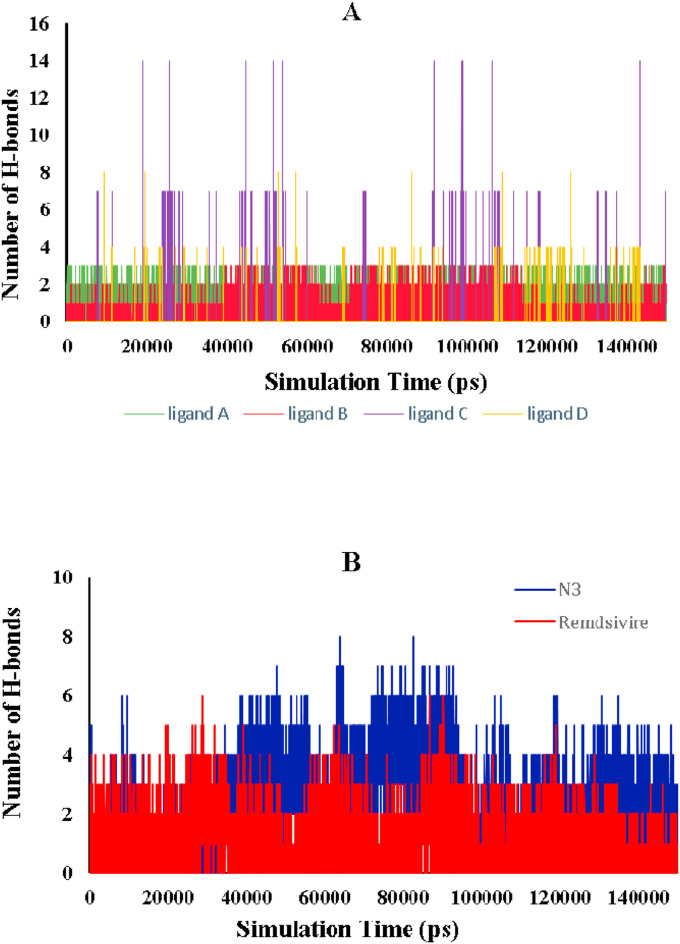
Hydrogen bonding is critical for determining the binding strength of ligands to proteins. The number of hydrogen bonds formed between the backbone of the protein and the ligands was calculated throughout the simulation time because they contribute to conformational stability. According to Fig. 7 (A & B), the average number of hydrogen bonds formed by ligands A, B, C, D, N3, and Remdesivir was 1.13, 0.71, 0.1, 0.09, 2.28, and 1.05, respectively. Fig. 7 (A) illustrates that in equilibrium simulation time, ligands A and B had a range of hydrogen bonds between 0 and 3, whereas ligands C and D exhibited changes in bonding within the active site of Mpro. As illustrated in Fig. 7 (B), the native co-crystal ligand N3 exhibited a range of hydrogen bonds ranging from 0 to 6 throughout the simulation time. The Remdesivir complex had fewer hydrogen bonds than N3, which follows a pattern almost identical to the ligand A complex. The docking and MD simulation analyses revealed that ligand A had the highest affinity for Mpro compared to the other selected ligand. As a result, all selected ligands formed fewer hydrogen bonds in the active site of Mpro than the native ligand.
Fig. 7.
(A) The total number of H-bonds formed during the simulation of the selected ligand with Mpro residue. (B) The total number of H-bonds formed during the simulation of N3 and Remdesivir with Mpro residue.
The binding free energy between selected ligands in the active site and the main protease was calculated using MM-PBSA. Table 2 summarizes the results of the MM-PBSA analysis for nonbonded interaction energies (van der Wall, electrostatic, polar solvation, and SASA) between selected ligands, N3 and Mpro, over a 150ns simulation time. More negative binding free energy values indicated a stronger interaction and increased affinity between receptor and ligands. The estimated binding free energy of docked ligands within Mpro was negative after a 150ns MD simulation, indicating that the ligands had a high affinity for and binding to the active site or receptor. Furthermore, decomposition of the binding energies revealed a more significant role for the vdW energy in the inhibitory effect of the selected ligands [49].
Table 2.
van der Waals, electrostatic, polar solvation, SASA, and binding energies of the selected ligands and native ligand (N3) into the binding pocket of Mpro (PDB ID: 6LU7).
| System | Energy (KJ/mol) ± SD |
|||||
|---|---|---|---|---|---|---|
| VdW | Elec | Polar solvation | SASA | Binding | ||
| Ligand A | −277.826 ± 32.473 | −4.478 ± 4.322 | 125.767 ± 34.384 | −28.070 ± 2.439 | −184.607 ± 27.904 | |
| Ligand B | −228.892 ± 20.955 | −11.573 ± 4.478 | 120.074 ± 43.335 | −23.942 ± 1.634 | −144.333 ± 43.092 | |
| Ligand C | −220.831 ± 25.535 | −8.841 ± 4.901 | 124.249 ± 23.515 | −22.764 ± 1.900 | −128.187 ± 14.640 | |
| Ligand D | −253.485 ± 13.226 | 5.447 ± 3.464 | 111.672 ± 12.354 | −24.114 ± 1.124 | −160.480 ± 14.472 | |
| N3 | −139.761 ± 18.610 | −741.750 ± 100.39 | 172.546 ± 110.099 | −18.484 ± 2.598 | −727.450 ± 55.857 | |
| Remdesivir | −241.243 ± 10.678 | −9.912 ± 4.345 | 104.723 ± 6.924 | −21.736 ± 1.173 | −168.168 ± 11.980 | |
Additionally, an analysis of the vdW interaction energy between ligands and receptors revealed a more negative pattern for ligand A than for the other ligands, indicating that ligand A has a higher binding affinity in the active site of Mpro. These results indicated that the N3 bound to the Mpro active site more efficiently than four selected ligands. According to the native ligand's (N3) negative binding energies, the complex in the active site was more advantageous than the four selected ligands. Moreover, decomposition of the binding energies revealed a greater contribution of the Elec energy to the inhibitory effect of N3. In comparison, the inhibitory effect of selected ligands depended on the Vdw energy required for binding to the active site residues of Mpro.
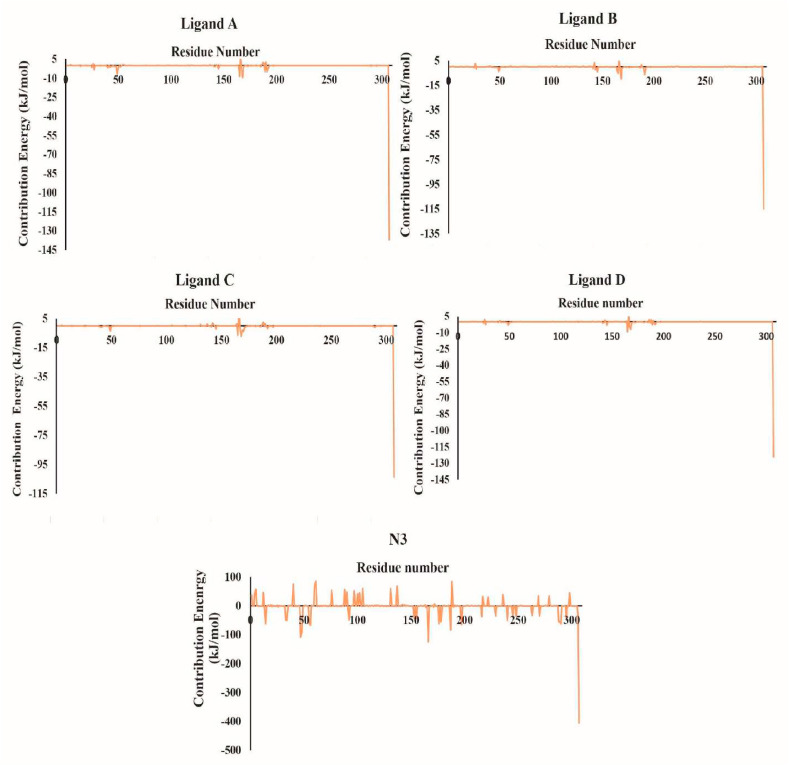
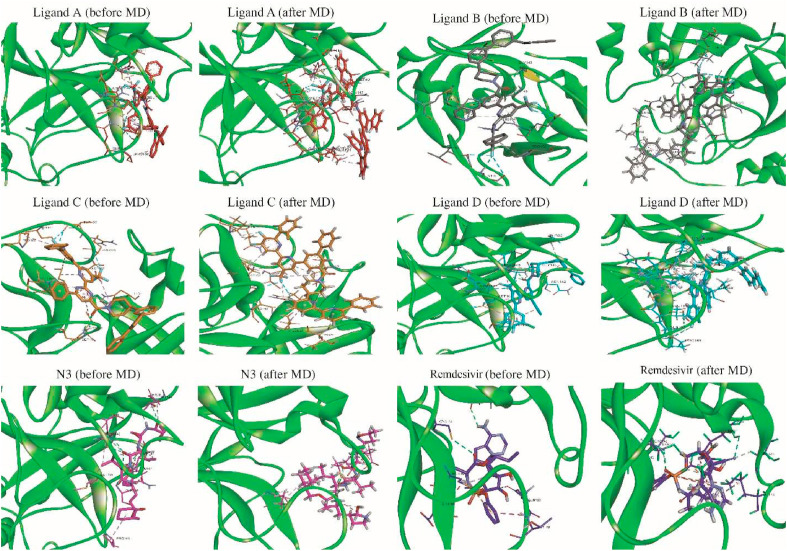
In addition, the total binding free energies between the four selected ligands and n3 and protein were decomposed into each residue to identify the key residues involved in the binding process, and the corresponding results are shown in Fig. 8 . Fig. 8 (ligands A, B, C, and D) shows that the significant beneficial energy contributions from residues SER1, LYS5, GLU14, THR26, LEU27, ARG40, MET49, CYS145, LEU141, MET165, PRO168, THR169, GLN189, THR190, GLN192, GLU290, CYS85, LYS100, GLY79, LEU30, and SER139 were prevalent in the systems studied with selected ligands. These amino acids interacted consistently with most of the four ligands chosen, and the findings corroborate the docking results. This demonstrates that the ligands make multiple contacts with critical residues in Mpro's active packet. Once the contributions of four selected ligands in the active packet are compared to the N3 system's residue contributions, some residues located and interacted in the active site of Mpro make very similar contributions. As illustrated in Fig. 8, the N3 study system contained significant favorable energy contributions from residues SER1, LYS5, LYS12, GLU14, ASP34, ARG40, ASP48, MET49, ARG76, LYS 90, ASP 92, LYS100, ARG131, GLY138, SER139, GLU166, GLU178, ASP187, GLN189, ASP197, ARG217, ASP229, VAL247, GLU240, 269LYS, GLU270, GLU288, and ARG298. Negatively correlated amino acids played a critical role in the interaction with ligands, and the common amino acids formed by decomposition residues were key residues in the active site. The residues that interact with specific ligands contribute a negligible amount, whereas the internal ligand (N3) contributes significantly more (N3). The fact that most residues in the active site contribute significantly to the overall binding energy indicates that they interact with N3 more favorably. The superimposed before and after structures of all simulated compounds from the MD simulations are shown in Fig. 9 .
Fig. 8.
Contribution of individual residues to the binding energy of each complex (ligands A, B, C, D, and N3).
Fig. 9.
Comparing the initial (a) and final (b) structures obtained from MD simulations.
3.4.1. Radius of gyration
The radius of gyration (Rg) in each simulation system was calculated to determine the structure's compactness. The lower degree of fluctuation and its consistency throughout the simulation indicates that the system is more compact and rigid. Thus, a stably folded protein is likely to maintain a relatively constant radius of gyration [50].
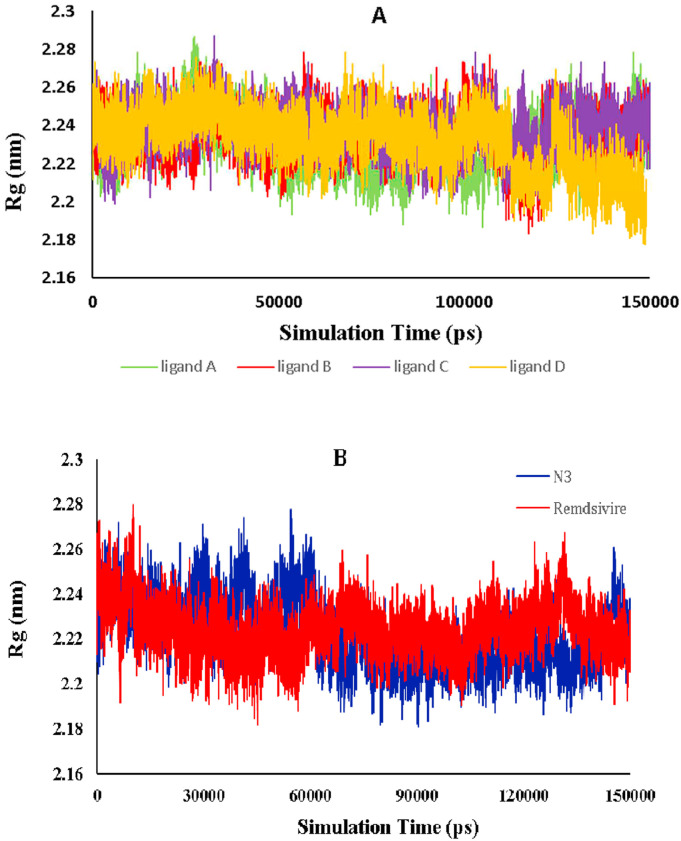
The radius of gyration for each system is plotted against simulation time in Fig. 10 to investigate the protein compactness. According to the relative curve, the N3 and Remdesivir complexes have the lowest Rg values, 2.222 and 2.224 nm, respectively, while ligand A, B, C, and D have Rg values of 2.235, 2.236, 2.242, and 2.231 nm, respectively. There was no significant difference between the Rg values obtained from the ligands-Mpro complex and those obtained from the references. The Rg of Mpro was found to be nearly stable in terms of fluctuation consistency across all simulations. This Rg value for the simulated systems indicated that the selected ligands were properly stabilized, formed compact structures, and bound to the SARS-CoV-2 protein in the same way that reference compounds do.
Fig. 10.
(A) The radius of the protein complex's gyration (Rg) profile in the presence of the selected ligands after 150000 ps MD simulations. (B) Radius of gyration (Rg) profile of the protein complex in the presence of N3 and Remdsivir after 150000 ps MD simulations. The plots display time (ps) on the x-axis and the gyration radius (nm) on the y-axis.
3.4.2. Principal component analysis (PCA) of molecular dynamics
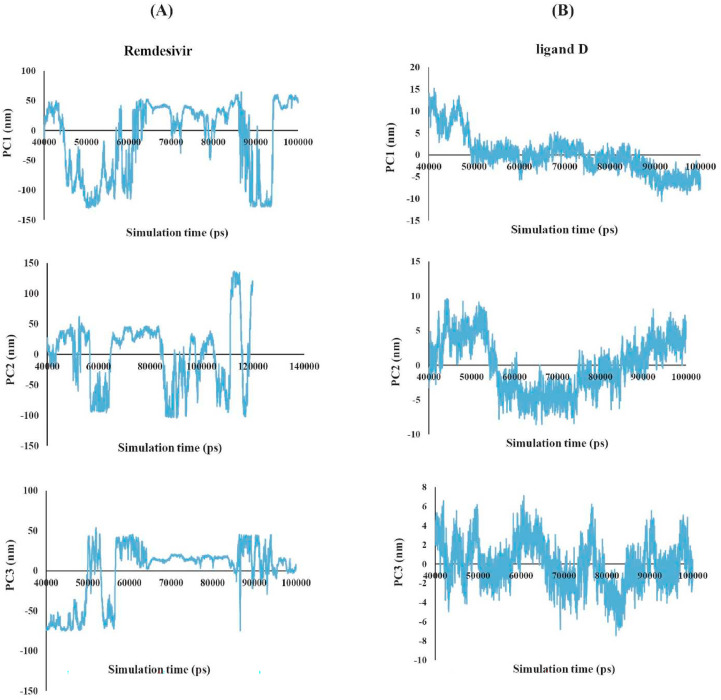
The last 60ns of simulation, where the tilt of the RMSD plot reached a stable value for the ligand 8 and Remdesivir were selected, and the cosine content of the first three principal components was computed for this sub-trajectory to assess the sufficiency of conformational sampling. Fig. 11 illustrates the values of the first three principal components for this time window, demonstrating sufficient conformational sampling.
Fig. 11.
(A) The projected trajectory of Remdesivir (positive reference) (left panel). (B) The projected trajectory of ligand 8 (right panel) for the last 60 ns subtrajectory.
4. Limitations and challenges
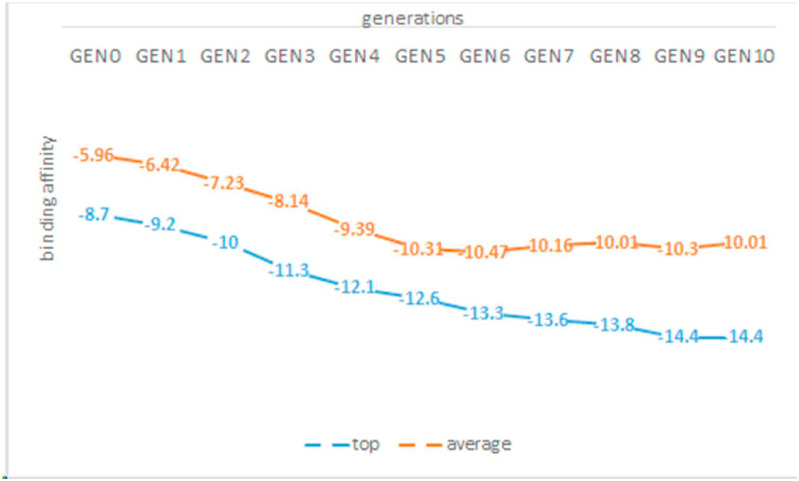
The study's limitations were the computational complexity and time constraints associated with calculating binding energy and performing simulations. Each generation stage of new compounds took approximately two to three days, and the tenth generation took approximately one month. Fig. 12 displays the minimum and average binding affinity of each generation's compounds. Given that these two values are decreasing, it is clear that improved compounds could be obtained by producing new generations. Simulating the combination of each of these ligands with the main proteases of SARS-CoV-2 is also time-consuming. Clustering operations were used to reduce this limitation to a manageable level, yielding 4158 ligands. However, only four ligands with the lowest binding affinity in each cluster were selected for the molecular dynamics simulation stage.
Fig. 12.
minimum and average binding affinity scores for the ten generations.
5. Conclusion
The current study introduced a novel pipeline for predicting drug-like compounds as COVID-19 inhibitors via a generative LSTM network. The network was fine-tuned for ten generations using a genetic algorithm, and computational screening techniques such as molecular docking and molecular dynamics simulation were used to identify the most potent inhibitory compounds against the SARS-CoV-2 main protease in comparison to the native ligand of Mpro, N3. The molecular docking simulation was used to predict the binding affinity of each cluster's representative compound at the active site of Mpro. The four top-ranked compounds, ligands A, B, C, and D, exhibited the highest affinity and strongest interactions with critical residues in the active site of the main N3 inhibitor, achieving a docking score of > −31.8 kJ/mol. MD simulations confirmed that four selected ligands were stable in the protein's active site and formed a significant number of hydrogen bond interactions with the main protease. In conclusion, based on docking binding affinity and molecular dynamics simulation trajectory analysis, the four selected ligands may be used as potential SARS-CoV-2 Mpro inhibitors to challenge the SARS-CoV-2 infection.
This study discovered several compounds that may be used to treat covid-19. While these are in-silico results, they must undergo in-vitro, in-vivo, and clinical trial phases to demonstrate their effectiveness in the real world.
Declaration of competing interest
None Declared.
Acknowledgments
This research has been partially funded by Shiraz University via Research Assistantship Award to Amir Hossein Arshia. The authors would like to thank Professor Foutse Khomh from Polytechnique Montreal, Canada and acknowledge CSC – IT Center for Science, Finland for providing computational resources.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2021.104967.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Wang C., Xiao X., Feng H., Hong Z., Li M., Tu N., et al. Ongoing COVID-19 pandemic: a concise but updated comprehensive review. Curr. Microbiol. 2021 May;78(5):1718–1729. doi: 10.1007/s00284-021-02413-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prevention Centers for Disease Control and. Novel Coronavirus. Information for Healthcare Professionals; Wuhan China: 2019. [Google Scholar]
- 3.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020 Feb 15;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elfiky A.A. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 2020 May 1;248:117477. doi: 10.1016/j.lfs.2020.117477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003 Jun 13;300(5626):1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 6.Boopathi S., Poma A.B., Kolandaivel P. Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. J. Biomol. Struct. Dyn. 2021 Jun;39(9):3409–3418. doi: 10.1080/07391102.2020.1758788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zumla A., Chan J.F.W., Azhar E.I., Hui D.S.C., Yuen K.-Y. Coronaviruses - drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016 May;15(5):327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chou K.-C. Structural bioinformatics and its impact to biomedical science. Curr. Med. Chem. 2004 Aug;11(16):2105–2134. doi: 10.2174/0929867043364667. [DOI] [PubMed] [Google Scholar]
- 9.Du Q., Wang S., Wei D., Sirois S., Chou K.-C. Molecular modeling and chemical modification for finding peptide inhibitor against severe acute respiratory syndrome coronavirus main proteinase. Anal. Biochem. 2005 Feb 15;337(2):262–270. doi: 10.1016/j.ab.2004.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020 Apr 24;368(6489):409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan H.C.S., Shan H., Dahoun T., Vogel H., Yuan S. Advancing drug discovery via artificial intelligence. Trends Pharmacol. Sci. 2019 Aug;40(8):592–604. doi: 10.1016/j.tips.2019.06.004. [DOI] [PubMed] [Google Scholar]
- 12.Encinar J.A., Menendez J.A. Potential drugs targeting early innate immune evasion of SARS-coronavirus 2 via 2’-O-methylation of viral RNA. Viruses. 2020 May 10;12(5) doi: 10.3390/v12050525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hijikata A., Shionyu-Mitsuyama C., Nakae S., Shionyu M., Ota M., Kanaya S., et al. Knowledge-based structural models of SARS-CoV-2 proteins and their complexes with potential drugs. FEBS Lett. 2020 May 25;594(12):1960–1973. doi: 10.1002/1873-3468.13806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Majumder R., Mandal M. Screening of plant-based natural compounds as a potential COVID-19 main protease inhibitor: an in silico docking and molecular dynamics simulation approach. J. Biomol. Struct. Dyn. 2020 Sep 8:1–16. doi: 10.1080/07391102.2020.1817787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhardwaj V.K., Singh R., Sharma J., Rajendran V., Purohit R., Kumar S. Identification of bioactive molecules from tea plant as SARS-CoV-2 main protease inhibitors. J. Biomol. Struct. Dyn. 2020 May 20:1–10. doi: 10.1080/07391102.2020.1766572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ton A.-T., Gentile F., Hsing M., Ban F., Cherkasov A. Rapid identification of potential inhibitors of SARS-CoV-2 main protease by deep docking of 1.3BillionCompounds. Mol Inform. 2020 Mar 23;39(8) doi: 10.1002/minf.202000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., et al. A trial of lopinavir-ritonavir in adults hospitalized with severe covid-19. N. Engl. J. Med. 2020 May 7;382(19):1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Popova M., Isayev O., Tropsha A. Deep reinforcement learning for de novo drug design. Sci Adv. 2018 Jul 25;4(7) doi: 10.1126/sciadv.aap7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhavoronkov A., Ivanenkov Y.A., Aliper A., Veselov M.S., Aladinskiy V.A., Aladinskaya A.V., et al. Deep learning enables rapid identification of potent DDR1 kinase inhibitors. Nat. Biotechnol. 2019 Sep 2;37(9):1038–1040. doi: 10.1038/s41587-019-0224-x. [DOI] [PubMed] [Google Scholar]
- 20.Stokes J.M., Yang K., Swanson K., Jin W., Cubillos-Ruiz A., Donghia N.M., et al. A deep learning approach to antibiotic discovery. Cell. 2020 Feb 20;180(4):688–702.e13. doi: 10.1016/j.cell.2020.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaulton A., Bellis L.J., Bento A.P., Chambers J., Davies M., Hersey A., et al. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012 Jan;40(Database issue):D1100–D1107. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Polykovskiy D., Zhebrak A., Sanchez-Lengeling B., Golovanov S., Tatanov O., Belyaev S., et al. Molecular sets (MOSES): a benchmarking platform for molecular generation models. Front. Pharmacol. 2020 Dec 18;11:565644. doi: 10.3389/fphar.2020.565644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010 Jan 30;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen H., Wei P., Huang C., Tan L., Liu Y., Lai L. Only one protomer is active in the dimer of SARS 3C-like proteinase. J. Biol. Chem. 2006 May 19;281(20):13894–13898. doi: 10.1074/jbc.M510745200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rahman M.M., Saha T., Islam K.J., Suman R.H., Biswas S., Rahat E.U., et al. Virtual screening, molecular dynamics and structure-activity relationship studies to identify potent approved drugs for Covid-19 treatment. J. Biomol. Struct. Dyn. 2020 Jul 21:1–11. doi: 10.1080/07391102.2020.1794974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A.E., Berendsen H.J.C. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005 Dec;26(16):1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 27.Chen J., Wang W., Sun H., Pang L., Bao H. Binding mechanism of inhibitors to p38α MAP kinase deciphered by using multiple replica Gaussian accelerated molecular dynamics and calculations of binding free energies. Comput. Biol. Med. 2021 Jul;134:104485. doi: 10.1016/j.compbiomed.2021.104485. [DOI] [PubMed] [Google Scholar]
- 28.Chen J., Wang X., Pang L., Zhang J.Z.H., Zhu T. Effect of mutations on binding of ligands to guanine riboswitch probed by free energy perturbation and molecular dynamics simulations. Nucleic Acids Res. 2019 Jul 26;47(13):6618–6631. doi: 10.1093/nar/gkz499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jorgensen W.L., Madura J.D., Swenson C.J. Optimized intermolecular potential functions for liquid hydrocarbons. J. Am. Chem. Soc. 1984 Oct;106(22):6638–6646. [Google Scholar]
- 30.Kräutler V., van Gunsteren W.F., Hünenberger P.H. A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J. Comput. Chem. 2001 Apr 15;22(5):501–508. [Google Scholar]
- 31.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996 Feb;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. 27. [DOI] [PubMed] [Google Scholar]
- 32.DeLano W.L. Pymol: an open-source molecular graphics tool. CCP4 Newsletter on protein crystallography. 2002;40(1):82–92. [Google Scholar]
- 33.Miller B.R., McGee T.D., Swails J.M., Homeyer N., Gohlke H., Roitberg A.E. MMPBSA.py: an efficient program for end-state free energy calculations. J. Chem. Theor. Comput. 2012 Sep 11;8(9):3314–3321. doi: 10.1021/ct300418h. [DOI] [PubMed] [Google Scholar]
- 34.Kumari R., Kumar R., Open Source Drug Discovery Consortium. Lynn A. g_mmpbsa--a GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014 Jul 28;54(7):1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- 35.Kollman P.A., Massova I., Reyes C., Kuhn B., Huo S., Chong L., et al. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc. Chem. Res. 2000 Dec;33(12):889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 36.Homeyer N., Gohlke H. Free energy calculations by the molecular mechanics Poisson−Boltzmann surface area method. Mol Inform. 2012 Feb;31(2):114–122. doi: 10.1002/minf.201100135. [DOI] [PubMed] [Google Scholar]
- 37.Wang J., Wolf R.M., Caldwell J.W., Kollman P.A., Case D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004 Jul 15;25(9):1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 38.Nayeem SkM., Sohail E.M., Ridhima G., Reddy M.S. Target SARS-CoV-2: computation of binding energies with drugs of dexamethasone/umifenovir by molecular dynamics using OPLS-AA force field. Res Biomed Eng. 2021 Jan 8 [Google Scholar]
- 39.Udhaya Kumar S., Thirumal Kumar D., Mandal P.D., Sankar S., Haldar R., Kamaraj B., et al. Comprehensive in silico screening and molecular dynamics studies of missense mutations in Sjogren-Larsson syndrome associated with the ALDH3A2 gene. Adv Protein Chem Struct Biol. 2020 Feb 4;120:349–377. doi: 10.1016/bs.apcsb.2019.11.004. [DOI] [PubMed] [Google Scholar]
- 40.Mohammadi M., Sakhteman A., Ahrari S., Hassanpour K., Hashemi S.E., Farnoosh G. Disulfide bridge formation to increase thermostability of DFPase enzyme: a computational study. Comput. Biol. Chem. 2018 Dec;77:272–278. doi: 10.1016/j.compbiolchem.2018.09.005. [DOI] [PubMed] [Google Scholar]
- 41.Abdel-Basset M., Hawash H., Elhoseny M., Chakrabortty R.K., Ryan M., DeepH-Dta Deep learning for predicting drug-target interactions: a case study of COVID-19 drug repurposing. IEEE Access. 2020;8:170433–170451. doi: 10.1109/ACCESS.2020.3024238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Majumdar S., Nandi S.K., Ghosal S., Ghosh B., Mallik W., Roy N.D., et al. Deep learning-based potential ligand prediction framework for COVID-19 with drug-target interaction model. Cognit Comput. 2021 Feb 2:1–13. doi: 10.1007/s12559-021-09840-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bung N., Krishnan S.R., Bulusu G., Roy A. De novo design of new chemical entities for SARS-CoV-2 using artificial intelligence. Future Med. Chem. 2021 Mar;13(6):575–585. doi: 10.4155/fmc-2020-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Santana M.V.S., Silva F.P., Jr. De novo design and bioactivity prediction of SARS-CoV-2 main protease inhibitors using recurrent neural network-based transfer learning. BMC Chemistry. 2021 Feb 2;15(1):8. doi: 10.1186/s13065-021-00737-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Talele T.T., Khedkar S.A., Rigby A.C. Successful applications of computer aided drug discovery: moving drugs from concept to the clinic. Curr. Top. Med. Chem. 2010;10(1):127–141. doi: 10.2174/156802610790232251. [DOI] [PubMed] [Google Scholar]
- 46.Bharadwaj S., Lee K.E., Dwivedi V.D., Yadava U., Panwar A., Lucas S.J., et al. Discovery of Ganoderma lucidum triterpenoids as potential inhibitors against Dengue virus NS2B-NS3 protease. Sci. Rep. 2019 Dec 13;9(1):19059. doi: 10.1038/s41598-019-55723-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mahmud S., Uddin M.A.R., Paul G.K., Shimu M.S.S., Islam S., Rahman E., et al. Virtual screening and molecular dynamics simulation study of plant-derived compounds to identify potential inhibitors of main protease from SARS-CoV-2. Briefings Bioinf. 2021 Mar 22;22(2):1402–1414. doi: 10.1093/bib/bbaa428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bharadwaj S., Rao A.K., Dwivedi V.D., Mishra S.K., Yadava U. Structure-based screening and validation of bioactive compounds as Zika virus methyltransferase (MTase) inhibitors through first-principle density functional theory, classical molecular simulation and QM/MM affinity estimation. J. Biomol. Struct. Dyn. 2021 Apr;39(7):2338–2351. doi: 10.1080/07391102.2020.1747545. [DOI] [PubMed] [Google Scholar]
- 49.Alamri M.A., Tahir Ul Qamar M., Mirza M.U., Bhadane R., Alqahtani S.M., Muneer I., et al. Pharmacoinformatics and molecular dynamics simulation studies reveal potential covalent and FDA-approved inhibitors of SARS-CoV-2 main protease 3CLpro. J. Biomol. Struct. Dyn. 2021 Aug;39(13):4936–4948. doi: 10.1080/07391102.2020.1782768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Islam R., Parves M.R., Paul A.S., Uddin N., Rahman M.S., Mamun A.A., et al. A molecular modeling approach to identify effective antiviral phytochemicals against the main protease of SARS-CoV-2. J. Biomol. Struct. Dyn. 2021 Jun;39(9):3213–3224. doi: 10.1080/07391102.2020.1761883. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.